Abstract
Proteus mirabilis, a gram-negative bacterium associated with complicated urinary tract infections, produces a metalloenzyme urease which hydrolyzes urea to ammonia and carbon dioxide. The apourease is comprised of three structural subunits, UreA, UreB, and UreC, assembled as a homotrimer of individual UreABC heterotrimers (UreABC)3. To become catalytically active, apourease acquires divalent nickel ions through a poorly understood process involving four accessory proteins, UreD, UreE, UreF, and UreG. While homologues of UreD, UreF, and UreG have been copurified with apourease, it remains unclear specifically how these polypeptides associate with the apourease or each other. To identify interactions among P. mirabilis accessory proteins, in vitro immunoprecipitation and in vivo yeast two-hybrid assays were employed. A complex containing accessory protein UreD and structural protein UreC was isolated by immunoprecipitation and characterized with immunoblots. This association occurs independently of coaccessory proteins UreE, UreF, and UreG and structural protein UreA. In a yeast two-hybrid screen, UreD was found to directly interact in vivo with coaccessory protein UreF. Unique homomultimeric interactions of UreD and UreF were also detected in vivo. To substantiate the study of urease proteins with a yeast two-hybrid assay, previously described UreE dimers and homomultimeric UreA interactions among apourease trimers were confirmed in vivo. Similarly, a known structural interaction involving UreA and UreC was also verified. This report suggests that in vivo, P. mirabilis UreD may be important for recruitment of UreF to the apourease and that crucial homomultimeric associations occur among these accessory proteins.
Urease (urea amidohydrolase; EC 3.5.1.5) is a nickel metalloenzyme which catalyzes the hydrolysis of urea into ammonia and carbamate (for a review, see reference 25). The biological role of urease varies from nitrogen recycling, as seen in many plants and soil-associated bacteria, to an essential virulence factor in several human pathogens (25). This study focused on urease produced by Proteus mirabilis, a gram-negative organism frequently associated with complicated urinary tract infections (26, 33). A hallmark of P. mirabilis infections is the formation of urinary stones. An increase in pH, arising from urease-mediated urea hydrolysis, culminates in precipitation of normally soluble ions in urine to form struvite and carbonate apatite stones (10, 16).
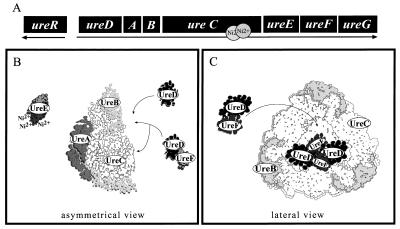
The urease gene cluster of P. mirabilis encodes three structural polypeptides, UreA, UreB, and UreC, which form the apoenzyme; four accessory polypeptides, UreD, UreE, UreF, and UreG; and an AraC-like positive transcriptional activator, UreR (see Fig. 7A) (18). Research published by this laboratory and others has amassed clues to the functional role played by the accessory proteins. For example, when ureolytic bacteria are grown in medium lacking nickel ions, the urease apoprotein is produced (21). Addition of nickel ions to purified apoprotein fails to generate active enzyme in standard purification or assay buffers (21, 31). Early genetic analyses of several ureolytic bacterial species revealed that it is possible to eliminate urease activity by disrupting genes encoding proteins other than the urease subunits (14, 17, 21, 25, 30). In our laboratory, independent in-frame mutations of ureD, ureF, and ureG led to the complete inactivation of P. mirabilis urease (14). Urease purified from homologous ure mutants in Klebsiella aerogenes has insufficient concentrations of the nickel cofactor to support enzymatic activity (21). Based on these observations, it is generally believed that urease accessory proteins facilitate nickel incorporation.
FIG. 7.
Hypothetical models of P. mirabilis urease interactions with structural and accessory proteins. (A) The 6,500-bp P. mirabilis urease gene cluster encodes eight proteins that comprise, regulate, and assemble the urease homoenzyme. Previously described UreA and UreE homomultimeric interactions were confirmed in vivo (Fig. 6) (15, 22, 38). Likewise, UreA and UreC structural interactions were also confirmed in vivo (Fig. 6) (15). (B) UreD associates with UreC in the context of the apourease independently of the UreA structural protein (Fig. 4); UreD was arbitrarily drawn contacting the apoenzyme face opposite UreA; there is no direct evidence for this structure. Although UreD and UreF interact in the absence of structural proteins (Fig. 6), UreD is still capable of associating with the apourease without coaccessory proteins such as UreF (Fig. 3). (C) Data reported here suggest that UreD is capable of homomultimeric interactions in vivo (Fig. 6). Based on the homotrimeric nature of the apourease, one explanation for our observation is that a single molecule of UreD associated with UreABC may interact with additional UreD molecules bound to adjacent UreABC heterotrimers. These interactions could stabilize overall the accessory protein interactions with the apourease and hypothetically coordinate nickel uptake among the three active sites of urease. A similar hypothesis could be applied to UreF; homomultimeric UreF interactions in vivo (Fig. 6) could occur between individual UreF molecules bound through UreD to adjacent UreABC heterotrimers. The three-dimensional structure of urease is inferred from the closely related urease of K. aerogenes (15).
Interestingly, accessory protein homologues UreD, UreF, and UreG of K. aerogenes have been copurified with the apourease (31, 32). In recombinant strains overproducing UreD, it was found to be associated with the urease apoprotein (31). Subsequent activation of the apourease was linked to UreD dissociation from the complex (31). Although the properties of UreD have only been examined in K. aerogenes, we speculated that it also serves as an apourease-specific chaperone in P. mirabilis, maintaining the optimal protein conformation to facilitate proper assembly of the metallocenter. It is worth noting that other accessory proteins do not appear to copurify with the apourease in a ureD mutant strain (32). Thus, we propose that UreD may be crucial for the recruitment and stabilization of other accessory proteins in complexes with the apourease.
The P. mirabilis UreE homologue possesses a histidine-rich motif at the carboxyl terminus (18). We exploited this feature to purify UreE protein in a single step with nickel affinity chromatography (38). While full-length UreE homologues have been reported to bind approximately six nickel ions per dimer (22), recent experiments have shown that UreE truncates, lacking the histidine-rich tail, retain some essential nickel-binding activity (3). It is postulated that nickel ions bound at the UreE dimer interface (see Fig. 1) may be important for transfer to the apourease (3, 7). Consistent with the role as a putative nickel donor, P. mirabilis ureE deletion mutants exhibit depressed urease activity in minimal medium that can be partially restored by adding higher concentrations of the metal ion (38). To date, UreE has not been demonstrated to interact with either apourease or coaccessory proteins.
FIG. 1.
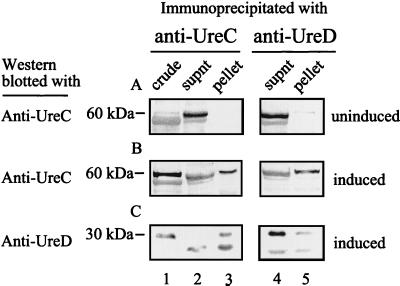
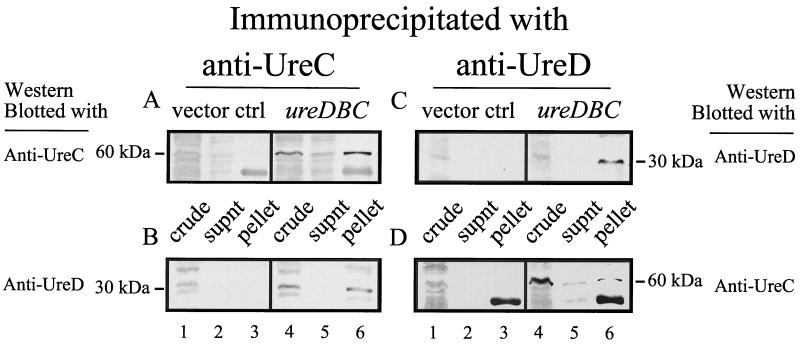
Immunoblots illustrating that UreC and UreD coimmunoprecipitate from lysates of E. coli DH5α carrying a recombinant P. mirabilis ure gene cluster (pMID1010). Soluble protein from uninduced (A) and urea (50 mM)-induced (B and C) E. coli DH5α(pMID1010) was immunoprecipitated with monoclonal anti-UreC (left column) and -UreD (right column) antibodies, separated by SDS–12% PAGE, and immunoblotted with polyclonal anti-UreC serum (A and B) and anti-UreD serum (C). The immunoblot is representative of three experiments. Lanes: 1, crude culture extracts of E. coli DH5α(pMID1010); 2, supernatants (supnt) from UreC immunoprecipitation reactions; 3, solubilized pellets from UreC immunoprecipitation reactions; 4, supernatants from UreD immunoprecipitation reactions; 5, solubilized pellets from UreD immunoprecipitation reactions.
