Abstract
Expression of the Vibrio vulnificus metalloprotease gene, vvp, was turned up rapidly when bacterial growth reached the late log phase. A similar pattern of expression has been found in the metalloprotease gene of Vibrio cholerae, and this has been shown to be regulated by a Vibrio harveyi LuxR-like transcriptional activator. To find out whether a LuxR homologue exists in V. vulnificus, a gene library of this organism was screened by colony hybridization using a probe derived from a sequence that is conserved in various luxR-like genes of vibrios. A gene containing a 618-bp open reading frame was identified and found to be identical to the smcR gene of V. vulnificus reported previously. An isogenic SmcR-deficient (RD) mutant was further constructed by an in vivo allelic exchange technique. This mutant exhibited an extremely low level of vvp transcription compared with that of the parent strain. On the other hand, the cytolysin gene, vvhA, was expressed at a higher level in the RD mutant than in the parent strain during the log phase of growth. These data suggested that SmcR might not only be a positive regulator of the protease gene but might also be involved in negative regulation of the cytolysin gene. Virulence of the RD mutant in either normal or iron-overloaded mice challenged by intraperitoneal injection was comparable to that of the parent strain, indicating that SmcR is not required for V. vulnificus virulence in mice.
Vibrio vulnificus, an opportunistic human pathogen, causes severe wound infection and primary septicemia (36, 38). This organism produces a few extracellular products implicated in bacterial virulence and pathogenesis, including cytolysin (12, 39), metalloprotease (11, 19), phospholipase (37), and siderophores (31). Only a single extracellular metalloprotease (designated Vvp) has been identified (11, 19). This protease has been shown to increase vascular permeability and edema through activating the Hageman factor-plasma kallikrein-kinin cascade (20, 21, 22) and to cause hypodermic hemorrhage in guinea pigs (23). It can also facilitate iron acquisition by the organism by digesting heme proteins, transferrin, and lactoferrin (24, 25). Based on these observations, Vvp was thought to be important in bacterial growth and disease development. Nevertheless, a Vvp-deficient mutant has been shown to be even more virulent than the wild-type strain, probably because of overexpression of the cytolysin in the absence of Vvp (30).
Vibrio cholerae hemagglutinin/protease (HA protease) (8) and Vibrio anguillarum EmpA (17), like Vvp of V. vulnificus, are members of the metalloprotease family in vibrios. The regulators of the genes of these two proteases have been identified: HapR for HA protease (10) and VanS for EmpA (D. Milton, U. Hope, M. Camara, and P. Williams, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. H-193, p. 366, 1999). Both HapR and VanS are members of the Vibrio harveyi LuxR family, with sequences and function similar to those of LuxR (10). The LuxR protein is a transcriptional activator which controls expression of the lux operon of V. harveyi at high bacterial cell density (34). Gene regulation by LuxR is a part of the quorum-sensing (cell density-dependent) regulatory system.
In this study, we investigated the transcription of vvp under various culture conditions, including different temperatures, osmotic pressures, iron levels, and oxygen levels. The results indicated that vvp was transcribed in a growth phase-dependent manner similar to that of the Pseudomonas aeruginosa elastase gene (lasB) (26) or the V. cholerae HA protease gene (hap) (10). Expression of LasB has been demonstrated to be regulated by a quorum-sensing regulatory system (26). A luxR homologue was identified in V. vulnificus by PCR with a pair of degenerate primers derived from sequences conserved in the genes of members of the V. harveyi LuxR family. The deduced amino acid sequence of this gene was identical to that of the smcR gene of V. vulnificus reported previously (15). We further demonstrated that SmcR is involved in the regulation of transcription of both vvp and the cytolysin gene by isolating and characterizing a mutant that was disrupted in smcR.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The V. vulnificus and Escherichia coli strains and the plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| V. vulnificus | ||
| YJ016 | Clinical isolate | Laboratory collection |
| YJ024 | Translucent mutant of YJ016 | 9 |
| CP080 | YJ016 Δvvp | 30 |
| CP154 | YJ016 smcR::pVR19 | This study |
| CP156 | YJ016 ΔsmcR | This study |
| CP194 | CP156/pVR21 | This study |
| E. coli | ||
| DH5α | supE44 lacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 7 |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpir Kmr Nalr | 16 |
| LLM1956 | DH5α/pRS205 | 14 |
| Plasmids | ||
| pRS205 | pUC19 with the lux operon of V. harveyi | 14 |
| pJRD215 | Shuttle vector; Kmr | 5 |
| pUC19 | Cloning vector; Apr | Laboratory collection |
| pCVD442 | Suicide vector; Apr | 6 |
| pVR11 | pBR322 inserted with a 3.3-kb fragment containing the intact smcR gene | This study |
| pVR18 | pVR11 with a 445-bp deletion in smcR (ΔsmcR) | This study |
| pVR19 | pCVD442 with the NheI-NheI fragment that contains ΔsmcR cloned from pVR18 | This study |
| pVR21 | pJRD215 with the NheI-NheI fragment that contains the intact smcR cloned from pVR11 | This study |
| pVR22 | pJRD215 with the NheI-NheI fragment that contains ΔsmcR cloned from pVR18 | This study |
Abbreviations: Apr, Kmr, and Nalr are resistance to ampicillin, kanamycin, and nalidixic acid, respectively.
Cultivation and storage of bacteria.
