Abstract
Background
Data regarding the influence of patatin-like phospholipase domain-containing 3 (PNPLA3) polymorphism for patients with liver cirrhosis (LC) are scarce.
Objective
This study assesses the role of the PNPLA3 polymorphism for the development of LC and its complications by the findings of genetic examinations.
Methods
Patients with LC caused by virus (n = 157), alcohol (n = 104), nonalcoholic fatty liver disease (NAFLD) (n = 106), or autoimmune disease (n = 33) and without LC (n = 128) were enrolled. LC was composed of the presence and absence of complications, such as variceal bleeding, hepatic ascites, and hepatic encephalopathy. To assess the role of the PNPLA3 polymorphism, odds ratio (OR) for the rs738409 variant was calculated for the patients between (i) with LC and without LC in the entire cohort and (ii) the presence and absence of complications in the patients with LC.
Results
There was a significant difference among the patients without LC and those with alcohol, NAFLD-related LC in the frequency of G alleles (p < 0.001, both). According to complications of LC, the OR for NAFLD-related cirrhosis significantly increased in the presence of the two mutated alleles (OR = 3.165; p = 0.046) when the wild type was used as the reference. However, there were no significant risks for the complications in the virus and alcohol-related cirrhosis unless there was a presence of G alleles.
Conclusion
The PNPLA3 polymorphism was associated with the risk of NAFLD-related LC and its complications.
Keywords: Patatin-like phospholipase domain-containing 3 genotype, Liver cirrhosis, Hepatic encephalopathy, Varix hemorrhage, Hepatic ascites
Introduction
Hepatic steatosis, which is defined as the presence of an intrahepatic triglyceride content >5% of the liver weight, is a common finding in obese people due to excessive fat and carbohydrate consumption [1]. Hepatic steatosis is subdivided into simple hepatic steatosis and steatohepatitis. Pathological findings of simple hepatic steatosis are microvesicular steatosis in hepatocytes without an inflammatory condition. Steatohepatitis, which is a more severe stage, is a condition of hepatocyte injury and degeneration characterized by neutrophilic infiltration, hepatocyte ballooning, and Mallory-Denk hyaline inclusions in the liver. Although the short-term prognosis in patients with the accumulation of fat in the liver is favorable, chronic liver damage is often caused by long-term exposure of hepatic steatosis, which is progressive with substantial risk of progression to advanced fibrosis and liver-related mortality [2].
The molecular mechanism from simple steatosis to steatohepatitis remains elusive. The most widely supported theory implicates an excessive accumulation of toxic lipids, mitochondrial structural abnormalities, and insulin resistance as the key mechanism leading to the increased free radical formation, hepatocellular injury, and fibrosis. Several potential oxidative stressors have been proposed to result in necroinflammation [3].
Especially, many researchers have suggested that the accumulation of triglycerides cause lipotoxicity, which increases cell damage induced by excessive oxidative stress [4]. Damaged hepatocytes from various factors release damage-endogenous-associated molecular patterns and directly cause hepatic stellate cell activation. Activated hepatic stellate cells synthesized accumulation of extracellular matrix protein leading to progressive fibrosis [5].
The most widely supported theory implicates a complex disorder with both environmental and genetic contributions. In environmental factors, patients with lipotoxicity damage typically present with insulin resistance linked to type 2 diabetes, dyslipidemia, hypertension, and cardiovascular disease [4]. According to genetic factors, the patatin-like phospholipase domain-containing 3 (PNPLA3) gene strongly influences hepatic steatosis [6, 7]. PNPLA3 is a multifunctional enzyme and could regulate metabolism of carbohydrates and lipids in the liver. Previously, the rs738409 C>G variant in exon 3b of the PNPLA3 gene was reported to influence the expression levels of the PNPLA3 protein [8]. The C to G substitution at this locus has led to reduce the lower PNPLA3 protein levels and enzyme activities. As a result, the G allele of rs738409 was strongly associated with increased liver steatosis and hepatic inflammation [9].
To date, there has been some research conducted to investigate whether or not the rs738409 polymorphism is a risk factor for fibrosis progression. Valenti et al. [10] showed that the rs738409 polymorphism was reported to associate with the severity of fibrosis and the hepatic inflammation for patients with nonalcoholic fatty liver disease (NAFLD). In alcohol-related liver disease (ALD), the rs738409 G allele increased the risk of mortality by fibrosis progression [11]. Furthermore, a significant association of the rs738409 G allele with steatosis and fibrosis was observed in viral hepatitis with histological damage [12]. Thus, the rs738409 polymorphism has major implications regarding the prognoses of patients with chronic liver disease regardless of the etiology.
Various degrees of liver fibrosis from liver steatosis ultimately lead to the occurrence of cirrhosis-related complications, such as the presence of hepatic ascites, hepatic encephalopathy, varix hemorrhage, and hepatocellular carcinoma. In patients with chronic liver disease, the presence or absence of complications are the most important factors to predict prognoses and determine treatment policy [13]. However, many subsequent PNPLA3 studies enrolled noncirrhotic patients. There are limited data on the PNPLA3 rs738409 polymorphism playing the role of fibrosis progression in cirrhotic patients. However, to our knowledge, there are as yet no data about any correlations between cirrhosis-related complications and the PNPLA3 rs738409 polymorphism. We therefore assessed the role of the PNPLA3 polymorphism for the development of LC and its complications by the findings of the genetic examinations.