Most urease accessory gene sequences do not exhibit extensive homology with other genes accessible in the GenBank database. However, UreG is an interesting exception; the predicted amino acid sequence shares similarities with a P-loop motif (PROSITE accession no. PDOC00017) which is characteristic of a variety of ATP- and GTP-binding proteins (21, 39). Equally important is the fact that the UreG amino acid sequence is somewhat related to the HypB protein (21). The hypB gene is part of the hydrogenase pleiotropic operon that is required for GTP-dependent activation of nickel-containing hydrogenases (23, 41). Limited research on the mechanism of UreG in K. aerogenes has been reported (28, 37). Complexes comprised of UreD, UreF, and UreG have been shown to bind nucleotide-linked resin and enhance apourease activation in vitro in a GTP-dependent manner; however, direct observations of UreG interactions with GTP are lacking (28, 37). Consistent with deoxynucleoside triphosphate requirements, P-loop variants of UreG have been correlated with reduced urease activity (28). Limited data suggest that UreG, as well as UreF, can bind to the UreD-apoprotein complex (32). Whether these associations are directly mediated by UreD is speculative and requires additional research.
Our objective was to identify how individual urease accessory proteins interact with the apourease and coaccessory proteins during the process of nickel incorporation. The significance of this work extends beyond understanding a crucial virulence factor of a widespread uropathogen. Findings reported here could be generalized to ureases produced by many other species (25), as well as have implications for how other metalloenzyme systems are activated. We conducted in vitro immunoprecipitation experiments and an in vivo yeast two-hybrid assay to screen for protein-protein interactions. In this study, we have identified new interactions involving P. mirabilis urease accessory proteins and confirmed previously described interactions among urease structural polypeptides.
MATERIALS AND METHODS
Strains and materials.
Recombinant DNA constructs reported here or elsewhere were maintained in Escherichia coli DH5α (Table 1). Cloned P. mirabilis urease genes were expressed in E. coli DH5α and BL21(DE3)(pLysS) for immunoprecipitation experiments. The Saccharomyces cerevisiae strains and plasmids used in the two-hybrid studies (Table 1) were described in detail by Ausubel et al. (1). P. mirabilis HI4320, a urease-positive clinical isolate, was the original source of the urease genes (17) used in this study and served as an apourease control in immunoprecipitation experiments (Table 1). Culture medium components were purchased from Bio 101, Inc. (La Jolla, Calif.), and Sigma (St. Louis, Mo.). Restriction endonucleases, DNA polymerases, and other DNA-modifying enzymes were obtained from either Gibco BRL (Rockville, Md.) or New England Biolabs (Beverly, Mass.). All immunological and chemical reagents, unless otherwise specified, were obtained from Sigma.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference(s) or source |
|---|---|---|
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA endA1 gyrA96 thi-1 relA1 | 35 |
| BL21(DE3)(pLysS) | B F−dcm ompT hsdS (rB mB) gal λ(DE3)(pLysS) Camr | 35 |
| Proteus mirabilis HI4320 | Urease-positive clinical isolate | 26 |
| Saccharomyces cerevisiae | ||
| EGY48 | his3 trp1 ura3 leu2 (4 lexA operators preceding leu2) | 8, 11 |
| EGY191 | his3 trp1 ura3 leu2 (1 lexA operator preceding leu2) | 8 |
| Cloning vectors | ||
| pBS SK+ | lacZ Ampr | Stratagene |
| pMALC-2 | malE Ampr | New England Biolabs |
| Recombinant P. mirabilis ure genes | ||
| pMID1010 | P. mirabilis ureDABCEFG Ampr | 17 |
| p1701.1 | P. mirabilis ureDABC Ampr | 38 |
| pDBC | P. mirabilis ureDBC Ampr | This study |
| Two-hybrid vectors | ||
| pEG202 | lexA his3 Ampr | 11 |
| pJG4-5 | gal1 B42 + HA epitope tag trp1 Ampr | 34 |
| pJK101 | gal1 2 lexA operators preceding lacZ ura3 Ampr | 4 |
| pSH18-34 | 8 lexA operators preceding lacZ ura3 Ampr | 42 |
| pJK103 | 1 lexA operator preceding lacZ ura3 Ampr | 20 |
Recombinant DNA techniques.
Recombinant DNA techniques, including restriction endonuclease digestion, DNA precipitation, agarose gel electrophoresis, T4 DNA ligation, and CaCl2 or lithium acetate DNA transformation, were performed in accordance with standard protocols (1, 19, 35). All PCR products and DNA restriction fragments were purified prior to ligation reactions by agarose gel electrophoresis and Qiaquick Gel Extraction (Qiagen, Valencia, Calif.) following the manufacturer's instructions. Plasmid DNA was isolated from yeast using a glass bead lysis procedure and phenol-chloroform extraction (1). Prior to restriction endonuclease analysis, yeast-extracted plasmids were amplified in E. coli DH5α. Plasmid DNA was isolated from bacterial cells either by rapid alkaline lysis (2) or on a large scale with Midi DNA purification columns (Qiagen) as described by the manufacturer.
Protein biochemistry techniques.
Qualitative and quantitative protein assays, such as sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), bicinchoninic acid (BCA) protein assay, immunoblotting with alkaline phosphatase conjugates, enzyme-linked immunosorbent assays (ELISA), and affinity chromatography, were performed in accordance with standard protocols (1) unless otherwise stated. Protease Factor Xa (New England Biolabs) was used in protein digests in accordance with the manufacturer's instructions.
Overexpression and purification of proteins UreC and UreD.
Using the primers described in Table 2, ureC and ureD were PCR amplified from pMID1010 and ligated blunt ended into the EcoRV site of pBS SK+. Isolated as an XhoI fragment, ureC was further subcloned into the corresponding site in vector pMALC-2 (New England Biolabs). The resulting plasmid encoded a malE fusion to the 5′ end of ureC; likewise, ureD was directionally subcloned as an EcoRI-XhoI fragment into pMALC-2 to produce a malE-ureD fusion. Fusion plasmids (pmalC and pmalD) were transformed into E. coli DH5α for overexpression and maintained under ampicillin selection at 100 μg/ml (1). Individual transformants were cultured in 100 ml of Luria broth under antibiotic selection at 37°C with aeration, induced in mid-exponential phase with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and incubated for an additional 2 h. Bacterial cells were collected by centrifugation (5,000 × g, 10 min, 4°C) and washed in 5 ml of cold amylose column buffer (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 10 mM β-mercaptoethanol, 1 mM EDTA). Washed bacterial pellets were resuspended in 5 ml of cold column buffer and ruptured by passage through a French press at 18,000 lb/in2. Unbroken cells and insoluble material were removed by centrifugation (14,000 × g, 30 min, 4°C). MalE-UreC and MalE-UreD fusion proteins were purified from the soluble fraction by passing lysate, diluted 1:5 in the above-described buffer, over a 1-ml amylose resin column (New England Biolabs) prepared in accordance with the manufacturer's instructions. Bound proteins were washed in 20 ml of the amylose column buffer, eluted in 30 ml of column buffer containing 10 mM maltose, and collected as 3-ml fractions (1). MalE fusions were identified in eluted fractions by BCA assay (Pierce, Rockville, Ill.), as well as in SDS–10% polyacrylamide gels stained with Coomassie brilliant blue (1). UreC and UreD were cleaved from MalE by digestion with protease Factor Xa (New England Biolabs) and separated by preparative SDS-PAGE (1). Proteins that were embedded in SDS–10% polyacrylamide gel were pooled and stored at −20°C for later use.
TABLE 2.
PCR primers used to amplify urease genes
| P. mirabilis gene | Forward primer | Reverse primer |
|---|---|---|
| ureA | 5′ AGCAGTAATGGAATTCACAC 3′ | 5′ GATCAGCTCGAGACCTACAC 3′ |
| ureB | 5′ ACTCGAATTCTGTGTAGGTA 3′ | 5′ TAAGCTCGAGGTGTGATAGT 3′ |
| ureC | 5′ ATGGAATTCACCTCACGTCA 3′ | 5′ CTCACTCGAGACGCTGGTTA 3′ |
| ureD | 5′ AAGTTGGAATTCGTGGGTATG 3′ | 5′ CCATCTCGAGTCCTAAAATAAAC 3′ |
| ureE | 5′ AACGGAATTCTTTACTCAG 3′ | 5′ TGCGAATTCTTGGATGATC 3′ |
| ureF | 5′ TGGGAATTCGATCATCAAAG 3′ | 5′ TGGGGAATTCTGGCGATCAC 3′ |
| ureG | 5′ TGAGAATTCGTGATGCAAGA 3′ | 5′ ACCCATACTCGAGCTCATAG 3′ |
Preparation of polyclonal antisera to UreD and MalE-UreC.
