Abstract
In Klebsiella pneumoniae, NifA-dependent transcription of nitrogen fixation (nif) genes is inhibited by NifL in response to molecular oxygen and combined nitrogen. We recently showed that K. pneumoniae NifL is a flavoprotein, which apparently senses oxygen through a redox-sensitive, conformational change. We have now studied the oxygen regulation of NifL activity in Escherichia coli and K. pneumoniae strains by monitoring its inhibition of NifA-mediated expression of K. pneumoniae ø(nifH′-′lacZ) fusions in different genetic backgrounds. Strains of both organisms carrying fnr null mutations failed to release NifL inhibition of NifA transcriptional activity under oxygen limitation: nif induction was similar to the induction under aerobic conditions. When the transcriptional regulator Fnr was synthesized from a plasmid, it was able to complement, i.e., to relieve NifL inhibition in the fnr mutant backgrounds. Hence, Fnr appears to be involved, directly or indirectly, in NifL-dependent oxygen regulation of nif gene expression in K. pneumoniae. The data indicate that in the absence of Fnr, NifL apparently does not receive the signal for anaerobiosis. We therefore hypothesize that in the absence of oxygen, Fnr, as the primary oxygen sensor, activates transcription of a gene or genes whose product or products function to relieve NifL inhibition by reducing the flavin adenine dinucleotide cofactor under oxygen-limiting conditions.
In diazotrophic proteobacteria, transcription of the nitrogen fixation (nif) genes is mediated by the nif-specific activator protein NifA, a member of a family of activators that functions with ς54 (2, 4). Both the expression and the activity of NifA can be regulated in response to the oxygen and/or combined nitrogen status of the cells; the mechanisms of the regulation differ with the organism. In Klebsiella pneumoniae and Azotobacter vinelandii, NifA transcriptional activity is regulated by a second regulatory protein, NifL. This negative regulator of the nif genes inhibits the transcriptional activation by NifA in response to combined nitrogen and/or external molecular oxygen. The translationally coupled synthesis of the two regulatory proteins, immunological studies, complex analyses, and studies using the two-hybrid system in Saccharomyces cerivisiae imply that the inhibition of NifA activity by NifL apparently occurs via direct protein-protein interaction (5, 11, 21, 26). The mechanism by which nitrogen is sensed in K. pneumoniae and A. vinelandii is currently the subject of extensive studies. Very recently, He et al. (10), and Jack et al. (15) provided evidence that in K. pneumoniae, the second PII protein, GlnK, is required for relief of NifL inhibition under nitrogen-limiting conditions. This indicates that GlnK regulates NifL inhibition of NifA in response to the nitrogen status of the cells by interacting with NifL or NifA.
In both organisms, K. pneumoniae and A. vinelandii, the negative regulator NifL is a flavoprotein with an N-terminally bound flavin adenine dinucleotide (FAD) as a prosthetic group (13, 19, 31). In vitro, the oxidized form of NifL inhibits NifA activity, whereas reduction of the FAD cofactor relieves NifL inhibition (13, 22). This indicates that NifL apparently acts as a redox switch in response to the environmental oxygen status and allows NifA activity, only under oxygen-limiting conditions. We recently showed that in vivo, the presence of iron is required to relieve inhibitory effects of NifL on transcriptional activation by NifA and, additionally, that iron is not present in NifL (31, 32). Therefore, we have postulated that an unidentified iron-containing protein may be the physiological reductant for NifL. This putative iron-containing protein is apparently not nif specific, since NifL function is regulated normally in response to cellular nitrogen and oxygen availability in Escherichia coli in the absence of nif proteins other than NifA (9).
The key question concerning the oxygen signal transduction in K. pneumoniae is whether NifL senses oxygen directly via a redox-induced conformational change, or whether oxygen is detected by a more general oxygen-sensing system, which then regulates NifL by inducing the oxidation or reduction of the flavin cofactor. One candidate for a general oxygen sensor is the transcriptional fumarate nitrate reductase regulator (Fnr) (35, 36), which in the case of E. coli Fnr, senses oxygen via an oxygen-labile iron-sulfur ([4Fe-4S]+2) cluster and is involved in signal transduction of the cellular redox state (7, 18, 25, 37). Recently we cloned and sequenced the fnr gene of K. pneumoniae and characterized the protein (6). Because the K. pneumoniae Fnr amino acid sequence is 98% identical to the E. coli Fnr and contains an iron-sulfur cluster, we have now tested the hypothesis that Fnr transduces the oxygen signal to NifL. We present evidence that in the absence of Fnr, NifL inhibits NifA activity under oxygen limitation, suggesting that Fnr is required for relief of NifL inhibition in K. pneumoniae under anaerobic conditions.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this work are listed in Table 1. Plasmid DNA was transformed into E. coli cells according to the method of Inoue et al. (14) and into K. pneumoniae cells by electroporation. Transduction by phage P1 was performed as described previously (33).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| NCM1529 | araD139Δ(argF-lacU)169 fthD5301 gyrA219 non-9 rpsL150 ptsF25 relA1 deoC1 trpDC700putPA1303::[Kanr-(nifH′-′lacZ)] (wild type) | 10 |
| NCM1528 | NCM1529/pNH3 | 10 |
| NCM1527 | NCM1529/pJES851 | 10 |
| RAS1 | NCM1529, but fnr::Tn10 | 6 |
| RAS2 | NCM1529/pRS107 | This study |
| RAS6 | RAS1/pRS107 | This study |
| RAS7 | RAS1/pNH3 | This study |
| RAS8 | RAS1/pJES851 | This study |
| RAS9 | RAS1/pNH3 and pRS79 | This study |
| RAS10 | RAS1/pNH3 and pRS120 | This study |
| RAS11 | RAS1/pNH3 and pMCL210 | This study |
| RAS12 | RAS1/pNH3 and pACYC184 | This study |
| RM101 | MC4100, but Δfnr | 30 |
| RAS13 | RM101, but [Kanr-(nifH′-′lacZ)] | This study |
| RAS21 | MC4100, but [Kanr-(nifH′-′lacZ)] | This study |
| RAS22 | RAS21/pNH3 | This study |
| RAS23 | RAS21/pJES851 | This study |
| RAS24 | RAS21/pRS107 | This study |
| RAS14 | RAS13/pNH3 | This study |
| EAS15 | RAS13/pJES851 | This study |
| RAS25 | RAS13/pRS107 | This study |
| RAS16 | RAS13/pNH3 and pRS120 | This study |
| RAS17 | RAS13/pNH3 and pACYC184 | This study |
| K. pneumoniae | ||
| M5al | Wild type | |
| UN4495 | φ(nifK-lacZ)5935 Δlac-4001 his D4226 Galr | 23 |
| RAS18 | φ(nifK-lacZ)5935 Δlac-4001 his D4226 Galrfnr::Ω | This study |
| RAS19 | RAS18/pRS137 | This study |
| RAS20 | RAS18/pACYC184 | This study |
| RAS26 | UN4495/pRS159 | This study |
| RAS27 | RAS18/pRS159 | This study |
| RAS28 | UN4495/pJES839 | 9 |
| RAS29 | RAS18/pJES839 | This study |
| RAS30 | UN4995Δ(nifLA)6293::Km/pJES839 | 32; this study |
| Plasmids | ||
| pNH3 | K. pneumoniae nifLA controlled by the tac promoter | 11 |
| pJES839 | pNH3, but additional tetracycline resistance cassette | 32 |
| pJES851 | K. pneumoniae nifA controlled by the tac promoter | 32 |
| pRS79 | E. coli fnr controlled by the lac promoter on pMCL210 | This study |
| pRS107 | K. pneumoniae nifLC184S/C187SnifA controlled by the tac promoter | This study |
| pRS159 | K. pneumoniae nifLC184S/C187SnifA controlled by the tac promoter | This study |
| pRS120 | E. coli fnr controlled by the tet promoter on pACYC184 | 6 |
| pRS127 | 2.1-kbp fragment in pBluescript SK+containing K. pneumoniae fnr | 6 |
| pRS137 | K. pneumoniae fnr controlled by the tet promoter on pACYC184 | 6 |
| pACYC184 | Low-copy vector | New England Biolabs |
| pMCL210 | Low-copy vector | 27 |
| pBluescript SK+ | Cloning vector | Stratagene, La Jolla, Calif. |
(i) E. coli strains.