Patients and Methods
Population
The LC Group
A multicenter study was conducted at six hospitals in Japan. Patients were required to cirrhotic patients based on laboratory results and imaging tests, showing an irregular surface in the liver, collateral portosystemic venous channels, ascites, hepatosplenomegaly, and/or esophageal (gastric) varices. Enrolled patients were categorized as present or absent regarding complications, which were defined by hepatic ascites, hepatic encephalopathy, and/or varix ruptures, which are life-threatening complications with LC [13]. A histological evaluation by liver biopsy was not performed because of the incidence of adverse events and sampling error staging inaccuracies [14]. LC in these patients was caused by viral infections with hepatitis B and C virus, ALD, NAFLD, autoimmune liver disease (AID) including primary biliary cholangitis, primary sclerosing cholangitis, and autoimmune hepatitis. The diagnosis of hepatitis B virus infection was based on the presence of hepatitis B surface antigen and/or polymerase chain reaction assays for direct determination of hepatitis B virus DNA. Hepatitis C virus (HCV) infection was based on the HCV-ab and/or HCV RNA tests detected level of 1.2 log IU/mL or lower. The ALD diagnosis was based on the intake of approximately 40–80 g/day (men) or 20–40 g/day (women) for longer than 10 years and the exclusion of other liver diseases. A definitive diagnosis of NAFLD requires histological features of steatohepatitis evidenced in a biopsy in the absence of significant alcohol consumption. However, the progression of liver fibrosis leads to the disappearance of steatosis, necroinflammation, ballooning degeneration, and Mallory bodies. We therefore diagnosed NASH associated with cirrhosis based on the clinical features. The patients with consumption of <20 g of ethanol per day and the appropriate exclusion of other liver diseases were categorized as NAFLD. Patients with severe kidney or heart disease or malignancies other than HCC were considered inappropriate for this study and were excluded.
The Non-LC Group
The control group was formed to access the frequency of the rs738409 polymorphism for non-LC. The control group subjects were recruited from Nippon Medical School Chiba Hokusoh Hospital in Tokyo, Japan. This group previously reported subclinical atherosclerosis in patients with NAFLD [15]. Patients in this control group were diagnosed by liver biopsy with deposition of fat in at least 5% of hepatocytes.
Endpoints
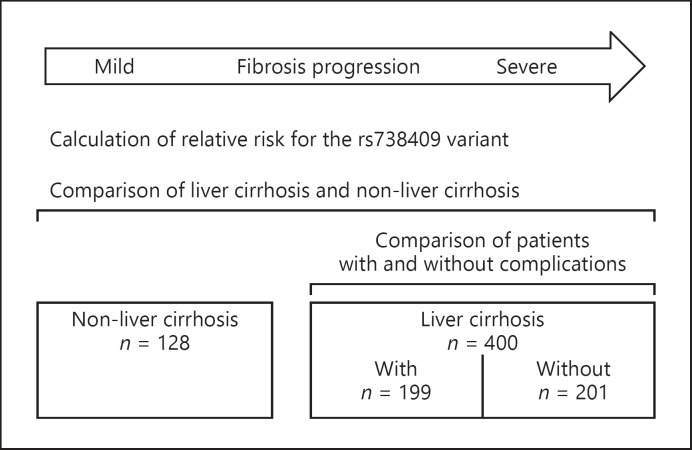
To assess the role of PNPLA3 polymorphism, odds ratio (OR) for the rs738409 variant was calculated for the patients between (i) with LC and without LC in the entire cohort and (ii) the presence or absence of complications in the patients with LC (Fig. 1). The secondary endpoint measurement was to determine whether or not the influence of another NAFLD-related gene (PNPLA3 rs2896019 and TM6SF2 rs58543926) polymorphism contributed to fibrosis progression.
Fig. 1.
Study design. The frequency of the PNPLA3 rs738409 variant was compared for the patients between those (i) with LC and without LC in the entire cohort and (ii) the presence or absence of complications in the patients with liver cirrhosis.
Genotyping for Polymorphisms in PNPLA3 and the Genetic Variants
Blood samples were exclusively collected in ethylenediaminetetraacetic acid tubes, and genomic DNA extraction from patient blood was used in an automated MagNA Pure Compact Instrument (Roche Diagnostics, Mannheim, Germany) with the MagNA Pure Compact Nucleic Acid Isolation Kit I (Roche Diagnostics) according to the manufacturer's instructions.
Genotyping for polymorphisms PNPLA3 (rs738409 and rs2896019) and TM6SF2 (rs58542926) was performed using TaqMan Single Nucleotide Polymorphism Genotyping Assays (Applied Biosystems, Waltham, MA, USA) and the LightCycler 96 System (Roche Diagnostics). The reaction consisted of 10 μL LightCycler FastStart Essential DNA Probes Master Mix (Roche Diagnostics), 1 μL of the primer-probe mix (20×), 7 μL of H2O (polymerase chain reaction grade), and 2 μL of DNA sample in a total reaction volume of 20 μL. Genotyping for the rs738409 variants was categorized as the CC genotype (wild type), the CG genotype (heterozygous polymorphism), and the GG genotype (homozygous polymorphism).
Baseline Characteristic Parameters
Demographic parameters, concomitant medication, and baseline laboratory data were obtained on the same day as the genotyping or within 1 month. Body mass index (BMI) was measured, and obesity was defined as a BMI >25.0 kg/m2. Diabetes diagnosis was based on the measurement of hemoglobin A1c level. Measurements of indirect biomarkers for the fibrosis-4 index and the albumin-bilirubin (ALBI) score were calculated [16]. Furthermore, we evaluated the reliability of the diagnosis of liver cirrhosis using the Bonacini cirrhosis discriminant score. A Bonacini cirrhosis discriminant score of >7 indicated a high probability, while <3 indicated a low probability. The Bonacini cirrhosis discriminant score was calculated using previously published formulas [17] (online suppl. Table 1; see www.karger.com/doi/10.1159/000521062 for all online suppl. material).
Statistical Analysis
Based on a previous study [8], the sample size was calculated to be approximately 400 patients (200 patients in each group with and those without complications) for this trial with a 95% confidence interval (CI) and a power of 80%. Data were expressed as the mean ± standard deviation. Genotype distribution was examined using the χ2 test. Patients with and those without complications were compared using the unpaired t test. Frequencies of the rs738409 polymorphism were analyzed in the Hardy-Weinberg equilibrium. The associations between genotypes and categorical variables were tested using logistic regression models. Differences with a p value of <0.05 were considered significant. Data were analyzed using the SPSS version 24.0 software package (IBM Corporation, Armonk, NY, USA). Statistical analysis was performed by the Statista Corporation, Kyoto, Japan.
