Abstract
The physical and genetic map of the Bradyrhizobium japonicum chromosome revealed that nitrogen fixation and nodulation genes are clustered. Because of the complex interactions between the bacterium and the plant, we expected this chromosomal sector to contain additional genes that are involved in the maintenance of an efficient symbiosis. Therefore, we determined the nucleotide sequence of a 410-kb region. The overall G+C nucleotide content was 59.1%. Using a minimum gene length of 150 nucleotides, 388 open reading frames (ORFs) were selected as coding regions. Thirty-five percent of the predicted proteins showed similarity to proteins of rhizobia. Sixteen percent were similar only to proteins of other bacteria. No database match was found for 29%. Repetitive DNA sequence-derived ORFs accounted for the rest. The sequenced region contained all nitrogen fixation genes and, apart from nodM, all nodulation genes that were known to exist in B. japonicum. We found several genes that seem to encode transport systems for ferric citrate, molybdate, or carbon sources. Some of them are preceded by −24/−12 promoter elements. A number of putative outer membrane proteins and cell wall-modifying enzymes as well as a type III secretion system might be involved in the interaction with the host.
Nodulation (nod) genes and nitrogen fixation (nif) genes are the key determinants in the interaction between rhizobia and their host plants (14, 47). However, other loci influence the efficiency of the interaction or change the host range. Sequencing of the symbiotic plasmid of Rhizobium sp. strain NGR234 revealed a gene cluster that encodes a type III secretion system (22). Secreted proteins are encoded within the same cluster (95). The closely related Sinorhizobium fredii carries a type III secretion system as well (51, 61). Mutations within the secretion systems of the two strains influence symbiosis in a host-dependent manner. Plant and animal pathogens use related systems to target proteins to host cells (35), but such proteins have not been identified in rhizobia.
During symbiosis, rhizobia exclusively rely on the carbon supply from the plant. Although bacteroids can utilize a wide range of carbon compounds, dicarboxylic acids are most likely the main carbon and energy source for bacteroids (45, 83). The main argument is that several strains that have a defect in the dicarboxylic acid transport system show a Fix− phenotype (7, 17, 19, 79, 94) or are at least strongly impaired in nitrogen fixation (37).
In our earlier work, we established a correlated physical and genetic map of the Bradyrhizobium japonicum genome (28, 53) and discovered that all known nod and nif genes were clustered within a chromosomal region of about 400 kb. Furthermore, we found that the G+C content of these genes was 58 mol% (76), considerably lower than the 61 to 65 mol% reported for the whole genome (43). Therefore, we concluded that the symbiotic genes have integrated into the chromosome after horizontal gene transfer from a different strain. In the absence of genomic rearrangements, this region might contain more genes involved in symbiosis. This possibility persuaded us to determine its nucleotide sequence. Here, we present the analysis of a 410-kb region and discuss the implications for symbiosis on the basis of selected examples.
MATERIALS AND METHODS
Bacterial strains.
Escherichia coli strains ED8767 (66) and JM101 (62) were used for propagation of the cosmid library and M13 clones, respectively. B. japonicum strain PSP7 (53) is a mutant derivative of B. japonicum USDA3I1b110 (hereinafter referred to as USDA110) that contains additional sites for PacI, SwaI, and PmeI within the tlpA locus. This strain served as the source for DNA from the symbiotic gene region.
Media and bacterial growth conditions.
E. coli strains were grown in Luria broth, 2× YT, or M9 minimal medium (85). B. japonicum was grown in peptone-salts-yeast extract medium (77).
Vectors.
M13mp18 (69) was digested with SmaI. Subsequently, a polylinker containing recognition sites for PacI, SwaI, and PmeI was inserted, yielding M13mp18Pme. M13mp18Pme was used for the preparation of single-stranded DNA for sequencing. The vector Lorist6 (25) was used for the construction of a cosmid library.
DNA protocols and construction of an overlapping cosmid library.
Standard procedures were used for DNA manipulations (85). DNA of strain PSP7 was restricted with SwaI. The DNA fragments were separated by pulsed-field gel electrophoresis as described previously (53). DNA of the 840-kb fragment that contained all of the known nif and nod genes was isolated by treatment of the gel slice with β-agarase I (Calbiochem, La Jolla, Calif.). The isolated DNA was digested partially with Sau3AI. DNA fragments in the size range 35 to 50 kb were isolated after pulsed-field gel electrophoresis by treatment with β-agarase, followed by further purification with phenol and a final ethanol precipitation. About 0.2 μg of DNA was ligated with 0.5 μg of vector DNA (Lorist6) that had been digested with BamHI and dephosphorylated. DNA was packaged (Gigapack II Gold packaging extract; Stratagene, La Jolla, Calif.) and transduced into E. coli ED8767. Selection for kanamycin resistance yielded more than 250 clones. Two hundred fifty clones were picked and cultivated in 1 ml of 2× YT. Aliquots of 100 μl were transferred into microtiter plates prefilled with 100 μl of 50% glycerol. The plates were sealed and stored. The rest of the culture was used for DNA isolation. DNA was transferred to nylon membranes and hybridized against 10 probes from the symbiotic gene region. This led to the selection of 99 clones. Restriction analysis finally resulted in an ordered cosmid library consisting of a minimal set of 16 partially overlapping cosmids encompassing the gene loci from nodVW to ndp.
DNA sequencing and assembly.
The minimal set of 16 cosmids was digested with a total of six different blunt-end-generating enzymes in independent reactions. From each reaction fragments of three or four different size ranges (with 500 bp being the smallest size) were isolated through agarose gel electrophoresis. Fragments were cloned into M13mp18Pme and sequenced (about 2,900 sequence reactions). Sequencing was done with dye terminator technology on a model 373A sequencer from Applied Biosystems. For assembly, the Genetics Computer Group software package was used. Single-stranded and double-stranded gaps were closed by sequencing of PCR-generated fragments (about 600 sequence reactions).
Nucleotide sequence analysis.