Polyclonal rabbit sera were generated using approximately 100 μg of purified UreD embedded in SDS-polyacrylamide. The gel was emulsified in Freund's complete adjuvant and used to immunize two New Zealand White rabbits by subcutaneous injection. Three 30-μg boosters were similarly administered at 3-week intervals with Freund's incomplete adjuvant. In an analogous manner, purified MalE-UreC was also used to immunize two New Zealand White rabbits. Specific UreD and MalE-UreC antibodies were isolated from rabbit antisera by affinity chromatography utilizing MalE-fusion proteins immobilized on an Aminolink column (Pierce) in accordance with the manufacturer's instructions. Bound antibodies were washed progressively with (i) 10 mM Tris-HCl (pH 7.5), (ii) 10 mM Tris-HCl (pH 7.5)–500 mM NaCl, (iii) 100 mM glycine (pH 2.5) collected in 1.0 M Tris-HCl (pH 8.0), (iv) 1.0 M Tris-HCl (pH 8.0), and (v) diethanolamine (pH 11.3) collected in 1.0 M Tris-HCl (pH 8.0). Specific antibodies that recognize the UreD and MalE-UreC proteins were found in the 100 mM glycine (pH 2.5) eluates neutralized with 10 mM Tris-HCl (pH 8.0).
Preparation of monoclonal antibodies to UreC and UreD.
Purified UreC and UreD proteins embedded in SDS-polyacrylamide were supplied to BioWorld Laboratories (Dublin, Ohio) for the production of monoclonal antibodies. Briefly, 12 BALB/c mice were injected with a homogeneous mixture of UreC and UreD, followed by four boosters at 2-week intervals. ELISA analysis of antisera identified two mice as having suitable ratios of MalE to MalE-UreC to MalE-UreD antibody titers, specifically, 1:20:52 and 1:5:2. Isolated murine spleen cells were fused with a select myeloma cell line (Bioworld) to generate hybridomas; in a similar manner, hybridoma supernatants were screened by ELISA against MalE, MalE-UreC, and MalE-UreD. Hybridomas, identified by the secretion of UreC- or UreD-specific antibodies, were used to produce ascites and expanded for frozen storage.
Construction of pDBC.
Using the primers described in Table 2, ureD was PCR amplified from the P. mirabilis gene cluster encoded by pMID1010. An 800-bp DNA fragment encoding ureD was subcloned into the EcoRV site of pBS SK+. The insertion of ureD was confirmed by restriction endonuclease mapping. The resulting vector (pDT7) encoded ureD under the control of a T7 promoter. Subsequently, ureBC was PCR amplified with the 5′ primer described for ureB and the 3′ primer outlined for ureC (Table 2). An ∼2.0-kb PCR product was subcloned into the SmaI site of pDT7. Restriction endonuclease mapping verified that the ureBC genes were transcribed in the same direction as ureD. This construct was transformed into E. coli BL21(DE3)(pLysS) for expression.
Coimmunoprecipitation of recombinant urease proteins.
P. mirabilis HI4320, E. coli DH5α, and E. coli BL21(DE3)(pLysS), transformed with recombinant P. mirabilis urease genes, were cultured in 100 ml of L broth under appropriate antibiotic selection with aeration at 37°C. P. mirabilis HI4320 and E. coli DH5α cultures were induced in early exponential-phase growth with 50 mM urea and allowed to incubate until growth reached late log phase. E. coli BL21(DE3)(pLysS) strains containing pBS SK+ and pDBC were induced in early exponential phase with 0.3 mM IPTG. Bacterial cells were harvested by centrifugation (5,000 × g, 10 min, 4°C), washed with 10 ml of 50 mM HEPES buffered at pH 7.5, and resuspended in 5 ml of 50 mM HEPES (pH 7.5). Washed cells were passed through a French press at 18,000 lb/in2, and lysates were cleared of debris by centrifugation (5,000 × g, 10 min, 4°C). Protein concentrations of the lysates were estimated by BCA assay (Pierce) in accordance with the manufacturer's instructions. Bacterial extracts (∼2 to 4 mg/ml) were gently mixed with 5 μl of anti-UreC or anti-UreD ascites in a final volume of 0.5 ml at 4°C for 2 h (12). Immunocomplexes were precipitated with 150 μl of protein A-Sepharose beads (Sigma) and washed in accordance with the manufacturer's instructions. Bound proteins were resuspended in 100 μl of Laemmli sample buffer and incubated at 100°C for 5 min (12). Denatured immunoprecipitated proteins (20 μl) were separated in an SDS–12% polyacrylamide gel and electroblotted onto a polyvinylidene difluoride membrane (Millipore) (1). Coimmunoprecipitated proteins were immunoblotted with affinity-purified rabbit antisera against MalE-UreC and UreD overnight at 4°C, washed, and treated with anti-rabbit immunoglobulin G conjugated to alkaline phosphatase (Sigma) in accordance with the manufacturer's instructions. Alkaline phosphatase conjugates were visualized with 5-bromo-4-chloro-3-indolylphosphate (BCIP)–Nitro Blue Tetrazolium (NBT).
PCR amplification.
P. mirabilis ure genes were individually PCR amplified from the recombinant ure cluster encoded by pMID1010 (17) using Vent DNA polymerase (Boehringer Mannheim, Indianapolis, Ind.) in an MJ Research Minicycler in accordance with the manufacturer's instructions. The DNA primers used to PCR amplify the ure genes are summarized in Table 2. Agarose-purified PCR products were ligated into pBS SK+, and constructs were confirmed by restriction endonuclease digestion. All of the PCR primers used in this study were designed to encode EcoRI and XhoI recognition sites that allowed directional cloning of the ure genes into pEG202 and pJG4-5.
Subcloning of ure genes into two-hybrid vectors.
Recombinant ure genes were isolated from pBS SK+ by restriction endonuclease digestion with EcoRI and XhoI, gel purified, and ligated into the corresponding restriction sites in pEG202 and pJG4-5. The resulting DNA constructs fused urease genes to the 3′ end of lexA and B42, respectively.
DNA sequencing.
The junctions of p202 and p45 derivatives encoding ureA, ureB, ureE, and ureG fusions in addition to p202C, p202D, and p202F were sequenced to confirm that each recombinant plasmid coded an in-frame fusion by the dideoxy-chain termination method (36) at the University of Maryland at Baltimore Biopolymer Core Facility (Applied Biosystems 373A automated DNA sequencer with the Big Dye Terminator Cycle Sequencing Kit). Fusion junctions of p45C, p45D, and p45F were sequenced using a [35S]dATP Sequenase kit (Amersham; Arlington Heights, Ill.) in accordance with the manufacturer's instructions. The sequencing primers used in this study were (i) pEG202-derived constructs (p202; 5′-TGTTGCCAGAAAATAGCGAG3′) and (ii) pJG4-5-derived constructs (p45; 5′ TGACTGGCTGAAATCGAATG3′).
Immunoblots to confirm expression of LexA- and B42-urease fusions.
S. cerevisiae EGY48 (p202- and p45-ure derivatives) were grown in 3 ml of synthetic defined dropout medium (Ura− His− Trp−) containing 2% (wt/vol) galactose overnight at 30°C with aeration. (Note that p45-ure fusions are regulated by the gal1 promoter.) Cultures were diluted 1:15 into 3 ml of fresh medium and grown for an additional 7 h. Each culture was harvested by adding 30 μl of polyethylene glycol 3350 (50% [wt/vol] stock solution) and centrifuged (5,000 × g, 10 min, 4°C). The resulting pellets were concentrated 20:1 in Tris-EDTA containing Laemmli sample buffer and then denatured at 100°C for 5 min (1). Lysates were separated by electrophoresis (Mighty Small II; Hoefer, San Francisco, Calif.) through an SDS–12% polyacrylamide gel in accordance with standard protocols using sample sizes that were ∼15% of the total lysate volume. Separated proteins were electroblotted to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, Mass.); afterwards, the membrane was blocked with 1% (wt/vol) bovine serum albumin–TTBS (100 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% [wt/vol] Tween 20) and washed with TTBS in accordance with standard protocols (1). Membranes blotted with EGY48(p202-ure) lysates were gently agitated overnight at 4°C in TTBS containing LexA antiserum diluted 1:5,000, whereas antihemagglutinin (anti-HA) monoclonal antibody (Boehringer Mannheim) was diluted 1:260 to treat blots of EGY48(p45-ure) lysates overnight at 4°C (1). Anti-rabbit immunoglobulin G and anti-mouse polyvalent immunoglobulin conjugated to alkaline phosphatase were used to identify primary antibodies bound to LexA and B42 derivatives, respectively, in accordance with the manufacturer's instructions. Alkaline phosphatase conjugates were visualized with BCIP-NBT.
Repression assay to confirm DNA-binding properties of LexA-urease fusions.
Yeast strains bearing p202-ure derivatives and pJK101 were grown at 30°C with aeration to mid-exponential phase in 5 ml of synthetic, defined-dropout medium (Ura− His−) containing 1% (wt/vol) raffinose–2% (wt/vol) galactose, which induces the lacZ reporter encoded by pJK101 (1). Cultures were collected by the addition of 50 μl of polyethylene glycol 3350 (50% [wt/vol] stock solution) and centrifugation at 5,000 × g for 10 min. Cell pellets were resuspended in an equal volume of Z buffer at pH 7.0 (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol]. β-Galactosidase activities produced by each strain were measured in triplicate with a standard chromogenic assay using o-nitrophenyl-β-d-galactopyranoside substrate (1). These activities are reported as percentages of the β-galactosidase activity produced by strain EGY48(pJK101), which was arbitrarily set at 100%.