Results
Patients
In this study, 528 patients were assessed from November 2019 through May 2021. Of those patients, LC and non-LC were made up of 400 and 128 patients, respectively. Table 1 summarizes the patients' demographics and other baseline characteristics. The t test confirmed that the ages of LC patients were significantly higher than those of non-LC patients (p < 0.001). The BMI of LC patients was significantly lower than that of non-LC patients (p < 0.001). Fasting blood sugar of LC patients was significantly higher than that of non-LC patients (p < 0.001). Total cholesterol of LC patients was significantly lower than that of non-LC patients (p < 0.001). In the Bonacini cirrhosis discriminant score, the frequency and population of high probability (>7) and low probability (<3) were 195 (48.8%) and 14 (3.5), respectively.
Table 1.
Patient baseline clinical characteristics
| LC |
Non-LC control | p value LC vs. non-LC | ||||||
|---|---|---|---|---|---|---|---|---|
| virus (HCV/HBV) | alcohol | NAFLD | AID (AIH/PBC) | total | ||||
| N | 157 (110/47) | 104 | 106 | 33 (14/19) | 400 | 128 | ||
| Age | years | 69.2±10.3 | 63.1±11.6 | 67.8±12.5 | 71.1±8.0 | 67.4±11.4 | 55.9±14.2 | <0.0001 |
| Gender: male | n (%) | 91 (58.0) | 81 (77.9) | 54 (50.9) | 7 (21.2) | 233 (58.3) | 67 (52.3) | 0.241 |
| BMI | kg/m2 | 23.4±4.07 | 22.9±3.26 | 26.2±5.06 | 22.4±3.53 | 23.9±4.35 | 28.8±4.52 | <0.0001 |
| Obesity | n (%) | 45 (28.8) | 27 (26.0) | 63 (59.4) | 6 (18.2) | 141 (35.3) | 106 (82.8) | <0.0001 |
| Liver cancer | n (%) | 73 (46.5) | 19 (18.3) | 20 (18.9) | 3 (9.1) | 115 (28.7) | − | |
| HCV SVR/non-SVR | n (%) | 79/27 | − | − | − | − | − | |
| Diabetes mellitus | n (%) | 50 (31.8) | 37 (35.6) | 58 (54.7) | 4 (12.1) | 149 (37.3) | 46 (35.9) | 0.789 |
| Liver fibrosis stage | (F0/F1/F2/F3) | − | − | − | − | − | 24/42/35/27 | |
| Complication | n (%) | 75 (47.8) | 57 (54.8) | 51 (48.1) | 16 (48.5) | 199 (49.8) | − | |
| HE/HA/VR | 30/55/11 | 39/40/19 | 32/41/11 | 7/8/6 | 108/144/47 | |||
| Child-Pugh score | 6.03±1.81 | 6.73±2.10 | 6.35±2.05 | 6.24±1.93 | 6.31±1.98 | − | ||
| FIB-4 index | 5.53±3.69 | 5.91±5.23 | 5.96±4.23 | 5.87±3.12 | 5.77±4.23 | − | ||
| ALBI score | −2.39±0.66 | −2.16±0.75 | −2.21±0.68 | −2.10±0.62 | −2.35±0.69 | − | ||
| Bonacini cirrhosis | ||||||||
| discriminant score | ≤3/4–6/≥7 | 3/75/69 | 5/54/58 | 2/42/49 | 4/20/19 | 14/191/195 | ||
| Fasting blood sugar | mg/dL | 132.1±53.4 | 141.2±62.3 | 140.4±53.1 | 114.1±33.4 | 135.4±54.9 | 113.1±27.5 | <0.0001 |
| Total cholesterol | mg/dL | 177.5±38.2 | 166.8±41.0 | 177.9±38.5 | 179.1±45.9 | 175.0±39.8 | 196.1±33.1 | <0.0001 |
Steatosis graded 0–3 based on the percentage of hepatocytes on biopsy specimens (0: <5%, 1: 5–33%, 2: 33–66%, 3: >66%). Fibrosis stage evaluated on biopsy: F0, no fibrosis; F1, perisinusoidal or periportal fibrosis; F2, perisinusoidal and portal/periportal fibrosis; F3, bridging fibrosis. HE, hepatic encephalopathy; HA, hepatic ascites; VR, varicose vein rupture.
Genotype Counts and Frequency of the rs738409 Allele in LC
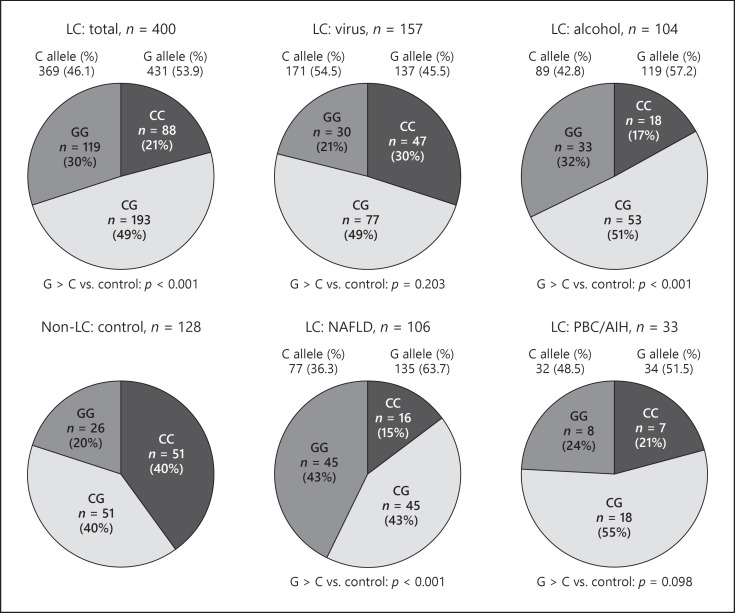
The frequencies of the PNPLA3 rs738409 CC, CG, and GG genotypes in LC were 22.0%, 48.2%, and 29.8%, respectively; and those in non-LC were 40.0%, 40.0%, and 20.0%, respectively. The highest frequency of the rs738409 G allele was 63.7% in NAFLD-related LC. The lowest frequency of the rs738409 G allele was 45.5% in virus-related LC.
