Abstract
We assessed the diagnostic potential of cardiovascular disease-associated microRNAs for the early prediction of gestational diabetes mellitus (GDM) in singleton pregnancies of Caucasian descent in the absence of other pregnancy-related complications. Whole peripheral venous blood samples were collected within 10 to 13 weeks of gestation. This retrospective study involved all pregnancies diagnosed with only GDM (n = 121) and 80 normal term pregnancies selected with regard to equality of sample storage time. Gene expression of 29 microRNAs was assessed using real-time RT-PCR. Upregulation of 11 microRNAs (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-23a-3p, miR-100-5p, miR-125b-5p, miR-126-3p, miR-181a-5p, miR-195-5p, miR-499a-5p, and miR-574-3p) was observed in pregnancies destinated to develop GDM. Combined screening of all 11 dysregulated microRNAs showed the highest accuracy for the early identification of pregnancies destinated to develop GDM. This screening identified 47.93% of GDM pregnancies at a 10.0% false positive rate (FPR). The predictive model for GDM based on aberrant microRNA expression profile was further improved via the implementation of clinical characteristics (maternal age and BMI at early stages of gestation and an infertility treatment by assisted reproductive technology). Following this, 69.17% of GDM pregnancies were identified at a 10.0% FPR. The effective prediction model specifically for severe GDM requiring administration of therapy involved using a combination of these three clinical characteristics and three microRNA biomarkers (miR-20a-5p, miR-20b-5p, and miR-195-5p). This model identified 78.95% of cases at a 10.0% FPR. The effective prediction model for GDM managed by diet only required the involvement of these three clinical characteristics and eight microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-100-5p, miR-125b-5p, miR-195-5p, miR-499a-5p, and miR-574-3p). With this, the model identified 50.50% of GDM pregnancies managed by diet only at a 10.0% FPR. When other clinical variables such as history of miscarriage, the presence of trombophilic gene mutations, positive first-trimester screening for preeclampsia and/or fetal growth restriction by the Fetal Medicine Foundation algorithm, and family history of diabetes mellitus in first-degree relatives were included in the GDM prediction model, the predictive power was further increased at a 10.0% FPR (72.50% GDM in total, 89.47% GDM requiring therapy, and 56.44% GDM managed by diet only). Cardiovascular disease-associated microRNAs represent promising early biomarkers to be implemented into routine first-trimester screening programs with a very good predictive potential for GDM.
Keywords: cardiovascular microRNAs, early pregnancy, gene expression, gestational diabetes mellitus, prediction, screening, whole peripheral venous blood
1. Introduction
Gestational diabetes mellitus (GDM), glucose intolerance in pregnancy [1,2,3], increases the risk of the onset of maternal pregnancy-related complications and neonatal morbidity. It also has long-term implications for both mother and child in form of risk of developing type 2 diabetes mellitus and cardiovascular diseases [1,4,5,6].
Several universal screening programs of GDM [1,2,7,8] have been implemented in the routine care of pregnant women. The first screening phase based on the monitoring of a fasting glucose is usually held at first visit during the first trimester of gestation and rules out patients with pre-existing diabetes and detects the occurrence of early GDM. The second screening phase is usually performed at 24–28 weeks of gestation in pregnancies with normal early screening with the oral glucose tolerance test (OGTT) and identifies the occurrence of GDM at the late second and early third pregnancy trimesters. If normal, the OGTT may be repeated again at 32 weeks of gestation [7].
As of now, several promising early predictive models for GDM have been established.
The initial logistic regression model based on the inclusion of maternal characteristics only (maternal age, weight, height, racial origin, family history of diabetes, use of ovulation drugs, birth weight, and previous history of GDM) showed a high accuracy for prediction of GDM at 11–13 weeks of gestation. It reached the following parameters: area under the curve (AUC) 0.823, 95% confidence interval (95% CI) 0.820–0.826, 55.0% sensitivity at a 10.0% false positive rate (FPR) [9]. A slightly older model for the prediction of GDM based on some of the above mentioned factors combined with serum concentrations of adiponectin and sex hormone binding globulin reached similar predictive results (AUC 0.842, 95% CI: 0.817–0.867, 58.6% at a 10.0% FPR) [10].
Similar data were reported by another research group which used a multivariate regression model for the early prediction of GDM. This model was also based on maternal clinical parameters such as age, body mass index (BMI), South/East Asian ethnicity, parity, family history of diabetes, and previous history of GDM (AUC 0.880, 95% CI: 0.850–0.920, 70.2% detection rate at a 10.0% FPR) [11]. Similarly, the same research group later introduced an improved first-trimester risk multivariate prediction model for GDM. This novel model incorporated family history of diabetes, previous history of GDM, South/East Asian ethnicity, parity, BMI, pregnancy-associated plasma protein A (PAPP-A), triglycerides, and lipocalin-2, and achieved a higher discrimination power (AUC 0.910, 95% CI: 0.890–0.960, 76.8% at a 10.0% FPR) [12].
Furthermore, reduced plasma levels of irisin in the first trimester of gestation were implemented into another model based on known risk factors (maternal age, BMI, gestational age at sampling, smoking, ethnicity, pre-existing hypertension or cardiovascular disease, family history of diabetes, physical activity, family history of diabetes, and blood levels of cholesterol, high-density lipoprotein cholesterol, triglycerides, insulin, fasting plasma glucose, and C-reactive protein). This improved the discrimination rate of predicting GDM in a Chinese population (AUC 0.809, 95% CI: 0.763–0.854) [13]. Another independent large-scale study performed in a Chinese population during the first trimester of pregnancy explored a total of 73 variables and also reached a high discriminative power for GDM (AUC 0.800) [14].
An additional non-invasive predictive model consisting of mean arterial blood pressure in the first trimester, age, ethnicity and previous history of GDM demonstrated relatively high predictive ability for a Singaporean population (AUC 0.820, 95% CI: 0.710–0.930), where UK NICE guidelines had poor GDM predictive outcome (AUC 0.600, 95% CI: 0.510–0.700) [15].
Additionally, metabolomics analyses performed on a Japanese population revealed novel promising metabolic biomarkers (serum glutamine, urine ethanolamine, and urine 1,3-diphosphoglycerate). Each biomarker individually demonstrated a high discrimination power for prediction of GDM during the first or early second trimesters of gestation (AUC over 0.800) [16].
First-trimester screening for GDM for an Israeli population reached very high discriminative power in both non-obese women (AUC 0.940, 95% CI: 0.850–0.990, 83.0% at a 10.0% FPR) and obese women (AUC 0.950, 95% CI: 0.880–0.990, 89% at a 10.0% FPR). These screening models were based on the combination of soluble cluster of differentiation 163 (sCD163), tumour necrosis factor alpha (TNFα), placental protein 13 (PP13), and PAPP-A or on the combination of BMI, insulin, sCD163, and TNFα [17].
The latest model was based on maternal clinical characteristics (age and pre-pregnancy BMI); maternal coagulation function (prothrombin time, international standardized ratio, activated partial thromboplastin time, fibrinogen, and thrombin time); and glycolipid metabolism indicators (fasting blood glucose, total cholesterol, triglycerides, low density lipoprotein cholesterol, small and dense low density lipoprotein cholesterol, apolipoprotein B, and apolipoprotein E). This model was applied to a Chinese population in the first trimester of gestation and reached a high clinical value for the prediction of GDM (AUC 0.892, 95% CI: 0.86–0.93) [18].
Previously, the potential usage of coagulation function examination variables such as prothrombin time and activated partial thromboplastin time as novel biomarkers for the prediction of GDM for a Chinese population at 19 weeks of gestation was demonstrated [19].
Similar results were reported for a Chinese population, when a mid-pregnancy risk prediction model for GDM was applied (AUC 0.911, 95% CI: 0.893–0.930). This model was based on maternal status in the combination with ultrasound and serological findings (age, pre-pregnancy BMI, family history of diabetes, polycystic ovary syndrome, previous history of GDM, high systolic pressure, glycosylated haemoglobin levels, triglyceride levels, total cholesterol levels, low density lipoprotein cholesterol levels, C-reactive protein levels, increased subcutaneous fat thickness, and visceral fat thickness) [20].
Similarly, a combined multivariate prediction model performed between 10 and 16 weeks of gestation in an Irish population also achieved a very high level of discrimination for the prediction of GDM (AUC 0.860, 95% CI: 0.774–0.945). This model was based on family history of diabetes, previous perinatal death, overall insulin resistant condition, ultrasound measurements of subcutaneous and visceral abdominal adipose tissue, 8-point skinfold thickness, mid-upper-arm circumference, and weight [21].
Interestingly, the latest study of Eidgahi et al. [22] presented a simplified GDM predictive model with a very good efficiency (AUC 0.83, 95% CI: 0.76–0.90) in an Irani population. This model was based on the mean values of basic indicators (haemoglobin, haematocrit, red blood cell count, and fasting blood glucose) obtained from repeated measures during the first and early second trimesters of gestation. They suggested that this GDM predictive model might be used mainly in poor and low-income countries.
Other models for the early prediction of GDM have not been as effective as the predictive models introduced above [23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40].
We focused on the exploration of gene expression profiles of selected cardiovascular disease-associated microRNAs in the whole peripheral venous blood of women during the early stages of gestation. The aim of the study was to assess the predictive potential for GDM in the absence of other pregnancy-related complications.
Previously, by searching the Medline database we identified a large number of microRNAs playing a role in pathogenesis of diabetes mellitus and cardiovascular/cerebrovascular diseases. Finally, we selected a shortlist of 29 microRNAs for the study which have been repeatedly demonstrated by numerous scientific teams to be involved in development and homeostasis of the cardiovascular system, angiogenesis, and adipogenesis. In addition, these microRNAs were reported to be associated with pathological conditions and diseases (vascular endothelial dysfunction and inflammation, hypoxia, hypertension and regulation of hypertension-related genes, obesity, dyslipidaemia, atherosclerosis and atherosclerotic plaque formation, insulin resistance, diabetes mellitus and diabetes-related complications, metabolic syndrome, cardiovascular diseases involving the blood vessels and/or the heart, chronic kidney disease, ischemia/reperfusion injury, cardiac regeneration, and cachexia) (Table 1) [41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220,221,222,223,224,225].
Table 1.
The role of studied microRNAs in the pathogenesis of diabetes mellitus and cardiovascular/cerebrovascular diseases.
| miRBase ID | Gene Location on Chromosome | Role in the Pathogenesis of Diabetes Mellitus and Cardiovascular/Cerebrovascular Diseases |
|---|---|---|
| hsa-miR-1-3p | 20q13.3 [41] 18q11.2 |
Acute myocardial infarction, heart ischemia, post-myocardial infarction complications, thoracic aortic aneurysm [43], diabetes mellitus [44,45], and vascular endothelial dysfunction [46] |
| hsa-miR-16-5p | 13q14.2 | Myocardial infarction [47,48], heart failure [49], acute coronary syndrome, cerebral ischaemic events [50], gestational diabetes mellitus [51,52,53], and diabetes mellitus [54,55,56] |
| hsa-miR-17-5p | 13q31.3 [57,58] | Cardiac development [59], ischemia/reperfusion-induced cardiac injury [60], kidney ischemia-reperfusion injury [61], diffuse myocardial fibrosis in hypertrophic cardiomyopathy [62], acute ischemic stroke [63], coronary artery disease [64], adipogenic differentiation [65], gestational diabetes mellitus [51,52], and diabetes mellitus [56,66] |
| hsa-miR-20a-5p | 13q31.3 [67] | Pulmonary hypertension [68], gestational diabetes mellitus [51,52,69], diabetic retinopathy [70], and diabetes with abdominal aortic aneurysm [71] |
| hsa-miR-20b-5p | Xq26.2 [67] | Hypertension-induced heart failure [72], insulin resistance [73], T2DM [74,75], and diabetic retinopathy [76] |
| hsa-miR-21-5p | 17q23.2 [77] | Homeostasis of the cardiovascular system [78], cardiac fibrosis and heart failure [79,80], thoracic aortic aneurysm [43], ascending aortic aneurysm [81], regulation of hypertension-related genes [82], myocardial infarction [83], insulin resistance [73], T2DM [84], T2DM with major cardiovascular events [85], T1DM [86,87,88], and diabetic nephropathy [89] |
| hsa-miR-23a-3p | 19p13.12 | Heart failure [90], coronary artery disease [91], cerebral ischemia-reperfusion [92], vascular endothelial dysfunction [46], small and large abdominal aortic aneurysm [93], obesity and insulin resistance [94] |
| hsa-miR-24-3p | 19p13.12 | Asymptomatic carotid stenosis [95], familial hypercholesterolemia and coronary artery disease [96], angina pectoris [97], ischemic dilated cardiomyopathy [98], small and large abdominal aortic aneurysm [93], myocardial ischemia/reperfusion [99,100], and diabetes mellitus [45,56,60,62] |
| hsa-miR-26a-5p | 3p22.2 [101] 12q14.1 |
Heart failure, cardiac hypertrophy, myocardial infarction [83,103,104], ischemia/reperfusion injury [105], pulmonary arterial hypertension [106], T1DM [107], and diabetic nephropathy [89] |
| hsa-miR-29a-3p | 7q32.3 | Ischemia/reperfusion-induced cardiac injury [108], cardiac cachexia, heart failure [109], atrial fibrillation [110], diffuse myocardial fibrosis in hypertrophic cardiomyopathy [62], coronary artery disease [111], pulmonary arterial hypertension [106], gestational diabetes mellitus [112], and diabetes mellitus [44,55,113,114] |
| hsa-miR-92a-3p | 13q31.3 Xq26.2 |
Mitral chordae tendineae rupture [115], children with rheumatic carditis [116], myocardial infarction [117], heart failure [118], coronary artery disease [119], and renal injury-associated atherosclerosis [120] |
| hsa-miR-100-5p | 11q24.1 | Failing human heart, idiopathic dilated cardiomyopathy, ischemic cardiomyopathy [98], regulation of hypertension-related genes [82], and T1DM [86] |
| hsa-miR-103a-3p | 5q34 [121] 20p13 |
Hypertension, hypoxia-induced pulmonary hypertension [123], myocardial ischemia/reperfusion injury, acute myocardial infarction [124], ischemic dilated cardiomyopathy [99], obesity, and regulation of insulin sensitivity [125], T1DM [126] |
| hsa-miR-125b-5p | 11q24.1 [126] 21q21.1 |
Acute ischemic stroke, acute myocardial infarction [128,129], ischemic dilated cardiomyopathy [98], ascending aortic aneurysm [81], gestational diabetes mellitus [130], T1DM [131,132], and T2DM [133] |
| hsa-miR-126-3p | 9q34.3 [134] | Acute myocardial infarction [104], thoracic aortic aneurysm [43], T2DM [85,135], T2DM with major cardiovascular events [85], and gestational diabetes mellitus [136] |
| hsa-miR-130b-3p | 22q11.21 | Hypertriglyceridemia [137,138], intracranial aneurysms [139], hyperacute cerebral infarction [140], T2DM [84,141,142], and gestational diabetes mellitus [136] |
| hsa-miR-133a-3p | 18q11.2 [143] 20q13.33 |
Heart failure, myocardial fibrosis in hypertrophic cardiomyopathy [62,145], arrhythmogenesis in the hypertrophic and failing hearts [146,147], coronary artery calcification [148], thoracic aortic aneurysm [43], ascending aortic aneurysm [81], and diabetes mellitus [41,45] |
| hsa-miR-143-3p | 5q33 | Intracranial aneurysms [149], coronary heart disease [150], myocardial infarction [151], myocardial hypertrophy [152], dilated cardiomyopathy [153], pulmonary arterial hypertension [154], acute ischemic stroke [127], and ascending aortic aneurysm [81], |
| hsa-miR-145-5p | 5q33 | Hypertension [155,156], dilated cardiomyopathy [157], myocardial infarction [158], stroke [159], acute cerebral ischemic/reperfusion [160], T2DM [56,161], T1DM [84], diabetic retinopathy [162], and gestational diabetes mellitus [163] |
| hsa-miR-146a-5p | 5q33.3 [164,165] | Angiogenesis [166], hypoxia, ischemia/reperfusion-induced cardiac injury [167], myocardial infarction [48], coronary atherosclerosis, coronary heart disease in patients with subclinical hypothyroidism [168], thoracic aortic aneurysm [43], acute ischemic stroke, acute cerebral ischemia [169], T2DM [56,84], T1DM [107], and diabetic nephropathy [89] |
| hsa-miR-155-5p | 21q21.3 | Thoracic aortic aneurysm [43], type 1 diabetes [125], gestational diabetes mellitus [53], adolescent obesity [170], diet-induced obesity and obesity resistance [171], atherosclerosis [172], hyperlipidemia-associated endotoxemia [173], coronary plaque rupture [174], children with cyanotic heart disease [175], chronic kidney disease and nocturnal hypertension [176], and atrial fibrillation [177] |
| hsa-miR-181a-5p | 1q32.1 [178] 9q33.3 |
Regulation of hypertension-related genes, atherosclerosis [178], metabolic syndrome, coronary artery disease [179], non-alcoholic fatty liver disease [180], ischaemic stroke, transient ischaemic attack, acute myocardial infarction [181,182], obesity and insulin resistance [94,178,179], T1DM [84,183], and T2DM [178,182] |
| hsa-miR-195-5p | 17p13.1 [184] | Cardiac hypertrophy, heart failure [185,186], abdominal aortic aneurysms [187], aortic stenosis [188], T2DM [161], and gestational diabetes mellitus [189] |
| hsa-miR-199a-5p | 1q24.3 19p13.2 |
T1DM, T2DM, gestational diabetes mellitus [190], diabetic retinopathy [191], cerebral ischemic injury [192], heart failure [193], hypertension [194,195], congenital heart disease [196], pulmonary artery hypertension [197], unstable angina [198], hypoxia in myocardium [196], and acute kidney injury [199] |
| hsa-miR-210-3p | 11p15.5 | Cardiac hypertrophy [200], acute kidney injury [201], myocardial infarction [202], and atherosclerosis [203] |
| hsa-miR-221-3p | Xp11.3 | Asymptomatic carotid stenosis [95], cardiac amyloidosis [204], heart failure [205], atherosclerosis [206,207], aortic stenosis [208], acute myocardial infarction [209], acute ischemic stroke [210], focal cerebral ischemia [211], pulmonary artery hypertension [212], and obesity [213] |
| hsa-miR-342-3p | 14q32.2 | Cardiac amyloidosis [204], obesity [214], T1DM [84,190,215], T2DM [190,216,217, gestational diabetes mellitus [190] and endothelial dysfunction [218] |
| hsa-miR-499a-5p | 20q11.22 | Myocardial infarction [48,219], hypoxia [220], cardiac regeneration [221], and vascular endothelial dysfunction [46] |
| hsa-miR-574-3p | 4p14 | Myocardial infarction [222], coronary artery disease [138], cardiac amyloidosis [204], stroke [223], and T2DM [142,224] |
T1DM: Diabetes mellitus type 1; T2DM: Diabetes mellitus type 2.
The epigenetic profiling of microRNAs (miR-1-3p, miR-16-5p, miR-17-5p, miR-20a-5p, miR-20b-5p, miR-21-5p, miR-23a-3p, miR-24-3p, miR-26a-5p, miR-29a-3p, miR-92a-3p, miR-100-5p, miR-103a-3p, miR-125b-5p, miR-126-3p, miR-130b-3p, miR-133a-3p, miR-143-3p, miR-145-5p, miR-146a-5p, miR-155-5p, miR-181a-5p, miR-195-5p, miR-199a-5p, miR-210-3p, miR-221-3p, miR-342-3p, miR-499a-5p, and miR-574-3p) was the subject of our interest (Table 1).
Up to now, no reports on microRNA gene profiling of the whole peripheral venous blood in early stages of gestation are at disposal in pregnancies with subsequent onset of GDM.
To our knowledge, only several studies have reported promising data on the early diagnosis of GDM during the first trimester of gestation via screening of circulating cardiovascular disease-associated microRNAs in maternal plasma or serum samples [112,130,226,227,228].
2. Results
2.1. Clinical Characteristics of GDM and Control Pregnancies
The clinical characteristics of GDM and control pregnancies are summarized in Table 2.
Table 2.
Clinical characteristics of the cases and controls.
