Abstract
To reveal the mechanisms underlying root adaptation to drought stress, we isolated and characterized an Arabidopsis mutant, dig5 (drought inhibition of lateral root growth 5), which exhibited increased sensitivity to the phytohormone abscisic acid (ABA) for the inhibition of lateral root growth. The dig5 mutant also had fewer lateral roots under normal conditions and the aerial parts were yellowish with a lower level of chlorophylls. The mutant seedlings also displayed phenotypes indicative of impaired auxin transport, such as abnormal root curling, leaf venation defects, absence of apical hook formation, and reduced hypocotyl elongation in darkness. Auxin transport assays with [3H]-labeled indole acetic acid (IAA) confirmed that dig5 roots were impaired in polar auxin transport. Map-based cloning and complementation assays indicated that the DIG5 locus encodes a chloroplast-localized tRNA adenosine deaminase arginine (TADA) that is involved in chloroplast protein translation. The levels of flavonoids, which are naturally occurring auxin transport inhibitors in plants, were significantly higher in dig5 roots than in the wild type roots. Further investigation showed that flavonoid biosynthetic genes were upregulated in dig5. Introduction of the flavonoid biosynthetic mutation transparent testa 4 (tt4) into dig5 restored the lateral root growth of dig5. Our study uncovers an important role of DIG5/TADA in retrogradely controlling flavonoid biosynthesis and lateral root development. We suggest that the DIG5-related signaling pathways, triggered likely by drought-induced chlorophyll breakdown and leaf senescence, may potentially help the plants to adapt to drought stress through optimizing the root system architecture.
Keywords: lateral roots, root system architecture, flavonoid, polar auxin transport, chloroplast, retrograde signaling, tRNA adenosine deaminase arginine
1. Introduction
The root system physically anchors a plant in soil and is responsible for acquiring water and nutrients to support plant growth. Roots can also sense the soil environment to adjust their own growth and physiology and are able to communicate the information to the above ground parts to initiate appropriate responses in the whole plant. The plant root system is thus highly versatile and its coordination with the shoot is critical for the entire plant to adapt to adverse conditions, such as soil water deficit (drought).
Drought is a common abiotic stress that plants encounter frequently. Even on well-irrigated agricultural land, periodic drought can occur on a daily basis. Upon encountering drought stress, plants can deploy both rapid and slow responses to deal with the stress. Rapid responses include closing stomata to minimize transpiration and activating a particular group of genes that are not expressed or expressed at low levels under normal conditions. The products of these stress-responsive genes may have diverse functions, such as facilitating osmotic adjustment and mitigating damages caused by stress. Slow responses are mostly related to developmental changes, such as altering stomata density, reinforcing leaf cuticles and waxes, promoting leaf senescence, and remodeling the root system [1,2,3,4]. The coordination of these various aspects of the drought stress response involves complex signal perception, transduction, and execution processes [3,5,6,7,8]. Understanding these signaling mechanisms is important for breeding crop tolerance to drought stress. Despite extensive research effort, many of the mechanisms underlying these diverse responses are still unclear. Major challenges, such as the complexity of drought stress as a signal, the difficulty in quantitatively and reproducibly applying drought treatments, and the lack of drought-specific responses in plants, have limited genetic studies of drought stress tolerance [5].
Root development has long been perceived as critical to plant drought stress tolerance, yet the particular traits of roots responsible for drought tolerance are rather obscure. Studies with model plants and analyses of quantitative trait loci (QTL) or genome-wide association studies (GWAS) in crop and other plants suggested that certain root traits may correlate with aspects of plant drought stress tolerance. These root traits include, for example, root biomass, diameter, length, surface area, root growth angle, presence of root hairs, and other properties, such as hydrotropism, anatomy, and hydraulic conductivity [9,10,11,12,13,14,15,16,17,18]. Yet many of these correlative relations lack strong genetic or other evidence to support their causal relationship. Furthermore, there are significant variations in the contributions of these traits to drought tolerance among different plant species.
While monocotyledonous plants have a fibrous root system, dicotyledonous plants such as Arabidopsis thaliana have a taproot system consisting of a main root (primary root) and multiple lateral roots growing out of the main root. Lateral roots develop from the pericycle cells adjacent to the xylem poles [19,20,21]. The development and growth of lateral roots are under the coordinated control of plant hormones, particularly auxin, and are strongly influenced by external conditions [21,22,23]. As a result, lateral root development is highly plastic and responsive to environmental conditions such as nutrient availability, soil mechanical property, and soil water status. Soil water deficit can dramatically affect root system architecture, including the development of lateral roots. Studies with Arabidopsis have indicated that osmotic stress and the phytohormone abscisic acid (ABA) can inhibit lateral root development [24,25,26]. We previously hypothesized that this response may be potentially linked to plant adaptation to drought and may underlie novel mechanisms of drought stress tolerance. We thus conducted genetic screens for dig (drought inhibition of lateral root growth) mutants that showed altered lateral root development in response to mannitol or ABA. Our study showed that there is a close link between this root response to drought or ABA and the whole plant drought tolerance [25]. We reasoned that reduced lateral root growth under drought stress may allow the plants to allocate more resources to grow deeper roots for water uptake, since water is often more available in deep soil [25]. Thus, the analysis of the lateral root response to drought may reveal novel determinants of plant drought tolerance.
In this study, we characterized a mutant isolated in our genetic screen, named dig5. The dig5 mutant plants showed increased inhibition by ABA of lateral root growth, although the mutant also had reduced lateral root growth under normal conditions. The dig5 mutant plants were yellowish with a reduced growth rate and a short stature. The mutant exhibited various phenotypes indicative of impaired auxin transport, which was confirmed by auxin transport assays. Further investigations showed that dig5 roots accumulated higher levels of flavonoids, secondary metabolites known to inhibit auxin transport. Introduction of the flavonoid biosynthesis mutation tt4 into dig5 restored lateral root growth of dig5. Map-based cloning found that the DIG5 locus encodes a chloroplast tRNA adenosine deaminase arginine (TADA) that edits the Arg (ACG) wobble position of tRNAArg to inosine (ICG). Our study discovered an important role of a plastidial protein in controlling lateral root development through regulating flavonoid synthesis and polar auxin transport. We suggest that the DIG5-related pathways may help plants to adapt to drought conditions.
2. Results
2.1. Isolation and Characterization of the dig5 Mutant
To identify genes important for drought tolerance, we screened for mutants defective in lateral root responses to osmotic stress or ABA. Seeds of Arabidopsis thaliana Col-gl1 (referred to as the wild type hereafter) were mutagenized and M2 seedlings were screened for mutants that showed more or less lateral root growth than the wild type under ABA or mannitol treatments. These mutants were referred to as dig (drought inhibition of growth of lateral roots) mutants [25] and one of these mutants, dig5, was characterized in this study.
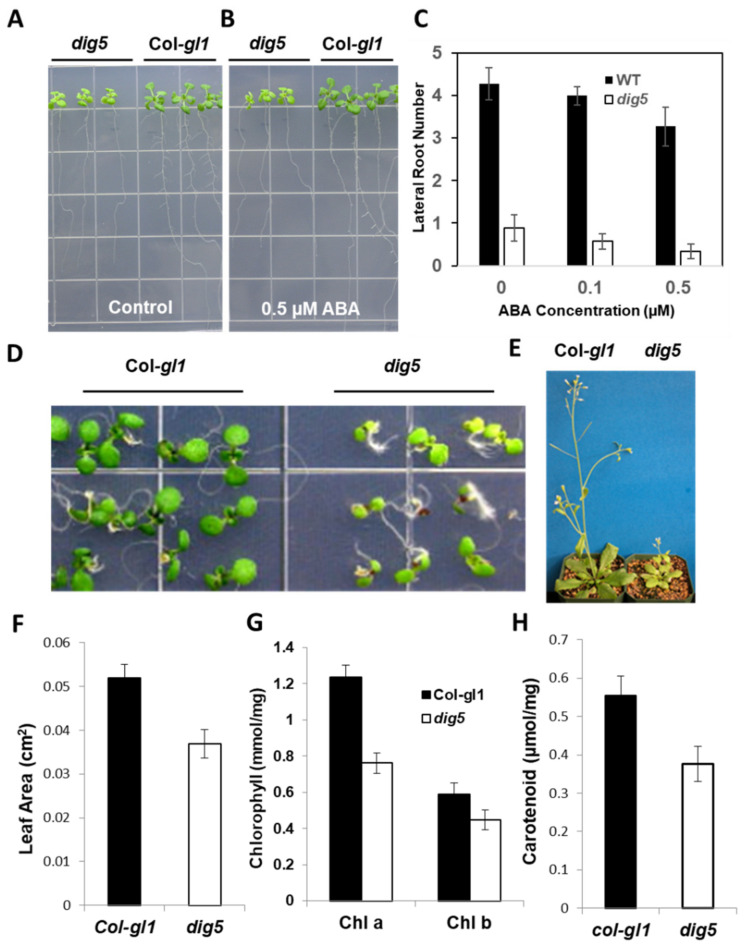
In the initial screen on 1/2 MS medium supplemented with 0.5 μM ABA, the wild type seedlings could still produce a reduced number of lateral roots compared with the control (without ABA), but the dig5 mutant had nearly no visible lateral roots. In subsequent re-screens, however, the number of lateral roots of dig5 was found to be fewer even on the control medium without ABA compared to wild type (Figure 1A). In media without ABA, the average number of visible lateral roots per plant in the wild type was 6 times that of dig5 (Supplementary Table S1). In the presence of ABA, while the lateral root number and length of both the wild type and dig5 decreased, these were decreased more in dig5, reflecting increased sensitivity to ABA inhibition of lateral root growth in the mutant (Figure 1A–C). On full-strength MS media (where lateral root development was suppressed compared with on 1/2 MS media), the primary root growth was inhibited more in dig5 than in the wild type by ABA or NaCl (Supplementary Figure S1A–C), suggesting a general increase in ABA sensitivity in the dig5 mutant.