All strains were grown at 37°C with vigorous aeration in Luria broth (LB), in which ampicillin (100 μg/ml), polymyxin B (50 U/ml), or tetracycline (15 μg/ml) was added as appropriate. They were maintained at −70°C in LB medium containing 17% glycerol.
DNA preparation and manipulation.
Plasmid DNA was extracted from the bacterial cells by the alkaline lysis method of Birnboim and Doly (3). Standard techniques were used to construct recombinant plasmids (28). DNA restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs. DNA fragments were purified from the agarose gels by using the GeneClean Glassmilk kit (Bio 101, Inc.).
PCR was performed with a thermocycler (GeneAmp PCR system 9600; Perkin-Elmer Cetus) as described previously (30). The degenerate primers used in PCR for amplifying the luxR homologue from the chromosome of V. vulnificus were 5′-GGTGGTCACGCG(A)GATATTG and 5′-CCGTGGAAT(C)AGG(A)T(C)TT(G/C)GCC. Nucleotide sequence was determined by an autosequencer (ABI Prism 377 DNA Sequencer; Applied Biosystems).
RNA slot, colony, and Southern hybridization.
Total RNA was prepared with an RNA isolation kit (RNeasy MiniKit; Qiagen GmbH, Hilden, Germany). Whole-cell DNA was prepared as described previously (30). Probes used in colony, Southern, and RNA slot hybridization were labeled with [α-32P]dCTP (Amersham Pharmacia Biotech Asia Pacific Ltd., Hong Kong, People's Republic of China) by random priming with a kit (Megaprime DNA labeling system; Amersham Pharmacia Biotech) using either the PCR products or fragments excised from the recombinant plasmids as the templates. The nylon membranes with DNA or RNA were prehybridized with the hybridization buffer (ExpressHyb Hybridization solution; Clontech Laboratories, Inc.) for 30 min at 68°C, hybridized for 1.5 h at the same temperature, washed, and visualized by autoradiography.
Primer extension.
Fifty micrograms of total RNA was used as the template in the primer extension reaction. A primer (5′-GCCACGACGAGCAAACACTTCCAG-3′) complementary to the coding strand 98 bp downstream of the start codon of smcR was end labeled with [γ-32P]ATP (Amersham Pharmacia Biotech). Primer extension was performed by the method of Wu et al. (40) with modifications. Superscript II RNase H− reverse transcriptase (GIBCO BRL Life Technologies, Inc.), instead of avian myeloblastosis virus (AMV) reverse transcriptase, was used and the reaction mixture was incubated at 50°C for 1.5 h. Plasmid pVR11 was primed with the same primer for a sequencing reaction by the dideoxy chain termination procedure of Sanger and Coulson (29) with the Sequenase 2.0 kit (United States Biochemicals) and [α-35S]dATP (Amersham Pharmacia Biotech). The primer extension product was then subjected to electrophoresis on a 7 M urea–6% polyacrylamide gel in parallel with the DNA sequencing products.
Bioluminescence assay.
The level of bioluminescence in a bacterial overnight culture was determined with a luminometer (Minilumate LB9506; Laboratorium Prof. Dr. Berthold GmbH & Co. KG, Bad Wildbad, Germany) and was expressed as relative light units (RLU) divided by the optical density at 600 nm (OD600) of the culture.
Construction of the smcR mutant.
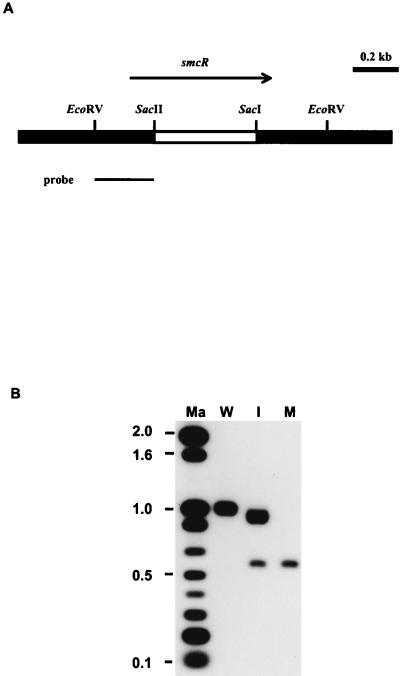
A deletion in smcR was introduced into the chromosome of V. vulnificus by in vivo allelic exchange. Plasmid pVR18 was first constructed by removing a fragment between the SacII and SacI sites in smcR (see Fig. 4) from pVR11. This resulted in a 445-bp deletion in smcR, which was confirmed by DNA sequence determination. The NheI-NheI fragment from pVR18 was cloned into the XhoI site in pCVD442 to create pVR19. Plasmid pCVD442 is a suicide vector containing the sacB gene, which allowed positive selection with sucrose for loss of the vector. Plasmid pVR19 was transferred from E. coli SM10λpir to V. vulnificus by conjugation. The transconjugants, which had pVR19 integrated in the chromosome via homologous recombination, were selected by ampicillin and polymyxin B and tested for sensitivity to 10% sucrose. Such transconjugants were obtained at a rate of 1 per 108 recipients. One of the sucrose-sensitive transconjugants was grown in LB containing 10% sucrose at 37°C overnight and then spread onto a 10% sucrose-containing LB plate for selecting the sucrose-resistant clones. The resultant strains were further tested for ampicillin sensitivity. Of the 200 sucrose-resistant colonies tested, 34 were ampicillin sensitive.