If not otherwise stated, the Genetics Computer Group software package was used. Blast similarity searches (2) were done at the National Center for Biotechnology Information (blast@ncbi.nlm.nih.gov). Open reading frames (ORFs) were annotated by the program Glimmer (12, 84). ATG, GTG, and TTG were allowed as start codons. With the sequences of ORFs that exhibited similarity to known genes, a new data set that was used to reanalyze the sequence was generated. Repeated sequences were included into this set only once. Analysis of the sequenced region resulted in the identification of a maximal number of 937 ORFs, which were assigned identification (id) numbers. All were screened for similarity to database entries. Software-assisted examination of many overlapping ORFs led to a considerable reduction in the number of putative genes. In general, only the ORFs with the highest coding probabilities were considered. ORFs with low coding probabilities were selected if they showed similarity to published sequences. This was the case for id56f2, id102, id725f2, id774R2, and id813r1. id56f2 (probability value, 64) exhibits high similarity to ferredoxins. id102 (frxA; probability value, 97) encodes a ferredoxin-like protein (16). Expression was demonstrated for id774r2 (hsfA; probability value, 64 [11]) and id813r1 (nrgB; probability value, 1 [68]). For id725f2 (nolM; probability value, 5 [59]), no data are available that could prove its functionality. If overlapping ORFs had the same coding probability but exhibited no similarity to other sequences, none of the ORFs was chosen. Because the smallest ORF with similarity to a published sequence encompassed 159 nucleotides, the lower limit was set to 150 nucleotides.
For the identification of potential promoters, the intergenic regions were compared by the program FastA with promoter sequences that are recognized by known regulators or sigma factors. The searched sequences were TTGAnCnnGATCAAnG (FixK consensus [20]) (where “n” represents any base), TGGCAC-n5-TTGCT/A (ς54 consensus [20]), (where “n5” represents any five bases), TTGACA-n17-TATAAT (ς70 consensus [56]), and ATCCA-n7-GATG-n6-ATCCAAACAATCGATTTTACCAATC (nod box; modified from reference 87).
Identification of transmembrane domains and N-terminal signal sequences.
Transmembrane domains were searched with TMpred (http://www.ch.embnet.org/software/TMPRED_form.html). Proteins supposed to contain at least one transmembrane domain were analyzed a second time with TopPred2 (http://www.sbc.su.se/∼erikw/toppred2). Likewise, the presence of N-terminal signal sequences (67) was investigated first by the program SPscan and second by Signal P (http://www.cbs.dtu.dk/services/SignalP/).
Nucleotide sequence accession number.
The nucleotide sequence has been deposited in the EMBL database under accession numbers AF322012 and AF322013.
RESULTS AND DISCUSSION
Evaluation of the sequence quality.
To analyze the quality on the nucleotide level, the new sequence data were compared with data from past projects. Within more than 10 kb, only two differences were found, and both were due to reading errors in the old sequences. In addition, more than 4 kb was sequenced a second time and independently by a company. Both sequences were identical.
In several cases, for example with hupD, hupH, and noeE, the sequence initially appeared to contain mistakes that caused frameshifts. In all cases, we could rely on high-quality sequence results, thus excluding the possibility of sequencing errors. In hupD it was possible to track the frameshift down to a single nucleotide. The corresponding region was reamplified directly from chromosomal DNA. Sequencing of the PCR fragment confirmed the frameshift. The same procedure was done for the verification of the noeE locus.
Based on these data, we expect that the error frequency is less than one mistake per 10,000 bp.
Coding capacity of the symbiotic region.
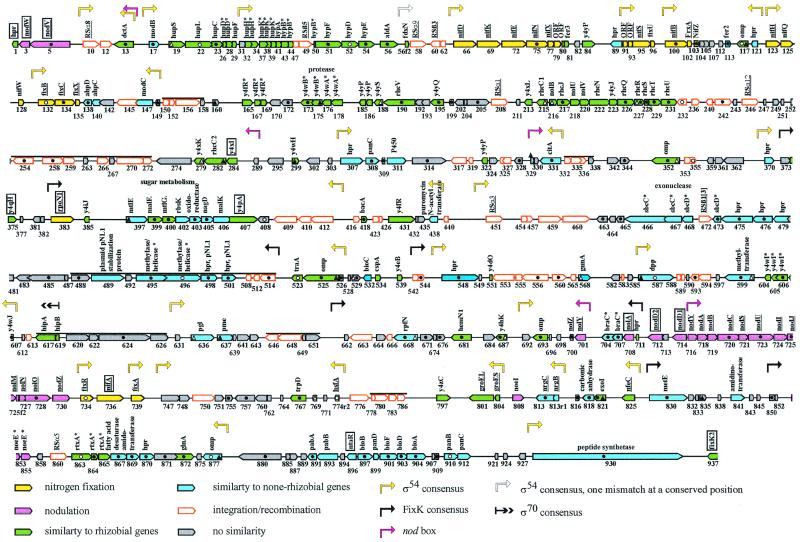
Analysis of the nucleotide sequence led to the selection of 388 ORFs, depicted in Fig. 1. Surprisingly, only 35% of the ORFs were similar to genes previously described for rhizobia (Table 1). Apart from having genes for nodulation, nitrogen fixation, and type III secretion, the symbiotic island of B. japonicum has little in common with the symbiotic plasmid of NGR234. This is remarkable because both strains have overlapping host ranges (they nodulate Macroptilium atropurpureum and Vigna radiata, for example). Obviously, the two strains follow partially different strategies for the establishment of an efficient symbiosis. This flexibility may be promoted by numerous insertion sequence (IS)-like elements that facilitate recombination events.
FIG. 1.
Map of the symbiotic gene region. Orientations and sizes of the ORFs are indicated. The number below each ORF corresponds to the id number in a table that is available at http://www.biologie.tu-dresden.de/genetik/molgen1.html. Gene designations of similar sequences are given. Genes or ORFs in strain USDA110 that have been described previously are underlined. Putative transcriptional regulators are boxed. nod boxes are indicated only if the three conserved regions are present. For the other promoter regions, a maximal deviation from the consensus sequence at two positions was accepted. With one indicated exception, no deviation at the highly conserved positions was allowed for −24/−12 promoter regions. Each line represents 52 kb. Filled triangles and circles within or next to a gene symbol indicate the presence of N-terminal signal sequences and transmembrane domains, respectively, as suggested by two programs with high probabilities. Open triangles and circles were used if these structural features were suggested with lower probability by one of the two programs. Bars above the gene symbols denote regions with a G+C content above 62%. The overall G +C content is 59.1%. Asterisks indicate neighboring ORFs whose deduced products have similarity to different parts of a single protein in other organisms.
TABLE 1.
