Abstract
Erwinia chrysanthemi exports degradative enzymes by using a type I protein secretion system. The proteases secreted by this system lack an N-terminal signal peptide but contain a C-terminal secretion signal. To explore the substrate specificity of this system, we have expressed the E. chrysanthemi transporter system (prtDEF genes) in Escherichia coli and tested the ability of this ABC transporter to export hybrid proteins carrying C-terminal fragments of E. chrysanthemi protease B. The C terminus contains six glycine-rich repeated motifs, followed by two repeats of the sequences DFLV and DIIV. Two types of hybrid proteins were assayed for transport, proteins with the 93-residue-protease-B C terminus containing one glycine-rich repeat and both hydrophobic terminal repeats and proteins with the 181-residue C terminus containing all repeat motifs. Although the shorter C terminus is unable to export the hybrids, the longer C terminus can promote the secretion of hybrid proteins with N termini as large as 424 amino acids, showing that the glycine-rich motifs are required for the efficient secretion of these hybrids. However, the secretion of hybrids occurs only if these proteins do not carry disulfide bonds in their mature structures. These latter results suggest that disulfide bond formation can occur prior to or during the secretion. Disulfide bonds may prevent type I secretion of hybrids. One simple hypothesis to explain these results is that the type I channel is too narrow to permit the export of proteins with secondary structures stabilized by disulfide bonds.
Nonpathogenic strains of Escherichia coli, such as the laboratory strain E. coli K-12, export few proteins to the external medium (53). The secretion of proteins in E. coli depends on a type II (Sec-dependent) mechanism in which unfolded proteins carrying an N-terminal signal sequence are transported across the inner membrane to the periplasm and then processed and folded in the periplasm prior to their translocation across the outer membrane. To initiate type II secretion, it is thought that a polypeptide and its signal peptide must be fully extended to cross the inner membrane (3). After crossing the inner membrane, proteins are folded in the periplasm in a process assisted by chaperones. In addition, the periplasm contains the enzymes required for the correct assembly of disulfide bonds to complete protein folding (5, 48).
In contrast, many gram-negative pathogens including enteropathogenic E. coli (38), Yersinia spp. (36), Salmonella enterica serovar Typhimurium (39), and Vibrio cholerae (35) export virulence factors required for host colonization and survival. These virulence factors are secreted by mechanisms that operate independently from the type II system, mechanisms that involve specialized membrane transport apparatuses (64). A subset of virulence factors is exported by a type I mechanism in which three proteins assemble to form a transmembrane structure that couples the export of protein substrates with ATP hydrolysis. These ABC transporters include the type I mechanisms involved in the secretion of a Serratia marcescens metalloprotease (43), E. coli beta-hemolysin (30, 61), Bordetella pertussis adenylate cyclase (29), Pseudomonas aeruginosa alkaline protease (23), and Pseudomonas fluorescens lipase (1). All of the substrates for these ABC transport systems have C-terminal signal sequences.
Many phytopathogens, including members of the genus Erwinia, also export proteins critical for virulence by using different secretion mechanisms. Among these, the secretion of metalloproteases in Erwinia chrysanthemi also proceeds by a type I mechanism (20). E. chrysanthemi secretes four proteases (19, 27, 28) to the external medium. Secretion requires the products of the prtD, prtE, and prtF genes, which also recognize a C-terminal signal sequence in their substrates (20). Like other ABC transporters, the products of the prtD, prtE, and prtF genes are thought to form a channel that allows the export of proteases directly from the cytoplasm to the external medium. The PrtD and PrtE proteins are associated with the inner membrane, and the PrtF protein is associated with the outer membrane (64). The large (ca. 60-kDa) PrtD protein has an N terminus with six transmembrane segments and a hydrophilic C terminus with a classical ATP binding motif (42). Studies with PrtD in vitro show that its P-type ATPase activity is inhibited by peptides carrying the C-terminal signal sequence, arguing that substrate translocation by this type I system requires the hydrolysis of ATP (18). PrtE, which belongs to the family of the membrane fusion proteins (MFP), has a short N terminus presumably anchored in the inner membrane, followed by a large hydrophilic periplasmic domain and a hydrophobic C terminus thought to interact with the outer membrane (22). It has been suggested that proteins in the MFP family may form transmembranous pores for their substrates that traverse the periplasmic space (22). The function of PrtF, associated with the outer membrane, is not well understood, as is the case for other ABC transporters (9).
Based on studies on the C-terminal secretion signal of E. chrysanthemi proteases, this signal has been located in the last 50 amino acids (20). Using an E. chrysanthemi protease, PrtG, it has been shown that the smallest C-terminal sequence allowing efficient secretion contains the last 29 amino acids of PrtG. This region contains two four-amino-acid motifs which are essential for protease secretion (28). The E. chrysanthemi C-terminal protease signal can promote the specific secretion of fused passenger polypeptides. However, the four-amino-acid terminal motifs are not sufficient to promote secretion. Studies on fusion protein secretion have revealed the role of a domain located just upstream from the C-terminal signal on most of the proteases and lipases secreted by the type I system. These proteins carry a domain of glycine-rich sequences (GGXGXD) that is repeated several times depending on the protein (66). It has been shown that these repeats play a critical role in the secretion of some polypeptide passengers, suggesting that they may act as internal chaperones (41).
Because E. chrysanthemi is closely related to E. coli, we are testing whether proteins expressed in E. coli hosts may be transported efficiently using the Erwinia chrysanthemi type I secretion system. Understanding the substrate requirements for the secretion of proteins expressed in E. coli by this system would facilitate the production and purification of recombinant proteins of medical relevance, as well as have other important biotechnological applications. In this work, we extend the results of previous work showing that the signal sequence required for the secretion of hybrid eukaryotic proteins carrying the C terminus of E. chrysanthemi protease B is larger than that required for the secretion of protease B hybrids made by fusions to prokaryotic proteins (41) and showing that similar rules appear to apply to the transport of hybrids made between eukaryotic proteins and protease B. Surprisingly, we find that the E. chrysanthemi PtrDEF ABC transport system cannot transport hybrid eukaryotic proteins that have disulfide bonds in their mature structures.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains used in this work are listed in Table 1. AD494(DE3) cells were from Novagen and were grown according to the vendor's instructions. E. coli JM105 and DH5α were from Pharmacia, and TOP10 was from Invitrogen. E. coli and Erwinia carotovora strains were grown in Luria-Bertani (LB) medium at 37 and 30°C, respectively, with constant shaking (200 rpm). Plasmids are listed in Table 2. Plasmids pRUW4 and pRUW500 (20) were kindly provided by P. Delepelaire. Isolation of recombinant plasmids was done by using the alkaline lysis method of Birnboim and Doly (10), with slight modifications. Bacterial cells carrying recombinant plasmids were grown in medium supplemented with the required antibiotics (100 μg of ampicillin/ml, 30 μg of chloramphenicol/ml, or 40 μg of kanamycin/ml).
TABLE 1.
List of bacterial strains used in this work
| Straina | Relevant properties | Source or reference |
|---|---|---|
| E. coli K-12 | Wild type | Our laboratory |
| E. coli B | Wild type | Our laboratory |
| E. coli C600 | supE44 hsdR thi-1 thr-1 leuB6 lacY1 tonA21 | Stratagene |
| E. coli DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Pharmacia Biotech |
| E. coli JM105 | supE endA sbcB15 hsdR4 rpsL thiΔ(lac-proAB) | 68 |
| E. coli TOP10 | mcrA Δ(mrr-hsdRMS-mcrBC) φ80ΔlacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL (Strr) endA1 nupG | Invitrogen Corp. |
| E. coli AD494(DE3) | Δara leu7967 ΔlacX74 ΔphoAPvuII phoR ΔmalF3 F−[Δlac+ (lacIq)pro] trxB::kan(DE3) | Novagen |
| E. carotovora subsp. carotovora | Ecc193 | A. K. Chatterjee |
Antibiotics were added to plasmid-transformed bacterial cultures or when required at the following concentrations: 40 μg of kanamycin/ml; 100 μg of ampicillin/ml; 30 μg of chloramphenicol/ml; 50 μg of tetracycline/ml.
TABLE 2.
List of plasmids used in this work
| Plasmid(s)a | Relevant properties | Source or reference |
|---|---|---|
| pSE280 (Apr), pSE420 (Apr) | Cloning and expression vectors | Invitrogen |
| pGEM-T, pGEMT Easy (Knr) | Vector for cloning of PCR fragments | Promega Corp. |
| pCR2.1 (Apr) | Vector for cloning of PCR fragments | Invitrogen |
| pRUW500 (Apr) | prtB (E. chrysanthemi protease B gene) | 20 |
| pRUW4 (Cmr) | prtD, prtE, and prtF genes | 20 |
| pACYC184 | Low-copy-number cloning vector | New England BioLabs |
| pKK233-2 (Apr) | IPTG-inducible expression vector | Pharmacia Biotech |
| pACYC-dsbC | E. chrysanthemi dsbC gene in pACYC184 | This work |
| pGFP | GFP gene in pUC19 | Clontech |
| pGFP-181 | GFP-protease B long hybrid gene in pSE420 | This work |
| pSE280-93CTPB | Short C-terminal protease B gene in pSE280 | This work |
| pSE420-93CTPB | Short C-terminal protease B gene in pSE420 | This work |
| pSE420-181CTPB | Long C-terminal protease B in pSE420 (Eco47III-HindIII) | This work |
| pSE420-181CTPB2 | Long C-terminal protease B in pSE420 (SmaI-HindIII) | This work |
| pECH42 | T. harzianum endochitinase cDNA | M. Gidekel |
| pCHIT-9, pCHIT-1 | Endochitinase gene in pSE420 | This work |
| pCHIT-93CTPB | Endochitinase-protease B short hybrid in pSE420 | This work |
| pCHIT-181CTPB | Endochitinase-protease B long hybrid in pSE420 | This work |
| pERY727 | Synthetic hEPO gene in pSVK10 | A. Venegas |
| phEPO-93CTPB | hEPO-protease B short hybrid gene in pSE420 | This work |
| phEPO-181CTPB | hEPO-protease B long hybrid gene in pSE420 | This work |
| pOmpC-181CTPB | S. typhi OmpC (205 amino acids) in pSE420-181CTPB-2 | This work |
| pOmpC-Bgl2 | A BglII insert of the S. typhi ompC gene in pUC19 | 69 |
| pβGAL-181CTPB | Initial codons of β-galactosidase plus polylinker-CTPB in pCR2.1 | This work |
| pKKTGH-23 | tGH cDNA in pKK233-2 | 49 |
| ptGH-93CTPB | tGH-protease B short hybrid gene in pSE420 | This work |
| ptGH-181CTPB | tGH-protease B long hybrid in pSE420 | This work |
Preparative amounts of plasmids were obtained by using QIAGEN columns according to vendor instructions.
