Abstract
Expression of the general stress regulon of Bacillus subtilis is controlled by the alternative transcription factor ςB, which is activated when cells encounter growth-limiting energy or environmental stresses. The RsbT serine-threonine kinase is required to convey environmental stress signals to ςB, and this kinase activity is magnified in vitro by the RsbR protein, a positive regulator important for full in vivo response to salt or heat stress. Previous genetic analysis suggested that RsbR function is redundant with other unidentified regulators. A search of the translated B. subtilis genome found six paralogous proteins with significant similarity to RsbR: YetI, YezB, YkoB, YojH, YqhA, and YtvA. Their possible regulatory roles were investigated using three different approaches. First, genetic analysis found that null mutations in four of the six paralogous genes have marked effects on the ςB environmental signaling pathway, either singly or in combination. The two exceptions were yetI and yezB, adjacent genes which appear to encode a split paralog. Second, biochemical analysis found that YkoB, YojH, and YqhA are specifically phosphorylated in vitro by the RsbT environmental signaling kinase, as had been previously shown for RsbR, which is phosphorylated on two threonine residues in its C-terminal region. Both residues are conserved in the three phosphorylated paralogs but are absent in the ones that were not substrates of RsbT: YetI and YezB, each of which bears only one of the conserved residues; and YtvA, which lacks both residues and instead possesses an N-terminal PAS domain. Third, analysis in the yeast two-hybrid system suggested that all six paralogs interact with each other and with the RsbR and RsbS environmental regulators. Our data indicate that (i) RsbR, YkoB, YojH, YqhA, and YtvA function in the environmental stress signaling pathway; (ii) YtvA acts as a positive regulator; and (iii) RsbR, YkoB, YojH, and YqhA collectively act as potent negative regulators whose loss increases ςB activity more than 400-fold in unstressed cells.
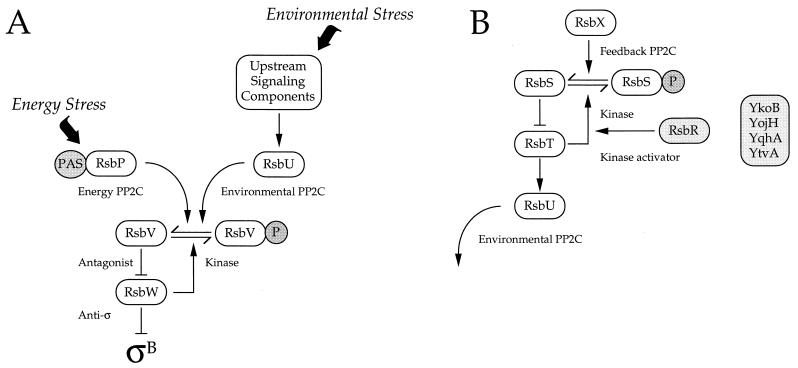
The general stress response of Bacillus subtilis is induced when cells encounter a variety of growth-limiting stresses, and this induction confers a multiple-stress resistance phenotype (17, 19, 32). Expression of the 200 or more genes comprising the general stress regulon is under control of the alternative transcription factor ςB, the activity of which is governed by a signal transduction pathway with two distinct branches. One branch is specific for energy stresses, such as carbon, phosphorus, or oxygen limitation, and the other is specific for environmental stresses, such as acid, ethanol, heat, or salt stress (23, 41, 46, 47). According to the model shown in Fig. 1A, the two branches converge on the common regulators RsbV and RsbW, which together control ςB activity by means of a partner-switching mechanism in which alternate interactions are determined by serine phosphorylation (2, 4, 10, 41, 44, 47).
FIG. 1.
Model of the ςB signal transduction network. (A) Two signaling pathways converge on phosphorylated RsbV (RsbV-P), the antagonist form found in unstressed cells. The energy-signaling pathway terminates with the RsbP PP2C phosphatase, which may employ its PAS domain to sense energy stress (41). In contrast, the environmental signaling pathway terminates with the RsbU PP2C phosphatase, which is activated by upstream elements (23, 47). When activated by stress, either the RsbP or the RsbU phosphatase removes the serine phosphate from RsbV-P (41, 44, 47). Dephosphorylated RsbV then binds the RsbW anti-ς factor, forcing it to release ςB, which can then activate its target general stress genes (2, 4, 10). (B) The upstream signaling elements which activate RsbU include the RsbS antagonist and the RsbT kinase, which are homologs of RsbV and RsbW, respectively (23, 47). Unphosphorylated RsbS is thought to be the antagonist form found in unstressed cells, and this form binds the RsbT kinase. Following environmental stress, RsbS is phosphorylated by RsbT, which is then released to bind and activate the RsbU phosphatase by direct protein-protein interaction (24, 47). Genetic and biochemical analyses indicate that RsbR (shaded) acts as a positive regulator of ςB activity by magnifying the activity of the RsbT kinase (1, 15). In contrast, the RsbX PP2C phosphatase appears to fulfill a feedback role by indirectly communicating the level of ςB protein in the cell (37, 43, 47). We show here that the four RsbR paralogs (shaded box) also act via the environmental signaling pathway.
The means by which energy and environmental stress signals enter their respective branches is presently unknown. Each branch terminates with a differentially regulated serine phosphatase, RsbP or RsbU (41, 47). The RsbP energy-signaling phosphatase contains a PAS domain in its amino-terminal half (41); PAS is an acronym for the proteins in which the domain was originally found (40). Many of the proteins containing this domain are involved in sensing fluctuations in redox, oxygen, or light, suggesting that RsbP alone could be sufficient for energy sensing and signal transduction. In contrast to the apparent simplicity of the energy branch, the RsbU environmental signaling phosphatase is known to be activated by upstream elements. Chief among these are RsbS and RsbT, which are homologs of RsbV and RsbW, respectively, and which are also thought to function by a partner-switching mechanism (23, 24, 47).
According to the model shown in Fig. 1B, in unstressed cells the unphosphorylated RsbS antagonist protein binds and sequesters the RsbT switch protein, holding the system in a nonsignaling state (47). However, when cells are subjected to environmental stress, RsbS is phosphorylated by the serine-threonine kinase activity of RsbT. RsbT is then released to switch partners and bind the RsbU phosphatase, which it activates by direct protein-protein interaction. The crux of this signaling mechanism is therefore the phosphorylation state of RsbS, which is controlled by the tension between the RsbT kinase and the RsbX phosphatase. Genetic analysis has shown that RsbX phosphatase activity is not required for the environmental stress response but instead provides part of a negative feedback loop that damps continued signaling (37, 43). In contrast, the RsbT kinase activity is essential (24). Modulation of the kinase activity of RsbT therefore provides one route by which environmental signals might enter the system.
The RsbR positive regulator has the biochemical and genetic properties expected for a modulator of RsbT kinase activity (1, 15). First, purified RsbR itself has no intrinsic kinase activity but greatly stimulates the kinase activity of RsbT toward RsbS in vitro. Second, complete absence of rsbR function disturbs only environmental signaling. However, in contrast to the other known Rsb regulators, loss of RsbR function causes an unusual partial phenotype that affects the response to some environmental stresses, such as salt and heat, but not others, such as ethanol. Moreover, even those responses that are affected by an rsbR null mutation are reduced only two- or threefold. One explanation for this partial phenotype is that RsbR function may be redundant with one or more additional regulators (1). The completion of the B. subtilis genome project (26) revealed six additional gene products with significant similarity to the RsbR protein, and these became prime candidates for the missing regulators. We report here that at least four of these newly discovered RsbR paralogs act collectively in the environmental stress signaling pathway as potent regulators of ςB activity.