E. coli NCM1529, which contains a ø(nifH′-′lacZ) fusion (9), and derivatives of NCM1529 were chosen to study NifA/NifL regulation in E. coli. The fnr::Tn10 allele was transferred from the fnr::Tn10 derivative of M182 (16) into NCM1529 by P1-mediated transduction with selection for tetracycline resistance, resulting in RAS1 (6). Strains RAS6, RAS7, RAS8, RAS9, RAS10, RAS11, and RAS12 contain plasmids pRS107, pNH3, pJES851, pNH3 plus pRS79, pNH3 plus pRS120, pNH3 plus pMCL210, and pNH3 plus pACYC184, respectively, in RAS1. To construct an independent second fnr null mutant, the [Kanr-(nifH′-′lacZ)] allele was transferred from strain NCM1529 by P1-mediated transduction into the independent fnr mutant strain RM101 (30) and into the parental strain MC4100 with selection for kanamycin resistance, resulting in RAS13 and RAS21, respectively. Strains RAS25, RAS14, RAS15, RAS16 and RAS17 contain plasmids pRS107, pNH3, pJES851, pNH3 plus pRS120, and pNH3 plus pACYC184, respectively, in RAS13.
(ii) Klebsiella strains.
K. pneumoniae strains M5al (wild type) and UN4495 [ø(nifK-lacZ)5935 Δlac-4001 his D4226 Galr] (23) were provided by Gary Roberts.
Construction of an fnr::Ω mutation.
Strain RAS18 was obtained by insertion of a kanamycin resistance cassette (28) into the fnr gene of K. pneumoniae UN4495 as detailed in the following steps. (i) The 2.1-kbp EcoRI-BamHI fragment, which carries the ogt-fnr-ydaA′- region of K. pneumoniae, was subcloned into pBluescript SK+ to produce pRS127. (ii) A 2.1-kb HindIII cassette containing an Ω interposon fragment with a kanamycin resistance gene derived from plasmid pHP45Ω (28) was cloned into the HindIII site of fnr in pRS127 to yield plasmid pRS142. (iii) A 2.9-kb PCR fragment carrying fnr::Ω was generated with pRS142 as a template and a set of primers which were homologues to the fnr flanking 5′ and 3′ regions, with additional BamHI synthetic restriction recognition sites (underlined) (5′ATATCAATGGATCCCTGAGCAGACTTATGATCC3′, sense primer; 5′CTTATATGGATCCAATGAAACAGGGGAGGA3′, antisense primer). The 2.9-kb PCR product was cloned into the BamHI site of the sacB-containing vector pKNG101 (17), creating plasmid pRS144. The correct insertion was analyzed by sequencing. (iv) pRS144 was transformed into K. pneumoniae UN4495, and recombinant strains (generated by means of a double crossover) were identified by the ability to grow on Luria-Bertani (LB) medium supplemented with 5% sucrose and resistance to kanamycin. The fnr::Ω mutation in strain RAS18 was confirmed by Southern blot analysis (29) and by PCR.
Strains RAS26 and RAS28 contain pRS159 and pJES839, respectively, in K. pneumoniae UN4495 and strains RAS19, RAS27 and RAS29 contain pRS137, pRS159 and pJES839, respectively, in RAS18.
(iii) Construction of plasmids.
Plasmid pRS107 contains the K. pneumoniae nifLC184S/C187SnifA operon under the control of the tac promoter, in which the Cys184 and Cys187 of nifL are changed to serine (Ser184-Ala-Asp-Ser187). It was constructed from pNH3 (11) by introducing the double mutation into nifL by site-directed mutagenesis. Site-directed mutagenesis was performed with the GeneEditor System (Promega) according to the protocol of the manufacturer. The double mutation was confirmed by sequencing. Plasmid pRS159 was constructed by inserting a tetracycline resistance cassette (32) into the ScaI site of plasmid pRS107. Plasmid pRS79 contains the E. coli fnr gene inserted into the BamHI-PstI site of pMCL210 (27) under the control of the lac promoter. pRS120 and pRS137 contain the E. coli fnr gene and K. pneumoniae fnr gene, respectively, inserted into the SalI-BamHI site of pACYC184 and thereby expressed from the tet promoter (6).
Growth.
K. pneumoniae and E. coli strains were grown under anaerobic conditions with N2 as the gas phase at 30°C in minimal medium (32) supplemented with 4 mM glutamine, 10 mM Na2CO3, 0.3 mM sulfide and 0.002% resazurin to monitor anaerobiosis. The medium was further supplemented with 0.004% histidine and with 0.4% sucrose as the sole carbon source for K. pneumoniae strains. For E. coli strains, the medium was supplemented with 0.1 mM tryptophan and 0.8% glucose as the carbon source. Precultures were grown overnight in closed bottles with N2 as the gas phase, in medium lacking sulfide and resazurin, but supplemented with 4 mM ammonium acetate in addition to glutamine; both ammonium and glutamine were completely utilized during growth of precultures. The cultures (25 ml) were grown in closed bottles with N2 as the gas phase at 30°C under strictly anaerobic conditions without shaking. Samples for monitoring growth at 600 nm and determining β-galactosidase activity were taken anaerobically. In E. coli strains carrying a plasmid encoding NifL and NifA (pNH3) (11) or NifLC184S/C187S and NifA (pRS107) or a plasmid encoding NifA alone (pJES851) (32), expression of nifLA, nifLC184S/C187SnifA, or nifA was induced from the tac promoter with 10 μM IPTG (isopropyl-β-d-thiogalactopyranoside).