Figure 1.
Phenotypes of the dig5 mutant. (A–C) Response of lateral roots of the wild-type Col-gl1 and dig5 seedlings to ABA. Three-day-old seedlings were transferred to the shown plate (A) without (control) or (B) with 0.5 μM ABA and grown for 3 days before taking the pictures. (C) Lateral root number per seedling of the wild type and dig5 shown in (A). Data are means ± SD from 8 to 9 seedlings. (D) Morphology of 7-day-old seedlings of the wild type and dig5 growing on agar plates. (E) Morphology of adult plants growing in soil for one and a half months. (F) Leaf area of the first two rosette leaves of 1-week-old wild type and dig5 mutants grown in agar media. (G) Chlorophyll contents of the wild type and dig5 mutant leaves of 3-week-old seedlings grown in soil. (H) Carotenoid content of leaves of 3-week-old wild type and dig5 seedlings grown in soil.
In addition to their reduced lateral root numbers, dig5 mutant seedlings had other phenotypes distinct from the wild type. The primary root of the mutant was shorter, at 60 percent that of the wild type (Figure 1A and Supplementary Table S1). The seedlings of dig5 were smaller in agar media (Figure 1D) and had a shorter stature when grown in soil (Figure 1E, Supplementary Table S1), which did not allow us to accurately compare their drought tolerance with the wild type due to the difference in their size. Another notable phenotype of the mutant was that the leaves were yellowish or pale green, either in MS media or in soil (Figure 1D,E). The contents of chlorophyll a, b, and carotenoids in dig5 were significantly lower than in the wild type (Figure 1G,H). Detailed morphometric analyses showed that there were significant differences in seedlings and in mature plants between the dig5 mutant and the wild type (Supplementary Table S1). For 8-day-old seedlings, the number of lateral roots in dig5 was about 83.5% fewer than that of the wild type. For adult plants growing in soil, the inflorescence stem length of mutant plants was only 28.6% that of the wild type, and the silique number and distance between siliques were 26.8% and 53.5% those of the wild type, respectively (Supplementary Table S1).
2.2. Auxin Responses of the dig5 Mutant
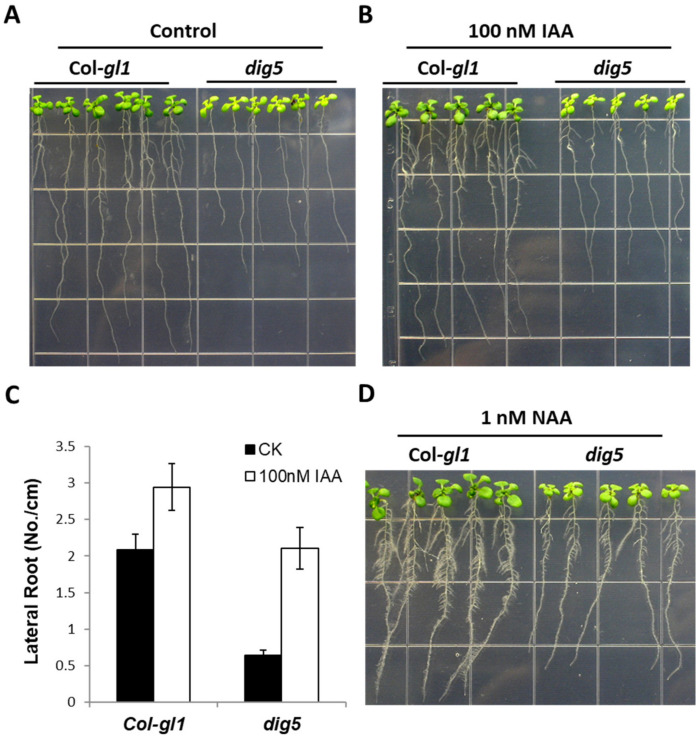
The reduced number of lateral roots in the dig5 mutant is striking. Since the phytohormone auxin plays critical roles in lateral root development [21], we examined whether the mutants were altered in auxin response. When growing in media containing the auxin indole-3-acetic acid (IAA), the number of lateral roots in both the wild type and dig5 increased (Figure 2A,B). Although the total number of lateral roots in dig5 was still lower than in the wild type, the increase in dig5 was about 165% higher than the increase in the wild type (Figure 2C). Similarly, the synthetic auxin analogs 1-naphthaleneacetic acid (NAA) and 2,4-dichlorophenoxyacetic acid (2,4-D) also significantly increased the number of lateral roots in dig5 (Figure 2D). These data indicated that dig5 was responsive to auxin and that exogenous auxin could partly rescue the defects in lateral root development in dig5.
Figure 2.
Lateral root phenotypes of dig5 mutant seedlings can be rescued by auxin. Lateral root growth of the wild type (Col-gl1) and dig5 without (A) or with (B) 100 nM IAA. (C) Lateral root density of seedlings shown in (A,B). Data are means and standard deviation (n = 18). (D) Morphology of seedlings of the wild type (Col-gl1) and dig5 on 1 nM NAA.
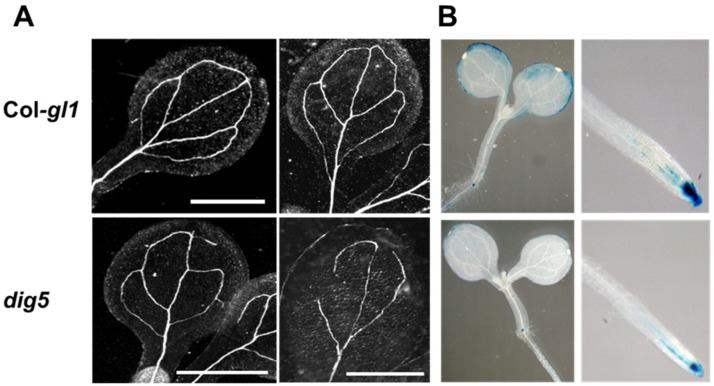
Auxins are also important in regulating the development of vasculature [27]. In contrast to the regular venation in the wild type leaves, dig5 leaves often had irregular, disrupted, and discontinuous veins (Figure 3A). These phenotypes suggested that dig5 mutants may have reduced levels of auxin or an impaired response to auxin. To test these possibilities, we introduced the auxin reporter gene DR5-GUS into the dig5 mutant through genetic crossing. Young seedlings of the resulting lines were stained for GUS expression (Figure 3B). The DR5-GUS expression was evident in leaves and roots of the wild type seedling, particularly at the root tip, in vascular tissues of the root, and the edge of leaves. In contrast, the DR5-GUS signal was significantly weaker in dig5 in both leaves and roots (Figure 3B). The nearly normal auxin response of the dig5 mutant in the lateral root development and the reduced auxin levels in the root suggest that the dig5 mutant may have a reduced auxin transport capability. We thus examined whether dig5 was defective in other processes that are affected by auxin transport.
Figure 3.
Leaf venation defects and reduced auxin levels in dig5 mutant seedlings. (A) Representative pictures of leaf venation in seedlings of wild type (Col-gl1) and dig5 mutant. Scale bars = 1 cm. (B) Expression of DR5-GUS reporter gene (GUS-staining) in wild type (Col-gl1) and dig5 seedlings.
2.3. Altered Root Curling, Hypocotyl Elongation and Apical Hook Formation in dig5
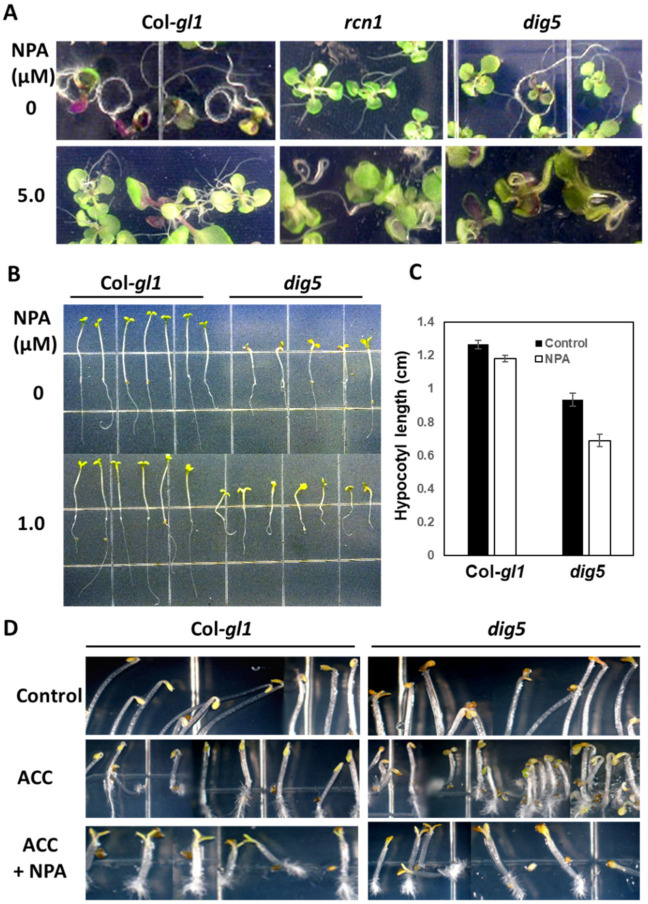
One of the phenotypes related to polar auxin transport in roots is root curling [28]. This phenotype can be seen with seedlings growing in a petri dish with agar media. When growing to the bottom of the agar plate, roots of the wild type seedlings will begin to curl in a circle-like pattern, presumably searching for the gravity vector. However, when the auxin transport inhibitor N-1-naphthylphthalamic acid (NPA) is added to the medium, roots grow in a straight line instead of curling. The auxin transport mutant roots curl in NPA 1 (rcn1) has an opposite phenotype to the wild type in that the roots do not curl in the absence of NPA but do curl strongly in the presence of NPA [28]. We examined the root curling behavior of dig5 seedlings and included the rcn1 mutant in the assay for comparison. In the control medium without NPA, the wild type roots curled as expected, but the dig5 mutant roots grew straight without or only slightly curling. In the presence of NPA, the wild type roots grew straight, but the dig5 roots curled (Figure 4A). Thus, the dig5 mutant was defective in the root curling response to NPA and behaved like the rcn1 mutant, consistent with the notion that dig5 may be defective in polar auxin transport.