MATERIALS AND METHODS
Bacterial strains and genetic methods.
Standard recombinant DNA methods were described by Sambrook et al. (33), and Escherichia coli DH5α (Bethesda Research Laboratories) was the host for all plasmid constructions. B. subtilis strains are shown in Table 1. To make the ykoBΔ1::kan null allele, we first removed a 711-bp NsiI-StuI fragment from within the ykoB coding region and substituted the 1,628-bp NsiI-StuI fragment containing the kanamycin resistance element from pDG792 (16). The resulting plasmid, pSA84, is a pUC19 derivative in which the kanamycin resistance cassette is flanked by the first 9 and the last 32 triplets of ykoB, together with additional contiguous DNA to permit recombination. Similar plasmids were constructed for the other four null alleles. These were pTG3, in which an spc spectinomycin resistance cassette (27) was inserted between triplets 21 and 829 of yqhA, and three plasmids bearing an ery erythromycin resistance cassette (20): pSA68, with ery between triplets 6 and 256 of ytvA; pSA79, with ery between triplet 2 of yetI and triplet 89 of the adjacent yezB; and pSA82, with ery replacing the region extending from triplet 4 of yojH through the yojH terminator. The pSA77 plasmid was used to carry an in-frame deletion of rsbT into strains bearing multiple mutations in the genes encoding rsbR and its paralogs. To avoid the possibility of correcting the rsbRΔ1 null allele, pSA77 was constructed by removing a 929-bp EcoRI fragment from pCK1, the plasmid bearing the in-frame deletion in rsbT (23).
TABLE 1.
B. subtilis strains
| Strain | Genotype | Reference or constructiona |
|---|---|---|
| PB2 | trpC2 | Wild-type Marburg strain |
| PB198 | amyE::ctc-lacZ trpC2 | 8 |
| PB427 | rsbRΔ1 trpC2 | 1 |
| PB491 | rsbRΔ1 amyE::ctc-lacZ trpC2 | 1 |
| PB520 | yqhAΔ1::spc amyE::ctc-lacZ trpC2 | pTG3→PB198 |
| PB521 | rsbRΔ1 yqhAΔ1::spc amyE::ctc-lacZ trpC2 | PB520→PB491 |
| PB528 | ykoBΔ1::kan amyE::ctc-lacZ trpC2 | pSA84→PB198 |
| PB529 | ykoBΔ1::kan yqhAΔ1::spc amyE::ctc-lacZ trpC2 | pSA84→PB520 |
| PB530 | rsbRΔ1 ykoBΔ1::kan yqhAΔ1::spc amyE::ctc-lacZ trpC2 | pSA84→PB521 |
| PB531 | rsbRΔ1 ykoBΔ1::kan amyE::ctc-lacZ trpC2 | pSA84→PB491 |
| PB545 | ykoBΔ1 ykoBΔ1::kan trpC2 | PB528→PB2 |
| PB545.1 | rsbRΔ1 ykoBΔ1::kan trpC2 | PB545→PB427 |
| PB551 | rsbRΔ1 ykoBΔ1::kan yqhAΔ1::spc trpC2 | pTG3→PB545.1 |
| PB565 | ytvAΔ1::ery amyE::ctc-lacZ trpC2 | pSA68→PB198 |
| PB566 | rsbRΔ1 ytvAΔ1::ery amyE::ctc-lacZ trpC2 | pSA68→PB491 |
| PB569 | yetI-yezBΔ1::ery amyE::ctc-lacZ trpC2 | pSA79→PB198 |
| PB574 | yojHΔ1::ery amyE::ctc-lacZ trpC2 | pSA82→PB198 |
| PB577 | rsbRΔ1 yojHΔ1::ery amyE::ctc-lacZ trpC2 | PB574→PB491 |
| PB578 | ykoBΔ1::kan ytvAΔ1::ery amyE::ctc-lacZ trpC2 | PB565→PB528 |
| PB579 | rsbRΔ1 rsbTΔ1 ykoBΔ1::kan yqhAΔ1::spc trpC2 | pSA77→PB551b |
| PB580 | rsbRΔ1 rsbTΔ1 ykoBΔ1::kan yqhAΔ1::spc amyE::ctc-lacZ trpC2 | pDH32-ctc (8)→PB579 |
| PB629 | rsbRΔ1 ykoBΔ1::kan yojHΔ1::ery yqhAΔ1::spc amyE::ctc-lacZ trpC2 | PB577→PB530 |
| PB639 | rsbRΔ1 yojHΔ1::ery yqhAΔ1::spc amyE::ctc-lacZ trpC2 | PB577→PB521 |
| PB653 | rsbRΔ1 ykoBΔ1::kan yojHΔ1::ery amyE::ctc-lacZ trpC2 | PB577→PB531 |
| PB654 | rsbRΔ1 rsbTΔ1 ykoBΔ1::kan yojHΔ1::ery yqhAΔ1::spc amyE::ctc-lacZ trpC2 | PB577→PB580 |
| 2A2 | B. subtilis W23 wild type | Bacillus Genetic Stock Center |
| 3A13 | B. subtilis var. amylosacchariticus wild type | Bacillus Genetic Stock Center |
Arrow indicates transformation from donor to recipient.
Two-step allele replacement (38).
β-Galactosidase accumulation assays.
For energy stress experiments, cells were grown to stationary phase in buffered Luria-Bertani medium (LB) lacking salt (7). For environmental stress experiments, cells were grown to early logarithmic phase in buffered LB lacking salt, at which point a chemical stressor was added to a final concentration of 0.3 M for salt or 4% (vol/vol) for ethanol. In all stress experiments, samples were collected at the times indicated and treated as described by Miller (28). Cells were washed with Z buffer and permeabilized using sodium dodecyl sulfate (SDS) and chloroform. Protein levels were determined using the Bio-Rad protein assay reagent (Bio-Rad Laboratories, Richmond, Calif.). Activity was defined as ΔA420 × 1,000 per minute per milligram of protein.
Construction of overexpression clones and purification of His-tagged proteins.
Overexpression clones which fused the rsbR, rsbS, rsbT, rsbV, and rsbW coding regions to a hexahistidine tag in the pET15b expression vector (Novagen, Madison, Wis.) were described previously (15, 24, 47). Here we constructed similar overexpression clones bearing the spoIIAA, spoIIAB, yetI, yezB, ykoB, yqhA, and ytvA reading frames (details of these and other plasmid constructions are available from the authors upon request). Hexahistidine-tagged proteins were purified from E. coli BL21(DE3)/pLysS extracts on nickel affinity columns according to the manufacturer's protocol (Novagen).
Kinase assays.
Kinase reactions were conducted as previously described (15). Reactions were initiated by adding 3 pmol of the kinase to be tested (RsbT, RsbW, or SpoIIAB) to 300-μl mixes containing 30 pmol of the purified protein substrate, 1 mM unlabeled ATP, and 150 μCi of [γ-32P]ATP in kinase assay buffer (50 mM Tris [pH 7.6], 50 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, and 0.1 mM EDTA). After 60 min of incubation at 37°C, labeled proteins were separated on SDS–13.5% polyacrylamide gels and visualized by exposure to X-ray film.
Yeast two-hybrid analysis.