Fnr phenotypes of RAS1, RAS13, and RAS18 and the respective complemented strains RAS9, RAS10, RAS16, and RAS19 were tested anaerobically by using glycerol and nitrate (0.5%) as the sole carbon and nitrogen sources, respectively, in minimal medium.
β-Galactosidase assay.
NifA-mediated activation of transcription from the nifHDK promoter in K. pneumoniae UN4495 and E. coli strains was monitored by measuring the differential rate of β-galactosidase synthesis during exponential growth (units per milliliter per optical density unit at 600 nm [OD600]) (32). Inhibitory effects of NifL on NifA activity were assessed by virtue of a decrease in nifH expression.
Western blot analysis.
Cells were grown anaerobically in minimal medium with glutamine as the nitrogen source; when the culture reached a turbidity of 0.4 to 0.7 at 660 nm, 1-ml samples of the exponentially growing cultures were harvested and concentrated 20-fold into sodium dodecyl sulfate (SDS) gel loading buffer (20). Samples were separated by SDS-polyacrylamide (12%) gel electrophoresis and transferred to nitrocellulose membranes as described previously (29). Membranes were exposed to polyclonal rabbit antisera directed against the NifL or NifA proteins of K. pneumoniae, and protein bands were detected with secondary antibodies directed against rabbit immunoglobulin G and coupled to horseradish peroxidase (Bio-Rad Laboratories). Purified NifA and NifL from K. pneumoniae and prestained protein markers (New England Biolabs, Frankfurt, Germany) were used as standards.
Nucleotide sequence accession number.
The sequence of K. pneumoniae fnr has been submitted to GenBank under accession no. AF220669.
RESULTS
We recently showed that in vivo iron is specifically required for nif induction in K. pneumoniae, and additionally, that iron is not present in NifL (31, 32). In order to examine whether oxygen is detected by a more general system rather than by NifL directly, we chose to examine the possible influence of Fnr on the nif induction in a heterologous E. coli system. We performed all experiments under nitrogen-limiting growth conditions to exclude NifA inhibition by NifL in response to the presence of ammonium. If Fnr is indeed the primary oxygen sensor, which transduces the oxygen signal to NifL, the iron requirement for the nif induction under oxygen-limiting conditions may be based on the iron requirement for the assembly of iron sulfur clusters of Fnr.
Studying the effect of Fnr on the nif induction in a heterologous E. coli system.
In order to study the effect of Fnr on nif regulation in response to oxygen, we chose a heterologous E. coli system. Strain NCM1529 carrying a chromosomal nifH′-′lacZ fusion was used as parental strain (9). NifL and NifA were induced independent of the Ntr system from plasmids which carried the K. pneumoniae nifLA (pNH3) and nifA (pJES851) genes under the control of the tac promoter. The two regulatory proteins were induced with 10 μM IPTG to levels at which NifL function is regulated normally in response to oxygen and combined nitrogen in E. coli in the absence of nif proteins other than NifA (9). To study the effect of an fnr null mutation on the regulation of NifL activity in response to oxygen, an fnr null allele (fnr::Tn10) was introduced by P1 transduction into the parental strain NCM1529 carrying the ø(nifH′-′lacZ) fusion as described in Materials and Methods, resulting in strain RAS1. After introducing nifLA and nifA on plasmids, the resulting strains were generally grown in mineral medium with glucose as the sole carbon source and under nitrogen limitation to exclude NifA inhibition by NifL in response to combined nitrogen. Determination of the doubling times of the different strains under anaerobic and aerobic conditions revealed no significant difference in growth rates for fnr mutant strains compared to the respective parental strains (Table 2). NifA-mediated activation of transcription from the nifH′ promoter in the different backgrounds was monitored by determining the differential rate of β-galactosidase synthesis during exponential growth. Inhibitory effects of NifL on NifA activity in strain RAS7 carrying the fnr null allele and carrying nifLA on a plasmid are detectable; they result in a decrease in nifH expression. Interestingly, under oxygen-limiting conditions, strain RAS7 showed a β-galactosidase synthesis rate from the nifH′ promoter of only 100 ± 10 U/ml/OD600 unit when nifLA was induced with 10 μM IPTG. This is in the range of the synthesis rate under aerobic conditions in the parental strain NCM1528 (60 ± 5 U/ml/OD600 unit) and equivalent to 3% of the synthesis rate under anaerobic conditions in NCM1528 (3,000 ± 100 U/ml/OD600 unit) (Table 2).
TABLE 2.