FIG. 4.
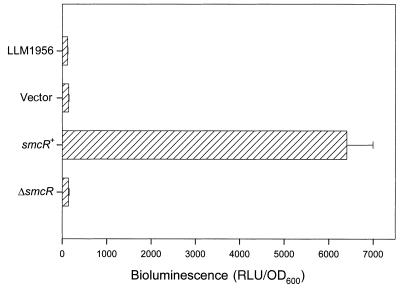
Cross activation of the lux operon by SmcR. Bacteria were grown overnight in LB at 37°C. The bacterial concentrations and bioluminescence (relative light units [RLU]) of the cultures were then determined. LLM1956, the reporter strain; vector, LLM1956 with pJRD215 as a negative control; smcR+, LLM1956 with pVR21 that carried the intact smcR gene; ΔsmcR, LLM1956 with pVR22 that carried ΔsmcR.
Protease and cytolysin assays.
The protease activity in the culture supernatant was determined as described by Kreger and Lockwood (12). The cytolysin activity in the culture supernatant was assayed as described previously (30) and was expressed as 100% × (OD545 of specimen/OD545 of complete hemolysis by Triton X-100).
Determination of bacterial concentration.
The bacterial concentration in the broth culture was estimated either by determining the OD600 value of the culture with a spectrophotometer (U-2000 Spectrophotometer; Hitachi Ltd., Tokyo, Japan), or by measuring the culture turbidity (expressed as Klett arbitrary units [kau]) with a turbidometer (Klett-Summerson photoelectric colorimeter; Klett Mfg. Co., Inc.).
Preconditioned medium assay.
Bacteria cultivated in LB overnight at 37°C were diluted 1:100 in fresh LB and grown for a given period. The culture supernatant was collected and sterilized by filtration through a 0.22-μm-pore-size membrane and was used as the preconditioned medium. The preconditioned media were prepared with an isogenic Δvvp mutant (30) to exclude the protease activity in them. A solution of 20× LB was added to the preconditioned medium to a final concentration of 0.5× to correct the nutrients when necessary. To test the autoinducer activity in the preconditioned medium, an overnight bacterial culture was diluted 1:500 in the preconditioned medium and grown at 37°C, and the protease activity in the culture supernatant was assayed at intervals.
Virulence assay.
Virulence of the mutant and parent strain were tested in either normal or iron-overloaded mice. C3H/HeN mice, 6 to 8 weeks old and purchased from the animal center of the College of Medicine, National Cheng-Kung University, were challenged by intraperitoneal (i.p.) injection of the bacterial suspension. Mice were made iron overloaded by injecting 25 mg of iron dextran (Sigma Chemical Co.) per mouse intramuscularly 2 h prior to challenge. A group of five mice was given 0.5 ml of a 10-fold serially diluted bacterial suspension in phosphate-buffered saline per mouse and mortality was recorded 48 h postinfection. The LD50 (the dose lethal to 50% of the mice) was calculated by the method of Reed and Muench (27).
Resistance to serum killing.
Forty microliters of bacterial suspension in phosphate-buffered saline was mixed with 160 μl of pooled serum from healthy volunteers, and the mixture was incubated with end-over-end rotation at 37°C for 30 min. The number of viable bacteria in the mixture was then determined by plate counts.
DNA database search.
The National Center for Biotechnology Information service was used to consult the GenBank database with the BLAST algorithm for searching the homologous sequences.
RESULTS
Regulation of vvp transcription.
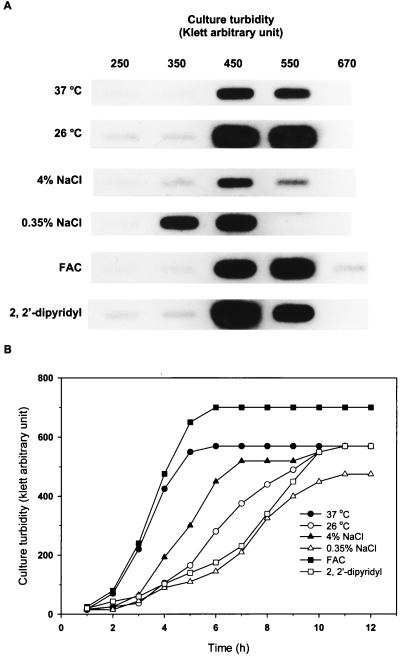
The transcription levels of vvp during growth under various conditions were examined by RNA slot blot analysis. Bacteria grown aerobically usually reached a maximal turbidity of 550 kau, except for those grown at low salt concentrations and high iron levels, the maximal turbidity for which was 450 and 670 kau, respectively. Transcription of vvp was higher at 26°C than at 37°C, higher at low salt than at high salt concentrations, and higher at low iron than at high iron levels (Fig. 1A). In every condition studied, transcription of vvp was turned up dramatically when the culture was just about to enter the stationary phase (Fig. 1A and B). V. vulnificus grew poorly anaerobically and only reached 90 kau (data not shown), and the transcription of vvp was undetectable.
FIG. 1.
(A) Transcriptional levels of vvp during growth under various culture conditions. Five micrograms of total RNA extracted from 109 bacteria of YJ016 was subjected to RNA slot blot hybridization with a 32P-labeled probe derived from the vvp gene. FAC, ferric ammonium citrate. The concentrations of ferric ammonium citrate and 2,2′-dipyridyl in LB medium were 0.1 mg/ml and 0.15 mM, respectively. All cultures (except for that cultured at 26°C) were grown at 37°C. (B) Growth curves of YJ016 cultivated under various conditions. The culture conditions were the same as those in panel A.
