Abstract
Synechocystis sp. strain PCC 6803 has five genes for putative Na+/H+ antiporters (designated nhaS1, nhaS2, nhaS3, nhaS4, and nhaS5). The deduced amino acid sequences of NhaS1 and NhaS2 are similar to that of NhaP, the Na+/H+ antiporter of Pseudomonas aeruginosa, whereas those of NhaS3, NhaS4, and NhaS5 resemble that of NapA, the Na+/H+ antiporter of Enterococcus hirae. We successfully induced the expression of nhaS1, nhaS3, and nhaS4 under control of an Na+-dependent promoter in Escherichia coli TO114, a strain that is deficient in Na+/H+ antiport activity. Inverted membrane vesicles prepared from TO114 nhaS1 and TO114 nhaS3 cells exhibited Na+(Li+)/H+ antiport activity. Kinetic analysis of this activity revealed that nhaS1 encodes a low-affinity Na+/H+ antiporter with a Km of 7.7 mM for Na+ ions and a Km of 2.5 mM for Li+ ions, while nhaS3 encodes a high-affinity Na+/H+ antiporter with a Km of 0.7 mM for Na+ ions and a Km of 0.01 mM for Li+ ions. Transformation of E. coli TO114 with the nhaS1 and nhaS3 genes increased cellular tolerance to high concentrations of Na+ and Li+ ions, as well as to depletion of K+ ions during cell growth. To our knowledge, this is the first functional characterization of Na+/H+ antiporters from a cyanobacterium. Inverted membrane vesicles prepared from TO114 nhaS4 cells did not have Na+/H+ antiport activity, and the cells themselves were as sensitive to Na+ and Li+ ions as the original TO114 cells. However, the TO114 nhaS4 cells were tolerant to depletion of K+ ions. Taking into account these results and the growth characteristics of Synechocystis mutants in which nhaS genes had been inactivated by targeted disruption, we discuss possible roles of NhaS1, NhaS3, and NhaS4 in Synechocystis.
High salinity is a major environmental factor that limits the growth and productivity of plants, eukaryotic microorganisms, and bacteria. Control of membrane permeability to Na+ ions and the counteracting K+ ions is the most important aspect of the acclimation of these organisms to high-salt conditions. Na+/H+ antiporters are membrane proteins that are essential for maintenance of the balance between Na+ and K+ ions in plant, fungal, and bacterial cells, in particular when the organism lacks primary Na+ pumps or when the Na+ pumps are not operative (8, 33).
Escherichia coli has at least three genes for Na+/H+ antiporters: nhaA (14, 23), nhaB (34), and chaA (19, 31). The presence of a primary Na+ pump has been suggested (4), but E. coli mutants deficient in all three of these genes are hypersensitive to Na+ and Li+ ions (31, 35). Saccharomyces cerevisiae has Na+-ATPases (17) and an Na+(K+)/H+ antiporter, Nha1, in the plasma membrane (5). In addition, it has been suggested that an Na+/H+ antiporter in yeast, designated Nhx1, functions to remove Na+ ions from the cytosol by sequestering these ions in a prevacuolar compartment (28, 29). In contrast, it is well established that high-affinity K+ channels that restrict the influx of Na+ ions determine the capacity of plant cells to tolerate high-salt stress (38). Moreover, Apse et al. (1) demonstrated that a vacuolar Na+/H+ antiporter in Arabidopsis thaliana, AtNHX1, which is homologous to Nhx1 of S. cerevisiae, also participates in the acclimation of A. thaliana to high-salt conditions. Shi et al. (40) proposed recently that SOS1 of A. thaliana, a homolog of Na+/H+ antiporters in plasma membranes, might play a role in Na+/K+ homeostasis.
We chose cyanobacteria as a model system for studies of the molecular mechanisms of the responses of plants to high-salt stress for the following reasons. (i) Cyanobacteria perform oxygenic photosynthesis using photosystems similar to those in plant chloroplasts. (ii) The structure and the lipid compositions of cyanobacterial membranes resemble those of chloroplasts of higher plants and algae (49). (iii) Cyanobacterial cells exhibit more obvious responses to salt stress than do plant cells, and they can be exposed directly to changes in external salt conditions, demonstrating a pronounced ability to acclimate to new conditions. (iv) Some strains of unicellular cyanobacteria, such as Synechocystis sp. strain PCC 6803 (hereafter “Synechocystis”) and Synechococcus sp. strain PCC 7942, are naturally transformable and can easily be modified by transformation and gene targeting (15). (v) The entire nucleotide sequence of the Synechocystis genome has been determined (22). Moreover, cyanobacteria themselves are unusual in that they contain thylakoid membranes in addition to the outer and cytoplasmic membranes. The thylakoid membranes provide sites for photosynthesis and a variety of metabolic pathways. The unusual structural and functional features of cyanobacterial cells led us to postulate that the systems that regulate ion fluxes across membranes in cyanobacterial cells might differ from those in other types of cells.
Cyanobacterial cells actively extrude Na+ ions via the actions of Na+/H+ antiporters. They maintain low intracellular concentrations of Na+ ions and relatively high intracellular concentrations of K+ ions (36). Therefore, they must have transport systems that discriminate between K+ and Na+ ions. When cyanobacterial cells are grown under high-salt conditions, the pH gradient-dependent (ΔpH-dependent) transport of Na+ ions across the cytoplasmic membrane is enhanced (7, 30). Respiratory activity and the activity of cytochrome c oxidase are also enhanced under high-salt conditions (13, 20, 26). These observations provide circumstantial evidence for the electron transport-driven extrusion of Na+ ions by an Na+/H+ antiporter in cyanobacterial cells. There have been extensive studies of the molecular aspects of salt-inducible proteins (2, 6) and of salt-regulated genes (3, 48). However, Na+/H+ antiporters and other transporters involved in the efflux of Na+ ions have not yet been identified in cyanobacteria.
In the present study, we attempted to identify the Na+/H+ antiporters in Synechocystis. This cyanobacterium has five putative genes for homologs of Na+/H+ antiporters (22). We used a mutant of E. coli that was deficient in Na+/H+ antiporters to characterize these cyanobacterial genes by functional complementation. We demonstrate here that Synechocystis has at least two genes that encode low-affinity and high-affinity Na+/H+ antiporters, respectively.
MATERIALS AND METHODS
Nomenclature of genes.
We refer to the putative genes for Na+/H+ antiporters in Synechocystis as nhaS1 (slr1727 in the designation system proposed by Kaneko et al. [22]), nhaS2 (sll0273), nhaS3 (sll0689), nhaS4 (slr1595), and nhaS5 (slr0415).
Bacterial strains and growth conditions.
E. coli TO114 (W3110 nhaA::Kmr nhaB::Emr chaA::Cmr) (31) was generously provided by H. Kobayashi (Chiba University, Chiba, Japan). It was used as the host for complementation tests with cyanobacterial genes. Cells were grown in modified Luria-Bertani medium (39) that consisted of 1.0% tryptone (Difco, Detroit, Mich.), 0.5% yeast extract (Difco), and 100 mM KCl (LBK medium; pH 6.8). For selection and growth of transformed cells, ampicillin was added to 50 μg ml−1.
The cyanobacterial strain Synechocystis sp. PCC 6803 was originally provided by J. G. K. Williams (DuPont de Nemours and Co., Wilmington, Del.). Cells were grown at 34°C in BG11 medium (41) supplemented with 20 mM HEPES, and the pH of the medium was adjusted to 7.5 with KOH. Cultures were supplied with illumination from incandescent lamps at 70 μE m−2 s−1 and aerated with air that contained 1% CO2. The growth of cells was monitored in terms of optical density at 730 nm (OD730).
