Abstract
We previously presented evidence that replicating but unsegregated nucleoids, along with the Min system, act as topological inhibitors to restrict assembly of the FtsZ ring (Z ring) to discrete sites in the cell. To test if nonreplicating nucleoids have similar exclusion effects, we examined Z rings in dnaA (temperature sensitive) mutants. Z rings were excluded from centrally localized nucleoids and were often observed at nucleoid edges. Cells with nonreplicating nucleoids formed filaments, some of which contained large nucleoid-free areas in which Z rings were positioned at regular intervals. Because MinE may protect FtsZ from the action of the MinC inhibitor in these nucleoid-free zones, we examined the localization of a MinE-green fluorescent protein (GFP) fusion with respect to Z rings and nucleoids. Like Z rings, MinE-GFP appeared to localize independently of nucleoid position, forming rings at regular intervals in nucleoid-free regions. Unlike FtsZ, however, MinE-GFP often localized on top of nucleoids, replicating or not, suggesting that MinE is relatively insensitive to the nucleoid inhibition effect. These data suggest that both replicating and nonreplicating nucleoids are capable of topologically excluding Z rings but not MinE.
Escherichia coli cells divide precisely at the midpoint of the long axis of the cell to produce two daughter cells of equal length. One of the key questions in bacterial cell biology is how the correct division plane is identified. We recently proposed a new model for division site selection in E. coli that integrated several previous hypotheses (39). One of these hypotheses, known as the nucleoid occlusion model, proposes that the nucleoid inhibits septation throughout the area of the cell that it occupies (37, 38). How might this occur? Recent evidence suggests that oriC and its associated components, such as SeqA, are centered with respect to the cell and coincide with future division sites (23, 24). We propose that when these components duplicate and migrate outward from midcell during replication and segregation, they may relieve the nucleoid-mediated inhibition at the vacated midcell site. The end result is that the Z ring, which is a ring structure composed of the tubulin-like protein FtsZ and is essential for initiation of septation (2, 15, 29), is allowed to assemble at the exact middle of the cell, followed by formation of the septum between the nucleoids.
The experimental evidence for nucleoid-mediated inhibition of Z rings includes (i) the tendency of Z rings and septa to form in nucleoid-free regions in various mutants (4, 21, 39) and (ii) the failure of Z rings to assemble at the middle of parC cells, defective in topoisomerase IV, that contain a large, centrally located unsegregated nucleoid (33). Instead of assembling at midcell, Z rings in such cells usually form on the edge of the unsegregated nucleoid mass. This seems to be the default site for FtsZ assembly when the primary site at midcell is blocked.
Nucleoid-mediated inhibition is only part of how division site positioning might be regulated. Surprisingly, Z rings are often present near the middle of anucleate cells produced by parC mutants as well as other mutants defective in chromosome segregation (33). The lower precision of Z-ring placement in these cells lacking nucleoids is consistent with the idea that normal nucleoid-mediated inhibition helps to guide FtsZ to the precise midcell location. However, the existence of Z rings near the center of these cells indicates that some factor other than the nucleoid must also play a role in guiding their localization. In support of this idea, Cook and Rothfield found that septa could form at locations far away from nucleoids in DNA replication mutants and that these septa were positioned at a fairly fixed distance from a cell pole (4). This result is consistent with our integrated model, in which the Min proteins are proposed to be the primary nucleoid-independent factors involved in guiding FtsZ to the correct cellular location.
The Min proteins consist of MinC, MinD, and MinE, encoded by the minB operon. Deletion of this operon results in the formation of septa at either midcell or the poles; polar septation events result in anucleate minicells (5, 6). MinC is an inhibitor of FtsZ self-assembly, whereas MinD enhances the activity of MinC by recruiting MinC to the membrane and driving a remarkable oscillation of the two proteins from one cell pole to the other (12, 13, 26, 28). MinE, on the other hand, localizes as a ring near midcell independently of FtsZ, although its localization is not as precise as that of FtsZ (27). The MinE ring is proposed to counteract MinCD inhibition in its vicinity, perhaps by physically excluding MinCD, allowing the Z ring to assemble safely nearby (32). It is unclear how MinE gets targeted to the cell center, although interestingly, this targeting requires MinD. One attractive model is that the MinD oscillation serves to mark the position equidistant between the two cell poles—perhaps the region of lowest average MinD concentration—guiding the MinE ring to this position.
How does the Min system fit into our integrated model? The presence of a MinE ring near the center of anucleate cells would explain how the Z ring can assemble near midcell in the absence of the guiding influence of the nucleoid. The presence of a MinE ring that locally counteracts MinCD inhibition would also explain how septa in nucleoid-free segments of filamentous DNA replication mutants are allowed to form. Without MinE, the midcell relief of nucleoid inhibition is not sufficient to suppress MinCD-mediated inhibition, explaining why minE mutants cannot divide (6). Likewise, without relief of nucleoid inhibition, the MinE ring at midcell is not sufficient to allow a Z ring to form there. The combined requirement of both the nucleoid and the Min system for Z-ring placement is best illustrated by our previous study, in which Z rings form promiscuously throughout a ΔminB cell, but only in nucleoid-free gaps (39). This finding prompted us to formulate our model, which proposes that there are two main negative topological regulators of Z-ring assembly in E. coli: the nucleoid and MinCD. These two components are proposed to be sufficient to define the division site, by preventing Z-ring assembly throughout the nucleoid-containing and nucleoid-free (polar) segments of the wild-type cell, respectively, until the inhibition at midcell is relieved. The mechanisms of nucleoid-mediated exclusion and its suppression are not yet known.
Some aspects of the model require refinement. For example, we made our conclusions from studies of a parC mutant, which contains very large nucleoids because they are replication competent despite being segregation defective. As a result, we could not rule out the possibility that such large nucleoids have nonspecific inhibitory effects on Z-ring assembly that are not typical of wild-type nucleoids. In addition, Z rings or septa have been shown clearly to transect, or guillotine, nucleoids in some mutants defective in chromosome replication or organization (4, 22, 33). This would seem to be a direct contradiction of the nucleoid occlusion portion of the model. However, such nucleoid cutting is not observed in wild-type cells, and it can be argued that in many cases where it is observed, such as mukB mutants, the structure of the nucleoid is grossly altered (30), which might suppress the inhibitory activity. Guillotining of nucleoids has also been proposed to occur in ftsK, dif, and xerCD mutants, which are unable to resolve dimeric chromosomes in a proportion of cells (11, 31). It is not yet clear what allows guillotining of these dimer chromosomes.