The possible interaction of the RsbR homologs with other proteins was tested by fusing the appropriate reading frame to the GAL4 DNA-binding domain in plasmid pGBT9 (Clontech, Palo Alto, Calif.). For the adjacent yetI and yezB genes, which appear to encode a split RsbR paralog, we coupled the two coding regions in frame to synthesize one full-length product fused to the GAL4 DNA-binding domain. These new paralog constructs were then paired in yeast SFY526 cells with another rsb or spoII reading frame fused to the GAL4 activation domain in plasmid pGAD424. Double transformants were selected on minimal medium plates. Transcriptional activation of the lacZ reporter gene in these double transformants was determined either qualitatively, using the colony lift filter assay from the Matchmaker Two-Hybrid System protocols (Clontech), or quantitatively, by assaying β-galactosidase activity following growth in minimal medium, as previously described (47).
RESULTS
Six RsbR paralogs are encoded by the B. subtilis genome.
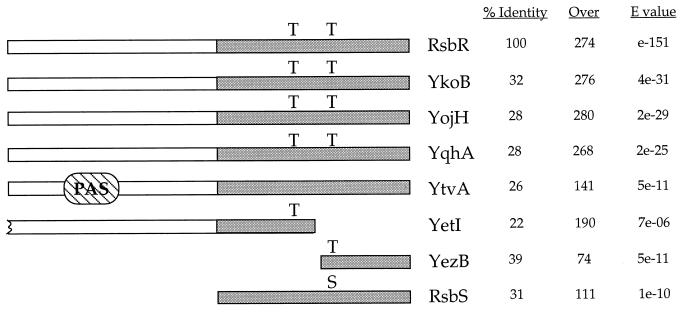
A Blast 2.0 search of the RsbR sequence against the translated B. subtilis genome found six clear RsbR paralogs. As shown in Fig. 2, the greatest sequence conservation is within their C-terminal halves, a region similar to the entire length of the smaller RsbS antagonist protein. Three of these proteins—YkoB, YojH, and YqhA—are the most similar to RsbR and include two characteristic threonine residues in their C-terminal regions. These conserved residues have been shown to be the sites at which RsbR is phosphorylated in vitro by the RsbT kinase, and both residues have been shown to be important for RsbR function in vivo (1, 15).
FIG. 2.
Sequence similarities between RsbR, its paralogs, and RsbS. The percent identical residues and the number of residues over which the similarity extends are shown in the first two columns, and the statistical significance of the Blast alignment is shown in the final column. E values of <1e-10 are considered probably significant, and those of <1e-20 are considered significant. The shaded regions indicate the sequences most highly conserved among the RsbR paralogs and the smaller RsbS antagonist protein. For RsbR, residues T171 and T205 are phosphorylated in vitro by the RsbT kinase and are important for RsbR function in vivo (15). These residues are conserved in YkoB, YojH, and YqhA, whereas the split paralogs YetI and YezB each carry one residue. In contrast, YtvA lacks the conserved threonines and instead bears a PAS domain in its N-terminal region. PAS domains are often found in proteins that monitor changes in redox potential, oxygen tension, or energy levels (40). The analysis reported here indicates that YtvA acts in the environmental stress-signaling branch.
A fourth protein, YtvA, also has significant similarity with RsbR but lacks the conserved threonine residues. Instead, YtvA has a PAS domain in its N-terminal region. PAS domains are found in a wide variety of intracellular proteins that monitor changes in redox potential, oxygen tension, and the overall energy level of the cell (40).
The fifth and sixth proteins, YetI and YezB, appear to have arisen from a frameshift mutation that split an ancestral RsbR paralog gene. As shown in Fig. 2, the translational stop for the yetI reading fame lies just after the triplet for the first conserved threonine residue, at which point the yezB frame begins, and this frame encodes the second conserved threonine. We first considered the possibility that the published genome sequence was incorrect over the yetI-yezB interval. However, our laboratory 168 strain proved to have an identical sequence through this region, as did two more divergent strains, B. subtilis W23 and B. subtilis var. amylosacchariticus (data not shown). We therefore conclude that yetI and yezB represent distinct reading frames in different B. subtilis strains.
Loss of YkoB or YtvA function affects ςB activation.
Three of the genes encoding RsbR paralogs appear to lie in monocistronic transcription units (ykoB, yqhA, and ytvA), and one is the downstream gene in an apparent two-gene operon (yojH). In order to determine the role of each paralog, we constructed a large insertion-deletion mutation in each gene and substituted these null alleles for the wild-type copies on the B. subtilis chromosome. In the case of the yetI-yezB reading frames, we constructed a single insertion-deletion mutation that removed most of both coding sequences. In order to assay the effect of these null mutations on ςB activity, each strain also carried at its amyE locus a single-copy transcriptional fusion between the ςB-dependent ctc promoter (21, 30) and a lacZ reporter gene.
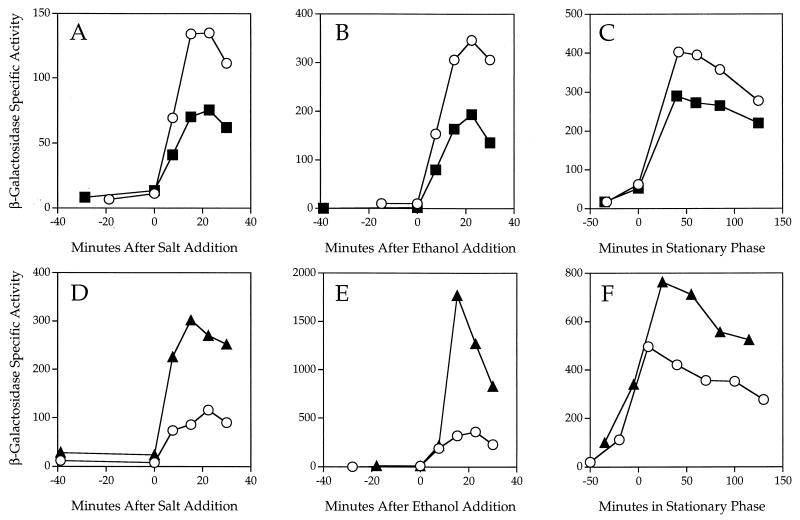
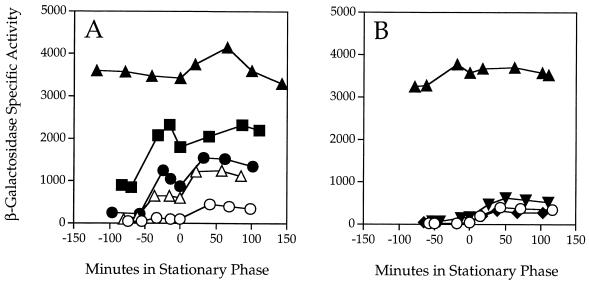
Two of the five single mutations, ytvA and ykoB, gave a clear ςB regulatory phenotype. As shown in Fig. 3A to C, the ytvA mutation resembled the original rsbR mutation in that it decreased response to salt stress about twofold. However, unlike the case with rsbR, loss of ytvA function equally affected response to ethanol stress and somewhat affected response to energy stress. These results indicate that YtvA is a positive regulator of ςB activity. The ykoB mutation manifested a different phenotype. As shown in Fig. 3D to F, loss of ykoB function increased ςB activity in response to salt and ethanol stress and also increased ςB activity in response to the energy stress associated with entry into stationary phase. These results indicate that YkoB is a negative regulator of ςB activity. Notably, the yojH, yqhA, and yetI-yezB single null alleles had no effect on ςB activity under any of the three conditions tested—salt, ethanol, or energy stress (data not shown). Moreover, none of the five null mutations had any obvious effect on sporulation timing or efficiency, suggesting that they do not strongly influence activity of ςF, the forespore-specific factor which provides the only other example of partner-switching regulation in B. subtilis (39 and references therein).