Similarity groups of proteins putatively encoded within the symbiotic island
| Group | Similarity to proteins | No. of proteins (% of totala) | Avg size (amino acids) |
|---|---|---|---|
| 1 | From rhizobia (excluding group 4) | 136 (35) | 259 |
| 2 | From organisms besides rhizobia (excluding group 4) | 64 (16) | 434 |
| 3 | No similarity | 114 (29) | 183 |
| 4 | Involved in transposition and recombination | 74 (19) | 255 |
| 5 | From Rhizobium sp. strain NGR234 (including group 4) | 120 (31) | NDb |
Rounded values.
ND, not determined.
Nodulation genes.
The nodulation gene cluster containing nodABC has been characterized previously (27, 59). Id808 has good similarity to NoeI of Rhizobium sp. strain NGR234. In NGR234, NoeI is involved in the 2-O-methylation of the Nod factor at the fucosyl residue (40). Because the Nod factor of B. japonicum also contains a methylated fucosyl residue (86), Id808 may be a homologue of NoeI.
Two deduced proteins of neighboring ORFs (Id853 and Id855) exhibit similarity to different parts of NoeE of Rhizobium sp. strain NGR234. NoeE was shown to have sulfotransferase activity (32, 75). However, no sulfatation of the Nod factor has been described for B. japonicum. The lack of a nod box-containing promoter region raises further doubt about the involvement of id853 and id855 in Nod factor biosynthesis.
The only nodulation gene that is known to exist in B. japonicum and that is missing within the sequenced region is nodM, which encodes a glucosamine synthase (5). In B. japonicum, nodM was found next to a region determining genotype-specific nodulation of soybean cultivars (58). Preliminary data suggest that this region is located about 100 kb downstream of nodVW (unpublished).
Nitrogen fixation and ferredoxin-like genes.
As with nodulation genes, most of the nitrogen fixation genes of B. japonicum had been studied before the onset of this project. The newly identified genes nifQ, nifW, nifZ, and fixU are known from other nitrogen-fixing bacteria, including rhizobia (47). NifW potentially in complex with NifZ may be required for oxygen protection of the nitrogenase (48, 54). The presence of NifQ is not surprising because it seems to be involved in the incorporation of molybdenum into the iron-molybdenum cofactor of nitrogenase (39). Nothing is known about the function of FixU, which has some similarity to NifT of Azotobacter vinelandii.
Interesting is the presence of several ferredoxin-like proteins that are encoded close to nif genes and that are present in other nitrogen-fixing bacteria as well. Carter et al. (10) purified a two-[4Fe-4S]-cluster-containing enzyme from a B. japonicum strain and suggested that it serves as an electron donor for nitrogenase. Based on the reported amino acid composition, the corresponding enzyme may be encoded by id56f2 (fdxN) in USDA110. A similar protein is encoded by frxA (id102). Both proteins contain the cysteine motifs (82) that are typical for ferredoxins with two [4Fe-4S] clusters. In S. meliloti, FdxN is essential for nitrogen fixation (50). In B. japonicum, a mutation of frxA does not significantly change symbiotic nitrogen fixation (16). Furthermore, a large deletion that encompasses fdxN (mutant strain E1-7d1) still fixes nitrogen (31). This finding suggests that both ferredoxins are able to substitute for each other.
The ferredoxin encoded by id81 is very similar to ferredoxin III from Rhodobacter capsulatus (44) and FdxB of NGR234 (22). In R. capsulatus, ferredoxin III probably does not serve as an electron donor to nitrogenase (44). Most similar to Id113 is a [2Fe-2S] ferredoxin of Clostridium pasteurianum, which was shown by cross-linking experiments to interact with the MoFe protein of nitrogenase (26).
Secretion of fixed nitrogen.
Fixed nitrogen might be translocated into the plant cytosol as ammonium (46, 93) or alanine (97). Recently, it was shown that bacteroids of Rhizobium leguminosarum secrete both alanine and ammonium (1). The product of id56 has high similarity to alanine dehydrogenases. Preliminary results suggest that the gene is transcribed under microaerobic conditions, supporting the idea that it may be involved in the formation of alanine that is then secreted.
Hydrogen metabolism.
Hydrogenase activity is widespread among Bradyrhizobium strains and has been described for USDA110 as well (9). Thus, it was not surprising to find the hup gene cluster within the symbiotic island. The genes encoding the small and large subunits of hydrogenase (id19 and id22) are highly similar to the corresponding genes of other bacteria. However, changes within the nucleotide sequence and the insertion of a repetitive element led to fragmentation and probably also to the deletion of several other genes. For example, hupG, -I and -J, which are present in R. leguminosarum (AC X52974) as well as in B. japonicum (23), are missing. hypB was split by the insertion of an IS-like element. It will be interesting to see if these changes still allow hydrogenase activity.
Type III secretion system.
More than 20 ORFs within a large cluster stretching from id185 to id289 are similar to genes of NGR234. In NGR234, several of these genes are known to be involved in the formation of a type III secretion system (22, 95). Also conserved is id284 (y4x1), whose product has similarity to transcriptional activators of the two-component regulatory family. The regulator seems to be under the control of a nod box-containing promoter sequence, raising the possibility that this cluster is active very early in symbiosis.
Several deduced proteins exhibit similarity to proteins of NGR234 but not to components of type III secretion systems from pathogenic bacteria. They may be part of a specialized transport complex. Alternatively, they might be secreted proteins. In NGR234 two secreted proteins, NolX and Y4xL, were identified (95). The corresponding genes are located downstream of rhcC1 together with six other ORFs. Interestingly, in B. japonicum these genes seem to be absent and only a short ORF (id213) has some similarity to the 5′ half of y4xL.
Candidates for secreted proteins are of course those that are encoded within the cluster. However, proteins encoded outside of this cluster may also be secreted. Id431 is a leucine-rich repeat protein that has good similarity to IpaH of Shigella flexneri and SspH1 of Salmonella enterica serovar Typhimurium; both were shown to be secreted (13, 63). Id165, Id167, and Id169 have some similarity to different parts of Id431. Id797 has weak similarity to proteins of the bean pathogen Pseudomonas syringae pathovar phaseolicola, one being the avirulence protein AvrPph3 (42) and the other being a virulence protein of unknown function that is encoded within the pathogenicity island (41).
Carbon metabolism.
id13 (dctA) encodes a protein that is highly similar to known C4-dicarboxylate permeases. The presence of a nod box as well as a −24/−12 promoter sequence suggests a complex regulation. Mutations in the two rpoN genes that encode ς54 do not affect the ability to grow on succinate or malate as a carbon source (52). There are several possibilities to explain this result. First, the identified dctA gene may be transcribed in a ς54-independent manner. Second, there might be more than one uptake system. The finding that the deletion mutant E1-7d1, in which dctA is also removed, is still able to fix nitrogen (31) supports the latter possibility. Two succinate uptake systems were reported to exist in B. japonicum 61-A-101 (36).