Background transcriptional activity of LexA- and B42-urease fusions.
LexA- and B42-urease protein fusions were assessed for endogenous transcriptional activity in S. cerevisiae strain EGY48(pSH18-34), which bears an integrated leu2 reporter and a plasmid-borne lacZ reporter. EGY48(p202-ure) derivatives were grown at 30°C for 5 or 6 days on the following synthetic, defined-dropout agar media: (i) Ura− His− Leu− medium with 2% (wt/vol) glucose (ii) Ura− His− Leu− medium with 1% (wt/vol) raffinose and 2% (wt/vol) galactose, (iii) Ura− His− medium with 2% (wt/vol) glucose and 2.5 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml; and (iv) Ura− His− medium with 1% (wt/vol) raffinose, 2% (wt/vol) galactose, and 2.5 mg of X-Gal per ml. Strains were scored daily for growth on leucine-deficient medium and the formation of blue patches on X-Gal-containing medium (1). Similarly, EGY48(p45-ure) derivatives were grown on the following supplemented dropout media: (i) Ura− Trp− Leu− medium containing 2% (wt/vol) glucose, (ii) Ura− Trp− Leu− medium with 1% (wt/vol) raffinose and 2% (wt/vol) galactose, (iii) Ura− Trp− medium containing 2% (wt/vol) glucose and 2.5 mg of X-Gal per ml, and (iv) Ura− Trp− medium containing 1% (wt/vol) raffinose, 2% (wt/vol) galactose, and 2.5 mg of X-Gal per ml. These strains were also scored for the same characteristics indicative of reporter activation. If within 1 or 2 days a fusion exhibited growth on leucine-deficient medium or blue patches on X-Gal-containing agar, the endogenous transcriptional activity was considered too strong for use in an interactive assay. As an alternative, constructs encoding transcriptionally active fusions were transformed into yeast strain EGY191(pJK103). EGY191 contains an integrated leu2 reporter downstream of a single LexA operator and is significantly less sensitive than strain EGY48, which contains four operators. Likewise, pJK103 has one LexA operator upstream of a lacZ reporter gene versus pSH18-34, which has eight operators (8, 20). Transformants were reevaluated as described above for transcriptional activating properties.
Screening for urease-protein interactions via two-hybrid system.
Fusions encoded by p202A, p202B, p202E, p202F, and p202G (i.e., LexA) were transformed into yeast strain EGY48(pSH18-34) and maintained on synthetic, defined-dropout agar medium (Ura− His− medium with 2% [wt/vol] glucose). In parallel, p202D was transformed into less-sensitive reporter strain EGY191(pJK103). Each p202-ure transformant was independently retransformed with constructs encoding B42-ure fusions (p45A, p45B, p45C, p45D, p45E, p45F, and p45G) and selected on dropout medium (Ura− His− Trp− medium with 2% [wt/vol] glucose). Transformants carrying dual-fusion plasmids were grown at 30°C for 5 or 6 days on the following synthetic, defined-dropout agar media: (i) Ura− His− Trp− Leu− medium with 2% [wt/vol] glucose, (ii) Ura− His− Trp− Leu− medium with 1% (wt/vol) raffinose and 2% (wt/vol) galactose, (iii) Ura− His− Trp− medium prepared with 2% glucose and 2.5 mg of X-Gal per ml, and (iv) Ura− His− Trp− medium prepared with 1% (wt/vol) raffinose, 2% (wt/vol) galactose, and 2.5 mg of X-Gal per ml. Interactive strains were scored each day for growth on leucine-deficient medium and the formation of blue patches on X-Gal-containing medium (1). Yeast strains maintaining vector controls were plated alongside each interactive strain for comparison.
RESULTS
Polyclonal and monoclonal antibodies against urease proteins.
Polyclonal and monoclonal antibodies were generated against two urease proteins. A translational fusion of UreC, the largest structural subunit, and the maltose-binding protein (MalE) was used for immunization of New Zealand White rabbits. Using a slightly different approach, an affinity-purified MalE-UreD fusion was digested with protease Factor Xa to release the UreD polypeptide. The products of the protease digests were separated in an SDS-polyacrylamide gel (data not shown), and isolated UreD polypeptide was used to immunize New Zealand White rabbits. Similar to the strategy employed for UreD polyclonal antisera production, purified MalE-UreC and MalE-UreD fusions were also cleaved by protease Factor Xa to generate UreC and UreD polypeptides. Purified proteins were also used for the commercial production and isolation of monoclonal antibodies.
Coimmunoprecipitation of accessory protein UreD with urease structural proteins.
Several metalloenzymes, dependent on accessory proteins for activation, require direct contact between the apoenzyme and an accessory protein in vitro prior to metal incorporation (5, 13, 32). To address whether analogous interactions occur with P. mirabilis urease, UreC was immunoprecipitated from lysates of E. coli DH5α expressing the P. mirabilis urease gene cluster encoded by pMID1010 and screened for coprecipitating accessory proteins. E. coli DH5α(pMID1010), regardless of whether it is cultured in modified M9 medium or Luria broth, produces similar amounts of active urease as measured by a phenol-hypochlorite urease assay (data not shown). However, immunoblotting of lysates with affinity-purified MalE-UreC antiserum revealed a greater quantity of UreC soluble protein from cultures grown in L broth (data not shown). If P. mirabilis urease is in an active state after associating with its accessory proteins, possibly a greater proportion of urease is in a prebound or bound state in lysates of L broth cultures as the result of nickel chelation by medium components.
Thus, lysates of E. coli DH5α(pMID1010) were prepared from L broth cultures, immunoprecipitated with monoclonal UreC antibodies, and analyzed by immunoblotting with affinity-purified antisera to MalE-UreC and UreD. Immunoblots confirmed that UreC is expressed and precipitated from lysates of E. coli DH5α(pMID1010) grown in the presence of 50 mM urea (Fig. 1B). MalE-UreC antiserum reacts with an expected 60-kDa protein band in the crude fraction (Fig. 1B, lane 1) and pellet (Fig. 1B, lane 3). A comparable protein band is nearly undetectable in lysates of uninduced cultures (Fig. 1A, lanes 1 and 3). Likewise, a 60-kDa protein is detected in neither the supernatant nor the pellet of control immunoprecipitations in which no lysate was added (data not shown). Consistent with our predictions, the accessory protein UreD coprecipitated with UreC, as shown in Fig. 1C. An appropriately sized 30-kDa protein band reacted strongly with UreD antiserum in the crude lysate and pellet of UreC-immunoprecipitated lysates from induced cultures (Fig. 1C, lanes 1 and 3). Similar protein bands were not detected in uninduced lysates and control immunoprecipitations comprised of monoclonal antibodies alone (data not shown).
In a complementary experiment, identical lysates were precipitated with monoclonal anti-UreD antibody. A faint, but distinct, 30-kDa protein band noted in the immunoblot (Fig. 1C, lane 5) verifies that UreD was present in the precipitate. Moreover, UreC was also detected in UreD-mediated immunoprecipitations of induced cultures (Fig. 1B, lane 5) and absent in uninduced cultures (Fig. 1A, lane 5). In these experiments, two additional protein bands were noted to cross-react with the polyclonal antisera; the light chain of monoclonal UreD antibodies is observed as a 25-kDa protein in Fig. 1C, and an unknown protein with mobility similar to that of UreC was seen in crude cultures and supernatants. Since this protein was not seen in the pellet fraction, it was regarded as a nonspecific reaction with an E. coli protein. Altogether, these results demonstrate that UreC, a structural component of urease, and the accessory protein UreD are both present in a multiprotein complex.
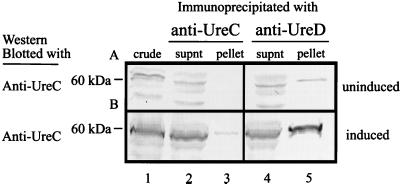
To assess whether a similar complex containing UreC and UreD polypeptides existed in P. mirabilis, lysates were prepared from wild-type strain HI4320 grown under inducing conditions (50 mM urea) in L broth and precipitated separately with monoclonal anti-UreC and -UreD antibodies. An immunoblot shows that the 60-kDa UreC protein is present in the pellets of both precipitation reactions (Fig. 2B, lanes 3 and 5), whereas UreC is barely detectable in precipitations of lysates from uninduced cultures (Fig. 2A, lanes 3 and 5). These observations corroborate experiments involving the recombinant urease; this implies that accessory protein UreD and urease structural protein UreC are present in a moderately stable protein complex in wild-type P. mirabilis strain HI4320. It is unclear what fraction of each protein is bound versus freely soluble protein in these lysates and whether this distribution contributes to a smaller quantity of UreC in the precipitate.
FIG. 2.