DNA manipulations.
Ligations and transformations were done using standard methods (54). Restriction fragments and linearized plasmids were purified by the GeneClean kit (Bio 101) or the Wizard kit (Promega). Vector DNAs were usually dephosphorylated with calf intestine alkaline phosphatase as described by Chaconas and van de Sande (13). Dephosphorylation reactions were stopped with phenol extraction, and dephosphorylated DNA was purified after chloroform-isoamyloalcohol (24:1) extraction followed by ethanol precipitation. Bacterial transformation of E. coli and Erwinia strains was done by electroporation (47). In some cases, E. coli cells were transformed by using the calcium chloride cell permeation method (67). For electroporation, a Bio-Rad gene pulser apparatus, model 2-89, coupled to a pulse controller was used. A high voltage (2,500 V) was applied to 40 μl of electrocompetent cells contained in a cuvette with a 0.2-cm electrode separation. Electrocompetent cells were prepared as described by Miller (47). Cells were recovered in SOC medium (54) at 30°C for 60 min, and electroporants were selected on LB agar plates with appropriate antibiotics. DNA sequencing was done by the dideoxy chain termination method (55), following the procedure of Chen and Seeburg (15) for double-stranded plasmid DNA templates. The Sequenase kit version 2.0 (Amersham Pharmacia Biotech) was used according to the instructions, with [α-35S]dATP as labeled substrate. Gels were exposed for 1 to 2 days to X-OmatAR Kodak film.
PCR amplifications.
PCRs were performed in thin-walled Eppendorf tubes, in a final volume of 100 μl containing 10 μl of 10× PCR buffer (200 mM Tris HCl [pH 8.4], 500 mM KCl), 3 μl of 50 mM MgCl2, 16 μl of 1.25 mM deoxynucleotide triphosphate, 50 to 75 pmol of each primer, and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer). Standard conditions were as follows: 30 to 35 cycles of 2 min at 94°C, 1 min at 55°C, and 1 min at 72°C, followed by a terminal elongation step at 72°C for 7 min. Primers for amplification of various fragments are listed in Table 3.
TABLE 3.
Oligonucleotide primer used in gene amplifications and hybrid constructions
| Name | 5′→3′ oligonucleotide sequencea | Restriction site(s) |
|---|---|---|
| DSBC-1 | ACC ACC ATG GAT GAC GCG GCG ATC AAA CAG GCG CTG | NcoI |
| DSBC-2 | AGC ACA AGC TTC CAT TAG CCG CCT GAT TTC AGC GAG | HindIII |
| PSE-2801 | CCG GCC CGT ATA ATG TGT GG | None |
| PSE-2802 | CAG ACC GCT TCT GCG TTC TG | None |
| POLYG | A TTA CTG CAG GCC GGC GGC ACC GAC ACC TTC GAC TTC TCC | PstI-NaeI |
| POLYG-2 | ATT ACC ATT GCA CCC GGG GGC ACC GAC ACC TTC GAC TTC TCC | SmaI |
| PRTB-4 | TC GCG AAG CTT TTA CAC AAT AAT ATC GGA TTG GCG | HindIII |
| CHIT-1 | AAC ACC ATG GCC CAG GCC ACT CTC ATT TCT GCA TCT CCT | NcoI |
| CHIT-4 | AAA CCG CGG TTA GCC GGC GTT GAG ACC GCT TCG GAT GTT ATC ATA CTG | SstI, NaeI |
| GFPNH-3 | C GCC AAG CTT GCA TGC CTG CAG GTC GAC TCT AGA | HindIII |
| GFPCT-4 | G GTA CCC GGG TTT GTA TAG TTC ATC CAT GCC ATG TG | SmaI |
| OC-1 | AG CCC CGG GCC ATG GAA ATT TAT AAT AAA GAC GGC AAC AAA TTA | NcoI |
| OC-51 | A ATA CCC GGG AGC GGT GTT GTT CTG ATC GGC AGT ACG TTT AG | SmaI |
| EPO-1 | T AGG ACC ATG GCG CCA CCA CGC CTG ATC TGT GAC AGC | NcoI |
| EPO-2 | T TAG GAT ATC TCG GTC CCC TGT CCG GCA GGC CTC CCC | EcoRV |
| TGH-21 | A TGA CCC ATG GAA AAC CAA CGT CTC TTC AAC ATC | NcoI |
| TGH-22 | G GCC GAT ATC CAG AGT GCA GTT GGC CTC CAG TGA | EcoRV |
Oligonucleotides were synthesized by Synthaid Biotechnology Inc., Ottawa, Canada. In all cases, signal peptide sequences were excluded in the primer design to avoid the use of a peptide signal-dependent secretion system during hybrid secretion evaluation. Restriction sites included in the oligonucleotides are underlined.
Construction of secretion vectors pSE420-93CTPB and pSE420-181CTPB.
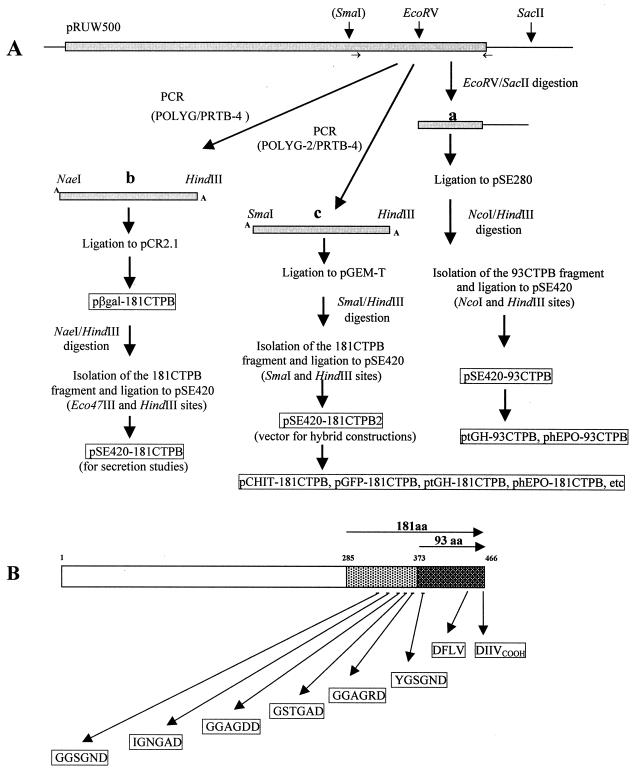
For the vector pSE420-93CTPB, a fragment containing the protease B coding region between residues Tyr 373 and Val 466 was released from plasmid pRUW500 by EcoRV and SacII digestions (Fig. 1) and inserted at the same sites in the pSE280 vector. Then, because of an additional EcoRV site present in pSE420, the 93CTPB region was transferred to pSE420 as a NcoI-HindIII fragment. The 181CTPB coding sequence was amplified by PCR from plasmid pRUW500 in two ways, using the primers POLYG and PRTB-4 (providing a NaeI site to be ligated in frame for the hybrid fusion at the Eco47III site of the vector) or using the pair POLYG-2 and PRTB-4 (providing an SmaI site for in-frame hybrid fusions) (see strategies in Fig. 1). Both amplified fragments were separately ligated to the pCR2.1 and the pGEM-T vectors. The first construction had the initial β-galactosidase residues in frame with the 181CTPB region and resulted in plasmid pβGAL-181CTPB. A fragment containing the 181CTPB region was isolated from pβGAL-181CTPB after digestion with NaeI and HindIII, and the same fragment was obtained from the pGEM-T construction after digestion with SmaI and HindIII. Each 181CTPB fragment was independently ligated into the vector pSE420 that had been previously digested with Eco47III and HindIII or SmaI and HindIII. The first construction, in which the plasmid once ligated has lost both the NaeI and the Eco47III sites, was used for secretion studies of the carboxyl terminus. The second construction, pSE420-181CTPB2, preserved intact the SmaI site after ligation, and this was utilized to generate the hybrids with different passenger proteins, as shown in Fig. 1.
FIG. 1.
Strategies for construction of the secretion vectors pSE420-93CTPB and pSE420-181CTPB and hybrid proteins and location of glycine-rich motifs in E. chrysanthemi protease B structure. (A) Relevant restriction endonuclease sites on the carboxyl terminus of the prtB gene cloned in pRUW500 and three strategies for construction of secretion vectors and hybrid genes are shown. The restriction fragment (a) corresponds to the 93CTPB coding region and was initially ligated into the EcoRV and SacII sites of pSE280 and then transferred to pSE420. The 181CTPB fragments (b and c) were obtained by PCR amplification as explained in the text. Relevant plasmid constructions are boxed. Details of hybrid constructions are given in Materials and Methods. The SmaI site shown in parentheses at the protease B gene was created by PCR to allow the in-frame ligation required for hybrid constructions. (B) Glycine-rich and hydrophobic motifs are displayed in the CTPB region. The vertical and diagonal arrows indicate the relative positions of the motifs (boxed sequences) along the amino acid protease B sequence. Numbers on the protease B graphic representation indicate amino acid positions. The 93- and 181-CTPB regions are shown as stippled boxes.
Construction of protease B hybrid genes.
To construct plasmids expressing endochitinase hybrids, the endochitinase gene was amplified by PCR using the ECh42 cDNA as the template (33) and primers CHIT-1 and CHIT-4 (Table 3). The use of primer CHIT-4 allowed the fusion in frame to the vector pSE420-181CTPB-2 after SmaI and NcoI digestions of the purified PCR fragment and ligation to the NcoI and SmaI sites of the vector. Ligation products were transformed into E. coli HB101 cells. Two pCHIT-181CTPB recombinant plasmids were selected for further studies. Plasmids expressing green fluorescent protein (GFP) hybrids were made using a similar strategy. A PCR fragment containing full-length GFP (238 codons) was amplified using template plasmid pGFP and primers GFP-NH3 and GFP-CT4. After NcoI and SmaI digestions, the fragment was ligated to pSE420-181CTPB2. The OmpC-181CTPB hybrid was constructed by amplifying the first 204 codons of the ompC coding region, using plasmid pOmpCBgl-2 as the template and primers OC-1 and OC-51. The PCR fragment was digested with NcoI and SmaI and ligated to pSE420-181CTPB2.