Despite the importance of C4-dicarboxylates in several systems (45), they may not be the only important energy and carbon sources for bacteroids of B. japonicum 110. ORFs id397 to id407 most likely encode a carbohydrate uptake system that includes a transcriptional regulator. There exists a pronounced similarity to proteins known to be involved in uptake of maltose or mannitol. It has been shown previously that the cytosol of soybean nodules contains a variety of usable carbohydrates besides sucrose, for example, maltose and trehalose (83, 90).
Additionally, B. japonicum is able to fix CO2 and ribulose-1, 5-bisphosphate carboxylase/oxygenase has been purified from chemolithoautotrophically grown cells (74). CO2 fixation may be supported by a β carbonic anhydrase that is probably encoded by id818. β carbonic anhydrases are ancient enzymes frequently found in prokaryotes and are required for efficient CO2 fixation (33, 88). Alternatively, other enzymes such as phosphoenolpyruvate carboxylase may utilize the generated HCO3−.
Genes that might be involved in metal ion uptake.
The product of id331 has similarity to citrate-proton symport proteins. Citrate is a prominent metabolite in soybean nodules and can be used as a carbon source by bacteroids (90). However, we favor the idea that the encoded protein is used for the import of Fe(III)-citrate. Fe(III)-citrate is probably the Fe transport form in soybean plants (92) and can be imported by bacteroids (65) and used by free-living cells of USDA110 (73). The presence of a −24/−12 promoter element indicates that the ORF may be regulated by ς54, which would guarantee its timely expression together with the nif genes. Compared to the well-known fec-encoded ABC-type ferric citrate uptake system (8), the suggested metal citrate-proton symport would have the advantage that it does not consume ATP.
A second element that is needed in increased amounts in nitrogen-fixing bacteroids is molybdenum. Indeed, the symbiotic island contains two ORFs that, based on similarities, may be involved in molybdenum uptake. Id17 is similar to ModB, a permease that has been shown to function in Mo transport. Id147 is similar to ModC and has the typical amino acid signature of ABC transporters. The finding that both of their ORFs are preceded by −24/−12 promoter sequences further supports their possible involvement in symbiosis.
Enzymatic functions that might interact with plant cell wall components.
Id568 shows weak similarity to glucuronidases. Similarity is higher between Id636 and polygalacturonases. All four conserved regions that have been proposed to contribute to the catalytic site of polygalacturonase from Erwinia carotovora (72) are present in Id636 as well, suggesting that this protein may have a similar function. A third protein (Id637) is similar to pectin methyl esterases of Arabidopsis thaliana. It is still disputed if such enzymatic functions contribute to the infection process. Early work suggested that pectic enzymes produced by the host play an important part in the infection process (18, 57), but these results have been questioned (38, 55). Later it was shown that rhizobia are indeed able to produce low levels of pectolytic enzymes (34); however, no genes have been identified so far. The fact that all three putative enzymes identified here are predicted to contain N-terminally located signal sequences underlines their potential extracellular function.
Outer membrane proteins.
Outer membrane proteins are the ideal candidates for mediating the interaction between bacteria and the host. Id117 has similarity to a family of outer membrane proteins that is characterized by eight amphipathic β strands (6). Id548 has weak similarity to OstA, a protein that contributes to the degree of organic solvent tolerance in E. coli (3). Four other proteins (Id352, Id525, Id693, and Id877) are similar to each other and to Omp31, a major outer membrane protein of Brucella melitensis that may serve as a porin (96).
Rhizobitoxine-related function.
Id863, Id864, and Id865 have high similarity to RtxA, which is involved in rhizobitoxine production in Bradyrhizobium elkanii (80, 81). Rhizobitoxine influences symbiosis with Glycine max, Vigna radiata, and Macroptilium atropurpureum, probably through inhibition of ethylene production (15, 71, 89, 99). Originally it was discovered due to its potential to induce chlorosis in soybeans (70). The identification of the corresponding genes in USDA110 was surprising because it was reported that this strain does not produce rhizobitoxine (24, 64). This may be due to differences in gene regulation or enzyme function. So far, the biosynthetic pathway for rhizobitoxine synthesis has not been elucidated, and the molecule potentially produced by USDA110 may have a different structure. Downstream of id865 and likely within the same operon, there are five additional ORFs (id863 to id872) that may be involved in the biosynthesis of a rhizobitoxine-like molecule.
The largest protein is probably a peptide synthetase.
The longest ORF found within the sequenced region may encode a protein of 3,310 amino acids. The protein has high similarity to peptide synthetases. Like other peptide synthetases, it has a characteristic order of conserved domains, and 4′-phosphopantetheine probably serves as carrier of acyl intermediates (reviewed in reference 49). Deduced from the conserved domain structure, Id930 may synthesize a tripeptide; however, the similarity to other synthetases does not reveal the potential composition of the product. Interesting is the presence of a −24/−12 promoter sequence that was shown to be functional (98). A gene cluster located a few kilobases upstream of id930 might be involved in the synthesis of the phosphopantetheine carrier.
Nineteen percent of the identified ORFs are related to genes involved in integration and recombination.
According to their similarities to published sequences, we grouped the integration- and recombination-related proteins, if appropriate, into the families that were summarized by Mahillon and Chandler (60). Most preponderant are members of the IS3, IS5, and IS21 families. Best conserved are the repeated sequences RSα that belong to the IS630 family. On the amino acid level, all six copies are identical in length and have at least 99.2% identical amino acid residues. Remarkably, all have the same orientation. The same orientation is also found for the six elements that share similarity with reverse transcriptases. However, peptide lengths and similarities are much more variant. Differences in the G+C contents, 65.8% for id266 and 58% for id776, suggest that they originate from independent sources.
In the face of the large amount of repeated sequences, it is astonishing that we have not encountered problems with strain stability so far. The only known gene that was interrupted by an IS-like element is hypB. However, deletions caused by recombination events between RSα elements can also occur either spontaneously (31) or after prolonged heat treatment (29).
Does the symbiotic region of B. japonicum result from an integration event?