Immunoblots depicting UreC and UreD coprecipitating from lysates of wild-type P. mirabilis HI4320. Soluble protein from strain HI4320, uninduced (A) and induced with 50 mM urea (B), was immunoprecipitated with anti-UreC (left column) and -UreD (right column) monoclonal antibodies, separated by SDS–12% PAGE, and immunoblotted with polyclonal anti-UreC serum. Lanes: 1, crude culture extracts of HI4320; 2, supernatants (supnt) from UreC immunoprecipitation reactions; 3, solubilized pellets from UreC immunoprecipitation reactions; 4, supernatants from UreD immunoprecipitation reactions; 5, solubilized pellets from UreD immunoprecipitation reactions.
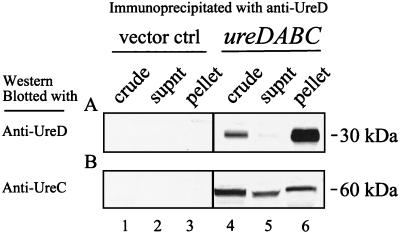
To establish whether urease coaccessory proteins such as UreE, UreF, and UreG are required for the formation of these structures, UreD was immunoprecipitated from lysates of E. coli DH5α carrying cloned P. mirabilis urease genes ureDABC (p1701.1) (38). Immunoblots establish that both UreD (Fig. 3A, lane 6) and UreC (Fig. 3B, lane 6) are among the pelleted proteins, whereas neither polypeptide is detected in lysates containing the pBS SK+ vector control (Fig. 3A and B, lanes 1 to 3). Thus, in the absence of other accessory proteins, UreD still associates with UreC.
FIG. 3.
Immunoblots demonstrating that recombinant UreC and UreD coprecipitate in the absence of coaccessory proteins. Cultures of E. coli DH5α(pBS SK+) (vector control) and DH5α(p1701.1) (encoding ureDABC) were induced with 50 mM urea. Soluble protein was immunoprecipitated with anti-UreD monoclonal antibodies, separated by SDS–12% PAGE, and immunoblotted with anti-UreD polyclonal serum (A) and anti-UreC polyclonal serum (B). Lanes: 1, crude culture extracts of E. coli DH5α(pBS SK+); 2, supernatants (supnt) collected from immunoprecipitation reactions of E. coli DH5α(pBS SK+); 3, solubilized pellets from immunoprecipitation reactions of E. coli DH5α(pBS SK+); 4, crude culture extracts of E. coli DH5α(p1701.1); 5, supernatants collected from E. coli DH5α(p1701.1) immunoprecipitation reactions; 6, solubilized pellets from E. coli DH5α(p1701.1) immunoprecipitation reactions.
As previously mentioned, K. aerogenes apourease has been stably purified from strains deficient in accessory proteins (21). Therefore, it is likely in a ureDABC-encoding strain (p1701.1) that UreC is associated with both the UreA and UreB costructural proteins. In light of this information, UreD may be involved in a structure that includes three urease structural proteins versus freely soluble UreC. Under these circumstances, it is possible that UreD is associated with UreC in an indirect manner or interacts directly with UreC only in the context of the apourease. To test these hypotheses, UreD was immunoprecipitated from lysates expressing only ureDBC and not ureA. If the association between UreD and UreC depends on UreA, then UreC should not be found in significant quantities among anti-UreD precipitates. Clearly, both UreD (Fig. 4C, lane 6) and UreC (Fig. 4D, lane 6) are immunoprecipitated by UreD antibodies. Experiments with UreC antibodies confirmed these results by coprecipitating UreC and UreD (Fig. 4A and B, lane 6). Unfortunately, neither UreC nor UreB has been expressed individually in a stable form, preventing similar experiments with ureB deletions (S. Heimer and H. Mobley, unpublished observation). These data imply that UreA is probably not necessary for UreD associations with UreC. It is uncertain whether UreB is directly or indirectly involved with the association.
FIG. 4.
Immunoblots revealing that UreC and UreD coprecipitate in the absence of UreA. Soluble protein from IPTG-induced cultures of E. coli BL21(DE3)(pLysS)(pBS SK+) and (pDBC) was immunoprecipitated with anti-UreC (left column) and anti-UreD (right column) monoclonal antibodies, separated by SDS–12% PAGE, and immunoblotted with anti-UreC polyclonal serum (A and D) and anti-UreD polyclonal serum (B and C). Lanes: 1, crude culture extracts of E. coli (pBS SK+); 2, supernatants (supnt) from immunoprecipitation reactions of E. coli (pBS SK+); 3, solubilized pellets from immunoprecipitation reactions of E. coli (pBS SK+); 4, crude culture extracts of E. coli (pDBC); 5, supernatants from E. coli (pDBC) immunoprecipitation reactions; 6, solubilized pellet from E. coli (pDBC) immunoprecipitation reactions. ctrl, control.
Expression of urease genes as lexA and b42 fusions.
To examine interactions of urease structural and accessory proteins in vivo, a yeast two-hybrid assay was employed that makes use of the DNA-binding domain of LexA and a transcriptional activation epitope called B42 (11). To prepare for two-hybrid screens, P. mirabilis urease genes ureDABCEFG were individually PCR amplified and cloned into pBS SK+.
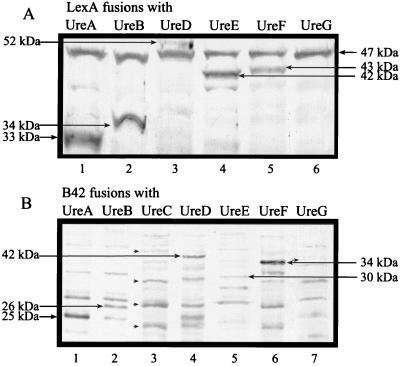
The derivatives of pEG202 and pJG4-5 (p202-ure and p45-ure, respectively) were transformed into S. cerevisiae strain EGY48, and the transformants were assayed for fusion protein expression by immunoblotting. On an anti-LexA immunoblot of whole lysates of various yeast transformants, unique protein bands of 33 and 34 kDa were observed (Fig. 5A, lanes 1 and 2) which are consistent with the expected sizes of LexA fusions with urease structural proteins UreA and UreB, respectively. Likewise, expression of urease accessory proteins UreD, UreE, and UreF as fusions with LexA were detected by anti-LexA immunoblotting (Fig. 5A, lanes 3, 4, and 5) as 52-, 42-, and 43-kDa protein bands, respectively. Accessory protein UreG fused to LexA is predicted to migrate as a 47-kDa protein. Unfortunately, fusion proteins in this size range cannot be identified conclusively due to a cross-reacting band recognized by the LexA antiserum. Several protein bands resembling truncates of a LexA-UreC fusion were detected in the lysate of EGY48(p202C) transformants; however, a full-length fusion product was not observed by Western blot analysis (data not shown).
FIG. 5.
Immunoblots confirming expression of LexA and B42 epitope fusions with urease proteins in S. cerevisiae. Protein extracts from S. cerevisiae EGY48 carrying p202-ure and p45-ure derivatives were separated by SDS-PAGE. (A) EGY48(p202-ure) lysates were reacted with anti-LexA polyclonal antibodies. Lanes contain lysates of EGY48 transformed with plasmids p202A (lane 1), p202B (lane 2), p202D (lane 3), p202E (lane 4), p202F (lane 5), and p202G (lane 6). (B) EGY48(p45-ure) lysates were reacted with anti-HA monoclonal antibodies. Lanes contain lysates of EGY48 transformed with plasmids p45A (lane 1), p45B (lane 2), p45C (lane 3), p45D (lane 4), p45E (lane 5), p45F (lane 6), and p45G (lane 7). Alkaline phosphatase-labeled immune complexes were visualized with the chromogenic substrate BCIP-NBT. Estimated molecular sizes are indicated. Truncated arrows denote possible truncated products of the B42-UreC fusion (panel B, lane 3) and the expected size of full-length B42-UreG (panel B, lane 7).
To identify B42 epitope fusions, whole lysates of EGY48(p45-ure) derivatives were immunoblotted with a commercially available anti-HA antibody which recognizes the HA epitope also encoded by pJG4-5 (1, 11). B42 epitope fusions and UreB were noted at 25 and 26 kDa (Fig. 5B, lanes 1 and 2, respectively), consistent with their predicted molecular weights. Accessory gene ureD, ureE, and ureF fusions with b42 were found to produce appropriately sized fusion proteins migrating at 42, 30, and 34 kDa (Fig. 5B, lanes 4, 5, and 6). Lysates of EGY48(p45C) produced several truncated B42-UreC polypeptides, but a full-length fusion product was not detectable (Fig. 5B, lane 3). Anti-HA immunoblotting also failed to detect a unique protein band of the size expected for a B42-UreG fusion (Fig. 5B, lane 7).
Thus, Western blotting has confirmed that structural proteins UreA and UreB are stably expressed as LexA and B42 epitope fusions in yeast, as well as accessory proteins UreD, UreE, and UreF. Structural protein UreC is probably expressed as both LexA and B42 epitope fusions; however, the stability of these fusions is questionable, as indicated by the various immunoreactive truncated products. Contrary to predictions made from DNA sequence analysis, accessory protein UreG was not detected as either a LexA or a B42 fusion.
Repression assay confirms DNA-binding activity of LexA fusions.