Effects of an fnr null allele on activity of the K. pneumoniae NifL protein in different E. coli backgrounds
| Strain | Relevant genotypea | Presence of oxygen | Expression of nifH′-′lacZ (U/ml/OD600 U)b | Doubling time (h) |
|---|---|---|---|---|
| NCM1528 | Wild type/Ptac-nifLA | − | 3,000 ± 100 | 5.0 |
| NCM1528 | Wild type/Ptac-nifLA | + | 60 ± 5 | 2.0 |
| NCM1527 | Wild type/Ptac-nifA | − | 5,300 ± 200 | 4.8 |
| NCM1527 | Wild type/Ptac-nifA | + | 5,118c | 2.1 |
| RAS2 | Wild type/Ptac-nifL mutant nifA | − | 2,950 ± 120 | 5.2 |
| RAS2 | Wild type/Ptac-nifL mutant nifA | + | 2,900 ± 50 | 2.0 |
| RAS8d | fnr mutant/Ptac-nifA | − | 4,800 ± 100 | 4.9 |
| RAS8d | fnr mutant/Ptac-nifA | + | 5,200 ± 200 | 2.2 |
| RAS6d | fnr mutant/Ptac-nifL mutant nifA | − | 2,800 ± 100 | 5.0 |
| RAS6d | fnr mutant/Ptac-nifL mutant nifA | + | 3,000 ± 200 | 2.0 |
| RAS7d | fnr mutant/Ptac-nifLA | − | 100 ± 10 | 5.0 |
| RAS7d | fnr mutant/Ptac-nifLA | + | 30 ± 3 | 2.0 |
| RAS9d | fnr mutant/Ptac-nifLA/Plac fnr | − | 3,000 ± 100 | 5.2 |
| RAS10d | fnr mutant/Ptac-nifLA/Ptet fnr | − | 2,870 ± 70 | 5.2 |
| RAS11d | fnr mutant/Ptac-nifLA/pMCL210 | − | 66 ± 5 | 5.5 |
| RAS12d | fnr mutant/Ptac-nifLA/pACYC184 | − | 70 ± 6 | 5.5 |
| RAS22 | Wild type/Ptac-nifLA | − | 3,500 ± 80 | 5.0 |
| RAS22 | Wild type/Ptac-nifLA | + | 70 ± 5 | 2.2 |
| RAS23 | Wild type/Ptac-nifA | − | 5,900 ± 250 | 5.1 |
| RAS23 | Wild type/Ptac-nifA | + | 5,725 ± 150 | 2.2 |
| RAS24 | Wild type/Ptac-nifL mutant nifA | − | 3,400 ± 200 | 4.9 |
| RAS24 | Wild type/Ptac-nifL mutant nifA | + | 2,800 ± 150 | 2.1 |
| RAS15e | fnr mutant/Ptac-nifA | − | 5,300 ± 200 | 5.6 |
| RAS15e | fnr mutant/Ptac-nifA | + | 5,130 ± 150 | 2.1 |
| RAS25e | fnr mutant/Ptac-nifL mutant nifA | − | 3,200 ± 200 | 5.0 |
| RAS25e | fnr mutant/Ptac-nifL mutant nifA | + | 3,400 ± 100 | 2.2 |
| RAS14e | fnr mutant/Ptac-nifLA | − | 160 ± 10 | 5.3 |
| RAS14e | fnr mutant/Ptac-nifLA | + | 40 ± 5 | 2.0 |
| RAS16e | fnr mutant/Ptac-nifLA/Ptet-fnr | − | 3,200 ± 100 | 5.2 |
| RAS17e | fnr mutant/Ptac-nifLA/pACYC184 | − | 190 ± 10 | 5.4 |
nifL mutant nifA, nifLC184S/C187SnifA (see Materials and Methods); Plac, Ptac, or Ptet, under the control of the lac, tac, or tet promoter, respectively.
Data are presented as mean values (± standard errors) of three independent experiments.
Determined by He et al. (9).
Strain contains the fnr null allele from M182 (fnr::Tn10) (16).
Strain contains the fnr null allele from RM101 (30).
In the case of NifA synthesis in the fnr mutant strain in the absence of NifL (RAS8), however, the β-galactosidase synthesis rate under anaerobic conditions was not significantly altered compared to the parental strain NCM1527 (4,800 ± 100 and 5,300 ± 200 U/ml/OD600 unit, respectively) and was not affected by oxygen (Table 2). This indicates that the observed Fnr effect is mediated by NifL towards NifA in RAS7. However, nif expression under anaerobic conditions by NifA induced from the tac promoter in the absence of NifL synthesis by using pJES851 (NCM1527) is significantly higher than that with plasmid pNH3 (NCM1528), in which NifA expression depends on NifL synthesis based on translational coupling in the nifLA operon (5). In addition, Western blot analysis showed that under our experimental conditions, the amounts of NifA synthesized in NCM1527 were approximately 30 to 40% higher than those synthesized in NCM1528 (data not shown). To rule out that nif expression in the fnr mutant using pJES851 (RAS8) is not due to this increase in NifA expression, we additionally constructed pRS107 containing nifLC184S/C187SnifA translationally coupled under the control of the tac promoter (see Materials and Methods). IPTG induction in NCM1529 containing pRS107 (RAS2) resulted in NifA expression comparable to that in NCM1528 (data not shown) and expression of NifLC184S/C187S, which completely lost its nitrogen and oxygen regulatory function (K. Klopprogge and R. A. Schmitz, unpublished observations). Determination of β-galactosidase synthesis rates showed that nif induction by NifA expressed from pRS107 in the absence of a functional NifL protein was again not affected by the fnr mutation (compare RAS2 with RAS6) and was in the range of nif induction in NCM1528 under anaerobic conditions (Table 2). These findings indicate that the fnr null allele does not affect NifA activity directly in the absence of functional NifL. In the presence of both regulatory proteins, however, NifL inhibits NifA activity under oxygen-limiting conditions when Fnr is absent, suggesting that the Fnr effect is mediated through NifL to NifA.
The finding that in the absence of Fnr NifL inhibits NifA activity under oxygen-limiting conditions to the same amount as under aerobic growth conditions indicates that NifL apparently does not receive the signal of anaerobiosis when Fnr is absent. To confirm this observation, we analyzed the nif induction under anaerobic conditions in a different fnr mutant strain (RAS13). After introduction of nifLA, nifA, and nifLC184S/C187SnifA on plasmids, the respective strains RAS14, RAS15, and RAS25 were grown under oxygen limitation. By determining the β-galactosidase synthesis rates from the nifH′ promoter in RAS14, we observed that in this independent fnr mutant strain, the nif induction was 160 ± 10 U/ml/OD600 unit, when nifLA was expressed under anaerobic conditions. This nif induction is again significantly lower than in the parental strain RAS22 (3,500 ± 80 U/ml/OD600 unit) and is in the range of aerobic nif induction in the parental strain (70 ± 5 U/ml/OD600 unit) (Table 2). Similar to RAS8 and RAS6, the β-galactosidase synthesis rate in the case of NifA synthesis in the absence of a functional NifL protein was not affected by the fnr mutation (RAS15 compared to RAS23 and RAS25 compared to RAS24).
The fnr null alleles do not affect the synthesis of NifL and NifA.
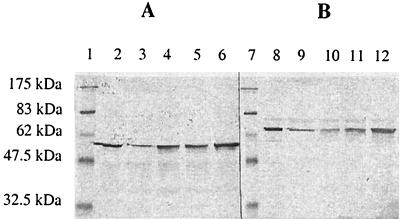
To demonstrate that the failure of the fnr mutant strains to express nifH under anaerobic conditions could not be accounted for by a decreased amount of NifA protein, we determined the amounts of NifA and NifL protein in the wild-type and fnr mutant strains by immunological means. As shown in Fig. 1, we observed no obvious differences in the amounts of the regulatory proteins of K. pneumoniae in the different fnr mutant backgrounds compared to those in the parental strains.
FIG. 1.