Plasmids hEPO-93CTPB and hEPO-181CTPB were constructed by amplifying a purified BglII restriction fragment obtained from plasmid pEry720 carrying a synthetic human erythropoietin (hEPO) gene (A. Venegas, unpublished results) as the PCR template and primers EPO-1 and EPO-2. The 518-bp PCR fragment was ligated to pGEM-T to make plasmid pGEM-hEPO-1, which was digested with NcoI and EcoRV to generate a fragment with the hEPO coding region lacking the coding sequences for the signal peptide and stop codon. This fragment was ligated into vectors pSE420-93CTPB and pSE420-181CTPB2 and introduced into E. coli C600. Construction of plasmids tGH-93CTPB and tGH-181CTPB was accomplished in a similar way as described for the construction of hEPO hybrids, using plasmid pKKTGH23 as the template and primers TGH-21 and TGH-22.
Cloning of the E. chrysanthemi dsbC gene.
The dsbC gene (57) was amplified without its signal peptide coding region from E. chrysanthemi chromosomal DNA prepared by the method of Grimberg et al. (31), using primers DSBC-1 and DSBC-2. A 1.4-kb amplified fragment was ligated into pGEM-T and introduced into DH5α cells by electroporation. An insert carrying dsbC was generated with NcoI and HindIII and then subcloned into plasmid pKK233-2, which carries the trc promoter. A BamHI-HindIII fragment from this construct was ligated into the BamHI and HindIII sites of low-copy-number plasmid pACYC184, and the recombinant plasmid was introduced into E. coli JM105 cells by electroporation and selection for Cmr Tcs recombinants. Expression of the DsbC protein was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), after induction of exponential cells with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), revealing an overexpressed protein with an apparent molecular mass of 42 kDa, as expected.
Detection of SH residues in hybrid proteins by covalent modification with iodoacetic acid.
Six milliliters of fresh overnight cultures was centrifuged at 2,000 × g for 5 min in an Eppendorf microcentrifuge. Cell pellets were suspended in double-distilled H2O (1/10 initial volume) and boiled for 5 min to inactivate disulfide bond formation enzymes. Cells were lysed by adding 300 μl of 25 mM Tris HCl (pH 8.0), 10 mM EDTA, and 50 mM glucose, containing 4 mg of lysozyme/ml, and vortexing, followed by incubation for 5 min at 25°C. Total proteins were precipitated with 10% trichloroacetic acid (TCA), and the pellets were washed twice with acetone, dried, and then suspended in 120 μl of 125 mM Tris HCl–6 M guanidinium chloride (pH 8.0) containing 20 mM iodoacetic acid and incubated for 10 min at 25°C. In order to remove iodoacetic acid remnants, a 50-μl aliquot was filtered through a 0.5-ml Sephadex G-25 minicolumn set on a disposable blue tip and previously equilibrated with 125 mM Tris HCl (pH 8.0). Proteins with modified SH residues change their electrophoretic mobility on an 8 M urea-polyacrylamide gel. The gel was prepared as described by Laemmli (40) but containing urea instead of SDS. No upper gel was used. An 8% polyacrylamide gel was run at 25°C using 150 V. The gel was made in 350 mM Tris HCl buffer (pH 8.9). Electrophoresis running buffer was 125 mM Tris-glycine (pH 9.0) without SDS or β-mercaptoethanol (β-ME). All samples were boiled for 5 min and some of them were reduced with 10 mM β-ME.
Purification of protease B and preparation of polyclonal antibodies.
The 52-kDa protease B protein from E. chrysanthemi cloned in plasmid pRUW500 (20) was expressed in E. coli C600 after 1 mM IPTG induction of an overnight culture for 6 h. The protein band was purified by SDS-PAGE (7% polyacrylamide), and an appropriate gel slice was electroeluted with a Bio-Rad electroeluter as described in the vendor instructions. Anti-protease B polyclonal antibody was obtained as described previously(16), using three 300-μg injections of protease B on rabbit pads. Sera from two rabbits were tested by an enzyme-linked immunosorbent assay, and each milliliter of a 1:10 dilution of serum in distilled water was adsorbed twice against a 5-ml extract of sonicated E. coli C600 cells previously immobilized on a 5-cm2 nitrocellulose filter.
Protein gel electrophoresis and Western blotting for detection of protease B hybrids.
Proteins with a molecular mass larger than 15 kDa were separated using SDS-PAGE (12 to 15% polyacrylamide) (40). Protein staining was done with 0.1% Coomassie blue R-250 in a 50% methanol–10% acetic acid solution for 1 to 2 h at 25°C. Gel destaining was done in 10% methanol–10% acetic acid at 25°C. Western blotting using anti-protease B polyclonal antibodies was performed by the method of Towbin et al. (63) with few modifications. After transfer, nitrocellulose filters were blocked with phosphate-buffered saline (PBS)–2% bovine serum albumin (BSA) for 2 h at 25°C. Prior to its use, anti-protease B antibody was preadsorbed with E. coli C600 total protein extract as described by Zaror (69). The antibody was added to a final dilution of 1:1,000 in PBS–2% BSA, and the filters were incubated for 1 h and then washed in PBS–0.1% Tween 20 three times for 5 min each. The filters were then incubated with anti-rabbit immunoglobulin G (diluted 1:1,000 in PBS–2% BSA) conjugated to horseradish peroxidase or alkaline phosphatase (Bio-Rad). For peroxidase reactions, the filters were developed in 25 ml of 50 mM Tris HCl (pH 7.4)–200 mM NaCl containing 15 mg of 4-chloro-1-naphthol (previously dissolved in 5 ml of methanol) and 60 μl of 30% hydrogen peroxide. For phosphatase reactions, the filters were incubated in 10 ml of a mixture of 100 mM Tris HCl (pH 9.5), 100 mM NaCl, 5 mM MgCl2, 66 μl of a 50-mg/ml concentration of nitroblue tetrazolium, and 33 μl of a 50-mg/ml concentration of 5-bromo-4-chloro-3-indolylphosphate. Western blot analyses of protease B hybrids were carried out using a 1:1,000 anti-protease B antibody dilution. To detect hybrids in supernatants, supernatants were concentrated 10-fold by precipitation with 10% TCA (32) prior to electrophoretic separation. In some cases, to standardize protein loading, the protein content of extracts was determined by the method of Bradford (11). Chemiluminescence assays were done with a Renaissance kit from NEN Life Science Products (Boston, Mass.).
RESULTS
Construction of plasmid vectors with E. chrysanthemi PrtB C-terminal signal sequences.
E. chrysanthemi secretes four proteases using its type I secretion system, proteases A, B, C, and G. These four proteases terminate with a similar 4-amino-acid motif, represented by the sequence Dhhh, in which h represents a hydrophobic amino acid. Initial studies have shown that this motif is critical for the secretion of protease G, because the addition of a single amino acid residue to the C terminus of protease G prevents its export by the type I mechanism (20, 28). Although fusion of the coding sequence for the 40 C-terminal residues of protease B to the coding region for the first 200 residues of amylomaltase results in a hybrid protein that is secreted in E. coli, other larger fusion proteins that carry only this motif are not secreted efficiently by the type I mechanism (20). In addition to this C-terminal motif, it has become clear that an additional glycine-rich motif, repeated immediately upstream of the C terminus of the E. chrysanthemi proteases, is required for the efficient type I transport of hybrids formed between heterologous proteins and the C termini of the secreted proteases (41). This additional motif has the consensus sequence GGXGXD, in which X represents any amino acid, and is repeated several times in all proteins secreted by a type I mechanism.
To explore the C-terminal requirements for type I secretion by the E. chrysanthemi system in greater detail, we constructed plasmid vectors carrying sequences encoding two different lengths of the C terminus of protease B. The PrtB sequences in these vectors correspond to the 93 or 181 C-terminal residues of protease B. Both sequences include the C-terminal signal essential for type I secretion, whereas only the latter vector encodes the glycine-rich repeats required for the secretion of larger, hybrid proteins. To construct these vectors, we obtained different portions of the 3′ end of the prtB gene and subcloned these products into plasmid expression vector pSE420, as described in Materials and Methods. These constructions allowed us to fuse part of the polylinker region of pSE420 in frame to the protease B C termini. This part of the polylinker region presumably is translated as a tract of 46 amino acid residues.
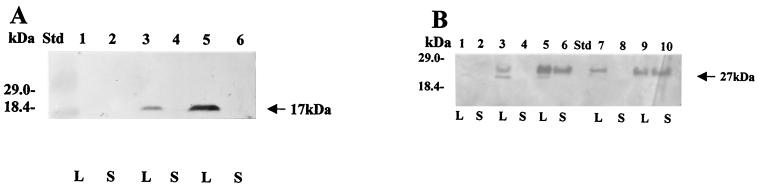
We tested the ability of the E. chrysanthemi type I secretion system to export the small fusion proteins expressed from plasmid vectors pSE420-93CTPB and pSE420-181CTPB in E. coli C600 cells. These proteins are predicted to be 139 and 229 amino acids in length, respectively. Western blot analysis of the proteins present in the total induced lysates of E coli C600 carrying these plasmids probed with anti-protease B antibodies reveals bands corresponding to fusion products with apparent molecular masses of 17 (Fig. 2A) and 27 (Fig. 2B) kDa, as expected. Plasmid pRUW4, which expresses the E. chrysanthemi prtD, prtE, and prtF genes, was introduced into these strains, and the ability of the PrtDEF transport system to secrete these proteins was tested by assaying for the presence of secreted proteins in cell supernatants. We note that the expression of proteins that are not secreted is higher in the presence of plasmid pRUW4 (Fig. 2A, compare lanes 3 and 5). At present, we have no explanation for this result. Figure 2B shows that only the 27-kDa fusion product (with the longer C-terminal signal sequence) is present in the supernatant fraction of cells (secretion scored 20 and 31% in two clones). No secretion was observed in the absence of plasmid pRUW4 (Fig. 2B, lanes 2 and 6). Similar results were obtained when the plasmid vectors were introduced into an E. carotovora Ecc193 host (data not shown). This result shows that the longer 181-amino-acid C terminus is essential for the efficient secretion of the short hybrid product formed between the polylinker sequence on expression plasmid pSE420 in an E. coli C600 host and thereby confirms that the glycine-rich repeats are required for the efficient export of hybrid passenger proteins.