Detection of autoinducer activity in culture supernatant of V. vulnificus for protease expression.
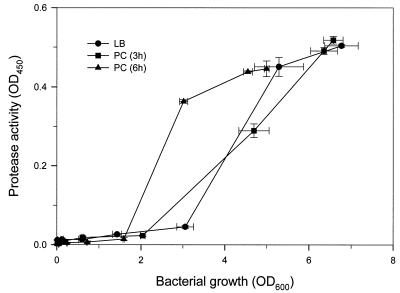
The kinetics of vvp transcription suggested that expression of this gene may be partly regulated by the quorum-sensing mechanism, in which the autoinducers secreted by the organism act as the signaling molecules. To test this, the culture supernatants collected from a 3-h (OD600 = 2.0) and a 6-h (OD600 = 6.6) culture were examined for autoinducer activity. V. vulnificus YJ016 grown in either preconditioned medium exhibited a similar growth rate as that of those grown in LB medium (data not shown). However, the bacteria grown in the preconditioned medium from a 6-h, but not a 3-h, culture expressed the protease activity at a lower bacterial cell density compared with those grown in LB (Fig. 2).
FIG. 2.
Effects of preconditioned media (PC) on expression of protease activity in culture supernatant. An overnight culture of V. vulnificus YJ016 was diluted 1:500 in LB, 3-h preconditioned medium, and 6-h preconditioned medium with 0.5× LB. The bacteria were then grown at 37°C, and the OD600 of the culture as well as the protease activity in the culture supernatant was determined every hour.
Cloning and nucleotide sequence determination of the luxR homologue in V. vulnificus.
To identify the V. harveyi luxR homologue in V. vulnificus, a gene library (4) of a clinical strain, YJ016, was screened by colony hybridization. The probe used was amplified from a chromosome with a pair of degenerate primers derived from sequences that are conserved in various luxR-like genes. One clone thus obtained was further used to determine the nucleotide sequence of the insert in the recombinant plasmid. Two complete and one partial open reading frames (ORFs) were identified. Sequence comparison of the two complete ORFs with those in the database of GenBank showed that they were highly homologous to the gene of dihydrolipoamide dehydrogenase (Lpd) of Vibrio parahaemolyticus and LuxR of V. harveyi. The incomplete ORF shared sequence homology with the N terminal of the hypoxanthine ribosyltransferase (Htp) of V. parahaemolyticus. The deduced amino acid sequence of the cloned luxR homologue (GenBank accession no. AY007308) was identical to that of the smcR gene of V. vulnificus published recently by McDougald et al. (15). Therefore, the gene we cloned was smcR. The amino acid sequence of SmcR was 93, 93, and 78% identical to LuxR of V. harveyi, OpaR of V. parahaemolyticus, and HapR of V. cholerae, respectively. The predicted molecular mass and isoelectric point of SmcR were 23.7 kDa and 5.81, respectively.
Determination of the tsp of smcR.
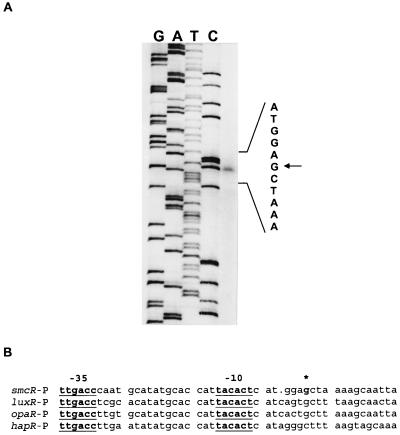
The transcription start point (tsp) of smcR was determined by primer extension (Fig. 3A). A G located 87 bp upstream of the start codon was identified. The putative promoter sequence (13) determined according to the tsp was TTGACC for the −35 and TACACT for the −10 sequences, which were separated by 16 bp. Alignment of the promoter regions of the luxR family members revealed identical promoter sequences among them (Fig. 3B).
FIG. 3.
(A) Determination of the tsp of smcR. Fifty micrograms of total RNA extracted from YJ016 was used in the primer extension reaction. Nucleotide sequences (GATC) of the noncoding strand up- and downstream of tsp obtained with the same oligoprimer are indicated. A G (indicated by an arrow) 87 bp upstream of the initiation codon was identified as the tsp. (B) Sequence alignment of the promoter regions of smcR, luxR, opaR, and hapR. The underlines indicate the promoter sequences (−10 and −35) and the asterisk denotes the tsp.
Transactivation of V. harveyi lux operon by SmcR.
An E. coli strain containing a functional but luxR-requiring V. harveyi lux operon in pRS205 was transformed with pJRD215, the vector; pVR21 that carried smcR; or pVR22 that carried ΔsmcR. Bioluminescence produced by the transformants was then measured. As shown in Fig. 4, a high level of bioluminescence was detected in the presence, but not in the absence, of SmcR, indicating that LuxR can be replaced by SmcR for activating the lux operon.
Isolation of V. vulnificus SmcR-deficient mutant.