Construction of plasmids for expression of nhaS genes in E. coli
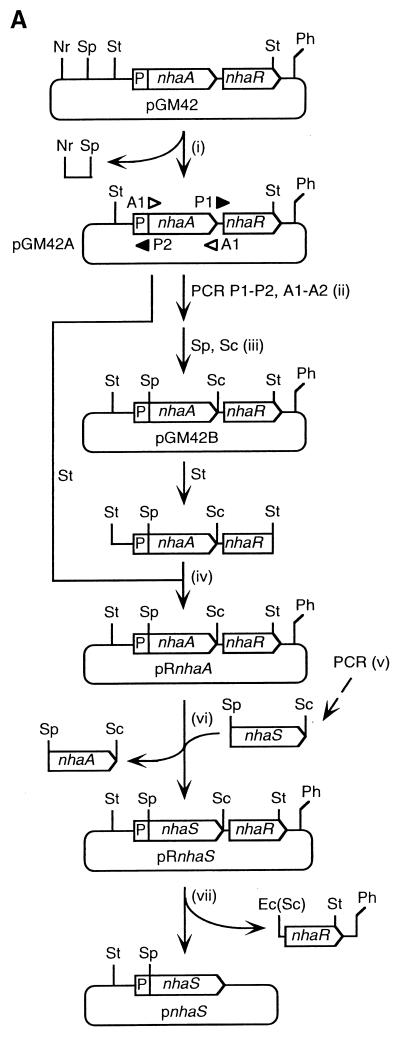
Plasmid pGM42 (14) was kindly provided by E. Padan (Hebrew University of Jerusalem, Jerusalem, Israel). This plasmid is a derivative of pBR322 and includes a 4.2-kbp segment of the chromosomal DNA of E. coli that contains the nhaA gene under control of the Na+-inducible promoter nhaAp plus the nhaR gene for the positive trans-acting regulator of the nhaA gene (11). Plasmids for expression of each of the five nhaS genes in E. coli were constructed from pGM42 as shown in Fig. 1A. (i) The SphI site in pGM42 was deleted to yield plasmid pGM42A by digestion with SphI and NruI, blunting with a DNA blunting kit (Takara Shuzo Co. Ltd., Tokyo, Japan), and self-ligation. (ii) A 6.7-kbp fragment that contained the nhaAp promoter, the nhaR gene, and the pBR322 backbone was amplified by PCR with pGM42A as template, the forward primer P1 (CGTCCATCAGTTTGAgAGctCGGTTTACCG, corresponding to nucleotides +1153 to +1182, counted from the site of initiation of translation of the nhaA gene, which was designated +1), and the reverse primer P2 (GATGCAGATGTTgCAtgcTTTATTTCTCTTTCAGG, complementary to nucleotides +13 to −19). (Italicized portions represent restriction sites for SphI [GCATGC] and SacI [GAGCTC]; nucleotides that differ from the ones in the template are lowercased.) The nhaA gene was also amplified with pGM42A as template, the forward primer A1 (CCTGAAAGAGAAATAAAgcaTGcAACATCTGCATC, corresponding to nucleotides −19 to +13, counted from the site of initiation of translation of the nhaA gene, which was designated +1), and the reverse primer A2 (CGGTAAACCGagCTcTCAAACTGATGGACG, complementary to nucleotides +1182 to +1153). (iii) The 6.7-kbp fragment and the nhaA gene were digested with SphI and SacI and ligated to yield plasmid pGM42B. (iv) A 2.2-kbp StuI-StuI fragment of pGM42A was replaced by the corresponding part of pGM42B. The resultant plasmid, designated pRnhaA, was identical to pGM42A except that it contained an SphI site at the site of initiation of translation of the nhaA gene (nucleotides −2 to +4) and a SacI site just downstream of the nhaA gene (nucleotides +1168 to +1173). (v) The various nhaS genes were amplified with the chromosomal DNA isolated from Synechocystis as template and the following synthetic oligonucleotides as primers: forward primer CAgCaTGcATACAGCGGTCAACGA (corresponding to nucleotides −4 to +20, counted from the site of initiation of translation of the nhaS1 gene, which was designated +1) and reverse primer aagagctcCTAGGATGGTTCGGCCACAT (complementary to nucleotides +1584 to +1565) for the nhaS1 gene, forward primer CTgCATGcCTTAAGCTCCCTGTGC (corresponding to nucleotides −4 to +19, counted from the site of initiation of translation of the nhaS2 gene, which was designated +1) and reverse primer TTgAGcTCGTCAGTCATCCTGCAGG (complementary to nucleotides +1632 to +1608) for the nhaS2 gene, forward primer ttgcATGcTTATGAACCCATTGCTCCCTC (corresponding to nucleotides +1 to +25, counted from the site of initiation of translation of the nhaS3 gene, which was designated +1) and reverse primer ttgagctcCTAATCTGGGGTGGGAACTG (complementary to nucleotides +1386 to +1367) for the nhaS3 gene, forward primer AAgcATGcACACCAATACTTTACTGCTAATT (corresponding to nucleotides −4 to +27, counted from the site of initiation of translation of the nhaS4 gene, which was designated +1) and reverse primer ttgaGcTcTTAATGGGCTGGGGCAGGAT (complementary to nucleotides +1237 to +1214) for the nhaS4 gene, and forward primer ttgcATGcATGGCCTATTCGCACCAATTC (corresponding to nucleotides +1 to +25, counted from the site of initiation of translation of the nhaS5 gene, which was designated +1) and reverse primer aagagctcCTAGGCGTAGGGATCGCCA (complementary to nucleotides +2097 to +2079) for the nhaS5 gene. (vi) The nhaA gene in pRnhaA was removed by digestion with SphI and SacI, and an amplified nhaS gene was inserted. The resultant plasmids were designated pRnhaS1, pRnhaS2, pRnhaS3, pRnhaS4, and pRnhaS5. (vii) To generate another set of plasmids that did not contain the nhaR gene, the plasmids pRnhaS1, pRnhaS2, pRnhaS3, pRnhaS4, pRnhaS5, and pRnhaA were further digested with Ecl136II (an isozyme of SacI) and PshA1 and self-ligated. The resultant plasmids were designated pnhaS1, pnhaS2, pnhaS4, pnhaS5, and pnhaA. We failed to generate pnhaS3. All the amplified fragments and the ligated junctions were verified by determination of nucleotide sequences.
FIG. 1.
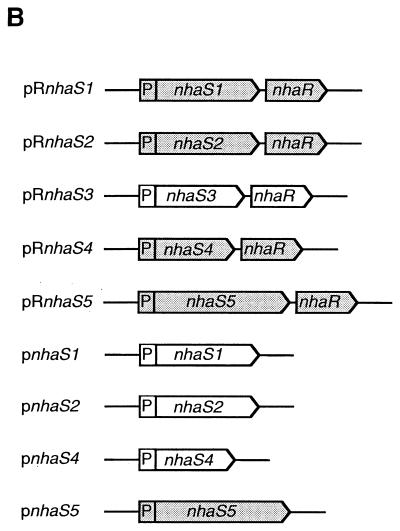
Plasmids used for expression of nhaS genes in E. coli TO114. (A) Construction of vector plasmids. Solid lines represent the pBR322 backbone and the flanking regions of the nhaA and nhaR genes of E. coli. P, Na+-inducible promoter of the nhaA gene (nhaAp). Restriction sites: Nr, NruI; Sp, SphI; St, StuI; Sc, SacI; Ph, PshAI; Ec, Ecl136II. For details, see Materials and Methods. (B) Plasmid characteristics. Plasmids pnhaS1, pnhaS2, pRnhaS3, and pnhaS4 were successfully introduced into TO114 cells (indicated by the absence of shading).
Isolation of RNA.