FIG. 2.
Expression and secretion of the carboxyl-terminal signals (93 and 181 amino acids) of the E. chrysanthemi protease B in E. coli C600. The hybrids were detected by SDS–12% PAGE followed by Western blot analysis. (A) The 93CTPB signal immunoblot revealed with alkaline phosphatase; (B) the 181CTPB signal immunoblot revealed by chemiluminescence with horseradish peroxidase. Cultures were induced with 1 mM IPTG for 4 h, when bacterial growth reached an optical density at 600 nm (OD600) of 0.5. Ten-microliter samples were applied to the gel from lysates (L) and supernatants (S). (A) Lanes: Std, prestained low-molecular-mass standard (GIBCO-BRL); 1 and 2, clone with pSE420 and pRUW4; 3 and 4, clone with pSE420-93CTPB; 5 and 6, clone with pSE420-93CTPB and pRUW4. Supernatants were concentrated 10 times with respect to the lysates by 10% TCA precipitation. (B) Lanes: 1 and 2, lysate and supernatant from control E. coli C600 cells; 3 and 4, lysate and supernatant from clone 8 (pSE420-181CTPB in E. coli C600); 5 and 6, lysate and supernatant from clone 8-1 (pSE420-181CTPB plus pRUW4 in E. coli C600); 7 and 8, lysate and supernatant from clone 4 (pSE420-181CTPB in E. coli C600); 9 and 10, lysate and supernatant from clone 4-1 (pSE420-181CTPB plus pRUW4 in E. coli C600). Std, migration of bands corresponding to low-molecular-mass standard (GIBCO-BRL). Supernatants were concentrated two times with respect to the lysates by 10% TCA precipitation.
The E. chrysanthemi type I secretion system permits the transport of hybrids between eukaryotic proteins and protease B.
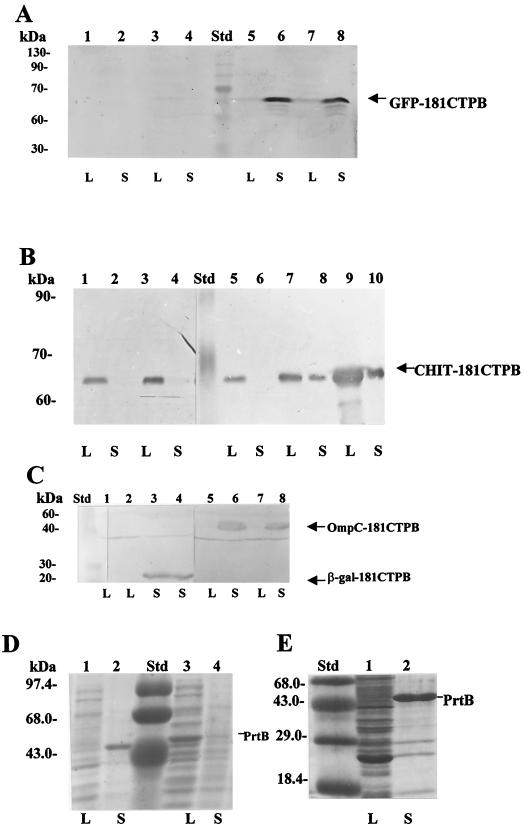
To test whether hybrids between eukaryotic passenger proteins and the C-terminal signal sequence of PrtB can be secreted in E. coli by the E. chrysanthemi type I system, plasmids to express hybrid proteins formed between a variety of eukaryotic proteins and the C terminus of protease B were constructed. These proteins include Trichoderma harzianum endochitinase (42 kDa), GFP from Aequorea victoria (28.5 kDa), (hEPO 20 kDa), and trout growth hormone (tGH, 22.6 kDa). For prokaryotic controls, we also constructed hybrids with segments of the E. coli lacZ (β-galactosidase segment, 3.6 kDa) and Salmonella typhi ompC porin genes (OmpC segment, 24.5 kDa) and included the protease B gene (full-length product, 56 kDa).
Each gene was cloned upstream of, and in frame with, the 181-amino-acid C-terminal segment of PrtB present in pSEB420-181CTPB2. Hybrid constructions were tested for secretion after overnight incubation at 37°C in LB broth containing 100 μg of ampicillin/ml and 50 μg of chloramphenicol/ml, followed by induction with 1 mM IPTG for 3 to 4 h. Figures 3A and 3B show secretion of GFP and chitinase hybrids, respectively. It was found that the GFP-181CTPB hybrid (55 kDa) was secreted in about 32 to 56% of the total amount of the synthesized hybrid depending on the particular clone, as shown in Fig. 3A (compare lanes 5 and 6 and and lanes 7 and 8). The chitinase-protease B hybrid (72 kDa) was modestly secreted to the medium at about 4 to 8% depending on the specific isolated clone (Fig. 3B, compare lanes 7 and 8 and lanes 9 and 10).
FIG. 3.
Expression of GFP- and endochitinase-181CTPB hybrids in E. coli. Protein hybrids were separated by SDS–12% PAGE; this was followed by Western blotting revealed with anti-protease B antibodies (diluted 1:1,000) and a second antibody (goat anti-rabbit immunoglobulin G coupled to peroxidase; diluted 1:1,000). Bacterial cultures harboring plasmids pGFP-181CTPB in E. coli HB101 cells and pCHIT-181CTPB in E. coli C600 were grown in LB medium with 100 μg of ampicillin/ml at 37°C until the OD600 was 0.5. The cultures were then left uninduced or were induced for 4 h with 1 mM IPTG. Some of the cultures were previously cotransformed with plasmid pRUW4, which carries the three genes that encode the secretion machinery. For total lysates (lanes L), 0.5 ml of cell culture was concentrated by centrifugation and lysed in 100 μl of electrophoresis buffer sample by heating at 100°C (40). The gel was loaded with 25 μl. The same volume of supernatant (lanes S) was precipitated with 10% TCA, and the pellets were washed twice with acetone, dissolved, and loaded as described for lysates. Std, GIBCO-BRL Benchmark molecular size standard in panels A through C and GIBCO-BRL high-molecular-weight standard in panels D and E. (A) GFP-181CTPB secretion in E. coli HB101. Lanes: 1 and 2, E. coli HB101 plus vector pSE420; 3 and 4, clone 6 (pGFP-181CTPB); 5 and 6, clone 6-1 (pGFP-181CTPB plus pRUW4); 7 and 8, clone 7-1 (pGFP-181CTPB plus pRUW4). (B) Endochitinase-181CTPB secretion in E. coli C600. Lanes: 1 and 2, clone CHIT-9 (pCHIT-181CTPB) uninduced; 3 and 4, clone CHIT-1 (pCHIT-181CTPB) uninduced; 5 and 6, clone pCHIT9-1 (pCHIT-181CTPB plus pRUW4) uninduced; 7 and 8, clone pCHIT1-1 (pCHIT-181CTPB plus pRUW4) induced with 1 mM IPTG; 9 and 10, clone pCHIT9-1 (pCHIT-181CTPB plus pRUW4) induced with 1 mM IPTG. (C) Secretion of β-galactosidase–181CTPB and OmpC-181CTPB hybrids in E. coli C600. All clones were induced for 3 h with 1 mM IPTG. Lanes: 1 and 2, lysates of two clones (pβgal-181CTPB plus pRUW4); 3 and 4, the corresponding supernatants; 5 and 6, clone OmpC-3 (pOmpC-181CTPB plus pRUW4); 7 and 8, clone OmpC-8 (pOmpC-181CTPB plus pRUW4). The supernatant proteins loaded into the gel were previously concentrated 10-fold by TCA precipitation with respect to the lysates. (D) Control of protease B secretion in E. coli DH5α cells grown overnight in LB medium and antibiotics as required and visualized by Coomassie blue staining. Lanes: 1 and 2, lysate and supernatant of clone containing pRUW500 plus pRUW4; 3 and 4, lysate and supernatant of clone containing only pRUW500. (E) Control of protease B secretion in E. coli C600 cells grown overnight in LB medium and visualized as described for panel D. Lanes: 1, lysate; 2, supernatant of clone containing pRUW500 plus pRUW4.
As expected, the β-galactosidase hybrid used as the control gave between 60 and 75% secretion for two clones (Fig. 3C). The OmpC-181CTPB hybrid (46 kDa) was also secreted to a substantial level (24 to 48%), even though this protein (when carrying its signal peptide and the entire coding sequence) is normally assembled in the outer membrane. For controls, protease B secretion assays in E. coli DH5α (Fig. 3D) as well as in C600 cells (Fig. 3E) were included. These results show that a variety of eukaryotic proteins can be secreted from an E. coli host by the E. chrysanthemi type I mechanism.
The E. chrysanthemi type I secretion system does not permit the transport of hybrids between eukaryotic proteins that form disulfide bridges in their native forms and protease B.