| Normal Term Pregnancies (n = 80) |
GDM Overall (n = 121) |
GDM Managed by Diet Only (n = 101) |
GDM Managed by Diet and Therapy (n = 20) |
p-Value 1 | p-Value 2 | p-Value 3 | |
|---|---|---|---|---|---|---|---|
| Maternal characteristics | |||||||
| Autoimmune diseases (SLE/APS/RA) | 0 (0%) | 1 (0.83%) | 1 (RA, 1.0%) | 0 (0%) | 0.672 OR: 2.004 95% CI: 0.081–49.814 |
0.593 OR: 2.403 95% CI: 0.096–59.786 |
0.497 OR: 3.927 95% CI: 0.076–203.916 |
| Other autoimmune diseases | 0 (0%) | 1 (0.83%) | 1 (vasculitis; 1.0%) | 0 (0%) | 0.672 OR: 2.004 95% CI: 0.081–49.814 |
0.593 OR: 2.403 95% CI: 0.096–59.786 |
0.497 OR: 3.927 95% CI: 0.076–203.916 |
| Any kind of autoimmune disease (SLE/APS/RA/other) | 0 (0%) | 2 (1.65%) | 2 (1.98%) | 0 (0%) | 0.435 OR: 3.368 95% CI: 0.160–71.088 |
0.369 OR: 4.045 95% CI: 0.191–85.468 |
0.497 OR: 3.927 95% CI: 0.076–203.916 |
| Trombophilic gene mutations | 0 (0%) | 11 (9.09%) | 9 (8.91%) | 2 (10.0%) | 0.052 OR: 16.756 95% CI: 0.973–288.513 |
0.055 OR: 16.535 95% CI: 0.947–288.589 |
0.050 OR: 21.757 95% CI: 1.002–472.533 |
| Family history of diabetes | |||||||
| First-degree relative with DM | 10 (12.50%) | 30 (24.79%) | 26 (25.74%) | 4 (20.0%) | 0.036 OR: 2.308 95% CI: 1.057–5.037 |
0.030 OR: 2.427 95% CI: 1.092–5.394 |
0.392 OR: 1.750 95% CI: 0.486–6.297 |
| Second-degree relative with DM | 21 (26.25%) | 44 (36.36%) | 36 (35.64%) | 8 (40.0%) | 0.135 OR: 1.605 95% CI: 0.863–2.986 |
0.178 OR: 1.556 95% CI: 0.818–2.961 |
0.230 OR: 1.873 95% CI: 0.673–5.215 |
| Parity | |||||||
| Nulliparous—no previous pregnancy | 40 (50.0%) | 54 (44.63%) | 46 (45.54%) | 8 (40.0%) | 0.455 OR: 0.806 95% CI: 0.458–1.419 |
0.551 OR: 0.836 95% CI: 0.465–1.505 |
0.425 OR: 0.667 95% CI: 0.246–1.805 |
| Parous—no prior GDM | 39 (48.75%) | 61 (50.41%) | 50 (49.50%) | 11 (55.0%) | 0.817 OR: 1.069 95% CI: 0.608–1.880 |
0.919 OR: 1.031 95% CI: 0.573–1.853 |
0.618 OR: 1.285 95% CI: 0.480–3.437 |
| Parous—prior GDM | 1 (1.25%) | 6 (4.96%) | 5 (4.95%) | 1 (5.0%) | |||
| History of macrosomia (FBW > 4000 g) | 4 (5.0%) | 2 (1.65%) | 1 (0.99%) | 1 (5.0%) | 0.194 OR: 0.319 95% CI: 0.057–1.786 |
0.141 OR: 0.190 95% CI: 0.021–1.735 |
1.0 OR: 1.000 95% CI: 0.106–9.471 |
| History of miscarriage spontaneous loss of a pregnancy before 22 weeks of gestation |
16 (20.0%) | 42 (34.71%) | 36 (35.64%) | 6 (30.0%) | 0.026 OR: 2.127 95% CI: 1.095–4.129 |
0.022 OR: 2.215 95% CI: 1.119–4.384 |
0.338 OR: 1.714 95% CI: 0.569–5.161 |
| History of perinatal death the death of a baby between 22 weeks of gestation (or weighing 500 g) and 7 days after birth |
0 (0%) | 4 (3.31%) | 3 (2.97%) | 1 (5.0%) | 0.224 OR: 6.166 95% CI: 0.327–116.113 |
0.251 OR: 5.721 95% CI: 0.291–112.387 |
0.128 OR: 12.385 95% CI: 0.486–315.805 |
| ART (IVF/ICSI/other) | 2 (2.5%) | 20 (16.53%) | 15 (14.85%) | 5 (25.0%) | 0.007 OR: 7.723 95% CI: 1.752–34.038 |
0.013 OR: 6.802 95% CI: 1.507–30.698 |
0.004 OR: 13.000 95% CI: 2.304–73.362 |
| Smoking during pregnancy | 2 (2.5%) | 6 (4.96%) | 4 (3.96%) | 2 (10.0%) | 0.392 OR: 2.035 95% CI: 0.108–10.343 |
0.589 OR: 1.608 95% CI: 0.287–9.012 |
0.156 OR: 4.333 95% CI: 0.572–32.859 |
| Pregnancy details (First trimester of gestation) | |||||||
| Maternal age (years) | 32 (25–42) | 33 (21–42) | 33 (21–42) | 32 (25–42) | 0.635 | 0.572 | 0.950 |
| Advanced maternal age (≥35 years old at early stages of gestation) | 18 (22.50%) | 49 (40.49%) | 42 (41.58%) | 7 (35.0%) | 0.009 OR: 2.618 95% CI: 1.238–4.437 |
0.007 OR: 2.675 95% CI: 1.271–4.731 |
0.252 OR: 1.144 95% CI: 0.644–5.343 |
| BMI (kg/m2) | 21.28 (17.16–29.76) | 24.24 (17.37–40.76) | 23.89 (17.37–40.76) | 26.55 (19.33–39.79) | <0.001 | <0.001 | <0.001 |
| BMI ≥ 30 kg/m2 | 0 (0%) | 25 (20.66%) | 17 (16.83%) | 8 (40%) | 0.009 OR: 42.544 95% CI: 2.550–709.837 |
0.015 OR: 33.343 95% CI: 1.972–563.719 |
0.002 OR: 109.480 95% CI: 5.941–2017.344 |
| Gestational age at sampling (weeks) | 10.29 (9.57–13.71) | 10.29 (9.43–13.57) | 10.29 (9.43–13.57) | 10.21 (9.43–12.71) | 0.737 | 0.548 | 0.521 |
| MAP (mmHg) | 88.75 (67.67–103.83) | 92.0 (72.83–127.58) | 91.96 (72.83–127.58) | 92.58 (82.85–101.92) | 0.051 | 0.083 | 0.022 |
| MAP (MoM) | 1.05 (0.84–1.25) | 1.05 (0.90–1.44) | 1.05 (0.90–1.44) | 1.07 (0.97–1.13) | 0.656 | 0.574 | 0.361 |
| Mean UtA-PI | 1.39 (0.56–2.43) | 1.35 (0.42–2.30) | 1.35 (0.42–2.30) | 1.25 (0.74–1.84) | 0.591 | 0.831 | 0.495 |
| Mean UtA-PI (MoM) | 0.90 (0.37–1.55) | 0.88 (0.26–1.48) | 0.89 (0.26–1.48) | 0.85 (0.52–1.26) | 0.539 | 0.710 | 0.402 |
| PIGF serum levels (pg/mL) | 27.1 (8.1–137.0) | 26.7 (9.2–71.0) | 26.8 (9.2–71.0) | 25.5 (14.5–46.0) | 0.420 | 0.377 | 0.375 |
| PIGF serum levels (MoM) | 1.04 (0.38–2.61) | 1.09 (0.44–2.0) | 1.06 (0.44–2.0) | 1.15 (0.62–1.59) | 0.934 | 0.690 | 0.065 |
| PAPP-A serum levels (IU/L) | 1.49 (0.48–15.69) | 1.28 (0.22–11.45) | 1.35 (0.22–11.45) | 1.0 (0.26–6.83) | 0.063 | 0.123 | 0.158 |
| PAPP-A serum levels (MoM) | 1.17 (0.37–3.18) | 1.05 (1.19–3.67) | 1.04 (0.28–3.02) | 1.43 (0.19–3.67) | 0.606 | 0.434 | 0.362 |
| Free b-hCG serum levels (μg/L) | 60.21 (9.9–200.6) | 50.25 (9.31–211.3) | 53.82 (9.31–211.3) | 32.62 (16.55–153.2) | 0.043 | 0.123 | 0.037 |
| Free b-hCG serum levels (MoM) | 1.02 (0.31–3.57) | 0.98 (0.18–4.54) | 1.0 (0.18–4.54) | 0.97 (0.33–2.74) | 0.317 | 0.437 | 0.446 |
| Screen positive for PE and/or FGR by FMF algorithm | 0 (0%) | 11 (9.09%) | 10 (9.90%) | 1 (5.0%) | 0.052 OR: 16.756 95% CI: 0.973–288.513 |
0.045 OR: 18.475 95% CI: 1.066–320.312 |
0.128 OR: 12.385 95% CI: 0.486–315.805 |
| Aspirin intake during pregnancy | 0 (0%) | 8 (6.61%) | 7 (6.93%) | 1 (5.0%) | 0.089 OR: 12.057 95% CI: 0.686–211.908 |
0.083 OR: 12.778 95% CI: 0.717–227.208 |
0.128 OR: 12.385 95% CI: 0.486–315.806 |
| Pregnancy details (At delivery) | |||||||
| BMI (kg/m2) | 26.66 (21.71–34.82) | 28.41 (20.11–49.31) | 28.24 (20.11–49.31) | 32.11 (23.23–44.98) | 0.004 | 0.042 | <0.001 |
| SBP (mmHg) | 122 (100–155) | 120 (90–160) | 121 (90–160) | 120 (100–140) | 0.823 | 0.950 | 0.330 |
| DBP (mmHg) | 76 (60–90) | 79 (57–109) | 79 (57–109) | 79 (60–89) | 0.898 | 0.945 | 0.816 |
| Gestational age at delivery (weeks) | 40.07 (37.57–42.0) | 39.14 (36.14–41.29) | 39.14 (36.14–41.29) | 38.93 (36.57–41.0) | <0.001 | <0.001 | 0.009 |
| Delivery at gestational age < 37 weeks | 0 (0%) | 6 (4.96%) | 4 (3.96%) 1 CS for vasculitis-associated adverse obstetric history 3 CS for abnormal CTG |
2 (10.0%) 1 CS for vasculitis-associated adverse obstetric history 1 CS for abnormal CTG |
0.135 OR: 9.061 95% CI: 0.503–163.118 |
0.181 OR: 7.431 95% CI: 0.394–140.092 |
0.050 OR: 21.757 95% CI: 1.002–472.533 |
| Polyhydramnios | 1 (1.25%) | 28 (23.14%) | 21 (20.79%) | 7 (35.0%) | 0.002 OR: 23.785 95% CI: 3.164–178.781 |
0.003 OR: 20.738 95% CI: 2.723–157.908 |
<0.001 OR: 42.538 95% CI: 4.828–374.768 |
| Fetal birth weight (grams) | 3470 (2920–4240) | 3370 (2430–4340) | 3310 (2430–4340) | 3625 (2950–4220) | 0.043 | 0.003 | 0.046 |
| LGA (FBW > 90th percentile) | 2 (2.5%) | 11 (9.09%) | 7 (6.93%) | 4 (20.0%) | 0.082 OR: 3.900 95% CI: 0.841–18.089 |
0.192 OR: 2.904 95% CI: 0.586–14.384 |
0.012 OR: 9.750 95% CI: 1.643–57.851 |
| Macrosomia (FBW > 4000g) | 5 (6.25%) | 10 (8.26%) | 8 (7.92%) | 2 (10.0%) | 0.596 OR: 1.351 95% CI: 0.444–4.112 |
0.666 OR: 1.290 95% CI: 0.405–4.108 |
0.560 OR: 1.667 95% CI: 0.299–9.295 |
| Fetal sex | |||||||
| Boy | 40 (50.0%) | 60 (49.59%) | 49 (48.51%) | 11 (55.0%) | 0.954 OR: 0.984 95% CI: 0.559–1.730 |
0.843 OR: 0.942 95% CI: 0.524–1.695 |
0.689 OR: 1.222 95% CI: 0.457–3.269 |
| Girl | 40 (50.0%) | 61 (50.41%) | 52 (51.49%) | 9 (45.0%) | |||
| Induced delivery | 8 (10.0%) 4 postterm pregnancy 1 polyhydramnios 1 suspicious CTG 2 programmed labour |
39 (32.23%) | 32 (31.68%) 29 term or postterm GDM pregnancy 2 suspicious CTG 1 hepatopathy |
7 (35.0%) 7 term or postterm GDM pregnancy |
<0.001 OR: 4.281 95% CI: 1.878–9.757 |
<0.001 OR: 4.174 95% CI: 1.798–9.689 |
0.008 OR: 4.846 95% CI: 1.498–15.674 |
| Mode of delivery | |||||||
| Vaginal | 69 (86.25%) | 66 (54.55%) | 58 (57.43%) | 8 (40.0%) | <0.001 OR: 5.227 95% CI: 2.519–10.848 |
<0.001 OR: 4.651 95% CI: 2.199–9.832 |
<0.001 OR: 9.409 95% CI: 3.139–28.205 |
| CS | 11 (13.75%) | 55 (45.45%) | 43 (42.57%) | 12 (60.0%) | |||
| Apgar score < 7, 5 min | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0.837 OR: 0.663 95% CI: 0.013–33.732 |
0.908 OR: 0.793 95% CI: 0.015–40.411 |
0.497 OR: 3.927 95% CI: 0.076–203.916 |
| Apgar score < 7, 10 min | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0.837 OR: 0.663 95% CI: 0.013–33.732 |
0.908 OR: 0.793 95% CI: 0.015–40.411 |
0.497 OR: 3.927 95% CI: 0.076–203.916 |
| Umbilical blood pH | 7.3 (7.29–7.38) | 7.3 (7.12–7.39) | 7.3 (7.29–7.30) | 0.981 | 0.796 |
Continuous variables, compared using the Mann–Whitney or Kruskal–Wallis test, are presented as median (range). Categorical variables, presented as number (percent), were compared using odds ratio test. p-value 1,2,3: the comparison among normal pregnancies and GDM pregnancies, the comparison among normal pregnancies and GDM pregnancies managed by diet only or GDM pregnancies managed by diet and therapy, respectively. GDM, gestational diabetes mellitus; BMI, body mass index; SBP; systolic blood pressure; DBP, diastolic blood pressure; SLE, systemic lupus erythematosus; APS, antiphospholipid syndrome; RA, rheumatoid arthritis; DM, diabetes mellitus; FBW, fetal birth weight; ART, assisted reproductive technology; IVF, in vitro fertilization; ICSI, intracytoplasmic sperm injection; MAP, mean arterial pressure; UtA-PI, uterine artery pulsatility index; PIGF, placental growth factor; PAPP-A, pregnancy-associated plasma protein-A; b-hCG, beta-subunit of human chorionic gonadotropin; PE, preeclampsia; FGR, fetal growth restriction; FMF, Fetal Medicine Foundation; LGA, large for gestational age; CS, caesarean section.
From the clinical characteristics of patients, it is obvious that maternal age (mainly advanced maternal age, ≥35 years), BMI (higher BMI values, BMI ≥ 30 kg/m2) at early stages of gestation, the necessity to undergo an infertility treatment by assisted reproductive technology, history of miscarriage, the presence of trombophilic gene mutations, positive first-trimester screening for preeclampsia and/or FGR by FMF algorithm, and family history of diabetes mellitus in first-degree relatives represent independent significant risk factors for the subsequent onset of GDM.
2.2. Dysregulation of Cardiovascular Disease-Associated MicroRNAs in Early Stages of Gestation in Pregnancies Destinated to Develop GDM
Initially, microRNA gene expression in peripheral blood leukocytes was compared in the early stages of gestation (within 10 to 13 weeks) between pregnancies destinated to develop GDM and term pregnancies with normal course of gestation (Figure 1). Afterwards, early microRNA gene expression was compared between pregnancies destinated to develop GDM and normal term pregnancies with respect to the treatment strategies (GDM pregnancies managed by diet only and GDM pregnancies requiring a combination of diet and administration of appropriate therapy).
Figure 1.
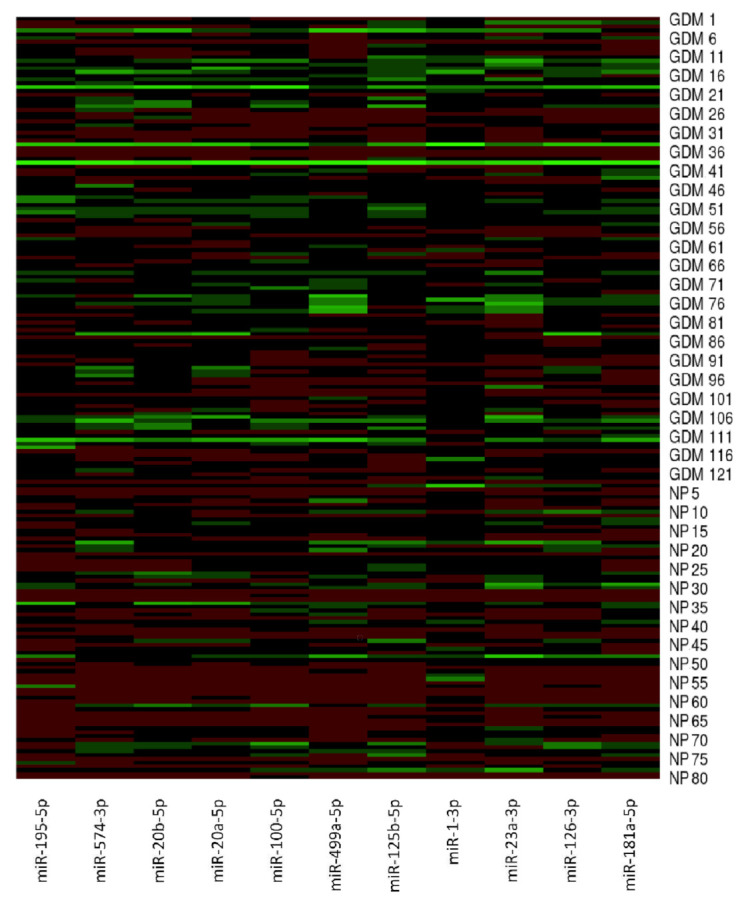
MicroRNA gene expression profile in early stages of gestation in pregnancies destinated to develop GDM and term pregnancies with normal course of gestation. MicroRNA gene expression data (2−∆∆Ct) are visualised using the heatmap. In this setting, each row represents a sample (GDM1–GDM121, NP1–NP80) and each column represents a microRNA gene. The colour and intensity of the boxes are used to represent changes of gene expression (2−∆∆Ct). Green colour indicates upregulation, and red colour indicates downregulation. GDM; gestational diabetes mellitus, NP; normal pregnancies.
Only the data that reached statistical significance after the application of Benjamini–Hochberg correction are discussed below (Supplementary Figures S1 and S2). To interpret the experimental data, new cutoff point p-values were set up. Significant results following the Benjamini–Hochberg correction are marked by asterisks for the appropriate significance levels (* for α = 0.05, ** for α = 0.01, and *** for α = 0.001). The data that were statistically non-significant after the application of Benjamini–Hochberg correction (Table 2 and Table 3) are also displayed (Supplementary Figures S3 and S4), but not discussed further.
Table 3.
MicroRNA expression profiles in peripheral blood leukocytes in early stages of gestation in pregnancies destinated to develop GDM and normal term pregnancies.
| Mann-Whitney Test Results GDM Overall (n = 121) vs. Normal Term Pregnancies (n = 80) |
|||
|---|---|---|---|
| Median (IQR) | Mean (SD) | p-Value | |
| miR-1-3p | 0.135 (0.071–0.254) vs. 0.075 (0.033–0.198) | 0.259 (0.525) vs. 0.176 (0.303) | p = 0.0028 ** |
| miR-16-5p | 1.216 (0.968–1.725) vs. 1.411 (0.890–1.980) | 1.495 (0.981) vs. 1.646 (1.129) | p = 0.5781 |
| miR-17-5p | 1.527 (1.181–2.311) vs. 1.384 (0.971–1.923) | 1.973 (1.473) vs. 1.748 (1.312) | p = 0.0538 |
| miR-20a-5p | 2.215 (1.493–3.398) vs. 1.576 (0.991–2.413) | 3.037 (3.068) vs. 1.909 (1.370) | p < 0.001 *** |
| miR-20b-5p | 2.662 (1.812–3.959) vs. 1.976 (1.111–2.675) | 3.706 (3.878) vs. 2.377 (2.291) | p < 0.001 *** |
| miR-21-5p | 0.344 (0.231–0.460) vs. 0.320 (0.167–0.538) | 0.433 (0.420) vs. 0.394 (0.219) | p = 0.2418 |
| miR-23a-3p | 0.239 (0.168–0.436) vs. 0.185 (0.103–0.376) | 0.367 (0.337) vs. 0.296 (0.329) | p = 0.0065 * |
| miR-24-3p | 0.292 (0.228–0.372) vs. 0.326 (0.196–0.468) | 0.331 (0.197) vs. 0.384 (0.284) | p = 0.5730 |
| miR-26a-5p | 0.699 (0.500–0.926) vs. 0.633 (0.410–1.066) | 0.837 (0.670) vs. 0.776 (0.521) | p = 0.3022 |
| miR-29a-3p | 0.405 (0.282–0.575) vs. 0.372 (0.221–0.545) | 0.510 (0.396) vs. 0.407 (0.245) | p = 0.0840 |
| miR-92a-3p | 2.179 (1.604–3.084) vs. 2.327 (1.188–3.743) | 2.702 (2.226) vs. 2.807 (2.132) | p = 0.9812 |
| miR-100-5p | 0.0023 (0.0013–0.0036) vs. 0.0013 (0.0006–0.0027) | 0.0030 (0.0039) vs. 0.0018 (0.0016) | p < 0.001 *** |
| miR-103a-3p | 1.565 (0.963–2.541) vs. 1.203 (0.815–2.425) | 2.121 (2.252) vs. 1.770 (1.466) | p = 0.1547 |
| miR-125b-5p | 0.0041 (0.0025–0.0057) vs. 0.0030 (0.0016–0.0054) | 0.0049 (0.0046) vs. 0.0036 (0.0027) | p = 0.0034 ** |
| miR-126-3p | 0.328 (0.231–0.509) vs. 0.272 (0.140–0.432) | 0.462 (0.551) vs. 0.336 (0.270) | p = 0.0137 * |
| miR-130b-3p | 0.745 (0.476–1.409) vs. 0.702 (0.407–1.157) | 1.075 (0.960) vs. 1.163 (2.425) | p = 0.2105 |
| miR-133a-3p | 0.109 (0.061–0.220) vs. 0.110 (0.550–0.233) | 0.193 (0.265) vs. 0.232 (0.483) | p = 0.8750 |
| miR-143-3p | 0.048 (0.030–0.880) vs. 0.038 (0.016–0.089) | 0.073 (0.086) vs. 0.058 (0.057) | p = 0.0260 |
| miR-145-5p | 0.176 (0.125–0.236) vs. 0.161 (0.980–0.243) | 0.209 (0.153) vs. 0.195 (0.143) | p = 0.2025 |
| miR-146a-5p | 1.224 (0.821–1.843) vs. 1.225 (0.578–1.765) | 1.658 (1.541) vs. 1.388 (1.096) | p = 0.1415 |
| miR-155-5p | 0.619 (0.434–0.778) vs. 0.607 (0.361–1.614) | 0.703 (0.523) vs 1.247 (1.439) | p = 0.2987 |
| miR-181a-5p | 0.250 (0.175–0.379) vs 0.181 (0.141–0.330) | 0.330 (0.318) vs 0.246 (0.184) | p = 0.0065 * |
| miR-195-5p | 0.267 (0.168–0.487) vs 0.106 (0.048–0.271) | 0.470 (0.690) vs 0.227 (0.364) | p < 0.001 *** |
| miR-199a-5p | 0.080 (0.037–0.159) vs 0.058 (0.023–0.111) | 0.136 (0.223) vs 0.096 (0.131) | p = 0.0288 |
| miR-210-3p | 0.102 (0.074–0.154) vs 0.138 (0.075–0.224) | 0.134 (0.105) vs 0.186 (0.180) | p = 0.0952 |
| miR-221-3p | 0.644 (0.448–0.969) vs 0.548 (0.293–0.906) | 0.815 (0.736) vs 0.693 (0.561) | p = 0.0947 |
| miR-342-3p | 3.069 (2.122–4.110) vs 2.542 (1.551–4.206) | 3.605 (2.724) vs 3.307 (2.383) | p = 0.1947 |
| miR-499a-5p | 0.460 (0.231–0.780) vs 0.269 (0.089–0.587) | 0.758 (1.070) vs 0.477 (0.566) | p < 0.001 *** |
| miR-574-3p | 0.275 (0.180–0.395) vs 0.181 (0.117–0.292) | 0.354 (0.332) vs 0.222 (0.156) | p < 0.001 *** |
MicroRNA gene expression is compared between groups using the Mann–Whitney test. Statistically significant results are marked in bold. Median (interquartile range, IQR) and mean (standard deviation, SD) fold values of relative gene expression of samples (2−∆∆Ct) are presented. Statistical significant data after Benjamini–Hochberg correction are marked by * for α = 0.05, ** for α = 0.01, and *** for α = 0.001.
Upregulation of miR-1-3p (p = 0.0028 **), miR-20a-5p (p < 0.001 ***), miR-20b-5p (p < 0.001 ***), miR-23a-3p (p = 0.0065 *), miR-100-5p (p < 0.001 ***), miR-125b-5p (p = 0.0034 **), miR-126-3p (p = 0.0137 *), miR-181a-5p (p = 0.0065 *), miR-195-5p (p < 0.001 ***), miR-499a-5p (p < 0.001 ***), and miR-574-3p (p < 0.001 ***) was detected during the first trimester of gestation in pregnancies destinated to develop GDM (Supplementary Figure S1, Table 3).
MiR-20a-5p (21.49%), miR-20b-5p (18.18%), miR-23a-3p (15.70%), miR-100-5p (20.66%), miR-125b-5p (14.88%), miR-126-3p (14.05%), miR-195-5p (19.83%) miR-499a-5p (14.88%), and miR-574-3p (23.14%) showed moderate sensitivities at a 10.0% FPR to distinguish between normal pregnancies and pregnancies destinated to develop GDM. In contrast, miR-1-3p (12.40%) and miR-181a-5p (10.74%) showed a low sensitivity to differentiate normal pregnancies and pregnancies with subsequent onset of GDM at a 10.0% FPR (Supplementary Figure S1). This means that the sensitivity in case of miR-1-3p and miR-181a-5p was similar to the false positive rate (10.0%) at which the expression data were assessed.
2.3. First-Trimester Combined MicroRNA Screening Is Able to Differentiate between Pregnancies Destinated to Develop GDM and Term Pregnancies with Normal Course of Gestation
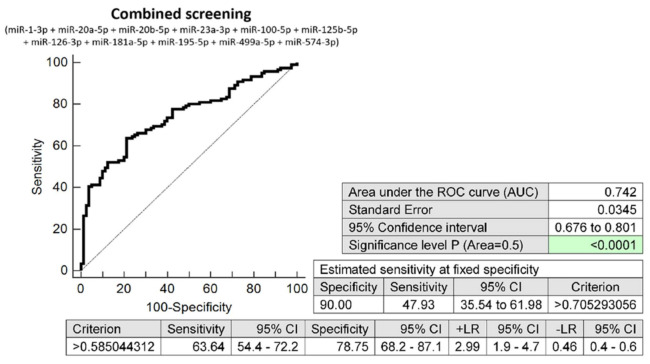
Despite the low sensitivities of miR-1-3p (12.40%) and miR-181a-5p (10.74%), the combined screening of all 11 dysregulated microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-23a-3p, miR-100-5p, miR-125b-5p, miR-126-3p, miR-181a-5p, miR-195-5p, miR-499a-5p, and miR-574-3p) showed the highest accuracy for the early identification of pregnancies destinated to develop GDM (AUC 0.742, p < 0.001, 63.64% sensitivity, 78.75% specificity, cut off >0.5850). This combined screening identified, in the early stages of gestation, 47.93% of pregnancies destinated to develop GDM at a 10.0% FPR (Figure 2).
Figure 2.
ROC analysis—the combination of microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-23a-3p, miR-100-5p, miR-125b-5p, miR-126-3p, miR-181a-5p, miR-195-5p, miR-499a-5p, and miR-574-3p). A total of 47.93% pregnancies destinated to develop GDM had an aberrant microRNA expression profile in the whole peripheral venous blood during the first trimester of gestation at a 10.0% FPR. This represents 58 out of 121 pregnancies correctly predicted to develop GDM and 8 out of 80 normal pregnancies predicted false positively to develop GDM.
2.4. The Very Good Accuracy of First-Trimester Combined Screening (MicroRNA Biomarkers and Selected Clinical Characteristics) to Differentiate between Pregnancies Destinated to Develop GDM and Term Pregnancies with Normal Course of Gestation
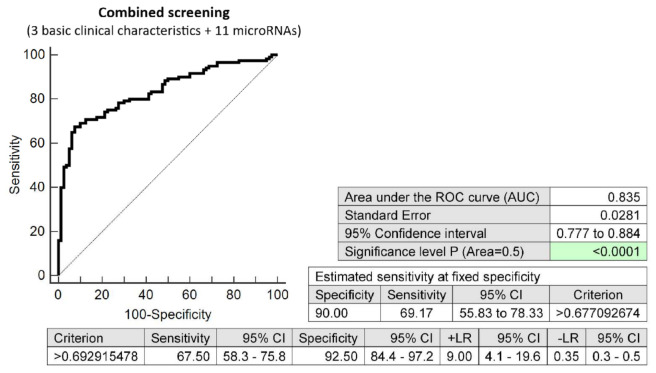
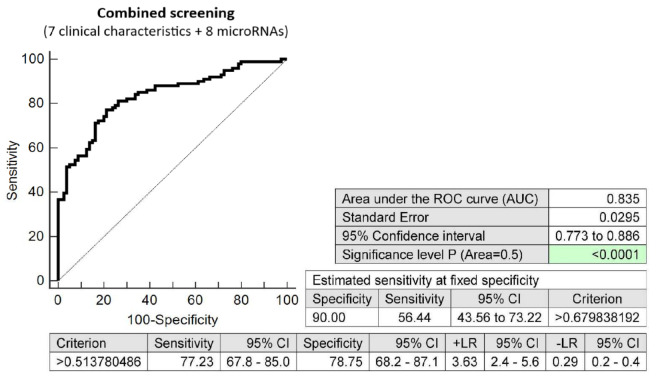
The effective screening based on the combination of minimal number of basic clinical characteristics (maternal age and BMI at early stages of gestation and an infertility treatment by assisted reproductive technology) and 11 dysregulated microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-23a-3p, miR-100-5p, miR-125b-5p, miR-126-3p, miR-181a-5p, miR-195-5p, miR-499a-5p, and miR-574-3p) showed relatively high accuracy for the early identification of pregnancies destinated to develop GDM (AUC 0.835, p < 0.001, 67.50% sensitivity, 92.50% specificity, cut off >0.6929). This combined screening identified, in the early stages of gestation, 69.17% of pregnancies destinated to develop GDM at a 10.0% FPR (Figure 3).