In the present study, we have sought to refine our integrated model in several ways. First, we have addressed the potential problem of high DNA concentrations by using a chromosome replication initiation mutant. We found that Z rings were still blocked from the central site when the nucleoid occupied this area and instead assembled at the edge of the nucleoid. In filaments arising after extensive incubation at the nonpermissive temperature, Z rings localized to discrete sites in the long nucleoid-free segments of these filaments in a Min-dependent fashion. Our results therefore suggest that small, nonreplicating nucleoids are able to inhibit local assembly of Z rings. Second, we investigated whether this unusual Z-ring positioning was a direct result of colocalization with, and thus protection by, MinE, and whether MinE localization was inhibited by nucleoids. We found that localization of a MinE-green fluorescent protein (GFP) fusion, unlike FtsZ, appears to be relatively insensitive to the postulated nucleoid-mediated inhibition.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
All strains used for this work are derivatives of E. coli K-12 and are listed in Table 1. The ΔminCDE dnaA46 double mutant WM1384 was constructed by P1-mediated cotransduction of tna::Tn10 and dnaA46 (14) into the ΔminCDE strain WM1032. Tetracycline-resistant, temperature-sensitive colonies which produced minicells were confirmed to be defective in DNA replication by staining with DAPI (4′,6-diamidino phenylindole).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| WM1270 | OFB24, thyA recA56 srl::Tn10 dnaA46 | 40 |
| PC5 | thyA47 dnaA5 | E. coli Stock Center |
| E177 | thyA6 dnaA177 | E. coli Stock Center |
| TX3772 | MG1655 ΔlacU169 | 34 |
| WM1179 | E177/pMK4 | This study |
| WM1032 | TX3772 ΔminCDE::kan | Laboratory collection |
| WM1033 | TX3772 parC281 | Laboratory collection |
| WM1081 | WM1033/pWM1079 | This study |
| WM1326 | WM1270/pWM1079 | This study |
| WM1384 | TX3772 ΔminCDE::kan dnaA46 tna::Tn10 | This study |
| Plasmids | ||
| pBAD33 | araBAD promoter vector, p15A origin, Cmr | 9 |
| pWM176 | Tetr, IncP cloning vector containing the tac promoter | 19 |
| pMK4 | E. coli ftsZ cloned downstream of tac promoter of pWM176 | 16 |
| pGBC | XbaI-BamHI fragment with gfp in pBCSK+/lacIq | 17 |
| pWM1005 | minC′DE cloned between SacI and XbaI of pGBC | This study |
| pWM1025 | pWM1005, with a linker inserted between minE and gfp | This study |
| pWM1079 | minDE-gfp in pBAD33 | This study |
To construct a minE-gfp fusion that also carries the minD gene, which is required for MinE localization, we first obtained the minC′DE fragment described previously (27) by PCR from E. coli DNA. We used the upstream primer CGGAGCTCAATCCCAGCAGA and the downstream primer TTTCTAGATTTCAGCTCTTCTGCT. Plasmid pWM1005 was constructed by cleaving the PCR fragment with SacI and XbaI and cloning in between the SacI and XbaI sites of pGBC to fuse gfp translationally with minE. A short linker was then inserted between minE and gfp as previously described (27) to generate pWM1025. Plasmid pWM1079 was constructed by inserting the SacI-KpnI fragment of pWM1025 containing the entire minC′DE-linker-gfp into SacI-KpnI-cleaved pBAD33. WM1081 was constructed by introducing the Cmr pWM1079 plasmid into the parC281 mutant WM1033, which is defective in topoisomerase IV function. WM1326 was constructed by introducing pWM1079 into the dnaA46 strain WM1270.
Unless otherwise noted, cells were grown overnight in Luria-Bertani (LB) medium with 0.5% NaCl at 30°C, diluted 200-fold in fresh LB, and grown at 30°C to early logarithmic phase. At this point, cells were shifted to 37 or 42°C and grown for several additional hours before they were analyzed. For normal growth of thyA strains, the medium was supplemented with 50 μg of thymine per ml. For induction of MinE-GFP synthesis, 0.2% l-arabinose was added, and FtsZ synthesis from pMK4 was induced by addition of 40 μM IPTG (isopropylthiogalactoside). For growth of the dnaA5 and dnaA177 strains in more defined medium, we used M9 salts supplemented with 0.2% glucose and 50 μg of thymine per ml, with or without 1% tryptone. For thymine starvation, thyA cells in logarithmic growth were washed and resuspended in fresh medium lacking thymine and grown at 42°C for an additional 3 to 4 h.
Cell fixation, staining, and microscopy.
Fixation of cells, staining with DAPI, and immunofluorescence microscopy (IFM) were performed as previously described (39). Rabbit polyclonal antibodies against purified FtsZ and purified GFP were raised at Cocalico Biologicals, Inc. (Reamstown, Pa.) and were affinity purified using Affi-Gel 15 (Bio-Rad Laboratories, Hercules, Calif.) according to the manufacturer's instructions (25). The affinity-purified antibodies were used as primary antibodies for IFM. Anti-rabbit immunoglobulin secondary antibodies conjugated to green Alexa 488 (Molecular Probes, Eugene, Oreg.) were used to visualize FtsZ- and GFP-tagged MinE.