Figure 4.
Abnormal root curling and apical hook formation of dig5 mutants. (A) Root curling of Col-gl1, rcn1, and dig5 seedlings in the absence or presence of 5 µM NPA. (B) Hypocotyl elongation of Col-gl1 and dig5 in the absence or presence of 1.0 µM NPA. (C) Hypocotyl length of seedlings in (B). Data are means and standard deviation from 18 seedlings. (D) Apical hook formation of seedlings under the control, ACC (10 µM), and ACC (10 µM) + NPA (1 µM) treatments. Seedlings were germinated and grown in darkness for 3 days before taking the pictures.
Inhibition of auxin transport by NPA also inhibits hypocotyl elongation of dark-grown seedlings. We measured the sensitivity of hypocotyl elongation to NPA in the dig5 mutant. Without NPA, dig5 seedlings had shorter hypocotyls than the wild type. NPA treatment inhibited the hypocotyl elongation of both wild type and dig5 seedlings, with the mutant hypocotyl inhibited by 27.3%, but the wild type by only 6.7% when compared with control treatments without NPA (Figure 4B,C).
Dark-grown Arabidopsis seedlings form apical hooks, and the hooks can be exaggerated by treating with ethylene or its precursors and can be reduced by treating with NPA. These responses are also mediated by auxin partly through auxin transport [28,29,30]. We examined apical hook formation in dig5 mutant seedlings. The wild type seedlings showed tight apical hooks on normal MS media, as expected, and exaggerated hooks were seen with addition of the ethylene precursor 1-aminocyclopropane-1-carboxylate (ACC) (Figure 4D). In contrast, the dig5 mutant seedlings grown under the same conditions did not form apical hooks or formed only partial hooks (Figure 4D). ACC also had limited enhancement on apical hook formation in dig5 seedlings (Figure 4D). As a control, hook formation in the wild type as well as in dig5 was blocked by NPA even in the presence of ACC (Figure 4D), consistent with the notion that polar auxin transport plays a dominant role in the apical hook formation.
2.4. Polar Auxin Transport Was Impaired in dig5
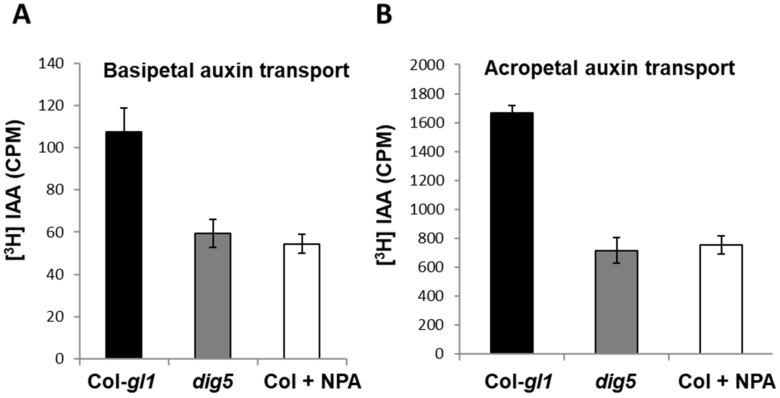
The above data all pointed to the notion that the dig5 mutant is likely impaired in polar auxin transport. To investigate the auxin transport capability in the dig5 mutant, we sought to measure the basipetal (toward the base) and acropetal (toward root tip) auxin transport in dig5 roots. We applied [3H]-labelled IAA to roots at either the root base or root tips and then measured the radioactivity at distal segments of the roots after a certain period of time using a method similar to that described [31]. Briefly, agar blocks containing 100 nM [3H] IAA with or without the auxin transport inhibitor NPA (100 μM) were applied to the root tips or root-shoot junctions, respectively. After incubation for 5 h (for basipetal) or 16 h (for acropetal) in the dark, the root samples were collected and the amount of radioactivity from the basipetal or acropetal transported [3H] IAA was determined using a liquid scintillation counter. As shown in Figure 5, both basipetal and acropetal IAA transport abilities in the dig5 mutant were significantly reduced compared to those in the wild type, and the transport rate in the mutant was similar to that of the NPA-treated wild type. This indicated that dig5 mutants were defective in both basipetal and acropetal auxin transport.
Figure 5.
Polar auxin transport in roots of dig5 and wild type seedlings. (A) Basipetal auxin transport. (B) Acropetal auxin transport. 100 nM [3H]-IAA with or without NPA (100 µM) was applied either to the root tip (A) or shoot-root junction (B) of seedlings on agar plates followed by incubation in the dark for 5 h (A) or 16 h (B). The distal segments of the roots were harvested to measure the radioactivity. Data were means and standard errors of 3 replicates, each with at least 10 seedlings.
2.5. Map-Based Cloning of the DIG5 Locus
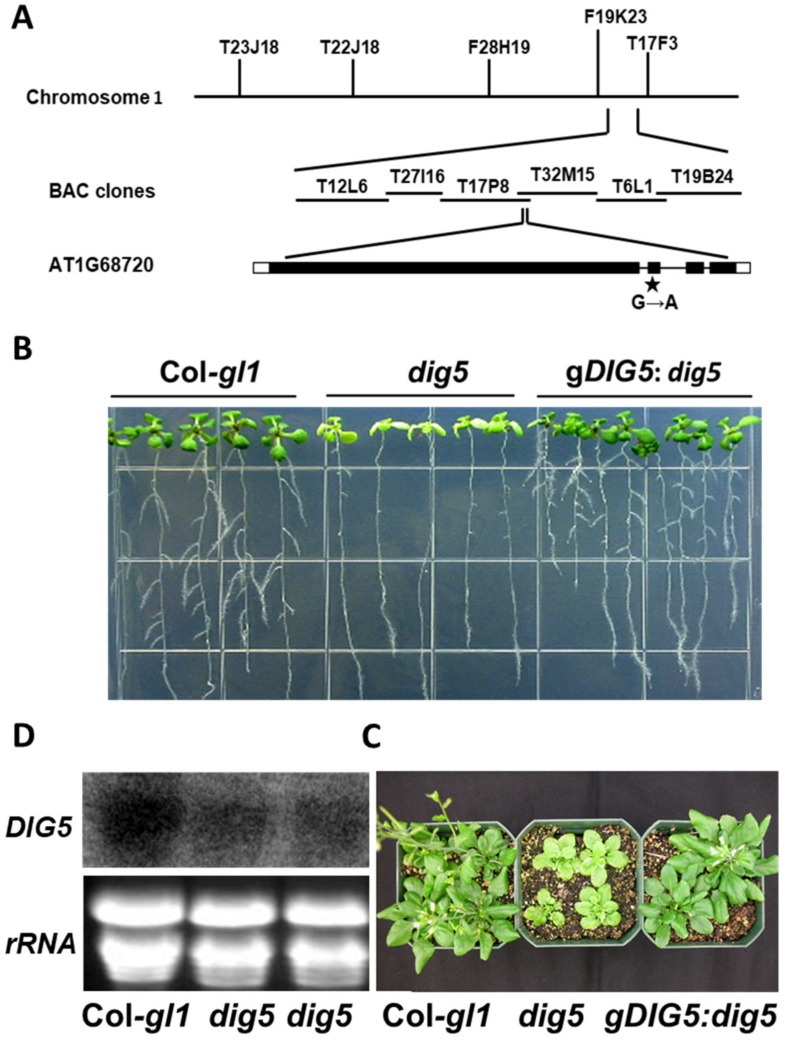
To determine the molecular nature the DIG5 locus, a map-based cloning strategy mapped the dig5 mutation to the lower arm of Chromosome I (Figure 6A), within an interval of about 105 kb. All the annotated genes in the mapped interval were PCR-amplified from the genomic DNA of the dig5 mutant and sequenced. This resulted in the identification of a single mutation in the interval. The G to A mutation at nucleotide 3511 from the predicted translational start site of the gene At1g68720 occurred at the junction between the first intron and the second exon. This mutation would alter the predicted splicing acceptor signal AG to AA and would thus shift the splicing acceptor to the immediately adjacent AG dinucleotide. Adoption of this new splicing acceptor site would result in removing this AG dinucleotide from the cDNA. RT-PCR amplification of the cDNA from dig5 and Col-gl1 and DNA sequencing confirmed this prediction. As a result, the mutation caused a frameshift and would produce 4 new amino acids before the transcript encountered a new pre-mature stop codon and terminate the open reading frame. RNA blotting of total RNA extracted from the wild type and dig5 showed that the transcript level of this gene in dig5 was lower than in the wild type (Figure 6D). This is likely caused by increased degradation of the pre-mature stop codon-containing mutant transcripts that trigger non-sense mediated decay (NMD).