To determine the roles of SmcR in the regulation of transcription of vvp and other genes in V. vulnificus, an isogenic ΔsmcR mutant (RD mutant) was isolated from a clinical V. vulnificus isolate, YJ016. A suicide vector, pVR19, which carried the smcR gene with a 445-bp deletion (Fig. 5), was constructed and used to isolate the RD mutant by an allelic exchange technique. Twelve randomly chosen sucrose-resistant, ampicillin-sensitive colonies were examined by PCR using a pair of primers complementary to sequences flanking the deletion, and seven of them were shown to contain the deletion (data not shown). The presence of the smcR deletion in the chromosome of one of the deletion-containing colonies, CP156, was further confirmed by Southern hybridization (Fig. 5).
FIG. 5.
(A) Restriction map of smcR and the flanking regions. The arrow indicates the direction of transcription. The extent of deletion (blank bar) in vvpR and the probe used in Southern hybridization are also depicted. (B) Detection of the deletion in smcR in the chromosome of V. vulnificus. Ten micrograms of genomic DNA was digested by EcoRV and then subjected to electrophoresis on a 1.2% agarose gel. Ma, molecular weight standards; W, V. vulnificus YJ016 (wild type); I, CP154 (YJ016 with pVR19 integrated in the chromosome); M, CP156 (ΔsmcR).
Expression of V. vulnificus metalloprotease and cytolysin in the RD mutant.
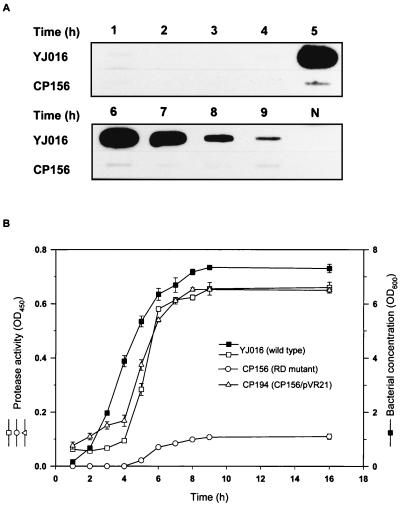
When examined by RNA slot blot hybridization with the gene-specific probes, transcription of vvp and the cytolysin gene, vvhA, in the RD mutant was different from that in the parent strain (Fig. 6 and 7), whereas transcription of the nuclease gene, vvn (41), was not affected by disrupting smcR (data not shown). As shown in Fig. 6A, the level of vvp transcription in the wild-type strain rose rapidly at 5 h of growth from basal to a high level, which was maintained for a few hours afterwards. However, transcription of vvp was greatly reduced in the RD mutant. Coinciding with the transcriptional level, the protease activity in the culture supernatant of the RD mutant is dramatically reduced compared to that of the parent strain (Fig. 6B). Such a low level of protease activity was also found in the RD mutant cultured under any of the other conditions (listed in Fig. 1) that have been used to examine the regulation of vvp expression in the parent strain (data not shown). Protease activity in the culture supernatant was restored to the wild-type level when pVR21, which carried the intact smcR, was introduced into the RD mutant (Fig. 6B).
FIG. 6.
Expression of Vvp by V. vulnificus strains. The bacteria were grown in LB at 37°C after a 1:100 dilution of an overnight culture. Total RNA (5 μg) was extracted from the bacteria harvested at intervals and examined for the transcriptional levels of the vvp gene by RNA slot blot hybridization (A). Bacterial cell density and the protease activity in the culture supernatant of each strain (n = 3) were determined at the same time (B). The growth curves of all strains were similar and only that of the wild-type strain is shown. N, E. coli total RNA used as a negative control.
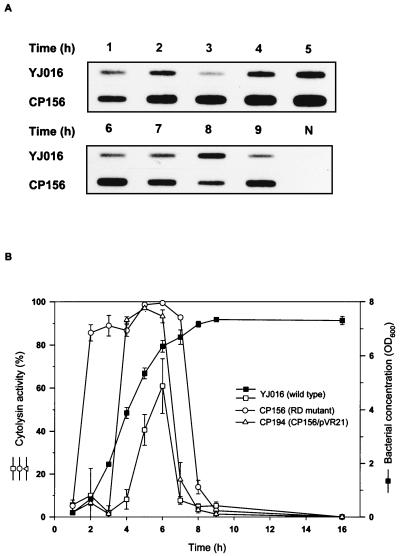
FIG. 7.
Expression of the cytolysin by V. vulnificus strains. Bacteria were grown in LB at 37°C after a 1:100 dilution of an overnight culture. Total RNA (5 μg) was extracted from the bacteria harvested at intervals and examined for the transcriptional levels of the vvhA gene by RNA slot blot hybridization (A). Bacterial cell density and the cytolysin activity in the culture supernatant of each strain (n = 3) were determined at the same time (B). The growth curves of all strains were similar and only that of the wild-type strain is shown. N, E. coli total RNA used as a negative control.
Transcription of vvhA, in contrast to that of vvp, was higher in the RD mutant than in the parent strain (Fig. 7A), particularly during the log phase of growth (2 to 6 h). The cytolysin activity in the culture supernatant not only was higher and detected earlier but also was sustained longer in the RD mutant than in the parent strain (Fig. 7B). Moreover, a biphasic expression was observed in the parent strain but not in the RD mutant for either the transcription of vvhA or the cytolysin activity in the culture supernatant. Introduction of pVR21 into the RD mutant resulted in greatly reduced cytolysin activity during the early log phase but not the late log phase and a slightly earlier decline of the cytolysin activity during the stationary phase (Fig. 7B).