In order to visualize nucleoids and GFP simultaneously in intact cells, we used blue Hoechst 33342 (Sigma Chemical Co., St. Louis, Mo.), which efficiently stains nucleoids in live E. coli cells. A 3-μl aliquot of cell culture was first attached to the surface of a 0.1% polylysine-coated cover glass. Then 20 μl of Hoechst dye (0.5 μg/ml in H2O) was added to stain nucleoids for several minutes. After staining, excess dye was removed, and the cover glass containing stained cells was placed on top of a slide containing a drop of antibleaching buffer (0.1% p-phenylenediamine and 80% glycerol in phosphate-buffered saline). Fixed samples were examined by fluorescence microscopy, and a DAPI filter was used to visualize the Hoechst staining. Red-green-blue (RGB) images were captured as previously described (39), with red and blue cables reversed in order to convert the blue DAPI and Hoechst images into red pseudocolor for better contrast. Distance and length measurements were made in Adobe Photoshop with the measurement tool. Data were then graphed with SigmaPlot (Jandel Scientific).
RESULTS
Deficient cell division in dnaA46 mutants.
DnaA protein is responsible for initiation of chromosome replication from oriC (3). To examine FtsZ localization in cells with nonreplicating chromosomes, we used a well-characterized temperature-sensitive allele of dnaA, dnaA46, present in strain WM1270 (20, 36). When shifted to the nonpermissive temperature of 42°C, dnaA(Ts) mutants stop initiating replication at oriC, and any active replication forks continue until they terminate. Assuming that it takes about 40 min to replicate the entire chromosome and that the amount of initiation at 42°C is negligible, essentially all replication should be stopped within 1 h after temperature shift.
We first examined WM1270 cells shifted to 42°C by DAPI staining to detect nucleoids and by IFM to detect Z rings. At 42°C, the cells continued to elongate for several hours, well after replication would have stopped, indicating that cell division is deficient. This cell division deficit was observed relatively early after the temperature shift, as short filaments containing two to four nucleoids were present in 26% of nucleoid-containing cells after 2 h at 42°C. Single Z rings were visible in the gaps between nucleoids in these cells (see below). Some of these Z rings were probably competent for subsequent septation, because the frequency of cells containing two or more nucleoids was reduced to 10% after 1 to 2 h of additional growth at 42°C. This suggested that the division problem was not caused by a deficiency of FtsZ in the cells as a result of lower gene dosage. In support of this idea, FtsZ levels normalized to total cell protein were found to be equivalent in WM1270 cells growing at either 42 or 28°C by immunoblotting (data not shown). Another possible cause of the division deficiency, induction of the SOS response and subsequent SulA-mediated inhibition of Z-ring assembly, was ruled out because WM1270 is recA; this is also consistent with the presence of Z rings in the filaments.
Continued growth and some cell division at 42°C resulted in the formation of short and long filaments, with long filaments being more common at later times. Both types of cells exhibited clear signs of chromosome replication arrest, with large areas of the cells devoid of DNA. Many of the short (4 to 8 μm) filaments contained one centrally located nucleoid. In contrast, other filaments contained a single nucleoid located acentrally or close to a cell pole (Fig. 1). Anucleate cells were also produced (Fig. 1), indicating that cell division, though delayed, was still occurring at regions distal from the nonreplicating nucleoid. A similar phenotype was also observed in a recA+ dnaA46 strain (data not shown).
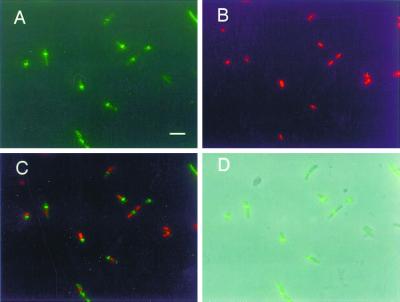
FIG. 1.
Asymmetric localization of FtsZ in a dnaA46 mutant. WM1270 cells shifted to 42°C for 3 h in LB plus thymine were fixed and immunostained for FtsZ. (A) FtsZ staining in green; (B) nucleoids stained with DAPI, pseudocolored red; (C) digital overlay of panels A and B; (D) overlay of panel A and a phase contrast image. Bar, 5 μm.
Exclusion of Z rings from the midcell site of short dnaA46 mutant filaments by a nonreplicating nucleoid.
How do such cells with acentral and polar nucleoids arise? We hypothesize that after temperature shift, the unreplicated nucleoid remains at midcell, blocking the normal division site. Z rings then form adjacent to the nucleoid, sometimes leading to septation. This type of nucleoid-inhibited septation is similar to what we previously observed in parC mutants (33). The end result is the generation of one daughter cell with a nucleoid near one pole and another cell without a nucleoid. Subsequent cell elongation will lead to a nucleoid that is neither adjacent to the cell pole nor at midcell.
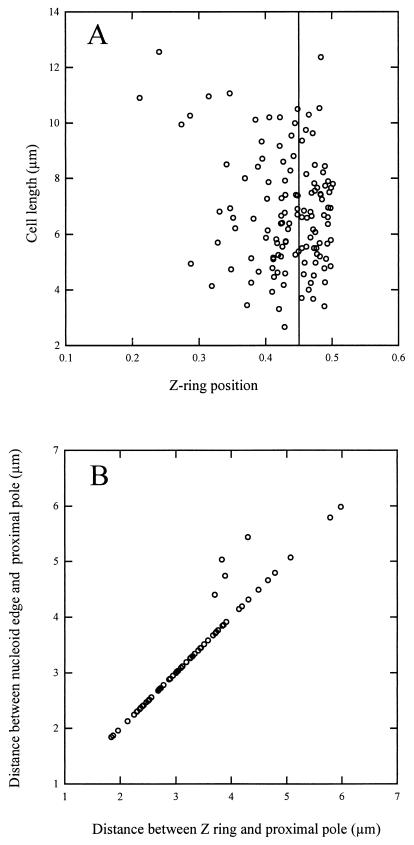
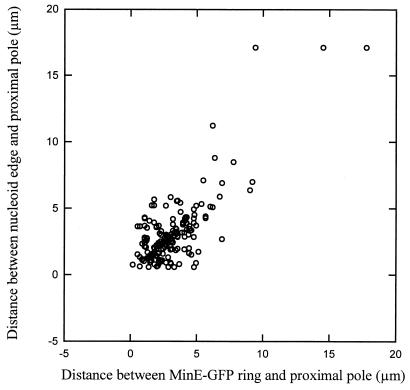
To test this hypothesis, we examined Z-ring localization with respect to the nucleoid in short filaments of WM1270 3 to 4 h after the temperature shift. The results indicate that most of these cells have a single Z ring, and, as expected, many Z rings were localized acentrally (Fig. 1). Whereas all Z rings were located at the midpoint of wild-type MG1655 cells in a range of 0.45 to 0.5 of the total cell length (0.5 is the midcell point, with the poles defined as 0 and 1, and the standard deviation from 0.5 was 0.013 for the 130 MG1655 cells counted), only about 45% of the Z rings were within this range in WM1270 cells (Fig. 2A). The scatter in the data suggests that Z rings did not localize to predetermined sites, such as potential division sites at the cell quarters. Importantly, in these short filaments, most Z rings (93%), including those at midcell, were positioned close to nucleoid edges (Fig. 2B). A small proportion (3.5%) of Z rings appeared to form on top of the nucleoid under our conditions, indicating that the apparent inhibition effect by the nucleoid is not absolute. In summary, these results are consistent with the idea that the majority of Z rings are excluded from their normal midcell position by unreplicated nucleoids. Similar results were observed in other strain backgrounds containing the dnaA46 mutation (data not shown).