Genotype counts and frequencies of rs738409 G alleles were compared between LC and non-LC (Table 2) (Fig. 2). ALD- and NAFLD-related LC were significantly different in the frequency of G alleles compared with non-LC (p < 0.001, both). However, there were no significant differences between virus-related LC and non-LC, and between AID-related LC and non-LC (p = 0.203 and p = 0.098, respectively).
Table 2.
Genotype counts and frequency of the rs738409 G allele for LC
| Etiology | N | rs738409 variant (genotype) |
p value G > C vs. control | rs738409 variant (allele) |
p value G > C vs. control | |||
|---|---|---|---|---|---|---|---|---|
| CC, n (%) | CG, n (%) | GG, n (%) | C allele, n (%) | G allele, n (%) | ||||
| Standard | ||||||||
| Total | 400 | 88 (22.0) | 193 (48.2) | 119 (29.8) | <0.0001 | 369 (46.1) | 431 (53.9) | <0.0001 |
| Virus | 157 | 47 (29.9) | 77 (49.0) | 33 (21.0) | 0.186 | 171 (54.5) | 143 (45.5) | 0.203 |
| Alcohol | 104 | 18 (17.3) | 53 (51.0) | 33 (31.7) | 0.001 | 89 (42.8) | 119 (57.2) | <0.0001 |
| NASH | 106 | 16 (15.1) | 45 (42.5) | 45 (42.5) | <0.0001 | 77 (36.3) | 135 (63.7) | <0.0001 |
| PBC/AIH | 33 | 7 (21.2) | 18 (54.5) | 8 (24.2) | 0.132 | 32 (48.5) | 34 (51.5) | 0.098 |
| Control | ||||||||
| NASH | 128 | 51 (39.8) | 51 (39.8) | 26 (20.3) | Reference | 153 (59.8) | 103 (40.2) | Reference |
Fig. 2.
Genotype counts and frequency of G allele rs738409 in LC and non-LC. The presence of liver cirrhosis was affected by the G allele of PNPLA3 rs738409 in the alcohol and NAFLD group (p < 0.001 and p < 0.001, respectively).
Genotype Counts and Frequency of the rs738409 G Alleles for LC-Related Complications
We evaluated the risk association between the G allele expressions and patients with complications in patients with LC (Table 3). In patients with NAFLD-related LC, although there were no differences in frequencies of G alleles (OR = 2.260; 95% CI = 0.922–5.544; p = 0.075), the OR for complications significantly increased in the presence of the two mutated alleles (OR = 3.165; 95% CI = 1.073–10.294; p = 0.046) when the CC genotype was used as the reference. In AID, there was a significant risk of complications in the presence of the two mutated alleles (p = 0.014, respectively). On the other hand, there were no significant risks or complications in patients with the virus- or alcohol-related LC unless there was a presence of G alleles.
Table 3.
Genotype counts and frequency of rs38409 G allele for cirrhosis-related complications
| Etiology | Complication present, n (%) | Complication absent, n (%) | OR | 95% CI | p value |
|---|---|---|---|---|---|
| Total | 199 (49.8) | 201 (50.3) | |||
| CC = 37 (18.6) | CC = 201 (25.4) | 1 | Reference | ||
| CG = 90 (45.2) | CG = 103 (51.2) | 1.204 | 0.724–2.004 | 0.474 | |
| GG = 72 (36.2) | GG = 47 (23.4) | 2.112 | 1.205–3.699 | 0.009 | |
| C allele = 164 (41.2) | C allele = 205 (51.0) | 1 | Reference | ||
| G allele = 234 (58.8) | G allele = 197 (49.0) | 1.401 | 0.916–2.144 | 0.120 | |
|
| |||||
| Virus | 75 (47.7) | 82 (52.3) | |||
| CC = 22 (29.3) | CC = 25 (30.5) | 1 | Reference | ||
| CG = 40 (53.3) | CG = 37 (45.1) | 1.229 | 0.594–2.541 | 0.579 | |
| GG = 13 (17.3) | GG = 20 (24.4) | 0.739 | 0.299–1.823 | 0.511 | |
| C allele = 84 (56) | C allele = 87 (53) | 1 | Reference | ||
| G allele = 66 (44) | G allele = 77 (47) | 0.904 | 0.458–1.786 | 0.771 | |
|
| |||||
| Alcohol | 57 (54.8) | 47 (45.2) | |||
| CC = 7 (12.3) | CC = 11 (23.4) | 1 | Reference | ||
| CG = 30 (52.6) | CG = 23 (48.9) | 2.050 | 0.688–6.110 | 0.198 | |
| GG = 20 (35.1) | GG = 13 (27.7) | 2.418 | 0.745–7.845 | 0.142 | |
| C allele = 44 (38.6) | C allele = 45 (47.9) | 1 | Reference | ||
| G allele = 70 (61.4) | G allele = 49 (52.1) | 1.371 | 0.592–3.176 | 0.460 | |
|
| |||||
| NAFLD | 51 (48.1) | 55 (51.9) | |||
| CC = 7 (13.7) | CC = 9 (16.4) | 1 | Reference | ||
| CG = 12 (23.5) | CG = 33 (60.0) | 0.468 | 0.142–1.534 | 0.210 | |
| GG = 32 (62.7) | GG = 13 (23.6) | 3.165 | 1.073–10.294 | 0.046 | |
| C allele = 26 (25.5) | C allele = 51 (46.4) | 1 | Reference | ||
| G allele = 76 (74.5) | G allele = 59 (53.6) | 2.260 | 0.922–5.544 | 0.075 | |
|
| |||||
| AID | 16 (48.5) | 17 (51.5) | |||
| CC = 1 (6.3) | CC = 6 (35.3) | 1 | Reference | ||
| CG = 8 (50) | CG = 10 (58.8) | 4.800 | 0.475–48.46 | 0.184 | |
| GG = 7 (43.8) | GG = 1 (5.9) | 42.00 | 2.136–825.7 | 0.014 | |
| C allele = 10 (31.3) | C allele = 22 (64.7) | 1 | Reference | ||
| G allele = 22 (68.8) | G allele = 12 (35.3) | 3.088 | 0.673–14.166 | 0.144 | |
Propensity Score Matching
We also evaluated the risk association between the G allele expressions and the progression of fibrosis by propensity score matching. Age and sex were chosen for the propensity score matching among the patients without LC and those with LC caused by the virus, alcohol, or NAFLD. After propensity score matching in non-LC patients and those with virus-related LC, the study population consisted of 162 patients (control: 82 patients, virus: 82 patients) (online suppl. Table 2). There was no difference between the control and the virus-related LC in the frequency of G alleles (p = 0.144). Regarding ALD, the study population consisted of 144 patients (control: 72 patients, ALD: 72 patients). There was a significant difference between the control and ALD in the frequency of G alleles (p = 0.001). However, there were no significant risks for complications in ALD unless there was a presence of G alleles. Regarding NAFLD, the study population consisted of 150 patients (control: 75 patients, NAFLD: 75 patients). There was a difference between the control and NAFLD in the frequency of G alleles (p < 0.001). Furthermore, the OR for complications significantly increased in the presence of the two mutated alleles (p < 0.001) when the CC genotype was used as the reference.