FIG. 3.
Effects of single null mutations on β-galactosidase activity of a ςB-dependent transcriptional fusion. ςB activity was measured indirectly, using a single-copy transcriptional fusion between the well-characterized ctc promoter (21, 30) and a lacZ reporter, with the PB198 wild-type control indicated by ○. At time zero, cells were subjected to either salt stress (A and D) or ethanol stress (B and E) in early logarithmic growth or to energy stress elicited by entry into stationary phase (C and F). (A to C) Assays using PB565 (ytvAΔ1::ery, ■); (D to F) assays using PB528 (ykoBΔ1::kan, ▴).
As described in the following two sections, analysis of strains carrying multiple mutations allowed us to establish the relationship among rsbR, ytvA, and ykoB and also led to the discovery of ςB regulatory phenotypes for the yojH and yqhA null alleles. In contrast, we were unable to find any combination of alleles which demonstrated a role for yetI-yezB under the conditions tested. If yetI or yezB in fact encodes a ςB regulator, its activity must be manifest only under specific circumstances.
Double-mutant analyses indicate that rsbR is dependent on ykoB and that ytvA acts independently.
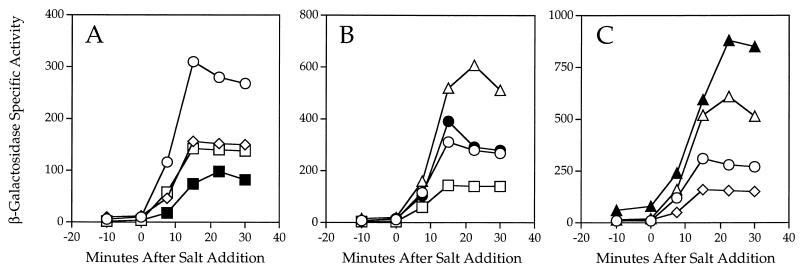
We made a series of double mutants in order to determine whether the RsbR, YtvA, and YkoB products act in the same signaling pathway or in different pathways that ultimately converge on ςB. As shown in Fig. 4A and B, combining the ytvA null allele with either rsbR or ykoB in a single strain produced additive phenotypes in response to salt stress. The ytvA-rsbR double null mutant had decreased response to salt stress compared with mutants carrying either single allele (Fig. 4A), whereas in the ytvA-ykoB double null, the increased response due to the ykoB null allele was essentially canceled by the decreased response due to the ytvA null allele (Fig. 4B). From these results, we conclude that YtvA functions independently of both RsbR and YkoB.
FIG. 4.
Double-mutant analyses indicate that rsbR is dependent on ykoB, whereas ytvA acts independently. A ctc-lacZ transcriptional fusion was used to measure ςB activity, with the PB198 wild-type control indicated by ○. At time zero, cells were subjected to salt stress in early logarithmic growth. (A) Effects of the ytvA null allele (PB565; ytvAΔ1::ery, □) and the rsbR null allele (PB491; rsbRΔ1, ◊) are additive in the double mutant (PB566; rsbRΔ1 ytvA1::ery, ■). (B) The ytvA null allele (PB565; ytvAΔ1::ery, □) and the ykoB null allele (PB528; ykoBΔ1::kan, ▵) have opposite effects on response to salt stress, and in the double mutant (PB578; ykoBΔ1::kan ytvAΔ1::ery, ●) the response is a blend of both phenotypes. (C) The ykoB null allele (PB528; ykoBΔ1::kan, ▵) and the rsbR null allele (PB491; rsbRΔ1, ◊) have opposite effects on response to salt stress, and in the double mutant (PB531; rsbRΔ1 ykoBΔ1::kan, ▴) the ykoB null allele fully masks the positive regulatory phenotype of the rsbR null allele. The seven strains shown were assayed together, and the results were distributed into the appropriate panels.
In contrast, the phenotype of the strain bearing the ykoB null allele together with an rsbR null allele was more similar to that of the ykoB single null mutant (Fig. 4C). We therefore conclude that RsbR is dependent upon YkoB function, either directly or indirectly.
Analyses of triple and quadruple mutants reveal that YojH and YqhA act in concert with RsbR and YkoB in the environmental signaling pathway.
The original rationale for investigating the RsbR paralogs was to determine whether they might perform redundant regulatory roles. Consistent with this notion, strains which contained multiple null alleles in fact manifested striking changes in ςB regulation. Loss of yqhA in a strain already lacking rsbR and ykoB caused a high basal level of ςB activity in unstressed cells and an earlier induction of ςB activity as cells approached stationary phase (Fig. 5A). The signal transduction network in this triple mutant still retained some capacity to activate ςB in response to environmental stress (imposed by salt addition; data not shown) or energy stress (imposed by entry into stationary phase). In contrast, loss of yojH in a strain lacking rsbR and ykoB produced only a slight increase in ςB activity over that seen in the rsbR-ykoB null parent (Fig. 5A).
FIG. 5.
Analyses of triple and quadruple mutants indicate that YojH and YqhA act in concert with RsbR and YkoB in the environmental signaling pathway. A ctc-lacZ transcriptional fusion was used to measure ςB activity in unstressed cells during logarithmic growth and also during entry into stationary phase. (A) PB198 (wild type, ○); PB531 (rsbRΔ1 ykoBΔ1::kan, ▵); PB653 (rsbRΔ1 ykoBΔ1::kan yojHΔ1::ery, ●); PB530 (rsbRΔ1 ykoBΔ1::kan yqhAΔ1::spc, ■); and quadruple mutant PB629 (rsbRΔ1 ykoBΔ1::kan yojHΔ1::ery yqhAΔ1::spc, ▴). (B) PB198 (wild type, ○); quadruple mutant PB629 (rsbRΔ1 ykoBΔ1::kan yojHΔ1::ery yqhAΔ1::spc, ▴); PB639 (rsbRΔ1 yojHΔ1::ery yqhAΔ1::spc, ▾); and PB654 (rsbRΔ1 rsbTΔ1 ykoBΔ1::kan yojHΔ1::ery yqhAΔ1::spc, ⧫)
However, the phenotype of a yojH null did become apparent in a quadruple mutant (Fig. 5A). The combined yojH, ykoB, yqhA, and rsbR null alleles elicited exceedingly high ςB activity in unstressed, early-logarithmic-phase cells (400- to 800-fold higher than the wild type, depending on the experiment). This high basal activity approaches the levels previously achieved only in the absence of the strong negative regulators RsbS, RsbW, and RsbX. Moreover, the high ςB activity of the quadruple mutant was markedly deleterious for growth, as had been previously observed for rsbS, rsbW, and rsbX null mutants (5, 8, 14, 21–23). From these experiments, we conclude that YojH and YqhA act as negative regulators and that this role is revealed in the absence of RsbR and YkoB.
In contrast to the significant effects noted for the yojH and yqhA null alleles (Fig. 5A), introduction of the yetI-yezB or ytvA null mutation into the rsbR-ykoB-yqhA triple null background had no added effect on ςB activation (data not shown). Since a ytvA null allele manifested a regulatory phenotype in an otherwise wild-type background (Fig. 3), and since YtvA acted independently of RsbR and YkoB (Fig. 4), we presume that it was loss of YqhA function that rendered the ytvA null allele silent in the triple mutant background. However, we cannot exclude the possibility that loss of two or more of the RsbR, YkoB, and YqhA regulators was in fact responsible for this silencing.