A number of novel genes that fit well into the symbiotic scheme have been identified. Despite the presence of a number of IS-related elements, there is no indication that recombination events led to an insertion of essential housekeeping genes from B. japonicum or to the translocation of essential symbiotic genes to a separate chromosomal location. In cases where genes are present whose functions are known to be required for the free-living bacterium, like groESL, and hemN, they represent nonessential copies of homologues that are present elsewhere in the genome as well (21, 21a). Several genes (id489 to id501) exhibit similarity to a region that was suggested to be involved in the maintenance of plasmid pNL1 of Sphingomonas aromaticivorans (78). Next to this region, a protein (Id523) that is similar to the N terminus of the nicking enzyme TraA is encoded. Genes id489 to id501 are also remarkable because of their high G+C contents of about 65.3%; the sequence stretches that contain the nif (id1 to id149) and nod (id668 to id748) genes have an average G+C content below 58%, suggesting that the symbiotic region is made of pieces from different origins.
So far we have no further evidence that an integration event took place. This is different from the situation in Mesorhizobium loti strain ICMP3153, in which the symbiotic island that can be transferred to other strains integrated into a phenylalanine-specific tRNA gene (91).
Perspectives.
The established nucleotide sequence significantly facilitates the design of future experiments. In many cases protein functions can be deduced from their similarity to known proteins and tested accordingly. For many ORFs, such similarities do not exist. Therefore, our work will also be directed towards mutational and gene expression analyses of the whole region. This will further increase our knowledge about genes required at different stages during nodule development. For a full understanding of the symbiotic process, however, a detailed knowledge of the complete genome is required.
ACKNOWLEDGMENTS
We thank Monika Weishaupt for technical assistance.
This work was supported by grant 0-20-918-94 from the ETH, Zurich, Switzerland.
REFERENCES
- 1.Allaway D, Lodwig E M, Crompton L A, Wood M, Parsons R, Wheeler T R, Poole P S. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol Microbiol. 2000;36:508–515. doi: 10.1046/j.1365-2958.2000.01884.x. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aono R, Negishi T, Nakajima H. Cloning of organic solvent tolerance gene ostA that determines n-hexane tolerance level in Escherichia coli. Appl Environ Microbiol. 1994;60:4624–4626. doi: 10.1128/aem.60.12.4624-4626.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arp D J. Hydrogen cycling in symbiotic bacteria. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman & Hall; 1992. pp. 432–460. [Google Scholar]
- 5.Baev N, Endre G, Petrovics G, Banfalvi Z, Kondorosi A. Six nodulation genes of nod box locus 4 in Rhizobium meliloti are involved in nodulation signal production: nodM codes for d-glucosamine synthetase. Mol Gen Genet. 1991;228:113–124. doi: 10.1007/BF00282455. [DOI] [PubMed] [Google Scholar]
- 6.Baldermann C, Lupas A, Lubieniecki J, Engelhardt H. The regulated outer membrane protein Omp21 from Comamonas acidovorans is identified as a member of a new family of eight-stranded β-sheet proteins by its sequence and properties. J Bacteriol. 1998;180:3741–3749. doi: 10.1128/jb.180.15.3741-3749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolton E, Higgisson B, Harrington A, O'Gara F. Dicarboxylic acid transport in Rhizobium meliloti: isolation of mutants and cloning of dicarboxylic acid transport genes. Arch Microbiol. 1986;144:142–146. [Google Scholar]
- 8.Braun V. Surface signaling: novel transcription initiation mechanism starting from the cell surface. Arch Microbiol. 1997;167:325–331. doi: 10.1007/s002030050451. [DOI] [PubMed] [Google Scholar]
- 9.Carter K R, Jennings N T, Hanus J, Evans H J. Hydrogen evolution and uptake by nodules of soybeans inoculated with different strains of Rhizobium japonicum. Can J Microbiol. 1978;24:307–311. doi: 10.1139/m78-051. [DOI] [PubMed] [Google Scholar]
- 10.Carter K R, Rawlings J, Orme-Johnson W H, Becker R R, Evans H J. Purification and characterization of a ferredoxin from Rhizobium japonicum bacteroids. J Biol Chem. 1980;255:4213–4223. [PubMed] [Google Scholar]
- 11.Chun J Y, Sexton G L, Roth L E, Stacey G. Identification and characterization of a novel Bradyrhizobium japonicum gene involved in host-specific nitrogen fixation. J Bacteriol. 1994;176:6717–6729. doi: 10.1128/jb.176.21.6717-6729.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delcher A L, Harmon D, Kasif S, White O, Salzberg S L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demers B, Sansonetti P J, Parsot C. Induction of type III secretion in Shigella flexneri is associated with differential control of transcription of genes encoding secreted proteins. EMBO J. 1998;17:2894–2903. doi: 10.1093/emboj/17.10.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Downie J A. Functions of rhizobial nodulation genes. In: Spaink H, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 387–402. [Google Scholar]
- 15.Duodu S, Bhuvaneswari T V, Stokkermans T J W, Peters N K. A positive role for rhizobitoxine in Rhizobium-legume symbiosis. Mol Plant-Microbe Interact. 1999;12:1082–1089. [Google Scholar]
- 16.Ebeling S, Noti J D, Hennecke H. Identification of a new Bradyrhizobium japonicum gene (frxA) encoding a ferredoxinlike protein. J Bacteriol. 1988;170:1999–2001. doi: 10.1128/jb.170.4.1999-2001.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.el Din A K. A succinate transport mutant of Bradyrhizobium japonicum forms ineffective nodules on soybeans. Can J Microbiol. 1992;38:230–234. doi: 10.1139/m92-039. [DOI] [PubMed] [Google Scholar]
- 18.Fåhraeus G, Ljunggren H. The possible significance of pectic enzymes in root hair infection by nodule bacteria. Physiol Plant. 1959;12:145–154. [Google Scholar]