Figure 6.
Map-based cloning of the DIG5 locus and complementation of the dig5 mutants. (A) Map-based cloning of the DIG5 locus. After delimiting the DIG5 locus to an interval of about 105 kb, DNA sequencing identified a single nucleotide mutation in the gene At1g68720. (B,C) Complementation of the dig5 mutants with the wild type genomic DNA of At1g68720. Shown are the wild type Col-gl1, dig5, and dig5 mutant transformed with the wild type DIG5 genomic DNA (gDIG5:dig5) on agar plates (B) or in soil (C). (D) Expression level of the DIG5 gene in the wild type and dig5 mutant seedlings. Shown are RNA blotting of total RNA probed with the radiolabeled DIG5 probe (upper panel) and loading control of ethidium bromide stained rRNA (lower panel).
To determine whether the phenotypes observed in dig5 were caused by the mutation in At1g68720, we made a construct consisting of the wild type At1g68720 genomic DNA sequence including its promoter and transferred the construct into the dig5 mutant via the Agrobacterium-mediated floral dip method [32]. Multiple independent stable transgenic plants were generated and tested for their phenotypes. It was found that the At1g68720 gene was able to rescue the leaf and the lateral root defects of the dig5 mutants on agar media (Figure 6B) and the yellowish, short stature and other phenotypes of dig5 mutants when grown in soil (Figure 6C). These data indicate that At1g68720 is the DIG5 gene and that the dig5 mutation was responsible for the mutant phenotypes observed.
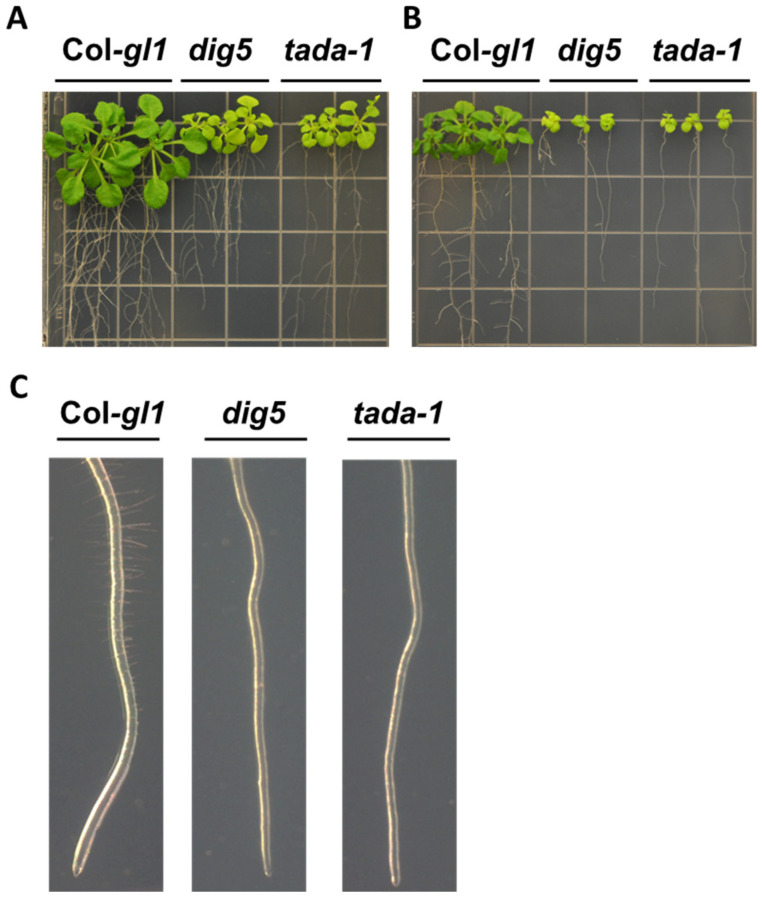
At the time we identified At1g68720 as DIG5, this gene was annotated as an unknown protein. The predicted protein contained a long N-terminus that showed no obvious sequence homology to other proteins, yet the C-terminus contained a short catalytic domain found in cytidine/deoxycytidylate deaminases that may participate in RNA editing in chloroplasts. Using the recombinant full-length protein or the truncated protein retaining the C-terminal catalytic domain, we failed in our attempt to detect cytosine deaminase activity. We also sequenced known RNA editing sites in related chloroplast and mitochondrial genes. However, no change in RNA editing in the mutant was detected. This gene was later reported to encode a tRNA adenosine deaminase arginine (TADA) that edited the adenosine at the wobble position of chloroplast tRNAArg(ACG) to inosine tRNAArg(ICG), and the mutant plants were found to accumulate lower levels of certain chloroplast proteins and to have reduced photosynthetic functions [33,34]. Phenotypes of yellowish leaves and a short stature similar to those of dig5 were also noted with the T-DNA knockout mutant tada [34]. After identifying the DIG5 locus, we obtained several T-DNA insertional alleles and an allele GK-119G08 (renamed as tada-1 for consistency) was used to compare their phenotypes with those of dig5. It was found that the tada mutant had phenotypes similar to those of dig5, such as reduced lateral root numbers (Figure 7A) and increased inhibition of primary roots by ABA (Supplementary Figure S1A,B). An additional phenotype was that both dig5 and tada-1 had significantly fewer root hairs than the wild type (Figure 7B,C). It is known that root hair development is also partly regulated by auxin transport [35]. Interestingly, the primary root of tada-1 was less inhibited by NaCl compared with dig5 (Supplementary Figure S1C), which likely was due to a possible leaky nature of the tada-1 mutation since the T-DNA was inserted at the N-terminus and might have spared some of transcripts with the intact C-terminal catalytic domain to produce a low level of the functional protein.
Figure 7.
The mutant lines dig5 and tada-1 had similar phenotypes. (A,B) Morphology of dig5 and tada-1 seedlings on 1/2 MS without (A) or with (B) 1.0 µM ABA. (C) Reduced numbers of root hairs of 10-day-old dig5 and tada-1 mutant roots compared with the wild type root.
2.6. Increased Flavonoid Contents in dig5 Roots
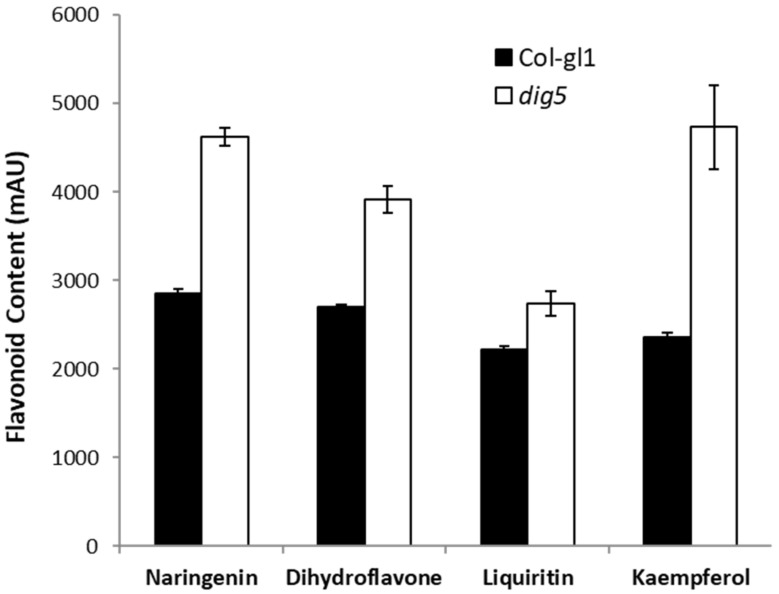
Our study showed that the dig5 mutant was defective in polar auxin transport. Since DIG5 did not encode a transporter but rather a putative enzyme (which we thought at the time we conducted the experiments), we hypothesized that there might be metabolite changes in dig5 that affect auxin transport. Plants have endogenous metabolites such as flavonoids that can regulate auxin transport under physiological conditions [36]. Indeed, flavonoids, such as quercetin, kaempherol, and apigenin, are endogenous inhibitors of polar auxin transport that can regulate auxin transport and impact root development [37,38]. We therefore examined whether there were any changes in the levels of these flavonoids in dig5. Roots of dig5 and wild type seedlings were harvested and extracted to measure flavonoids using an HPLC method. Our data showed that the levels of flavonoids, including naringenin, kaempherol, dihydroflavone, and liquiritin were all higher in dig5 than in the wild type. Particularly, the level of kaempherol in dig5 was twice that of the wild type (Figure 8).
Figure 8.