Figure 3.
ROC analysis—the combination of 3 basic clinical characteristics (maternal age and BMI values at early stages of gestation and an infertility treatment by assisted reproductive technology) and 11 dysregulated microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-23a-3p, miR-100-5p, miR-125b-5p, miR-126-3p, miR-181a-5p, miR-195-5p, miR-499a-5p, and miR-574-3p). At a 10.0% FPR, 69.17% of pregnancies destinated to develop GDM were identified during the first trimester of gestation. This represents 84 out of 121 pregnancies correctly predicted to develop GDM and 8 out of 80 normal pregnancies predicted false positively to develop GDM.
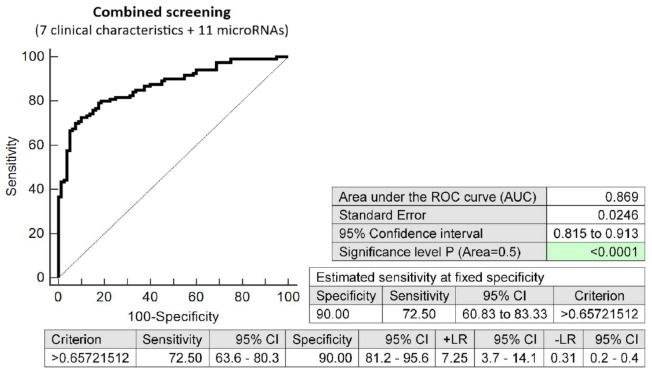
The screening based on the combination of seven clinical characteristics (maternal age and BMI at early stages of gestation, an infertility treatment by assisted reproductive technology, history of miscarriage, the presence of trombophilic gene mutations, positive first-trimester screening for preeclampsia and/or FGR by FMF algorithm, and family history of diabetes mellitus in first-degree relatives) and 11 dysregulated microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-23a-3p, miR-100-5p, miR-125b-5p, miR-126-3p, miR-181a-5p, miR-195-5p, miR-499a-5p, and miR-574-3p) showed the highest possible accuracy for the early identification of pregnancies destinated to develop GDM (AUC 0.869, p < 0.001, 72.50% sensitivity, 90.0% specificity, cut off >0.6572). This combined screening identified, in the early stages of gestation, 72.50% of pregnancies destinated to develop GDM at a 10.0% FPR (Figure 4).
Figure 4.
ROC analysis—the combination of 7 clinical characteristics (maternal age and BMI at early stages of gestation, an infertility treatment by assisted reproductive technology, history of miscarriage, the presence of trombophilic gene mutations, positive first-trimester screening for PE and/or FGR by FMF algorithm, and family history of diabetes mellitus in first-degree relatives) and 11 dysregulated microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-23a-3p, miR-100-5p, miR-125b-5p, miR-126-3p, miR-181a-5p, miR-195-5p, miR-499a-5p, and miR-574-3p). At a 10.0% FPR, 72.50% of pregnancies destinated to develop GDM were identified during the first trimester of gestation. This represents 88 out of 121 pregnancies correctly predicted to develop GDM and 8 out of 80 normal pregnancies predicted false positively to develop GDM.
2.5. Dysregulation of Cardiovascular Disease-Associated MicroRNAs in Pregnancies Destinated to Develop GDM with Respect to the Treatment Strategies (Diet Only and a Combination of Diet and Administration of Appropriate Therapy)
Concurrently, upregulation of miR-20a-5p (p = 0.0015 **, p = 0.0098 *), miR-20b-5p (p < 0.001 ***, p = 0.0054 **), and miR-195-5p (p < 0.001 ***, p < 0.001 ***) was observed in both groups of pregnancies destinated to develop GDM, irrespective of the treatment strategies (diet only or a combination of diet and therapy).
In addition, upregulation of miR-1-3p (p = 0.0045 *), miR-100-5p (p = 0.0010 **), miR-125b-5p (p = 0.0109 *), miR-499-5p (p = 0.0043 *), and miR-574-3p (p < 0.001 ***) was observed in only the group of pregnancies destinated to develop GDM, which was managed well by diet only (Supplementary Figure S2, Table 4).
Table 4.
MicroRNA expression profiles in peripheral blood leukocytes in early stages of gestation in pregnancies destinated to develop GDM with respect to the treatment strategies and normal term pregnancies.
| Kruskal–Wallis Test Results GDM Managed by Diet Only (n = 101) vs. Normal Term Pregnancies (n = 80) GDM Managed by Diet and Therapy (n = 20) vs. Normal Term Pregnancies (n = 80) |
|||
|---|---|---|---|
| Median (IQR) | Mean (SD) | p-Value | |
| miR-1-3p | 0.141 (0.075–0.274) vs. 0.075 (0.033–0.198) 0.099 (0.071–0.175) vs. 0.075 (0.033–0.198) |
0.278 (0.568) vs. 0.176 (0.303) 0.162 (0.190) vs. 0.176 (0.303) |
p = 0.0045 * p = 1.000 |
| miR-16-5p | 1.216 (0.981–1. 785) vs. 1.411 (0.890–1.980) 1.268 (0.923–2.007) vs. 1.411 (0.890–1.980) |
1.469 (0.976) vs. 1.646 (1.129) 1.626 (1.019) vs. 1.646 (1.129) |
p = 1.000 p = 1.000 |
| miR-17-5p | 1.480 (1.166–2.267) vs. 1.384 (0.971–1.923) 1.893 (1.346–2.362) vs. 1.384 (0.971–1.923) |
1.950 (1.553) vs. 1.748 (1.312) 2.085 (0.996) vs. 1.748 (1.312) |
p = 0.3822 p = 0.1019 |
| miR-20a-5p | 2.144 (1.486–3.398) vs. 1.576 (0.991–2.413) 2.598 (1.787–3.384) vs. 1.576 (0.991–2.413) |
3.019 (3.220) vs. 1.909 (1.370) 3.130 (2.204) vs. 1.909 (1.370) |
p = 0.0015 **
p = 0.0098 * |
| miR-20b-5p | 2.577 (1.784–3.719) vs. 1.976 (1.111–2.675) 3.072 (2.085–5.484) vs. 1.976 (1.111–2.675) |
3.678 (4.112) vs. 2.377 (2.291) 3.850 (2.439) vs. 2.377 (2.291) |
p < 0.001 ***
p = 0.0054 ** |
| miR-21-5p | 0.339 (0.222–0.460) vs. 0.320 (0.167–0.538) 0.352 (0.260–0.464) vs. 0.320 (0.167–0.538) |
0.426 (0.436) vs. 0.394 (0.219) 0.472 (0.332) vs. 0.394 (0.219) |
p = 1.000 p = 0.4483 |
| miR-23a-3p | 0.229 (0.160–0.444) vs. 0.185 (0.103–0.376) 0.299 (0.219–0.344) vs. 0.185 (0.103–0.376) |
0.364 (0.346) vs. 0.296 (0.329) 0.383 (0.293) vs. 0.296 (0.329) |
p = 0.0627 p = 0.0371 |
| miR-24-3p | 0.292 (0.222–0.370) vs. 0.326 (0.196–0.468) 0.301 (0.241–0.377) vs. 0.326 (0.196–0.468) |
0.330 (0.206) vs. 0.384 (0.284) 0.339 (0.147) vs. 0.384 (0.284) |
p = 1.000 p = 1.000 |
| miR-26a-5p | 0.729 (0.497–0.938) vs. 0.633 (0.410–1.066) 0.658 (0.560–0.917) vs. 0.633 (0.410–1.066) |
0.841 (0.705) vs. 0.776 (0.521) 0.815 (0.462) vs. 0.776 (0.521) |
p = 0.9599 p = 1.000 |
| miR-29a-3p | 0.404 (0.276–0.571) vs. 0.372 (0.221–0.545) 0.435 (0.358–0.666) vs. 0.372 (0.221–0.545) |
0.486 (0.377) vs. 0.407 (0.245) 0.630 (0.471) vs. 0.407 (0.245) |
p = 0.5656 p = 0.1198 |
| miR-92a-3p | 2.171 (1.604–3.036) vs. 2.327 (1.188–3.743) 2.258 (1.603–3.681) vs. 2.327 (1.188–3.743) |
2.647 (2.217) vs. 2.807 (2.132) 2.979 (2.3086) vs. 2.807 (2.132) |
p = 1.000 p = 1.000 |
| miR-100-5p | 0.0024 (0.0013–0.0036) vs. 0.0013 (0.0006–0.0027) 0.0014 (0.0012–0.0037) vs. 0.0013 (0.0006–0.0027) |
0.0031 (0.0041) vs. 0.0018 (0.0016) 0.0028 (0.0025) vs. 0.0018 (0.0016) |
p = 0.0010 ** p = 0.2898 |
| miR-103a-3p | 1.531 (0.949–2.533) vs. 1.203 (0.815–2.425) 1.618 (1.234–2.554) vs. 1.203 (0.815–2.425) |
2.085 (2.294) vs. 1.770 (1.466) 2.304 (2.075) vs. 1.770 (1.466) |
p = 0.7368 p = 0.4354 |
| miR-125b-5p | 0.0041 (0.0026–0.0057) vs. 0.0030 (0.0016–0.0054) 0.0038 (0.0021–0.0055) vs. 0.0030 (0.0016–0.0054) |
0.0050 (0.0048) vs. 0.0036 (0.0027) 0.0045 (0.0029) vs. 0.0036 (0.0027) |
p = 0.0109 * p = 0.4855 |
| miR-126-3p | 0.332 (0.219–0.500) vs. 0.272 (0.140–0.432) 0.324 (0.280–0.546) vs. 0.272 (0.140–0.432) |
0.470 (0.595) vs. 0.336 (0.270) 0.418 (0.228) vs. 0.336 (0.270) |
p = 0.0842 p = 0.1516 |
| miR-130b-3p | 0.707 (0.453–1.315) vs. 0.702 (0.407–1.157) 1.087 (0.577–1.481) vs. 0.702 (0.407–1.157) |
1.051 (0.995) vs. 1.163 (2.425) 1.194 (0.769) vs. 1.163 (2.425) |
p = 1.000 p = 0.1983 |
| miR-133a-3p | 0.118 (0.066–0.228) vs. 0.110 (0.550–0.233) 0.071 (0.055–0.105) vs. 0.110 (0.550–0.233) |
0.209 (0.283) vs. 0.232 (0.483) 0.113 (0.109) vs. 0.232 (0.483) |
p = 1.000 p = 0.4015 |
| miR-143-3p | 0.048 (0.029–0.087) vs. 0.038 (0.016–0.089) 0.049 (0.033–0.090) vs. 0.038 (0.016–0.089) |
0.072 (0.088) vs. 0.058 (0.057) 0.078 (0.077) vs. 0.058 (0.057) |
p = 0.1327 p = 0.2766 |
| miR-145-5p | 0.176 (0.122–0.235) vs. 0.161 (0.980–0.243) 0.171 (0.131–0.242) vs. 0.161 (0.980–0.243) |
0.210 (0.162) vs. 0.195 (0.143) 0.200 (0.100) vs. 0.195 (0.143) |
p = 0.6997 p = 1.000 |
| miR-146a-5p | 1.116 (0.800–1.798) vs. 1.225 (0.578–1.765) 1.451 (1.167–2.129) vs. 1.225 (0.578–1.765) |
1.634 (1.621) vs. 1.388 (1.096) 1.780 (1.068) vs. 1.388 (1.096) |
p = 0.8676 p = 0.1619 |
| miR-155-5p | 0.624 (0.432–0.820) vs. 0.607 (0.361–1.614) 0.566 (0.448–0.695) vs. 0.607 (0.361–1.614) |
0.701 (0.516) vs. 1.247 (1.439) 0.710 (0.573) vs. 1.247 (1.439) |
p = 1.000 p = 1.000 |
| miR-181a-5p | 0.246 (0.175–0.375) vs. 0.181 (0.141–0.330) 0.260 (0.190–0.393) vs. 0.181 (0.141–0.330) |
0.331 (0.336) vs. 0.246 (0.184) 0.326 (0.208) vs. 0.246 (0.184) |
p = 0.0399 p = 0.1367 |
| miR-195-5p | 0.269 (0.154–0.487) vs. 0.106 (0.048–0.271) 0.246 (0.210–0.522) vs. 0.106 (0.048–0.271) |
0.460 (0.707) vs. 0.227 (0.364) 0.520 (0.609) vs. 0.227 (0.364) |
p < 0.001 ***
p < 0.001 *** |
| miR-199a-5p | 0.073 (0.033–0.139) vs. 0.058 (0.023–0.111) 0.088 (0.052–0.163) vs. 0.058 (0.023–0.111) |
0.134 (0.233) vs. 0.096 (0.131) 0.148 (0.165) vs. 0.096 (0.131) |
p = 0.1575 p = 0.1701 |
| miR-210-3p | 0.102 (0.074–0.154) vs. 0.138 (0.075–0.224) 0.099 (0.075–0.155) vs. 0.138 (0.075–0.224) |
0.134 (0.109) vs. 0.186 (0.180) 0.131 (0.080) vs. 0.186 (0.180) |
p = 0.2982 p = 1.000 |
| miR-221-3p | 0.644 (0.448–0.948) vs. 0.548 (0.293–0.906) 0.616 (0.459–1.032) vs. 0.548 (0.293–0.906) |
0.819 (0.776) vs. 0.693 (0.561) 0.796 (0.503) vs. 0.693 (0.561) |
p = 0.3698 p = 0.7241 |
| miR-342-3p | 3.093 (2.070–3.955) vs. 2.542 (1.551–4.206) 2.884 (2.159–4.844) vs. 2.542 (1.551–4.206) |
3.555 (2.756) vs. 3.307 (2.383) 3.858 (2.610) vs. 3.307 (2.383) |
p = 0.6912 p = 1.000 |
| miR-499a-5p | 0.459 (0.218–0.881) vs. 0.269 (0.089–0.587) 0.472 (0.285–0.611) vs. 0.269 (0.089–0.587) |
0.771 (1.104) vs. 0.477 (0.566) 0.692 (0.902) vs. 0.477 (0.566) |
p= 0.0043 * p = 0.1765 |
| miR-574-3p | 0.275 (0.182–0.392) vs. 0.181 (0.117–0.292) 0.279 (0.178–0.485) vs. 0.181 (0.117–0.292) |
0.350 (0.339) vs. 0.222 (0.156) 0.375 (0.301) vs. 0.222 (0.156) |
p < 0.001 *** p = 0.0356 |
MicroRNA gene expression is compared between individual groups using Kruskal–Wallis test. Statistically significant results are marked in bold. Median (interquartile range, IQR) and mean (standard deviation, SD) values of relative fold gene expression of samples (2−∆∆Ct) are presented. Statistical significant data after Benjamini–Hochberg correction are marked by * for α = 0.05, ** for α = 0.01, and *** for α = 0.001.
Sensitivities at a 10.0% FPR were reported for miR-20a-5p (21.78%, 20.0%), miR-20b-5p (15.84%, 30.0%), and miR-195-5p (18.81%, 25.0%) in pregnancies destinated to develop GDM requiring management by diet only or a combination of diet and administration of appropriate therapy.
Sensitivities at a 10.0% FPR were reported for miR-1-3p (13.86%), miR-100-5p (19.80%), miR-125b-5p (14.85%), miR-499a-5p (15.84%), and miR-574-3p (21.78%) in pregnancies destinated to develop GDM requiring diet only (Supplementary Figure S2).
2.6. First-Trimester Combined MicroRNA Screening Is Able to Differentiate between Pregnancies Destinated to Develop GDM Requiring a Combination of Diet and Administration of Appropriate Therapy and Term Pregnancies with Normal Course of Gestation
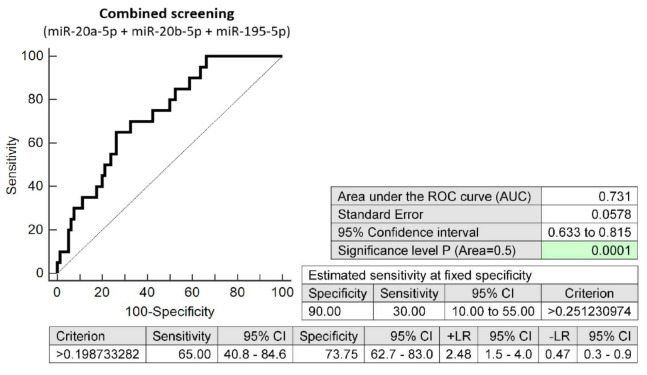
The combined screening of three microRNA biomarkers (miR-20a-5p, miR-20b-5p and miR-195-5p) in early stages of gestation was able to detect aberrant microRNA expression profile in 30.0% pregnancies destinated to develop GDM requiring a combination of diet and administration of appropriate therapy at a 10.0% FPR (AUC 0.731, p < 0.001, 65.0% sensitivity, 73.75% specificity, cut off >0.1987) (Figure 5).
Figure 5.
ROC analysis—the combination of microRNA biomarkers (miR-20a-5p, miR-20b-5p and miR-195-5p). A total of 30.0% pregnancies destinated to develop GDM requiring a combination of diet and administration of appropriate therapy had aberrant microRNA expression profile in the whole peripheral venous blood during the first trimester of gestation at a 10.0% FPR. This represents 6 out of 20 pregnancies correctly predicted to develop GDM and 8 out of 80 normal pregnancies predicted false positively to develop GDM.
2.7. The Very High Accuracy of First-Trimester Combined Screening (MicroRNA Biomarkers and Selected Clinical Characteristics) to Differentiate between Pregnancies Destinated to Develop GDM Requiring a Combination of Diet and Administration of Appropriate Therapy and Term Pregnancies with Normal Course of Gestation
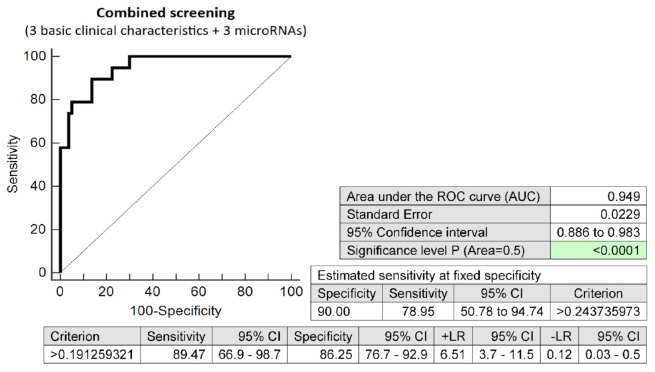
The effective screening based on the combination of minimal number of basic clinical characteristics (maternal age and BMI at early stages of gestation, and an infertility treatment by assisted reproductive technology) and three dysregulated microRNA biomarkers (miR-20a-5p, miR-20b-5p, and miR-195-5p) showed very high accuracy for the early identification of pregnancies destinated to develop GDM requiring a combination of diet and administration of appropriate therapy (AUC 0.949, p < 0.001, 89.47% sensitivity, 86.25% specificity, cut off >0.1912). The screening identified 78.95% of cases at a 10.0% FPR in the early stages of gestation (Figure 6).
Figure 6.
ROC analysis—the combination of 3 basic clinical characteristics (maternal age and BMI values at early stages of gestation and an infertility treatment by assisted reproductive technology) and 3 dysregulated microRNA biomarkers (miR-20a-5p, miR-20b-5p, and miR-195-5p). At a 10.0% FPR, 78.95% pregnancies destinated to develop GDM requiring a combination of diet and administration of appropriate therapy were identified during the first trimester of gestation. This represents 16 out of 20 pregnancies correctly predicted to develop GDM and 8 out of 80 normal pregnancies predicted false positively to develop GDM.
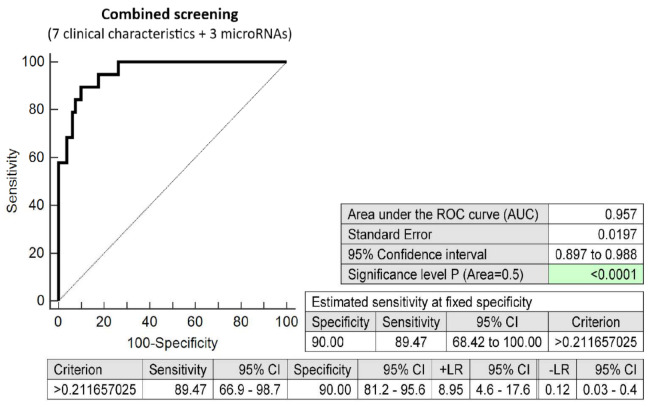
The screening based on the combination of seven clinical characteristics (maternal age and BMI at early stages of gestation, an infertility treatment by assisted reproductive technology, history of miscarriage, the presence of trombophilic gene mutations, positive first-trimester screening for preeclampsia and/or FGR by FMF algorithm, family history of diabetes mellitus in first-degree relatives) and three dysregulated microRNA biomarkers (miR-20a-5p, miR-20b-5p, and miR-195-5p) showed the highest possible accuracy for the early identification of pregnancies destinated to develop GDM requiring a combination of diet and administration of appropriate therapy (AUC 0.957, p < 0.001, 89.47% sensitivity, 90.0% specificity, cutoff >0.2116). This screen identified 89.47% of cases in the early stages of gestation at a 10.0% FPR (Figure 7).
Figure 7.
ROC analysis—the combination of 7 clinical characteristics (maternal age and BMI at early stages of gestation, an infertility treatment by assisted reproductive technology, history of miscarriage, the presence of trombophilic gene mutations, positive first-trimester screening for PE and/or FGR by FMF algorithm, and a family history of diabetes mellitus in first-degree relatives) and 3 dysregulated microRNA biomarkers (miR-20a-5p, miR-20b-5p, and miR-195-5p). At a 10.0% FPR, 89.47% pregnancies destinated to develop GDM requiring a combination of diet and administration of appropriate therapy were identified during the first trimester of gestation. This represents 18 out of 20 pregnancies correctly predicted to develop GDM and 8 out of 80 normal pregnancies predicted false positively to develop GDM.
2.8. First-Trimester Combined MicroRNA Screening Is Able to Differentiate between Pregnancies Destinated to Develop GDM Managed by Diet Only and Normal Term Pregnancies
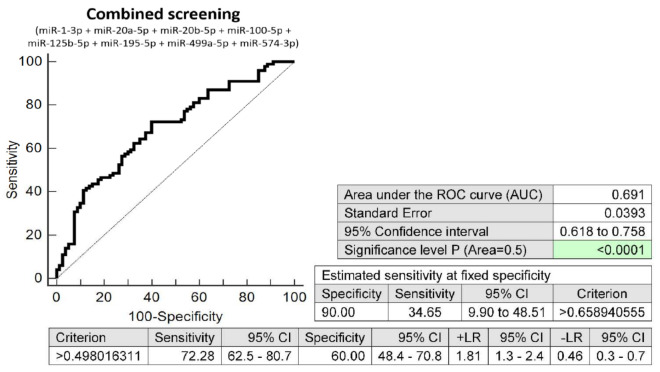
The combined screening of eight microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-100-5p, miR-125b-5p, miR-195-5p, miR-499a-5p, and miR-574-3p) was able to detect, in the early stages of gestation, an aberrant microRNA expression profile in 34.65% of pregnancies destinated to develop GDM managed by diet only at a 10.0% FPR (AUC 0.691, p < 0.001, 72.28% sensitivity, 60.0% specificity, cut off >0.4980) (Figure 8).
Figure 8.
ROC analysis—the combination of microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-100-5p, miR-125b-5p, miR-195-5p, miR-499a-5p, and miR-574-3p). A total of 34.65% pregnancies destinated to develop GDM on diet only had an aberrant microRNA expression profile in the whole peripheral venous blood during the first trimester of gestation at a 10.0% FPR. This represents 35 out of 101 pregnancies correctly predicted to develop GDM and 8 out of 80 normal pregnancies predicted false positively to develop GDM.