- 19.Finan T M, Wood J M, Jordan D C. Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J Bacteriol. 1983;154:1403–1413. doi: 10.1128/jb.154.3.1403-1413.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer H M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer H M, Babst M, Kaspar T, Acuña G, Arigoni F, Hennecke H. One member of a gro-ESL-like chaperonin multigene family in Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 1993;12:2901–2912. doi: 10.1002/j.1460-2075.1993.tb05952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Fischer H-M, Velasco L, Delgado M J, Bedmar E J, Schären S, Zingg D, Göttfert M, Hennecke H. One of two hemN genes in Bradyrhizobium japonicum is functional during anaerobic growth and in symbiosis. J Bacteriol. 2001;183:1300–1311. doi: 10.1128/JB.183.4.1300-1311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 23.Fu C, Maier R J. Organization of the hydrogenase gene cluster from Bradyrhizobium japonicum: sequences and analysis of five more hydrogenase-related genes. Gene. 1994;145:91–96. doi: 10.1016/0378-1119(94)90328-x. [DOI] [PubMed] [Google Scholar]
- 24.Fuhrmann J. Symbiotic effectiveness of indigenous soybean bradyrhizobia as related to serological, morphological, rhizobitoxine, and hydrogenase phenotypes. Appl Environ Microbiol. 1990;56:224–229. doi: 10.1128/aem.56.1.224-229.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson T J, Rosenthal A, Waterston R H. Lorist6, a cosmid vector with BamHI, NotI, ScaI and HindIII cloning sites and altered neomycin phosphotransferase gene expression. Gene. 1987;53:283–286. doi: 10.1016/0378-1119(87)90017-5. [DOI] [PubMed] [Google Scholar]
- 26.Golinelli M P, Gagnon J, Meyer J. Specific interaction of the [2Fe-2S] ferredoxin from Clostridium pasteurianum with the nitrogenase MoFe protein. Biochemistry. 1997;36:11797–11803. doi: 10.1021/bi970528p. [DOI] [PubMed] [Google Scholar]
- 27.Göttfert M. Regulation and function of rhizobial nodulation genes. FEMS Microbiol Rev. 1993;10:39–63. doi: 10.1111/j.1574-6968.1993.tb05863.x. [DOI] [PubMed] [Google Scholar]
- 28.Göttfert M, Kündig C, Hennecke H. Bradyrhizobium japonicum strain 3I1b110. In: de Bruijn F J, Lupski J R, Weinstock G M, editors. Bacterial genomes: physical structures and analysis. New York, N.Y: Chapman & Hall; 1998. pp. 625–628. [Google Scholar]
- 29.Hahn M. Genomstruktur von Bradyrhizobium japonicum: gemeinsames Vorkommen von repetitiven Sequenzen und Genen für die Wurzelknöllchensymbiose. Ph.D. thesis. Zurich, Switzerland: Eidgenössische Technische Hochschule; 1986. [Google Scholar]
- 30.Hahn M, Hennecke H. Mapping of a Bradyrhizobium japonicum DNA region carrying genes for symbiosis and an asymmetric accumulation of reiterated sequences. Appl Environ Microbiol. 1987;53:2247–2252. doi: 10.1128/aem.53.9.2247-2252.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahn M, Meyer L, Studer D, Regensburger B, Hennecke H. Insertion and deletion mutations within the nif region of Rhizobium japonicum. Plant Mol Biol. 1984;3:159–168. doi: 10.1007/BF00016063. [DOI] [PubMed] [Google Scholar]
- 32.Hanin M, Jabbouri S, Quesada-Vincens D, Freiberg C, Perret X, Promé J C, Broughton W J, Fellay R. Sulphation of Rhizobium sp. NGR234 Nod factors is dependent on noeE, a new host-specificity gene. Mol Microbiol. 1997;24:1119–1129. doi: 10.1046/j.1365-2958.1997.3981777.x. [DOI] [PubMed] [Google Scholar]
- 33.Hewett-Emmett D, Tashian R E. Functional diversity, conservation, and convergence in the evolution of the α, β, and γ carbonic anhydrase gene families. Mol Phylogenet Evol. 1996;5:50–77. doi: 10.1006/mpev.1996.0006. [DOI] [PubMed] [Google Scholar]
- 34.Hubbell D H, Morales V M, Umali-Garcia M. Pectolytic enzymes in Rhizobium. Appl Environ Microbiol. 1978;35:210–213. doi: 10.1128/aem.35.1.210-213.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Humbeck C, Werner D. Two succinate uptake systems in Bradyrhizobium japonicum. Curr Microbiol. 1987;14:259–262. [Google Scholar]
- 37.Humbeck C, Werner D. Delayed nodule development in a succinate transport mutant of Bradyrhizobium japonicum. J Plant Physiol. 1989;134:276–283. [Google Scholar]
- 38.Hunter W J, Elkan G H. Role of pectic and cellulolytic enzymes in the invasion of the soybean by Rhizobium japonicum. Can J Microbiol. 1975;21:1254–1258. doi: 10.1139/m75-187. [DOI] [PubMed] [Google Scholar]
- 39.Imperial J, Ugalde R A, Shah V K, Brill W J. Role of the nifQ gene product in the incorporation of molybdenum into nitrogenase in Klebsiella pneumoniae. J Bacteriol. 1984;158:187–194. doi: 10.1128/jb.158.1.187-194.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jabbouri S, Relic B, Hanin M, Kamalaprija P, Burger U, Promé D, Promé J C, Broughton W J. nolO and noel (HsnIII) of Rhizobium sp. NGR234 are involved in 3-O-carbamoylation and 2-O-methylation of Nod factors. J Biol Chem. 1998;273:12047–12055. doi: 10.1074/jbc.273.20.12047. [DOI] [PubMed] [Google Scholar]
- 41.Jackson R W, Athanassopoulos E, Tsiamis G, Mansfield J W, Sesma A, Arnold D L, Gibbon M J, Murillo J, Taylor J D, Vivian A. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc Natl Acad Sci USA. 1999;96:10875–10880. doi: 10.1073/pnas.96.19.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenner C, Hitchin E, Mansfield J, Walters K, Betteridge P, Teverson D, Taylor J. Gene-for-gene interactions between Pseudomonas syringae pv. phaseolicola and Phaseolus. Mol Plant-Microbe Interact. 1991;4:553–562. [PubMed] [Google Scholar]
- 43.Jordan D C. Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing, root nodule bacteria from leguminous plants. Int J Syst Bacteriol. 1982;32:136–139. [Google Scholar]
- 44.Jouanneau Y, Meyer C, Gaillard J, Forest E, Gagnon J. Purification and characterization of a novel dimeric ferredoxin (FdIII) from Rhodobacter capsulatus. J Biol Chem. 1993;268:10636–10644. [PubMed] [Google Scholar]