Flavonoid levels in dig5 roots. Flavonoids in roots of 2-week-old dig5 and wild type Col-gl1 seedlings were extracted and quantified with HPLC (mAU, milli Absorbance Unit).
2.7. The Expression of Flavonoid Biosynthetic Genes Was Enhanced in dig5
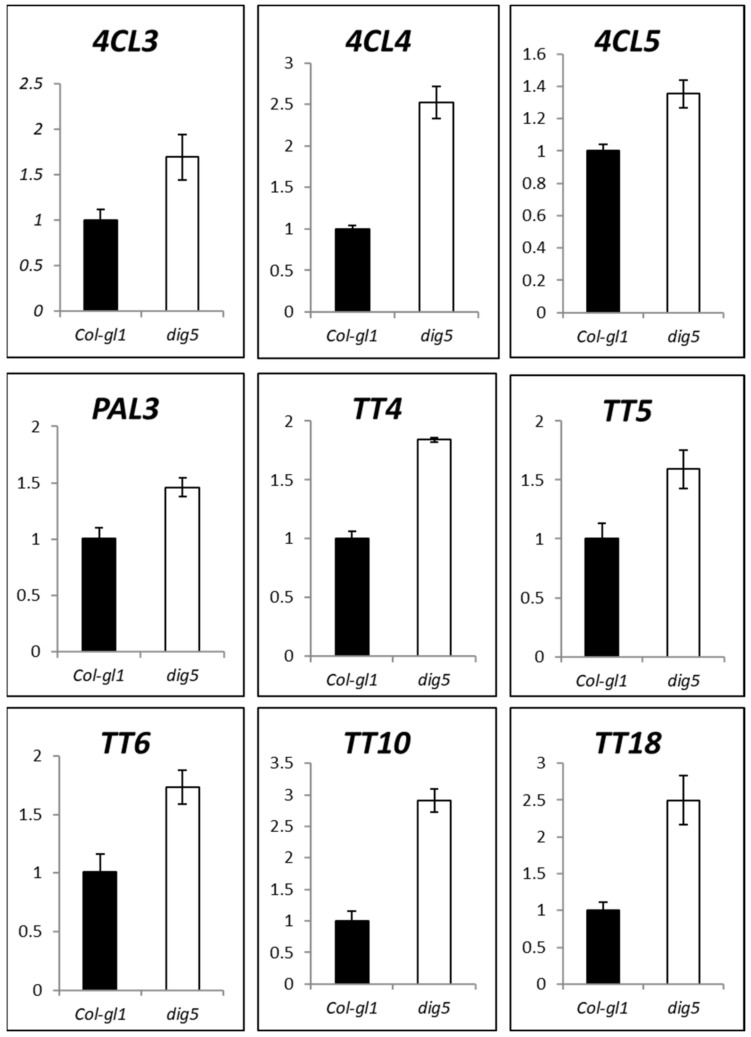
The fact that dig5 mutants had higher contents of flavonoids suggests that there may be increased biosynthesis of these metabolites. To test this possibility, we performed quantitative real-time PCR to measure the expression of major flavonoid biosynthesis genes in the roots of dig5 in comparison with the wild type. The flavonoid biosynthesis pathways are well studied in Arabidopsis and other plants, through genetic and biochemistry approaches [39,40,41]. Flavonoids are synthesized from phenylalanine through the phenylpropanoid pathway where the first enzyme of the pathway phenylalanine ammonia-lyase (PAL) converts phenylalanine into cinnamate, which is then converted to ρ-coumarate. ρ-coumarate is further catalyzed by 4-coumarate coenzyme A ligase (4CL) to form 4-coumarate coenzyme A (CoA). Chalcone synthase (CHS/TT4) then combines 4-coumarate CoA with malonyl-CoA to produce naringenin chalcones. Chalcone isomerase (CHI/TT5) isomerizes chalcones into naringenin, which is catalyzed by flavanone 3-hydroxylase (F3H/TT6) into dihydroflavones. In the late part of the pathway, flavanol synthase (FLS) converts various dihydrofavones into common flavanols, such as kaempferol and quercetin [40]. Our quantitative real-time PCR analyses found that there was significantly increased expression of major genes in the flavonoid biosynthesis pathway: PAL3, 4CL3, 4CL4, 4CL5, TT4, TT5, TT6, TT10, and TT18 (Figure 9). The higher expression of these genes in dig5 may contribute to increased syntheses of flavonoids in the mutant.
Figure 9.
Expression of flavonoid biosynthetic genes in dig5. Shown are quantitative real-time PCR detection of transcript levels in roots of 2-week-old dig5 and wild type (Col-gl1) seedlings. 4CL3/4CL4/4CL5 encode 4-coumarate:CoA ligase 3/4/5; PAL3 encodes phenylalanine ammonia-lyase 3; TT4 encodes chalcone synthase (CHS); TT5(At3g55120) encodes chalcone isomerase (CHI); TT6 (At3g51240) encodes flavanone 3-hydroxylase; TT10 (At5g48100) encodes a laccase-like polyphenol oxidase; TT18 (At4g22880) encodes anthocyanidin synthase (ANS).
2.8. Genetically Blocking Flavonoid Synthesis Rescued dig5 Phenotypes
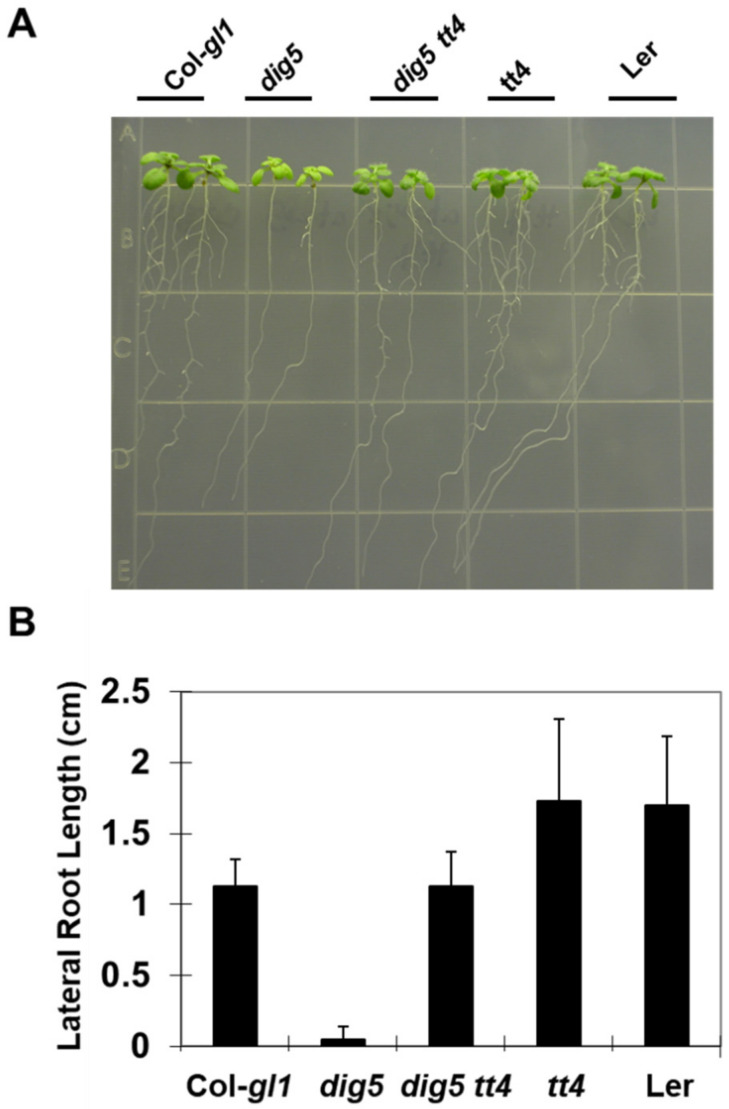
In the dig5 mutant, the enhanced expression of flavonoid biosynthetic genes and the higher levels of flavonoids in roots of dig5 are consistent with the impaired auxin transport and the reduced lateral root development in the mutant. To determine whether the increased flavonoid content was the cause of these phenotypes, we made a double mutant between dig5 and transparent testa 4 (the tt4-1 allele) by crossing the single mutants. TT4 encodes a chalcone synthase, which catalyzes the first committed step in the flavonoid pathway. The tt4 mutant has very low levels of all major flavonoids [37,41,42]. The dig5 tt4 double mutant along with the single mutants and Col-gl1 (background of dig5) and Landsberg-erecta (Ler) (background of tt4) were germinated and one-week-old seedlings were transferred to MS medium to grow for 10 days (Figure 10A,B). While dig5 had no or very few lateral roots, the dig5 tt4 double mutant had a lateral root phenotype close to that of the wild type Col-gl1. Measurement of the total lateral root length per seedling showed that tt4 suppressed the dig5 lateral root phenotype, suggesting that TT4 acts downstream of DIG5 in controlling lateral root development and that higher levels of flavonoids may underlie the lateral root defects in the dig5 mutant.
Figure 10.
Loss-of-function mutation of TT4 rescued the development defects in lateral roots of dig5 mutant seedlings. The dig5 tt4 double mutant was generated by genetic crossing. (A) Morphology of 2-week-old seedlings grown on 1/2 MS medium. (B) Total lateral root length per plants. Data are means and standard deviation from 6 seedlings.