To verify that the LexA DNA-binding domain retained its sequence-specific recognition and DNA-binding activity as a fusion in S. cerevisiae, EGY48(p202-ure) derivatives were transformed with pJK101. This construct is designed such that lexA operators, derived from colE1, interrupt the gal1 promoter upstream of lacZ (1, 4). If a LexA fusion binds specifically to these sites, galactose-inducible lacZ transcription will be repressed. In this study, β-galactosidase activity was measured in triplicate for each transformant and reported as a percentage of the activity detected in the positive control, EGY48(pJK101). All strains cotransformed with p202-ure derivatives of the accessory genes and pJK101 produced less than 20% of the β-galactosidase activity measured in EGY48(pJK101). Likewise, structural protein LexA-UreA and LexA-UreB fusions also repressed lacZ expression to less than 5.0% of uninhibited reporter levels. Surprisingly, p202C and pJK101 cotransformants had high β-galactosidase activity (∼360%), suggesting that LexA-UreC fusions activate transcription of the lacZ reporter upon binding to recognition sites. These repression assays demonstrate that urease structural and accessory proteins do not interfere with the DNA-binding functions of LexA fusions. However, the LexA-UreC fusion has enhanced transcriptional activating activity and thus cannot be used for interactive studies with B42 fusions.
Background transcriptional activity of LexA and B42 fusions.
The two-hybrid system used in this study relies on two reporter genes, leu2 and lacZ, to assess whether LexA and B42 epitope fusions interact in vivo (1, 11). These reporter genes are expressed when a transcriptionally active complex of fusion proteins occupies the LexA-binding sites upstream of their respective promoters. In S. cerevisiae strain EGY48, the upstream activating sequence of leu2 has been with replaced with four LexA operators from colE1 (1, 8, 11). This strain has been transformed with the reporter plasmid pSH18-34, which encodes gal1-lacZ downstream of eight LexA operators (1). It is essential that individual urease protein fusions with either LexA or B42 do not modulate expression of these reporter genes in a direct or indirect manner. EGY48(pSH18-34) was independently transformed with p202-ure (i.e., LexA) and p45-ure (i.e., B42) derivatives and streaked onto synthetic, defined-dropout agar medium that was either deficient in leucine or contained 2.5 mg of X-Gal per ml (1). Experiments were performed in the presence of 2% glucose or 2% galactose to regulate the expression of B42 fusions. During 5 days of 30°C incubation, strains were visually scored for growth and color. Background reporter activities were considered significant if growth occurred on leucine-deficient medium and blue patches formed on X-Gal-containing medium within 2 days of incubation. LexA fusions with structural protein UreC and accessory protein UreD were associated with significantly high levels of reporter activities in the presence of both glucose and galactose (data not shown). Within 2 days of incubation, the B42-UreC fusion also exhibited high reporter background levels in yeast strain EGY48(pSH18-34) (data not shown). To compensate for the high background levels, alternate reporter systems were tested which were expected to be less sensitive. These reporters were S. cerevisiae strain EGY191, which contains only one LexA operator upstream of the leu2 gene, and pJK103 encoding gal1-lacZ preceded by one LexA operator (1, 20). Unfortunately, alternate p202C transformants continued to generate high levels of reporter activity and were omitted from further studies (data not shown).
Most fusions produced a low yet detectable level of background activity that appeared in 3 to 4 days of incubation. These reporter activities were considered to be modest, and we did not anticipate interference with the detection of specific interactions. This characteristic was noted in strain EGY48(pSH18-34) cotransformed with ureA, ureE, and ureF fused with lexA and B42 (data not shown). Comparable results were observed with p202D or p45C when expressed in EGY191(pJK103) in the presence of galactose (data not shown). Screens for urease-protein interactions were conducted with strains expressing this phenotype.
Two-hybrid analysis of structural protein interactions.
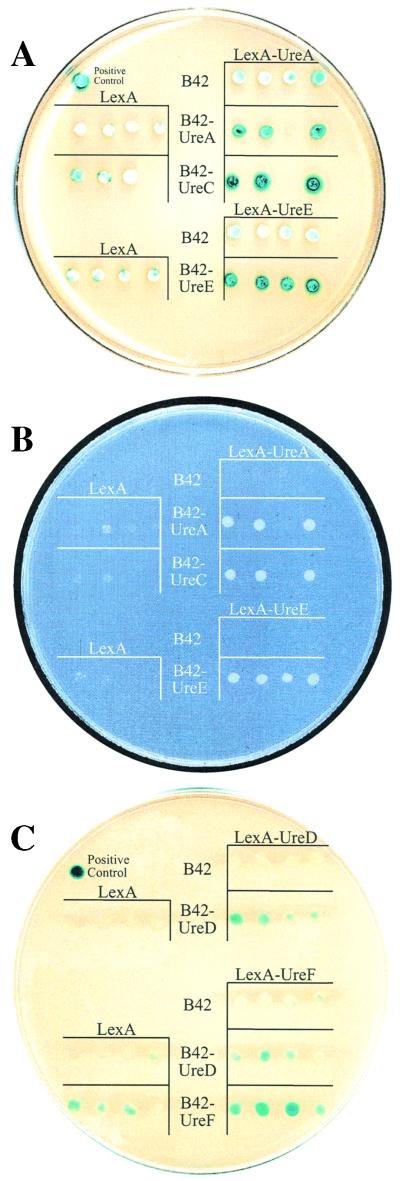
Fusions of lexA with ureA and ureB were individually cotransformed into EGY48(pSH18-34) with B42 fusions to ureA, ureB, and ureC. These cotransformants were patched onto a synthetic, defined-dropout agar medium that was deficient in leucine or contained 2.5 mg of X-Gal per ml. Each growth experiment was performed in the presence of 2% glucose or 2% galactose. Strains were visually scored for growth and color over a 5-day period of incubation at 30°C. Striking reporter activities were observed in EGY48(pSH18-34) strains coexpressing LexA-UreA and B42 fusions of UreA and UreC within 2 to 3 days of inoculation in the presence of galactose. Strains producing LexA-UreA and B42-UreA or B42-UreC (Fig. 6A, right column, rows 2 and 3, respectively) synthesize significantly more β-galactosidase than do control strains producing LexA-UreA and only B42 (right column, row 1), B42-UreA and LexA (left column, row 2), or B42-UreC and LexA (left column, row 3). Comparable observations were made with the leu2 reporter shown in Fig. 6B. These results were not observed in the absence of galactose (data not shown). Altogether, these results suggest that UreA interacts in vivo with other UreA and UreC polypeptides, which is consistent with X-ray diffraction studies of K. aerogenes urease crystals (15). Reporter activation was not detected among strains carrying ureB fusions (data not shown).
FIG. 6.
Urease-protein interactions identified in vivo with a yeast two-hybrid assay. EGY48(pSH18-34) bearing p202A, p202E, or p202F was transformed with p45A, p45C, p45D, p45E, or p45F. Likewise, p202D EGY191(pJK103) bearing was transformed with p45-ura derivatives. Four independent transformants were grown at 30°C for 3 to 4 days on synthetic, defined-dropout agar medium (Ura− His− Trp− medium prepared with 1% [wt/vol] raffinose, 2% [wt/vol] galactose, and 2.5 mg of X-Gal per ml [A and C] or Ura− His− Trp− Leu− medium prepared with 1% [wt/vol] raffinose and 2% [wt/vol] galactose [B]). Interactive strains were scored for the formation of blue colonies (A and C) and growth on leucine-deficient medium (1) (B). Yeast strains maintaining vector controls were plated alongside each interactive strain for comparison. Representative plates are shown.
Two-hybrid analysis of accessory protein interactions.
To identify novel interactions between urease accessory proteins, p202E, p202F, and p202G were independently cotransformed into EGY48(pSH18-34) with B42 fusions of ureD, ureE, ureF, and ureG. Cotransformants were patched onto a synthetic, defined-dropout agar medium that was deficient in leucine or contained 2.5 mg of X-Gal per ml in the presence of 2% glucose or 2% galactose. During incubation at 30°C, strains were visually scored for growth and color. P. mirabilis UreE dimers, which were initially characterized by size exclusion chromatography (38), were identified in vivo in this two-hybrid screen. EGY48(pSH18-34) cotransformed with p202E and p45E grew vigorously on medium deficient in leucine (Fig. 6B, right column, row 5) and generated high levels of β-galactosidase activity (Fig. 6A, right column, row 5) in the presence of galactose compared to strains containing vector control p202E (right column, row 4) or p45E (left column, row 3). LexA-UreF fusions also produced significant amounts of β-galactosidase activity when coexpressed with B42-UreD and B42-UreF (Fig. 6C, right column, rows 4 and 5, respectively) compared with LexA (left column, row 3) and B42 (right column, row 3) controls in the presence of galactose. Protein interactions involving the LexA-UreD fusion were examined with less sensitive reporters (EGY191 and pJK103) due to high background activity. When UreD was coexpressed as a LexA and B42 fusion in the presence of galactose, levels of β-galactosidase activity suggestive of in vivo protein interactions were observed (Fig. 6C, right column, row 2). Controls for B42-UreD and LexA-UreD are also shown (left column, row 1, and right column, row 1, respectively). The reporter activities described here were not detectable in the absence of galactose; hence, these activities are not the result of non-specific interactions but require both LexA and B42 fusions.