FIG. 2.
Distribution of Z-ring positions in the dnaA46 mutant. A total of 129 cells equivalent to those used in Fig. 1 were used to measure positioning of Z rings and nucleoid edges relative to the cell poles. A total of 96% of the cells examined contained only one Z ring. (A) The x axis represents the distance between the Z ring and the proximal pole, with 0.0 defined as the pole and 0.5 as the midcell. The solid line at the 0.45 position represents the border of the normal range of FtsZ localization (0.45 to 0.50) in 130 wild-type MG1655 cells counted. (B) Distances between the nucleoid edge and the proximal pole are plotted versus the distances between the Z ring and the proximal pole; 93% of the Z rings examined localized as close to the edge of the nucleoid as could be measured.
Z-ring localization in other dnaA(Ts) mutants.
To rule out the possibility that the effects on Z-ring positioning resulted from a peculiarity of the dnaA46 allele, we investigated FtsZ localization in two other dnaA mutants, PC5 (dnaA5) and E177 (dnaA177). The dnaA5 mutant had been used in a previous study to analyze septal position relative to the nucleoid and cell pole (4). Both PC5 and E177, like WM1270, require thymine for growth. In LB, M9 tryptone medium, or M9 minimal medium, all containing thymine, we frequently found nucleoids of extended size (over 4 μm in length) at the nonpermissive temperature. This was different from what we observed in the dnaA46 mutant and surprising, because these mutants cannot form colonies at the nonpermissive temperature and should not be able to initiate replication under these conditions. This prompted us to suspect either that residual replication occurred under our experimental conditions or that the nucleoids were not replicating but instead were decondensed. These effects were also observed in other strain backgrounds containing these two alleles (data not shown).
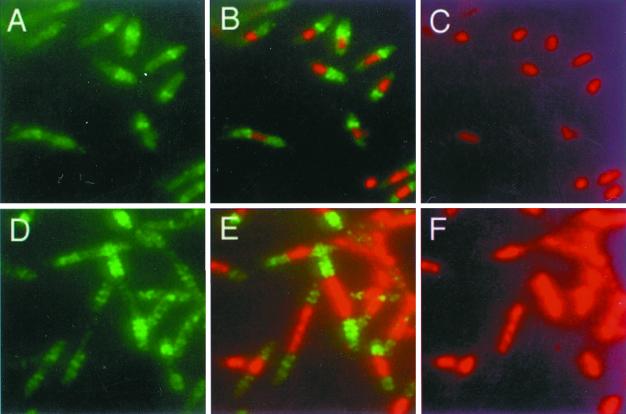
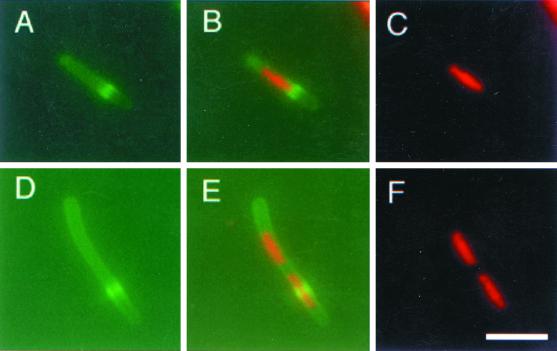
To circumvent this problem, we arrested DNA replication by removing thymine from the medium, thus depleting cellular thymine pools. Nucleoids in thymine-depleted PC5 and E177 cells were significantly smaller than those in cells with thymine, as expected. Interestingly, IFM analysis of these cells revealed no FtsZ localization at all, probably because thymine starvation of recA+ cells induces the SOS response and prevents Z-ring assembly via the action of SulA. To overcome these effects, we introduced plasmid pMK4, which expresses ftsZ under tac promoter control, into the dnaA177 mutant E177. In the absence of IPTG, some additional FtsZ was synthesized in these cells, resulting in the formation of acentral rings at nucleoid edges (Fig. 3A to C). These results are similar to what we found in the dnaA46 mutant. When higher levels of FtsZ were induced by addition of IPTG, multiple Z rings appeared in the short filaments, and they usually assembled in nucleoid-free regions (Fig. 3D to F). This suggests that nonreplicating nucleoids are able to exclude Z rings even when FtsZ levels are relatively high.
FIG. 3.
Z rings in the dnaA177 mutant. To counteract any potential SOS response in the recA+ dnaA177 cells after replication arrest, FtsZ synthesis was increased with a plasmid expressing ftsZ under tac promoter control (WM1179). WM1179 cells were grown in LB plus thymine to early log phase at 30°C. Cells were then washed three times and resuspended in LB without thymine, containing no IPTG (A to C) or 40 μM IPTG to induce extra FtsZ synthesis (D to F), and grown at 42°C for 3 h. (A and D) FtsZ staining in green; (C and F), nucleoids stained with DAPI, pseudocolored red; (B and E), digital overlays of panels A plus C and panels D plus F, respectively.
Z rings localize at regular intervals in long nucleoid-free regions of dnaA mutant filaments.