3. Discussion
The plant root system is of critical importance for plants fighting drought stress. Roots not only mine water from soil, but can also coordinate other processes of the whole plant to manage the water economy. Complicated mechanisms are expected to exist in plants that regulate these processes, yet few of these mechanisms are well elucidated. A consensus is that a robust root system that can forage water from deeper soil (also known as a drought avoidance strategy) would enhance plant tolerance to drought stress. In this regard, allocation of limited resources to grow deeper roots will be of adaptative advantage to the plants. Reduced lateral root growth of Arabidopsis in response to osmotic stress and ABA is thus considered to be one of such adaptative strategy [25,43]. This altered growth habit underlies an important aspect of the plasticity of root developmental adaptation to the environment. Although the exact mechanisms for this adaptive response are not very clear, the involvement of phytohormones, particularly auxin and ABA, in the process has been implicated. The identification of the DIG5 locus in this study highlights further an unexpected route of regulation of lateral root development.
Auxin control of lateral root development has been well documented [20,21,44,45]. Nonetheless, the interaction between auxin and drought or ABA in regulating the root response to drought is less well understood. Drought stress may alter auxin biosynthesis, catabolism, transport, and signaling to affect root development. For instance, drought or ABA enhances the production of the microRNA miR393, which targets the transcripts of auxin receptor genes, such as TIR1 and AFB2, for cleavage and degradation. In transgenic plants expressing miR393-resistant versions of TIR1 or AFB2, these mutant transcripts are no longer cleaved and neither ABA nor osmotic stress (generated by polyethylene glycol) inhibits the elongation of lateral roots [46]. Thus, auxin signaling is important for regulating lateral root growth during plant response to drought stress.
Although auxin can be synthesized locally, long distance polar auxin transport is important for organogenesis including lateral root development [45]. Several classes of auxin transporters are described, among which the PIN family of auxin efflux carriers are of particular importance and have been extensively investigated [47]. The functionality of these carriers is determined by their transport activity and polar localization, both of which are regulated by various processes such as phosphorylation [48]. Besides these posttranslational modifications, cellular metabolites appear to regulate the transporter activities as well. Among these metabolites, flavonoids have been widely reported to inhibit auxin transport [36].
Flavonoids are synthesized through the phenylpropanoid pathway. The first committed step in the flavonoid pathway is catalyzed by chalcone synthase (CHS) encoded by the TT4 (transparent testa 4) gene in Arabidopsis. The other major steps of the pathway have also been defined genetically with other tt mutants, such as tt5, tt6, tt7, etc. [40,41]. Mutations in these genes can significantly decrease flavonoid accumulation. In dig5 mutant roots, levels of several flavonoids increased significantly, particularly kaempferol, which is a major flavonoid that accumulates in Arabidopsis roots [40,49]. This increased flavonoid accumulation was likely caused by the increased expression of flavonoid biosynthetic genes. Indeed, the expression of major genes in the early steps in the pathway increased significantly (Figure 8). Another possibility is that more substrates of the pathway may have channeled into flavonoid production.
Flavonoids have important functions in processes, such as UV tolerance, oxidative stress response, defense response, and abiotic stress, as well as bacterium–host interactions [38,50,51,52]. The involvement of flavonoids in regulating auxin transport has long been known [36,38,42], and the discovery of synthetic auxin transport inhibitors such as NPA that mimic flavonoids makes chemical regulation of auxin transport much easier. Several targets of NPA or flavonoids in auxin transport have been proposed [53]. Recent studies suggested that the PIN family of polar auxin efflux carriers are the major targets. Flavanols and NPA stabilize PIN dimers, thus inhibiting the transport activity of these auxin efflux carriers [54,55]. Recent structure analyses shed light on how NPA may affect auxin transport, e.g., by competing for the IAA-binding site of PIN transporters [56,57]. In dig5, multiple phenotypes indicative of auxin transport defects were observed. These include reduced lateral root growth, abnormal root curling, loose apical hook formation, and reduced number of root hairs, etc. Measurement of auxin polar transport in roots indeed showed that both acropetal auxin transport and basipetal auxin transport were significantly reduced in the dig5 mutant (Figure 8). We therefore suggest that increased accumulation of flavonoids in dig5 plants impairs auxin transport, which in turn leads to reduced lateral root growth and the other phenotypes related to defects in auxin transport. Consistently, introduction of the tt4 mutation (which abolishes flavonoid production) into the dig5 mutant restored the lateral root growth to the level of the wild type (Figure 10), indicating an important role of DIG5 in maintaining flavonoid homeostasis in plants.
Our map-based cloning uncovered that DIG5 encodes a protein that was subsequently identified as tRNA adenosine deaminase Arginine (TADA) [34]. TADA catalyzes the deamination of the adenosine at the wobble position of tRNAArg (ACG) to inosine in tRNAArg (ICG) in chloroplasts/plastids. The tada mutation reduced chloroplast translation efficiency and led to reduced levels of certain plastid-encoded proteins and impaired photosynthesis functions [34,58]. These affected proteins, however, are not directly involved in the phenylpropanoid biosynthesis pathway. Thus, the defect in plastid protein homeostasis may generate signal(s) that affect the expression of nuclear genes involved in flavonoid biosynthesis. Retrograde signaling from chloroplasts or mitochondria has been shown to be common in regulating nuclear gene expression [59]. Mutants of several chloroplast genes were reported to affect lateral root development. For instance, the Arabidopsis mutants of the chloroplast-localized protein FIERY1 (FRY1) [60] had fewer lateral roots and were more sensitive to ABA inhibition of lateral root growth [61,62]. The plastidial glycolate and glycerate transporter mutant plgg1 [63] and the NADPH thioredoxin reductase C mutant [64] also had reduced lateral roots. That chloroplast retrograde signaling inhibits lateral root development is likely of adaptive advantage to the plant. Drought stress is known to cause chlorophyll breakdown and leaf senescence, which, as shown in the current study, may trigger the production of flavonoids to inhibit horizontal root growth and to promote roots to grow deeper to increase drought tolerance of the plant. This possibility can be tested in future studies.
How chloroplast defects may trigger flavonoid production and inhibit lateral root development is not yet clear. In the current study, we speculate that the defects in chloroplast translation caused by lack of DIG5/TADA protein may lead to an imbalance in the chloroplast of, e.g., reactive oxygen species, which in turn activates the expression of flavonoid genes in the nucleus in order to maintain organellar redox homeostasis in the dig5 mutant. The increased accumulation of flavonoids would result in reduced auxin transport and impaired lateral root development. The exact mechanisms through which chloroplast protein homeostasis triggers the retrograde signaling to activate nuclear flavonoid biosynthetic genes warrant further investigation.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 carrying the glabrous1(gl1) mutation (referred to as Col-gl1 or the wild type) was used to conduct mutagenesis with ethyl methanesulfonate. Mutant screening of the M2 generation for altered lateral root growth responses under treatment of mannitol or ABA was described previously [25]. Isolated mutants were re-screened and backcrossed to Col-gl twice before being used for further characterization. For general growth assays, unless otherwise stated, seeds were surface sterilized and planted on 1/2 MS media (1.2% agar and 3% Sucrose). The plates were then incubated at 4 °C for 3 d before being placed vertically under constant white light at 22 °C for germination and seedling growth.
4.2. Root Growth, Hypocotyl Elongation, Apical Hook Formation and Auxin Responses
For root growth assays, 5-day-old seedlings were individually transferred from 1/2 MS medium plates with a pair of forceps to the treatment medium plates and incubated in an incubator for a specified period before taking photographs or measurements.
For root curling assay, the surface-sterilized seeds were sown on 1/2 × MS medium agar plates without or with 5 μM NPA supplement. After cold treatment, plates were incubated for 2 days in the light and then 5 days in the dark (all at 22 °C) before taking pictures from the back of the plates. For hypocotyl elongation measurements, seedlings were germinated and grown on the surface of the medium in a vertical position for 5 days at 22 °C in the dark. For apical hypocotyl hook observation, plates were wrapped with aluminum foil and incubated at 22 °C in a vertical position for 3 days before taking pictures. For the hook formation assays, NPA was used at 1 µM and ACC was at 10 µM.
To test the auxin responses, IAA, NAA or 2,4-D was added into the medium separately with the final concentration at 100 nM, 10 nM, and 1 nM, respectively. The phenotypes of roots were observed, and the pictures were taken with a digital camera.
Length of lateral roots and primary roots were measured with the digital images of seedlings using the Image J software as described [25]. Number of visible lateral roots per seedling was counted and the density was calculated by dividing with the length of the primary root.
4.3. Chlorophyll a and b and Carotenoid Measurements
For pigment measurements, fresh leaves of 3-week-old seedlings grown in soil were ground in absolute ethanol. After centrifugation, the supernatant was used to measure the absorbance at 663, 645, and 480 nm. Pigment contents were calculated as described [65].
4.4. Leaf Venation Observation
Fresh leaf tissues were fixed in saturated chloral hydrate. The samples were then mounted on glass slides with coverslips, the leaf veins were visualized, and the pictures were taken with differential interference contrast (DIC) images using a Nikon SMZ1500 microscope and a Qimaging Retiga cooled camera (Burnaby, BC, Canada).