E. coli cells were grown in LBK medium to the early exponential phase of growth (OD600, 0.4). Aliquots of the culture were withdrawn, mixed immediately with an equal volume of ice-cold ethanol that contained 5% (wt/vol) phenol, and centrifuged at 3,000 × g for 10 min. Each pellet was washed with 50 mM Tris-HCl (pH 8.0) and 100 mM EDTA and then resuspended in 600 μl of 50 mM Tris-HCl (pH 8.0)–5 mM EDTA–0.25% sodium dodecyl sulfate (SDS). The suspension was mixed with 600 μl of acid phenol (a mixture of 50% phenol, 48% chloroform, and 2% isoamyl alcohol [vol/vol], buffered with an equal volume of 50 mM sodium acetate, pH 5.2), and the mixture was incubated at 65°C for 5 min to disrupt the cells. Total nucleic acids were extracted three times with acid phenol and precipitated in ethanol. Total RNA was separated from DNA by precipitation twice in LiCl and stored at −80°C.
DNA probes.
The DNA fragments used for the preparation of probes for Northern blotting analysis were generated by excision from the nhaS genes that had been amplified by PCR as described above: nhaS1 (with HincII, nucleotides +13 to +648), nhaS2 (with SphI and NcoI, nucleotides +1 to +545), nhaS3 (with SphI and EcoRI, nucleotides +1 to +652), and nhaS4 (with SphI and BstEII, nucleotides +1 to +576). The resultant DNA fragments were labeled with [α-32P]dCTP using a BcaBEST labeling kit (Takara Shuzo).
Northern blotting.
Fifteen micrograms of total RNA was fractionated by electrophoresis on a 1.2% agarose gel that contained 6.3% formaldehyde in 3-(N-morpholino)propanesulfonic acid buffer, pH 7.0 (39), and bands of RNA were transferred to a nylon membrane (NEN Life Science Products, Boston, Mass.). The membrane was baked at 80°C for 2 h and then incubated for 2 h at 65°C in a solution of 0.5 M sodium phosphate buffer (pH 7.2), 5% SDS, 5× Denhardt's reagent (39), and 100 μg of denatured salmon sperm DNA ml−1. Then the DNA probe was added (2 × 105 cpm ml−1), and hybridization was allowed to proceed for 16 h at 65°C. After a 1-h wash at 55°C in a solution of 0.05 M sodium phosphate buffer (pH 7.2) and 0.5% SDS, the membrane was exposed to an X-ray film (Eastman Kodak Company, Rochester, N.Y.).
Measurement of Na+/H+ antiport activities of IMVs.
Cells were grown in LBK medium to the middle of the exponential phase of growth (OD600, 1.5). Inverted membrane vesicles (IMVs) were prepared with a French pressure cell (SLM Instruments, Inc., Urbana, Ill.) as described previously (37). The Na+/H+ antiport activities of IMVs were estimated from the extent of the collapse of a preformed proton gradient, with acridine orange as the pH indicator, essentially as described previously (14). The assay solution consisted of 140 mM choline chloride, 5 mM MgCl2, 1 μM acridine orange, and 10 mM Tris titrated with 2-(N-morpholino)ethanesulfonic acid (MES; pH 8.5). In some cases choline chloride was replaced by 140 mM KCl. An aliquot corresponding to 20 μg of vesicle protein was added to 2 ml of the assay solution that was being stirred in a cuvette. Fluorescence from acridine orange was monitored in a fluorometer (model RF-5000; Shimadzu, Kyoto, Japan). The wavelength of excitation light was 495 nm, and fluorescence was monitored at 530 nm. Addition of Tris-d-lactate to a final concentration of 2 mM energized the IMVs and resulted in quenching of the fluorescence. Subsequent addition of NaCl or LiCl resulted in restoration of fluorescence. The initial rate of this restoration, as measured during the 2-s interval that followed the addition of NaCl or LiCl at various concentrations was taken as the Na+/H+ antiport activity, which was expressed in arbitrary units (fluorescence units s−1 mg of protein−1). IMVs from pBR322+ cells (negative control) had low Na+/H+ antiport activity, which was taken as the background activity. For calculations of kinetic parameters, the Na+/H+ antiport activity of IMVs prepared from pBR322+ cells was subtracted from the activity of IMVs prepared from nhaA+, nhaS1+, and nhaS3+ cells.
Evaluation of the sensitivity of cell growth to salt stress.
Transformed cells that had been grown in LBK medium were spread on plates prepared with 1.0% tryptone, 0.5% yeast extract, and 1.5% agar (Difco; LBn solid medium) that had been supplemented with various concentrations of NaCl or LiCl, in addition to KCl, for evaluation of the sensitivity of cell growth to high concentrations of Na+ and Li+ ions. For evaluation of the sensitivity of cell growth to depletion of K+ ions, we used plates of LBn solid medium that had been supplemented with various concentrations of KCl. LBn solid medium by itself contained 20 mM Na+ ions and 5 mM K+ ions. Formation of colonies was examined after incubation for 24 h at 37°C.
Targeted mutagenesis of the nhaS genes in Synechocystis.
Plasmid pAM1573, which contained a chloramphenicol resistance (Cmr) gene cartridge, and plasmid pAM1303, which contained a spectinomycin resistance (Spr) gene cartridge, were kindly provided by S. S. Golden (Texas A&M University, College Station, Tex.). The nhaS genes that had been amplified by PCR, as described above, were subcloned into the TA cloning site of plasmid pT7Blue (Novagen, Madison, Wis.). For construction of a plasmid with a disrupted nhaS1 gene, the region between the BbsI and StuI sites of the nhaS1 gene in pT7Blue was removed and the ends of the cleaved plasmid were blunted with the DNA blunting kit. The cleaved and blunted plasmid was ligated with a kanamycin resistance (Kmr) gene cartridge, which had been excised by SmaI from plasmid pUC-KIXX (Pharmacia, Uppsala, Sweden). The resultant plasmid was designated pnhaS1::Kmr.
For construction of a plasmid with a disrupted nhaS2 gene, the region between the BstEII and HpaI sites of the nhaS2 gene in pT7Blue was removed and the ends of the cleaved plasmid were blunted with the DNA blunting kit. The cleaved and blunted plasmid was ligated with a Cmr gene cartridge, which had been excised by BstEII and HindIII from pAM1573 and blunted with the DNA blunting kit. The resultant plasmid was designated pnhaS2::Cmr. A plasmid with a disrupted nhaS3 gene was constructed by inserting the Kmr gene cartridge, which had been excised from pUC-KIXX with SmaI, into the EcoRV site of the nhaS3 gene in pT7Blue. The resultant plasmid was designated pnhaS3::Kmr. For construction of a plasmid with a disrupted nhaS4 gene, the region between the BstEII and BbsI sites of the nhaS4 gene in pT7Blue was removed and the ends of the cleaved plasmid were blunted with the DNA blunting kit. The cleaved and blunted plasmid was ligated with the Cmr gene cartridge, which had been excised by BstEII and HindIII from pAM1573 and blunted with the DNA blunting kit. The resultant plasmid was designated pnhaS4::Cmr. A disrupted nhaS5 gene was constructed by replacing the region between the two BalI sites in the nhaS5 gene in pT7Blue by the Spr gene cartridge, which had been excised by EcoRV and SmaI from pAM1303. The resultant plasmid was designated pnhaS5::Spr.
Wild-type cells of Synechocystis were transformed with the individual plasmids to generate ΔnhaS cells, as described previously (44). For the construction of the double mutants ΔnhaS1ΔnhaS2 and ΔnhaS4ΔnhaS5, we transformed ΔnhaS1 cells with pnhaS2::Cmr and ΔnhaS4 cells with pnhaS5::Spr, respectively. For selection of mutant cells, kanamycin, spectinomycin, and chloramphenicol were included in the medium at 25, 15, and 15 μg/ml, respectively. Disruption with the antibiotic resistance cartridges of the nhaS genes on all copies of the chromosome was examined by PCR (44).
Concentrations of proteins in IMVs.
The concentrations of proteins in IMVs were determined as described elsewhere (9).
RESULTS
Expression of nhaS genes in E. coli TO114.