Using a similar strategy, EGY48(pSH18-34) cotransformants of p202A and p202B combined with p45D, p45E, p45F, or p45G were screened for interactions. Five days following inoculation, significant levels of reporter activity were lacking in these strains (data not shown). Thus, direct interactions between these urease structural and accessory proteins were not detectable in vivo.
In summary, UreD and UreF proteins were found in vivo to form homomultimeric structures using a yeast two-hybrid assay. The accessory protein UreD was observed to associate in vivo with coaccessory protein UreF. Finally, the formation of UreE protein homomultimers was confirmed in vivo.
DISCUSSION
A variety of mechanisms for the synthesis and assembly of metalloenzymes have been described in the literature. In several instances, accessory (i.e., nonstructural) proteins are required for the production of catalytically active metalloenzymes (24). These accessory proteins are postulated to serve various functions, including cofactor synthesis, metal ion chelators or donors, and deoxynucleoside triphosphatases, as well as chaperones and proteases of structural proteins. It is likely that some accessory proteins require physical contact with their respective apoenzyme or other coaccessory proteins to fulfill these putative roles and facilitate proper metal ion incorporation. One group of multipolypeptide complexes has been observed in studies of K. aerogenes urease involving three accessory proteins (UreD, UreF, and UreG) and the apourease (27, 31, 32). The interactions stabilizing these protein complexes weaken in the presence of nickel, and the accessory proteins dissociate from the apourease as it becomes enzymatically active (31, 32). Similar metal ion-dependent interactions have been reported between K. pneumoniae apodinitrogenase and NifY protein (13), as well as Streptomyces apotyrosinase and MelC1 (5).
To identify whether analogous and heretofore unrecognized interactions occur between P. mirabilis apourease and accessory proteins, monoclonal antibodies to UreC and UreD were used to precipitate protein complexes from P. mirabilis and E. coli DH5α expressing cloned urease genes. UreC and UreD proteins coprecipitated from strains encoding the entire P. mirabilis ure gene cluster (Fig. 1 and 2). These observations are consistent with the hypothesis that UreD of P. mirabilis interacts with the apourease similar to the K. aerogenes homologue (31). Comparable results from immunoprecipitation experiments with E. coli DH5α carrying only ureDABC imply that the association between UreC and UreD is not mediated by and does not require other coaccessory proteins (Fig. 3 and 7A). Corroborating studies with homologues in K. aerogenes have also been reported (32).
It has been shown that the apourease can be purified from urease accessory gene mutants, and this protein can be partially reactivated with the subsequent addition of nickel under certain conditions (21, 31). Thus, it is generally accepted that the accessory proteins are not involved in apourease assembly. We speculate that UreD associations with UreC in vivo probably do not precede apourease formation. However, it remains unclear whether UreD interaction occurs exclusively within the context of the apourease such that other structural proteins are required to stabilize it. Immunoprecipitation experiments using lysates of E. coli DH5α expressing only P. mirabilis ureDBC indicate that UreA is not necessary for UreC and UreD to coprecipitate (Fig. 4 and 7B). To determine whether UreC alone is sufficient for UreD interaction in vivo, we expressed UreC as an amino-terminal fusion with LexA and a B42 epitope in a yeast two-hybrid system. Unfortunately, UreC fusions affected growth rates and promoted high levels of transcription in yeast, which interfered with the detection of most reporter gene activities (an exception was seen in UreA-UreC interactions [Fig. 6]). Since ureC was unsuitable for most uses in a two-hybrid assay and was difficult to express by itself recombinantly in E. coli DH5α, we are still uncertain whether this structural protein directly interacts with UreD. An equally important factor to address is the role played by the structural protein UreB in UreD interactions (40).
We postulate that P. mirabilis UreD may function in the recruitment or stabilization of other coaccessory protein associations with the apourease, in addition to acting as a chaperone similar to its homologue in K. aerogenes (31). In agreement with that hypothesis, we have identified a new interaction between UreD and another accessory protein, UreF, in a yeast two-hybrid screen for in vivo protein associations (Fig. 7B and C). UreF homologues have been also described as putative chaperones that prevent the binding of nickel ions to the noncarbamylated apourease (27). In a two-hybrid assay involving LexA-UreF coexpressed with B42-UreD, reporter activities were detected in the presence of galactose, which are indicative of specific in vivo interactions (Fig. 6C). A direct interaction of this kind is also consistent with two prior observations involving urease accessory protein homologues. In one case, the apourease could not be purified in a form associated with UreF from a ureD mutant strain (32). The second observation was the loss of immunoblot detection of UreD bound to apourease in native gels in the presence of UreF (27). Unfortunately, we were unable to confirm UreF and UreD interactions with the two-hybrid system when the prior accessory protein was fused to the B42 epitope and the latter was expressed as a LexA fusion (data not shown). The transcriptional activity of the LexA-UreD fusion may have obscured the detection of interactions that generate weaker reporter activity (data not shown).
The homotrimeric nature of P. mirabilis urease poses an interesting activation barrier; namely, each of the three active sites must acquire and properly coordinate two nickel ions to achieve maximum catalytic activity. It is known that UreD homologues associate with apourease as multimers, varying from one to three molecules; it is assumed that each molecule is associated with a separate trimer (32). Furthermore, one could speculate that P. mirabilis apourease could be bound simultaneously by three distinct accessory protein complexes, comprised of UreD, UreF, and UreG, at each of the active sites. Hypothetically, accessory proteins bound to one trimer of the apourease could interact with another complex of accessory proteins anchored to an adjacent trimer. Similar to models of cooperactivity, perhaps, conformational changes associated with nickel incorporation could be communicated from one accessory protein complex to another, affecting the overall energy required to generate fully active holourease (29). Simpler still, initial accessory protein interactions with the apourease may stabilize additional accessory complexes assembling at other active sites through direct protein contact. In this study, two novel interactions were identified in two-hybrid screens, i.e., UreD and UreF self-interactions, which could be explained sterically by such an arrangement. This observation emphasizes the likelihood, for example, that UreD can form important associations in vivo with other molecules of UreD. It is not known whether UreD multimers are found within a distinct accessory protein complex associated with the apourease. However, it is feasible that UreD multimers form between individual proteins bound to different trimers of P. mirabilis apourease (Fig. 7B). Similar arguments could be made for the UreF self-interactions detected in vivo (Fig. 7B). If contacts did occur between accessory proteins bound at different trimers, it could serve as a mechanism by which to coordinate nickel uptake in urease. To our knowledge, there has been no discussion in the literature of the possibility of cooperative nickel or substrate binding among the three active sites.
It is worth noting that other biologically relevant multimers formed by P. mirabilis urease proteins were detected in vivo with a two-hybrid assay (Fig. 7B). Specifically, the accessory protein UreE, coexpressed in yeast as both LexA and B42 epitope fusions, yielded reporter activity suggesting homomultimer formation (Fig. 6). UreE homologues are probably the best studied of the urease accessory proteins. This protein is involved in nickel ion chelation within the cell and regulation of nickel transfer to the apourease (6, 7). UreE has been purified from P. mirabilis in a single step on a nickel affinity column and migrated as a dimer on a gel filtration column (38). In a similar fashion, UreA was found with the two-hybrid system to self-interact in vivo (Fig. 6 A and B). X-ray crystallography of K. aerogenes urease indicates that three UreA homologues are arranged in triad symmetry on the same face of the apourease (Fig. 7B) (15). This arrangement is stabilized by interactions between adjacent UreA subunits in addition to direct associations of UreA with the structural protein UreC. Also seen in Fig. 6A and B is evidence supporting the interaction between structural proteins UreA and UreC in vivo. None of these interactive strains produced significant reporter activity in the absence of galactose, which argues against nonspecific reporter activation. Confirming these biologically significant interactions among P. mirabilis urease proteins with a yeast two-hybrid system, including the formation of homomultimers, we believe, validates this method of detection for the purpose of our study.
We were unable to detect any interactions with UreB and UreG using a two-hybrid system that has been suggested of homologues elsewhere (15, 21, 28). Immunoblotting (Fig. 5A and B) and in vivo DNA-binding assays suggest that UreB fusions are synthesized in yeast; however, the conformation of these fusions could be significantly different than that of the wild type, preventing protein interactions. For similar reasons, other possible interactions may have escaped detection as well. Strangely, UreG fusions were not detected in either anti-HA or anti-LexA immunoblots although the DNA constructs were found to encode in-frame fusions. While protein instability seems a reasonable explanation, p202G conferred LexA DNA-binding activity on transformed yeast.