2.9. The Very Good Accuracy of First-Trimester Combined Screening (MicroRNA Biomarkers and Selected Clinical Characteristics) to Differentiate between Pregnancies Destinated to Develop GDM Managed by Diet Only and Term Pregnancies with Normal Course of Gestation
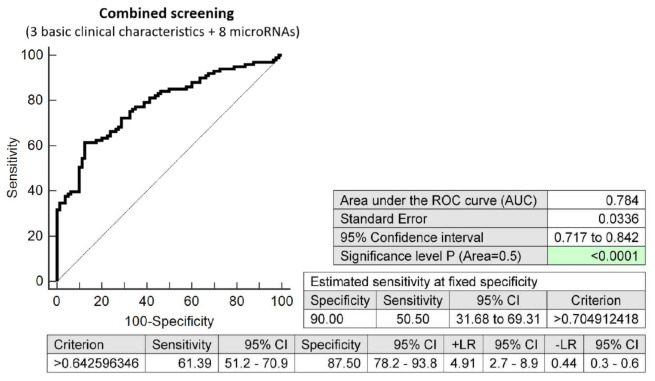
The effective screening based on the combination of a minimal number of basic clinical characteristics (maternal age and BMI at early stages of gestation and an infertility treatment by assisted reproductive technology) and eight dysregulated microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-100-5p, miR-125b-5p miR-195-5p, miR-499a-5p, and miR-574-3p) showed relatively good accuracy for the early identification of pregnancies destinated to develop GDM managed by diet only (AUC 0.784, p < 0.001, 61.39 sensitivity, 87.50% specificity, cut off >0.6425). This screening identified 50.50% of cases during the early stages of gestation at a 10.0% FPR (Figure 9).
Figure 9.
ROC analysis—the combination of 3 basic clinical characteristics (maternal age and BMI values at early stages of gestation and an infertility treatment by assisted reproductive technology) and 8 dysregulated microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-100-5p, miR-125b-5p, miR-195-5p, miR-499a-5p, and miR-574-3p). At a 10.0% FPR, 50.50% pregnancies destinated to develop GDM managed by diet only were identified during the first trimester of gestation. This represents 51 out of 101 pregnancies correctly predicted to develop GDM and 8 out of 80 normal pregnancies predicted false positively to develop GDM.
The screening based on the combination of seven clinical characteristics (maternal age and BMI at early stages of gestation, an infertility treatment by assisted reproductive technology, history of miscarriage, the presence of trombophilic gene mutations, positive first-trimester screening for preeclampsia and/or FGR by FMF algorithm, and family history of diabetes mellitus in first-degree relatives) and eight dysregulated microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-100-5p, miR-125b-5p, miR-195-5p, miR-499a-5p, and miR-574-3p) showed the highest possible accuracy for the early identification of pregnancies destinated to develop GDM managed by diet only (AUC 0.835, p < 0.001, 77.23% sensitivity, 78.75% specificity, cut off >0.5137. This combined screening identified, in the early stages of gestation, 56.44% of pregnancies destinated to develop GDM managed by diet only at a 10.0% FPR (Figure 10).
Figure 10.
ROC analysis—the combination of 7 clinical characteristics (maternal age and BMI at early stages of gestation, an infertility treatment by assisted reproductive technology, history of miscarriage, the presence of trombophilic gene mutations, positive first-trimester screening for PE and/or FGR by FMF algorithm, and family history of diabetes mellitus in first-degree relatives) and 8 dysregulated microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-100-5p, miR-125b-5p, miR-195-5p, miR-499a-5p, and miR-574-3p). At a 10.0% FPR, 56.44% of pregnancies destinated to develop GDM managed by diet only were identified during the first trimester of gestation. This represents 57 out of 101 pregnancies correctly predicted to develop GDM and 8 out of 80 normal pregnancies predicted false positively to develop GDM.
2.10. Information on MicroRNA-Gene-Biological Pathways Interactions
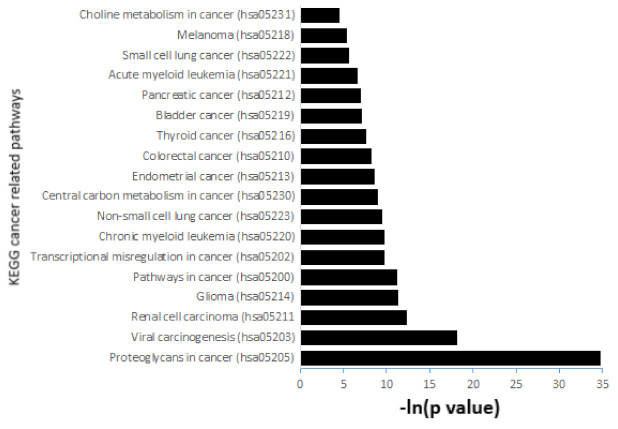
The KEGG pathway enrichment analysis of 11 microRNAs dysregulated in early stages of gestation in pregnancies destinated to develop GDM revealed a total of 62 pathways, where at least 18 (29.03%) pathways were cancer related. The cancer-related pathways with the highest −ln(p-values) were proteoglycans in cancer (hsa05205; 34.738), viral carcinogenesis (hsa05203; 18.144), renal cell carcinoma (hsa05211; 12.364), glioma (hsa05214; 11.400), and pathways in cancer (hsa05200; 11.269).
Other cancer-related pathways showed slightly lower −ln(p-values): transcriptional misregulation in cancer (hsa05202; 9.818), chronic myeloid leukaemia (hsa05220; 9.818), non-small cell lung cancer (hsa05223; 9.492), central carbon metabolism in cancer (hsa05230; 9.047), endometrial cancer (hsa05213; 8.698), colorectal cancer (hsa05210; 8.296), thyroid cancer (hsa05216; 7.630), bladder cancer (hsa05219; 7.099), pancreatic cancer (hsa05212; 6.996), acute myeloid leukaemia (hsa05221; 6.648), small cell lung cancer (hsa05222; 5.661), melanoma (hsa05218; 5.424), and choline metabolism in cancer (hsa05231; 4.536) (Figure 11).
Figure 11.
The KEGG pathway enrichment analysis of 11 microRNAs dysregulated in early pregnancies destinated to develop GDM. The analysis revealed a total of 62 pathways, where at least 18 (29.03%) pathways were cancer related. The results were expressed as –ln of the p-value (−ln(p-value)).
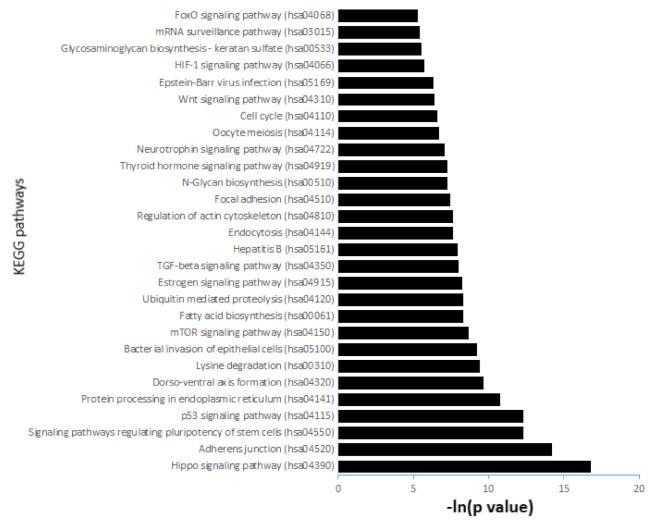
The other pathways with the highest −ln(p-values) have been shown to play a role in physiological processes and besides the pathogenesis of cancer. These are Hippo signalling pathway (hsa04390; 16.800), adherens junction (hsa04520; 14.198), signalling pathways regulating pluripotency of stem cells (hsa04550; 12.276), p53 signalling pathway (hsa04115; 12.276), and protein processing in endoplasmatic reticulum (hsa04141; 10.769) (Figure 12).
Figure 12.
The KEGG pathway enrichment analysis of 11 microRNAs dysregulated in early pregnancies destinated to develop GDM. The analysis revealed a total of 62 various pathways, where a majority of pathways have been shown to play a role in physiological processes and besides to the pathogenesis of cancer. The results were expressed as –ln of the p-value (−ln(p-value)).
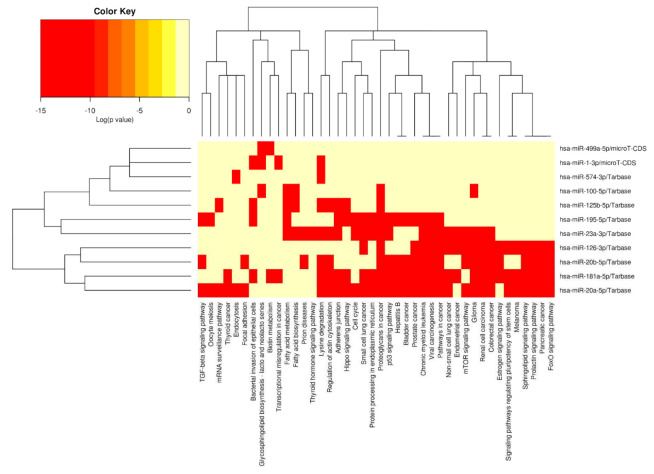
The microRNA/KEGG pathway heatmap and hierarchical clustering demonstrated the level of involvement of particular microRNAs in various biological pathways (Figure 13).
Figure 13.
The microRNA/KEGG pathway heatmap and hierarchical clustering in early pregnancies destinated to develop GDM. The heatmap represents the level of involvement of particular microRNAs in various biological pathways. The results were expressed as log of the p-value (log(p-value)).
3. Discussion
Gene expression of 29 preselected cardiovascular disease-associated microRNAs was compared between pregnancies destinated to develop GDM and normal term pregnancies in the whole peripheral venous blood during the first trimester of gestation. The study was held within the framework of routine screening to assess the risk for a wide array of major fetal chromosomal and non-chromosomal defects as well as other pregnancy-related complications such as PE and/or FGR.
Upregulation of 11 cardiovascular disease-associated microRNAs (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-23a-3p, miR-100-5p, miR-125b-5p, miR-126-3p, miR-181a-5p, miR-195-5p, miR-499a-5p, and miR-574-3p) was detected during the early stages of gestation in the entire group of pregnancies destinated to develop GDM.
To our knowledge, several studies have reported promising data on the early diagnosis of GDM during the first trimester of gestation via screening of circulating microRNAs in maternal plasma/serum or peripheral blood samples. Our study produced similar findings to Yoffe et al. [226], Lamadrid-Romero et al. [130], and Legare et al. [227].
Yoffe et al. validated two upregulated microRNAs (miR-23a and miR-223) as potential plasma biomarkers for early prediction of GDM (after the ninth gestational week and before completion of the 12th week of gestation) in women diagnosed with GDM via a 75 g OGTT performed at 22–24 weeks of gestation [226].
The study of Lamadrid-Romero et al. [130] reported higher miR-125b-5p expression levels in first-trimester serum samples in GDM pregnancies when compared with the control group. On the other hand, the study of Zhang et al. [229] reported downregulation of miR-125b in circulating plasma exosomes in patients with confirmed diagnosis of GDM within 26–40 weeks of pregnancy. Nevertheless, microRNA expression profile may differ between free circulating microRNAs and circulating exosomes; therefore, these findings are not necessarily contradictory results.
Our data may also support the data presented by Tagoma et al. [190], who observed upregulation of miR-100-5p and miR-195-5p in maternal plasma samples collected during the late second and early third pregnancy trimesters in patients who had a positive glucose tolerance test between 23 and 31 weeks of gestation, in which case miR-195-5p showed the highest fold upregulation, similar to our first-trimester study. Our data and the data of Tagoma et al. [190] are also consistent with the data of Wang et al. [230], who also observed increased expression levels of miR-195-5p in serum samples of GDM patients at 25 weeks of gestation.
Concerning miR-20a-5p, our first-trimester data may support the data of Zhu et al. [51] and Cao et al. [52]. Zhu et al. [51] observed upregulation of miR-20a-5p in peripheral blood samples of women at 16–19 weeks of pregnancy, whereas GDM was diagnosed via a 50 g glucose challenge test at 24–28 weeks of pregnancy. Cao et al. [52] observed upregulation of miR-20a-5p in plasma samples derived from patients at the time of diagnosis of GDM determined at 24–28 gestational weeks via performance of 50 g glucose challenge test and 75 g OGTT test.
Nevertheless, our data are inconsistent with the results of other researchers concerning miR-16-5p and miR-17-5p [51,52,228,231]. While in our study, first-trimester whole peripheral blood levels did not differ between pregnancies destinated to develop GDM and control groups, the expression levels of miR-16-5p and miR-17-5p have been reported to be significantly increased in patients with a diagnosis of GDM confirmed at 24–28 gestational weeks [52]. Similarly, Zhu et al. [51], Sorensen et al. [231], and Juchnicka et al. [228] presented similar findings to Cao et al. [52]. Zhu et al. [5] was able to observe upregulation of miR-16-5p and miR-17-5p in peripheral blood samples of women with subsequent onset of GDM at 16–19 weeks of pregnancy. Similarly, Sorensen et al. [231] observed elevated serum levels of miR-16-5p even in the earlier stages of gestation (mean 15th gestational week) in women destinated to develop GDM. Juchnicka et al. [228] showed upregulation of miR-16-5p in first-trimester serum samples of normoglycemic women that developed GDM within the 24–26 gestational weeks.
In addition, Zhao et al. [232] and Sorensen et al. [231] identified miR-29a and miR-29a-3p as other potentially predictive circulating GDM biomarkers. Unfortunately, they did not show any dysregulation when first-trimester expression levels were compared between pregnancies destinated to develop GDM and the control group in our study.
Parallelly, our data concerning miR-155-5p are inconsistent with the study of Wander et al. [112], who observed a positive association between early–mid-pregnancy plasma miR-155-5p levels and occurrence of GDM.
With regard to miR-1-3p, our study produced supportive findings to the study of Kennedy et al. [233], in which they reported increased levels of miR-1-3p in serum extracellular vesicles in patients with confirmed GDM diagnoses within 26–28 gestational weeks that subsequently delivered large-for-gestational-age new-borns (LGA) when compared with appropriately grown-for-gestational-age new-borns (AGA). Nevertheless, our data concerning miR-133a-3p and miR-145-5p are inconsistent with the study of Kennedy et al. [233]. While they observed reduced levels of miR-145-5p and increased levels of miR-133a-3p in GDM pregnancies delivering LGA new-borns, we did not detect any changes in the gene expression of miR-133a-3p and miR-145-5p during the early stages of gestation in pregnancies destinated to develop GDM.
Similarly, our data concerning miR-143-3p and miR-221-3p did not confirm the data of Legare et al. [227], that implemented these first-trimester dysregulated plasmatic microRNAs into the Lasso regression model for prediction of insulin sensitivity estimated by the Matsuda index at the end of the second trimester of pregnancy. However, our data concerning miR-100-5p concurred with Legare et al. [227], who also observed increased levels of miR-100-5p in plasma samples in the early stages of gestation in pregnancies that subsequently developed GDM.
In addition, other studies have introduced a whole range of other circulating microRNAs which were not subject of interest in our study as biomarkers with predictive or diagnostic potential for GDM. These are the following: let-7b-3p [227], miR-10b-5p [227], miR-16-1-3p [227] miR-19a and miR-19b [234], miR-21-3p [53,112], miR-33a-5p [235], miR-130a-3p [227], miR-132 [232], miR-134-5p [231], miR-141-3p [227], miR-142-3p [228], miR-144 [229], miR-144-3p [228], miR-200a-3p [227], miR-205-5p [227], miR-215-5p [227], miR-218-5p [227], miR-222 [232], miR-330-3p [236], miR-338-3p [227], miR-340 [237], miR-375 [227], miR-429 [227], miR-483-5p [227], miR-499a-3p [233], miR-503 [238], miR-512-3p [227], miR-515-5p [227], miR-516a-5p [227], miR-516b-5p [227], miR-517a-3p [227], miR-517b-3p [227], miR-518e-3p [227], miR-518e-5p [227], miR-519a-5p [227], miR-519b-5p [227], miR-519c-5p [227], miR-519d-5p [227], miR-520a-3p [227], miR-520d-3p [227], miR-522-5p [227], miR-523-5p [227], miR-524-3p [227], miR-582-5p [227], miR-873-5p [227], miR-877-5p [227], miR-1283 [227], miR-1323 [239], miR-2116-3p [227], miR-3183 [227], and miR-4772-5p [227].
The current study revealed that aberrant gene expression of miR-1-3p, miR-20a-5p, miR-20b-5p, miR-23a-3p, miR-100-5p, miR-125b-5p, miR-126-3p, miR-181a-5p, miR-195-5p, miR-499a-5p, and miR-574-3p expression is present during the early stages of gestation in pregnancies destinated to develop GDM.
During the first trimester of gestation, we have also recently observed an aberrant expression profile of these cardiovascular disease-associated microRNAs in pregnancies with chronic hypertension (miR-1-3p, miR-20a-5p, and miR-126-3p) and in normotensive pregnancies with subsequent onset of PE (miR-20a-5p, miR-126-3p, miR-181a-5p, and miR-574-3p), FGR (miR-20a-5p, miR-100-5p, miR-181a-5p, miR-195-5p, and miR-574-3p), SGA (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-126-3p, miR-181a-5p, and miR-499a-5p), and/or preterm delivery (miR-20b-5p) [240,241,242].
Parallelly, not long ago we observed the upregulation of 11 microRNAs (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-23a-3p, miR-100-5p, miR-125b-5p, miR-126-3p, miR-181a-5p, miR-195-5p, miR-499a-5p, and miR-574-3p) in the whole peripheral blood samples of mothers with a history of GDM [243]. At the same time, the upregulation of multiple other cardiovascular disease-associated microRNAs (miR-16-5p, miR-17-5p, miR-21-5p, miR-24-3p, miR-26a-5p, miR-29a-3p, miR-103a-3p, miR-130b-3p, miR-133a-3p, miR-143-3p, miR-145-5p, miR-146a-5p, miR-199a-5p, miR-221-3p, and miR-342-3p) was identified postpartum in mothers with a history of GDM [243], which had not yet been present in the early stages of gestation, and probably appeared later with the onset of GDM.
Existing data suggest that dysregulated microRNAs in early pregnancies destinated to develop GDM play a role, not only in the pathogenesis of cardiovascular and cerebrovascular diseases, but also in the pathogenesis of cancer. Since women with a history of GDM were reported to have a higher risk of developing both cardiovascular diseases [244,245,246,247,248] and cancer [249,250,251,252,253,254,255,256], cardiovascular risk assessment [243] together with cancer screening [249] should be implemented into the routine preventive programmes of women with a previous occurrence of GDM.
4. Materials and Methods
4.1. Patients Cohort
Within the framework of the retrospective case-control study held at the Institute for the Care of Mother and Child, Prague, Czech Republic, within the period 11/2012–5/2018, the whole peripheral venous blood samples were collected at 10–13 gestational weeks from a total of 4187 singleton pregnancies of Caucasian descent. Finally, 3028 out of 4187 pregnancies had complete medical records from the first trimester of gestation until the time of delivery. Out of these 3028 pregnancies, 121 women were consecutively confirmed to only have GDM, where 101 GDM pregnancies were managed by diet only and 20 GDM pregnancies were managed by the combination of diet and therapy (15 patients required insulin administration and metformin was prescribed for 5 patients). GDM was rarely diagnosed during the first trimester of gestation—only in four patients. Otherwise, the onset of GDM was confirmed in majority of patients (n = 117) within 24–28 gestational weeks.
Gestational diabetes mellitus was defined as any degree of glucose intolerance with the first onset during gestation [2,3,257]. The International Association of Diabetes and Pregnancy Study Groups’ (IADPSG) recommendations on the diagnosis and classification of hyperglycaemia in pregnancy were followed, and universal early testing was performed in all pregnancies [2]. The first screening phase, during the first trimester of gestation, detected patients with overt diabetes (fasting plasma glucose level ≥ 7.0 mmol/L) and patients with GDM (fasting plasma glucose level ≥ 5.1 mmol/L–<7.0 mmol/L). The second screening phase, 2 h 75 g OGTT at 24–28 weeks of gestation, was performed for all patients not previously found to have overt diabetes or GDM and identified GDM if fasting plasma glucose level was ≥5.1 mmol/L, 1 h plasma glucose was ≥10.0 mmol/L, or 2 h plasma glucose was ≥8.5 mmol/L [2].
Patients newly diagnosed with diabetes mellitus, patients with the occurrence of chronic hypertension, and those carrying growth-restricted or small-for-gestational-age fetuses, or fetuses with anomalies or chromosomal abnormalities were intentionally excluded from the study. Likewise, patients concurrently demonstrating other pregnancy-related complications such as gestational hypertension, preeclampsia, HELLP syndrome, in utero infections, spontaneous preterm birth, preterm prelabour rupture of membranes, fetal demise in utero, or stillbirth were also excluded from the study.
The control group was selected with regard to the uniformity of gestational age at sampling and storage times of biological samples, and included 80 women with normal courses of gestation that delivered healthy infants after the completion of 37 weeks of gestation with a weight above 2500 g.
No woman had a history of any cardiovascular disease (a positive anamnesis of cardiac remodelling, cardiac hypertrophy, heart failure, or acute myocardial infarction). All pregnant women had normal clinical findings (electrocardiography and echocardiography).
4.2. Processing of Samples
Homogenized leukocyte lysates were prepared from 200 µL maternal whole peripheral venous blood samples immediately after collection using a QIAamp RNA Blood Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Firstly, lysis of erythrocytes was performed using EL buffer. Then, the pelleted leukocytes were stored in a mixture of RLT buffer and β-mercaptoethanol (β-ME) at −80 °C for several months until further processing.
Subsequently, a mirVana microRNA Isolation kit (Ambion, Austin, TX, USA) was used to isolate the RNA fraction highly enriched for small RNAs from whole peripheral blood leukocyte lysates.
Concentration and quality of RNA was assessed using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The A(260/280) absorbance ratio of isolated RNA samples was 1.8–2.1, demonstrating that the RNA samples were pure and could be used for further analysis. The concentration of the isolated RNA ranged within 2.0–10.0 ng/μL.
Real-time RT-PCR analyses were performed regularly every six months to process the collection of frozen samples derived from GDM and normal term pregnancies. The gene expression levels of 29 cardiovascular disease-associated microRNAs (miR-1-3p, miR-16-5p, miR-17-5p, miR-20a-5p, miR-20b-5p, miR-21-5p, miR-23a-3p, miR-24-3p, miR-26a-5p, miR-29a-3p, miR-92a-3p, miR-100-5p, miR-103a-3p, miR-125b-5p, miR-126-3p, miR-130b-3p, miR-133a-3p, miR-143-3p, miR-145-5p, miR-146-5p, miR-155-5p, miR-181a-5p, miR-195-5p, miR-199a-5p, miR-210-3p, miR-221-3p, miR-342-3p, miR-499a-5p, and miR-574-3p) (Table 5) was determined.
Table 5.