We transformed E. coli TO114 cells with plasmids that contained individual nhaS genes (Fig. 1B). The resultant transformed cells were grown on solid LBK medium supplemented with 50 μg of ampicillin ml−1. We obtained colonies only when cells had been transformed with pnhaS1, pnhaS2, pRnhaS3, or pnhaS4. Transformation with plasmids pRnhaS1, pRnhaS2, pRnhaS4, pRnhaS5, and pnhaS5 failed to yield colonies under our selection conditions. Thus, transformation of cells with the nhaS5 gene was unsuccessful. We also obtained TO114 cells that harbored pBR322, pRnhaA, or pnhaA.
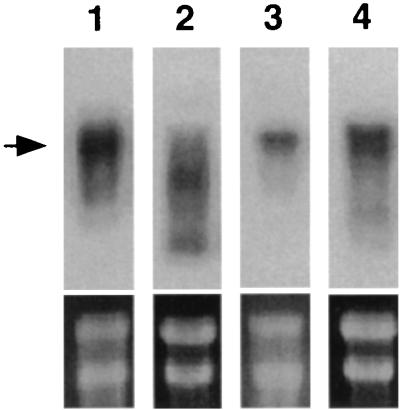
We attempted to determine the levels of products of nhaS genes in membrane fractions of transformed E. coli cells by SDS-polyacrylamide gel electrophoresis and silver staining. However, we failed to detect bands that corresponded unequivocally to the Synechocystis proteins either before or after induction by NaCl. Thus, to evaluate whether nhaS genes were at least transcribed in E. coli under control of the nhaAp promoter, we performed Northern blotting of total RNA extracted from pnhaS1/TO114 (nhaS1+), pnhaS2/TO114 (nhaS2+), pRnhaS3/TO114 (nhaS3+), and pnhaS4/TO114 (nhaS4+) cells that had been grown in LBK medium, using probes derived from each nhaS gene (Fig. 2). Transcripts of the nhaS1, nhaS3, and nhaS4 genes accumulated in nhaS1+, nhaS3+, and nhaS4+ cells, respectively. In contrast, most transcripts of the nhaS2 gene in nhaS2+ cells were shorter than the expected length of nhaS2 mRNA. These transcripts might be degradation products of the nhaS2 mRNA.
FIG. 2.
Northern blotting analysis of the expression of nhaS genes in transformed E. coli TO114 cells. Total RNA was extracted from cells that had been grown in LBK medium. Results are shown for nhaS1 transcripts in nhaS1+ cells (lane 1), nhaS2 transcripts in nhaS2+ cells (lane 2), nhaS3 transcripts in nhaS3+ cells (lane 3), and nhaS4 transcripts in nhaS4+ cells (lane 4). The positions of the expected transcripts are indicated (arrow). The lower panels show bands that correspond to 16S and 23S rRNAs on each gel, as revealed after staining with ethidium bromide prior to blotting. Three independent experiments yielded essentially the same results.
We also examined changes in the levels of transcripts upon an increase in the concentration of NaCl in the medium to 200 mM (data not shown). During exposure to 200 mM NaCl, the level of nhaS3 transcripts in nhaS3+ cells increased gradually over the course of 40 min, while the levels of transcripts of nhaS1, nhaS2, and nhaS4 in nhaS1+, nhaS2+, and nhaS4+ cells, respectively, did not change significantly. This result was probably due to the presence of the Na+-dependent regulatory gene nhaR in the construct for expression of the nhaS3 gene (Fig. 1B), which might have promoted transcription of the nhaS3 gene under high-salt conditions (11).
Na+/H+ antiport activities of IMVs.
We measured the Na+/H+ antiport activity of IMVs prepared from transformed cells as the Na+-mediated and Li+-mediated net efflux of protons, which we monitored by observing changes in the fluorescence of acridine orange. Since IMVs from pnhaA/TO114 and pRnhaA/TO114 cells had almost the same Na+/H+ antiport activity (data not shown), we used pRnhaA/TO114 cells in further experiments as the positive control, referring to them as nhaA+ cells. We used pBR322/TO114 (pBR322+) cells as the negative control.
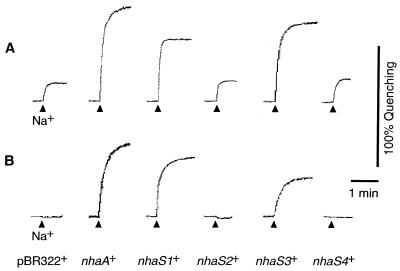
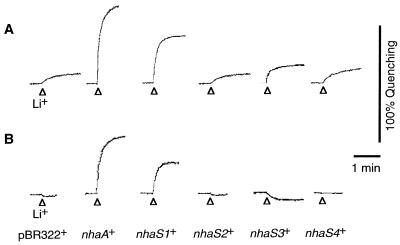
Figure 3 shows profiles of Na+/H+ antiport activity after addition of 5 mM NaCl in the presence of 140 mM choline chloride under K+-free conditions (Fig. 3A) or in the presence of 140 mM KCl (Fig. 3B; K+-rich conditions). Under K+-free conditions, IMVs from pBR322+ cells had low Na+/H+ antiport activity (Fig. 3A). Such activity might have been due to a nonspecific monovalent cation/H+ antiport system (35) that did not transport Na+ ions under K+-rich conditions (Fig. 3B). IMVs from nhaS1+ and nhaS3+ cells had significant Na+/H+ antiport activity under K+-rich conditions, as did the IMVs from nhaA+ cells (Fig. 3B). These results clearly demonstrated that the Na+/H+ antiport activity had been transferred to the host E. coli cells by transformation with the nhaS1 and nhaS3 genes. The IMVs prepared from nhaS2+ and nhaS4+ cells did not have Na+/H+ antiport activity under K+-rich conditions.
FIG. 3.
Activities of the Na+/H+ antiport system in IMVs prepared from transformed cells. IMVs were prepared from cells that had been grown in LBK medium. Activity was assayed in a solution that consisted of 5 mM MgCl2, 1 μM acridine orange, and 10 mM Tris titrated with MES (pH 8.5) and supplemented with 140 mM choline chloride (A) or 140 mM KCl (B). Arrowheads, time at which 5 mM NaCl was added to the assay solution.
Figure 4 shows profiles of Li+/H+ antiport activity, as determined upon addition of 5 mM LiCl under K+-free conditions (Fig. 4A) and under K+-rich conditions (Fig. 4B). The IMVs prepared from nhaS1+ cells had high Li+/H+ antiport activity under K+-rich conditions (Fig. 4B), demonstrating that Li+/H+-antiport activity had also been transferred to the host E. coli cells by transformation with the nhaS1 gene. IMVs prepared from nhaS3+ cells did not have Li+/H+ antiport activity under K+-rich conditions (Fig. 4B), but they had considerably higher Li+/H+ antiport activity under K+-free conditions than IMVs prepared from pBR322+ cells (Fig. 4A). These results indicated that Li+/H+ antiport activity had been transferred to the host E. coli cells upon transformation with the nhaS3 gene and that the activity was strongly inhibited by the presence of K+ ions in the assay solution.
FIG. 4.
Activities of the Li+/H+ antiport system in IMVs prepared from transformed cells. Experiments were carried out as described in the legend to Fig. 3 except that 5 mM LiCl was added to the assay solution instead of 5 mM NaCl.
Table 1 shows the kinetic parameters of the Na+/H+ antiport activity of IMVs prepared from nhaA+, nhaS1+, and nhaS3+ cells and assayed under K+-free conditions. For both Na+ and Li+ ions, the activity of IMVs from nhaS1+ cells gave larger values of Km than the activity of IMVs from nhaS3+ cells. The activity of IMVs from nhaS3+ cells revealed a strikingly high affinity for Li+ ions. The Km of the activity of IMVs from nhaA+ cells for Na+ ions was of the same magnitude as the value reported previously for purified NhaA (i.e., 0.1 mM at pH 8.6) (43).
TABLE 1.