4.5. Auxin Transport Assays
The basipetal and acropetal auxin transport capabilities of seedlings were measured according to the method described [31], with modifications. The growth medium contained 0.8% agar, 1 × MS, and 1.5% sucrose, at pH 5.7. Seeds were germinated, and seedlings were grown on vertically placed agar petri dishes for about 5 days until the roots reached the length of 1.0–1.5 cm. Seedlings were transferred to treatment plates with root tips aligned. 3H-IAA (American Radiolabeled Chemical, St. Louis, MO, USA) at 100 nM was mixed with 1% (v/v) agar either with or without 100 μM NPA and was applied to localized areas of roots for transport assays. For basipetal auxin transport, the radiolabeled IAA-containing agar was applied to just touch the root tip. Plates remained vertically oriented for 5 h in the dark to minimize IAA breakdown. For acropetal transport assay, 10 μM cold IAA was added as it could increase 3H-IAA transport. The 3H-IAA agar block was placed at the root-shoot junction of the seedlings. Plates were incubated in the dark for 16 h with seedlings upside-down. Individual 2- or 5-mm segments were cut and rinsed and placed into 2.5 mL of scintillation fluid. The radioactivity was measured using a liquid scintillation counter. Three independent experiments were conducted, each with at least 10 individual seedlings.
4.6. HPLC Detection of the Flavonoid Content in Roots
Roots of seedlings vertically growing on agar petri dishes for 2 weeks were harvested and grounded immediately in liquid N2. The samples were then extracted with 80% (v/v) methanol at 1 g/10 mL at room temperature. After centrifugation (4000× g, 5 min) and filtration (0.45 μm syringe filter), the samples were re-extracted with ethyl acetate. Extracts were concentrated to dryness with an Eppendorf vacufuge at 45 °C and dissolved in 80% methanol for HPLC analysis. The extracts were analyzed on an Agilent 1100 series HPLC system (Agilent, Santa Clara, CA, USA) using authentic standards as internal controls as described [66].
4.7. Constructs and Arabidopsis Transformation
The full-length cDNA of DIG5 was amplified and cloned into the Ecor I and Sal I sites of the pZP vector. The genomic sequence of At1g68720 was also amplified from Col-0 plants and used in the complementation assay. The constructs were introduced into Agrobacterium tumefaciens strain GV3101 and then transformed into the dig5 mutant using the floral dip method [32]. The transgenic plants were screened on a MS agar medium containing 50 mg/L kanamycin.
4.8. Quantitative Real-Time PCR
Total RNAs were extracted from 10-day-old seedlings using the RNeasy Plant Mini kit (Qiagen), and RT reactions were conducted with SuperScript III First-Strand Synthesis SuperMix (Invitrogen) using random hexamers for the first strand synthesis. After 10 times dilution, 1 µL of the diluted solution was used as a template in a 10-µL reaction with 2 × SYBR Green SuperMix, ROX (Invitrogen). The quantitative RT-PCRs were performed in triplicate using the ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). UBQ3 was used as internal control. Primer sequences are listed in Supplementary Table S2.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231810642/s1.
Author Contributions
Conceptualization: W.L. and L.X.; methodology, W.L., T.C., Y.L., Q.T.L., R.W. and L.X., with most experiments performed by W.L.; data curation, W.L., T.C. and L.X.; writing—draft preparation, W.L. and L.X.; writing—review and editing, everyone reviewed, edited and approved the manuscript; supervision, H.L. and L.X.; funding acquisition, L.X. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was supported by the Donald Danforth Plant Science Center (2002#1200) and by the Research Council of HKBU (SGT2/1920/SCI-006) (to L.X.).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yoshida T., Fernie A.R., Shinozaki K., Takahashi F. Long-distance stress and developmental signals associated with abscisic acid signaling in environmental responses. Plant J. 2021;105:477–488. doi: 10.1111/tpj.15101. [DOI] [PubMed] [Google Scholar]
- 2.Zhao P., Liu P., Shao J., Li C., Wang B., Guo X., Yan B., Xia Y., Peng M. Analysis of different strategies adapted by two cassava cultivars in response to drought stress: Ensuring survival or continuing growth. J. Exp. Bot. 2015;66:1477–1488. doi: 10.1093/jxb/eru507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong Z., Xiong L., Shi H., Yang S., Herrera-Estrella L.R., Xu G., Chao D.Y., Li J., Wang P.Y., Qin F., et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020;63:635–674. doi: 10.1007/s11427-020-1683-x. [DOI] [PubMed] [Google Scholar]
- 4.Basu S., Ramegowda V., Kumar A., Pereira A. Plant adaptation to drought stress. F1000Research. 2016;5:1554. doi: 10.12688/f1000research.7678.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong L., Schumaker K.S., Zhu J.K. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14:S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shinozaki K., Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 7.Waadt R., Seller C.A., Hsu P.K., Takahashi Y., Munemasa S., Schroeder J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022 doi: 10.1038/s41580-022-00479-6. online ahead of print . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaves M.M., Oliveira M.M. Mechanisms underlying plant resilience to water deficits: Prospects for water-saving agriculture. J. Exp. Bot. 2004;55:2365–2384. doi: 10.1093/jxb/erh269. [DOI] [PubMed] [Google Scholar]
- 9.Oyiga B.C., Palczak J., Wojciechowski T., Lynch J.P., Naz A.A., Léon J., Ballvora A. Genetic components of root architecture and anatomy adjustments to water-deficit stress in spring barley. Plant Cell Environ. 2020;43:692–711. doi: 10.1111/pce.13683. [DOI] [PubMed] [Google Scholar]
- 10.Xiong R., Liu S., Considine M.J., Siddique K.H.M., Lam H.M., Chen Y. Root system architecture, physiological and transcriptional traits of soybean (Glycine max L.) in response to water deficit: A review. Physiol. Plant. 2021;172:405–418. doi: 10.1111/ppl.13201. [DOI] [PubMed] [Google Scholar]
- 11.Li C., Li L., Reynolds M.P., Wang J., Chang X., Mao X., Jing R. Recognizing the hidden half in wheat: Root system attributes associated with drought tolerance. J. Exp. Bot. 2021;72:5117–5133. doi: 10.1093/jxb/erab124. [DOI] [PubMed] [Google Scholar]
- 12.Ober E.S., Alahmad S., Cockram J., Forestan C., Hickey L.T., Kant J., Maccaferri M., Marr E., Milner M., Pinto F., et al. Wheat root systems as a breeding target for climate resilience. Theor. Appl. Genet. 2021;134:1645–1662. doi: 10.1007/s00122-021-03819-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halder T., Liu H., Chen Y., Yan G., Siddique K.H.M. Identification of candidate genes for root traits using genotype-phenotype association analysis of near-isogenic lines in hexaploid Wheat (Triticum aestivum L.) Int. J. Mol. Sci. 2021;22:3579. doi: 10.3390/ijms22073579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma J., Zhao D., Tang X., Yuan M., Zhang D., Xu M., Duan Y., Ren H., Zeng Q., Wu J., et al. Genome-wide association study on root system architecture and identification of candidate genes in wheat (Triticum aestivum L.) Int. J. Mol. Sci. 2022;23:1843. doi: 10.3390/ijms23031843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X., Guo Z., Lv Y., Cen X., Ding X., Wu H., Li X., Huang J., Xiong L. Genetic control of the root system in rice under normal and drought stress conditions by genome-wide association study. PLoS Genet. 2017;13:e1006889. doi: 10.1371/journal.pgen.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comas L.H., Becker S.R., Cruz V.M., Byrne P.F., Dierig D.A. Root traits contributing to plant productivity under drought. Front. Plant Sci. 2013;4:442. doi: 10.3389/fpls.2013.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S., Begum N., An T., Zhao T., Xu B., Zhang S., Deng X., Lam H.M., Nguyen H.T., Siddique K.H.M., et al. Characterization of root system architecture traits in diverse soybean genotypes using a semi-hydroponic system. Plants. 2021;10:2781. doi: 10.3390/plants10122781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai G., Ahmed M.A., Abdalla M., Carminati A. Root hydraulic phenotypes impacting water uptake in drying soils. Plant Cell Environ. 2022;45:650–663. doi: 10.1111/pce.14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nibau C., Gibbs D.J., Coates J.C. Branching out in new directions: The control of root architecture by lateral root formation. New Phytol. 2008;179:595–614. doi: 10.1111/j.1469-8137.2008.02472.x. [DOI] [PubMed] [Google Scholar]
- 20.Petricka J.J., Winter C.M., Benfey P.N. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 2012;63:563–590. doi: 10.1146/annurev-arplant-042811-105501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavenus J., Goh T., Roberts I., Guyomarc’h S., Lucas M., De Smet I., Fukaki H., Beeckman T., Bennett M., Laplaze L. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends. Plant Sci. 2013;18:450–458. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Satbhai S.B., Ristova D., Busch W. Underground tuning: Quantitative regulation of root growth. J. Exp. Bot. 2015;66:1099–1112. doi: 10.1093/jxb/eru529. [DOI] [PubMed] [Google Scholar]
- 23.Waidmann S., Sarkel E., Kleine-Vehn J. Same same, but different: Growth responses of primary and lateral roots. J. Exp. Bot. 2020;71:2397–2411. doi: 10.1093/jxb/eraa027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Smet I., Signora L., Beeckman T., Inzé D., Foyer C.H., Zhang H. An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 2003;33:543–555. doi: 10.1046/j.1365-313X.2003.01652.x. [DOI] [PubMed] [Google Scholar]