Amounts of NifA and NifL in wild-type and fnr mutant strains of E. coli. Cultures were grown at 30°C in minimal medium under anaerobic conditions with 4 mM glutamine as a limiting nitrogen source. The strains carried K. pneumoniae NifL and NifA under the control of the tac promoter on pNH3. Expression of NifL and NifA was induced with 10 μM IPTG in the wild-type strain (lanes 2 and 8), in fnr null allele strains RAS7 (lanes 3 and 9) and RAS14 (lanes 5 and 11), and in complemented strains RAS10 (lanes 4 and 10) and RAS16 (lanes 6 and 12). The amounts of NifL (A) and NifA (B) were determined by Western blotting. Prestained broad-range protein markers (lanes 1 and 7) were purchased from New England Biolabs.
Fnr is required for release of NifL inhibition of NifA activity under anaerobic conditions in the heterologous E. coli system.
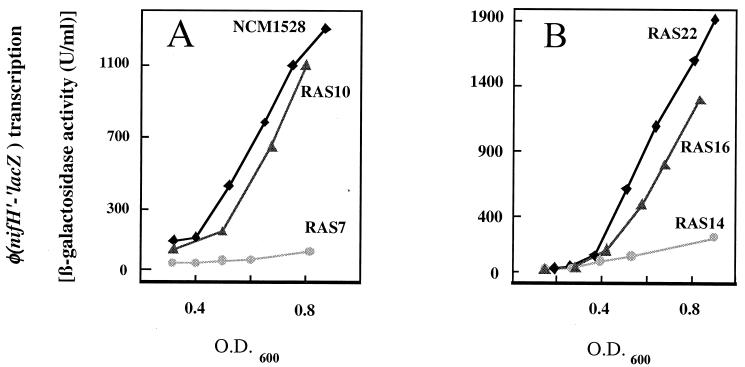
To determine if constitutive expression of fnr is able to restore nif induction in the fnr mutant strains, we expressed E. coli fnr from the tet promoter (pRS120) or the lac promoter (pRS79) in addition to the nifLA operon. Expression of Fnr in trans from either promoter resulted in complementation with a restoration of anaerobic growth on nitrate and glycerol (data not shown). It further resulted in relief of NifL inhibition of NifA activity under oxygen-limiting conditions. This restoration of nif induction was achieved in both strains carrying independent chromosomal fnr null alleles (RAS10 and RAS16, respectively) and is displayed graphically in Fig. 2. The nif induction under anaerobic conditions in both mutant strains was restored to the induction level of the parental strains (NCM1528 and RAS22, respectively) by expressing E. coli fnr from promoter Ptet on pACYC184 or promoter Plac on pMCL210, whereas the vectors pACYC184 and pMCL210 alone did not restore nif induction (Table 2). These results and the finding that Fnr affects NifA only in the presence of NifL (see above) strongly indicate that in the heterologous E. coli system, Fnr is required for release of NifL inhibition of NifA activity under anaerobic conditions.
FIG. 2.
Effects of fnr null alleles on expression of a ø(nifH′-′lacZ) fusion in heterologous E. coli strains carrying K. pneumoniae nifLA on a plasmid. The activity of β-galactosidase was plotted as a function of OD600 for cultures grown at 30°C in minimal medium under anaerobic conditions with 4 mM glutamine as a limiting nitrogen source. Differential rates of transcription from the nifH promoter, which reflect NifA activity, were determined from the slopes of these plots. All strains carried a single copy of a ø(nifH′-′lacZ) fusion at the trp locus (9) and plasmid pNH3 encoding NifL and NifA under the control of the tac promoter. (A) fnr null allele transduced from M182 (fnr::Tn10): wild-type NCM1528 (diamonds), the respective fnr null allele in NCM1528 (RAS7) (circles), and the complemented respective fnr mutant by constitutive expression of E. coli fnr on pACYC184 (RAS10) (triangles) are shown. (B) fnr null allele from RM101: wild-type RAS22 (diamonds), the respective fnr null allele in RAS22 (RAS14) (circles), and the complemented respective fnr mutant by constitutive expression of E. coli fnr on pACYC184 (RAS16) (triangles) are shown.
The wild-type strain (NCM1528) grown in the presence of 10 mM ammonium showed nif inductions of approximately 3 ± 1 U/ml/OD600 unit independent of oxygen availability (data not shown). This induction level is significantly lower than the nif induction observed in the fnr mutant strains (RAS7 and RAS14) under oxygen- and nitrogen-limiting growth conditions (100 ± 10 and 160 ± 10 U/ml/OD600 unit, respectively). These data suggest that Fnr is required for the oxygen signal transduction to NifL rather than for the ammonium signal transduction. They further indicate that in the absence of Fnr, NifL apparently does not receive the signal for absence of oxygen and therefore inhibits NifA activity under anaerobic conditions.
Studying the effect of Fnr on the nif induction in K. pneumoniae
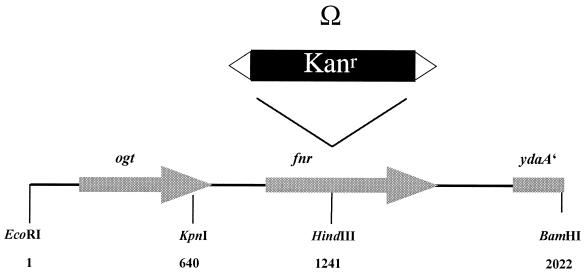
In order to confirm the requirement of Fnr for relief of NifL inhibition under anaerobic conditions in the heterologous E. coli system, we constructed a chromosomal fnr null allele in K. pneumoniae. We used K. pneumoniae strain UN4495 carrying nifLA and a nifK-lacZ fusion on the chromosome, which allows monitoring of NifA-mediated transcription from the nifHDK promoter by measuring the differential rate of β-galactosidase synthesis (32). The fnr deletion was constructed on a plasmid by inserting an Ω interposon fragment with a kanamycin resistance gene into K. pneumoniae fnr (Fig. 3), which was then introduced into the chromosome by marker exchange using the sac system (see Materials and Methods). The disruption of the fnr gene was confirmed by PCR and Southern blot analysis (data not shown).
FIG. 3.
Map of the cloned EcoRI-BamHI fragment (pRS127) showing the site of insertion of the Ω interposon fragment with a kanamycin resistance gene derived from plasmid pHP45Ω (28) in K. pneumoniae fnr. The Ω interposon fragment is flanked by short inverted repeats, including strong transcription termination signals. The sequence of the EcoRI-BamHI fragment has been submitted to GenBank under accession no. F220669.
