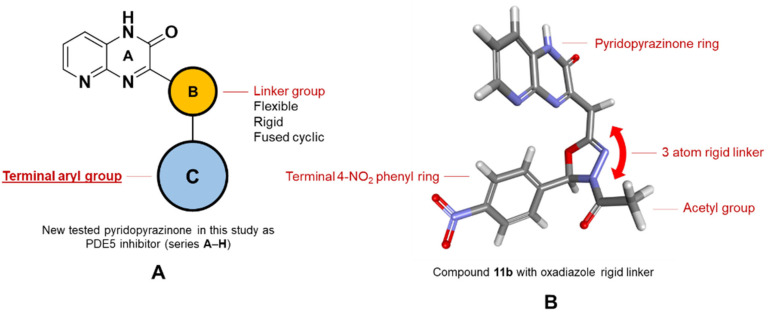
Figure 10.
The proposed structure activity relationship (SAR) of the new pyridopyrazinone-based derivatives (series A–H) into PDE5 protein (PDB ID: 3HC8). (A) pyridopyrazinone ring favors to be anchored into the H pocket close to M-loop and affords hydrophobic interactions as well as HB interaction. The central linker group acts as a hinge between the 2 binding motifs. The flexible spacer with optimum length (5 atoms) will allow this hinge to bring the 2 binding motifs inside the binding pocket. The rigid spacer with five-membered ring is rigid enough to keep the angular orientation between the 2 binding motifs inside the active site. The fused cyclic spacer creates an unfavorable strain between the 2 binding motifs. The terminal aryl motif with HBD/HBA groups will enhance the binding through HB to Gln 817 in the Q pocket. Bulky aryl groups (series C) will deteriorate the residence of the 2 binding motifs together into the active site. (B) the stable conformation of compound 11b inside PDE5 binding site. The oxadiazole rigid ring affords a stable hinge (3 atoms length) between the 2 binding motifs. The terminal NO2 group affords a strong HB interaction with Gln 817 in Q pocket (2.69 Å). The acetyl substitution swims in the solvent-exposed area.