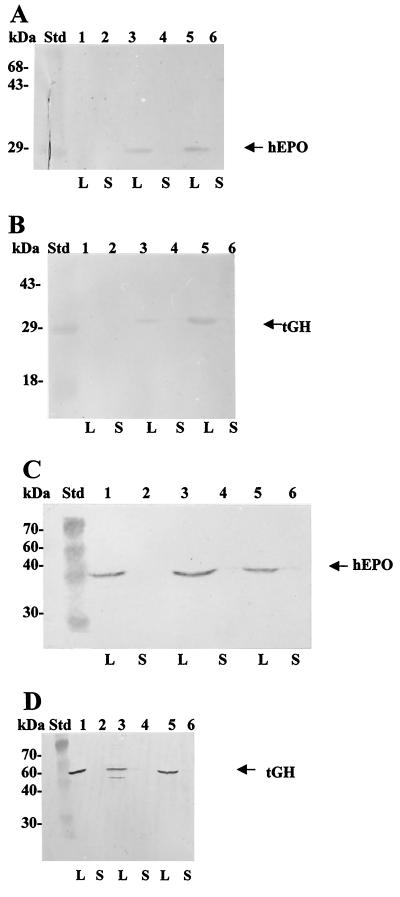
Two of the hybrid proteins we tested are not secreted by the E. chrysanthemi type I system. The hybrids with hEPO and tGH were assayed with both the 93CTPB and 181CTPB secretion signals. Both of these proteins are unique among the proteins that have been tested for secretion by a type I system because they are capable of forming disulfide bonds in their native structures. The hEPO hormone contains two disulfide bonds, and because its active form is highly glycosylated (34), it has not been expressed previously in E. coli. In contrast, tGH, with a molecular mass of 22 kDa, contains two disulfide bonds, is not glycosylated, and has been expressed intracellularly in E. coli (49). Both tGH-93CTPB and hEPO-93CTPB hybrids appeared in E. coli C600 lysates as the corresponding 32- and 35-kDa bands after separation by SDS-PAGE and Western blot assays. Thus, hybrids with both proteins are expressed in E. coli. However, these proteins were absent in supernatant fractions of cells that also carry plasmid pRUW4, which encodes the PrtDEF ABC transporter (Fig. 4A and B). In spite of positive secretion results with the other eukaryotic hybrids, the hEPO-181CTPB and tGH-181CTPB hybrids are not secreted when expressed in host E. coli DH5α cells carrying pRUW4 (Fig. 4C and D). The same results were obtained when the 93CTPB or 181CTPB hybrid contructions and plasmid pRUW4 were transferred to E. coli HB101 (results not shown). These results argue that the PrtDEF type I secretion system cannot facilitate the secretion of passenger proteins that can form disulfide bonds. A summary of the results obtained for secretion of different hybrid constructions by the type I system in E. coli is shown in Table 4.
FIG. 4.
Expression and secretion assays of hEPO and tGH hybrids in E. coli C600. The hybrids were visualized by SDS–12% PAGE followed by a Western blot assay revealed with anti-protease B polyclonal antibodies. Cultures were induced for 4 h at 37°C with 1 mM IPTG after the bacterial cultures reached an OD600 of 1.0. Ten-microliter samples of lysates (L) and supernatants (S) that were concentrated 10 times with respect to the lysates by 10% TCA precipitation were loaded in the gel. Std, Benchmark prestained protein ladder. (A) Expression of hybrid hEPO-93CTPB. Lanes: 1 and 2, clone harboring pSE420 plus pRUW4; 3 and 4, clone with phEPO-93CTPB; 5 and 6, clone with phEPO-93CTPB plus pRUW4. (B) Expression of hybrid tGH2-93CTPB. Lanes: 1 and 2, clone with pSE420 plus pRUW4; 3 and 4, clone with ptGH2-93CTPB; 5 and 6, clone with ptGH2-93CTPB plus pRUW4. The arrows indicate the electrophoretic migration of the protein bands expected in each case. (C) Expression of hybrid hEPO-181CTPB. Lanes: 1 and 2, clone 3-1 (phEPO-181CTPB plus pRUW4 in E. coli DH5α); 3 and 4, clone 12-1 (phEPO-181CTPB plus pRUW4 in E. coli DH5α); 5 and 6, clone 4-1 (phEPO-181CTPB plus pRUW4 in E. coli DH5α). (D) Lanes: 1 and 2, clone 6-1 (ptGH-181CTPB plus pRUW4 in E. coli DH5α); 3 and 4, clone 5-1 (ptGH-181CTPB plus pRUW4 in E. coli DH5α); 5 and 6, clone 2-1 (ptGH-181CTPB plus pRUW4 in E. coli DH5α).
TABLE 4.
Secretion of hybrid constructions in E. coli
| Plasmid-signal | Promoter-hybrid gene structurea | Sizes of hybrid proteins (split in moieties)b | Total sizec (in kDa) | No. of SH groups | Secretion (%)d of E. coli |
|---|---|---|---|---|---|
| pSE420-93CTP | 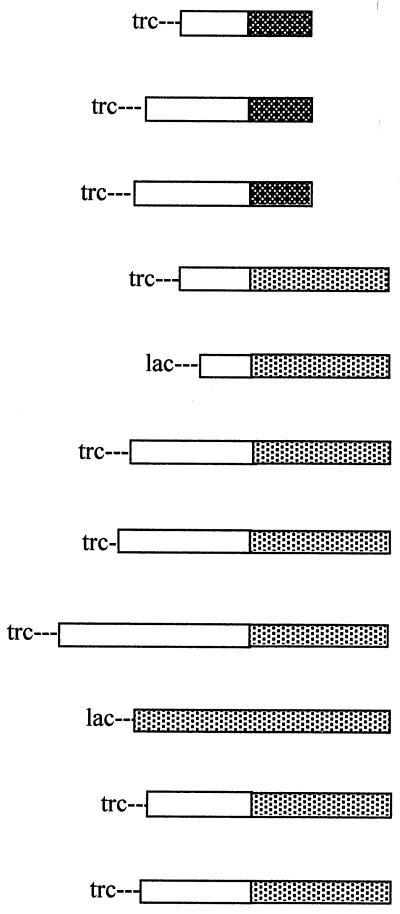 |
46/93 | 16.7 | 1 | 0 |
| phEPO-93CTPB | 167/93 | 31.2 | 4 | 0 | |
| ptGH-93CTPB | 189/93 | 33.8 | 4 | 0 | |
| pSE420-181CTPB | 48/181 | 27.5 | 1 | 27–31 | |
| p-βGal-181CTPB | 30/181 | 25.3 | 0 | 60–75 | |
| pOmpC-181CTPB | 204/181 | 46.0 | 0 | 24–48 | |
| pGFP-181CTPB | 238/181 | 55.3 | 2e | 32–56 | |
| pCHIT-181CTPB | 424/181 | 72.6 | 0 | 4–8 | |
| pRUW500 (PrtB) | 466f | 56.0 | 0 | 98–100 | |
| phEPO-181CTPB | 167/181 | 41.7 | 4 | 0 | |
| ptGH-181CTPB | 189/181 | 44.4 | 4 | 0 |
Hybrid size structures represented by horizontal bars are not drawn to scale.
Hybrid sizes are displayed in amino acid residues as separated contributions of the passenger protein and the corresponding CTPB region.
Molecular mass was estimated considering an average mass of 120 Da per amino acid residue.
Data were obtained for two independently isolated clones, and values were not averaged. All secretion assays required introduction of plasmid pRUW4 by electroporation. Secretion was determined by scanning Western blot bands of hybrids detected with anti-protease B antibodies and expressed as a percentage of the supernatant fraction compared to total amount of the synthesized hybrid. Data were processed by the Image J V1.12 program (Wayne Rasband, National Institutes of Health).
Not a disulfide bond.
The entire E. chrysanthemi protease B gene as a positive secretion control (not a hybrid).
Cytoplasmic enhanced expression of E. chrysanthemi disulfide isomerase C (DsbC) in E. coli does not improve the secretion of proteins carrying disulfide bonds.
For some mechanisms of secretion, it has been proposed that a passenger protein must be completely folded to permit translocation through the secretion channel or outer membrane (52, 25). In E. coli, the majority of intramolecular disulfide bonds are formed in the periplasm (59), and it is believed that intermediates translocated by the E. chrysanthemi type I secretion system are not in contact with the periplasm during secretion (64). Because the bacterial cytoplasm is a reducing environment, and most disulfide bonds are formed in the periplasm (53), we reasoned that proteins secreted by this pathway do not have the opportunity to form disulfide bridges. If such proteins must be folded prior to transport, the failure to form disulfide bonds may account for their failure to be transported by the type I mechanism. Therefore, we tested whether the formation of disulfide bonds in these proteins prior to transport might increase their efficiency of transport in two different ways. First, we expressed the hEPO and the tGHII hybrid proteins in a trxB mutant deficient in thioredoxin production to increase the oxidizing potential of the cytoplasm, and thereby to favor cytoplasmic disulfide bond formation (59). Second, we expressed the hEPO and the tGHII hybrids in a strain of E. coli in which a soluble form of the dsbC gene product (disulfide isomerase) is also expressed in the cytoplasm, to promote the enzyme-catalyzed formation of disulfide bonds prior to secretion.
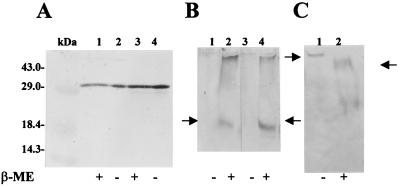
Figure 5 shows the results of a Western blot assay for expression of the tGH-93CTPB hybrid in total lysates of the trxB-deficient E. coli host strain AD494, with or without β-ME included in the sample prior to the protein separation by electrophoresis. We did not visualize any change in the electrophoretic mobility of this protein under the two conditions, indicating that the absence of thioredoxin does not enhance the formation of disulfide bonds in the hybrid protein. We suspect that this is due to the fact that disulfide bonds can form in this hybrid protein in the E. coli cytoplasm both in the presence and absence of thioredoxin (see Discussion). Similar results were obtained with the hEPO-93CTPB hybrid and both constructions carrying the 181CTPB region.
FIG. 5.