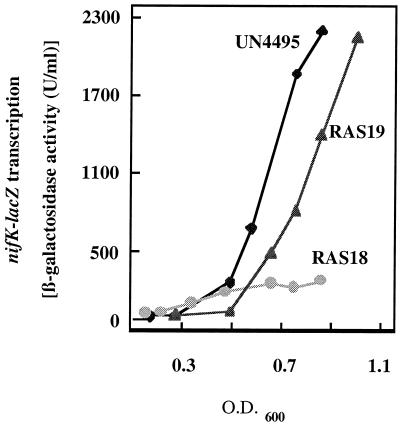
Klebsiella strains with the exception of RAS26 and RAS27 were generally grown in minimal medium under nitrogen limitation to exclude NifA inhibition by NifL in response to ammonium. The fnr::Ω mutation in K. pneumoniae UN4495 did not result in a significant growth rate reduction, but did reduce the nif induction under oxygen-limiting conditions to 10% of the nif induction in the parental strain. The observed induction level of the K. pneumoniae fnr mutant strain (RAS18) under anaerobic conditions (400 ± 20 U/ml/OD600 unit) again is in the same range as the nif induction in the presence of oxygen in the parental K. pneumoniae strain (220 ± 20 U/ml/OD600 unit) (Table 3). Determination of NifA and NifL proteins in the fnr mutant strain revealed no differences in the amount of the regulatory proteins compared to those of the parental strain (data not shown), indicating that the failure to express nifH could not be accounted for by a decrease in NifA expression. Normal NifL/NifA-dependent regulation was restored by introduction of the K. pneumoniae fnr gene expressed from the tet promoter on pRS137 into the fnr mutant (Fig. 4). nif induction in the complemented mutant (RAS19) was determined to be 3,800 ± 50 U/ml/OD600 unit, whereas the low-copy vector pACYC184 alone did not result in complementation (RAS20). These findings in the native background again suggest that Fnr is required for nif expression in K. pneumoniae under anaerobic conditions.
TABLE 3.
Effects of an fnr::Ω mutation on NifL activity in K. pneumoniae UN4495
| Strain | Relevant genotypea | Nitrogen source | Presence of oxygen | Expression of nifH′-′lacZ (U/ml/OD600 U)b | Doubling time (h) |
|---|---|---|---|---|---|
| UN 4495 | Wild type | Glutamine | − | 4,400 ± 100 | 3.5 |
| UN 4495 | Wild type | Glutamine | + | 220 ± 10 | 2.0 |
| RAS18 | fnr mutant | Glutamine | − | 400 ± 20 | 4.0 |
| RAS18 | fnr mutant | Glutamine | + | 100 ± 10 | 2.2 |
| RAS19 | fnr mutant/Ptet-fnrc | Glutamine | − | 3,800 ± 50 | 3.8 |
| RAS20 | fnr mutant/pACYC184 | Glutamine | − | 660 ± 30 | 4.2 |
| RAS26 | Wild type/Ptac-nifL mutant nifA | Ammoniumd | − | 2,350 ± 100 | 3.7 |
| RAS26 | Wild type/Ptac-nifL mutant nifA | Ammoniumd | + | 2,100 ± 100 | 1.7 |
| RAS27 | fnr mutant/Ptac-nifL mutant nifA | Ammoniumd | − | 2,200 ± 50 | 4.1 |
| RAS27 | fnr mutant/Ptac-nifL mutant nifA | Ammoniumd | + | 2,150 ± 150 | 1.6 |
| RAS28 | Wild type/Ptac-nifLA | Glutamine | − | 2,400 ± 30 | 4.0 |
| RAS28 | Wild type/Ptac-nifLA | Glutamine | + | 160 ± 5 | 1.6 |
| RAS29 | fnr mutant/Ptac-nifLA | Glutamine | − | 430 ± 30 | 3.6 |
| RAS29 | fnr mutant/Ptac-nifLA | Glutamine | + | 310 ± 30 | 1.6 |
| RAS30 | 4495ΔnifLA/Ptac-nifLA | Glutamine | − | 2,450 ± 30 | 4.1 |
nifL mutant nifA, nifLC184S/C187S nifA (see Materials and Methods); Ptac, under the control of the tac promoter.
Data are presented as mean values (± standard errors) of three independent experiments.
K. pneumoniae fnr is expressed under the control of the tet promoter (Ptet).
Grown in the presence of 10 mM ammonium to repress chromosomal nifLA induction.
FIG. 4.
Effects of an fnr null allele on expression of an nifK-lacZ fusion in K. pneumoniae strain UN4495. The activity of β-galactosidase was plotted as a function of the OD600 for cultures grown at 30°C in minimal medium under anaerobic conditions with 4 mM glutamine as a limiting nitrogen source. Differential rates of transcription from the nifHDK promoter were determined from the slopes of these plots. Wild-type UN4495 (diamonds), the fnr mutant strain of UN4495 (RAS18) (circles), and the complemented respective fnr mutant by constitutive expression of K. pneumoniae fnr on pACYC184 (RAS19) (triangles) are shown.
In order to confirm our finding in the heterologous E. coli system that Fnr is required to relieve NifL inhibition of NifA activity under anaerobic conditions, we studied the effect of the fnr null allele on NifA in Klebsiella. Plasmid pRS159 carrying nifLC184S/C187SnifA translationally coupled under the control of the tac promoter was introduced into K. pneumoniae UN4495 and the corresponding fnr mutant strain RAS18. Because growth in minimal medium in the presence of 10 mM ammonium results in repression of the chromosomal nifLA operon, under nitrogen sufficiency, only nifLC184S/C187SnifA from pRS159 is induced, resulting in the synthesis of NifA and a nonfunctional NifL protein (see above). Determination of β-galactosidase synthesis rates under those conditions in the fnr mutant strain (RAS27) and the parental strain (RAS26) showed that the absence of Fnr under anaerobic conditions does not affect NifA activity in the absence of a functional NifL protein (2,200 ± 50 and 2,350 ± 100 U/ml/OD600 unit, respectively) (Table 3). These results indicate that the Fnr effect on nif regulation observed in the native background is based on the Fnr requirement for relief of NifL inhibition under oxygen-limiting growth conditions. Based on our findings, we hypothesize that in K. pneumoniae, Fnr is the primary oxygen sensor for the nif regulation, which transduces the signal directly or indirectly to NifL.