The Influence of rs738409 GG Genotypes for the Basal Characteristics
A correlation between rs738409 polymorphism and baseline clinical characteristics in patients with LC was observed (Table 4). The BMI in the CC + CG group was significantly lower than that in the GG group (p = 0.004). The platelet count was higher in the CG + CC group than that in the GG group (p = 0.007). Hemoglobin A1c was lower in the CG + CC group than that in the GG group (p = 0.019). However, there were no associations between the rs738409 polymorphisms and the total cholesterol of other hepatic enzymes.
Table 4.
Effect of the rs738409 polymorphism on clinical characteristics
| CC + CG | GG | p value | |
|---|---|---|---|
| n | 281 | 119 | |
| Age, years | 67.7±11.6 | 66.8±10.9 | 0.476 |
| BMI, kg/m2 | 23.5±4.2 | 24.9±4.6 | 0.004 |
| White blood cells, µl | 4,719±1,883 | 4,255±1,404 | 0.007 |
| Hemoglobin, g/dL | 12.6±2.3 | 12.6±2.1 | 0.803 |
| Platelets, ×104/µl | 11.9±5.6 | 10.3±5.7 | 0.007 |
| Prothrombin time, % | 1.2±0.3 | 1.21±0.3 | 0.548 |
| Serum albumin, g/dL | 3.7±0.7 | 3.6±0.7 | 0.376 |
| Blood urea nitrogen, mg/dL | 17.6±9.5 | 16.8±8.2 | 0.419 |
| Serum creatinine, mg/dL | 0.9±0.3 | 0.91±0.4 | 0.767 |
| Aspartate aminotransferase, IU/L | 39.3±25.7 | 42.4±23.7 | 0.256 |
| Alanine aminotransferase, IU/L | 27.7±18.4 | 30.3±19.0 | 0.241 |
| Total bilirubin, mg/dL | 1.3±1.1 | 1.5±1.2 | 0.144 |
| Hemoglobin A1c, % | 5.9±1.1 | 6.3±1.5 | 0.019 |
| Total cholesterol, mg/dL | 173.3±41.1 | 178.2±36.5 | 0.316 |
Multivariate Analyses of Factors Affecting Complications in NAFLD-Related LC
The relationship between the complications of NALFL-related LC and baseline characteristic parameters using a logistic regression model was calculated (Table 5). In the univariate analysis, rs738409 polymorphisms and the ALBI score were identified as independent factors. Multivariate analysis confirmed that rs738409 polymorphisms and the ALBI score were independent factors that predicted complications of LC (OR = 5.750; 95% CI = 2.229–14.83, p < 0.001 and OR = 9.375; 95% CI = 2.969–29.60, p < 0.001).
Table 5.
Multivariate analyses of factors affecting LC-related complications in patients caused by NAFLD
| Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|
| OR (95% CI) | p value | OR (95% CI) | p value | ||
| rs738409 genotype | |||||
| 1: CC + CG | |||||
| 2: GG | 5.441 (2.344–12.63) | <0.001 | 5.750 (2.229–14.83) | <0.001 | |
| TM6Sf2 CC versus TT + CT | |||||
| 1: CT + TT | |||||
| 2: TT | 1.108 (0.467–2.629) | 0.816 | |||
| Age <65 versus ≥65 | |||||
| 1: <65 | |||||
| 2: >65 | 1.222 (0.555–2.695) | 0.174 | |||
| BMI | |||||
| 1: <22 | 2.812 (1.054–7.503) | 0.039 | |||
| 2: ≥22 | |||||
| DM | |||||
| 1: − | |||||
| 2: + | 1.607 (0.743–3.477) | 0.228 | |||
| AST | |||||
| 1: <35 | |||||
| 2: ≥35 | 2.119 (0.429–2.840) | 0.410 | |||
| MELD score | |||||
| 1: <9 | |||||
| 2: ≥9 | 1.939 (0.719–3.001) | 0.518 | |||
| Platelets <10 versus ≥10 | |||||
| 1: <10×104/µL | 2.002 (0.923–4.344) | 0.079 | |||
| 2: ≥10×104/µL | |||||
| ALBI score | |||||
| 1: ≤–2.60 | 8.871 (3.062–25.69) | <0.001 | 9.375 (2.969–29.60) | <0.001 | |
| 2: >−2.60 | |||||
Other Single-Nucleotide Polymorphisms Linked to NAFLD-Related Genes and Complications
We evaluated risk alleles in the NAFLD-related genotypes (PNPLA3 rs2896019 and TM6SF2 rs58542926) for the complication of LC in the patients with LC. The frequency of the GG, GT, and TT genotypes in PNPLA3 rs2896019 was 29.5%, 47.0%, and 22.3%, respectively. The rate of concordance with the G1 to the G2 alleles of the PNPLA3 rs2896019 and rs738409 gene polymorphisms was 97.3%. Therefore, there were a significant risk association and complications when comparing the TT to GG + GT genotypes in the entire cohort of the LC patients (OR = 1.864; 95% CI = 1.203–2.889; p = 0.005).