As mentioned above, when WM1270 was grown for extended periods at the nonpermissive temperature of 42°C, many cells became long filaments because of an overall delay in septation. Most of these long filaments contained far fewer nucleoids per cell length than replication-proficient cells, resulting in long anucleate regions up to 20 μm in length. IFM analysis revealed that the anucleate regions often contained regularly spaced Z rings. Interestingly, the distance between the rings was significantly longer than the normal 2 to 3 μm seen in filaments with normal nucleoids, and this distance varied from cell to cell (Fig. 4). This result suggests that in DNA replication mutants, Z rings can localize at discrete intervals in a regular pattern in the absence of the nucleoid.
FIG. 4.
Periodic localization of Z rings in anucleate segments of long dnaA46 filaments. WM1270 cells shifted to 42°C for 4 h in LB plus thymine were fixed and immunostained for FtsZ. The left panel shows FtsZ staining in green, and the right panel shows nucleoids stained with DAPI, pseudocolored red. The middle panel is a digital overlay of the left and right panels. Bar, 5 μm.
Involvement of the Min system in regular Z-ring localization in the absence of the nucleoid.
What factors might guide Z rings to assemble at regular intervals in the extensive nucleoid-free spaces in these dnaA mutants? Because the Min system is intact in these cells, our integrated model would predict that MinCD inhibits Z rings from assembling throughout most of the nucleoid-free areas except within zones protected by MinE.
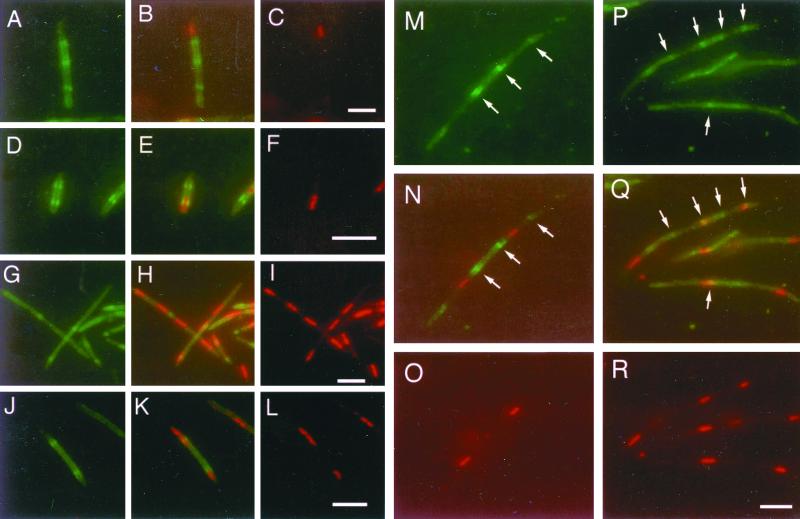
We therefore investigated whether MinE rings localized in patterns similar to FtsZ in these cells. We used a MinE-GFP fusion to report localization of MinE in the dnaA46 mutant. Such fusions appear to be fully functional (27). WM1326 is WM1270 (dnaA46) containing a plasmid (pWM1079) coexpressing MinE-GFP and MinD, which is required for localization of MinE, under arabinose control. Because the MinE-GFP fluorescent signal was unstable at temperatures above 40°C in WM1326, the mutant was shifted instead from 30 to 37°C to observe MinE-GFP localization. At 37°C, WM1326 still failed to form colonies, and nucleoid and Z-ring patterns were equivalent to those observed at 42°C. To detect both MinE-GFP and nucleoids with DAPI in the same cells, we fixed the cells and immunostained for MinE-GFP using a polyclonal anti-GFP antibody. Cell fixation was necessary for efficient DAPI staining, and both MinE-GFP and nucleoid staining signals were stable and clear under these conditions. To use an independent method, we also used the membrane-permeating DNA stain Hoechst 33342 and MinE-GFP to detect both nucleoids and MinE localization directly in live cells. With either technique, we observed that MinE-GFP localized in a pattern roughly similar to that of Z rings in nucleoid-free segments of these mutant filaments (Fig. 5M to R, arrows, and data not shown). This result suggests that MinE may be required for regular Z-ring localization in the anucleate portions of the filaments.
FIG. 5.
MinE-GFP localization in the dnaA46 mutant. WM1326 (MinDE-GFP under pBAD control in dnaA46) cells growing at 30°C in LB plus thymine were shifted to 37°C. This temperature, like 42°C, is an effective nonpermissive temperature for this mutant. After 0 and 1 h of growth at 37°C, l-arabinose was added to induce MinE-GFP synthesis. Samples were taken after 3 to 4 h of incubation at 37°C. (A to L) MinE-GFP rings in short filaments. (M to R) MinE-GFP rings in longer filaments. (A, D, G, J, M, and P) MinE-GFP immunostaining with anti-GFP and Alexa 488 (green). (C, F, I, L, O and R) Nucleoids stained with DAPI, pseudocolored red. (B, E, H, K, N, and Q) Digital overlays of MinE-GFP and nucleoid staining, corresponding to panels A plus C, D plus F, G plus I, J plus L, M plus O, and P plus R, respectively. Arrows in panels M, N, P, and Q highlight the regular MinE-GFP bands in longer filaments. Bars, 5 μm; the bar in panel R pertains to panels M to R.
To test further the requirement of the Min system for Z-ring localization in nucleoid-free segments of dnaA filaments, the dnaA46 mutation was introduced into the ΔminCDE strain WM1032, producing the double dnaA46 ΔminCDE mutant WM1384. Cells lacking the Min system are normally a mixture of short multinucleate filaments and minicells, because each polar division event occurs at the expense of a midcell division (1). As expected, prior to shifting to the nonpermissive temperature, many of the WM1384 cells already were filamentous and multinucleate, and cells continued to elongate after the temperature shift in spite of the replication arrest. This resulted in filaments with multiple nonreplicating nucleoids and some long anucleate regions, as well as small anucleate cells. In IFM experiments, dramatic clusters of Z rings localized within nucleoid-free gaps (Fig. 6). We did not observe transection of nucleoids by any Z rings (Fig. 6). This promiscuous and seemingly random assembly of Z rings in nucleoid-free regions of ΔminCDE cells is similar to our previous results with parC mutants (39) and further supports the idea that the Min system limits Z-ring assembly to discrete locations in these nucleoid-free segments. These results also support the idea that nonreplicating nucleoids can inhibit FtsZ localization in ΔminCDE strains as efficiently as unsegregated but replicating nucleoids.