Characteristics of microRNAs involved in the study.
| Assay Name | ID | NCBI Location Chromosome | Sequence |
|---|---|---|---|
| hsa-miR-1 | hsa-miR-1-3p | Chr.20: 62554306–62554376 [+] | 5′-UGGAAUGUAAAGAAGUAUGUAU-3′ |
| hsa-miR-16 | hsa-miR-16-5p | Chr.13: 50048973–50049061 [−] | 5′-UAGCAGCACGUAAAUAUUGGCG- 3′ |
| hsa-miR-17 | hsa-miR-17-5p | Chr.13: 91350605–91350688 [+] | 5′-CAAAGUGCUUACAGUGCAGGUAG-3′ |
| hsa-miR-20a | hsa-miR-20a-5p | Chr.13: 91351065–91351135 [+] | 5′-UAAAGUGCUUAUAGUGCAGGUAG-3′ |
| hsa-miR-20b | hsa-miR-20b-5p | Chr.X: 134169809–134169877 [−] | 5′-CAAAGUGCUCAUAGUGCAGGUAG-3′ |
| hsa-miR-21 | hsa-miR-21-5p | Chr.17: 59841266–59841337 [+] | 5′-UAGCUUAUCAGACUGAUGUUGA-3′ |
| hsa-miR-23a | hsa-miR-23a-3p | Chr.19: 13836587–13836659 [−] | 5′-AUCACAUUGCCAGGGAUUUCC-3′ |
| hsa-miR-24 | hsa-miR-24-3p | Chr.9: 95086021–95086088 [+] | 5′-UGGCUCAGUUCAGCAGGAACAG-3′ |
| hsa-miR-26a | hsa-miR-26a-5p | Chr.3: 37969404–37969480 [+] | 5′-UUCAAGUAAUCCAGGAUAGGCU-3′ |
| hsa-miR-29a | hsa-miR-29a-3p | Chr.7: 130876747–130876810 [−] | 5′-UAGCACCAUCUGAAAUCGGUUA-3′ |
| hsa-miR-92a | hsa-miR-92a-3p | Chr.13: 91351314–91351391 [+] | 5′-UAUUGCACUUGUCCCGGCCUGU-3′ |
| hsa-miR-100 | hsa-miR-100-5p | Chr.11: 122152229–122152308 [−] | 5′-AACCCGUAGAUCCGAACUUGUG-3′ |
| hsa-miR-103 | hsa-miR-103a-3p | Chr.5: 168560896–168560973 [−] | 5′-AGCAGCAUUGUACAGGGCUAUGA-3′ |
| hsa-miR-125b | hsa-miR-125b-5p | Chr.11: 122099757–122099844 [−] | 5′-UCCCUGAGACCCUAACUUGUGA-3′ |
| hsa-miR-126 | hsa-miR-126-3p | Chr.9: 136670602–136670686 [+] | 5′-UCGUACCGUGAGUAAUAAUGCG-3′ |
| hsa-miR-130b | hsa-miR-130b-3p | Chr.22: 21653304–21653385 [+] | 5′-CAGUGCAAUGAUGAAAGGGCAU-3′ |
| hsa-miR-133a | hsa-miR-133a-3p | Chr.18: 21825698–21825785 [−] | 5′-UUUGGUCCCCUUCAACCAGCUG-3′ |
| hsa-miR-143 | hsa-miR-143-3p | Chr.5: 149428918–149429023 [+] | 5′-UGAGAUGAAGCACUGUAGCUC-3′ |
| hsa-miR-145 | hsa-miR-145-5p | Chr.5: 149430646–149430733 [+] | 5′-GUCCAGUUUUCCCAGGAAUCCCU-3′ |
| hsa-miR-146a | hsa-miR-146a-5p | Chr.5: 160485352–160485450 [+] | 5′-UGAGAACUGAAUUCCAUGGGUU-3′ |
| hsa-miR-155 | hsa-miR-155-5p | Chr.21: 25573980–25574044 [+] | 5′-UUAAUGCUAAUCGUGAUAGGGGU-3′ |
| hsa-miR-181a | hsa-miR-181a-5p | Chr.1: 198859044–198859153 [−] | 5′-AACAUUCAACGCUGUCGGUGAGU-3′ |
| hsa-miR-195 | hsa-miR-195-5p | Chr.17: 7017615–7017701 [−] | 5′-UAGCAGCACAGAAAUAUUGGC-3′ |
| hsa-miR-199a | hsa-miR-199a-5p | Chr.19: 10817426–10817496 [−] | 5′-CCCAGUGUUCAGACUACCUGUUC-3′ |
| hsa-miR-210 | hsa-miR-210-3p | Chr.11: 568089–568198 [−] | 5′-CUGUGCGUGUGACAGCGGCUGA-3′ |
| hsa-miR-221 | hsa-miR-221-3p | Chr.X: 45746157–45746266 [−] | 5′-AGCUACAUUGUCUGCUGGGUUUC-3′ |
| hsa-miR-342-3p | hsa-miR-342-3p | Chr.14: 100109655–100109753 [+] | 5′-UCUCACACAGAAAUCGCACCCGU-3′ |
| mmu-miR-499 | hsa-miR-499a-5p | Chr.20: 34990376–34990497 [+] | 5′-UUAAGACUUGCAGUGAUGUUU-3′ |
| hsa-miR-574-3p | hsa-miR-574-3p | Chr.4: 38868032–38868127 [+] | 5′-CACGCUCAUGCACACACCCACA-3′ |
| RNU58A | 664243 | Chr.18: 49491283–49491347 [−] | 5′-CTGCAGTGATGACTTTCTTGGGACACCTTTGGA TTTACCGTGAAAATTAATAAATTCTGAGCAGC-3′ |
| RNU38B | 568914 | Chr.1: 44778390–44778458 [+] | 5′-CCAGTTCTGCTACTGACAGTAAGTGAAGATAA AGTGTGTCTGAGGAGA-3′ |
mRNAs of the appropriate microRNAs were reverse transcribed into cDNA using a TaqMan MicroRNA assays containing miRNA-specific stem loop primers and a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Branchburg, NJ, USA). The total reaction volumes were 10 µL. Furthermore, 3 µL of cDNA was mixed with the components of TaqMan MicroRNA assays (specific primers and the TaqMan MGB probes) and the components of the TaqMan Universal PCR Master Mix (Applied Biosystems, Branchburg, NJ, USA). The total reaction volumes were 15 µL. Reverse transcription and real-time qPCR were performed on a 7500 Real-Time PCR System using the TaqMan PCR conditions described in the TaqMan guidelines. The reverse transcription thermal cycling parameters were the following: 30 min at 16 °C, 30 min at 42 °C, 5 min at 85 °C, and then held at 4 °C. The real-time qPCR thermal cycling parameters were the following: 2 min at 50 °C, 10 min at 95 °C, then 50 cycles at 95 °C for 15 s, and 60 °C for 1 min.
Assessment of microRNA gene expression was performed using the comparative Ct method [258]. The geometric mean of the Ct values of selected endogenous controls (RNU58A and RNU38B) was used as a normalization factor [259] to normalize microRNA gene expression. Selection and validation of endogenous controls for microRNA expression studies in whole peripheral blood samples affected by pregnancy-related complications has already been described in one of our previous studies [260]. In brief, the expression of 20 candidate endogenous controls (HY3, RNU6B, RNU19, RNU24, RNU38B, RNU43, RNU44, RNU48, RNU49, RNU58A, RNU58B, RNU66, RPL21, U6 snRNA, U18, U47, U54, U75, Z30, and cel-miR-39) was investigated using NormFinder (NormFinder v.5, Aarhus University Hospital, Aarhus, Denmark) [261]. RNU58A and RNU38B were identified as the most stable small nucleolar RNAs (ncRNA) and equally expressed in patients with normal and abnormal courses of gestation. Therefore, these ncRNA were selected as the most suitable endogenous controls for the normalization of microRNA qPCR expression studies performed on whole peripheral blood samples affected by pregnancy-related complications.
4.3. Statistical Analysis
Initially, power analysis was used to determine the sample size required to detect an effect of a given size with a given degree of confidence (G * Power Version 3.1.9.6, Franz Faul, University of Kiel, Kiel, Germany). A total of 51 cases and 51 controls were required to achieve a power of 0.805 and a total of 70 cases and 70 controls were required to achieve a power of 0.902.
With respect to non-normal data distribution, unpaired nonparametric tests were used for subsequent statistical analyses. Initially, microRNA gene expression was compared between GDM and normal term pregnancies using the Mann–Whitney test. Subsequently, microRNA gene expression was compared between particular groups with respect to the treatment strategies using the Kruskal–Wallis one-way analysis of variance. Afterwards, a post-hoc test for comparison between groups and the Benjamini–Hochberg correction were applied [262] (Table 6 and Table 7).
Table 6.
Benjamini–Hochberg correction: comparison of microRNA gene expression between GDM and normal term pregnancies.
| K | i | Alpha = 0.05 | Alpha = 0.01 | Alpha = 0.001 |
|---|---|---|---|---|
| 2 | 0.05 | 0.01 | 0.001 | |
| 1 | 0.025 | 0.005 | 0.001 |
Table 7.
Benjamini–Hochberg correction for multiple comparisons: comparison of microRNA gene expression between GDM and normal term pregnancies with respect to the treatment strategies (GDM pregnancies managed by diet only vs. GDM pregnancies managed by diet and therapy vs. normal term pregnancies).
| K | i | Alpha = 0.05 | Alpha = 0.01 | Alpha = 0.001 |
|---|---|---|---|---|
| 3 | 0.05 | 0.01 | 0.001 | |
| 1 | 0.017 | 0.003 | 0.000 | |
| 2 | 0.033 | 0.007 | 0.001 | |
| 3 | 0.050 | 0.010 | 0.001 |
Boxplots display the median, the 75th and 25th percentiles (the upper and lower limits of the boxes), the maximum and minimum values that are no more than 1.5 times the span of the interquartile range (the upper and lower whiskers), outliers (circles), and extremes (asterisks). Statistica software (version 9.0; StatSoft, Inc., Tulsa, OK, USA) was used to produce the boxplots.
Receivers operating characteristic (ROC) curve analyses state the areas under the curves (AUC), p-values, the best cutoff point-related sensitivities, specificities, positive and negative likelihood ratios (LR+, LR−) together with the 95% CI (confidence interval). Furthermore, estimated specificities at fixed sensitivities and estimated sensitivities at fixed specificities are stated (MedCalc Software bvba, Ostend, Belgium). Sensitivities at a 90.0% specificity corresponding to a 10.0% false positive rate (FPR) were selected for data presentation. To select the optimal microRNA combinations, logistic regression with subsequent ROC curve analyses were applied (MedCalc Software bvba, Ostend, Belgium).
4.4. Information on MicroRNA-Gene-Biological Pathways Interactions
The DIANA miRPath v.3 database (DIANA TOOLS-mirPath v.3 (uth.gr)) and genes union mode were used as an a priori analysis method to perform KEGG pathway enrichment analysis to investigate the regulatory mechanisms of the microRNAs dysregulated in the early stages of gestation in the whole peripheral blood of mothers destinated to develop GDM. The results of this enrichment analysis were expressed as –ln of the p-value (−ln(p-value)). Preferentially, the database of experimentally verified microRNA targets (Tarbase v7.0) was used. In case that Tarbase v7.0 database did not provide a sufficient list of experimentally verified microRNA targets, the target prediction algorithm (microT-CDS v5.0) was used as an alternative.
In addition, the pathways/categories union mode, an a posteriori analysis method, was applied with the aim to identify merged p-values for each pathway significantly enriched with the gene targets of microRNAs dysregulated in early pregnancies destinated to develop GDM. Furthermore, the targeted pathway clusters/heatmap mode was applied to obtain the microRNA/KEGG pathway heatmap with hierarchical clustering.
5. Conclusions
Overall, we observed aberrant expression profiles of 11 microRNAs in the whole peripheral venous blood during the first trimester of gestation in pregnancies destinated to develop GDM. We confirmed the observations of other researchers that miR-23a-3p, miR-100-5p, and miR-125b-5p may serve as microRNA biomarkers with early predictive potential for GDM. In addition, novel microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-126-3p, miR-181a-5p, miR-195-5p, miR-499a-5p, and miR-574-3p) were identified, with the potential to predict GDM during the early stages of gestation.
Combined screening of all 11 dysregulated microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-23a-3p, miR-100-5p, miR-125b-5p, miR-126-3p, miR-181a-5p, miR-195-5p, miR-499a-5p, and miR-574-3p) showed the highest accuracy for the early identification of pregnancies destinated to develop GDM irrespective of the severity of the disease. This screening identified, in the early stages of gestation, 47.93% of pregnancies destinated to develop GDM at a 10.0% FPR.
The predictive model for GDM based on microRNA aberrant expression profile was further improved via the implementation of a minimal number of basic clinical characteristics (maternal age and BMI at early stages of gestation and an infertility treatment by assisted reproductive technology). Following this, 69.17% of pregnancies destinated to develop GDM were identified during the early stages of gestation at a 10.0% FPR.
The simplified prediction model for severe GDM (requiring management of diet and administration of appropriate therapy) using the combination of three basic clinical characteristics and three dysregulated microRNA biomarkers (miR-20a-5p, miR-20b-5p, and miR-195-5p) was able to identify 78.95% of cases at a 10.0% FPR during the early stages of gestation.
Parallelly, the simplified prediction model for GDM with a milder course (managed well by diet only) was more complex and required the involvement of three basic clinical characteristics and eight dysregulated microRNA biomarkers (miR-1-3p, miR-20a-5p, miR-20b-5p, miR-100-5p, miR-125b-5p, miR-195-5p, miR-499a-5p, and miR-574-3p). Following this, the model was able to identify 50.50% of cases at a 10.0% FPR during the early stages of gestation.
The implementation of additional clinical variables into the final GDM predictive model is feasible; however, it depends on the availability of the clinical data, which differs between various health care providers.
The screening based on the combination of seven clinical characteristics (maternal age and BMI at early stages of gestation, an infertility treatment by assisted reproductive technology, history of miscarriage, the presence of trombophilic gene mutations, positive first-trimester screening for preeclampsia and/or FGR by FMF algorithm, and family history of diabetes mellitus in first-degree relatives) and microRNA biomarkers showed the highest possible accuracy for the early identification of pregnancies destinated to develop GDM either regardless or with regard to the severity of the disease. The screening was able to identify, in the early stages of gestation, 72.50% of GDM cases in total—89.47% of GDM cases requiring management by diet and administration of appropriate therapy and 56.44% GDM cases managed well by diet only—at a 10.0% FPR. Nevertheless, we prefer to leave the first-trimester GDM screening simplified as much as possible.
The implementation of a novel first-trimester GDM predictive model based on the combination of basic maternal clinical characteristics and aberrant microRNA expression profile into routine screening programmes may significantly improve the care of pregnancies at risk of the development of GDM. In pregnancies identified to be destinated to develop GDM, effective dietary counselling may be already provided during the early stages of gestation, and a healthy-eating plan naturally rich in nutrients and low in fat and calories may be developed to control blood glucose, manage weight, and control heart disease risk factors, such as a high blood pressure and high blood fats. This preventive measure may contribute to lowering the incidence of GDM overall and may also contribute to a reduction in the number of severe GDM cases that require the administration of an appropriate therapy. This may also contribute to a decrease in the occurrence of other pregnancy-related complications such as gestational hypertension, preeclampsia, and fetal growth restriction.
Since women with a history of GDM have an increased risk of developing diabetes (predominantly type 2 diabetes) and cardiovascular diseases later in life, the implementation of effective early screening programme for GDM alongside subsequent preventive measures into early prenatal care may contribute to a subsequent decrease in the occurrence of diabetes and cardiovascular diseases in young and middle-aged mothers. This would also have a large impact on the offspring descending from GDM-affected pregnancies. Accumulating data suggest that exposure to hyperglycemia in utero, as occurs in gestational diabetes mellitus, may expose the offspring to short-term and long-term adverse effects.
The cost of the implementation of the novel first-trimester GDM predictive model based on the combination of basic maternal clinical characteristics and aberrant microRNA expression profile into routine screening programmes is minimal when compared to the costs related to prenatal, peripartal, postpartal, neonatal, postnatal, and lifelong healthcare. In this manner, a significant reduction in healthcare cost can be achieved.
Large-scale follow-up studies need to be performed to verify diagnostic potential of cardiovascular disease-associated microRNA biomarkers to predict the subsequent occurrence of GDM.
Any changes to the epigenome, including the dysregulation of cardiovascular microRNAs induced during the early stages of gestation in pregnancies complicated by GDM, may predispose mothers to later development of diabetes mellitus and cardiovascular/cerebrovascular diseases. This hypothesis may also be supported by our previous finding that epigenetic changes (upregulation of serious cardiovascular microRNAs) appeared in a proportion of women with a history of GDM throughout postpartal life [243].
6. Patents
National patent application—Industrial Property Office, Czech Republic (Patent n. PV 2022-335).
Acknowledgments
We would like to thank the staff of the Institute for the Care of Mother and Child for assistance with the collection of the patients’ biological samples.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231810635/s1.
Author Contributions
Conceptualization, I.H. and L.K.; methodology, I.H. and K.K.; software, I.H., K.K. and L.K.; validation, I.H., K.K. and L.K.; formal analysis, I.H., K.K.; investigation, K.K.; resources, I.H. and L.K.; data curation, I.H. and K.K.; writing—original draft preparation, I.H.; writing—review and editing, I.H. and K.K.; visualization, K.K.; supervision, I.H. and L.K.; project administration, I.H. and L.K.; funding acquisition, I.H. and L.K. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The approval for the study was initially obtained from the Ethics Committee of the Third Faculty of Medicine, Charles University (Implication of placental specific microRNAs in maternal circulation for diagnosis and prediction of pregnancy-related complications, date of approval: 7 April 2011). Ongoing approvals for the study were obtained from the Ethics Committee of the Third Faculty of Medicine, Charles University (Long-term monitoring of complex cardiovascular profiles in mother, foetus, and offspring descending from pregnancy-related-complications; date of approval: 27 March 2014) and the Ethics Committee of the Institute for the Care of the Mother and Child, Charles University (Long-term monitoring of complex cardiovascular profiles in mother, foetus, and offspring descending from pregnancy-related-complications; date of approval: 28 May 2015; number of approval: 1/4/2015). This informed consent is very complex and involves consent for the collection of peripheral blood samples at the beginning of pregnancy. In addition, in case of the onset of pregnancy-related complications, it also involves consent for the collection of peripheral blood samples at the time of the onset of pregnancy-related complications and the collection of a piece of placenta sample during the childbirth. All procedures were in compliance with the Helsinki Declaration of 1975, as revised in 2000.
Informed Consent Statement
Informed consent for the study was obtained from patients during the first trimester of gestation when the collection of peripheral blood samples for the first-trimester screening was held. Informed consent was signed by all pregnant women involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to rights reserved by funding supporters.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the Charles University research program Cooperatio—Mother and Childhood Care (no. 207035) and research grant SVV (no. 260529). All rights reserved.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Committee on Practice Bulletins—Obstetrics ACOG Practice Bulletin No. 190: Gestational Diabetes Mellitus. Obstet. Gynecol. 2018;131:e49–e64. doi: 10.1097/AOG.0000000000002501. [DOI] [PubMed] [Google Scholar]
- 2.International Association of Diabetes and Pregnancy Study Groups Consensus Panel. Metzger B.E., Gabbe S.G., Persson B., Buchanan T.A., Catalano P.A., Damm P., Dyer A.R., Leiva A.D., Hod M., et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care. 2010;33:676–682. doi: 10.2337/dc10-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Diabetes Association Diagnosis and classification of diabetes mellitus (Position Statement) Diabetes Care. 2009;32:S62–S67. doi: 10.2337/dc09-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.England L.J., Dietz P.M., Njoroge T., Callaghan W.M., Bruce C., Buus R.M., Williamson D.F. Preventing type 2 diabetes: Public health implications for women with a history of gestational diabetes mellitus. Am. J. Obstet. Gynecol. 2009;200:365.e1–365.e8. doi: 10.1016/j.ajog.2008.06.031. [DOI] [PubMed] [Google Scholar]
- 5.O’Sullivan J.B. Body weight and subsequent diabetes mellitus. JAMA. 1982;248:949–952. doi: 10.1001/jama.1982.03330080031024. [DOI] [PubMed] [Google Scholar]
- 6.Kim C., Newton K.M., Knopp R.H. Gestational diabetes and the incidence of type 2 diabetes: A systematic review. Diabetes Care. 2002;25:1862–1868. doi: 10.2337/diacare.25.10.1862. [DOI] [PubMed] [Google Scholar]
- 7.International Diabetes Federation IDF GDM Model of Care. [(accessed on 16 May 2022)]. Available online: https://www.idf.org/e-library/guidelines/77-idf-gdm-model-of-care-implementation-protocol-guidelines-for-healthcare-professionals.html.