Kinetic parameters under K+-free conditions, of Na+/H+ antiport activities of IMVs from transformed cellsa
| Cells | Na+
|
Li+
|
||
|---|---|---|---|---|
| Vmaxb | Km (mM) | Vmaxb | Km (mM) | |
| nhaA+ | 42 | 0.2 | 42 | 0.3 |
| nhaS1+ | 120 | 7.7 | 40 | 2.5 |
| nhaS3+ | 22 | 0.7 | 5 | 0.01 |
The activities of IMVs from pBR322+ cells (Km = 1.5 mM and Vmax = 12 U s−1 mg of protein−1 for Na+ ions; Km = 0.7 mM and Vmax = 1.5 U s−1 mg of protein−1 for Li+ ions) were taken as background activities, as described in Materials and Methods. Two independent experiments yielded essentially the same results.
In fluorescence units s−1 mg of protein−1.
Sensitivity of cell growth to high concentrations of Na+ and Li+ ions.
Table 2 shows the maximum concentrations of Na+ and Li+ ions that allowed growth of transformed cells on solid LBn medium prepared with 5, 25, 105, or 305 mM K+ ions. In the presence of 105 mM K+ ions, growth of pBR322+ cells was inhibited at 120 mM Na+ ions and at 3 mM Li+ ions, and it was completely arrested at 170 mM Na+ ions and at 5 mM Li+ ions. In contrast, nhaS1+ and nhaS3+ cells were able to grow at 570 mM Na+ ions and 10 mM Li+ ions and at 420 mM Na+ ions and 70 mM Li+ ions, respectively, in the presence of 105 mM K+ ions. These results were consistent with the restored Na+/H+ antiport activity in the membranes isolated from the respective cell lines. nhaS1+ and nhaS3+ cells retained their high tolerance to Na+ and Li+ ions when the concentration of K+ ions was decreased to 5 mM.
TABLE 2.
Effects of K+ ions on the maximum concentrations of Na+ and Li+ ions that allowed growth of transformed cells on solid LBn medium
| Concn of K+ in the medium (mM) | Maximum concn for growth
(mM)a
|
|
|---|---|---|
| Na+ | Li+ | |
| pBR322+ | ||
| 5 | <20 | 0 |
| 25 | 60 | 2 |
| 105 | 120 | 3 |
| 305 | 120 | 3 |
| nhaA+ | ||
| 5 | 1,070 | 400 |
| 25 | 1,070 | 400 |
| 105 | 970 | 350 |
| 305 | 770 | 300 |
| nhaS1+ | ||
| 5 | 570 | 15 |
| 25 | 570 | 15 |
| 105 | 570 | 10 |
| 305 | 420 | 5 |
| nhaS2+ | ||
| 5 | <20 | 0 |
| 25 | 60 | 2 |
| 105 | 120 | 3 |
| 305 | 120 | 3 |
| nhaS3+ | ||
| 5 | 370 | 40 |
| 25 | 370 | 40 |
| 105 | 420 | 70 |
| 305 | 320 | 90 |
| nhaS4+ | ||
| 5 | <20 | 0 |
| 25 | 60 | 2 |
| 105 | 120 | 3 |
| 305 | 120 | 3 |
Three independent experiments yielded essentially similar results.
Both nhaS2+ and nhaS4+ cells were as sensitive as pBR322+ cells to Na+ and Li+ ions, as expected from the absence under K+-rich conditions of Na+/H+ antiport activity of the IMVs prepared from such cells. This sensitivity of pBR322+, nhaS2+, and nhaS4+ cells to Na+ and Li+ ions decreased as the concentration of K+ ions in the medium was increased from 5 to 105 mM. As described below, this dependence on K+ ions seemed to reflect the absence of Na+/H+ antiport activity in the membranes.
Sensitivity of cell growth to depletion of K+ ions.
pBR322+ cells did not grow in the presence of 5 mM K+ ions, the background level, even in the absence of additional Na+ and Li+ ions (Table 2), an observation that was consistent with previous reports on a ΔnhaAΔnhaB strain of E. coli (16, 47) and was probably due to the inability of these cells to maintain intracellular concentrations of Na+ ions at an appropriate level when the ratio of K+ ions to Na+ ions in the medium was low (16). To elucidate the effect of transformation on the sensitivity to depletion of K+ ions, we examined the growth of transformed cells at various concentrations of K+ ions. pBR322+ cells required at least 20 mM K+ ions; nhaS1+ and nhaS3+ cells grew at 5 mM K+ ions, the background level, as did nhaA+ cells. nhaS4+ cells also exhibited a lower requirement for K+ ions (6 mM) than that of pBR322+ cells. In contrast, the requirement of nhaS2+ cells for K+ ions did not differ significantly from that of pBR322+ cells.
Disruption of nhaS genes in Synechocystis.
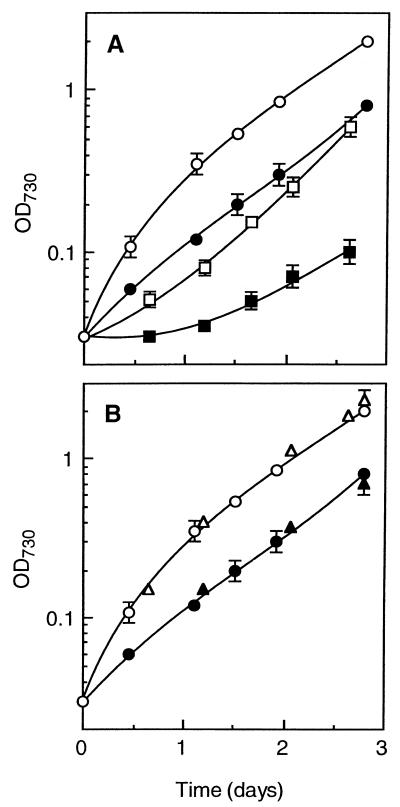
We created single and double mutants of Synechocystis in which individual nhaS genes were disrupted by insertion of an antibiotic resistance gene cartridge. We verified the disruption of the nhaS1, nhaS2, nhaS4, and nhaS5 genes on all copies of the chromosomal DNA by PCR. We failed to disrupt the nhaS3 gene under any conditions tested. In our efforts to disrupt the nhaS3 gene we used the following media: BG11 medium that contained 18 mM Na+ ions (pH 7.5), a low-sodium medium in which all the sodium salts of BG11 medium had been replaced by potassium salts (this medium was estimated to contain 50 μM Na+ ions from the extent of contamination by Na+ ions of the potassium salts [Wako Pure Chemical Industries, Ltd., Oaska, Japan] that we used), and media prepared by adding different concentrations of NaCl (100 μM to 100 mM) to the low-salt medium. The single mutants that we did obtain did not show any phenotypic changes in terms of sensitivity to high concentrations of NaCl (data not shown). ΔnhaS1ΔnhaS2 cells grew more slowly than wild-type cells both in BG11 medium and in a high-salt medium prepared by adding NaCl to 0.5 M to BG11 medium (Fig. 5A). The retardation of growth of ΔnhaS1ΔnhaS2 cells, compared to the growth of wild-type cells, appeared to be greater in the presence of 0.5 M NaCl than in its absence. In contrast, ΔnhaS4ΔnhaS5 cells grew as rapidly as wild-type cells regardless of the presence or absence of 0.5 M NaCl (Fig. 5B).
FIG. 5.
Growth curves for Synechocystis in BG11 medium that contained 18 mM Na+ ions (open symbols) or in high-salt medium prepared by increasing the concentration of NaCl in BG11 medium to 0.5 M (closed symbols). (A) Wild-type cells (circles) and mutant cells with disrupted nhaS1 and nhaS2 genes (ΔnhaS1ΔnhaS2) (squares). (B) Wild-type cells (circles) and mutant cells with disrupted nhaS4 and nhaS5 genes (ΔnhaS4ΔnhaS5) (triangles). The results were obtained from three independent determinations for each line of cells.
DISCUSSION
Homologs of eukaryotic and prokaryotic Na+/H+ antiporters in Synechocystis.