Of the four alleles whose loss causes extremely high ςB activity (Fig. 5A), only loss of rsbR or ykoB elicited a regulatory phenotype when present in an otherwise wild-type genetic background. As shown in Fig. 4C, the phenotype of an rsbR single null mutant indicated that RsbR acts as a positive regulator of ςB activity, whereas the phenotype of a ykoB single null indicated that YkoB acts as a negative regulator. We therefore hypothesized that loss of YkoB function was key to the striking phenotype of the quadruple mutant. To test this hypothesis, we compared ςB activity in unstressed cells of two different strains: the quadruple mutant and a related triple mutant in which YkoB function had been restored. As shown in Fig. 5B, ςB activity in the triple mutant was close to that of the wild type in unstressed cells. Because restoration of YkoB function almost completely reversed the phenotype of the quadruple null mutant, we conclude that YkoB function is critical to this phenotype. We further conclude that the combined loss of RsbR, YojH, and YqhA functions greatly exacerbates loss of YkoB function.
We then wished to establish at what point in the ςB signal transduction network the newly discovered RsbR paralogs might act. Loss of YkoB function in the single null mutant affected ςB activation in response to both environmental and energy stress (Fig. 3D to F), and loss of YkoB function in the quadruple mutant elicited such a high basal level of ςB activity that response to both classes of stresses was compromised (Fig. 5A). To distinguish if the RsbR paralogs functioned in the environmental signaling branch, the energy-signaling branch, or the common part of the signal transduction network, we introduced into the quadruple mutant an in-frame deletion in rsbT. The RsbT positive regulator is required for environmental signaling but is not important for energy signaling (15, 24, 47). As shown in Fig. 5B, ςB activity in the quintuple mutant was similar to that in the wild type in unstressed cells. Because loss of RsbT function completely reversed the strong phenotype of the quadruple null mutant, we conclude that RsbR, YkoB, YojH, and YqhA act upstream of RsbT in the environmental signaling branch of the ςB regulatory network.
Purified RsbR paralogs are specifically phosphorylated by the RsbT kinase.
RsbT in the environmental signaling branch and RsbW in the common part of the ςB pathway are related serine-threonine kinases with different substrate specificities (47), and it is known that RsbR is phosphorylated in vitro by RsbT but not by RsbW (15). We therefore purified His-tagged versions of the six RsbR paralogs to determine whether they could be phosphorylated by either kinase.
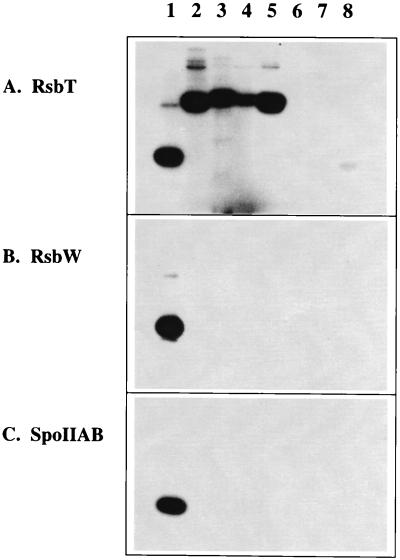
As shown in Fig. 6A, the RsbT kinase transferred the labeled γ-phosphate from ATP to RsbR, YkoB, YojH, and YqhA as readily as it did to its cognate antagonist protein, RsbS. These four paralogs all retain the two conserved threonine residues that are known to be important for RsbR function in vitro and in vivo (see Fig. 2). The presumably split paralogs YetI and YezB, each of which contains only one of the threonine residues, were labeled poorly or not at all, and YtvA, which lacks both residues, was likewise unlabeled. In contrast, the RsbW kinase did not transfer label to any of the RsbR paralogs. A control reaction indicated that the RsbW kinase was indeed active and could readily label its cognate antagonist protein, RsbV (Fig. 6B, lane 1).
FIG. 6.
Purified RsbR, YkoB, YojH, and YqhA are phosphorylated by the RsbT environmental signaling kinase. Purified proteins were incubated with [γ-32P]ATP in kinase assay buffer, as described in the text. After 60 min at 37°C, the reaction mixtures were subjected to SDS-PAGE and autoradiography. Reactions included the kinase of interest, either RsbT (A), RsbW (B), or SpoIIAB (C), and a target protein: lane 1, the cognate antagonist protein of the kinase, either RsbS (A), RsbV (B), or SpoIIAA (C); lane 2, RsbR; lane 3, YkoB; lane 4, YqhA; lane 5, YojH; lane 6, YtvA; lane 7, YetI; lane 8, YezB. In lanes 1 to 6, the faint bands of lower mobility than the main band are presumed to be dimers and multimers of the target protein.
RsbT and RsbW belong to a class of serine-threonine kinases that are closely related to bacterial histidine kinases (13, 18, 23, 29, 47). The only other such kinase in B. subtilis is SpoIIAB, which regulates activity of the forespore-specific ςF factor by a partner-switching mechanism similar to the one that regulates ςB (3, 9, 13, 29, 34). We therefore tested whether the SpoIIAB kinase could label any of the RsbR paralogs in vitro. As was the case for RsbW, SpoIIAB did not transfer label to any of the paralogs but readily labeled its cognate antagonist protein, SpoIIAA (Fig. 6C, lane 1). We conclude that RsbR, YkoB, YojH, and YqhA are specifically phosphorylated by the RsbT kinase in vitro. These in vitro experiments lend strong support to the genetic analysis, which indicated that the RsbR paralogs function in the environmental signaling branch.
RsbR paralogs interact with each other and with RsbS of the environmental signaling branch.
Reversible protein-protein interactions controlled by serine or threonine phosphorylation are hallmarks of the partner-switching mechanism that regulates ςB activity. These interactions have been detected in cell extracts by immune precipitations (4), sizing columns (10, 11), and affinity chromatography (2, 12); between purified proteins by chemical cross-linking (2, 3, 47) and by nondenaturing gel electrophoresis (9); and also in the yeast two-hybrid system (1, 45, 47). We therefore used the yeast two-hybrid system to determine whether the RsbR paralogs interacted with any of the previously described ςB regulators.
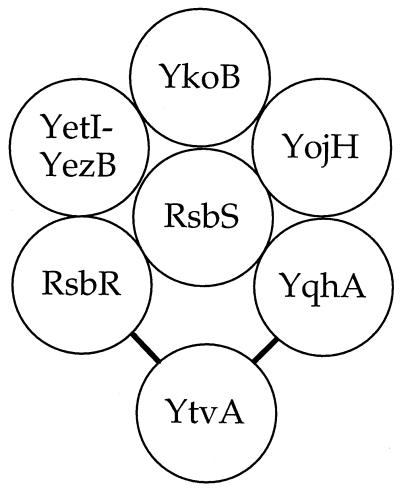
As shown in Table 2, each of the new paralogs was able to activate transcription in the yeast two-hybrid system when paired with either RsbR or RsbS (their homologous regulators in the environmental signaling branch) or with each other. The exception was YtvA, which was only able to activate the system when paired with RsbR or YqhA. In contrast, none of the RsbR paralogs could activate transcription when paired with other Rsb regulators, including the RsbT kinase and the RsbU phosphatase from the environmental branch, the RsbV antagonist protein and the RsbW kinase from the common pathway, and the RsbX feedback phosphatase, nor could they activate transcription when paired with the SpoIIAA antagonist protein or the SpoIIAB kinase from the related partner-switching pathway that governs ςF activity. The absence of detectable interaction between the RsbR paralogs and either RsbV or SpoIIAA is particularly striking. Because RsbS, RsbV, and SpoIIAA are homologous antagonist proteins (22, 23) which bear significant similarity to the C-terminal portions of the RsbR paralogs, this result suggests that the interactions detected among the paralogs and RsbS were specific and not simply a reflection of similar primary structures. In contrast, given the lack of a discernible two-hybrid activation when any RsbR paralog was paired with the RsbT kinase, we infer that the phosphorylation events noted in Fig. 6 involve a transient rather than a long-lived interaction. The positive interactions detected in the two-hybrid system are summarized in Fig. 7.