- 8.National Institute for Health and Care Excellence (NICE) Diabetes in Pregnancy: Management from Preconception to the Postnatal Period. [(accessed on 16 May 2022)]. Available online: https://www.guidelines.co.uk/diabetes/nice-diabetes-in-pregnancy-guideline/252595.article. [PubMed]
- 9.Syngelaki A., Pastides A., Kotecha R., Wright A., Akolekar R., Nicolaides K.H. First-Trimester Screening for Gestational Diabetes Mellitus Based on Maternal Characteristics and History. Fetal Diagn. Ther. 2015;38:14–21. doi: 10.1159/000369970. [DOI] [PubMed] [Google Scholar]
- 10.Nanda S., Savvidou M., Syngelaki A., Akolekar R., Nicolaides K.H. Prediction of gestational diabetes mellitus by maternal factors and biomarkers at 11 to 13 weeks. Prenat. Diagn. 2011;31:135–141. doi: 10.1002/pd.2636. [DOI] [PubMed] [Google Scholar]
- 11.Sweeting A.N., Appelblom H., Ross G.P., Wong J., Kouru H., Williams P.F., Sairanen M., Hyett J.A. First trimester prediction of gestational diabetes mellitus: A clinical model based on maternal demographic parameters. Diabetes Res. Clin. Pract. 2017;127:44–50. doi: 10.1016/j.diabres.2017.02.036. [DOI] [PubMed] [Google Scholar]
- 12.Sweeting A.N., Wong J., Appelblom H., Ross G.P., Kouru H., Williams P.F., Sairanen M., Hyett J.A. A Novel Early Pregnancy Risk Prediction Model for Gestational Diabetes Mellitus. Fetal Diagn. Ther. 2019;45:76–84. doi: 10.1159/000486853. [DOI] [PubMed] [Google Scholar]
- 13.Wang P., Ma H.H., Hou X.Z., Song L.L., Song X.L., Zhang J.F. Reduced plasma level of irisin in first trimester as a risk factor for the development of gestational diabetes mellitus. Diabetes Res. Clin. Pract. 2018;142:130–138. doi: 10.1016/j.diabres.2018.05.038. [DOI] [PubMed] [Google Scholar]
- 14.Wu Y.T., Zhang C.J., Mol B.W., Kawai A., Li C., Chen L., Wang Y., Sheng J.Z., Fan J.X., Shi Y., et al. Early Prediction of Gestational Diabetes Mellitus in the Chinese Population via Advanced Machine Learning. J. Clin. Endocrinol. Metab. 2021;106:e1191–e1205. doi: 10.1210/clinem/dgaa899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar M., Chen L., Tan K., Ang L.T., Ho C., Wong G., Soh S.E., Tan K.H., Chan J.K.Y., Godfrey K.M., et al. Population-centric risk prediction modeling for gestational diabetes mellitus: A machine learning approach. Diabetes Res. Clin. Pract. 2022;185:109237. doi: 10.1016/j.diabres.2022.109237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakurai K., Eguchi A., Watanabe M., Yamamoto M., Ishikawa K., Mori C. Exploration of predictive metabolic factors for gestational diabetes mellitus in Japanese women using metabolomic analysis. J. Diabetes Investig. 2019;10:513–520. doi: 10.1111/jdi.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tenenbaum-Gavish K., Sharabi-Nov A., Binyamin D., Møller H.J., Danon D., Rothman L., Hadar E., Idelson A., Vogel I., Koren O., et al. First trimester biomarkers for prediction of gestational diabetes mellitus. Placenta. 2020;101:80–89. doi: 10.1016/j.placenta.2020.08.020. [DOI] [PubMed] [Google Scholar]
- 18.Zheng Y., Hou W., Xiao J., Huang H., Quan W., Chen Y. Application Value of Predictive Model Based on Maternal Coagulation Function and Glycolipid Metabolism Indicators in Early Diagnosis of Gestational Diabetes Mellitus. Front. Public Health. 2022;10:850191. doi: 10.3389/fpubh.2022.850191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong Y., Lin L., Chen Y., Salerno S., Li Y., Zeng X., Li H. Prediction of gestational diabetes mellitus in the first 19 weeks of pregnancy using machine learning techniques. J. Matern. Fetal Neonatal Med. 2022;35:2457–2463. doi: 10.1080/14767058.2020.1786517. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y.Z., Zhou L., Tian L., Li X., Zhang G., Qin J.Y., Zhang D.D., Fang H. A mid-pregnancy risk prediction model for gestational diabetes mellitus based on the maternal status in combination with ultrasound and serological findings. Exp. Ther. Med. 2020;20:293–300. doi: 10.3892/etm.2020.8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cremona A., O’Gorman C.S., Ismail K.I., Hayes K., Donnelly A.E., Hamilton J., Cotter A. A risk-prediction model using parameters of maternal body composition to identify gestational diabetes mellitus in early pregnancy. Clin. Nutr. ESPEN. 2021;45:312–321. doi: 10.1016/j.clnesp.2021.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Shaarbaf Eidgahi E., Nasiri M., Kariman N., Safavi Ardebili N., Salehi M., Kazemi M., Zayeri F. Diagnostic accuracy of first and early second trimester multiple biomarkers for prediction of gestational diabetes mellitus: A multivariate longitudinal approach. BMC Pregnancy Childbirth. 2022;22:13. doi: 10.1186/s12884-021-04348-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lappharat S., Rothmanee P., Jandee K., Suksai M., Liabsuetrakul T. A model for predicting gestational diabetes mellitus in early pregnancy: A prospective study in Thailand. Obstet. Gynecol. Sci. 2022;65:156–165. doi: 10.5468/ogs.21250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X., Zheng X., Yan J., Xu R., Xu M., Zheng L., Xu L., Lin Z. The Clinical Values of Afamin, Triglyceride and PLR in Predicting Risk of Gestational Diabetes During Early Pregnancy. Front. Endocrinol. 2021;12:723650. doi: 10.3389/fendo.2021.723650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Lv B., Chen X., Pan Y., Chen K., Zhang Y., Li Q., Wei L., Liu Y. An early model to predict the risk of gestational diabetes mellitus in the absence of blood examination indexes: Application in primary health care centres. BMC Pregnancy Childbirth. 2021;21:814. doi: 10.1186/s12884-021-04295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang M., Zhang H., Zhang J., Huang K., Zhao J., Hu J., Lu C., Shao J., Weng J., Yang Y., et al. A Novel Nomogram for Predicting Gestational Diabetes Mellitus During Early Pregnancy. Front. Endocrinol. 2021;12:779210. doi: 10.3389/fendo.2021.779210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buerger O., Elger T., Varthaliti A., Syngelaki A., Wright A., Nicolaides K.H. First-Trimester Screening for Gestational Diabetes Mellitus in Twin Pregnancies. J. Clin. Med. 2021;10:3814. doi: 10.3390/jcm10173814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snyder B.M., Baer R.J., Oltman S.P., Robinson J.G., Breheny P.J., Saftlas A.F., Bao W., Greiner A.L., Carter K.D., Rand L., et al. Early pregnancy prediction of gestational diabetes mellitus risk using prenatal screening biomarkers in nulliparous women. Diabetes Res. Clin. Pract. 2020;163:108139. doi: 10.1016/j.diabres.2020.108139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., Zhao X., Huo L., Yuan N., Sun J., Du J., Nan M., Ji L. Risk prediction model of gestational diabetes mellitus based on nomogram in a Chinese population cohort study. Sci. Rep. 2020;10:21223. doi: 10.1038/s41598-020-78164-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Hoorn F., Koster M., Naaktgeboren C.A., Groenendaal F., Kwee A., Lamain-de Ruiter M., Franx A., Bekker M.N. Prognostic models versus single risk factor approach in first-trimester selective screening for gestational diabetes mellitus: A prospective population-based multicentre cohort study. BJOG. 2021;128:645–654. doi: 10.1111/1471-0528.16446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benhalima K., Van Crombrugge P., Moyson C., Verhaeghe J., Vandeginste S., Verlaenen H., Vercammen C., Maes T., Dufraimont E., De Block C., et al. Estimating the risk of gestational diabetes mellitus based on the 2013 WHO criteria: A prediction model based on clinical and biochemical variables in early pregnancy. Acta Diabetol. 2020;57:661–671. doi: 10.1007/s00592-019-01469-5. [DOI] [PubMed] [Google Scholar]
- 32.Meertens L., Smits L., van Kuijk S., Aardenburg R., van Dooren I., Langenveld J., Zwaan I.M., Spaanderman M., Scheepers H. External validation and clinical usefulness of first-trimester prediction models for small- and large-for-gestational-age infants: A prospective cohort study. BJOG. 2019;126:472–484. doi: 10.1111/1471-0528.15516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan Y., Hu J., Zhong S. The joint prediction model of pBMI and eFBG in predicting gestational diabetes mellitus. J. Int. Med. Res. 2020;48:300060519889199. doi: 10.1177/0300060519889199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng T., Ye W., Wang X., Li X., Zhang J., Little J., Zhou L., Zhang L. A simple model to predict risk of gestational diabetes mellitus from 8 to 20 weeks of gestation in Chinese women. BMC Pregnancy Childbirth. 2019;19:252. doi: 10.1186/s12884-019-2374-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamain-de Ruiter M., Kwee A., Naaktgeboren C.A., de Groot I., Evers I.M., Groenendaal F., Hering Y.R., Huisjes A.J., Kirpestein C., Monincx W.M., et al. External validation of prognostic models to predict risk of gestational diabetes mellitus in one Dutch cohort: Prospective multicentre cohort study. BMJ. 2016;354:i4338. doi: 10.1136/bmj.i4338. [DOI] [PubMed] [Google Scholar]
- 36.Ravnsborg T., Andersen L.L., Trabjerg N.D., Rasmussen L.M., Jensen D.M., Overgaard M. First-trimester multimarker prediction of gestational diabetes mellitus using targeted mass spectrometry. Diabetologia. 2016;59:970–979. doi: 10.1007/s00125-016-3869-8. [DOI] [PubMed] [Google Scholar]
- 37.Hassiakos D., Eleftheriades M., Papastefanou I., Lambrinoudaki I., Kappou D., Lavranos D., Akalestos A., Aravantinos L., Pervanidou P., Chrousos G. Increased Maternal Serum Interleukin-6 Concentrations at 11 to 14 Weeks of Gestation in Low Risk Pregnancies Complicated with Gestational Diabetes Mellitus: Development of a Prediction Model. Horm. Metab. Res. 2016;48:35–41. doi: 10.1055/s-0034-1395659. [DOI] [PubMed] [Google Scholar]
- 38.Alptekin H., Çizmecioğlu A., Işık H., Cengiz T., Yildiz M., Iyisoy M.S. Predicting gestational diabetes mellitus during the first trimester using anthropometric measurements and HOMA-IR. J. Endocrinol. Investig. 2016;39:577–583. doi: 10.1007/s40618-015-0427-z. [DOI] [PubMed] [Google Scholar]
- 39.Papastefanou I., Eleftheriades M., Kappou D., Lambrinoudaki I., Lavranos D., Pervanidou P., Sotiriadis A., Hassiakos D., Chrousos G.P. Maternal serum osteocalcin at 11–14 weeks of gestation in gestational diabetes mellitus. Eur. J. Clin. Investig. 2015;45:1025–1031. doi: 10.1111/eci.12500. [DOI] [PubMed] [Google Scholar]
- 40.Eleftheriades M., Papastefanou I., Lambrinoudaki I., Kappou D., Lavranos D., Akalestos A., Souka A.P., Pervanidou P., Hassiakos D., Chrousos G.P. Elevated placental growth factor concentrations at 11–14 weeks of gestation to predict gestational diabetes mellitus. Metabolism. 2014;63:1419–1425. doi: 10.1016/j.metabol.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 41.Li J., Dong X., Wang Z., Wu J. MicroRNA-1 in Cardiac Diseases and Cancers. Korean J. Physiol. Pharmacol. 2014;18:359–363. doi: 10.4196/kjpp.2014.18.5.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y.Q., Zhang M.F., Wen H.Y., Hu C.L., Liu R., Wei H.Y., Ai C.M., Wang G., Liao X.X., Li X. Comparing the diagnostic values of circulating microRNAs and cardiac troponin T in patients with acute myocardial infarction. Clinics. 2013;68:75–80. doi: 10.6061/clinics/2013(01)OA12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gasiulė S., Stankevičius V., Patamsytė V., Ražanskas R., Zukovas G., Kapustina Z., Zaliaduonytė D., Benetis R., Lesauskaitė V., Vilkaitis G. Tissue-Specific miRNAs Regulate the Development of Thoracic Aortic Aneurysm: The Emerging Role of KLF4 Network. J. Clin. Med. 2019;8:1609. doi: 10.3390/jcm8101609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerlinger-Romero F., Yonamine C.Y., Junior D.C., Esteves J.V., Machado U.F. Dysregulation between TRIM63/FBXO32 expression and soleus muscle wasting in diabetic rats: Potential role of miR-1-3p, -29a/b-3p, and -133a/b-3p. Mol. Cell. Biochem. 2017;427:187–199. doi: 10.1007/s11010-016-2910-z. [DOI] [PubMed] [Google Scholar]
- 45.Kokkinopoulou I., Maratou E., Mitrou P., Boutati E., Sideris D.C., Fragoulis E.G., Christodoulou M.I. Decreased expression of microRNAs targeting type-2 diabetes susceptibility genes in peripheral blood of patients and predisposed individuals. Endocrine. 2019;66:226–239. doi: 10.1007/s12020-019-02062-0. [DOI] [PubMed] [Google Scholar]
- 46.Hromadnikova I., Kotlabova K., Dvorakova L., Krofta L. Evaluation of Vascular Endothelial Function in Young and Middle-Aged Women with Respect to a History of Pregnancy, Pregnancy-Related Complications, Classical Cardiovascular Risk Factors, and Epigenetics. Int. J. Mol. Sci. 2020;21:430. doi: 10.3390/ijms21020430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X., Shang Y., Dai S., Wu W., Yi F., Cheng L. MicroRNA-16-5p aggravates myocardial infarction injury by targeting expression of insulin receptor substrates 1 and mediating myocardial apoptosis and angiogenesis. Curr. Neurovasc. Res. 2019 doi: 10.2174/1567202617666191223142743. Epub ahead of print . [DOI] [PubMed] [Google Scholar]
- 48.O’Sullivan J.F., Neylon A., McGorrian C., Blake G.J. miRNA-93-5p and other miRNAs as predictors of coronary artery disease and STEMI. Int. J. Cardiol. 2016;224:310–316. doi: 10.1016/j.ijcard.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 49.Vegter E.L., Schmitter D., Hagemeijer Y., Ovchinnikova E.S., van der Harst P., Teerlink J.R., O’Connor C.M., Metra M., Davison B.A., Bloomfield D., et al. Use of biomarkers to establish potential role and function of circulating microRNAs in acute heart failure. Int. J. Cardiol. 2016;224:231–239. doi: 10.1016/j.ijcard.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 50.Gacoń J., Badacz R., Stępień E., Karch I., Enguita F.J., Żmudka K., Przewłocki T., Kabłak-Ziembicka A. Diagnostic and prognostic micro-RNAs in ischaemic stroke due to carotid artery stenosis and in acute coronary syndrome: A four-year prospective study. Kardiol. Pol. 2018;76:362–369. doi: 10.5603/KP.a2017.0243. [DOI] [PubMed] [Google Scholar]
- 51.Zhu Y., Tian F., Li H., Zhou Y., Lu J., Ge Q. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int. J. Gynaecol. Obstet. 2015;130:49–53. doi: 10.1016/j.ijgo.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 52.Cao Y.L., Jia Y.J., Xing B.H., Shi D.D., Dong X.J. Plasma microRNA-16-5p, -17-5p and -20a-5p: Novel diagnostic biomarkers for gestational diabetes mellitus. J. Obstet. Gynaecol. Res. 2017;43:974–981. doi: 10.1111/jog.13317. [DOI] [PubMed] [Google Scholar]
- 53.Hocaoglu M., Demirer S., Senturk H., Turgut A., Komurcu-Bayrak E. Differential expression of candidate circulating microRNAs in maternal blood leukocytes of the patients with preeclampsia and gestational diabetes mellitus. Pregnancy Hypertens. 2019;17:5–11. doi: 10.1016/j.preghy.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 54.Duan Y.R., Chen B.P., Chen F., Yang S.X., Zhu C.Y., Ma Y.L., Li Y., Shi J. Exosomal microRNA-16-5p from human urine-derived stem cells ameliorates diabetic nephropathy through protection of podocyte. J. Cell Mol. Med. 2019. Epub ahead of print . [DOI] [PMC free article] [PubMed]
- 55.Assmann T.S., Recamonde-Mendoza M., Costa A.R., Puñales M., Tschiedel B., Canani L.H., Bauer A.C., Crispim D. Circulating miRNAs in diabetic kidney disease: Case-control study and in silico analyses. Acta Diabetol. 2019;56:55–65. doi: 10.1007/s00592-018-1216-x. [DOI] [PubMed] [Google Scholar]
- 56.Alicka M., Major P., Wysocki M., Marycz K. Adipose-Derived Mesenchymal Stem Cells Isolated from Patients with Type 2 Diabetes Show Reduced “Stemness” through an Altered Secretome Profile, Impaired Anti-Oxidative Protection, and Mitochondrial Dynamics Deterioration. J. Clin. Med. 2019;8:765. doi: 10.3390/jcm8060765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mogilyansky E., Rigoutsos I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou L., Qi R.Q., Liu M., Xu Y.P., Li G., Weiland M., Kaplan D.H., Mi Q.S. microRNA miR-17-92 cluster is highly expressed in epidermal Langerhans cells but not required for its development. Genes Immun. 2014;15:57–61. doi: 10.1038/gene.2013.61. [DOI] [PubMed] [Google Scholar]
- 59.Danielson L.S., Park D.S., Rotllan N., Chamorro-Jorganes A., Guijarro M.V., Fernandez-Hernando C., Fishman G.I., Phoon C.K., Hernando E. Cardiovascular dysregulation of miR-17-92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB J. 2013;27:1460–1467. doi: 10.1096/fj.12-221994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du W., Pan Z., Chen X., Wang L., Zhang Y., Li S., Liang H., Xu C., Zhang Y., Wu Y., et al. By targeting Stat3 microRNA-17-5p promotes cardiomyocyte apoptosis in response to ischemia followed by reperfusion. Cell. Physiol. Biochem. 2014;34:955–965. doi: 10.1159/000366312. [DOI] [PubMed] [Google Scholar]
- 61.Kaucsár T., Révész C., Godó M., Krenács T., Albert M., Szalay C.I., Rosivall L., Benyó Z., Bátkai S., Thum T., et al. Activation of the miR-17 family and miR-21 during murine kidney ischemia-reperfusion injury. Nucleic Acid Ther. 2013;23:344–354. doi: 10.1089/nat.2013.0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang L., Ellims A.H., Moore X.L., White D.A., Taylor A.J., Chin-Dusting J., Dart A.M. Circulating microRNAs as biomarkers for diffuse myocardial fibrosis in patients with hypertrophic cardiomyopathy. J. Transl. Med. 2015;13:314. doi: 10.1186/s12967-015-0672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu J., Du K., Lu X. Elevated expressions of serum miR-15a, miR-16, and miR-17-5p are associated with acute ischemic stroke. Int. J. Clin. Exp. Med. 2015;8:21071–21079. [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J., Xu L., Hu Q., Yang S., Zhang B., Jiang H. MiR-17-5p as circulating biomarkers for the severity of coronary atherosclerosis in coronary artery disease. Int. J. Cardiol. 2015;197:123–124. doi: 10.1016/j.ijcard.2015.06.037. [DOI] [PubMed] [Google Scholar]
- 65.Tian L., Song Z., Shao W., Du W.W., Zhao L.R., Zeng K., Yang B.B., Jin T. Curcumin represses mouse 3T3-L1 cell adipogenic differentiation via inhibiting miR-17-5p and stimulating the Wnt signalling pathway effector Tcf7l. Cell Death Dis. 2017;8:e2559. doi: 10.1038/cddis.2016.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen T.C., Sung M.L., Kuo H.C., Chien S.J., Yen C.K., Chen C.N. Differential regulation of human aortic smooth muscle cell proliferation by monocyte-derived macrophages from diabetic patients. PLoS ONE. 2014;9:e113752. doi: 10.1371/journal.pone.0113752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mendell J.T. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brock M., Samillan V.J., Trenkmann M., Schwarzwald C., Ulrich S., Gay R.E., Gassmann M., Ostergaard L., Gay S., Speich R., et al. AntagomiR directed against miR-20a restores functional BMPR2 signalling and prevents vascular remodelling in hypoxia-induced pulmonary hypertension. Eur. Heart J. 2014;35:3203–3211. doi: 10.1093/eurheartj/ehs060. [DOI] [PubMed] [Google Scholar]
- 69.Pheiffer C., Dias S., Rheeder P., Adam S. Decreased Expression of Circulating miR-20a-5p in South African Women with Gestational Diabetes Mellitus. Mol. Diagn. Ther. 2018;22:345–352. doi: 10.1007/s40291-018-0325-0. [DOI] [PubMed] [Google Scholar]
- 70.Platania C.B.M., Maisto R., Trotta M.C., D’Amico M., Rossi S., Gesualdo C., D’Amico G., Balta C., Herman H., Hermenean A., et al. Retinal and circulating miRNA expression patterns in diabetic retinopathy: An in silico and in vivo approach. Br. J. Pharmacol. 2019;176:2179–2194. doi: 10.1111/bph.14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lareyre F., Clément M., Moratal C., Loyer X., Jean-Baptiste E., Hassen-Khodja R., Chinetti G., Mallat Z., Raffort J. Differential micro-RNA expression in diabetic patients with abdominal aortic aneurysm. Biochimie. 2019;162:1–7. doi: 10.1016/j.biochi.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 72.Dickinson B.A., Semus H.M., Montgomery R.L., Stack C., Latimer P.A., Lewton S.M., Lynch J.M., Hullinger T.G., Seto A.G., van Rooij E. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur. J. Heart Fail. 2013;15:650–659. doi: 10.1093/eurjhf/hft018. [DOI] [PubMed] [Google Scholar]
- 73.Flowers E., Aouizerat B.E., Abbasi F., Lamendola C., Grove K.M., Fukuoka Y., Reaven G.M. Circulating microRNA-320a and microRNA-486 predict thiazolidinedione response: Moving towards precision health for diabetes prevention. Metabolism. 2015;64:1051–1059. doi: 10.1016/j.metabol.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katayama M., Wiklander O.P.B., Fritz T., Caidahl K., El-Andaloussi S., Zierath J.R., Krook A. Circulating Exosomal miR-20b-5p Is Elevated in Type 2 Diabetes and Could Impair Insulin Action in Human Skeletal Muscle. Diabetes. 2019;68:515–526. doi: 10.2337/db18-0470. [DOI] [PubMed] [Google Scholar]
- 75.Xiong Y., Chen L., Yan C., Zhou W., Endo Y., Liu J., Hu L., Hu Y., Mi B., Liu G. Circulating Exosomal miR-20b-5p Inhibition Restores Wnt9b Signaling and Reverses Diabetes-Associated Impaired Wound Healing. Small. 2020;16:e1904044. doi: 10.1002/smll.201904044. [DOI] [PubMed] [Google Scholar]
- 76.Zhu K., Hu X., Chen H., Li F., Yin N., Liu A.L., Shan K., Qin Y.W., Huang X., Chang Q., et al. Downregulation of circRNA DMNT3B contributes to diabetic retinal vascular dysfunction through targeting miR-20b-5p and BAMBI. EBioMedicine. 2019;49:341–353. doi: 10.1016/j.ebiom.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sekar D., Venugopal B., Sekar P., Ramalingam K. Role of microRNA 21 in diabetes and associated/related diseases. Gene. 2016;582:14–18. doi: 10.1016/j.gene.2016.01.039. [DOI] [PubMed] [Google Scholar]
- 78.Suárez Y., Fernández-Hernando C., Pober J.S., Sessa W.C. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 79.Dong S., Ma W., Hao B., Hu F., Yan L., Yan X., Wang Y., Chen Z., Wang Z. microRNA-21 promotes cardiac fibrosis and development of heart failure with preserved left ventricular ejection fraction by up-regulating Bcl-2. Int. J. Clin. Exp. Pathol. 2014;7:565–574. [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J., Xing Q., Zhou X., Li J., Li Y., Zhang L., Zhou Q., Tang B. Circulating miRNA 21 is a promising biomarker for heart failure. Mol. Med. Rep. 2017;16:7766–7774. doi: 10.3892/mmr.2017.7575. [DOI] [PubMed] [Google Scholar]
- 81.Licholai S., Blaż M., Kapelak B., Sanak M. Unbiased Profile of MicroRNA Expression in Ascending Aortic Aneurysm Tissue Appoints Molecular Pathways Contributing to the Pathology. Ann. Thorac. Surg. 2016;102:1245–1252. doi: 10.1016/j.athoracsur.2016.03.061. [DOI] [PubMed] [Google Scholar]
- 82.Kriegel A.J., Baker M.A., Liu Y., Liu P., Cowley A.W., Jr., Liang M. Endogenous microRNAs in human microvascular endothelial cells regulate mRNAs encoded by hypertension-related genes. Hypertension. 2015;66:793–799. doi: 10.1161/HYPERTENSIONAHA.115.05645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Velle-Forbord T., Eidlaug M., Debik J., Sæther J.C., Follestad T., Nauman J., Gigante B., Røsjø H., Omland T., Langaas M., et al. Circulating microRNAs as predictive biomarkers of myocardial infarction: Evidence from the HUNT study. Atherosclerosis. 2019;289:1–7. doi: 10.1016/j.atherosclerosis.2019.07.024. [DOI] [PubMed] [Google Scholar]
- 84.Demirsoy İ.H., Ertural D.Y., Balci Ş., Çınkır Ü., Sezer K., Tamer L., Aras N. Profiles of Circulating MiRNAs Following Metformin Treatment in Patients with Type 2 Diabetes. J. Med. Biochem. 2018;37:499–506. doi: 10.2478/jomb-2018-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olivieri F., Spazzafumo L., Bonafè M., Recchioni R., Prattichizzo F., Marcheselli F., Micolucci L., Mensà E., Giuliani A., Santini G., et al. MiR-21-5p and miR-126a-3p levels in plasma and circulating angiogenic cells: Relationship with type 2 diabetes complications. Oncotarget. 2015;6:35372–35382. doi: 10.18632/oncotarget.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Assmann T.S., Recamonde-Mendoza M., De Souza B.M., Crispim D. MicroRNA expression profiles and type 1 diabetes mellitus: Systematic review and bioinformatic analysis. Endocr. Connect. 2017;6:773–790. doi: 10.1530/EC-17-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lakhter A.J., Pratt R.E., Moore R.E., Doucette K.K., Maier B.F., DiMeglio L.A., Sims E.K. Beta cell extracellular vesicle miR-21-5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia. 2018;61:1124–1134. doi: 10.1007/s00125-018-4559-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grieco G.E., Cataldo D., Ceccarelli E., Nigi L., Catalano G., Brusco N., Mancarella F., Ventriglia G., Fondelli C., Guarino E., et al. Serum Levels of miR-148a and miR-21-5p Are Increased in Type 1 Diabetic Patients and Correlated with Markers of Bone Strength and Metabolism. Noncoding RNA. 2018;4:37. doi: 10.3390/ncrna4040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gholaminejad A., Abdul Tehrani H., Gholami Fesharaki M. Identification of candidate microRNA biomarkers in diabetic nephropathy: A meta-analysis of profiling studies. J. Nephrol. 2018;31:813–831. doi: 10.1007/s40620-018-0511-5. [DOI] [PubMed] [Google Scholar]