In summary, we have demonstrated that, in P. mirabilis, accessory protein UreD is present in a protein complex containing the structural protein UreC. This association occurs independent of other coaccessory proteins and the structural protein UreA. It remains unclear whether UreD interacts directly with UreC or if UreB plays a role in this association. We have provided evidence for direct interaction between UreD and UreF, a coaccessory protein. This finding is consistent with our hypothesis that UreD also recruits and/or stabilizes other P. mirabilis accessory proteins in structures involving the apourease. In this study, unique homomultimers were observed among UreD and UreF proteins. We propose that these interactions may be important in vivo to stabilize multiaccessory protein complexes with the apourease and could play a role in coordinating nickel incorporation among different active sites. Lastly, we have confirmed homomultimeric protein interactions, previously described in Klebsiella, such as that of UreA and UreE in addition to the UreA-UreC association in vivo, which validates the use of two-hybrid technology in this study.
ACKNOWLEDGMENTS
We thank Roger Brent (Boston, Mass.), who generously supplied the S. cerevisiae strains and plasmids necessary for the two-hybrid experiments; Erica Golemis for supplying the LexA polyclonal antiserum and discussion of the two-hybrid experiments; Christopher Coker and David McGee for their critiques of the manuscript; and Christopher Coker, John Fulkerson, and Xin Li for numerous technical suggestions.
This work was supported in part by Public Health Service grant AI23328 from the National Institutes of Health.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1987. [Google Scholar]
- 2.Birnboim H C, Doly J. Rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brayman T G, Hausinger R P. Purification, characterization, and functional analysis of a truncated Klebsiella aerogenes UreE urease accessory protein lacking the histidine-rich carboxyl terminus. J Bacteriol. 1996;178:5410–5416. doi: 10.1128/jb.178.18.5410-5416.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brent R, Ptashne M. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature. 1984;321:612–615. doi: 10.1038/312612a0. [DOI] [PubMed] [Google Scholar]
- 5.Chen L Y, Chen M Y, Leu W M, Tsai T Y, Lee Y H. Copper transfer and activation of the Streptomyces apotyrosinase are mediated through a complex formation between apotyrosinase and its trans-activator MelC1. J Biol Chem. 1992;267:20100–20107. [PubMed] [Google Scholar]
- 6.Colpas G J, Brayman T G, Ming L-J, Hausinger R P. Identification of metal-binding residues in the Klebsiella aerogenes urease nickel metallochaperone, UreE. Biochemistry. 1999;38:4078–4088. doi: 10.1021/bi982435t. [DOI] [PubMed] [Google Scholar]
- 7.Colpas G J, Hausinger R P. In vivo and in vitro kinetics of metal transfer by the Klebsiella aerogenes urease nickel metallochaperone, UreE. J Biol Chem. 2000;275:10731–10737. doi: 10.1074/jbc.275.15.10731. [DOI] [PubMed] [Google Scholar]
- 8.Estojak J, Brent R, Golemis E A. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Field S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 10.Griffith D P, Musher D M, Itin C. Urease: the primary cause of infection-induced urinary stones. Investig Urol. 1976;13:346–350. [PubMed] [Google Scholar]
- 11.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 12.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 13.Homer M J, Paustian T D, Shah V K, Roberts G P. The nifY product of Klebsiella pneumoniae is associated with apodinitrogenase and dissociates upon activation with the iron-molybdenum cofactor. J Bacteriol. 1993;175:4907–4910. doi: 10.1128/jb.175.15.4907-4910.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Island M D, Mobley H L T. Proteus mirabilis urease: linker-insertion mutagenesis of the positive activator and accessory genes. J Bacteriol. 1995;177:5653–5660. doi: 10.1128/jb.177.19.5653-5660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jabri E, Carr M B, Hausinger R P, Karplus P A. The crystal structure of urease from Klebsiella aerogenes. Science. 1995;268:998–1004. [PubMed] [Google Scholar]
- 16.Johnson D E, Russell R G, Lockatell C V, Zulty J C, Warren J W, Mobley H L T. Contribution of Proteus mirabilis urease to persistence, urolithiasis, and acute pyelonephritis in a mouse model of ascending urinary tract infection. Infect Immun. 1993;61:2748–2754. doi: 10.1128/iai.61.7.2748-2754.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones B D, Mobley H L T. Proteus mirabilis urease: genetic organization, regulation, and expression of structural gene. J Bacteriol. 1988;170:3342–3349. doi: 10.1128/jb.170.8.3342-3349.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones B D, Mobley H L T. Proteus mirabilis urease: nucleotide sequence determination and comparison with jack bean urease. J Bacteriol. 1989;171:6414–6422. doi: 10.1128/jb.171.12.6414-6422.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 20.Karmens J, Richardson P, Mosialo G, Brent R, Gilmore T. Oncogenic transformation by vRel requires an amino-terminal activation domain. Mol Cell Biol. 1990;10:2840–2847. doi: 10.1128/mcb.10.6.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee M H, Mulrooney S B, Renner M J, Markowicz Y, Hausinger R P. Klebsiella aerogenes urease gene cluster: sequence of ureD and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. J Bacteriol. 1992;174:4324–4330. doi: 10.1128/jb.174.13.4324-4330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M H, Pankratz S, Wang S, Scott R A, Finnegan M G, Johnson M K, Ippolito J A, Christianson D W, Hausinger R P. Purification and characterization of Klebsiella aerogenes UreE protein: a nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 1993;2:1042–1052. doi: 10.1002/pro.5560020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maier T, Jacobi A, Sauter M, Böck A. The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J Bacteriol. 1993;175:630–635. doi: 10.1128/jb.175.3.630-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maroney M J. Structure/function relationships in nickel metallobiochemistry. Curr Opin Chem Biol. 1999;3:188–199. doi: 10.1016/S1367-5931(99)80032-5. [DOI] [PubMed] [Google Scholar]
- 25.Mobley H L T, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mobley H L T, Warren J W. Urease-positive bacteriuria and obstruction of long-term urinary catheters. J Clin Microbiol. 1987;25:2216–2217. doi: 10.1128/jcm.25.11.2216-2217.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moncrief M C, Hausinger R P. Purification and activation properties of UreD-UreF-urease apoprotein complexes. J Bacteriol. 1996;178:5417–5421. doi: 10.1128/jb.178.18.5417-5421.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moncrief M C, Hausinger R P. Characterization of UreG, identification of a UreD-UreF-UreG complex, and evidence suggesting that a nucleotide-binding site in UreG is required for in vivo metallocenter assembly of Klebsiella aerogenes urease. J Bacteriol. 1997;179:4081–4086. doi: 10.1128/jb.179.13.4081-4086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monod J, Wyman J, Changeux J-P. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 30.Mulrooney S B, Hausinger R P. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol. 1990;172:5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park I-S, Carr M B, Hausinger R P. In vitro activation of urease apoprotein and role of UreD as a chaperone required for nickel metallocenter assembly. Proc Natl Acad Sci USA. 1994;91:3233–3237. doi: 10.1073/pnas.91.8.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park I-S, Hausinger R P. Evidence for the presence of urease apoprotein complexes containing UreD, UreF, and UreG in cells that are competent for in vivo enzyme activation. J Bacteriol. 1995;177:1947–1951. doi: 10.1128/jb.177.8.1947-1951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubins R H, Tolkoff-Rubins N E, Cotran R S. Urinary tract infection, pyelonephritis, and reflux nephropathy. In: Brenner B M, Rector F C, editors. The kidney. Philadelphia, Pa: The W.B. Saunders Co.; 1986. pp. 1085–1141. [Google Scholar]
- 34.Ruden D M, Ma J, Li Y, Wood K, Ptashne M. Generating yeast transcriptional activators containing no yeast protein sequences. Nature. 1991;35:250–252. doi: 10.1038/350250a0. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soriano A, Hausinger R P. GTP-dependent activation of urease apoprotein in complex with the UreD, UreF, and UreG accessory proteins. Proc Natl Acad Sci USA. 1999;96:11140–11144. doi: 10.1073/pnas.96.20.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sriwanthana B, Island M D, Maneval D, Mobley H L T. Single-step purification of Proteus mirabilis urease accessory protein UreE, a protein with a naturally occurring histidine tail, by nickel chelate affinity chromatography. J Bacteriol. 1994;176:6836–6841. doi: 10.1128/jb.176.22.6836-6841.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sriwanthana B, Island M D, Mobley H L T. Sequence of the Proteus mirabilis urease accessory gene ureG. Gene. 1993;129:103–106. doi: 10.1016/0378-1119(93)90703-6. [DOI] [PubMed] [Google Scholar]
- 40.Taha T S M, Brayman T G, Karplus A, Hausinger R P. Urease nickel metallocenter structure and assembly. In: Winkelmann G, Carrano C J, editors. Transition metals in microbial metabolism. Amsterdam, The Netherlands: Harwood Academic Publishers; 1997. pp. 391–413. [Google Scholar]
- 41.Waugh R, Boxer D H. Pleiotropic hydrogenase mutants of Escherichia coli K-12: growth in the presence of nickel can restore hydrogenase activity. Biochimie. 1986;68:157–166. doi: 10.1016/s0300-9084(86)81080-x. [DOI] [PubMed] [Google Scholar]
- 42.West R W J, Yocum R R, Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activator sequence UASG. Mol Cell Biol. 1984;4:2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]