TABLE 2.
Interaction of RsbR paralogs with other partner-switching regulators
| Fusion protein expressed from pGAD424 | Interactiona with fusion
protein expressed from pGBT9
|
||||
|---|---|---|---|---|---|
| YkoB | YojH | YqhA | YtvA | YetI-YezB | |
| RsbR | ++ | ++ | ++ | ++ | ++ |
| YkoB | ++ | ++ | ++ | 0 | ++ |
| YojH | ND | ND | ND | 0 | ++ |
| YqhA | ++ | + | ++ | ++ | ++ |
| YtvA | ND | 0 | ND | ND | 0 |
| RsbS | ++ | ++ | ++ | 0 | ++ |
| RsbT | 0 | 0 | 0 | 0 | 0 |
| RsbU | 0 | 0 | 0 | 0 | 0 |
| RsbV | 0 | 0 | 0 | 0 | 0 |
| RsbW | 0 | 0 | 0 | 0 | 0 |
| RsbX | 0 | 0 | 0 | 0 | ND |
| SigB | 0 | 0 | 0 | 0 | ND |
| SpoIIAA | 0 | 0 | 0 | 0 | 0 |
| SpoIIAB | 0 | 0 | 0 | 0 | 0 |
++, strong blue color in colony lift assay; +, weak blue color; 0, no blue color detected; ND, not done.
FIG. 7.
Cartoon summarizing the interactions among RsbR, its paralogs, and RsbS. As shown in Table 2, RsbR, YkoB, YojH, YqhA, and the fused YetI-YezB proteins interacted with each other and with the environmental signaling antagonist protein RsbS when paired in the yeast two-hybrid system. In contrast, YtvA interacted only with RsbR and YqhA. None of the newly characterized RsbR paralogs interacted with the downstream RsbT or RsbU regulator from the environmental signaling pathway, with RsbV or RsbW from the common part of the ςB regulatory pathway, or with SpoIIAA or SpoIIAB from the ςF regulatory pathway.
In addition to the qualitative plate assays shown in Table 2, we also conducted β-galactosidase assays to provide a more quantitative estimate of the interactions of YkoB, which was found to be a key negative regulator of ςB activity (Fig. 5B). The reciprocal comparisons shown in Table 3 give similar values and closely parallel the results of the plate assay. However, assuming that each of the tested regulators was equally stable in yeast cells and had equal access to the yeast nucleus, these results suggest that the interaction between YkoB and RsbS was not as strong as that between RsbR and RsbS. In sum, the results of the two-hybrid analysis provide supporting evidence that the RsbR paralogs are associated with the environmental signaling branch.
TABLE 3.
Activation of the yeast two-hybrid system by YkoB paired with RsbR, RsbS, or RsbV
| Fusion protein expressed from pGAD424 | Interaction with Fusion protein expressed
from pGBT9 (sp act)a
|
||
|---|---|---|---|
| YkoB [6]b | RsbR [10] | RsbV [14] | |
| YkoB | NDc | 173 | 6 |
| RsbR | 331 | ND | 11 |
| RsbS | 445 | 2,731 | 3 |
| RsbV | 4 | 4 | 3 |
Average β-galactosidase specific activity (103 ΔA420 min−1 mg−1) for four independent transformants of each pairwise combination. A larger value indicates a stronger interaction.
The basal activation elicited by the protein of interest fused to the DNA binding domain carried by pGBT9 and tested in the absence of a companion pGAD424 plasmid is shown in brackets. The basal activation caused by RsbS in pGBT9 was too high for it to be used in pairwise combinations.
ND, not determined.
DISCUSSION
The data presented here indicate that four of the six new RsbR paralogs are regulators of ςB activity. The exceptions are YetI and YezB, products of an apparently split paralog gene which may function under different conditions that we have tested here. Of the four new paralogs which do not affect ςB regulation, the contribution of YtvA appears to be relatively modest. By contrast, the extraordinary increase in ςB activity caused by the combined loss of RsbR, YkoB, YojH, and YqhA functions indicates that these gene products collectively play an important role in ςB regulation. What is that role, and by what mechanism is it accomplished?
With regard to the mechanism of their action, the available genetic data do not allow us to establish a clear regulatory hierarchy for the RsbR paralogs. Instead, we see an apparent redundancy as phenotypes are progressively revealed in multiply mutant backgrounds. This is in marked contrast to the other regulators in the ςB signal transduction network, which act in a linear dependent pathway (5, 8, 23, 42). One explanation for these results is that the RsbR paralogs might function in a complex, as suggested by the results of our two-hybrid analysis. In such a complex, loss of an RsbR paralog could have a primary effect on its own activity and a secondary effect on the structure of the complex. Consistent with this notion, the RsbR and RsbS regulators are indeed found in a large complex in unstressed B. subtilis cells (11, 36), but there is presently no evidence that the new RsbR paralogs also associate in vivo.
The present genetic analysis confirms and extends earlier work which indicated that RsbR has both positive and negative regulatory roles. Moreover, these dual roles are consistent with an interaction of RsbR and its paralogs, either pairwise or in a complex. For example, the phenotype of a null rsbR mutant indicates that RsbR functions as a positive regulator of ςB activity (1). In contrast, the phenotypes of single and double null mutants indicate that YkoB functions as a negative regulator (Fig. 3D to F) which acts downstream from RsbR in the signal transduction pathway (Fig. 4C). Notably, we also find that loss of rsbR function greatly magnifies the loss of ykoB function in either a triple or quadruple mutant (Fig. 5A). This result suggests that in addition to its positive regulatory role, RsbR also supplements the negative action of YkoB in wild-type cells. These results may explain the opposite phenotypes elicited by rsbR point mutations in which the triplet for the key threonine residue T205 was altered to encode either an alanine or an aspartate (1). The T205D alteration, which presumably mimics the threonine in its phosphorylated state, decreases activation of ςB in response to salt stress even more than does the complete loss of RsbR function in an rsbR null mutant. In contrast, the T205A alteration, which presumably cannot be phosphorylated, increases activation of ςB in response to salt stress even more than does wild-type RsbR. These point mutant phenotypes were taken as evidence that RsbR could exhibit positive or negative regulatory activities depending on the phosphorylation state of T205 (1). The only known biochemical activity of RsbR—its ability to increase the activity of the RsbT serine kinase toward the RsbS antagonist in vitro—is consistent with a positive regulatory role for RsbR (15). On the basis of the results reported here, we can now suggest that one negative role of RsbR is associated with its influence on YkoB function.