- 90.Long B., Gan T.Y., Zhang R.C., Zhang Y.H. miR-23a Regulates Cardiomyocyte Apoptosis by Targeting Manganese Superoxide Dismutase. Mol. Cells. 2017;40:542–549. doi: 10.14348/molcells.2017.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang S., He W., Wang C. MiR-23a Regulates the Vasculogenesis of Coronary Artery Disease by Targeting Epidermal Growth Factor Receptor. Cardiovasc. Ther. 2016;34:199–208. doi: 10.1111/1755-5922.12187. [DOI] [PubMed] [Google Scholar]
- 92.Cong X., Li Y., Lu N., Dai Y., Zhang H., Zhao X., Liu Y. Resveratrol attenuates the inflammatory reaction induced by ischemia/reperfusion in the rat heart. Mol. Med. Rep. 2014;9:2528–2532. doi: 10.3892/mmr.2014.2090. [DOI] [PubMed] [Google Scholar]
- 93.Černá V., Ostašov P., Pitule P., Moláček J., Třeška V., Pešta M. The Expression Profile of MicroRNAs in Small and Large Abdominal Aortic Aneurysms. Cardiol. Res. Pract. 2019;2019:8645840. doi: 10.1155/2019/8645840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lozano-Bartolomé J., Llauradó G., Portero-Otin M., Altuna-Coy A., Rojo-Martínez G., Vendrell J., Jorba R., Rodríguez-Gallego E., Chacón M.R. Altered Expression of miR-181a-5p and miR-23a-3p Is Associated With Obesity and TNFα-Induced Insulin Resistance. J. Clin. Endocrinol. Metab. 2018;103:1447–1458. doi: 10.1210/jc.2017-01909. [DOI] [PubMed] [Google Scholar]
- 95.Dolz S., Górriz D., Tembl J.I., Sánchez D., Fortea G., Parkhutik V., Lago A. Circulating MicroRNAs as Novel Biomarkers of Stenosis Progression in Asymptomatic Carotid Stenosis. Stroke. 2017;48:10–16. doi: 10.1161/STROKEAHA.116.013650. [DOI] [PubMed] [Google Scholar]
- 96.De Gonzalo-Calvo D., Cenarro A., Garlaschelli K., Pellegatta F., Vilades D., Nasarre L., Camino-Lopez S., Crespo J., Carreras F., Leta R., et al. Translating the microRNA signature of microvesicles derived from human coronary artery smooth muscle cells in patients with familial hypercholesterolemia and coronary artery disease. J. Mol. Cell. Cardiol. 2017;106:55–67. doi: 10.1016/j.yjmcc.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 97.Gecys D., Tatarunas V., Veikutiene A., Lesauskaite V. New potential modulators of CYP4F2 enzyme activity in angina pectoris: Hsa-miR-24-3p and hsa-miR-34a-5p. Biomarkers. 2020;25:40–47. doi: 10.1080/1354750X.2019.1690580. [DOI] [PubMed] [Google Scholar]
- 98.Onrat S.T., Onrat E., Ercan Onay E., Yalım Z., Avşar A. The Genetic Determination of the Differentiation between Ischemic Dilated Cardiomyopathy and Idiopathic Dilated Cardiomyopathy. Genet. Test. Mol. Biomark. 2018;22:644–651. doi: 10.1089/gtmb.2018.0188. [DOI] [PubMed] [Google Scholar]
- 99.Tan H., Qi J., Fan B.Y., Zhang J., Su F.F., Wang H.T. MicroRNA-24-3p Attenuates Myocardial Ischemia/Reperfusion Injury by Suppressing RIPK1 Expression in Mice. Cell. Physiol. Biochem. 2018;51:46–62. doi: 10.1159/000495161. [DOI] [PubMed] [Google Scholar]
- 100.Xiao X., Lu Z., Lin V., May A., Shaw D.H., Wang Z., Che B., Tran K., Du H., Shaw P.X. MicroRNA miR-24-3p Reduces Apoptosis and Regulates Keap1-Nrf2 Pathway in Mouse Cardiomyocytes Responding to Ischemia/Reperfusion Injury. Oxidative Med. Cell. Longev. 2018;2018:7042105. doi: 10.1155/2018/7042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gao J., Liu Q.G. The role of miR-26 in tumors and normal tissues. Oncol. Lett. 2011;2:1019–1023. doi: 10.3892/ol.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zheng L., Lin S., Lv C. MiR-26a-5p regulates cardiac fibroblasts collagen expression by targeting ULK1. Sci Rep. 2018;8:2104. doi: 10.1038/s41598-018-20561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bye A., Røsjø H., Nauman J., Silva G.J., Follestad T., Omland T., Wisløff U. Circulating microRNAs predict future fatal myocardial infarction in healthy individuals—The HUNT study. J. Mol. Cell. Cardiol. 2016;97:162–168. doi: 10.1016/j.yjmcc.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 104.Hsu A., Chen S.J., Chang Y.S., Chen H.C., Chu P.H. Systemic approach to identify serum microRNAs as potential biomarkers for acute myocardial infarction. BioMed Res. Int. 2014;2014:418628. doi: 10.1155/2014/418628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xing X., Guo S., Zhang G., Liu Y., Bi S., Wang X., Lu Q. miR-26a-5p protects against myocardial ischemia/reperfusion injury by regulating the PTEN/PI3K/AKT signaling pathway. Braz. J. Med. Biol. Res. 2020;53:e9106. doi: 10.1590/1414-431x20199106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chouvarine P., Geldner J., Giagnorio R., Legchenko E., Bertram H., Hansmann G. Trans-Right-Ventricle and Transpulmonary MicroRNA Gradients in Human Pulmonary Arterial Hypertension. Pediatr. Crit. Care Med. 2019 doi: 10.1097/PCC.0000000000002207. Epub ahead of print . [DOI] [PubMed] [Google Scholar]
- 107.Garavelli S., Bruzzaniti S., Tagliabue E., Prattichizzo F., Di Silvestre D., Perna F., La Sala L., Ceriello A., Mozzillo E., Fattorusso V., et al. Blood Co-Circulating Extracellular microRNAs and Immune Cell Subsets Associate with Type 1 Diabetes Severity. Int. J. Mol. Sci. 2020;21:477. doi: 10.3390/ijms21020477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ye Y., Hu Z., Lin Y., Zhang C., Perez-Polo J.R. Downregulation of microRNA-29 by antisense inhibitors and a PPAR-gamma agonist protects against myocardial ischaemia-reperfusion injury. Cardiovasc. Res. 2010;87:535–544. doi: 10.1093/cvr/cvq053. [DOI] [PubMed] [Google Scholar]
- 109.Moraes L.N., Fernandez G.J., Vechetti-Júnior I.J., Freire P.P., Souza R.W.A., Villacis R.A.R., Rogatto S.R., Reis P.P., Dal-Pai-Silva M., Carvalho R.F. Integration of miRNA and mRNA expression profiles reveals microRNA-regulated networks during muscle wasting in cardiac cachexia. Sci. Rep. 2017;7:6998. doi: 10.1038/s41598-017-07236-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao Y., Yuan Y., Qiu C. Underexpression of CACNA1C Caused by Overexpression of microRNA-29a Underlies the Pathogenesis of Atrial Fibrillation. Med. Sci. Monit. 2016;22:2175–2181. doi: 10.12659/MSM.896191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang L., Zhang Y., Xue S., Ding H., Wang Y., Qi H., Wang Y., Zhu W., Li P. Clinical significance of circulating microRNAs as diagnostic biomarkers for coronary artery disease. J. Cell. Mol. Med. 2020;24:1146–1150. doi: 10.1111/jcmm.14802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wander P.L., Boyko E.J., Hevner K., Parikh V.J., Tadesse M.G., Sorensen T.K., Williams M.A., Enquobahrie D.A. Circulating early- and mid-pregnancy microRNAs and risk of gestational diabetes. Diabetes Res. Clin. Pract. 2017;132:1–9. doi: 10.1016/j.diabres.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kong L., Zhu J., Han W., Jiang X., Xu M., Zhao Y., Dong Q., Pang Z., Guan Q., Gao L., et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A clinical study. Acta Diabetol. 2011;48:61–69. doi: 10.1007/s00592-010-0226-0. [DOI] [PubMed] [Google Scholar]
- 114.Widlansky M.E., Jensen D.M., Wang J., Liu Y., Geurts A.M., Kriegel A.J., Liu P., Ying R., Zhang G., Casati M., et al. miR-29 contributes to normal endothelial function and can restore it in cardiometabolic disorders. EMBO Mol. Med. 2018;10:e8046. doi: 10.15252/emmm.201708046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bulent Vatan M., Kalaycı Yigin A., Akdemir R., Tarik Agac M., Akif Cakar M., Aksoy M., Tatli E., Kilic H., Gunduz H., Guzel D., et al. Altered Plasma MicroRNA Expression in Patients with Mitral Chordae Tendineae Rupture. J. Heart Valve Dis. 2016;25:580–588. [PubMed] [Google Scholar]
- 116.Gumus G., Giray D., Bobusoglu O., Tamer L., Karpuz D., Hallioglu O. MicroRNA values in children with rheumatic carditis: A preliminary study. Rheumatol. Int. 2018;38:1199–1205. doi: 10.1007/s00296-018-4069-2. [DOI] [PubMed] [Google Scholar]
- 117.Rogg E.M., Abplanalp W.T., Bischof C., John D., Schulz M.H., Krishnan J., Fischer A., Poluzzi C., Schaefer L., Bonauer A., et al. Analysis of Cell Type-Specific Effects of MicroRNA-92a Provides Novel Insights Into Target Regulation and Mechanism of Action. Circulation. 2018;138:2545–2558. doi: 10.1161/CIRCULATIONAHA.118.034598. [DOI] [PubMed] [Google Scholar]
- 118.Marques F.Z., Vizi D., Khammy O., Mariani J.A., Kaye D.M. The transcardiac gradient of cardio-microRNAs in the failing heart. Eur. J. Heart Fail. 2016;18:1000–1008. doi: 10.1002/ejhf.517. [DOI] [PubMed] [Google Scholar]
- 119.Liu Y., Li Q., Hosen M.R., Zietzer A., Flender A., Levermann P., Schmitz T., Frühwald D., Goody P., Nickenig G., et al. Atherosclerotic Conditions Promote the Packaging of Functional MicroRNA-92a-3p Into Endothelial Microvesicles. Circ. Res. 2019;124:575–587. doi: 10.1161/CIRCRESAHA.118.314010. [DOI] [PubMed] [Google Scholar]
- 120.Wiese C.B., Zhong J., Xu Z.Q., Zhang Y., Ramirez Solano M.A., Zhu W., Linton M.F., Sheng Q., Kon V., Vickers K.C. Dual inhibition of endothelial miR-92a-3p and miR-489-3p reduces renal injury-associated atherosclerosis. Atherosclerosis. 2019;282:121–131. doi: 10.1016/j.atherosclerosis.2019.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Moncini S., Salvi A., Zuccotti P., Viero G., Quattrone A., Barlati S., De Petro G., Venturin M., Riva P. The role of miR-103 and miR-107 in regulation of CDK5R1 expression and in cellular migration. PLoS ONE. 2011;6:e20038. doi: 10.1371/journal.pone.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huang L., Li L., Chen X., Zhang H., Shi Z. MiR-103a targeting Piezo1 is involved in acute myocardial infarction through regulating endothelium function. Cardiol. J. 2016;23:556–562. doi: 10.5603/CJ.a2016.0056. [DOI] [PubMed] [Google Scholar]
- 123.Deng B., Du J., Hu R., Wang A.P., Wu W.H., Hu C.P., Li Y.J., Li X.H. MicroRNA-103/107 is involved in hypoxia-induced proliferation of pulmonary arterial smooth muscle cells by targeting HIF-1β. Life Sci. 2016;147:117–124. doi: 10.1016/j.lfs.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 124.Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., Heim M.H., Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 125.Assmann T.S., Recamonde-Mendoza M., Puñales M., Tschiedel B., Canani L.H., Crispim D. MicroRNA expression profile in plasma from type 1 diabetic patients: Case-control study and bioinformatic analysis. Diabetes Res. Clin. Pract. 2018;141:35–46. doi: 10.1016/j.diabres.2018.03.044. [DOI] [PubMed] [Google Scholar]
- 126.Shaham L., Binder V., Gefen N., Borkhardt A., Izraeli S. MiR-125 in normal and malignant hematopoiesis. Leukemia. 2012;26:2011–2018. doi: 10.1038/leu.2012.90. [DOI] [PubMed] [Google Scholar]
- 127.Tiedt S., Prestel M., Malik R., Schieferdecker N., Duering M., Kautzky V., Stoycheva I., Böck J., Northoff B.H., Klein M., et al. RNA-Seq Identifies Circulating miR-125a-5p, miR-125b-5p, and miR-143-3p as Potential Biomarkers for Acute Ischemic Stroke. Circ. Res. 2017;121:970–980. doi: 10.1161/CIRCRESAHA.117.311572. [DOI] [PubMed] [Google Scholar]
- 128.Jia K., Shi P., Han X., Chen T., Tang H., Wang J. Diagnostic value of miR-30d-5p and miR-125b-5p in acute myocardial infarction. Mol. Med. Rep. 2016;14:184–194. doi: 10.3892/mmr.2016.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bayoumi A.S., Park K.M., Wang Y., Teoh J.P., Aonuma T., Tang Y., Su H., Weintraub N.L., Kim I.M. A carvedilol-responsive microRNA, miR-125b-5p protects the heart from acute myocardial infarction by repressing pro-apoptotic bak1 and klf13 in cardiomyocytes. J. Mol. Cell. Cardiol. 2018;114:72–82. doi: 10.1016/j.yjmcc.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lamadrid-Romero M., Solís K.H., Cruz-Reséndiz M.S., Pérez J.E., Díaz N.F., Flores-Herrera H., García-López G., Perichart O., Reyes-Muñoz E., Arenas-Huertero F., et al. Central nervous system development-related microRNAs levels increase in the serum of gestational diabetic women during the first trimester of pregnancy. Neurosci. Res. 2018;130:8–22. doi: 10.1016/j.neures.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 131.Satake E., Pezzolesi M.G., Md Dom Z.I., Smiles A.M., Niewczas M.A., Krolewski A.S. Circulating miRNA Profiles Associated With Hyperglycemia in Patients With Type 1 Diabetes. Diabetes. 2018;67:1013–1023. doi: 10.2337/db17-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Samandari N., Mirza A.H., Kaur S., Hougaard P., Nielsen L.B., Fredheim S., Mortensen H.B., Pociot F. Influence of Disease Duration on Circulating Levels of miRNAs in Children and Adolescents with New Onset Type 1 Diabetes. Noncoding RNA. 2018;4:35. doi: 10.3390/ncrna4040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yu C.Y., Yang C.Y., Rui Z.L. MicroRNA-125b-5p improves pancreatic β-cell function through inhibiting JNK signaling pathway by targeting DACT1 in mice with type 2 diabetes mellitus. Life Sci. 2019;224:67–75. doi: 10.1016/j.lfs.2019.01.031. [DOI] [PubMed] [Google Scholar]
- 134.Wu X.J., Zhao Z.F., Kang X.J., Wang H.J., Zhao J., Pu X.M. MicroRNA-126-3p suppresses cell proliferation by targeting PIK3R2 in Kaposi’s sarcoma cells. Oncotarget. 2016;7:36614–36621. doi: 10.18632/oncotarget.9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Matsha T.E., Kengne A.P., Hector S., Mbu D.L., Yako Y.Y., Erasmus R.T. MicroRNA profiling and their pathways in South African individuals with prediabetes and newly diagnosed type 2 diabetes mellitus. Oncotarget. 2018;9:30485–30498. doi: 10.18632/oncotarget.25271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lan X., Wu L., Wu N., Chen Q., Li Y., Du X., Wei C., Feng L., Li Y., Osoro E.K., et al. Long Noncoding RNA lnc-HC Regulates PPARγ-Mediated Hepatic Lipid Metabolism through miR-130b-3p. Mol. Ther. Nucleic Acids. 2019;18:954–965. doi: 10.1016/j.omtn.2019.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tryggestad J.B., Vishwanath A., Jiang S., Mallappa A., Teague A.M., Takahashi Y., Thompson D.M., Chernausek S.D. Influence of gestational diabetes mellitus on human umbilical vein endothelial cell miRNA. Clin. Sci. 2016;130:1955–1967. doi: 10.1042/CS20160305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang J., Jazii F.R., Haghighi M.M., Alvares D., Liu L., Khosraviani N., Adeli K. miR-130b is a potent stimulator of hepatic very-low-density lipoprotein assembly and secretion via marked induction of microsomal triglyceride transfer protein. Am. J. Physiol. Endocrinol. Metab. 2020;318:E262–E275. doi: 10.1152/ajpendo.00276.2019. [DOI] [PubMed] [Google Scholar]
- 139.Li P., Zhang Q., Wu X., Yang X., Zhang Y., Li Y., Jiang F. Circulating microRNAs serve as novel biological markers for intracranial aneurysms. J. Am. Heart Assoc. 2014;3:e000972. doi: 10.1161/JAHA.114.000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tian C., Li Z., Yang Z., Huang Q., Liu J., Hong B. Plasma MicroRNA-16 Is a Biomarker for Diagnosis, Stratification, and Prognosis of Hyperacute Cerebral Infarction. PLoS ONE. 2016;11:e0166688. doi: 10.1371/journal.pone.0166688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Prabu P., Rome S., Sathishkumar C., Aravind S., Mahalingam B., Shanthirani C.S., Gastebois C., Villard A., Mohan V., Balasubramanyam M. Circulating MiRNAs of ‘Asian Indian Phenotype’ Identified in Subjects with Impaired Glucose Tolerance and Patients with Type 2 Diabetes. PLoS ONE. 2015;10:e0128372. doi: 10.1371/journal.pone.0128372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Feng T., Li K., Zheng P., Wang Y., Lv Y., Shen L., Chen Y., Xue Z., Li B., Jin L., et al. Weighted Gene Coexpression Network Analysis Identified MicroRNA Coexpression Modules and Related Pathways in Type 2 Diabetes Mellitus. Oxidative Med. Cell. Longev. 2019;2019:9567641. doi: 10.1155/2019/9567641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liang H.W., Yang X., Wen D.Y., Gao L., Zhang X.Y., Ye Z.H., Luo J., Li Z.Y., He Y., Pang Y.Y., et al. Utility of miR 133a 3p as a diagnostic indicator for hepatocellular carcinoma: An investigation combined with GEO, TCGA, meta analysis and bioinformatics. Mol. Med. Rep. 2018;17:1469–1484. doi: 10.3892/mmr.2017.8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.van Rooij E., Olson E.N. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J. Clin. Investig. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang J., Xu R., Lin F., Zhang S., Zhang G., Hu S., Zheng Z. MicroRNA: Novel regulators involved in the remodeling and reverse remodeling of the heart. Cardiology. 2009;113:81–88. doi: 10.1159/000172616. [DOI] [PubMed] [Google Scholar]
- 146.Kukreja R.C., Yin C., Salloum F.N. MicroRNAs: New players in cardiac injury and protection. Mol. Pharmacol. 2011;80:558–564. doi: 10.1124/mol.111.073528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Duisters R.F., Tijsen A.J., Schroen B., Leenders J.J., Lentink V., van der Made I., Herias V., van Leeuwen R.E., Schellings M.W., Barenbrug P., et al. miR-133 and miR-30 regulate connective tissue growth factor: Implications for a role of microRNAs in myocardial matrix remodeling. Circ. Res. 2009;104:170–178. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- 148.Liu W., Ling S., Sun W., Liu T., Li Y., Zhong G., Zhao D., Zhang P., Song J., Jin X., et al. Circulating microRNAs correlated with the level of coronary artery calcification in symptomatic patients. Sci. Rep. 2015;5:16099. doi: 10.1038/srep16099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jiang Y., Zhang M., He H., Chen J., Zeng H., Li J., Duan R. MicroRNA/mRNA profiling and regulatory network of intracranial aneurysm. BMC Med. Genom. 2013;6:36. doi: 10.1186/1755-8794-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu H., Xiong W., Liu F., Lin F., He J., Liu C., Lin Y., Dong S. Significant role and mechanism of microRNA-143-3p/KLLN axis in the development of coronary heart disease. Am. J. Transl. Res. 2019;11:3610–3619. [PMC free article] [PubMed] [Google Scholar]
- 151.Li C., Li J., Xue K., Zhang J., Wang C., Zhang Q., Chen X., Gao C., Yu X., Sun L. MicroRNA-143-3p promotes human cardiac fibrosis via targeting sprouty3 after myocardial infarction. J. Mol. Cell. Cardiol. 2019;129:281–292. doi: 10.1016/j.yjmcc.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 152.Yu B., Zhao Y., Zhang H., Xie D., Nie W., Shi K. Inhibition of microRNA-143-3p attenuates myocardial hypertrophy by inhibiting inflammatory response. Cell Biol. Int. 2018;42:1584–1593. doi: 10.1002/cbin.11053. [DOI] [PubMed] [Google Scholar]
- 153.Jiao M., You H.Z., Yang X.Y., Yuan H., Li Y.L., Liu W.X., Jin M., Du J. Circulating microRNA signature for the diagnosis of childhood dilated cardiomyopathy. Sci. Rep. 2018;8:724. doi: 10.1038/s41598-017-19138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Deng L., Blanco F.J., Stevens H., Lu R., Caudrillier A., McBride M., McClure J.D., Grant J., Thomas M., Frid M., et al. MicroRNA-143 Activation Regulates Smooth Muscle and Endothelial Cell Crosstalk in Pulmonary Arterial Hypertension. Circ. Res. 2015;117:870–883. doi: 10.1161/CIRCRESAHA.115.306806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shi L., Tian C., Sun L., Cao F., Meng Z. The lncRNA TUG1/miR-145-5p/FGF10 regulates proliferation and migration in VSMCs of hypertension. Biochem. Biophys. Res. Commun. 2018;501:688–695. doi: 10.1016/j.bbrc.2018.05.049. [DOI] [PubMed] [Google Scholar]
- 156.Yang X., Niu X., Xiao Y., Lin K., Chen X. MiRNA expression profiles in healthy OSAHS and OSAHS with arterial hypertension: Potential diagnostic and early warning markers. Respir. Res. 2018;19:194. doi: 10.1186/s12931-018-0894-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Toro R., Blasco-Turrión S., Morales-Ponce F.J., Gonzalez P., Martínez-Camblor P., López-Granados A., Brugada R., Campuzano O., Pérez-Serra A., Rosa Longobardo F., et al. Plasma microRNAs as biomarkers for Lamin A/C-related dilated cardiomyopathy. J. Mol. Med. 2018;96:845–856. doi: 10.1007/s00109-018-1666-1. [DOI] [PubMed] [Google Scholar]
- 158.Yuan M., Zhang L., You F., Zhou J., Ma Y., Yang F., Tao L. MiR-145-5p regulates hypoxia-induced inflammatory response and apoptosis in cardiomyocytes by targeting CD40. Mol. Cell. Biochem. 2017;431:123–131. doi: 10.1007/s11010-017-2982-4. [DOI] [PubMed] [Google Scholar]
- 159.Wu G., Tan J., Li J., Sun X., Du L., Tao S. miRNA-145-5p induces apoptosis after ischemia-reperfusion by targeting dual specificity phosphatase J. Cell. Physiol. 2019;234:16281–16289. doi: 10.1002/jcp.28291. [DOI] [PubMed] [Google Scholar]
- 160.Xie X., Peng L., Zhu J., Zhou Y., Li L., Chen Y., Yu S., Zhao Y. miR-145-5p/Nurr1/TNF-α Signaling-Induced Microglia Activation Regulates Neuron Injury of Acute Cerebral Ischemic/Reperfusion in Rats. Front. Mol. Neurosci. 2017;10:383. doi: 10.3389/fnmol.2017.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nunez Lopez Y.O., Retnakaran R., Zinman B., Pratley R.E., Seyhan A.A. Predicting and understanding the response to short-term intensive insulin therapy in people with early type 2 diabetes. Mol. Metab. 2019;20:63–78. doi: 10.1016/j.molmet.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang J., Cui C., Xu H. Downregulation of miR-145-5p elevates retinal ganglion cell survival to delay diabetic retinopathy progress by targeting FGF5. Biosci. Biotechnol. Biochem. 2019;83:1655–1662. doi: 10.1080/09168451.2019.1630251. [DOI] [PubMed] [Google Scholar]
- 163.Zamanian Azodi M., Rezaei-Tavirani M., Rezaei-Tavirani M., Robati R.M. Gestational Diabetes Mellitus Regulatory Network Identifies hsa-miR-145-5p and hsa-miR-875-5p as Potential Biomarkers. Int. J. Endocrinol. Metab. 2019;17:e86640. doi: 10.5812/ijem.86640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Paterson M.R., Kriegel A.J. MiR-146a/b: A family with shared seeds and different roots. Physiol. Genom. 2017;49:243–252. doi: 10.1152/physiolgenomics.00133.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang X., Ye Z.H., Liang H.W., Ren F.H., Li P., Dang Y.W., Chen G. Down-regulation of miR-146a-5p and its potential targets in hepatocellular carcinoma validated by a TCGA- and GEO-based study. FEBS Open Bio. 2017;7:504–521. doi: 10.1002/2211-5463.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang X., Ha T., Liu L., Zou J., Zhang X., Kalbfleisch J., Gao X., Williams D., Li C. Increased expression of microRNA-146a decreases myocardial ischaemia/reperfusion injury. Cardiovasc. Res. 2013;97:432–442. doi: 10.1093/cvr/cvs356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Quan X., Ji Y., Zhang C., Guo X., Zhang Y., Jia S., Ma W., Fan Y., Wang C. Circulating MiR-146a May be a Potential Biomarker of Coronary Heart Disease in Patients with Subclinical Hypothyroidism. Cell. Physiol. Biochem. 2018;45:226–236. doi: 10.1159/000486769. [DOI] [PubMed] [Google Scholar]
- 169.Li S.H., Chen L., Pang X.M., Su S.Y., Zhou X., Chen C.Y., Huang L.G., Li J.P., Liu J.L. Decreased miR-146a expression in acute ischemic stroke directly targets the Fbxl10 mRNA and is involved in modulating apoptosis. Neurochem. Int. 2017;107:156–167. doi: 10.1016/j.neuint.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 170.Barberio M.D., Kasselman L.J., Playford M.P., Epstein S.B., Renna H.A., Goldberg M., DeLeon J., Voloshyna I., Barlev A., Salama M., et al. Cholesterol efflux alterations in adolescent obesity: Role of adipose-derived extracellular vesical microRNAs. J. Transl. Med. 2019;17:232. doi: 10.1186/s12967-019-1980-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gaudet A.D., Fonken L.K., Gushchina L.V., Aubrecht T.G., Maurya S.K., Periasamy M., Nelson R.J., Popovich P.G. miR-155 Deletion in Female Mice Prevents Diet-Induced Obesity. Sci. Rep. 2016;6:22862. doi: 10.1038/srep22862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chen L., Zheng S.Y., Yang C.Q., Ma B.M., Jiang D. MiR-155-5p inhibits the proliferation and migration of VSMCs and HUVECs in atherosclerosis by targeting AKT1. Eur. Rev. Med. Pharmacol. Sci. 2019;23:2223–2233. doi: 10.26355/eurrev_201903_17270. [DOI] [PubMed] [Google Scholar]
- 173.Zhu M., Wei Y., Geißler C., Abschlag K., Corbalán Campos J., Hristov M., Möllmann J., Lehrke M., Karshovska E., Schober A. Hyperlipidemia-Induced MicroRNA-155-5p Improves β-Cell Function by Targeting Mafb. Diabetes. 2017;66:3072–3084. doi: 10.2337/db17-0313. [DOI] [PubMed] [Google Scholar]
- 174.Li S., Lee C., Song J., Lu C., Liu J., Cui Y., Liang H., Cao C., Zhang F., Chen H. Circulating microRNAs as potential biomarkers for coronary plaque rupture. Oncotarget. 2017;8:48145–48156. doi: 10.18632/oncotarget.18308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mukai N., Nakayama Y., Murakami S., Tanahashi T., Sessler D.I., Ishii S., Ogawa S., Tokuhira N., Mizobe T., Sawa T., et al. Potential contribution of erythrocyte microRNA to secondary erythrocytosis and thrombocytopenia in congenital heart disease. Pediatr. Res. 2018;83:866–873. doi: 10.1038/pr.2017.327. [DOI] [PubMed] [Google Scholar]