- 25.Xiong L., Wang R.G., Mao G., Koczan J.M. Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic Acid. Plant Physiol. 2006;142:1065–1074. doi: 10.1104/pp.106.084632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deak K.I., Malamy J. Osmotic regulation of root system architecture. Plant J. 2005;43:17–28. doi: 10.1111/j.1365-313X.2005.02425.x. [DOI] [PubMed] [Google Scholar]
- 27.Scarpella E., Marcos D., Friml J., Berleth T. Control of leaf vascular patterning by polar auxin transport. Genes. Dev. 2006;20:1015–1027. doi: 10.1101/gad.1402406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garbers C., DeLong A., Deruere J., Bernasconi P., Soll D. A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 1996;15:2115–2124. doi: 10.1002/j.1460-2075.1996.tb00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., Guo H. On hormonal regulation of the dynamic apical hook development. New Phytol. 2019;222:1230–1234. doi: 10.1111/nph.15626. [DOI] [PubMed] [Google Scholar]
- 30.Beziat C., Kleine-Vehn J. The road to auxin-dependent growth repression and promotion in apical hooks. Curr. Biol. 2018;28:R519–R525. doi: 10.1016/j.cub.2018.01.069. [DOI] [PubMed] [Google Scholar]
- 31.Rashotte A.M., Brady S.R., Reed R.C., Ante S.J., Muday G.K. Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiol. 2000;122:481–490. doi: 10.1104/pp.122.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 33.Karcher D., Bock R. Identification of the chloroplast adenosine-to-inosine tRNA editing enzyme. RNA. 2009;15:1251–1257. doi: 10.1261/rna.1600609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delannoy E., Le Ret M., Faivre-Nitschke E., Estavillo G.M., Bergdoll M., Taylor N.L., Pogson B.J., Small I., Imbault P., Gualberto J.M. Arabidopsis tRNA adenosine deaminase arginine edits the wobble nucleotide of chloroplast tRNAArg(ACG) and is essential for efficient chloroplast translation. Plant Cell. 2009;21:2058–2071. doi: 10.1105/tpc.109.066654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee R.D., Cho H.T. Auxin, the organizer of the hormonal/environmental signals for root hair growth. Front. Plant Sci. 2013;4:448. doi: 10.3389/fpls.2013.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown D.E., Rashotte A.M., Murphy A.S., Normanly J., Tague B.W., Peer W.A., Taiz L., Muday G.K. Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 2001;126:524–535. doi: 10.1104/pp.126.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buer C.S., Muday G.K. The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell. 2004;16:1191–1205. doi: 10.1105/tpc.020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buer C.S., Imin N., Djordjevic M.A. Flavonoids: New roles for old molecules. J. Integ. Plant Biol. 2010;52:98–111. doi: 10.1111/j.1744-7909.2010.00905.x. [DOI] [PubMed] [Google Scholar]
- 39.Liu W., Feng Y., Yu S., Fan Z., Li X., Li J., Yin H. The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 2021;22:12824. doi: 10.3390/ijms222312824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito K., Yonekura-Sakakibara K., Nakabayashi R., Higashi Y., Yamazaki M., Tohge T., Fernie A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013;72:21–34. doi: 10.1016/j.plaphy.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Shirley B.W., Kubasek W.L., Storz G., Bruggemann E., Koornneef M., Ausubel F.M., Goodman H.M. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995;8:659–671. doi: 10.1046/j.1365-313X.1995.08050659.x. [DOI] [PubMed] [Google Scholar]
- 42.Peer W.A., Bandyopadhyay A., Blakeslee J.J., Makam S.N., Chen R.J., Masson P.H., Murphy A.S. Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell. 2004;16:1898–1911. doi: 10.1105/tpc.021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan G.A., Declerck M., Sorin C.L., Hartmann C., Crespi M., Lelandais-Brière C. MicroRNAs as regulators of root development and architecture. Plant Mol. Biol. 2011;77:47–58. doi: 10.1007/s11103-011-9793-x. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y., Ma Y., Zhao D., Tang Z., Zhang T., Zhang K., Dong J., Zhang H. Genetic regulation of lateral root development. Plant Signal Behav. 2022:2081397. doi: 10.1080/15592324.2022.2081397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Overvoorde P., Fukaki H., Beeckman T. Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2010;2:a001537. doi: 10.1101/cshperspect.a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H., Li Z., Xiong L. A plant microRNA regulates the adaptation of roots to drought stress. FEBS Lett. 2012;586:1742–1747. doi: 10.1016/j.febslet.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 47.Michniewicz M., Brewer P.B., Friml J. Polar auxin transport and asymmetric auxin distribution. Arab. Book. 2007;5:e0108. doi: 10.1199/tab.0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barbosa I.C.R., Hammes U.Z., Schwechheimer C. Activation and polarity control of PIN-FORMED auxin Transporters by phosphorylation. Trends. Plant Sci. 2018;23:523–538. doi: 10.1016/j.tplants.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Yonekura-Sakakibara K., Tohge T., Matsuda F., Nakabayashi R., Takayama H., Niida R., Watanabe-Takahashi A., Inoue E., Saito K. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell. 2008;20:2160–2176. doi: 10.1105/tpc.108.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor L.P., Grotewold E. Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 2005;8:317–323. doi: 10.1016/j.pbi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 51.Falcone Ferreyra M.L., Rius S., Casati P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012;3:222. doi: 10.3389/fpls.2012.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002;5:218–223. doi: 10.1016/S1369-5266(02)00256-X. [DOI] [PubMed] [Google Scholar]
- 53.Teale W., Palme K. Naphthylphthalamic acid and the mechanism of polar auxin transport. J. Exp. Bot. 2018;69:303–312. doi: 10.1093/jxb/erx323. [DOI] [PubMed] [Google Scholar]
- 54.Teale W.D., Pasternak T., Dal Bosco C., Dovzhenko A., Kratzat K., Bildl W., Schworer M., Falk T., Ruperti B., Schaefer J.V., et al. Flavonol-mediated stabilization of PIN efflux complexes regulates polar auxin transport. EMBO J. 2021;40:e104416. doi: 10.15252/embj.2020104416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abas L., Kolb M., Stadlmann J., Janacek D.P., Lukic K., Schwechheimer C., Sazanov L.A., Mach L., Friml J., Hammes U.Z. Naphthylphthalamic acid associates with and inhibits PIN auxin transporters. Proc. Natl. Acad. Sci. USA. 2021;118:e2020857118. doi: 10.1073/pnas.2020857118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Su N., Zhu A., Tao X., Ding Z.J., Chang S., Ye F., Zhang Y., Zhao C., Chen Q., Wang J., et al. Structures and mechanisms of the Arabidopsis auxin transporter PIN3. Nature. 2022 doi: 10.1038/s41586-022-05142-w. online ahead of print . [DOI] [PubMed] [Google Scholar]
- 57.Yang Z., Xia J., Hong J., Zhang C., Wei H., Ying W., Sun C., Sun L., Mao Y., Gao Y., et al. Structural insights into auxin recognition and efflux by Arabidopsis PIN1. Nature. 2022 doi: 10.1038/s41586-022-05143-9. online ahead of print . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strand D.D., Livingston A.K., Satoh-Cruz M., Koepke T., Enlow H.M., Fisher N., Froehlich J.E., Cruz J.A., Minhas D., Hixson K.K., et al. Defects in the expression of chloroplast proteins leads to H(2)O(2) accumulation and activation of cyclic electron flow around photosystem I. Front. Plant Sci. 2017;7:2073. doi: 10.3389/fpls.2016.02073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nott A., Jung H.S., Koussevitzky S., Chory J. Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- 60.Chen H., Zhang B., Hicks L.M., Xiong L. A nucleotide metabolite controls stress-responsive gene expression and plant development. PLoS ONE. 2011;6:e26661. doi: 10.1371/journal.pone.0026661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen H., Xiong L. The bifunctional abiotic stress signalling regulator and endogenous RNA silencing suppressor FIERY1 is required for lateral root formation. Plant Cell Environ. 2010;33:2180–2190. doi: 10.1111/j.1365-3040.2010.02218.x. [DOI] [PubMed] [Google Scholar]
- 62.Chen H., Xiong L. Genetic interaction of two abscisic acid signaling regulators, HY5 and FIERY1, in mediating lateral root formation. Plant Signal Behav. 2011;6:123–125. doi: 10.4161/psb.6.1.14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dong H., Bai L., Chang J., Song C.P. Chloroplast protein PLGG1 is involved in abscisic acid-regulated lateral root development and stomatal movement in Arabidopsis. Biochem. Biophys. Res. Commun. 2018;495:280–285. doi: 10.1016/j.bbrc.2017.10.113. [DOI] [PubMed] [Google Scholar]
- 64.Kirchsteiger K., Ferrández J., Pascual M.B., González M., Cejudo F.J. NADPH thioredoxin reductase C is localized in plastids of photosynthetic and nonphotosynthetic tissues and is involved in lateral root formation in Arabidopsis. Plant Cell. 2012;24:1534–1548. doi: 10.1105/tpc.111.092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hendry G.A.F., Grime J.P. Methods in Comparative Plant Ecology: A Laboratory Manual. Chapman & Hall; London, UK: New York, NY, USA: 1993. [Google Scholar]
- 66.Zhang J., Subramanian S., Zhang Y., Yu O. Flavone synthases from Medicago truncatula are flavanone-2-hydroxylases and are important for nodulation. Plant Physiol. 2007;144:741–751. doi: 10.1104/pp.106.095018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.