Other phenotypes of the RD mutant.
Opacity of the colonies of the RD mutant was between that of the opaque parent strain and a translucent mutant, YJ024 (data not shown). The virulence of the RD mutant, represented by the LD50 value, in either normal or iron-overloaded mice was comparable to that of the parent strain (Table 2). The RD mutant was also as resistant as its parent strain to the human serum killing effect (P > 0.05) (Table 2). The translucent mutant, which served as a control, was weakly virulent in mice and was very sensitive to human serum (Table 2).
TABLE 2.
Virulence of V. vulnificus strains in mice and their resistance to human serum
| Straina | LD50 (CFU/mouse) for:
|
Bacterial resistance to human serum (%)b
|
||
|---|---|---|---|---|
| Normal mouse | Iron-overloaded mouse | Untreated | Heat inactivated | |
| YJ016 | 1.0 × 106 | <1.0 × 101 | 25.6 ± 11.7 | 35.9 ± 4.4 |
| CP156 | 2.0 × 106 | <1.0 × 101 | 46.3 ± 11.6 | 38.9 ± 9.6 |
| YJ024 | 3.2 × 107 | 9.5 × 105 | 0 ± 0 | 53.8 ± 3.8 |
YJ016, wild-type strain; CP156, RD mutant; YJ024, translucent mutant of YJ016.
Bacterial resistance to human serum, either untreated or heated at 56°C for 30 min to inactivate the complement, is expressed as 100% × (the number of viable bacteria after treatment/the number of viable bacteria before treatment). Values represent means ± standard deviations.
DISCUSSION
Transcription of vvp, which encodes the metalloprotease of V. vulnificus, was shown to be affected by a variety of culture conditions, such as temperature, iron levels, and salt concentrations, indicating that multiple factors may be involved in the regulation of vvp. Nevertheless, in every case studied, transcription of vvp was turned up dramatically in the late log growth phase, like that of many other bacterial protease genes (10, 26). Some protease genes with such expression patterns have been shown to be regulated by the quorum-sensing, or cell density-dependent, regulatory systems (2).
Two different families of quorum-sensing regulatory systems, represented by those of Vibrio fischeri and V. harveyi, have been found widely distributed in the gram-negative bacteria. The quorum-sensing system in V. fischeri that regulates the genes involved in the production of bioluminescence is composed of a signaling molecule (the autoinducer) and a positive regulator, which is activated upon binding with the autoinducer (2, 33). The quorum-sensing system in V. harveyi, which is also involved in the regulation of the bioluminescence genes, is more complicated and is composed of two autoinducers (AI1 and AI2), the autoinducer receptors, a signal transducer, and a negative regulator (1, 2). Although the transcriptional activator, LuxR, of V. harveyi does not respond to stimulation by the autoinducers, it is required for the activation of transcription of the target genes (34).
Existence of a V. harveyi quorum-sensing system-like regulatory mechanism in V. vulnificus has been proposed based on the detection of an AI2-like activity in the culture supernatant (B. L. Bassler, personal communication) and the identification of a LuxR homologue (15). A LuxS-like AI2 synthase has also been identified recently (S. Y. Kim et al., Abstr. 100th Gen. Meet. Am. Soc. Microbiol. 2000, abstr. B-248, p. 97, 2000). We detected an autoinducer activity in a high-cell-density (OD600 = 6.6) culture supernatant, but not in that of a low-cell-density (OD600 = 2.0) culture, for the expression of protease at a lower bacterial cell density. This suggested that vvp may be partly regulated by the quorum-sensing mechanism. The LuxR homologue of V. vulnificus, SmcR, was shown in this study to complement a LuxR-deficient E. coli in activating the bioluminescence genes of V. harveyi. The functional similarity between LuxR and SmcR also implies that SmcR may be involved in a quorum-sensing regulatory system. We further demonstrated that SmcR is required for vvp expression because the transcription of vvp was greatly reduced in an SmcR-deficient V. vulnificus mutant grown under a variety of culture conditions.
The V. harveyi luxR family members identified so far, including smcR, are not only highly homologous in the encoding sequences but are also identical in the promoter regions (the −10 and −35 sequences). In addition, each member has been shown to be capable of transactivating the lux operon of V. harveyi, suggesting that they may employ a common mechanism in activating the target genes. Two LuxR-binding sites (LuxR boxes) have been identified in the promoter of luxC, a target gene of luxR, by footprint analysis (18, 35). However, the LuxR boxes were not found in the promoters of two putative target genes of the LuxR homologues: hap of V. cholerae (10) and vvp of V. vulnificus (4). Therefore, the LuxR homologues may recognize specific binding sequences that share a low level of homology with each other. Alternatively, the hap and vvp genes may each be regulated indirectly rather than directly by the LuxR homologue via another regulatory factor. A study of the interaction of SmcR and HapR with the promoter regions of vvp and hap, respectively, is required for distinguishing between the two possibilities.