- 45.Kahn M L, McDermott T R, Udvardi M K. Carbon and nitrogen metabolism in rhizobia. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 461–485. [Google Scholar]
- 46.Kaiser B N, Finnegan P M, Tyerman S D, Whitehead L F, Bergersen F J, Day D A, Udvardi M K. Characterization of an ammonium transport protein from the peribacteroid membrane of soybean nodules. Science. 1998;281:1202–1206. doi: 10.1126/science.281.5380.1202. [DOI] [PubMed] [Google Scholar]
- 47.Kaminski P A, Batut J, Boistard P. A survey of symbiotic nitrogen fixation by rhizobia. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 431–460. [Google Scholar]
- 48.Kim S, Burgess B K. Evidence for the direct interaction of the nifW gene product with the MoFe protein. J Biol Chem. 1996;271:9764–9770. doi: 10.1074/jbc.271.16.9764. [DOI] [PubMed] [Google Scholar]
- 49.Kleinkauf H, von Döhren H. A nonribosomal system of peptide biosynthesis. Eur J Biochem. 1996;236:335–351. doi: 10.1111/j.1432-1033.1996.00335.x. [DOI] [PubMed] [Google Scholar]
- 50.Klipp W, Reiländer H, Schlüter A, Krey R, Pühler A. The Rhizobium meliloti fdxN gene encoding a ferredoxin-like protein is necessary for nitrogen fixation and is cotranscribed with nifA and nifB. Mol Gen Genet. 1989;216:293–302. doi: 10.1007/BF00334368. [DOI] [PubMed] [Google Scholar]
- 51.Kovacs L G, Balatti P A, Krishnan H B, Pueppke S G. Transcriptional organization and expression of noIXWBTUV, a locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol Microbiol. 1995;17:923–933. doi: 10.1111/j.1365-2958.1995.mmi_17050923.x. [DOI] [PubMed] [Google Scholar]
- 52.Kullik I, Fritsche S, Knobel H, Sanjuan J, Hennecke H, Fischer H M. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the ς54 gene (rpoN) J Bacteriol. 1991;173:1125–1138. doi: 10.1128/jb.173.3.1125-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kündig C, Hennecke H, Göttfert M. Correlated physical and genetic map of the Bradyrhizobium japonicum 110 genome. J Bacteriol. 1993;175:613–622. doi: 10.1128/jb.175.3.613-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S H, Pulakat L, Parker K C, Gavini N. Genetic analysis on the NifW by utilizing the yeast two-hybrid system revealed that the NifW of Azotobacter vinelandii interacts with the NifZ to form higher-order complexes. Biochem Biophys Res Commun. 1998;244:498–504. doi: 10.1006/bbrc.1998.8119. [DOI] [PubMed] [Google Scholar]
- 55.Lillich T T, Elkan G H. Evidence countering the role of polygalacturonase in invasion of root hairs of leguminous plants by Rhizobium spp. Can J Microbiol. 1968;14:617–625. doi: 10.1139/m68-104. [DOI] [PubMed] [Google Scholar]
- 56.Lisser S, Margalit H. Compilation of E. coli messenger RNA promoter sequences. Nucleic Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ljunggren H, Fåhraeus G. The role of polygalacturonase in root-hair invasion by nodule bacteria. J Gen Microbiol. 1961;26:521–528. doi: 10.1099/00221287-26-3-521. [DOI] [PubMed] [Google Scholar]
- 58.Lohrke S M, Day B, Kolli V S, Hancock R, Yuen J P, de Souza M L, Stacey G, Carlson R, Tong Z, Hur H-G, Orf J H, Sadowsky M J. The Bradyrhizobium japonicum noeD gene: a negatively acting, genotype-specific nodulation gene for soybean. Mol Plant-Microbe Interact. 1998;11:476–488. doi: 10.1094/MPMI.1998.11.6.476. [DOI] [PubMed] [Google Scholar]
- 59.Luka S, Sanjuan J, Carlson R W, Stacey G. nolMNO genes of Bradyrhizobium japonicum are co-transcribed with nodYABCSUIJ, and nolO is involved in the synthesis of the lipo-oligosaccharide nodulation signals. J Biol Chem. 1993;268:27053–27059. [PubMed] [Google Scholar]
- 60.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meinhardt L W, Krishnan H B, Balatti P A, Pueppke S G. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol Microbiol. 1993;9:17–29. doi: 10.1111/j.1365-2958.1993.tb01665.x. [DOI] [PubMed] [Google Scholar]
- 62.Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- 63.Miao E A, Scherer C A, Tsolis R M, Kingsley R A, Adams L G, Bäumler A J, Miller S I. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol Microbiol. 1999;34:850–864. doi: 10.1046/j.1365-2958.1999.01651.x. [DOI] [PubMed] [Google Scholar]
- 64.Minamisawa K, Seki T, Onodera S, Kubota M, Asami T. Genetic relatedness of Bradyrhizobium japonicum field isolates as revealed by repeated sequences and various other characteristics. Appl Environ Microbiol. 1992;58:2832–2839. doi: 10.1128/aem.58.9.2832-2839.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moreau S, Meyer J M, Puppo A. Uptake of iron by symbiosomes and bacteroids from soybean nodules. FEBS Lett. 1995;361:225–228. doi: 10.1016/0014-5793(95)00155-3. [DOI] [PubMed] [Google Scholar]
- 66.Murray N E, Brammar W J, Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977;150:53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- 67.Nielsen H, Brunak S, von Heijne G. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 1999;12:3–9. doi: 10.1093/protein/12.1.3. [DOI] [PubMed] [Google Scholar]
- 68.Nienaber A, Huber A, Göttfert M, Hennecke H, Fischer H M. Three new NifA-regulated genes in the Bradyrhizobium japonicum symbiotic gene region discovered by competitive DNA-RNA hybridization. J Bacteriol. 2000;182:1472–1480. doi: 10.1128/jb.182.6.1472-1480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 70.Owens L D, Wright D A. Rhizobial-induced chlorosis in soybeans: isolation, production, and varietal specificity of the toxin. Plant Physiol. 1965;40:927–930. doi: 10.1104/pp.40.5.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Owens L D, Liebermann M, Kunishi A. Inhibition of ethylene production by rhizobitoxine. Plant Physiol. 1971;48:1–4. doi: 10.1104/pp.48.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pickersgill R, Smith D, Worboys K, Jenkins J. Crystal structure of polygalacturonase from Erwinia carotovora ssp. carotovora. J Biol Chem. 1998;273:24660–24664. doi: 10.1074/jbc.273.38.24660. [DOI] [PubMed] [Google Scholar]