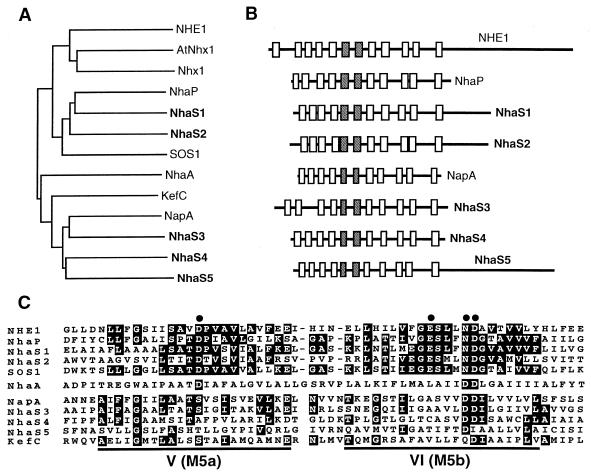
Phylogenetic analysis (Fig. 6A) revealed that NhaS1 and NhaS2 are related to isoforms of NHE found in vertebrates (NHE1 to -6 and βNHE) and to NHE-like Na+/H+ antiporters in plant, fungal, and bacterial cells. NhaS1 and NhaS2 appeared to be most similar to NhaP, an Na+/H+ antiporter of Pseudomonas aeruginosa (46), and to SOS1, a putative Na+/H+ antiporter in A. thaliana (40). NhaS3, NhaS4, and NhaS5 resembled NapA, an Na+/H+ antiporter in Enterococcus hirae (42, 51), as well as KefC, a putative K+/H+ antiporter in E. coli (27). Genes for homologs of both NHE-like (“eukaryotic”) and NapA-like (“prokaryotic”) Na+/H+ antiporters have been found in many eubacteria, archaea, and eukaryotes, suggesting that the two types of Na+/H+ antiporter might have been selected early in evolution. NHE-like and NapA-like Na+/H+ antiporters appear to have distinct properties. The isoforms of NHE catalyze the electroneutral exchange of Na+ ions for protons, being activated by internal protons (50). It has been proposed that Nhx1 of S. cerevisiae, an NHE-like Na+/H+ antiporter, might be activated by decreases in cytoplasmic pH (28). The isoforms of NHE have rather high Km values for Na+ ions, which range from 4.7 to 59 mM (32). A high Km (7 mM) for Na+ ions was also reported for vacuoles of A. thaliana that overexpressed the AtNHX1 gene (1). In contrast, it was reported that NapA has a relatively low Km (1.0 mM) for Na+ ions (42). The present study of the expression in E. coli of cyanobacterial genes from Synechocystis provides the first example, to our knowledge, of the functional identification of the two types of Na+/H+ antiporter in a single organism. A. thaliana has a number of genes for putative NHE-like and NapA-like Na+/H+ antiporters. They might be localized in different tissues and membranes.
FIG. 6.
Relationships of NhaS proteins to other Na+/H+ antiporters and related proteins. (A) Phylogenetic relationships as determined with the CLUSTAL W multiple sequence alignment algorithm (45). (B) Secondary structures predicted from hydropathy profiles, as determined by the algorithm of Kyte and Doolittle (24). Putative transmembrane segments are boxed, and segments that exhibit the strongest homology are shaded. (C) Alignment of amino acid sequences of two strongly homologous segments—the putative fifth (V) and sixth (VI) segments—of NhaS proteins, which correspond, respectively, to the sixth (M5a) and seventh (M5b) segments of NHE1. Identical residues are shaded, and conserved Glu and Asp residues are indicated by dots. NHE1, human P19634; AtNhx1, A. thaliana AAD16946; Nhx1, S. cerevisiae NP 010744; NhaP, P. aeruginosa BAA31695; SOS1, A. thaliana AAF76139; NhaA, NhaB, and KefC, E. coli C64722, G64864, and QQECRD, respectively; NapA, Enterococcus hirae A42111.
Each of the NhaS proteins appears to contain 11 transmembrane segments (Fig. 6B). NhaS1, NhaS2, and NhaS5 include a large hydrophilic extension at the carboxyl terminus, as do the NHE isoforms and the NHE-like Na+/H+ antiporters in eukaryotic cells. In the various isoforms of NHE, the carboxy-terminal extension mediates the response of the antiporter to various stimuli (50). Therefore, the carboxy-terminal extensions of NhaS1, NhaS2, and NhaS5 might each also play a role in the regulation of the activity.
The strongest homology was found within the putative fifth and sixth transmembrane segments of the NhaS proteins and the corresponding regions of the Na+/H+ antiporters from other organisms (Fig. 6C). This region is the most strongly conserved among the NHE isoforms and includes several acidic residues, the importance of which has been demonstrated both for human NHE1 (12) and for NhaA of E. coli (18). Some of these residues are also conserved in the NhaS proteins.
Functional expression of the nhaS1, nhaS3, and nhaS4 genes in E. coli under the control of the nhaAp promoter.
The nhaS genes were expressed at very low levels in wild-type Synechocystis (unpublished results). This observation suggested that expression of each nhaS gene in E. coli from its own promoter would not result in a sufficient level of product. However, overproduction of proteins that contain several transmembrane segments might be expected to have detrimental effects on host cells. In wild-type E. coli, NhaA is a membrane-bound protein that is present at a low level (less than 0.2% of the total membrane proteins [43]). When this protein was overexpressed under the control of the strongly inducible tac promoter, cell growth ceased (43). Therefore, we chose to use the nhaAp promoter for expression of the various nhaS genes in E. coli at appropriate levels.
Expression in E. coli TO114 of the nhaS1 and nhaS3 genes under control of the nhaAp promoter resulted in production of functional Na+/H+ antiporters. In contrast, the expression of the nhaS4 gene did not result in expression of detectable Na+/H+ antiport activity in the transformed host cells. This failure might have been due to an insufficient level of the expressed protein, which, in turn, would have resulted in the inability of nhaS4+ cells to acquire Na+/H+ antiport activity. Alternatively, NhaS4 might not function as an efficient system for extrusion of Na+ ions.
Transcripts of the nhaS2 gene appeared to be degraded in the absence of NaCl. The instability of the heterologous transcripts might have been related to inefficient translation, due in turn to the presence of codons that are used at low frequencies in E. coli (21). Inefficient translation can increase the susceptibility of transcripts to RNases (10). However, this situation does not appear to have been operative in the present case because the proportion of such unusual codons in nhaS2 transcripts was not much higher than that in the transcripts of the other nhaS genes (unpublished data). It has been suggested that NhaS2 might be required for the uptake of Na+ ions in Synechocystis (25). The instability of nhaS2 transcripts might have been a consequence of the disturbed balance of ions in the transformed E. coli cells.
Our failure to introduce the nhaS5 gene into TO114 cells suggests that the introduction of this gene under the control of the nhaAp promoter might have had a detrimental effect on the host cells, even when expression was not induced by high concentrations of Na+ ions.
NhaS1 and NhaS3 are low-affinity and high-affinity Na+/H+ antiporters, respectively.
The kinetic properties of the Na+/H+ antiport system in IMVs prepared from nhaS1+ cells (Table 1) indicated that the expressed protein, NhaS1, had low affinity for Na+ ions (Km, 7.7 mM) and for Li+ ions (Km, 2.5 mM). The Km for Na+ ions is close to that reported for AtNhx1 of A. thaliana (1). The lower Km of NhaS1 for Li+ ions than for Na+ ions suggests that Li+ ions might be a better substrate than Na+ ions. However, transformation with the nhaS1 gene had only a minimal effect on the tolerance of the host cells to Li+ ions, while it dramatically increased the tolerance of host cells to Na+ ions (Table 2). This result suggests that the Li+/H+ antiport activity of NhaS1 might not have any physiological relevance.