In terms of primary sequence, RsbR, YkoB, YojH, and YqhA are the most similar members of this new regulatory family (Fig. 2), and they behave in a similar ways in the three different analyses reported here. In contrast, YtvA is more divergent in both its sequence and behavior. First, YtvA was the only positive regulator identified among the new RsbR paralogs (Fig. 3), and it acted independently of RsbR and YkoB (Fig. 4A and B). Second, it was the only full-length paralog that was not phosphorylated by the RsbT kinase in vitro (Fig. 6). And third, it was the only paralog which did not interact with RbsS and each of the other paralogs in the yeast two-hybrid system (Table 2). Instead, YtvA appeared to interact only with RsbR and YqhA.
The inability of RsbT to phosphorylate YtvA likely reflects the absence of two conserved threonine residues in its C-terminal region. As shown in Fig. 2, these threonines are present in the RsbR, YkoB, YojH, and YqhA paralogs, which are phosphorylated by RsbT, and the in vitro activity of RsbR is known to be controlled by the phosphorylation state of these residues (15). This difference suggests that YtvA responds to an input signal distinct from the one conveyed by the RsbT kinase. Consistent with this notion, YtvA contains a PAS domain in its N-terminal region. PAS domains are usually found in proteins that sense changes in redox, oxygen, or light (40), and indeed, the RsbP phosphatase in the energy-signaling branch contains a PAS domain thought to be important for its in vivo function (41). In bacteria, such domains are commonly associated with a chromophore that confers specificity for the parameter sensed (40). Intriguingly, we found that the hexahistidine-tagged YtvA purified from E. coli had both the yellow color and the spectrum characteristic of a flavin-containing chromophore (data not shown). Moreover, chloroform extraction did not remove this yellow color from YtvA, suggesting a covalent attachment. Although the presence of a PAS domain and potential chromophore hint that YtvA acts in the energy-signaling branch, we infer from the phenotype of the ytvA null mutation that it, like the other RsbR paralogs, acts in the environmental signaling branch (Fig. 3). This inference is supported by the observation that the rsbR-ykoB-yqhA triple null mutations are together epistatic to the ytvA null mutation (data not shown). The environmental signal that might be monitored by the PAS domain in YtvA is presently unknown.
What is the physiological role of the RsbR, YkoB, YojH, YqhA, and YtvA regulators? We can imagine two roles: the transmission of environmental stress signals to ςB and the coordination of ςB activity with other stress response pathways. These roles are not mutually exclusive.
With regard to the transmission of environmental signals, the data presented here indicate that four of the regulators collectively act via the environmental signaling branch of the network, with the complete reversal of the rsbR-ykoB-yojH-yqhA mutant phenotype by loss of rsbT function providing the most convincing evidence. It is therefore attractive to consider that these four regulators convey signals of environmental stress to the established members of the environment-signaling branch, RsbS and RsbT. Whether RsbR, YkoB, YojH, and YqhA are solely responsible for environmental stress transmission might be tested by determining if the system retains the ability to respond to stress in their absence. However, because the high ςB activity elicited by the quadruple mutant approaches the maximum seen when ςB is largely unregulated, as is the case in an rsbS or rsbW null mutant, it is not straightforward to determine whether the quadruple mutant still responds to environmental stress.
With regard to coordination of ςB activity with other stress pathways, these four regulators may serve to tie expression of the ςB regulon to the expanding web of stress response systems that protect B. subtilis cells. These other responses include chemotaxis and motility, the synthesis of degradative enzymes and antibiotics, the development of natural competence for DNA uptake, and the sporulation process. The regulatory interactions among these other systems are becoming increasingly well established (31), but thus far there is no evidence for their coordination with the general stress response. Because the cell devotes considerable resources to the general stress response, including about 5% of its genetic coding capacity (32) and between 25 and 35% of its protein synthetic capacity under growth-limiting conditions (6), it is likely that its action is integrated with other stress systems. In this regard, the essential GTP-binding protein Obg was found to be required for environmental stress activation of the general stress response (35), and in subsequent analysis, Obg appeared to be physically associated with the ribosome (36). Scott and her colleagues (36) interpreted these results to suggest that the ribosome functions as the sensor of environmental stress and communicates these stress signals to ςB via Obg. However, another interpretation is that a signal of sufficient translational capacity is required for the cell to activate ςB in response to environmental stress. Whether the transmission of such a signal also involves the RsbR paralogs remains to be tested.
Whatever their physiological role, RsbR and its six paralogs define a new family of ςB regulators, the larger antagonist family, with RsbR as its prototype. The C-terminal regions of these larger antagonist proteins are similar to the entire lengths of the smaller antagonist family members, defined by RsbS and RsbV of the ςB regulatory network and by SpoIIAA of the ςF network. However, each of the larger antagonist proteins also possesses an N-terminal extension of unknown function. Moreover, although homologs of the smaller antagonist family are surprisingly widespread among eubacteria and are found even among organisms that are not known to possess partner-switching regulation (25), the distribution of the larger antagonist proteins appears to be confined to members of the gram-positive lineage. A Blast 2.0 search found members of the larger antagonist family in Bacillus licheniformis (GenBank accession AAC29504; e value of e-101), Deinococcus radiodurans (AAF12578; 8e-32), Mycobacterium avium (unfinished fragment; 4e-28), and Streptomyces coelicolor (CAB92870; 1e-27), organisms whose genomes also encode clear orthologs of a variety of partner-switching regulators. Thus, the physiological roles and the molecular mechanisms by which these larger antagonist proteins function in B. subtilis are likely to be conserved across a broad spectrum of gram-positive bacteria.
ACKNOWLEDGMENTS
We thank Yee Peing Chia for her assistance in constructing the ykoB null mutant, Ronald Zeigler of the Bacillus Genetic Stock Center for furnishing plasmid pDG792 and B. subtilis strains, and Valley Stewart for his helpful discussions and comments on the manuscript.