- 73.Plessner O, Klapatch T, Guerinot M L. Siderophore utilization by Bradyrhizobium japonicum. Appl Environ Microbiol. 1993;59:1688–1690. doi: 10.1128/aem.59.5.1688-1690.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Purohit K, Becker R R, Evans H J. d-Ribulose-1, 5-bisphosphate carboxylase/oxygenase from chemolithotrophically grown Rhizobium japonicum. Biochim Biophys Acta. 1982;715:230–239. [Google Scholar]
- 75.Quesada-Vincens D, Hanin M, Broughton W J, Jabbouri S. In vitro sulfotransferase activity of NoeE, a nodulation protein of Rhizobium sp. NGR234. Mol Plant-Microbe Interact. 1998;11:592–600. doi: 10.1094/MPMI.1998.11.7.592. [DOI] [PubMed] [Google Scholar]
- 76.Ramseier T M, Göttfert M. Codon usage and G+C content in Bradyrhizobium japonicum genes are not uniform. Arch Microbiol. 1991;156:270–276. doi: 10.1007/BF00262997. [DOI] [PubMed] [Google Scholar]
- 77.Regensburger B, Hennecke H. RNA polymerase from Rhizobium japonicum. Arch Microbiol. 1983;135:103–109. doi: 10.1007/BF00408017. [DOI] [PubMed] [Google Scholar]
- 78.Romine M F, Stillwell L C, Wong K K, Thurston S J, Sisk E C, Sensen C, Gaasterland T, Fredrickson J K, Saffer J D. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J Bacteriol. 1999;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ronson C W, Lyttleton P, Robertson J G. C4-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens. Proc Natl Acad Sci USA. 1981;78:4284–4288. doi: 10.1073/pnas.78.7.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ruan X, Zhang C, Peters N K. Bradyrhizobium japonicum rhizobitoxine genes and putative enzyme functions: expression requires a translational frameshift. Proc Natl Acad Sci USA. 1993;90:2641–2645. doi: 10.1073/pnas.90.7.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ruan X, Zhang C, Peters N K. Bradyrhizobium japonicum rhizobitoxine genes and putative enzyme functions—expression requires a translational frameshift. Proc Natl Acad Sci USA. 1993;90:12055. doi: 10.1073/pnas.90.7.2641. . (Erratum, 90:2641.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saeki K, Tokuda K, Fukuyama K, Matsubara H, Nadanami K, Go M, Itoh S. Site-specific mutagenesis of Rhodobacter capsulatus ferredoxin I, FdxN, that functions in nitrogen fixation. J Biol Chem. 1996;271:31399–31406. doi: 10.1074/jbc.271.49.31399. [DOI] [PubMed] [Google Scholar]
- 83.Salminen S O, Streeter J G. Uptake and metabolism of carbohydrates by Bradyrhizobium japonicum bacteroids. Plant Physiol. 1987;83:535–540. doi: 10.1104/pp.83.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Salzberg S L, Delcher A L, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 86.Sanjuan J, Carlson R W, Spaink H P, Bhat U R, Barbour W M, Glushka J, Stacey G. A 2-O-methylfucose moiety is present in the lipo-oligosaccharide nodulation signal of Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 1992;89:8789–8793. doi: 10.1073/pnas.89.18.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schlaman H-R M, Phillips D-A, Kondorosi E. Genetic organization and transcriptional regulation of rhizobial nodulation genes. In: Spaink H, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 361–386. [Google Scholar]
- 88.Smith K S, Jakubzick C, Whittam T S, Ferry J G. Carbonic anhydrase is an ancient enzyme widespread in prokaryotes. Proc Natl Acad Sci USA. 1999;96:15184–15189. doi: 10.1073/pnas.96.26.15184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stokkermans T J W, Sanjuan J, Ruan X, Stacey G, Peters N K. Bradyrhizobium japonicum rhizobitoxine mutants with altered host-range on Rj4 soybeans. Mol Plant-Microbe Interact. 1992;5:504–512. [Google Scholar]
- 90.Streeter J G. Carbohydrate, organic acid, and amino acid composition of bacteroids and cytosol from soybean nodules. Plant Physiol. 1987;85:768–773. doi: 10.1104/pp.85.3.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sullivan J T, Ronson C W. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci USA. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tiffin L O. Translocation of iron citrate and phosphorus in xylem exudate of soybean. Plant Physiol. 1970;45:280–283. doi: 10.1104/pp.45.3.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tyerman S D, Whitehead L F, Day D A. A channel-like transporter for NH4+ on the symbiotic interface of N2-fixing plants. Nature. 1995;378:629–632. [Google Scholar]
- 94.van Slooten J C, Bhuvanasvari T V, Bardin S, Stanley J. Two C4-dicarboxylate transport systems in Rhizobium sp. NGR234: rhizobial dicarboxylate transport is essential for nitrogen fixation in tropical legume symbioses. Mol Plant-Microbe Interact. 1992;5:179–186. doi: 10.1094/mpmi-5-179. [DOI] [PubMed] [Google Scholar]
- 95.Viprey V, Del Greco A, Golinowski W, Broughton W J, Perret X. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol Microbiol. 1998;28:1381–1389. doi: 10.1046/j.1365-2958.1998.00920.x. [DOI] [PubMed] [Google Scholar]
- 96.Vizcaino N, Cloeckaert A, Zygmunt M S, Dubray G. Cloning, nucleotide sequence, and expression of the Brucella melitensis omp31 gene coding for an immunogenic major outer membrane protein. Infect Immun. 1996;64:3744–3751. doi: 10.1128/iai.64.9.3744-3751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Waters J K, Hughes B L, Purcell L C, Gerhardt K O, Mawhinney T P, Emerich D W. Alanine, not ammonia, is excreted from N2-fixing soybean nodule bacteroids. Proc Natl Acad Sci USA. 1998;95:12038–12042. doi: 10.1073/pnas.95.20.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weidenhaupt M, Fischer H M, Acuña G, Sanjuan J, Hennecke H. Use of a promoter-probe vector system in the cloning of a new NifA-dependent promoter (ndp) from Bradyrhizobium japonicum. Gene. 1993;129:33–40. doi: 10.1016/0378-1119(93)90693-w. [DOI] [PubMed] [Google Scholar]
- 99.Yuhashi K I, Ichikawa N, Ezura H, Akao S, Minakawa Y, Nukui N, Yasuta T, Minamisawa K. Rhizobitoxine production by Bradyrhizobium elkanii enhances nodulation and competitiveness on Macroptilium atropurpureum. Appl Environ Microbiol. 2000;66:2658–2663. doi: 10.1128/aem.66.6.2658-2663.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]