- 176.Klimczak D., Kuch M., Pilecki T., Żochowska D., Wirkowska A., Pączek L. Plasma microRNA-155-5p is increased among patients with chronic kidney disease and nocturnal hypertension. J. Am. Soc. Hypertens. 2017;11:831–841.e4. doi: 10.1016/j.jash.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 177.Wang M., Sun L., Ding W., Cai S., Zhao Q. Ablation alleviates atrial fibrillation by regulating the signaling pathways of endothelial nitric oxide synthase/nitric oxide via miR-155-5p and miR-24-3p. J. Cell. Biochem. 2019;120:4451–4462. doi: 10.1002/jcb.27733. [DOI] [PubMed] [Google Scholar]
- 178.Sun X., Sit A., Feinberg M.W. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc. Med. 2014;24:105–112. doi: 10.1016/j.tcm.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hulsmans M., Sinnaeve P., Van der Schueren B., Mathieu C., Janssens S., Holvoet P. Decreased miR-181a expression in monocytes of obese patients is associated with the occurrence of metabolic syndrome and coronary artery disease. J. Clin. Endocrinol. Metab. 2012;97:E1213–E1218. doi: 10.1210/jc.2012-1008. [DOI] [PubMed] [Google Scholar]
- 180.Du X., Yang Y., Xu C., Peng Z., Zhang M., Lei L., Gao W., Dong Y., Shi Z., Sun X., et al. Upregulation of miR-181a impairs hepatic glucose and lipid homeostasis. Oncotarget. 2017;8:91362–91378. doi: 10.18632/oncotarget.20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wu J., Fan C.L., Ma L.J., Liu T., Wang C., Song J.X., Lv Q.S., Pan H., Zhang C.N., Wang J.J. Distinctive expression signatures of serum microRNAs in ischaemic stroke and transient ischaemic attack patients. Thromb. Haemost. 2017;117:992–1001. doi: 10.1160/TH16-08-0606. [DOI] [PubMed] [Google Scholar]
- 182.Zhu J., Yao K., Wang Q., Guo J., Shi H., Ma L., Liu H., Gao W., Zou Y., Ge J. Circulating miR-181a as a Potential Novel Biomarker for Diagnosis of Acute Myocardial Infarction. Cell. Physiol. Biochem. 2016;40:1591–1602. doi: 10.1159/000453209. [DOI] [PubMed] [Google Scholar]
- 183.Nabih E.S., Andrawes N.G. The Association Between Circulating Levels of miRNA-181a and Pancreatic Beta Cells Dysfunction via SMAD7 in Type 1 Diabetic Children and Adolescents. J. Clin. Lab. Anal. 2016;30:727–731. doi: 10.1002/jcla.21928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.He J.F., Luo Y.M., Wan X.H., Jiang D. Biogenesis of MiRNA-195 and its role in biogenesis, the cell cycle, and apoptosis. J. Biochem. Mol. Toxicol. 2011;25:404–408. doi: 10.1002/jbt.20396. [DOI] [PubMed] [Google Scholar]
- 185.van Rooij E., Sutherland L.B., Liu N., Williams A.H., McAnally J., Gerard R.D., Richardson J.A., Olson E.N. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.You X.Y., Huang J.H., Liu B., Liu S.J., Zhong Y., Liu S.M. HMGA1 is a new target of miR-195 involving isoprenaline-induced cardiomyocyte hypertrophy. Biochemistry. 2014;79:538–544. doi: 10.1134/S0006297914060078. [DOI] [PubMed] [Google Scholar]
- 187.Zampetaki A., Attia R., Mayr U., Gomes R.S., Phinikaridou A., Yin X., Langley S.R., Willeit P., Lu R., Fanshawe B., et al. Role of miR-195 in aortic aneurysmal disease. Circ. Res. 2014;115:857–866. doi: 10.1161/CIRCRESAHA.115.304361. [DOI] [PubMed] [Google Scholar]
- 188.Du J., Zheng R., Xiao F., Zhang S., He K., Zhang J., Shao Y. Downregulated MicroRNA-195 in the Bicuspid Aortic Valve Promotes Calcification of Valve Interstitial Cells via Targeting SMAD7. Cell. Physiol. Biochem. 2017;44:884–896. doi: 10.1159/000485356. [DOI] [PubMed] [Google Scholar]
- 189.Collares C.V., Evangelista A.F., Xavier D.J., Rassi D.M., Arns T., Foss-Freitas M.C., Foss M.C., Puthier D., Sakamoto-Hojo E.T., Passos G.A., et al. Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients. BMC Res. Notes. 2013;6:491. doi: 10.1186/1756-0500-6-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tagoma A., Alnek K., Kirss A., Uibo R., Haller-Kikkatalo K. MicroRNA profiling of second trimester maternal plasma shows upregulation of miR-195-5p in patients with gestational diabetes. Gene. 2018;672:137–142. doi: 10.1016/j.gene.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 191.Massaro J.D., Polli C.D., Costa e Silva M., Alves C.C., Passos G.A., Sakamoto-Hojo E.T., de Holanda Miranda W.R., Bispo Cezar N.J., Rassi D.M., Crispim F., et al. Post-transcriptional markers associated with clinical complications in Type 1 and Type 2 diabetes mellitus. Mol. Cell. Endocrinol. 2019;490:1–14. doi: 10.1016/j.mce.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 192.Li M., Luan L., Liu Q., Liu Y., Lan X., Li Z., Liu W. MiRNA-199a-5p Protects Against Cerebral Ischemic Injury by Down-Regulating DDR1 in Rats. World Neurosurg. 2019;131:e486–e494. doi: 10.1016/j.wneu.2019.07.203. [DOI] [PubMed] [Google Scholar]
- 193.Yan M., Yang S., Meng F., Zhao Z., Tian Z., Yang P. MicroRNA 199a-5p induces apoptosis by targeting JunB. Sci. Rep. 2018;8:6699. doi: 10.1038/s41598-018-24932-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lynch S.M., Ward M., McNulty H., Angel C.Z., Horigan G., Strain J.J., Purvis J., Tackett M., McKenna D.J. Serum levels of miR-199a-5p correlates with blood pressure in premature cardiovascular disease patients homozygous for the MTHFR 677C > T polymorphism. Genomics. 2020;112:669–676. doi: 10.1016/j.ygeno.2019.04.019. [DOI] [PubMed] [Google Scholar]
- 195.Tian X., Yu C., Shi L., Li D., Chen X., Xia D., Zhou J., Xu W., Ma C., Gu L., et al. MicroRNA-199a-5p aggravates primary hypertension by damaging vascular endothelial cells through inhibition of autophagy and promotion of apoptosis. Exp. Ther. Med. 2018;16:595–602. doi: 10.3892/etm.2018.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhou Y., Pang B., Xiao Y., Zhou S., He B., Zhang F., Liu W., Peng H., Li P. The protective microRNA-199a-5p-mediated unfolded protein response in hypoxic cardiomyocytes is regulated by STAT3 pathway. J. Physiol. Biochem. 2019;75:73–81. doi: 10.1007/s13105-018-0657-6. [DOI] [PubMed] [Google Scholar]
- 197.Liu Y., Liu G., Zhang H., Wang J. MiRNA-199a-5p influences pulmonary artery hypertension via downregulating Smad3. Biochem. Biophys. Res. Commun. 2016;473:859–866. doi: 10.1016/j.bbrc.2016.03.140. [DOI] [PubMed] [Google Scholar]
- 198.Wang J., Yu G. A Systems Biology Approach to Characterize Biomarkers for Blood Stasis Syndrome of Unstable Angina Patients by Integrating MicroRNA and Messenger RNA Expression Profiling. Evid.-Based Complement. Altern. Med. 2013;2013:510208. doi: 10.1155/2013/510208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yu L., Gu T., Shi E., Wang Y., Fang Q., Wang C. Dysregulation of renal microRNA expression after deep hypothermic circulatory arrest in rats. Eur. J. Cardiothorac Surg. 2016;49:1725–1731. doi: 10.1093/ejcts/ezv460. [DOI] [PubMed] [Google Scholar]
- 200.Hirt M.N., Werner T., Indenbirken D., Alawi M., Demin P., Kunze A.C., Stenzig J., Starbatty J., Hansen A., Fiedler J., et al. Deciphering the microRNA signature of pathological cardiac hypertrophy by engineered heart tissue- and sequencing-technology. J. Mol. Cell. Cardiol. 2015;81:1–9. doi: 10.1016/j.yjmcc.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 201.Aguado-Fraile E., Ramos E., Conde E., Rodríguez M., Martín-Gómez L., Lietor A., Candela Á., Ponte B., Liaño F., García-Bermejo M.L. A Pilot Study Identifying a Set of microRNAs As Precise Diagnostic Biomarkers of Acute Kidney Injury. PLoS ONE. 2015;10:e0127175. doi: 10.1371/journal.pone.0127175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ma H., Chen P., Sang C., Huang D., Geng Q., Wang L. Modulation of apoptosis-related microRNAs following myocardial infarction in fat-1 transgenic mice vs wild-type mice. J. Cell. Mol. Med. 2018;22:5698–5707. doi: 10.1111/jcmm.13846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Qiao X.R., Wang L., Liu M., Tian Y., Chen T. MiR-210-3p attenuates lipid accumulation and inflammation in atherosclerosis by repressing IGF2. Biosci. Biotechnol. Biochem. 2020;84:321–329. doi: 10.1080/09168451.2019.1685370. [DOI] [PubMed] [Google Scholar]
- 204.Derda A.A., Pfanne A., Bwangär C., Schimmel K., Kennel P.J., Xiao K., Schulze P.C., Bauersachs J., Thum T. Blood-based microRNA profiling in patients with cardiac amyloidosis. PLoS ONE. 2018;13:e0204235. doi: 10.1371/journal.pone.0204235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Verjans R., Peters T., Beaumont F.J., van Leeuwen R., van Herwaarden T., Verhesen W., Munts C., Bijnen M., Henkens M., Diez J., et al. MicroRNA-221/222 Family Counteracts Myocardial Fibrosis in Pressure Overload-Induced Heart Failure. Hypertension. 2018;71:280–288. doi: 10.1161/HYPERTENSIONAHA.117.10094. [DOI] [PubMed] [Google Scholar]
- 206.Zhuang X., Li R., Maimaitijiang A., Liu R., Yan F., Hu H., Gao X., Shi H. miR-221-3p inhibits oxidized low-density lipoprotein induced oxidative stress and apoptosis via targeting a disintegrin and metalloprotease-22. J. Cell. Biochem. 2019;120:6304–6314. doi: 10.1002/jcb.27917. [DOI] [PubMed] [Google Scholar]
- 207.Pereira-da-Silva T., Coutinho Cruz M., Carrusca C., Cruz Ferreira R., Napoleão P., Mota Carmo M. Circulating microRNA profiles in different arterial territories of stable atherosclerotic disease: A systematic review. Am. J. Cardiovasc. Dis. 2018;8:1–13. [PMC free article] [PubMed] [Google Scholar]
- 208.Coffey S., Williams M.J., Phillips L.V., Galvin I.F., Bunton R.W., Jones G.T. Integrated microRNA and messenger RNA analysis in aortic stenosis. Sci. Rep. 2016;6:36904. doi: 10.1038/srep36904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Coskunpinar E., Cakmak H.A., Kalkan A.K., Tiryakioglu N.O., Erturk M., Ongen Z. Circulating miR-221-3p as a novel marker for early prediction of acute myocardial infarction. Gene. 2016;591:90–96. doi: 10.1016/j.gene.2016.06.059. [DOI] [PubMed] [Google Scholar]
- 210.Sørensen S.S., Nygaard A.B., Nielsen M.Y., Jensen K., Christensen T. miRNA expression profiles in cerebrospinal fluid and blood of patients with acute ischemic stroke. Transl. Stroke Res. 2014;5:711–718. doi: 10.1007/s12975-014-0364-8. [DOI] [PubMed] [Google Scholar]
- 211.Gusar V.A., Timofeeva A.V., Zhanin I.S., Shram S.I., Pinelis V.G. Estimation of Time-Dependent microRNA Expression Patterns in Brain Tissue, Leukocytes, and Blood Plasma of Rats under Photochemically Induced Focal Cerebral Ischemia. Mol. Biol. 2017;51:683–695. doi: 10.1134/S0026893317040100. [DOI] [PubMed] [Google Scholar]
- 212.Nie X., Chen Y., Tan J., Dai Y., Mao W., Qin G., Ye S., Sun J., Yang Z., Chen J. MicroRNA-221-3p promotes pulmonary artery smooth muscle cells proliferation by targeting AXIN2 during pulmonary arterial hypertension. Vascul. Pharmacol. 2019;116:24–35. doi: 10.1016/j.vph.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 213.Villard A., Marchand L., Thivolet C., Rome S. Diagnostic Value of Cell-free Circulating MicroRNAs for Obesity and Type 2 Diabetes: A Meta-analysis. J. Mol. Biomark. Diagn. 2015;6:251. doi: 10.4172/2155-9929.1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang L., Xu L., Xu M., Liu G., Xing J., Sun C., Ding H. Obesity-Associated MiR-342-3p Promotes Adipogenesis of Mesenchymal Stem Cells by Suppressing CtBP2 and Releasing C/EBPα from CtBP2 Binding. Cell. Physiol. Biochem. 2015;35:2285–2298. doi: 10.1159/000374032. [DOI] [PubMed] [Google Scholar]
- 215.Hezova R., Slaby O., Faltejskova P., Mikulkova Z., Buresova I., Raja K.R., Hodek J., Ovesna J., Michalek J. microRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell. Immunol. 2010;260:70–74. doi: 10.1016/j.cellimm.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 216.Eissa S., Matboli M., Bekhet M.M. Clinical verification of a novel urinary microRNA panal: 133b, -342 and -30 as biomarkers for diabetic nephropathy identified by bioinformatics analysis. Biomed. Pharmacother. 2016;83:92–99. doi: 10.1016/j.biopha.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 217.Cheng S., Cui Y., Fan L., Mu X., Hua Y. T2DM inhibition of endothelial miR-342-3p facilitates angiogenic dysfunction via repression of FGF11 signaling. Biochem. Biophys. Res. Commun. 2018;503:71–78. doi: 10.1016/j.bbrc.2018.05.179. [DOI] [PubMed] [Google Scholar]
- 218.Khalyfa A., Kheirandish-Gozal L., Bhattacharjee R., Khalyfa A.A., Gozal D. Circulating microRNAs as Potential Biomarkers of Endothelial Dysfunction in Obese Children. Chest. 2016;149:786–800. doi: 10.1378/chest.15-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hoekstra M. MicroRNA-499-5p: A therapeutic target in the context of cardiovascular disease. Ann. Transl. Med. 2016;4:539. doi: 10.21037/atm.2016.11.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhao L., Wang B., Zhang W., Sun L. Effect of miR-499a-5p on damage of cardiomyocyte induced by hypoxia-reoxygenation via downregulating CD38 protein. J. Cell. Biochem. 2020;121:996–1004. doi: 10.1002/jcb.29334. [DOI] [PubMed] [Google Scholar]
- 221.Neshati V., Mollazadeh S., Fazly Bazzaz B.S., de Vries A.A.F., Mojarrad M., Naderi-Meshkin H., Neshati Z., Mirahmadi M., Kerachian M.A. MicroRNA-499a-5p Promotes Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells to Cardiomyocytes. Appl. Biochem. Biotechnol. 2018;186:245–255. doi: 10.1007/s12010-018-2734-2. [DOI] [PubMed] [Google Scholar]
- 222.Boštjančič E., Zidar N., Glavač D. MicroRNAs and cardiac sarcoplasmic reticulum calcium ATPase-2 in human myocardial infarction: Expression and bioinformatic analysis. BMC Genom. 2012;13:552. doi: 10.1186/1471-2164-13-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Salinas J., Lin H., Aparico H.J., Huan T., Liu C., Rong J., Beiser A., Himali J.J., Freedman J.E., Larson M.G., et al. Whole blood microRNA expression associated with stroke: Results from the Framingham Heart Study. PLoS ONE. 2019;14:e0219261. doi: 10.1371/journal.pone.0219261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Baldeón Rojas L., Weigelt K., de Wit H., Ozcan B., van Oudenaren A., Sempértegui F., Sijbrands E., Grosse L., van Zonneveld A.J., Drexhage H.A., et al. Study on inflammation-related genes and microRNAs, with special emphasis on the vascular repair factor HGF and miR-574-3p, in monocytes and serum of patients with T2D. Diabetol. Metab. Syndr. 2016;8:6. doi: 10.1186/s13098-015-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hromadnikova I., Kotlabova K., Dvorakova L., Krofta L., Sirc J. Substantially Altered Expression Profile of Diabetes/Cardiovascular/Cerebrovascular Disease Associated microRNAs in Children Descending from Pregnancy Complicated by Gestational Diabetes Mellitus-One of Several Possible Reasons for an Increased Cardiovascular Risk. Cells. 2020;9:1557. doi: 10.3390/cells9061557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Yoffe L., Polsky A., Gilam A., Raff C., Mecacci F., Ognibene A., Crispi F., Gratacós E., Kanety H., Mazaki-Tovi S., et al. Early diagnosis of gestational diabetes mellitus using circulating microRNAs. Eur. J. Endocrinol. 2019;181:565–577. doi: 10.1530/EJE-19-0206. [DOI] [PubMed] [Google Scholar]
- 227.Légaré C., Desgagné V., Poirier C., Thibeault K., White F., Clément A.A., Scott M.S., Jacques P.É., Perron P., Guérin R., et al. First trimester plasma microRNAs levels predict Matsuda Index-estimated insulin sensitivity between 24th and 29th week of pregnancy. BMJ Open Diabetes Res. Care. 2022;10:e002703. doi: 10.1136/bmjdrc-2021-002703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Juchnicka I., Kuźmicki M., Niemira M., Bielska A., Sidorkiewicz I., Zbucka-Krętowska M., Krętowski A.J., Szamatowicz J. miRNAs as Predictive Factors in Early Diagnosis of Gestational Diabetes Mellitus. Front. Endocrinol. 2022;13:839344. doi: 10.3389/fendo.2022.839344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zhang L., Zhang T., Sun D., Cheng G., Ren H., Hong H., Chen L., Jiao X., Du Y., Zou Y., et al. Diagnostic value of dysregulated microribonucleic acids in the placenta and circulating exosomes in gestational diabetes mellitus. J. Diabetes Investig. 2021;12:1490–1500. doi: 10.1111/jdi.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wang J., Pan Y., Dai F., Wang F., Qiu H., Huang X. Serum miR-195-5p is upregulated in gestational diabetes mellitus. J. Clin. Lab. Anal. 2020;34:e23325. doi: 10.1002/jcla.23325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sørensen A.E., van Poppel M.N.M., Desoye G., Damm P., Simmons D., Jensen D.M., Dalgaard L.T., The DALI Core Investigator Group The Predictive Value of miR-16, -29a and -134 for Early Identification of Gestational Diabetes: A Nested Analysis of the DALI Cohort. Cells. 2021;10:170. doi: 10.3390/cells10010170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zhao C., Dong J., Jiang T., Shi Z., Yu B., Zhu Y., Chen D., Xu J., Huo R., Dai J., et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS ONE. 2011;6:e23925. doi: 10.1371/journal.pone.0023925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kennedy M., Cartland S., Saravanan P., Simpson N., Scott E., Forbes K. miR-1-3p and miR-133-3p are altered in maternal serum EVs and placenta in pregnancies complicated by gestational diabetes with large-for-gestational age babies. Endocr. Abstr. 2019;65:349. doi: 10.1530/endoabs.65.P349. [DOI] [Google Scholar]
- 234.Wang F., Zhang X., Zhou H. Role of cell free microRNA-19a and microRNA-19b in gestational diabetes mellitus patients. 3 Biotech. 2019;9:406. doi: 10.1007/s13205-019-1952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Feng Y., Qu X., Chen Y., Feng Q., Zhang Y., Hu J., Li X. MicroRNA-33a-5p sponges to inhibit pancreatic β-cell function in gestational diabetes mellitus LncRNA DANCR. Reprod. Biol. Endocrinol. 2020;18:61. doi: 10.1186/s12958-020-00618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sebastiani G., Guarino E., Grieco G.E., Formichi C., Delli Poggi C., Ceccarelli E., Dotta F. Circulating microRNA (miRNA) Expression Profiling in Plasma of Patients with Gestational Diabetes Mellitus Reveals Upregulation of miRNA miR-330-3p. Front. Endocrinol. 2017;8:345. doi: 10.3389/fendo.2017.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Stirm L., Huypens P., Sass S., Batra R., Fritsche L., Brucker S., Abele H., Hennige A.M., Theis F., Beckers J., et al. Maternal whole blood cell miRNA-340 is elevated in gestational diabetes and inversely regulated by glucose and insulin. Sci. Rep. 2018;8:1366. doi: 10.1038/s41598-018-19200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Xu K., Bian D., Hao L., Huang F., Xu M., Qin J., Liu Y. microRNA-503 contribute to pancreatic beta cell dysfunction by targeting the mTOR pathway in gestational diabetes mellitus. EXCLI J. 2017;16:1177–1187. doi: 10.17179/excli2017-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Liu L., Zhang J., Liu Y. MicroRNA-1323 serves as a biomarker in gestational diabetes mellitus and aggravates high glucose-induced inhibition of trophoblast cell viability by suppressing TP53INP1. Exp. Ther. Med. 2021;21:230. doi: 10.3892/etm.2021.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Hromadnikova I., Kotlabova K., Krofta L. Cardiovascular Disease-Associated MicroRNA Dysregulation during the First Trimester of Gestation in Women with Chronic Hypertension and Normotensive Women Subsequently Developing Gestational Hypertension or Preeclampsia with or without Fetal Growth Restriction. Biomedicines. 2022;10:256. doi: 10.3390/biomedicines10020256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hromadnikova I., Kotlabova K., Krofta L. First-Trimester Screening for Fetal Growth Restriction and Small-for-Gestational-Age Pregnancies without Preeclampsia Using Cardiovascular Disease-Associated MicroRNA Biomarkers. Biomedicines. 2022;10:718. doi: 10.3390/biomedicines10030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Hromadnikova I., Kotlabova K., Krofta L. First Trimester Prediction of Preterm Delivery in the Absence of Other Pregnancy-Related Complications Using Cardiovascular-Disease Associated MicroRNA Biomarkers. Int. J. Mol. Sci. 2022;23:3951. doi: 10.3390/ijms23073951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hromadnikova I., Kotlabova K., Dvorakova L., Krofta L. Diabetes Mellitus and Cardiovascular Risk Assessment in Mothers with a History of Gestational Diabetes Mellitus Based on Postpartal Expression Profile of MicroRNAs Associated with Diabetes Mellitus and Cardiovascular and Cerebrovascular Diseases. Int. J. Mol. Sci. 2020;21:2437. doi: 10.3390/ijms21072437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Shah B.R., Retnakaran R., Booth G.L. Increased risk of cardiovascular disease in young women following gestational diabetes mellitus. Diabetes Care. 2008;31:1668–1669. doi: 10.2337/dc08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kramer C.K., Campbell S., Retnakaran R. Gestational diabetes and the risk of cardiovascular disease in women: A systematic review and meta-analysis. Diabetologia. 2019;62:905–914. doi: 10.1007/s00125-019-4840-2. [DOI] [PubMed] [Google Scholar]
- 246.Yu Y., Soohoo M., Sørensen H.T., Li J., Arah O.A. Gestational Diabetes Mellitus and the Risks of Overall and Type-Specific Cardiovascular Diseases: A Population- and Sibling-Matched Cohort Study. Diabetes Care. 2022;45:151–159. doi: 10.2337/dc21-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Bentley-Lewis R. Late cardiovascular consequences of gestational diabetes mellitus. Semin. Reprod. Med. 2009;27:322–329. doi: 10.1055/s-0029-1225260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Shostrom D.C.V., Sun Y., Oleson J.J., Snetselaar L.G., Bao W. History of Gestational Diabetes Mellitus in Relation to Cardiovascular Disease and Cardiovascular Risk Factors in US Women. Front. Endocrinol. 2017;8:144. doi: 10.3389/fendo.2017.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Peng Y.S., Lin J.R., Cheng B.H., Ho C., Lin Y.H., Shen C.H., Tsai M.H. Incidence and relative risk for developing cancers in women with gestational diabetes mellitus: A nationwide cohort study in Taiwan. BMJ Open. 2019;9:e024583. doi: 10.1136/bmjopen-2018-024583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Han K.T., Cho G.J., Kim E.H. Evaluation of the Association between Gestational Diabetes Mellitus at First Pregnancy and Cancer within 10 Years Postpartum Using National Health Insurance Data in South Korea. Int. J. Environ. Res. Public Health. 2018;15:2646. doi: 10.3390/ijerph15122646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Liu Y., Chen X., Sheng J., Sun X., Chen G.Q., Zhao M., Chen Q. Complications of Pregnancy and the Risk of Developing Endometrial or Ovarian Cancer: A Case-Control Study. Front. Endocrinol. 2021;12:642928. doi: 10.3389/fendo.2021.642928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Simon J., Goueslard K., Arveux P., Bechraoui-Quantin S., Petit J.M., Quantin C. Increased Risk of Hospitalization for Pancreatic Cancer in the First 8 Years after a Gestational Diabetes Mellitus regardless of Subsequent Type 2 Diabetes: A Nationwide Population-Based Study. Cancers. 2021;13:308. doi: 10.3390/cancers13020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Fuchs O., Sheiner E., Meirovitz M., Davidson E., Sergienko R., Kessous R. The association between a history of gestational diabetes mellitus and future risk for female malignancies. Arch. Gynecol. Obstet. 2017;295:731–736. doi: 10.1007/s00404-016-4275-7. [DOI] [PubMed] [Google Scholar]
- 254.Dawson S.I. Long-term risk of malignant neoplasm associated with gestational glucose intolerance. Cancer. 2004;100:149–155. doi: 10.1002/cncr.20013. [DOI] [PubMed] [Google Scholar]
- 255.Perrin M.C., Terry M.B., Kleinhaus K., Deutsch L., Yanetz R., Tiram E., Calderon-Margalit R., Friedlander Y., Paltiel O., Harlap S. Gestational diabetes and the risk of breast cancer among women in the Jerusalem Perinatal Study. Breast Cancer Res. Treat. 2008;108:129–135. doi: 10.1007/s10549-007-9585-9. [DOI] [PubMed] [Google Scholar]
- 256.Perrin M.C., Terry M.B., Kleinhaus K., Deutsch L., Yanetz R., Tiram E., Calderon R., Friedlander Y., Paltiel O., Harlap S. Gestational diabetes as a risk factor for pancreatic cancer: A prospective cohort study. BMC Med. 2007;5:25. doi: 10.1186/1741-7015-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Metzger B.E., Coustan D.R. Summary and recommendations of the Fourth International Workshop-Conference on Gestational Diabetes Mellitus. The Organizing Committee. Diabetes Care. 1998;21:B161–B167. [PubMed] [Google Scholar]
- 258.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 259.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of re-al-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Hromadnikova I., Kotlabova K., Hympanova L., Krofta L. Gestational hypertension, preeclampsia and intrauterine growth restriction induce dysregulation of cardiovascular and cerebrovascular disease associated microRNAs in maternal whole peripheral blood. Thromb. Res. 2016;137:126–140. doi: 10.1016/j.thromres.2015.11.032. [DOI] [PubMed] [Google Scholar]
- 261.Andersen C.L., Jensen J.L., Ørntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon can-cer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 262.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to rights reserved by funding supporters.