FIG. 6.
Z-ring clusters in the dnaA46 ΔminCDE double mutant. WM1384 cells shifted to 42°C for 4 h in LB were fixed and immunostained for FtsZ. Many filaments appear to be multinucleate because many ΔminCDE cells were already multinucleate filaments prior to the temperature shift. (A and D) FtsZ staining in green; (C and F), nucleoids stained with DAPI, pseudocolored red; (B and E), digital overlays of panels A plus C and panels D plus F, respectively.
MinE-GFP localization is relatively insensitive to inhibition by the nucleoid and is independent of nucleoid position.
The apparent inhibition of Z-ring assembly by the nucleoid and the similar patterns of MinE and FtsZ localization prompted us to ask whether the nucleoid also inhibits MinE localization in its vicinity. To address this question, we first investigated the localization of MinE-GFP in short dnaA46 mutant filaments. WM1326, the dnaA46 mutant carrying MinE-GFP under arabinose control (pWM1079), was shifted to the nonpermissive temperature of 37°C, and MinE-GFP expression was induced by adding 0.2% l-arabinose. In IFM experiments, we found that MinE-GFP rings, in addition to localizing independently of nucleoids, often formed on top of them (Fig. 5E, H, and K). Staining of FtsZ in parallel samples showed, as expected, that Z rings were excluded from the nucleoid (data not shown). This suggests that, unlike the Z ring, the MinE ring appears to be relatively insensitive to nucleoid-mediated inhibition.
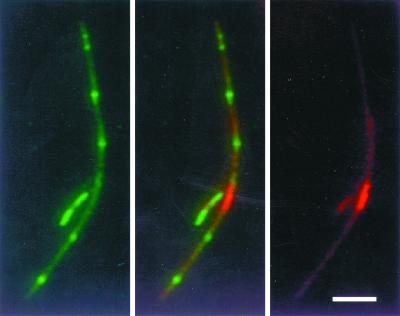
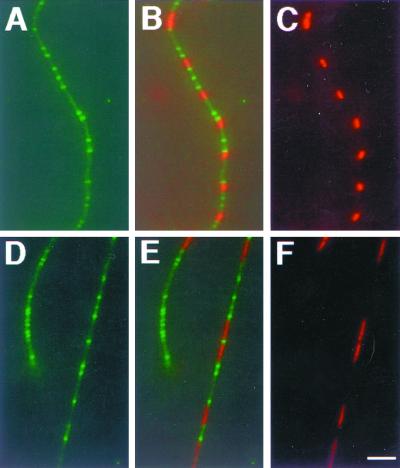
To test this idea more rigorously, we investigated whether large parC mutant nucleoids containing multiple unsegregated chromosomes might be more likely to exert a negative effect on MinE ring formation. WM1081 is a parC mutant containing the arabinose-inducible MinE-GFP. As was done with the dnaA mutants, we obtained stable MinE-GFP localization by shifting to a lower nonpermissive temperature (37°C) for parC, which still exhibited the segregation defect at this temperature. MinE-GFP and nucleoids were visualized simultaneously in unfixed cells by using Hoechst 33342. As in the dnaA46 mutant, MinE-GFP rings localized both on top of unsegregated nucleoids (Fig. 7D to F) and in the polar nucleoid-free regions of these short filaments (Fig. 7A to C). Among 190 cells examined, 22% exhibited clear transection of the nucleoid by the MinE-GFP ring (Fig. 8). The results with the parC mutant therefore agree with those with the dnaA46 mutant. Our data suggest that MinE localization is independent of and relatively insensitive to (i) unsegregated but replicating nucleoids and (ii) nonreplicating nucleoids.
FIG. 7.
MinE-GFP localization in the parC mutant. WM1081 cells were grown at 30°C to early logarithmic phase. Then 0.2% l-arabinose was added, and the culture was shifted to 37°C for 2 h. Live cells with MinE-GFP fluorescence were then stained with Hoechst 33342 in order to visualize MinE-GFP and nucleoids simultaneously. (A and D) MinE-GFP; (C and F) nucleoids stained with Hoechst 33342; (B and E) digital overlays of MinE-GFP and Hoechst 33342 staining, corresponding to panels A plus C and D plus F, respectively.
FIG. 8.
Relative positions of MinE-GFP and the nucleoid with respect to the cell pole. WM1081 (parC) cultures treated as described in the legend to Fig. 7 were used to measure distances between the MinE-GFP ring and the pole and the nucleoid edge to the pole. A total of 190 cells containing MinE-GFP rings were counted.
We also examined the localization of Z rings in this WM1081 (parC) strain by IFM under the same conditions used to visualize MinE rings. Similar to our previous results (33), essentially all Z rings were positioned at nucleoid edges, and cutting of unsegregated nucleoids by a Z ring was not detected (data not shown). The results suggest that the unsegregated chromosomes in this mutant retained the capacity to inhibit Z-ring formation, causing the rings to be acentral, but often allowed central localization of MinE rings.
DISCUSSION
In this study, we present evidence suggesting that a nonreplicating nucleoid is able to exclude the Z ring, an essential component of the division septum. This evidence complements and extends our previous studies, in which Z rings formed throughout nucleoid-free portions of the cell and in small gaps between nucleoids, but did not form on top of a clearly unsegregated nucleoid mass (33, 39). The unsegregated nucleoid masses in the parC mutant were very large, because they were replication proficient despite being segregation deficient. Therefore, the possibility remained that Z rings are not normally excluded from normal-sized nucleoids, only from the large parC nucleoids. In the present study, we show that Z rings are almost always excluded from small, nonreplicating nucleoids. These results suggest that nucleoid-mediated inhibition of FtsZ is broadly significant and is not restricted to a particular type of mutant nucleoid. The fact that MinE-GFP rings often form on top of both parC and dnaA mutant nucleoids suggests that the inhibitory effect of an unsegregated nucleoid on FtsZ is fairly specific, not merely a steric effect on any protein that normally would localize in the nucleoid zone.