DISCUSSION
Our goal is to determine how K. pneumoniae NifL perceives the oxygen status of the cells in order to regulate NifA activity in response to environmental oxygen. The main question concerning the oxygen signal transduction is whether NifL senses oxygen directly via a redox-induced conformational change, or whether oxygen is detected by a more general system. After receiving the oxygen signal, directly or indirectly, the redox state of the flavoprotein NifL is thought to influence the ability of NifL to modulate the NifA activity in response to environmental oxygen and to allow NifA activity only in the absence of oxygen (13, 22, 31). We recently showed that iron is specifically required for nif induction, but is not present in NifL (31, 32). To determine whether this iron requirement for nif induction could be accounted for by the role of Fnr in transducing the oxygen signal to NifL, we determined the effect of an fnr null allele on nif regulation. Using different genetic backgrounds and independent fnr null alleles, we were able to show that the absence of Fnr affects the nif regulation dramatically. The nif induction in the absence of Fnr was low, similar to the nif induction under aerobic conditions, even though cells were growing under oxygen limitation. Normal nif regulation was achieved in the mutant strains by introduction of a low-copy vector expressing fnr constitutively (Fig. 2 and 4). These data indicate that Fnr is required to relieve NifL inhibition of NifA activity under anaerobic conditions, and this appears to account for the iron requirement of nif induction (32). Therefore, in addition to the rhizobial homologous Fnr proteins, FnrN and FixK, which are known to be involved in regulation of nitrogen fixation in the symbiotic bacteria (8; see reference 4 and references cited therein), in K. pneumoniae, the transcriptional activator Fnr is apparently also involved in regulation of nitrogen fixation. These results are in contrast to the report of Hill (12), that redox regulation of nif expression in a heterologous E. coli strain is independent of the E. coli fnr gene product. This discrepancy may be due to experimental differences. We determined NifA-mediated transcriptional activation by measuring differential rates of β-galactosidase expression from a chromosomal nifK-lacZ fusion in order to monitor nif induction. In contrast, Hill determined acetylene reduction by nitrogenase after growing heterologous E. coli fnr mutant strains carrying the Nif+ plasmid pRD1 under derepressing conditions. Also, because plasmid pRD1 contains in addition to the nif genes nonidentified K. pneumoniae genes (3), we cannot completely rule out that K. pneumoniae fnr is encoded on the plasmid. Apart from these experimental differences concerning the heterologous E. coli systems, we confirmed the Fnr requirement for the nif regulation in the native-genetic-background K. pneumoniae.
We further showed that the general oxygen sensor Fnr is required for relief of NifL inhibition under anaerobic growth conditions and that the presence of ammonium results in significantly lower nif inductions in the wild-type strain than those observed in fnr mutant strains under nitrogen and oxygen limitation. Both of these findings suggest that the oxygen signal is not detected by NifL directly but by Fnr, which transduces the signal—directly or indirectly—to NifL. However, at this state of experimental data, we cannot completely rule out that the Fnr requirement might be due to some Fnr-dependent metabolic signals not directly related to the lack of oxygen. If Fnr is indeed the primary oxygen sensor for the nif regulation in K. pneumoniae, how the oxygen signal is transmitted to NifL remains to be explained. Fnr either is transducing the oxygen signal by directly interacting with NifL in the absence of oxygen, or under anaerobic conditions, Fnr is activating the transcription of a gene or genes whose product or products mediate the signal to NifL. Because Fnr is a transcriptional activator and can be excluded as the physiological electron donor for NifL reduction, it is more reasonable that under anaerobic conditions, Fnr transduces the signal by transcriptional activation.
Hypothetical model for oxygen signal transduction.
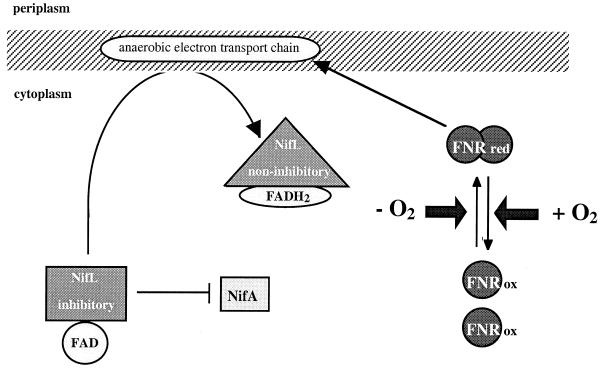
In K. pneumoniae, as in A. vinelandii, the redox state of the flavoprotein NifL is thought to influence its ability to modulate the NifA activity in response to the oxygen levels. However, the physiological electron donors for NifL have not yet been identified (19, 22). If the redox state of the flavoproteins is indeed responsible for mediating the oxygen signal to NifA, one could postulate that by reducing the cofactor of NifL, the physiological electron donor is transducing the oxygen signal to NifL. Thus, the physiological electron donor for the NifL reduction may be a component of the oxygen signal transduction. Because one can exclude Fnr as the physiological electron donor for NifL reduction in the absence of oxygen, one has to postulate another downstream signal transductant following Fnr. We therefore hypothesize that in the absence of oxygen, Fnr activates transcription of a gene or genes whose product or products function to relieve NifL inhibition by reducing the FAD cofactor of NifL. Attractive hypothetical candidates for the physiological electron donor for NifL are components of the anaerobic electron transport system (Fig. 5), particularly the electron transport system to fumarate, whose transcription under anaerobic conditions is directly dependent on Fnr activation (1, 24, 34, 38). Preliminary data, which indicate that K. pneumoniae NifL under anaerobic conditions is membrane associated, whereas in the presence of oxygen NifL is in the cytosolic fraction, support this model (Klopprogge and Schmitz, unpublished). Studies of the anaerobic electron transport system components as potential physiological electron donors for NifL are in process.
FIG. 5.
Hypothetical model of oxygen signal transduction in K. pneumoniae. red, reduced; ox, oxidized.
ACKNOWLEDGMENTS
We thank Gerhard Gottschalk for generous support and helpful discussions; Andrea Shauger for critical reading of the manuscript, and G. Unden for providing the fnr deletion strains RM101 and M182(fnr::Tn10).