Expression of the cytolysin of V. vulnificus has been shown previously to be regulated by Vvp at the posttranslational level (30). We further found in this study that transcription of the cytolysin gene, vvhA, was affected by disrupting smcR. Transcription of vvhA was increased during the log phase, suggesting that SmcR may be involved in negative regulation of vvhA expression. In the RD mutant, the production of Vvp is greatly reduced and the transcription of vvhA is increased. Consequently, the cytolysin activity in the culture supernatant is detected earlier and reaches a maximal activity higher than that of the parent strain, as demonstrated in this study. However, in contrast to the prolonged high cytolysin activity detected in the Vvp-deficient mutant (30), this high level of cytolysin activity was sustained for about 5 h and then declined to undetectable levels at 8 h of growth in the RD mutant. Decline of the cytolysin activity in the RD mutant may be caused by the low level of protease expressed.
The role of SmcR in the regulation of Vvp or cytolysin expression was confirmed by complementing the RD mutant with SmcR expressed from a plasmid. The pattern of Vvp expression in the reconstituted strain, as represented by the protease activity detected in the culture supernatant, was similar to that in the wild-type strain. However, the pattern of cytolysin expression in the reconstituted strain was different from that in the wild-type strain. The biphasic expression was restored by complementation with SmcR, but the maximal cytolysin activity in the reconstituted strain was much higher than that in the wild-type strain. We do not know currently what caused this partial repression of cytolysin expression in the reconstituted strain. Nevertheless, our results imply that the regulation of vvhA expression by SmcR may occur via a mechanism that is more complex than that involved in the regulation of vvp by SmcR.
Various colony morphological changes have been found to be associated with the LuxR homologues in a number of vibrios. Disruption of hapR in V. cholerae resulted in a rugose phenotype (10), while expression of OpaR in a translucent strain of V. parahaemolyticus brought about opaque colonies (14). Morphological change of the colony was also observed in the RD mutant of V. vulnificus: the colonies of the RD mutant were less opaque than those of the parent strain. Such a morphological change could be caused by alterations of the bacterial cell surface compositions. Translucent variants are sometimes obtained from the opaque V. vulnificus strains, and the variation in opacity has been found to be accompanied by variation of capsular polysaccharide and bacterial virulence in mice (32). In contrast to the opaque strains, the translucent variants, including YJ024 used in this study, usually contain less or no capsular polysaccharide and are much less virulent in iron-overloaded mice (32). Although the RD mutant exhibited an intermediate phenotype between the parent strain and the translucent mutant in colonial opacity, it was as virulent as the parent strain in the iron-overloaded mice. Moreover, it showed a wild-type level of resistance to human serum killing activity. Therefore, disruption of smcR either did not affect the amount of the capsular polysaccharide or, if it had any effect on capsular synthesis, the effect was not sufficient to result in reduction of bacterial virulence in mice or resistance to human serum.
In conclusion, our data demonstrated that the V. harveyi LuxR homologue, SmcR, positively controls the transcription of the metalloprotease gene and may also be involved in the negative regulation of the cytolysin gene. The target genes of SmcR remain to be identified.
ACKNOWLEDGMENTS
This work was partly supported by grants DOH 88-HR-606 from the National Health Institute and NSC 89-2320-B-006-018 from the National Science Council, Taiwan, Republic of China.
REFERENCES
- 1.Bassler B L, Wright M, Silverman M R. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequences and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 2.Bassler B L. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol. 1999;2:582–587. doi: 10.1016/s1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng J C, Shao C P, Hor L I. Cloning and nucleotide sequencing of the protease gene of Vibrio vulnificus. Gene. 1996;183:255–257. doi: 10.1016/s0378-1119(96)00488-x. [DOI] [PubMed] [Google Scholar]
- 5.Davison J, Heusterspreute M, Chevalier N, Ha-Thi V, Brunel F. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene. 1987;51:275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- 6.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 8.Hase C C, Finkelstein R A. Cloning and nucleotide sequence of the Vibrio cholerae hemagglutinin/protease (HA/protease) gene and construction of an HA/protease-negative strain. J Bacteriol. 1991;173:3311–3317. doi: 10.1128/jb.173.11.3311-3317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hor L I, Chang Y K, Chang C C, Lei H Y, Ou J T. Mechanism of high susceptibility of iron-overloaded mouse to Vibrio vulnificus infection. Microbiol Immunol. 2000;44:871–878. doi: 10.1111/j.1348-0421.2000.tb02577.x. [DOI] [PubMed] [Google Scholar]
- 10.Jobling M G, Holmes R K. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol. 1997;26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 11.Kothary M H, Kreger A S. Purification and characterization of an elastolytic protease of Vibrio vulnificus. J Gen Microbiol. 1987;133:1783–1791. doi: 10.1099/00221287-133-7-1783. [DOI] [PubMed] [Google Scholar]