Despite the evidence for nucleoid-mediated exclusion of Z rings, the nature of the exclusion is not clear. A recent population study in wild-type cells suggested that Z rings form at about the time of termination of chromosomal replication (7). As chromosome segregation is probably an ongoing process, it is reasonable to propose that relief of nucleoid-mediated inhibition would occur when chromosomes were initially being pulled apart, which might occur at the time of termination. Proper timing of this relief of inhibition would prevent assembly of the division machinery until after chromosome replication is complete. Moreover, proper positioning of the relief of inhibition at the precise center of the cell would help to prevent scission of the symmetrically segregating chromosomes by the septum. Although MinE counteracts MinCD inhibition at midcell to allow Z-ring assembly, MinE localization is not precise; therefore, we postulate that it is the task of the nucleoid inhibition mechanism and its subsequent suppression of segregation to provide a more precise site for Z-ring assembly. When a nucleoid is not nearby, our data suggest that the Min system may be sufficient to allow Z rings to form in discrete zones that correspond roughly to MinE rings.
In a recent study, Cook and Rothfield analyzed the positions of septa in mutant filaments defective in DNA replication (4). They found that most septa formed approximately 4 to 6 μm from the poles, usually within nucleoid-free regions of these cells, and that the position of the often distant nucleoids had no effect on the position of these septa. This result is consistent with our findings. The ability of septa to form in a range might be explained by protection of this region by a nearby MinE ring; the imprecision of septal positioning within this 4- to 6-μm range would be consistent with the imprecision of MinE localization. However, another aspect of their study differed from ours, namely, that large nucleoids near the pole in dnaA5 ftsA and dnaB sfiA mutants were transected by septa at higher frequencies than the 3.5% that we observed. The dnaA5 ftsA mutant is defective in both DNA replication initiation and a post-FtsZ step in septation at 42°C, necessitating a return to the permissive temperature of 30°C in order to form visible septa. It is possible that replication reinitiation under these conditions might have resulted in the suppression of nucleoid-mediated inhibition of Z rings. We also found that nucleoids of both dnaA5 and dnaA177 cells at 42°C were significantly larger than those of dnaA46 mutant cells at 42°C and that these effects were independent of strain background. This suggests that differences in nucleoid structure in dnaA5 and dnaA177 mutants compared to dnaA46 mutants might lead to different effects on Z rings. Another possible explanation is that the nucleoid may not be properly segregated away from the septum in certain mutants. While Z rings initially form at the nucleoid edge, subsequent extension of the nucleoid during a delay in septation might result in occasional trapping of segments at the edges of nucleoids by the septum. Clearly, further studies are needed to refine our model for division site placement and to define more precisely the nucleoid-mediated inhibition mechanism and the effects of cell physiology on its proper functioning.
The apparent nucleoid independence of MinE localization suggests that, unlike FtsZ, MinE uses cues other than the nucleoid for its proper targeting. The dependence of MinE localization on the MinD oscillator suggests that MinE rings form in the area of the cell with the lowest average concentration of MinD. This area in wild-type cells should be around midcell, because MinD dwells mostly at each cell pole and only appears to migrate transiently through the midcell area (28). This model is also consistent with the imprecision of MinE localization (27), in that if MinE dynamically senses MinD concentration during its oscillation in real time, MinE may also oscillate in a small range around midcell (18) and thus may seldom be precisely at midcell.
In the parC mutant, 22% of the MinE rings formed on top of an unsegregated nucleoid, a position where Z-ring assembly is normally prevented. Because Z rings often form at the edge of these unsegregated nucleoids, it is therefore possible that in some of these cells with MinE rings on top of nucleoids, Z rings might assemble at a significant distance away from MinE. How might MinE protect FtsZ from MinCD if this occurs? Perhaps FtsZ, once above its critical concentration for assembly, integrates the level of inhibition from both the nucleoid and MinCD and nucleates a ring at the site with the lowest total inhibitory activity. In cells with unsegregated nucleoids with active inhibition mechanisms, this spot would automatically be in the nucleoid-free area. When the MinE ring is on top of the nucleoid and not in this area, the inhibition of MinCD by MinE may occur via a gradient of inhibition that is lowest at the poles and highest near midcell. We speculate that Z rings form at the edge of the nucleoid because this is farthest away from the maximum inhibitory activity of MinCD and still not in the nucleoid space. In the dnaA mutant filaments, MinCD and the nucleoid block most parts of the cell except for regularly spaced zones occupied by MinE rings, which are available sites for Z-ring assembly. The problem of how MinD oscillates and how MinE localization depends on this oscillation must be addressed in the future in order to refine further our integrated division site placement model. Additional insights about the regulation of Z-ring placement will come from other model systems that lack MinE and whose nucleoids exert somewhat different effects on Z-ring assembly and activity (10, 35).
The formation of filaments after inhibition of DNA replication indicates that there is a deficiency in cell division under these conditions, although production of cells with fewer nucleoids or no nucleoids implies that division still occurs, just at a lower rate. The fact that most of these filaments contain Z rings suggests that the deficiency is not caused by the lack of Z rings, but instead by a block downstream from Z-ring formation that results in unused Z rings. The nature of this block is not yet clear. The Z rings in most cases appear normal in morphology, indicating that if they are structurally defective, the defect must be subtle. Another possibility is that a later cell division protein, such as FtsA, may be limiting, perhaps because of the lower gene dosage relative to cell volume in these filaments. Limitation of FtsA can have a major effect on cell division even in the presence of sufficient FtsZ (1).
After this paper was submitted, Gullbrand and Nordström reported the effects of nonreplicating nucleoids on Z-ring positioning (8). They observed both cell division deficiencies and anucleate cell production in a dnaA46 mutant and also found that Z rings were located on both sides of the centrally located nucleoids. These results were similar to ours. Work is currently in progress to understand the mechanism of nucleoid occlusion and the reasons for the division delay in dnaA mutants.
ACKNOWLEDGMENTS
We thank Jon Kaguni, Jill Zielstra-Ryalls, and Mary Berlyn at the E. coli Genetic Stock Center for sending us dnaA mutant strains and Shelley Sazer for suggesting the use of Hoechst for staining nucleoids of live cells.