This work was supported by the Deutsche Forschungsgemeinschaft (SCHM1052/4–3) and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Ackrell B A. Progress in understanding structure-function relationships in respiratory chain complex II. FEBS Lett. 2000;466:1–5. doi: 10.1016/s0014-5793(99)01749-4. [DOI] [PubMed] [Google Scholar]
- 2.Dixon R. The oxygen-responsive NifL-NifA complex: a novel two-component regulatory system controlling nitrogenase synthesis in gamma-proteobacteria. Arch Microbiol. 1998;169:371–380. doi: 10.1007/s002030050585. [DOI] [PubMed] [Google Scholar]
- 3.Dixon R A, Cannon F, Kondorosi A. Construction of a P plasmid carrying nitrogen fixation genes from Klebsiella pneumoniae. Nature. 1976;260:268–271. doi: 10.1038/260268a0. [DOI] [PubMed] [Google Scholar]
- 4.Fischer H-M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govantes F, Andujar E, Santero E. Mechanism of translational coupling in the nifLA operon of Klebsiella pneumoniae. EMBO J. 1998;17:2368–2377. doi: 10.1093/emboj/17.8.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grabbe R, Kuhn A, Schmitz R A. Cloning, sequencing and characterization of Fnr from Klebsiella pneumoniae. 2000. Antonie Leeuwenhoek, in press. [DOI] [PubMed] [Google Scholar]
- 7.Green J, Bennett B, Jordan P, Ralph E T, Thomson A J, Guest J R. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem J. 1996;316:887–892. doi: 10.1042/bj3160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutierrez D, Hernando Y, Palacios J M, Imperial J, Ruiz-Argueso T. FnrN controls symbiotic nitrogen fixation and hydrogenase activities in Rhizobium leguminosarum biovar viciae UPM791. J Bacteriol. 1997;179:5264–5270. doi: 10.1128/jb.179.17.5264-5270.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He L, Soupene E, Kustu S. NtrC is required for control of Klebsiella pneumoniae NifL activity. J Bacteriol. 1997;179:7446–7455. doi: 10.1128/jb.179.23.7446-7455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He L, Soupene E, Ninfa A, Kustu S. Physiological role for the GlnK protein of enteric bacteria: relief of NifL inhibition under nitrogen-limiting conditions. J Bacteriol. 1998;180:6661–6667. doi: 10.1128/jb.180.24.6661-6667.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson N, Austin S, Dixon R A. Role of metal ions in negative regulation of nitrogen fixation by the nifL gene product from Klebsiella pneumoniae. Mol Gen Genet. 1989;216:484–491. [Google Scholar]
- 12.Hill S. Redox regulation of enteric nif expression is independent of the fnr gene product. FEMS Microbiol Lett. 1985;29:5–9. [Google Scholar]
- 13.Hill S, Austin S, Eydmann T, Jones T, Dixon R. Azotobacter vinelandii NifL is a flavoprotein that modulates transcriptional activation of nitrogen-fixation genes via a redox-sensitive switch. Proc Natl Acad Sci USA. 1996;93:2143–2148. doi: 10.1073/pnas.93.5.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;9:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 15.Jack R, DeZamaroczy M, Merrick M. The signal transduction protein GlnK is required for NifL-dependent nitrogen control of nif gene expression in Klebsiella pneumoniae. J Bacteriol. 1999;181:1156–1162. doi: 10.1128/jb.181.4.1156-1162.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayaraman P-S, Gaston K L, Cole J A, Busby S J W. The nirB promoter of Escherichia coli: location of nucleotide sequences essential for regulation by oxygen, the FNR protein and nitrite. Mol Microbiol. 1988;2:527–530. doi: 10.1111/j.1365-2958.1988.tb00059.x. [DOI] [PubMed] [Google Scholar]
- 17.Kaniga K, Delor I, Cornelis G R. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 18.Khoroshilova N, Popescu C, Munck E, Beinert H, Kiley P J. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc Natl Acad Sci USA. 1997;94:6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klopprogge K, Schmitz R A. NifL of Klebsiella pneumoniae: redox characterization in relation to the nitrogen source. Biochem Biophys Acta. 1999;1431:462–470. doi: 10.1016/s0167-4838(99)00075-8. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lei S, Pulakat L, Gavini N. Genetic analysis of nif regulatory genes by utilizing the yeast two-hybrid system detected formation of a NifL-NifA complex that is implicated in regulated expression of nif genes. J Bacteriol. 1999;181:6535–6539. doi: 10.1128/jb.181.20.6535-6539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macheroux P, Hill S, Austin S, Eydmann T, Jones T, Kim S O, Poole R, Dixon R. Electron donation to the flavoprotein NifL, a redox-sensing transcriptional regulator. Biochem J. 1998;332:413–419. doi: 10.1042/bj3320413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacNeil D, Zhu J, Brill W J. Regulation of nitrogen fixation in Klebsiella pneumoniae: isolation and characterization of strains with nif-lac fusions. J Bacteriol. 1981;145:348–357. doi: 10.1128/jb.145.1.348-357.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manodori A, Cecchini G, Schröder I, Gunsalus R P, Werth M T, Johnson M K. [3Fe-4S] to [4Fe-4S] cluster conversion in Escherichia coli fumarate reductase by site-directed mutagenesis. Biochemistry. 1992;31:2703–2712. doi: 10.1021/bi00125a010. [DOI] [PubMed] [Google Scholar]
- 25.Melville S B, Gunsalus R P. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J Biol Chem. 1990;265:18733–18736. [PubMed] [Google Scholar]
- 26.Money T, Jones T, Dixon R, Austin S. Isolation and properties of the complex between the enhancer binding protein NifA and the sensor NifL. J Bacteriol. 1999;181:4461–4468. doi: 10.1128/jb.181.15.4461-4468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakano Y, Yoshida Y, Yamashita Y, Koga T. Construction of a series of pACYC-derived plasmid vectors. Gene. 1995;162:157–158. doi: 10.1016/0378-1119(95)00320-6. [DOI] [PubMed] [Google Scholar]
- 28.Prentki P, Kirsch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Sawers G, Suppmann B. Anaerobic induction of pyruvate formate-lyase gene expression is mediated by the ArcA and FNR proteins. J Bacteriol. 1992;174:3474–3478. doi: 10.1128/jb.174.11.3474-3478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitz R A. NifL of Klebsiella pneumoniae carries an N-terminally bound FAD cofactor, which is not directly required for the inhibitory function of NifL. FEMS Microbiol Lett. 1997;1577:313–318. doi: 10.1111/j.1574-6968.1997.tb12791.x. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz R A, He L, Kustu S. Iron is required to relieve inhibitory effects of NifL on transcriptional activation by NifA in Klebsiella pneumoniae. J Bacteriol. 1996;178:4679–4687. doi: 10.1128/jb.178.15.4679-4687.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silhavy T J, Bermann M, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. pp. 107–112. [Google Scholar]
- 34.Skotnicki L M, Rolfe B G. Pathways of energy metabolism required for phenotypic expression of nif+Kp genes in Escherichia coli. Aust J Biol Sci. 1979;32:637–649. doi: 10.1071/bi9790637. [DOI] [PubMed] [Google Scholar]
- 35.Spiro S. The FNR family of transcriptional regulators. Antonie Leeuwenhoek. 1994;66:23–36. doi: 10.1007/BF00871630. [DOI] [PubMed] [Google Scholar]
- 36.Spiro S, Guest J R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990;6:399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- 37.Unden G, Schirawski J. The oxygen-responsive transcriptional regulator FNR of Escherichia coli: the search for signals and reactions. Mol Microbiol. 1997;25:205–210. doi: 10.1046/j.1365-2958.1997.4731841.x. [DOI] [PubMed] [Google Scholar]
- 38.VanHellemond J J, Tielens A G. Expression and functional properties of fumarate reductase. Biochem J. 1994;304:321–331. doi: 10.1042/bj3040321. [DOI] [PMC free article] [PubMed] [Google Scholar]