- 12.Kreger A, Lockwood D. Detection of extracellular toxin(s) produced by Vibrio vulnificus. Infect Immun. 1981;33:583–590. doi: 10.1128/iai.33.2.583-590.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lisser S, Margalit H. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarter L L. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio paraheamolyticus. J Bacteriol. 1998;180:3166–3173. doi: 10.1128/jb.180.12.3166-3173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDougald D, Rice S A, Kjelleberg S. The marine pathogen Vibrio vulnificus encodes a putative homologue of the Vibrio harveyi regulatory gene, luxR: a genetic and phylogenetic comparison. Gene. 2000;248:213–221. doi: 10.1016/s0378-1119(00)00117-7. [DOI] [PubMed] [Google Scholar]
- 16.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutants: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milton D L, Norqvist A, Wolf-Watz H. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J Bacteriol. 1992;174:7235–7244. doi: 10.1128/jb.174.22.7235-7244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyamoto C M, Smith E E, Swartzman E, Cao J G, Graham F, Meighen E A. Proximal and distal sites bind LuxR independently and activate expression of the Vibrio harveyi lux operon. Mol Microbiol. 1994;14:255–262. doi: 10.1111/j.1365-2958.1994.tb01286.x. [DOI] [PubMed] [Google Scholar]
- 19.Miyoshi N, Shimizu C, Miyoshi S, Shinoda S. Purification and characterization of Vibrio vulnificus protease. Microbiol Immunol. 1987;31:13–25. doi: 10.1111/j.1348-0421.1987.tb03064.x. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi N, Miyoshi S, Sugiyama K, Suzuki Y, Furuta H, Shinoda S. Activation of the plasma kallikrein-kinin system by Vibrio vulnificus protease. Infect Immun. 1987;55:1936–1939. doi: 10.1128/iai.55.8.1936-1939.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyoshi S, Shinoda S. Role of the protease in the permeability enhancement by Vibrio vulnificus. Microbiol Immunol. 1988;32:1025–1032. doi: 10.1111/j.1348-0421.1988.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 22.Miyoshi S, Shinoda S. Activation mechanism of human Hageman factor-plasma kallikrein-kinin system by Vibrio vulnificus metalloprotease. FEBS Lett. 1992;308:315–319. doi: 10.1016/0014-5793(92)81301-2. [DOI] [PubMed] [Google Scholar]
- 23.Miyoshi S, Nakazawa H, Kawata K, Tomochika K, Tobe K, Shinoda S. Characterization of the hemorrhagic reaction caused by Vibrio vulnificus metalloprotease, a member of the thermolysin family. Infect Immun. 1998;66:4851–4855. doi: 10.1128/iai.66.10.4851-4855.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishina Y, Miyoshi S, Nagase A, Shinoda S. Significant role of an exocellular protease in utilization of heme by Vibrio vulnificus. Infect Immun. 1992;60:2128–2132. doi: 10.1128/iai.60.5.2128-2132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okujo N, Akiyama T, Miyoshi S, Shinoda S, Yamamoto S. Involvement of vulnibactin and exocellular protease in utilization of transferrin- and lactoferrin-bound iron by Vibrio vulnificus. Microbiol Immunol. 1996;40:595–598. doi: 10.1111/j.1348-0421.1996.tb01114.x. [DOI] [PubMed] [Google Scholar]
- 26.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence gene requires cell-to-cell communication. Science. 1993;260:1127–1137. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 27.Reed L J, Muench H. A simple method of estimating the fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sanger F, Coulson A R. A rapid method for determining sequences in DNA primer synthesis with DNA polymerase. J Mol Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 30.Shao C P, Hor L I. Metalloprotease is not essential for Vibrio vulnificus in mice. Infect Immun. 2000;68:3569–3573. doi: 10.1128/iai.68.6.3569-3573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson L M, Oliver J D. Siderophore production by Vibrio vulnificus. Infect Immun. 1983;41:644–649. doi: 10.1128/iai.41.2.644-649.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simpson L M, White V K, Zane S F, Oliver J D. Correlation between virulence and colony morphology in Vibrio vulnificus. Infect Immun. 1987;55:269–272. doi: 10.1128/iai.55.1.269-272.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sitnikov D M, Schineller J B, Baldwin T O. Transcriptional regulation of bioluminescence genes from Vibrio fischeri. Mol Microbiol. 1995;17:801–812. doi: 10.1111/j.1365-2958.1995.mmi_17050801.x. [DOI] [PubMed] [Google Scholar]
- 34.Swartzman E, Silverman M, Meighen E A. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J Bacteriol. 1992;174:7490–7493. doi: 10.1128/jb.174.22.7490-7493.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swartzman E, Meighen E A. Purification and characterization of a poly(dA-dT) lux-specific DNA-binding protein from Vibrio harveyi and identification as LuxR. J Biol Chem. 1993;268:16706–16716. [PubMed] [Google Scholar]
- 36.Tacket C O, Brenner F, Blake P A. Clinical features and an epidemiological study of Vibrio vulnificus infections. J Infect Dis. 1984;149:558–561. doi: 10.1093/infdis/149.4.558. [DOI] [PubMed] [Google Scholar]
- 37.Teata J, Daniel L W, Kreger A S. Extracellular phospholipase A2 and lysophospholipase produced by Vibrio vulnificus. Infect Immun. 1984;45:458–463. doi: 10.1128/iai.45.2.458-463.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warnock E W, MacMath T L. Primary Vibrio vulnificus septicemia. J Emerg Med. 1993;11:153–156. doi: 10.1016/0736-4679(93)90510-e. [DOI] [PubMed] [Google Scholar]
- 39.Wright A C, Morris J G., Jr The extracellular cytolysin of Vibrio vulnificus: inactivation and relationship to virulence in mice. Infect Immun. 1991;59:192–197. doi: 10.1128/iai.59.1.192-197.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J J, Howard M G, Piggot P J. Regulation of transcription of the Bacillus subtilis spoIIA locus. J Bacteriol. 1989;171:692–698. doi: 10.1128/jb.171.2.692-698.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, S. I., S. K. Lo, C. P. Shao, H. W. Tsai, and L. I. Hor. Cloning and characterization of a periplasmic nuclease of Vibrio vulnificus and its role in preventing uptake of foreign DNA. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]