The frequency of the CC, CT, and TT genotypes in the TM6SF2 rs58542926 gene polymorphism was 81.7%, 16.1%, and 2.3%, respectively. There were no significant risks or complications when comparing the CC to CT + TT genotypes in the entire cohort of LC patients.
Discussion
PNPLA3 and TM6SF2 polymorphisms were identified as major genetic determinants for hepatic steatosis by the genome-wide association studies [18, 19]. In this study, the PNPLA3 rs738409 polymorphism had a clinical significance to evaluate the risk factor for the progression of fibrosis due to hepatic steatosis in LC patients. However, the frequency of the TM6SF2 rs58542926 T allele in the Japanese people with LC was relatively low so that we could not accurately evaluate the association between rs58542926 and fibrosis progression.
Two main results in this study are especially noteworthy. First, genotype counts and the frequency of the rs738409 G alleles in patients with ALD- and NAFLD-related LC were higher than those without LC. This result further clarifies the revelation that some researchers had previously reported that the G allele of the rs738409 variant significantly influences not only liver fat accumulation but also a heightened susceptibility of developing severe hepatitis, therefore, having a greater risk of fibrosis progression in non-LC patients [20, 21, 22]. These results suggest that the rs738409 genotype might have a causal role in disease severity regardless of etiology. However, the influence on the virus seems to be lower than that in NAFLD. In this cohort, the OR for the virus-related patients was lower than that in NAFLD, and liver fat accumulation from two mutated alleles of the rs738409 genotype had an insignificant effect in patients with LC caused by virus. The established factors associated with the progression of fibrosis in viral hepatitis were male, duration of infection, and long-term excessive alcohol consumption [23]. Second, the rs738409 GG genotype (homozygous polymorphism) was a risk factor for complications in NAFLD-related cirrhosis. Several studies revealed that the OR for the association between the rs738409 genotype and liver damage significantly increased in the GG genotype when the CC genotype was used as the reference [7, 24]. The frequency of G allies was correlated with the degree of steatosis, leading to lipotoxicity and inflammatory damage to the major parenchymal cells in the liver [25, 26, 27].
For all of these reasons, patients with the rs738409 GG genotype must be provided with an appropriate treatment therapy to prevent fibrosis progression, particularly in the early stages of LC. Especially, severely obese patients with homozygous polymorphism require medical care. The pathogenesis of NAFLD involves a complex interaction among the genetic and environmental factors, including the oxidative, endoplasmic reticulum stress, insulin resistance, lipotoxicity, and higher triglycerides in severely obese patients [28, 29, 30].
Previously, Nishioji et al. [31] reported the correlation between the PNPLA3 polymorphism and weight gain on NAFLD in Japanese people. The frequencies of the PNPLA3 rs738409 CC, CG, and GG genotypes in patients of a relatively normal weight without NAFLD were 28.7%, 52.6%, and 18.7%, respectively; and overweight patients with NAFLD were 14.3%, 54.3%, and 31.4%, respectively [31].
Thus, genetic modifiers might be one of many factors and strong interaction with the environment for the pathogenesis of triglyceride accumulation in hepatocytes. Therefore, we recommend to analyze the PNPLA3 genotype for the patients with the environmental factors, such diabetes and dyslipidemia. In the present study, the frequency of diabetes and obesity was higher in the GG genotype of the rs738409 than in the CC genotype in this cohort. The dyslipidemia should be carefully interpreted. The rs738409 variant would not correlated with the levels of total cholesterol unless the risk alleles play a role in hepatic triglyceride hydrolysis. The reason for this discrepancy could be the severalty of liver injury. The liver plays a central role in the secretion, degradation, synthesis, and storage of lipids. LC does not function properly due to the slow and gradual exposure of chronic liver damage over months or years [32, 33]. As a result, the level of total cholesterol tends to decrease with the length of LC.
To our knowledge, this is the first report about the influence of the PNPLA3 on the development of LC and its complications by the findings of genetic examinations. However, the diagnosis of liver cirrhosis, which was crucial for the study, was not complete in the clinical study. There are inherent errors associated with the use of a liver biopsy as a gold standard including selection bias, sampling error, estimation of fibrosis, and interobserver variability. A biopsy specimen only samples an estimated 1/50,000th of the entire liver mass, which can result in an underestimation of the prevalence of cirrhosis [14]. Therefore, we evaluated the reliability of our data using the Bonacini cirrhosis discriminant scores, which is the best way to predict cirrhosis in patients with suspected liver disease [34]. In the present study, the frequency and population of the Bonacini cirrhosis discriminant score <3 were 14 (3.5%). In a meta-analysis, a combination of simple laboratory tests with the Bonacini cirrhosis discriminant score is the most frequently studied, reliable, and informative indirect marker of fibrosis [17]. The clinical data strongly suggest the presence of cirrhosis, and the results reflect the patient's management.
This study has several limitations. First, this clinical study enrolled 400 patients with LC whose etiologies were not uniform. Noteworthy, the number of patients with AID was relatively small. Because there were significant risks and complications in AID, further research is warranted. Second, the diagnosis of NAFLD in this study did not meet the classical criteria for steatohepatitis because the diagnosis of NAFLD was not based on liver biopsy in the LC group. Third, there were correlations between HbA1c and the PNPLA3 rs738409 G allele frequency. However, the HbA1c test can be unreliable in chronic liver disease. Especially, liver cirrhosis may falsely decrease the HbA1c level because of anemia and/or splenomegaly due to portal hypertension. Finally, genomic DNA in this clinical study was extracted from ethnically homogeneous Japanese people. Therefore, reexamination of the data for other and more diverse ethnic groups would be desirable.