This work was supported by grants from the Texas Advanced Technology Program (011618-0231-1999) and the National Institutes of Health (1R01-GM61074-01).
REFERENCES
- 1.Begg K, Nikolaichik Y, Crossland N, Donachie W D. Roles of FtsA and FtsZ in activation of division sites. J Bacteriol. 1998;180:881–884. doi: 10.1128/jb.180.4.881-884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bramhill D. Bacterial cell division. Annu Rev Cell Dev Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 3.Bramhill D, Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988;54:915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- 4.Cook W R, Rothfield L I. Nucleoid-independent identification of cell division sites in Escherichia coli. J Bacteriol. 1999;181:1900–1905. doi: 10.1128/jb.181.6.1900-1905.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Boer P A, Cook W R, Rothfield L I. Bacterial cell division. Annu Rev Genet. 1990;24:249–274. doi: 10.1146/annurev.ge.24.120190.001341. [DOI] [PubMed] [Google Scholar]
- 6.de Boer P A, Crossley R E, Rothfield L I. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J Bacteriol. 1992;174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.den Blaauwen T, Buddelmeijer N, Aarsman M E, Hameete C M, Nanninga N. Timing of FtsZ assembly in Escherichia coli. J Bacteriol. 1999;181:5167–5175. doi: 10.1128/jb.181.17.5167-5175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gullbrand B, Nordström K. FtsZ ring formation without subsequent cell division after replication runout in Escherichia coli. Mol Microbiol. 2000;36:1349–1359. doi: 10.1046/j.1365-2958.2000.01949.x. [DOI] [PubMed] [Google Scholar]
- 9.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harry E J, Rodwell J, Wake R G. Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Mol Microbiol. 1999;33:33–40. doi: 10.1046/j.1365-2958.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 11.Hendricks E C, Szerlong H, Hill T, Kuempel P. Cell division, guillotining of dimer chromosomes and SOS induction in resolution mutants (dif, xerC and xerD) of Escherichia coli. Mol Microbiol. 2000;36:973–981. doi: 10.1046/j.1365-2958.2000.01920.x. [DOI] [PubMed] [Google Scholar]
- 12.Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- 13.Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci USA. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang D S, Kaguni J M. Purification and characterization of the dnaA46 gene product. J Biol Chem. 1988;263:10625–10632. [PubMed] [Google Scholar]
- 15.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 16.Ma X, Ehrhardt D W, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma X, Sun Q, Wang R, Singh G, Jonietz E L, Margolin W. Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J Bacteriol. 1997;179:6788–6797. doi: 10.1128/jb.179.21.6788-6797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margolin W. Themes and variations in prokaryotic cell division. FEMS Microbiol Rev. 2000;24:531–548. doi: 10.1111/j.1574-6976.2000.tb00554.x. [DOI] [PubMed] [Google Scholar]
- 19.Margolin W, Long S R. Rhizobium meliloti contains a novel second copy of the cell division gene ftsZ. J Bacteriol. 1994;176:2033–2043. doi: 10.1128/jb.176.7.2033-2043.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messer W, Weigel C. Initiation of chromosome replication. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 1579–1601. [Google Scholar]
- 21.Mulder E, Woldringh C L. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol. 1989;171:4303–4314. doi: 10.1128/jb.171.8.4303-4314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14:212–223. [PMC free article] [PubMed] [Google Scholar]
- 24.Onogi T, Niki H, Yamazoe M, Hiraga S. The assembly and migration of SeqA-Gfp fusion in living cells of Escherichia coli. Mol Microbiol. 1999;31:1775–1782. doi: 10.1046/j.1365-2958.1999.01313.x. [DOI] [PubMed] [Google Scholar]
- 25.Pringle J R, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 26.Raskin D M, de Boer P A. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raskin D M, de Boer P A. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 28.Raskin D M, de Boer P A. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothfield L, Justice S, Garcia-Lara J. Bacterial cell division. Annu Rev Genet. 1999;33:423–448. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- 30.Sawitzke J A, Austin S. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc Natl Acad Sci USA. 2000;97:1671–1676. doi: 10.1073/pnas.030528397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steiner W, Liu G, Donachie W D, Kuempel P. The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol Microbiol. 1999;31:579–583. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan S M, Maddock J R. Bacterial division: finding the dividing line. Curr Biol. 2000;10:249–252. doi: 10.1016/s0960-9822(00)00376-6. [DOI] [PubMed] [Google Scholar]
- 33.Sun Q, Yu X-C, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol Microbiol. 1998;29:491–504. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- 34.Tsui H C, Feng G, Winkler M E. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J Bacteriol. 1997;179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward D, Newton A. Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus. Mol Microbiol. 1997;26:897–910. doi: 10.1046/j.1365-2958.1997.6242005.x. [DOI] [PubMed] [Google Scholar]
- 36.Wechsler J, Gross J. Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol Gen Genet. 1971;113:273–84. doi: 10.1007/BF00339547. [DOI] [PubMed] [Google Scholar]
- 37.Woldringh C L, Mulder E, Huls P G, Vischer N. Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol. 1991;142:309–20. doi: 10.1016/0923-2508(91)90046-d. [DOI] [PubMed] [Google Scholar]
- 38.Woldringh C L, Mulder E, Valkenburg J A, Wientjes F B, Zaritsky A, Nanninga N. Role of the nucleoid in the toporegulation of division. Res Microbiol. 1990;141:39–49. doi: 10.1016/0923-2508(90)90096-9. [DOI] [PubMed] [Google Scholar]
- 39.Yu X-C, Margolin W. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol. 1999;32:315–326. doi: 10.1046/j.1365-2958.1999.01351.x. [DOI] [PubMed] [Google Scholar]
- 40.Zeilstra-Ryalls J, Fayet O, Georgopoulos C. In vivo protein folding: suppressor analysis of mutations in the groES cochaperone gene of Escherichia coli. FASEB J. 1996;10:148–152. doi: 10.1096/fasebj.10.1.8566535. [DOI] [PubMed] [Google Scholar]