The Na+/H+ antiport system in IMVs prepared from nhaS3+ cells had high affinity for Na+ ions (Km, 0.7 mM) and extremely high affinity for Li+ ions (Km, 0.01 mM). These results suggest that Li+ ions might be a better substrate of NhaS3 than Na+ ions. The Km of NhaS3 for Na+ ions was similar to the value obtained for NapA of Enterococcus hirae that was expressed in E. coli (i.e., 1.0 mM [42]). However, the Km of NhaS3 for Li+ ions was much smaller than that reported for NapA (i.e., 0.1 mM [42]). K+ ions in the assay solution significantly inhibited the Li+/H+ antiport activity (Fig. 4). However, the tolerance of nhaS3+ cells to Li+ ions increased as the concentration of K+ ions in the medium was increased (Table 2), suggesting that K+ ions in the medium might have had a positive rather than a negative effect on the extrusion in vivo of Li+ ions by NhaS3. There might be a direct interaction between K+ ions and NhaS3. For example, extracellular K+ ions might activate the extrusion of Li+ ions by NhaS3.
Possible roles of NhaS1 and NhaS3 in Synechocystis.
The existence of high-affinity and low-affinity Na+/H+ antiporters in Synechocystis is consistent with the ability of this organism to acclimate to a wide range of extracellular concentrations of Na+ ions. The low affinity of NhaS1 for Na+ ions suggests that this Na+/H+ antiporter might be able to function at relatively high concentrations of Na+ ions. However, disruption of the nhaS1 gene did not cause any phenotypic changes in the tolerance to high salt, suggesting that other Na+/H+ antiporters might complement the function of NhaS1. Disruption of both the nhaS1 and the nhaS2 genes resulted in retardation of growth in the standard BG11 medium. Moreover, retardation of the growth of ΔnhaS1ΔnhaS2 cells appeared to be enhanced by high salt. These results suggest that the functions of NhaS1 and NhaS2, homologs of eukaryotic Na+/H+ antiporters, might complement one another and that both might be involved in the tolerance of Synechocystis to high-salt stress.
Synechocystis requires a very low concentration of Na+ ions for optimal growth. Wild-type cells grow more slowly in low-sodium medium (50 μM Na+) than in the standard BG11 medium (18 mM Na+). The nhaS3 gene is essential for the viability of Synechocystis even in the low-sodium medium at close to neutral pH. This requirement for the nhaS3 gene is very specific: all other Na+/H+ antiporters characterized to date in heterotrophic bacteria have been shown to be dispensable under such conditions. In contrast, disruption of both the nhaS4 and the nhaS5 genes had no effects on phenotypes in terms of high-salt tolerance, an observation that suggests that NhaS4 and NhaS5 might make little contribution to tolerance to high-salt stress. The high affinity of NhaS3 for Na+ ions and for Li+ ions indicates that NhaS3 is able to transport Na+ and Li+ ions at low concentrations. Therefore, NhaS3 might function in monitoring changes in intracellular concentrations of ions and might be involved in the appropriate adjustment of such concentrations.
It remains to be determined whether the various Na+/H+ antiporters are localized on the plasma membrane, on the thylakoid membrane, or on both. Their locations should help us to clarify their physiological roles in Synechocystis. Furthermore, we cannot exclude the possibility that NhaS2, NhaS4, and NhaS5 are also Na+/H+ antiporters. The nhaS4 gene reversed the inability of TO114 cells to grow under K+-depleted conditions, as did the nhaS1 and nhaS3 genes, an observation that suggests that the nhaS4 gene might encode a membrane-bound protein that transports K+ and/or Na+ ions.
ACKNOWLEDGMENTS
We express appreciation to E. Padan, H. Kobayashi, and S. S. Golden for gifts of plasmids and bacterial strains. We are also grateful to M. Hagemann and M. L. Verkhovskaya for helpful suggestions.
This work was supported in part by a grant-in-aid for specially promoted research (grant 08102011 to N.M.) from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Apse M P, Aharon G S, Snedden W A, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 2.Apte S K, Bhagwat A A. Salinity-stress-induced proteins in two nitrogen-fixing Anabaenastrains differentially tolerant to salt. J Bacteriol. 1989;171:909–915. doi: 10.1128/jb.171.2.909-915.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apte S K, Haselkorn R. Cloning of salinity stress-induced genes from the salt-tolerant nitrogen-fixing cyanobacterium Anabaena torulosa. Plant Mol Biol. 1990;15:723–733. doi: 10.1007/BF00016122. [DOI] [PubMed] [Google Scholar]
- 4.Avetisyan A V, Bogachev A V, Murtasina R A, Skulachev V P. ATP-driven Na+ transport and Na+-dependent ATP synthesis in Escherichia coli grown at low ΔμH+ FEBS Lett. 1992;317:267–270. doi: 10.1016/0014-5793(93)81290-g. [DOI] [PubMed] [Google Scholar]
- 5.Bañuelos M A, Sychrová H, Bleykasten-Grosshans C, Souciet J-L, Potier S. The Nha1 antiporter of Saccharomyces cerevisiaemediates sodium and potassium efflux. Microbiology. 1998;144:2749–2758. doi: 10.1099/00221287-144-10-2749. [DOI] [PubMed] [Google Scholar]
- 6.Bhagwat A A, Apte S K. Comparative analysis of proteins induced by heat shock, salinity, and osmotic stress in the nitrogen-fixing cyanobacterium Anabaenasp. strain L-31. J Bacteriol. 1989;171:5187–5189. doi: 10.1128/jb.171.9.5187-5189.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumwald E, Wolosin J M, Packer L. Na+/H+ exchange in the cyanobacterium Synechococcus6311. Biochem Biophys Res Commun. 1984;122:452–459. doi: 10.1016/0006-291x(84)90497-2. [DOI] [PubMed] [Google Scholar]
- 8.Blumwald E, Aharon G S, Apse M P. Sodium transport in plant cells. Biochim Biophys Acta. 2000;1465:140–151. doi: 10.1016/s0005-2736(00)00135-8. [DOI] [PubMed] [Google Scholar]
- 9.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Braun F, Le Derout J, Régnier P. Ribosomes inhibit an RNase E cleavage which induces the decay of the rpsO mRNA of Escherichia coli. EMBO J. 1998;17:4790–4797. doi: 10.1093/emboj/17.16.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carmel O, Rahav-Manor O, Dover N, Shaanan B, Padan E. The Na+-specific interaction between the LysR-type regulator, NhaR, and the nhaA gene encoding the Na+/H+ antiporter of Escherichia coli. EMBO J. 1997;16:5922–5929. doi: 10.1093/emboj/16.19.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fafournoux P, Nöel J, Pouysségur J. Evidence that Na+/H+exchanger isoforms NHE1 and NHE3 exist as stable dimers in membranes with a high degree of specificity for homodimers. J Biol Chem. 1994;269:2589–2596. [PubMed] [Google Scholar]
- 13.Fry I V, Huflejt M, Erber W W A, Peschek G A, Packer L. The role of respiration during adaptation of the freshwater cyanobacterium Synechococcus6311 to salinity. Arch Biochem Biophys. 1986;244:686–691. doi: 10.1016/0003-9861(86)90637-5. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg E B, Arbel T, Chen J, Karpel R, Mackie G A, Schuldiner S, Padan E. Characterization of a Na+/H+ antiporter gene of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:2615–2619. doi: 10.1073/pnas.84.9.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golden S S, Brusslan J, Haselkorn R. Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 1987;153:215–231. doi: 10.1016/0076-6879(87)53055-5. [DOI] [PubMed] [Google Scholar]