Conclusions
The high frequency of the PNPLA3 rs738409 G allele was associated with the risk of NAFLD-related LC and its complications.
Statement of Ethics
The research protocol for this study conformed to the provisions of the Declaration of Helsinki and was approved by the Institutional Review Board Ethics Committee of the Kitasato University School of Medicine (Number: G19-06), Yokohama City University Graduate School of Medicine (Number: A200528001), Nippon Medical School Chiba Hokusoh Hospital (Approval Number: 603), and the Tokushukai Medical Group (Number: TGE01428-024). This is registered in the UMIN Clinical Trials Registry as UMIN000039573. All patients were informed about the significance of gene analysis and gave written informed consent.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
This study was partly supported by the Research Program on Hepatitis from the Japan AMED (Agency for Medical Research and Development) Grant No.: JP19fk0210048 and JP20fk0210048.
Author Contributions
Shao X., Uojima H., Arai T., Ogawa Y., Setsu T., Atsukawa M., Furuichi Y., Arase Y., Horio K., Hidaka H., Nakazawa T., Kako M., Kagawa T., Iwakiri K., Nakajima A., Terai S., Tanaka Y., and Koizumi W. contributed equally to this work. Uojima H. and Shao X. collected and analyzed the data. Shao X. drafted the manuscript. Hidaka H. designed and supervised the study. Arai T., Ogawa Y., Setsu T., Atsukawa M., Furuichi Y., Arase Y., Horio K., Hidaka H., Nakazawa T., Kako M., Kagawa T., Iwakiri K., Nakajima A., Terai S., Tanaka Y., and Koizumi W. offered technical or material support. All authors discussed the results and approved the final manuscript for publication.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgment
We thank Robert E. Brandt, Founder, CEO, and CME of MedEd Japan, for editing and formatting the manuscript.
References
- 1.Matteoni CA, Younossi ZM, Gramlich T, Boparai N, Liu YC, McCullough AJ. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology. 1999;116((6)):1413–9. doi: 10.1016/s0016-5085(99)70506-8. [DOI] [PubMed] [Google Scholar]
- 2.Nassir F, Rector RS, Hammoud GM, Ibdah JA. Pathogenesis and prevention of hepatic steatosis. Gastroenterol Hepatol. 2015;11((3)):167–75. [PMC free article] [PubMed] [Google Scholar]
- 3.Kakisaka K, Suzuki Y, Fujiwara Y, Abe T, Yonezawa M, Kuroda H, et al. Evaluation of ballooned hepatocytes as a risk factor for future progression of fibrosis in patients with non-alcoholic fatty liver disease. J Gastroenterol. 2018;53((12)):1285–91. doi: 10.1007/s00535-018-1468-9. [DOI] [PubMed] [Google Scholar]
- 4.Parthasarathy G, Revelo X, Malhi H. Pathogenesis of nonalcoholic steatohepatitis: an overview. Hepatol Commun. 2020;4((4)):478–92. doi: 10.1002/hep4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gjorgjieva M, Sobolewski C, Dolicka D, Correia de Sousa M, Foti M. miRNAs and NAFLD: from pathophysiology to therapy. Gut. 2019 Nov;68((11)):2065–79. doi: 10.1136/gutjnl-2018-318146. [DOI] [PubMed] [Google Scholar]
- 6.Eslam M, Valenti L, Romeo S. Genetics and epigenetics of NAFLD and NASH: clinical impact. J Hepatol. 2018;68:268–79. doi: 10.1016/j.jhep.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Trépo E, Romeo S, Zucman-Rossi J, Nahon P. PNPLA3 gene in liver diseases. J Hepatol. 2016;65((2)):399–412. doi: 10.1016/j.jhep.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 8.Friedrich K, Wannhoff A, Kattner S, Brune M, Hov JR, Weiss KH, et al. PNPLA3 in end-stage liver disease: alcohol consumption, hepatocellular carcinoma development, and transplantation-free survival. J Gastroenterol Hepatol. 2014;29((7)):1477–84. doi: 10.1111/jgh.12540. [DOI] [PubMed] [Google Scholar]
- 9.Dai G, Liu P, Li X, Zhou X, He S. Association between PNPLA3 rs738409 polymorphism and nonalcoholic fatty liver disease (NAFLD) susceptibility and severity: a meta-analysis. Medicine. 2019;98((7)):e14324. doi: 10.1097/MD.0000000000014324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valenti L, Al-Serri A, Daly AK, Galmozzi E, Rametta R, Dongiovanni P, et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51((4)):1209–17. doi: 10.1002/hep.23622. [DOI] [PubMed] [Google Scholar]
- 11.Friedrich K, Wannhoff A, Kattner S, Brune M, Hov JR, Weiss KH, et al. PNPLA3 in end-stage liver disease: alcohol consumption, hepatocellular carcinoma development, and transplantation-free survival. J Gastroenterol Hepatol. 2014;29((7)):1477–84. doi: 10.1111/jgh.12540. [DOI] [PubMed] [Google Scholar]
- 12.Yasui K, Kawaguchi T, Shima T, Mitsuyoshi H, Seki K, Sendo R, et al. Effect of PNPLA3 rs738409 variant (I148 M) on hepatic steatosis, necroinflammation, and fibrosis in Japanese patients with chronic hepatitis C. J Gastroenterol. 2015;50((8)):887–93. doi: 10.1007/s00535-014-1018-z. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Tsao G, Abraldes JG, Berzigotti A, Bosch J. Portal hypertensive bleeding in cirrhosis: risk stratification, diagnosis, and management: 2016 practice guidance by the American Association for the Study of Liver Diseases. Hepatology. 2017;65((1)):310–35. doi: 10.1002/hep.28906. [DOI] [PubMed] [Google Scholar]