This research was supported by Public Health Service grant GM42077 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Akbar S, Kang C M, Gaidenko T A, Price C W. Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis. Mol Microbiol. 1997;24:567–578. doi: 10.1046/j.1365-2958.1997.3631732.x. [DOI] [PubMed] [Google Scholar]
- 2.Alper S, Dufour A, Garsin D A, Duncan L, Losick R. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol. 1996;260:165–177. doi: 10.1006/jmbi.1996.0390. [DOI] [PubMed] [Google Scholar]
- 3.Alper S, Duncan L, Losick R. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell. 1994;77:195–205. doi: 10.1016/0092-8674(94)90312-3. [DOI] [PubMed] [Google Scholar]
- 4.Benson A K, Haldenwang W G. Bacillus subtilis ςBis regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci USA. 1993;90:2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson A K, Haldenwang W G. Characterization of a regulatory network that controls ςB expression in Bacillus subtilis. J Bacteriol. 1992;174:749–757. doi: 10.1128/jb.174.3.749-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhardt J, Volker U, Volker A, Antelmann H, Schmid R, Mach H, Hecker M. Specific and general stress proteins in Bacillus subtilis—a two-dimensional protein electrophoresis study. Microbiology. 1997;143:999–1017. doi: 10.1099/00221287-143-3-999. [DOI] [PubMed] [Google Scholar]
- 7.Boylan S A, Redfield A R, Brody M S, Price C W. Stress-induced activation of the ςB transcription factor of Bacillus subtilis. J Bacteriol. 1993;175:7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boylan S A, Rutherford A, Thomas S M, Price C W. Activation of Bacillus subtilis transcription factor ςBby a regulatory pathway responsive to stationary-phase signals. J Bacteriol. 1992;174:3695–3706. doi: 10.1128/jb.174.11.3695-3706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diederich B, Wilkinson J F, Magnin T, Najafi M, Erringston J, Yudkin M D. Role of interactions between SpoIIAA and SpoIIAB in regulating cell-specific transcription factor ςF of Bacillus subtilis. Genes Dev. 1994;8:2653–2663. doi: 10.1101/gad.8.21.2653. [DOI] [PubMed] [Google Scholar]
- 10.Dufour A, Haldenwang W G. Interactions between a Bacillus subtilisanti-sigma factor (RsbW) and its antagonist (RsbV) J Bacteriol. 1994;176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dufour A, Voelker U, Voelker A, Haldenwang W G. Relative levels and fractionation properties of Bacillus subtilis ςBand its regulators during balanced growth and stress. J Bacteriol. 1996;178:3701–3709. doi: 10.1128/jb.178.13.3701-9sigma.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duncan L, Alper S, Losick R. SpoIIAA governs the release of the cell-type specific transcription factor ςFfrom its anti-ς factor SpoIIAB. J Mol Biol. 1996;260:147–164. doi: 10.1006/jmbi.1996.0389. [DOI] [PubMed] [Google Scholar]
- 13.Duncan L, Losick R. SpoIIAB is an anti-sigma factor that binds to and inhibits transcription by regulatory protein ςF from Bacillus subtilis. Proc Natl Acad Sci USA. 1993;90:2325–2329. doi: 10.1073/pnas.90.6.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan M L, Kalman S S, Thomas S M, Price C W. Gene encoding the 37,000-dalton minor sigma factor of Bacillus subtilisRNA polymerase: isolation, nucleotide sequence, chromosomal locus, and cryptic function. J Bacteriol. 1987;169:771–778. doi: 10.1128/jb.169.2.771-778.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaidenko T A, Yang X, Lee Y M, Price C W. Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the environmental stress signaling pathway of Bacillus subtilis. J Mol Biol. 1999;288:29–39. doi: 10.1006/jmbi.1999.2665. [DOI] [PubMed] [Google Scholar]
- 16.Guerout-Fleury A M, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 17.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris R A, Popov K M, Zhao Y, Kedishvili N Y, Shimomura Y, Crabb D W. A new family of protein kinases—the mitochondrial protein kinases. Adv Enzyme Regul. 1995;35:147–162. doi: 10.1016/0065-2571(94)00020-4. [DOI] [PubMed] [Google Scholar]
- 19.Hecker M, Volker U. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the ςBregulon. Mol Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 20.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982;150:804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igo M, Lampe M, Ray C, Schafer W, Moran C P, Jr, Losick R. Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis. J Bacteriol. 1987;169:3464–3469. doi: 10.1128/jb.169.8.3464-3469.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalman S, Duncan M L, Thomas S M, Price C W. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilisRNA polymerase. J Bacteriol. 1990;172:5575–5585. doi: 10.1128/jb.172.10.5575-5585.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang C M, Brody M S, Akbar S, Yang X, Price C W. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor ςBin response to environmental stress. J Bacteriol. 1996;178:3846–3853. doi: 10.1128/jb.178.13.3846-3853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang C M, Vijay K, Price C W. Serine kinase activity of a Bacillus subtilisswitch protein is required to transduce environmental stress signals but not to activate its target PP2C phosphatase. Mol Microbiol. 1998;30:189–196. doi: 10.1046/j.1365-2958.1998.01052.x. [DOI] [PubMed] [Google Scholar]
- 25.Koonin E V, Aravind L, Galperin M Y. A comparative-genomic view of the microbial stress response. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: American Society for Microbiology; 2000. pp. 417–444. [Google Scholar]
- 26.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 27.LeBlanc D J, Lee L N, Inamine J M. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:1804–1810. doi: 10.1128/aac.35.9.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 29.Min K T, Hilditch C M, Diederich B, Errington J, Yudkin M D. ςF, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-sigma factor that is also a protein kinase. Cell. 1993;74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- 30.Moran C P, Jr, Johnson W C, Losick R. Close contacts between sigma 37-RNA polymerase and a Bacillus subtilischromosomal promoter. J Mol Biol. 1982;162:709–713. doi: 10.1016/0022-2836(82)90399-0. [DOI] [PubMed] [Google Scholar]
- 31.Msadek T. When the going gets tough: survival strategies and environmental signaling networks in Bacillus subtilis. Trends Microbiol. 1999;7:201–207. doi: 10.1016/s0966-842x(99)01479-1. [DOI] [PubMed] [Google Scholar]
- 32.Price C W. Protective function and regulation of the general stress response in Bacillus subtilis and related gram-positive bacteria. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: American Society for Microbiology; 2000. pp. 179–197. [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Schmidt R, Margolis P, Duncan L, Coppolecchia R, Moran C P, Jr, Losick R. Control of developmental transcription factor ςF by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:9221–9225. doi: 10.1073/pnas.87.23.9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott J M, Haldenwang W G. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor ςB. J Bacteriol. 1999;181:4653–4660. doi: 10.1128/jb.181.15.4653-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott J M, Ju J, Mitchell T, Haldenwang W G. The Bacillus subtilis GTP binding protein Obg and regulators of the ςBstress response transcription factor cofractionate with ribosomes. J Bacteriol. 2000;182:2771–2777. doi: 10.1128/jb.182.10.2771-2777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smirnova N, Scott J, Voelker U, Haldenwang W G. Isolation and characterization of Bacillus subtilis sigBoperon mutations that suppress the loss of the negative regulator RsbX. J Bacteriol. 1998;180:3671–3680. doi: 10.1128/jb.180.14.3671-3680.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stahl M L, Ferrari E. Replacement of the Bacillus subtilissubtilisin structural gene with an in vitro-derived deletion mutation. J Bacteriol. 1984;158:411–418. doi: 10.1128/jb.158.2.411-418.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 40.Taylor B L, Zhulin I B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vijay K, Brody M S, Fredlund E, Price C W. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the ςB transcription factor of Bacillus subtilis. Mol Microbiol. 2000;35:180–185. doi: 10.1046/j.1365-2958.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- 42.Voelker U, Dufour A, Haldenwang W G. The Bacillus subtilis rsbU gene product is necessary for RsbX-dependent regulation of ςB. J Bacteriol. 1995;177:114–122. doi: 10.1128/jb.177.1.114-122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voelker U, Luo T, Smirnova N, Haldenwang W. Stress activation of Bacillus subtilis ςB can occur in the absence of the ςBnegative regulator RsbX. J Bacteriol. 1997;179:1980–1984. doi: 10.1128/jb.179.6.1980-1984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voelker U, Voelker A, Haldenwang W G. Reactivation of the Bacillus subtilis anti-ςBantagonist, RsbV, by stress- or starvation-induced phosphatase activities. J Bacteriol. 1996;178:5456–5463. doi: 10.1128/jb.178.18.5456-5463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voelker U, Voelker A, Haldenwang W G. The yeast two-hybrid system detects interactions between Bacillus subtilis ςBregulators. J Bacteriol. 1996;178:7020–7023. doi: 10.1128/jb.178.23.7020-7023.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang W G. Separate mechanisms activate ςB of Bacillus subtilisin response to environmental and metabolic stresses. J Bacteriol. 1995;177:3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X, Kang C M, Brody M S, Price C W. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]