- 16.Harel-Bronstein M, Dibrov P, Olami Y, Pinner E, Schuldiner S, Padan E. MH1, a second-site revertant of an Escherichia coli mutant lacking Na+/H+ antiporters (ΔnhaAΔnhaB), regains Na+ resistance and a capacity to excrete Na+ in a ΔμH+-independent fashion. J Biol Chem. 1995;270:3816–3822. doi: 10.1074/jbc.270.8.3816. [DOI] [PubMed] [Google Scholar]
- 17.Haro R, Garciadeblas B, Rodríguez-Navarro A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- 18.Inoue H, Noumi T, Tsuchiya T, Kanazawa H. Essential aspartic acid residues, Asp-133, Asp-163 and Asp-164, in the transmembrane helices of a Na+/H+ antiporter (NhaA) from Escherichia coli. FEBS Lett. 1995;363:264–268. doi: 10.1016/0014-5793(95)00331-3. [DOI] [PubMed] [Google Scholar]
- 19.Ivey D M, Guffanti A A, Zemsky J, Pinner E, Karpel R, Padan E, Schuldiner S, Krulwich T A. Cloning and characterization of a putative Ca2+/H+ antiporter gene from Escherichia coli upon functional complementation of Na+/H+antiporter-deficient strains by the overexpressed gene. J Biol Chem. 1993;268:11296–11303. [PubMed] [Google Scholar]
- 20.Jeanjean R, Matthijs H C P, Onana B, Havaux M, Joset F. Exposure of the cyanobacterium SynechocystisPCC6803 to salt stress induces concerted changes in respiration and photosynthesis. Plant Cell Physiol. 1993;34:1073–1079. [Google Scholar]
- 21.Kane J F. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystissp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 23.Karpel R, Olami Y, Taglicht D, Schuldiner S, Padan E. Sequencing of the gene ant which affects the Na+/H+ antiporter activity in Escherichia coli. J Biol Chem. 1988;263:10408–10414. [PubMed] [Google Scholar]
- 24.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 25.Mikkat S, Milkowski C, Hagemann M. A putative Na+/H+ antiporter encoded by sll0273 of the cyanobacterium Synechocystis sp. strain PCC 6803 is essential for growth at high Na+/K+ratios. Plant Cell Environ. 2000;23:549–559. [Google Scholar]
- 26.Molitor V, Erber W, Peschek G A. Increased levels of cytochrome oxidase and sodium-proton antiporter in the plasma membrane of Anacystis nidulansafter growth in sodium-enriched media. FEBS Lett. 1986;204:251–256. [Google Scholar]
- 27.Munro A W, Ritchie G Y, Lamb A J, Douglas R M, Booth I R. The cloning and DNA sequence of the gene for the glutathione-regulated potassium-efflux system KefC of Escherichia coli. Mol Microbiol. 1991;5:607–616. doi: 10.1111/j.1365-2958.1991.tb00731.x. [DOI] [PubMed] [Google Scholar]
- 28.Nass R, Cunningham K W, Rao R. Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhanced by mutations in the plasma membrane H+-ATPase. J Biol Chem. 1997;272:26145–26152. doi: 10.1074/jbc.272.42.26145. [DOI] [PubMed] [Google Scholar]
- 29.Nass R, Rao R. Novel localization of a Na+/H+exchanger in a late endosomal compartment of yeast. J Biol Chem. 1998;273:21054–21060. doi: 10.1074/jbc.273.33.21054. [DOI] [PubMed] [Google Scholar]
- 30.Nitschmann W H, Packer L. NMR studies on Na+ transport in SynechococcusPCC 6311. Arch Biochem Biophys. 1992;294:347–352. doi: 10.1016/0003-9861(92)90694-r. [DOI] [PubMed] [Google Scholar]
- 31.Ohyama T, Igarashi K, Kobayashi H. Physiological role of the chaA gene in sodium and calcium circulations at a high pH in Escherichia coli. J Bacteriol. 1994;176:4311–4315. doi: 10.1128/jb.176.14.4311-4315.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orlowski J. Heterologous expression and functional properties of amiloride high affinity (NHE-1) and low affinity (NHE-3) isoforms of the rat Na+/H+exchanger. J Biol Chem. 1993;268:16369–16377. [PubMed] [Google Scholar]
- 33.Padan E, Schuldiner S. Molecular physiology of Na+/H+ antiporters, key transporters in circulation of Na+ and H+in cells. Biochim Biophys Acta. 1994;1185:129–151. doi: 10.1016/0005-2728(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 34.Pinner E, Padan E, Schuldiner S. Cloning, sequencing, and expression of the nhaB gene, encoding a Na+/H+ antiporter in Escherichia coli. J Biol Chem. 1992;267:11064–11068. [PubMed] [Google Scholar]
- 35.Pinner E, Kotler Y, Padan E, Schuldiner S. Physiological role of NhaB, a specific Na+/H+ antiporter in Escherichia coli. J Biol Chem. 1993;268:1729–1734. [PubMed] [Google Scholar]
- 36.Reed R H, Richardson D L, Stewart W D P. Na+ uptake and extrusion in the cyanobacterium Synechocystis PCC6714 in response to hypersaline treatment. Evidence for transient changes in plasmalemma Na+permeability. Biochim Biophys Acta. 1985;814:347–355. [Google Scholar]
- 37.Rosen B P. Ion extrusion systems in Escherichia coli. Methods Enzymol. 1986;125:328–336. doi: 10.1016/s0076-6879(86)25028-4. [DOI] [PubMed] [Google Scholar]
- 38.Rubio F, Gassmann W, Schroeder J I. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science. 1995;270:1660–1663. doi: 10.1126/science.270.5242.1660. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Shi H, Ishitani M, Kim C, Zhu J-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+antiporter. Proc Natl Acad Sci USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanier R Y, Kunisawa R, Mandel M, Cohen-Bazire G. Purification and properties of unicellular blue-green algae (order Chroococcales) Bacteriol Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strausak D, Waser M, Solioz M. Functional expression of the Enterococcus hirae NaH-antiporter in Escherichia coli. J Biol Chem. 1993;268:26334–26337. [PubMed] [Google Scholar]
- 43.Taglicht D, Padan E, Schuldiner S. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J Biol Chem. 1991;266:11289–11294. [PubMed] [Google Scholar]
- 44.Tasaka Y, Gombos Z, Nishiyama Y, Mohanty P, Ohba T, Ohki K, Murata N. Targeted mutagenesis of acyl-lipid desaturases in Synechocystis: evidence for the important roles of polyunsaturated membrane lipids in growth, respiration and photosynthesis. EMBO J. 1996;15:6416–6425. [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Utsugi J, Inaba K, Kuroda T, Tsuda M, Tsuchiya T. Cloning and sequencing of a novel Na+/H+ antiporter gene from Pseudomonas aeruginosa. Biochim Biophys Acta. 1998;1398:330–334. doi: 10.1016/s0167-4781(98)00058-x. [DOI] [PubMed] [Google Scholar]
- 47.Verkhovskaya M L, Barquera B, Verkhovsky M I, Wikström M. The Na+ and K+ transport deficiency of an E. colimutant lacking the NhaA and NhaB proteins is apparent and caused by impaired osmoregulation. FEBS Lett. 1998;439:271–274. doi: 10.1016/s0014-5793(98)01380-5. [DOI] [PubMed] [Google Scholar]
- 48.Vinnemeier J, Hagemann M. Identification of salt-regulated genes in the genome of the cyanobacterium Synechocystissp. strain PCC 6803 by subtractive RNA hybridization. Arch Microbiol. 1999;172:377–386. doi: 10.1007/s002030050774. [DOI] [PubMed] [Google Scholar]
- 49.Wada H, Murata N. Membrane lipids in cyanobacteria. In: Siegenthaler P-A, Murata N, editors. Lipids in photosynthesis: structure, function and genetics. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 65–81. [Google Scholar]
- 50.Wakabayashi S, Shigekawa M, Pouyssegur J. Molecular physiology of vertebrate Na+/H+exchangers. Physiol Rev. 1997;77:51–74. doi: 10.1152/physrev.1997.77.1.51. [DOI] [PubMed] [Google Scholar]
- 51.Waser M, Hess-Bienz D, Davies K, Solioz M. Cloning and disruption of a putative NaH-antiporter gene of Enterococcus hirae. J Biol Chem. 1992;267:5396–5400. [PubMed] [Google Scholar]