(A) Western blot assay carried out in bacterial lysates to detect cytoplasmic formation of disulfide bonds in the tHG-93CTPB hybrid expressed in a trxB mutant. All E. coli AD494 and DH5α strains carrying both ptGH-93CTPB and pRUW4 plasmids were grown overnight and induced with 1 mM IPTG for 3 h. Bacterial cells (1 ml) were centrifuged in a microcentrifuge, and pellets were lysed by boiling for 5 min in 100 μl of Laemmli sample buffer (40). Aliquots of 30 μl were loaded in an SDS–12% polyacrylamide gel. Lanes: Std, GIBCO-BRL molecular mass standard; 1, DH5α/ptGH-93CTPB plus 25 mM β-ME; 2, DH5α/ptGH-93CTPB without β-ME; 3, AD494/ptGH-93CTPB plus 25 mM β-ME; 4, AD494/ptGH-93CTPB without β-ME. (B) Detection of change in electrophoretic mobility of the tGH-181CTPB hybrid in DH5α cell extracts after treatment with β-ME. Proteins were resolved by 8 M urea–8% polyacrylamide gel electrophoresis (see Materials and Methods), and the hybrid (arrows) was detected by Western blotting as described in Materials and Methods. Lanes: 1, ptGH-181CTPB without β-ME; 2, ptGH-181CTPB with 10 mM β-ME; 3, ptGH-181CTPB plus pRUW4, without β-ME added; 4, ptGH-181CTPB plus pRUW4 with 10 mM β-ME. Loaded samples contained 10 μl of cell extracts and 10 μl of Laemmli sample buffer without SDS or β-ME. Hybrids loaded on lanes 1 and 3 did not enter the gel and were lost prior to or during transfer to the nitrocellulose filter. (C) Detection of change in electrophoretic mobility of hEPO-181 hybrid in DH5α cells overexpressing the dsbC gene by β-ME treatment of extracts previously blocked with 20 mM iodoacetic acid. Reaction with iodoacetic acid was done as described in Materials and Methods. Proteins were resolved by 8 M urea–8% polyacrylamide gel electrophoresis, and the hybrid (arrows) was detected by a chemiluminescence assay (see Materials and Methods). Loaded samples contained 10 μl of cell extracts and 10 μl of Laemmli sample buffer without SDS or β-ME. Lanes: 1, phEPO-181CTPB plus pRUW4 plus pACYC-dsbC, without β-ME added; 2, phEPO-181CTPB plus pRUW4 plus pACYC-dsbC with 10 mM β-ME.
To overexpress a soluble form of DsbC in the cytoplasm, we cloned a version of the E. chrysanthemi dsbC gene without the coding region for its signal sequence in plasmid pACYC184 as described in Materials and Methods and expressed this truncated form of dsbC from a strong IPTG-inducible trc promoter. The plasmid pACYC-dsbC was transferred by electroporation to an E. coli DH5α host carrying plasmids phEPO-181CTPB and pRUW4. When we assayed for the expression and secretion of the hEPO hybrid as described previously, we found that overexpression of DsbC activity in the cytoplasm did not improve secretion of the hEPO-181CTPB hybrid (data not shown). In this case, the redox state of the hybrids was evaluated by modification of the SH residues by iodoacetic acid (see Materials and Methods). An electrophoretic mobility change for the EPO hybrid was noticed when the cell extract previously treated with 20 mM iodoacetic acid was reduced by the addition of β-ME, showing that the sample treated with the reducing agent entered the gel and the untreated sample did not (Fig. 5C). This suggests that the hEPO hybrid has been previously oxidized.
DISCUSSION
The main goal of the work we have described in this report has been to test whether the E. chrysanthemi type I secretion system permits the efficient secretion of eukaryotic proteins in an E. coli host. The secretion of eukaryotic proteins to the external medium using this system will facilitate and simplify the purification of proteins for basic scientific, medical, and industrial purposes. Our demonstration that a subset of eukaryotic proteins can be produced and excreted by this system in E. coli provides yet another method for the large-scale purification of eukaryotic protein hybrids in a prokaryotic host. Such hybrids, once purified, can be processed proteolytically to remove the secretion signal from the hybrid after exportation, if necessary.
Consistent with the results of previous studies with prokaryotic fusion proteins, we found that a 93-residue signal sequence from E. chrysanthemi PrtB is not sufficient to allow secretion of the small hybrid resulting from the construction of the secretion vector pSE420-93CTPB but that a larger hybrid with a 181-residue signal sequence is sufficient to allow secretion of the product of vector pSE420-181CTPB. Similar results were obtained with fusions of the same C termini to an endochitinase and GFP (Table 4). Thus, as for previous hybrids constructed between prokaryotic proteins and the C terminus of PrtB, the secretion of hybrids with eukaryotic proteins, in general, depends not only on the C-terminal DFLV and DIIV motifs of PrtB but also on the presence of six short repeats of residues rich in glycines, three of them matching precisely the consensus sequence GGXGXD. These motifs have been defined as additional components of the secretion signal (41). This feature is also present in a Serratia sp. metalloprotease, S. marcescens lipase, Rhizobium leguminosarum NodO, Proteus mirabilis metalloprotease, P. aeruginosa alkaline protease, and hemolysin, all secreted by a type I mechanism.
Although some hybrids with eukaryotic proteins are secreted by the E. chrysanthemi type I mechanism, others are not, including those with hEPO and tGH. The proteins that are not secreted by this mechanism have the common feature that they form intramolecular disulfide bonds in their native structures. This result is striking because it calls attention to the fact that none of the 22 known proteins secreted by type I mechanisms are capable of forming intramolecular disulfide bonds (Table 5). Of these proteins, 20 of 22 have no cysteine residues and only NodO and PllktA have single cysteine residues. Given the average size of the proteins secreted by a type I mechanism, and the frequency of occurrence of cysteine residues in proteins, this conspicuous absence of disulfide bonds in these 22 proteins cannot be due to mere coincidence. Furthermore, we have shown that fusions with GFP are secreted by the type I apparatus. GFP has two cysteine residues that do not participate in disulfide bond formation because they are sufficiently distant from one another on the surface of the GFP monomer to exclude their direct intramolecular interaction (51). Thus, the simplest hypothesis that can account for our results is that the formation of disulfide bonds, or the potential to form disulfide bonds, precludes the secretion of a subset of proteins by the E. chrysanthemi type I mechanism.
TABLE 5.
List of proteins secreted by type I secretion system and content of cysteine residues
| Secreted protein (gene) | Microorganism | Components of type I secretion system | No. of cysteine residues | Reference |
|---|---|---|---|---|
| Protease AB374 (prtA) | Erwinia chrysanthemi | PrtD, PrtE, PrtF | 0 | 27 |
| Protease B (prtB) | Erwinia chrysanthemi | PrtD, PrtE, PrtF | 0 | 19 |
| Protease CE374 (prtC) | Erwinia chrysanthemi | PrtD, PrtE, PrtF | 0 | 20 |
| Protease CEc16 (prtC) | Erwinia chrysanthemi | PrtD, PrtE, PrtF | 0 | 17 |
| Protease G (prtG) | Erwinia chrysanthemi | PrtD, PrtE, PrtF | 0 | 28 |
| Protease A (prtA) | Erwinia amylovora | PrtD, PrtE, PrtF | 0 | 70 |
| Protease W (prtW) | Erwinia carotovora | Not yet identified | 0 | 46 |
| Adenylate cyclase (cyaA) | Bordetella pertussis | CyaB, CyaD, CyaE | 0 | 29 |
| Alpha-hemolysin (hlyA) | Escherichia coli | HlyB, HlyD, TolC | 0 | 61 |
| Alpha-hemolysin (hlyA) | Escherichia coli | HlyB, HlyD, TolC | 0 | 60 |
| Leukotoxin (pllktA) | Pasteurella haemolytica | PllktB, PllktD | 1 | 14 |
| Metalloprotease (prt) | Serratia sp. strain E-15 | PrtD, PrtE | 0 | 50 |
| Hb binding protein (hasA) | Serratia marcescens | PrtDSM, PrtESMa | 0 | 44 |
| Protease SM (prtSM) | Serratia marcescens | PrtDSM, PrtESM, PrtFSM | 0 | 12 |
| S-layer protein (rsaA) | Caulobacter crescentus | RsaD, RsaE | 0 | 4 |
| Lipase A (lipA) | Serratia marcescens Sr41 | LipB, LipC, LipD | 0 | 2 |
| Lipase A (lipA) | Pseudomonas strain LS107d2 | Not reported yet | 0 | 37 |
| Thermostable lipase (tliA) | Pseudomonas fluorescens | tLiD, tLiE, tLiF | 0 | 1 |
| Protease A (prtA) | Pseudomonas fluorescens | PrtD, PrtE, PrtF | 0 | 1 |
| Alkaline protease (aprA) | Pseudomonas aeruginosa | AprD, AprE, AprF | 0 | 23 |
| Metalloprotease (zapA) | Proteus mirabilis | PrtD, PrtE, PrtF | 0 | 65 |
| Nodulating protein (nodO) | Rhizobium leguminosarum | Not yet reported | 1 | 24 |
PrtDSM and PrtESM components of the HasA secretion machinery have been renamed as HasB and HasD, respectively, and HasF, a TolC analog, has been cloned (8).
Initially, we considered the latter hypothesis because the E. coli cytoplasm is a reducing environment and most disulfide bond formation in E. coli occurs in the periplasm, due to the enzymatic activity of the Dsb proteins. According to this hypothesis, the potential to form disulfide bonds would preclude the secretion of a subset of proteins by the E. chrysanthemi type I mechanism, presumably because these proteins would need to be folded prior to transport. By this hypothesis, then, the lack of secretion for hEPO and tGH would be due to the incomplete folding of these hybrids because of the lack of disulfide bonds in their structure. However, we find that this is unlikely to be the case because overexpression of a soluble form of active DsbC in the cytoplasm does not increase the efficiency of export of proteins with disulfide bonds in their mature structures. This strategy of overexpressing members of the DsbC family to catalyze the formation of disulfide bonds in the cytoplasm has been used successfully to improve the production of apo-retinol-binding protein (56), mouse urokinase, and human tissue plasminogen activator (6). Given that we can detect cytoplasmic activity of the expressed E. chrysanthemi DsbC protein without its signal peptide, we presume that this protein can function to stimulate disulfide bond formation in the cytoplasm like its homologs. It is quite clear that the hEPO-181CTPB and tGH-181CTPB hybrids are sufficiently small to be secreted by the type I mechanism, because the endochitinase hybrid with 605 amino acid residues, almost twice the size of these proteins, is secreted to the same extent.