- 14.Afdhal NH, Nunes D. Evaluation of liver fibrosis: a concise review. Am J Gastroenterol. 2004;99((6)):1160–74. doi: 10.1111/j.1572-0241.2004.30110.x. [DOI] [PubMed] [Google Scholar]
- 15.Arai T, Atsukawa M, Tsubota A, Kawano T, Koeda M, Yoshida Y, et al. Factors influencing subclinical atherosclerosis in patients with biopsy-proven nonalcoholic fatty liver disease. PLoS One. 2019;14:e0224184. doi: 10.1371/journal.pone.0224184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiraoka A, Kumada T, Michitaka K, Toyoda H, Tada T, Ueki H, et al. Usefulness of albumin-bilirubin grade for evaluation of prognosis of 2584 Japanese patients with hepatocellular carcinoma. J Gastroenterol Hepatol. 2016;31((5)):1031–6. doi: 10.1111/jgh.13250. [DOI] [PubMed] [Google Scholar]
- 17.Bonacini M, Hadi G, Govindarajan S, Lindsay KL. Utility of a discriminant score for diagnosing advanced fibrosis or cirrhosis in patients with chronic hepatitis C virus infection. Am J Gastroenterol. 1997;92((8)):1302–4. [PubMed] [Google Scholar]
- 18.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53((6)):1883–94. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 19.Raksayot M, Chuaypen N, Khlaiphuengsin A, Pinjaroen N, Treeprasertsuk S, Poovorawan Y, et al. Independent and additive effects of PNPLA3 and TM6SF2 polymorphisms on the development of non-B, non-C hepatocellular carcinoma. J Gastroenterol. 2019;54((5)):427–36. doi: 10.1007/s00535-018-01533-x. [DOI] [PubMed] [Google Scholar]
- 20.Arriazu E, Ruiz de Galarreta M, Cubero FJ, Varela-Rey M, Pérez de Obanos MP, Leung TM, et al. Extracellular matrix and liver disease. Antioxid Redox Signal. 2014;21((7)):1078–97. doi: 10.1089/ars.2013.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Z, Guo X, Zhang G, Liang L, Nong B. Correlation between PNPLA3 rs738409 polymorphism and hepatocellular carcinoma: a meta-analysis of 10,330 subjects. Int J Biol Markers. 2019;34((2)):117–22. doi: 10.1177/1724600818812471. [DOI] [PubMed] [Google Scholar]
- 22.Seko Y, Yamaguchi K, Mizuno N, Okuda K, Takemura M, Taketani H, et al. Combination of PNPLA3 and TLL1 polymorphism can predict advanced fibrosis in Japanese patients with nonalcoholic fatty liver disease. J Gastroenterol. 2018;53((3)):438–48. doi: 10.1007/s00535-017-1372-8. [DOI] [PubMed] [Google Scholar]
- 23.Sircana A, Paschetta E, Saba F, Molinaro F, Musso G. Recent insight into the role of fibrosis in nonalcoholic steatohepatitis-related hepatocellular carcinoma. Int J Mol Sci. 2019;20((7)):1745. doi: 10.3390/ijms20071745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueyama M, Nishida N, Korenaga M, Korenaga K, Kumagai E, Yanai H, et al. The impact of PNPLA3 and JAZF1 on hepatocellular carcinoma in non-viral hepatitis patients with type 2 diabetes mellitus. J Gastroenterol. 2016;51((4)):370–9. doi: 10.1007/s00535-015-1116-6. [DOI] [PubMed] [Google Scholar]
- 26.Sookoian S, Pirola CJ. Genetics of nonalcoholic fatty liver disease: from pathogenesis to therapeutics. Semin Liver Dis. 2019;39((2)):124–40. doi: 10.1055/s-0039-1679920. [DOI] [PubMed] [Google Scholar]
- 27.Lake AC, Sun Y, Li JL, Kim JE, Johnson JW, Li D, et al. Expression, regulation, and triglyceride hydrolase activity of adiponutrin family members. J Lipid Res. 2005;46((11)):2477–87. doi: 10.1194/jlr.M500290-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Chrostek L, Supronowicz L, Panasiuk A, Cylwik B, Gruszewska E, Flisiak R. The effect of the severity of liver cirrhosis on the level of lipids and lipoproteins. Clin Exp Med. 2014;14((4)):417–21. doi: 10.1007/s10238-013-0262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Working Group; Association of Pathologists for Guidebook of NASH and NAFLD, 2015: The Japan Society of Hepatology Pathological findings of NASH and NAFLD: for guidebook of NASH and NAFLD, 2015: the Japan Society of Hepatology. Hepatol Res. 2017;47((1)):3–10. doi: 10.1111/hepr.12847. [DOI] [PubMed] [Google Scholar]
- 30.Povsic M, Wong OY, Perry R, Bottomley J. A structured literature review of the epidemiology and disease burden of non-alcoholic steatohepatitis (NASH) Adv Ther. 2019;36((7)):1574–94. doi: 10.1007/s12325-019-00960-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishioji K, Mochizuki N, Kobayashi M, Kamaguchi M, Sumida Y, Nishimura T, et al. The impact of PNPLA3 rs738409 genetic polymorphism and weight gain ≥10 kg after age 20 on non-alcoholic fatty liver disease in non-obese Japanese individuals. PLoS One. 2015;10((10)):e0140427. doi: 10.1371/journal.pone.0140427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parthasarathy G, Revelo X, Malhi H. Pathogenesis of nonalcoholic steatohepatitis: an overview. Hepatol Commun. 2020;4((4)):478–92. doi: 10.1002/hep4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chrostek L, Supronowicz L, Panasiuk A, Cylwik B, Gruszewska E, Flisiak R. The effect of the severity of liver cirrhosis on the level of lipids and lipoproteins. Clin Exp Med. 2014;14((4)):417–21. doi: 10.1007/s10238-013-0262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Udell JA, Wang CS, Tinmouth J, FitzGerald JM, Ayas NT, Simel DL, et al. Does this patient with liver disease have cirrhosis? JAMA. 2012;307((8)):832–42. doi: 10.1001/jama.2012.186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.