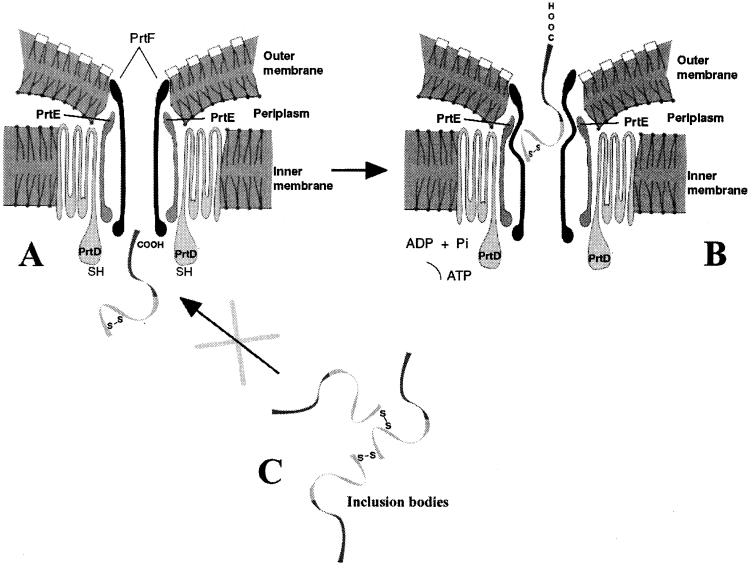
Thus, it appears that the formation of disulfide bonds per se excludes the export of a subset of eukaryotic proteins. At some step during the secretion process, these hybrid proteins can form disulfide bonds, and once formed, disulfide bonds block secretion. It is inviting to speculate that this is the case for two reasons (see a proposed model in Fig. 6). First, because the signal sequence essential for type I secretion is C-terminal, it is likely that transport of type I passenger proteins is initiated with this signal sequence, as it is with the signal sequences of proteins secreted by a type II mechanism. We might imagine that the C terminus of type I passenger proteins is first threaded through the export channel, followed by the N-terminal remainder of the protein (Fig. 6A). A priori, an optimal export channel will have a small diameter, to minimize the exchange of ions between the extracellular and intracellular environments during transport. Thus, it is reasonable to assume that type I passenger proteins, like type II proteins transported to the periplasm, are transported in an unfolded form to be accommodated by an optimally small diameter of the type I secretion channel. Because SecB is required for the S. marcescens HasA protein to be secreted via a type I system (21), it is likely that the passenger protein must be in an unfolded state as is the case for type II mechanisms.
FIG. 6.
Proposed model to explain blockage of secretion for hybrids containing disulfide bonds. (A) The hybrid carrying a preformed disulfide bond approaches the channel entrance, is recognized by the secretion machinery (PrtD), but cannot go across the channel. (B) The hybrid is stacked inside the channel during secretion. Disulfide bonds may be formed at this stage. (C) The hybrid has formed inclusion bodies by intermolecular S—S bonds and cannot reach the channel or enter it.
This argument provides a simple explanation for why proteins capable of forming disulfide bonds cannot be transported by a type I mechanism. If they form disulfide bonds at some step during transport, either prior to interaction with PrtD or in contact with the periplasm, they may acquire a secondary structure that cannot be unfolded during the process of secretion, a structure that is too large to fit through the type I channel and which therefore blocks the channel (Fig. 6B). It is unlikely that passenger proteins secreted by a type I mechanism can form disulfide bonds in the periplasm. This is because a hybrid formed by E. coli alkaline phosphatase (PhoA), which forms two intramolecular disulfide bonds in the periplasm dependent on DsbC, and the last 60-amino-acid region of the alpha-hemolysin carboxyl terminus is efficiently secreted in E. coli by the Hly type I transporter, even though no enzymatic activity has been reported (26). Thus, we suspect that, unlike many prokaryotic proteins, some eukaryotic proteins with intramolecular disulfide bonds can form these bonds in the reducing environment of the E. coli cytoplasm. We have found direct evidence that our hEPO hybrid protein does form disulfide bonds in E. coli, because, when overproduced in E. coli, a fraction of the hEPO hybrid protein aggregates by forming disulfide bonds that are sensitive to reduction with β-ME (Fig. 5C). Also, the folding of growth hormone occurs independently of the formation of at least one of its two disulfide bridges, to position two cysteine side chains sufficiently close to one another to permit spontaneous, base-catalyzed disulfide bond formation (62). In addition, formation of prochymosin inclusion bodies in E. coli cytoplasm by cross-linking of disulfide bonds has been reported (58). Formation of inclusion bodies in the cytoplasm could also restrict secretion of hybrid proteins by the type I secretion system (Fig. 6C).
Currently, we are testing the hypothesis that the formation of intramolecular disulfide bonds in the cytoplasm by passenger proteins can block their secretion by a type I mechanism directly, by determining whether E. coli strains with the E. chrysanthemi type I secretion system are capable of exporting proteins that form disulfide bonds in their mature structures dependent on the activity of cytoplasmically expressed DsbC.
ACKNOWLEDGMENTS
This work was funded by the Comisión Nacional Científica y Tecnológica de Chile (grant FONDECYT 1971010) and Fondo de Desarrollo Innovativo (CORFO, Santiago, Chile; grant FDI AT-1).
E. carotovora subsp. carotovora Ecc193 was kindly provided by A. K. Chatterjee. We gratefully acknowledge P. Delepelaire for providing plasmids pRUW4 and pRUW500 and P. Youderian for critical reading of the manuscript.
REFERENCES
- 1.Ahn J, Pan J, Rhee J. Identification of the tliDEF ABC transporter specific for lipase in Pseudomonas fluorescens SIK W1. J Bacteriol. 1999;181:1847–1852. doi: 10.1128/jb.181.6.1847-1852.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akatsuka H, Kawai E, Omori K, Shibatani T. The three genes lipB, lipC, and lipD involved in the extracellular secretion of Serratia marcescens lipase which lacks an N-terminal signal peptide. J Bacteriol. 1995;177:6381–6389. doi: 10.1128/jb.177.22.6381-6389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arkowitz R, Joly J, Wickner W. Translocation can drive the unfolding of a preprotein domain. EMBO J. 1993;12:243–253. doi: 10.1002/j.1460-2075.1993.tb05650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awram P, Smith J. The Caulobacter crescentus paracrystalline S-layer protein is secreted by an ABC transporter type I secretion apparatus. J Bacteriol. 1998;180:3062–3069. doi: 10.1128/jb.180.12.3062-3069.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardwell J. Building bridges: disulfide bond formation in the cell. Mol Microbiol. 1994;14:199–205. doi: 10.1111/j.1365-2958.1994.tb01281.x. [DOI] [PubMed] [Google Scholar]
- 6.Bessette P, Aslund F, Beckwith J, Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci USA. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binet R, Wandersman C. Protein secretion by hybrid bacterial ABC-transporters: specific functions of the membrane ATPase and the membrane fusion protein. EMBO J. 1995;14:2298–2306. doi: 10.1002/j.1460-2075.1995.tb07224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binet R, Wandersman C. Cloning of the Serratia marcescens hasF gene encoding the Has ABC exporter outer membrane component: a TolC analog. Mol Microbiol. 1996;22:265–273. doi: 10.1046/j.1365-2958.1996.00103.x. [DOI] [PubMed] [Google Scholar]
- 9.Binet R, Létoffé S, Ghigo J, Delepelaire P, Wandersman C. Protein secretion by Gram negative bacterial ABC export: a review. Gene. 1997;192:7–11. doi: 10.1016/s0378-1119(96)00829-3. [DOI] [PubMed] [Google Scholar]
- 10.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 12.Braunagel S C, Benedik M J. The metalloprotease gene of Serratia marcescens strain SM6. Mol Gen Genet. 1990;222:446–451. doi: 10.1007/BF00633854. [DOI] [PubMed] [Google Scholar]
- 13.Chaconas G, van de Sande J. 5′-32P labeling of RNA and DNA restriction fragments. Methods Enzymol. 1990;65:75–85. doi: 10.1016/s0076-6879(80)65012-5. [DOI] [PubMed] [Google Scholar]
- 14.Chang Y, Ma D, Shi J, Chengappa M. Molecular characterization of a leukotoxin gene from a Pasteurella haemolytica-like organism, encoding a new member of the RTX toxin family. Infect Immun. 1993;61:2089–2095. doi: 10.1128/iai.61.5.2089-2095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen E, Seeburg P. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985;4:165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- 16.Coligan J. In: Current protocols in immunology. 2nd ed. Coligan J, Kruisbeck A, Margulies D, Shevach E, Strober W, editors. New York, N.Y: Wiley Liss Inc.; 1994. pp. 1.6.1–1.7.8. [Google Scholar]
- 17.Dahler G S, Barras F, Keen N T. Protease C cloning of genes encoding extracellular metalloproteases from Erwinia chrysanthemi EC16. J Bacteriol. 1990;172:5803–5815. doi: 10.1128/jb.172.10.5803-5815.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delepelaire P. PrtD, the integral membrane ATP-binding cassette component of the Erwinia chrysanthemi metalloprotease secretion system, exhibits a secretion signal-regulated ATPase activity. J Biol Chem. 1994;269:27952–27957. [PubMed] [Google Scholar]
- 19.Delepelaire P, Wandersman C. Protease secretion by Erwinia chrysanthemi. Proteases B and C are synthesized and secreted as zymogens without a signal peptide. J Biol Chem. 1989;264:9083–9089. [PubMed] [Google Scholar]
- 20.Delapelaire P, Wandersman C. Protein secretion in gram-negative bacteria. The extracellular metalloprotease B from Erwinia chrysanthemi contains a C-terminal secretion signal analogous to that of Escherichia coli α-hemolysin. J Biol Chem. 1990;265:17118–17125. [PubMed] [Google Scholar]
- 21.Delepelaire P, Wandersman C. The SecB chaperone is involved in the secretion of the Serratia marcescens HasA protein through an ABC transporter. EMBO J. 1998;17:936–944. doi: 10.1093/emboj/17.4.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinh T, Paulsen I, Saier M. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J Bacteriol. 1994;176:3825–3831. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duong F, Lazdunski A, Cami B, Murgier M. Sequence of a cluster of genes controlling synthesis and secretion of alkaline protease in Pseudomonas aeruginosa: relationships to other secretory pathways. Gene. 1992;121:47–54. doi: 10.1016/0378-1119(92)90160-q. [DOI] [PubMed] [Google Scholar]
- 24.Economou A, Hamilton W, Johnston A, Downie J. The Rhizobium nodulation gene nodO encodes a Ca2(+)-binding protein that is exported without N-terminal cleavage and is homologous to haemolysin and related proteins. EMBO J. 1990;9:349–354. doi: 10.1002/j.1460-2075.1990.tb08117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujishige A, Smith K, Silen J, Agard D. Correct folding of α-lytic protease is required for extracellular secretion from Escherichia coli. J Cell Biol. 1992;118:33–42. doi: 10.1083/jcb.118.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentschev I, Hess J, Goebel W. Change in the cellular localization of alkaline phosphatase by alteration of its carboxy-terminal sequence. Mol Gen Genet. 1990;222:211–216. doi: 10.1007/BF00633820. [DOI] [PubMed] [Google Scholar]
- 27.Ghigo J, Wandersman C. Cloning, nucleotide sequence and characterization of the gene encoding the Erwinia chrysanthemi B374 PrtA metalloprotease: a third metalloprotease secreted via a C-terminal secretion signal. Mol Gen Genet. 1992;236:135–144. doi: 10.1007/BF00279652. [DOI] [PubMed] [Google Scholar]
- 28.Ghigo J, Wandersman C. A carboxyl-terminal four-amino acid motif is required for secretion of the metalloprotease PtrG through the Erwinia chrysanthemi protease secretion pathway. J Biol Chem. 1994;269:8979–8985. [PubMed] [Google Scholar]
- 29.Glasser P, Sakamoto H, Bellalou J, Ullman A, Danchin A. Secretion of cyclolysin, the calmodulin-sensitive adenylate cyclase-haemolysin bifunctional protein of Bordetella pertussis. EMBO J. 1988;7:3997–4004. doi: 10.1002/j.1460-2075.1988.tb03288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray L, Kenny B, Mackman N, High R, Holland I. A novel C-terminal signal sequence targets Escherichia coli haemolysin directly to the medium. J Cell Sci. 1989;11(Suppl.):45–57. doi: 10.1242/jcs.1989.supplement_11.4. [DOI] [PubMed] [Google Scholar]
- 31.Grimberg J, Maguire S, Belluscio L. A simple method for the preparation of plasmid and chromosomal Escherichia coli DNA. Nucleic Acids Res. 1989;17:8893. doi: 10.1093/nar/17.21.8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hames B. Trichloroacetic acid precipitation. In: Bollag D, Edelstein S, editors. Protein methods. New York, N.Y: Wiley Liss Inc.; 1981. pp. 71–73. [Google Scholar]
- 33.Hayes C K, Klemsdal S, Lorito M, Di Pietro A, Peterbauer C, Nakas J P, Tronsmo A, Harman G E. Isolation and sequence of an endochitinase-encoding gene from a cDNA library of Trichoderma harzianum. Gene. 1994;138:143–148. doi: 10.1016/0378-1119(94)90797-8. [DOI] [PubMed] [Google Scholar]
- 34.Higuchi M, Oh-eda M, Kuboniwa H, Tomonoh K, Shimonaka Y, Ochi N. Role of sugar chains in the expression of the biological activity of human erythropoietin. J Biol Chem. 1992;267:7703–7709. [PubMed] [Google Scholar]
- 35.Hirst T, Holmgren J. Conformation of protein secreted across bacterial outer membranes: a study of enterotoxin translocation from Vibrio cholerae. Proc Natl Acad Sci USA. 1987;84:7418–7422. doi: 10.1073/pnas.84.21.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huek C. Type III protein secretion systems in bacterial pathogens in plants and animals. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson L A, Beacham I R, MacRae I C, Free M L. Degradation of triglycerides by a pseudomonad isolated from milk: molecular analysis of a lipase-encoding gene and its expression in Escherichia coli. Appl Environ Microbiol. 1992;58:1776–1779. doi: 10.1128/aem.58.5.1776-1779.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kenny B, Finlay B. Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc Natl Acad Sci USA. 1995;92:7991–7995. doi: 10.1073/pnas.92.17.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán J, Aizawa S. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 40.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 41.Létoffé S, Wandersman C. Secretion of Cya-PrtB and Hly-PrtB fusion proteins in Escherichia coli: involvement of the glycine-rich repeat domain of Erwinia chrysanthemi protease B. J Bacteriol. 1992;174:4920–4927. doi: 10.1128/jb.174.15.4920-4927.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Létoffé S, Delapelaire P, Wandersman C. Protease secretion by Erwinia chrysanthemi: the specific secretion functions are analogous to those of Escherichia coli α-hemolysin. EMBO J. 1990;9:1375–1382. doi: 10.1002/j.1460-2075.1990.tb08252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Létoffé S, Delepelaire P, Wandersman C. Cloning and expression in Escherichia coli of the Serratia marcescens metalloprotease gene: secretion of the protease from E. coli in the presence of the Erwinia chrysanthemi protease functions. J Bacteriol. 1991;173:2160–2166. doi: 10.1128/jb.173.7.2160-2166.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Létoffé S, Ghigo J, Wandersman C. Secretion of the Serratia marcescens HasA protein by an ABC transporter. J Bacteriol. 1994;176:5372–5377. doi: 10.1128/jb.176.17.5372-5377.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Létoffé S, Ghigo J, Wandersman C. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc Natl Acad Sci USA. 1994;91:9876–9880. doi: 10.1073/pnas.91.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marits R, Köiv V, Laasik E, Mäe A. Isolation of an extracellular protease gene of Erwinia carotovora subsp. carotovora strain SCC3193 by transposon mutagenesis and the role of protease in phytopathogenicity. Microbiology. 1999;145:1959–1966. doi: 10.1099/13500872-145-8-1959. [DOI] [PubMed] [Google Scholar]
- 47.Miller J. Bacterial transformation by electroporation. Methods Enzymol. 1994;235:375–385. doi: 10.1016/0076-6879(94)35156-2. [DOI] [PubMed] [Google Scholar]
- 48.Missiakas D, Georgopoulos C, Raina S. The Escherichia coli dsbC (xprA) gene encodes a periplasmic protein involved in disulfide bond formation. EMBO J. 1994;13:2013–2020. doi: 10.1002/j.1460-2075.1994.tb06471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müller I. Expression of trout growth hormone tGHII in Escherichia coli. Biochemist professional title thesis. Concepción, Chile: University of Concepción; 1991. [Google Scholar]
- 50.Nakahama K, Yoshimura K, Marumoto R, Kikuchi M, Lee I S, Hase T, Matsubara H. Cloning and sequencing of Serratia protease gene. Nucleic Acids Res. 1986;14:5843–5855. doi: 10.1093/nar/14.14.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ormö M, Cubitt A B, Kallio K, Gross L A, Tsien R Y, Remington S J. Crystal structure of the aequorea victoria green fluorescent protein. Science. 1996;273:1392–1295. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 52.Pugsley A. Translocation of a folded protein across the outer membrane in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:12058–12062. doi: 10.1073/pnas.89.24.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pugsley A. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt A, Bloss I, Skerra A. Improved folding of apo-retinol-binding protein in the periplasm of Escherichia coli: positive influences of dsbC coexpression and of an amino acid exchange in the vitamin A binding site. Protein Eng. 1998;11:601–607. doi: 10.1093/protein/11.7.601. [DOI] [PubMed] [Google Scholar]
- 57.Shevchik V, Condemine G, Robert-Baudouy J. Characterization of DsbC, a periplasmic protein of Erwinia carotovora and Escherichia coli with disulfide isomerase activity. EMBO J. 1994;13:2007–2012. doi: 10.1002/j.1460-2075.1994.tb06470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shoemaker J M, Brasnett A H, Marston F A. Examination of calf prochymosin accumulation in Escherichia coli: disulphide linkages are a structural component of prochymosin-containing inclusion bodies. EMBO J. 1985;4:775–780. doi: 10.1002/j.1460-2075.1985.tb03696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stewart E, Aslund F, Beckwith J. Disulfide bond formation in Escherichia coli cytoplasm and in vivo role reversal for the thioredoxins. EMBO J. 1998;17:5543–5550. doi: 10.1093/emboj/17.19.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taneike, I., N. Wakisaka-Saito, Y. Harada, H.-M. Zhang, and T. Yamamoto. The enterohemorrhagic Escherichia coli (EHEC)-hemolysin genes of a Shiga toxin 1 (Stx1)- and Stx2-producing, serotype O128 Escherichia coli strain with a greatest hemolytic activity. Acta Med. Biol., in press.
- 61.Thomas W, Wagner S, Welch R. A heterologous membrane protein domain fused to the C-terminal ATP-binding domain of HlyB can export Escherichia coli hemolysin. J Bacteriol. 1992;174:6771–6779. doi: 10.1128/jb.174.21.6771-6779.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tokunaga T, Tanaka T, Ikehara M, Ohtsuka E. Synthesis and expression of a human growth hormone (somatotropin) gene mutated to change cysteine-165 to alanine. Eur J Biochem. 1985;153:445–449. doi: 10.1111/j.1432-1033.1985.tb09322.x. [DOI] [PubMed] [Google Scholar]
- 63.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wandersman C. Secretion across the bacterial outer membrane. Trends Genet. 1992;8:317–322. doi: 10.1016/0168-9525(92)90264-5. [DOI] [PubMed] [Google Scholar]
- 65.Wassif C, Cheek D, Belas R. Molecular analysis of a metalloprotease from Proteus mirabilis. J Bacteriol. 1995;177:5790–5798. doi: 10.1128/jb.177.20.5790-5798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Welch R A. Pore-forming cytolysins of gram-negative bacteria. Mol Microbiol. 1991;5:521–528. doi: 10.1111/j.1365-2958.1991.tb00723.x. [DOI] [PubMed] [Google Scholar]
- 67.Weston A, Brow M, Perkins H, Saunders J, Humphreys G. Transformation of Escherichia coli with plasmid deoxyribonucleic acid: calcium-induced binding of deoxyribonucleic acid to whole cells and to isolated membrane fractions. J Bacteriol. 1981;145:780–787. doi: 10.1128/jb.145.2.780-787.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 69.Zaror M. Isolation and characterization of the Salmonella typhi ompC gene. Structural and functional studies of the OmpC porin. Ph.D. thesis. Santiago: Catholic University of Chile; 1989. [Google Scholar]
- 70.Zhang Y, Bak D D, Heid H, Geider K. Molecular characterization of a protease secreted by Erwinia amylovora. J Mol Biol. 1999;289:1239–1251. doi: 10.1006/jmbi.1999.2846. [DOI] [PubMed] [Google Scholar]