Abstract
Sesquiterpenes are common small-molecule natural products with a wide range of promising applications and are biosynthesized by sesquiterpene synthase (STS). Basidiomycetes are valuable and important biological resources. To date, hundreds of related sesquiterpenoids have been discovered in basidiomycetes, and the biosynthetic pathways of some of these compounds have been elucidated. This review summarizes 122 STSs and 2 fusion enzymes STSs identified from 26 species of basidiomycetes over the past 20 years. The biological functions of enzymes and compound structures are described, and related research is discussed.
Keywords: basidiomycetes, sesquiterpene, biosynthesis, sesquiterpene synthase
1. Introduction
Fungi are widely distributed in various ecosystems of the Earth. Based on high-throughput sequencing methods, approximately 5.1 million species of fungi exist in nature, but only approximately 100,000 species have been discovered [1]. Basidiomycota (commonly known as basidiomycetes) is one of the major phyla of the fungal kingdom, with more than 31,000 species identified [2]. Basidiomycetes are divided into three subphyla: rusts (Puccinomycotina), smuts (Ustilagomycotina), and mushrooms (Agaricomycotina), with several taxonomic ranks below them. Sesquiterpenes are among the most structurally diverse natural products and have many applications in various industries. They contain C15 polymers composed of three isoprene units and derivatives with diverse chemical skeletons. Fungi are rich in sesquiterpenoid natural products, many of which have good biological activities, including antibacterial, antifungal, anti-inflammatory, antitumor, vascular-relaxing, immunosuppressant, and cytotoxic activities. They can be used as lead compounds for new drugs [3,4,5,6,7,8,9,10]; especially in basidiomycetes, sesquiterpenes have various pharmacological activities [11].
Basidiomycetes often produce large fruiting bodies to disperse spores; however, these fruiting bodies are constantly threatened by other organisms that feed on them [12]. As a result, basidiomycetes have evolved a number of protective strategies against threats from other organisms, one of which is the production of toxins. Basidiomycetes produce toxic sesquiterpenes, mainly as protoilludane skeleton, to protect against predators [11]. In addition, basidiomycetes often form symbiotic relationships with roots and their hosts, providing plant hormones [13,14]. For example, basidiomycetes in the genus Lactarius produce modified lactarane and protoilludane-derived sesquiterpenes that promote plant growth [15,16,17]. Sesquiterpenes isolated from basidiomycetes also exhibit pharmacological activity. For instance, hydroxymethylacylfulvene (HMAF) is a semisynthetic antitumor agent based on the naturally occurring illudin S from the mushroom Omphalotus olearius [18]. It is currently in human clinical trials because of its anti-cancer properties [19,20]. Phellinignin A and 11,12-epoxy-12β-hydroxy-1-tremulen-5-one isolated from the genus Phellinus igniarius showed high cytotoxicity to HL-60, SMMC-7721, and SW480 cancer cells [21]. 10β,12-Dihydroxy-tremulene isolated from Phellinus igniarius showed good vasodilatory activity in the experiment [9].
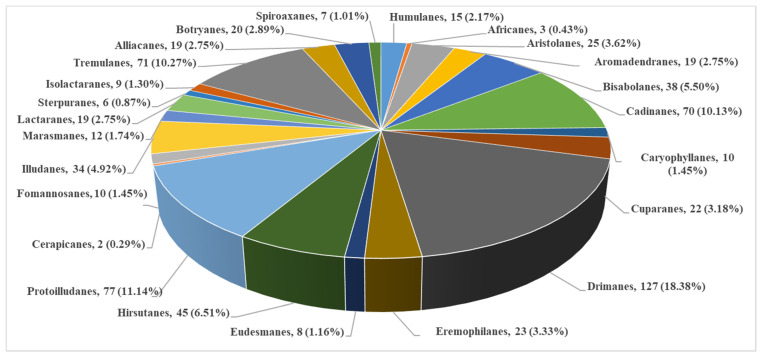
So far, approximately one thousand sesquiterpenoids have been reportedly obtained from basidiomycetes (Supplementary Table S1) [22,23]. Drimanes, protoilludanes, illudanes, hirstutanes, cadinanes, and tremulanes sesquiterpene skeletons are the main skeleton types of sesquiterpenoids in basidiomycetes, comprising approximately 60% of the total population (Figure 1, Supplementary Table S2. There are 79 genera of basidiomycetes that produce sesquiterpenes, and Lactarius, Xylaria, Armillaria, Phellinus, Granulobasidium, and Conocybe are the main sources (Supplementary Table S1). According to the reported genomic data, the average number of sesquiterpene genes per strain in basidiomycetes is 12, which is much higher than the average 3.5 sesquiterpene genes in ascomycetes [24]. These results suggest that basidiomycetes produce more sesquiterpenes. However, most of the compounds produced by these potentially functional genes are unknown and require further clarification.
Figure 1.
Basidiomycota sesquiterpenes classified by a skeleton.
2. Cyclization Mode of STSs
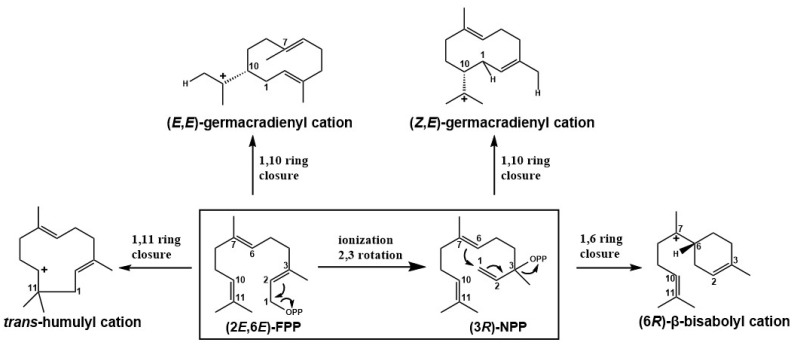
The sesquiterpene biosynthetic pathway is divided into two steps [25]. The first step is a coupling reaction that connects isoprene precursors, dimethylallyl dipyrophosphate (DMAPP) and isoprenyl dipyrophosphate (IPP), from geranyl pyrophosphate (GPP) in a head-to-tail manner, and then condenses with another molecule of IPP to generate farnesyl pyrophosphate (FPP), which is a sesquiterpenoid biosynthetic precursor and substrate for STS [26]. As the second step, FPP generates different sesquiterpene carbon skeletons through irregular coupling reactions. Typical STS contains conserved D(D/E)XXD and NSE/DTE motifs, and these amino acid residues play important roles in coordinating the stabilization of divalent metal ions at the active site for defocusing the catalytic reaction of phosphoric acid. Cyclization is initiated by the metal-ion-induced departure of inorganic pyrophosphate (PPi) to form allyl cations, facilitating the structural shift and catalyzing cyclization closure [27,28]. For cyclic sesquiterpenes, this step can be further divided into two. FPP undergoes one or more cyclizations to form intermediates, which are then converted to sesquiterpene skeletal end-products under the action of STS. The reaction mechanism is divided into four categories [29,30,31] (Figure 2 and Figure 3)—Clade I: After (2E,6E)-FPP is deionized by pyrophosphate, it electrophilically attacks the double bond at the other end and forms a 10-membered ring carbon-positive intermediate E, E-germacradienyl cation through a 1,10 cyclization reaction; Clade II: FPP is first ionized and isomerized to form (3R)-nerolidyl diphosphate ((3R)-NPP), deionized by pyrophosphate, and electrophilically attacks the double bond to form a 10-membered ring of the carbon-positive intermediate Z, E-germacradienyl cation through 1,10 cyclization; Clade III: (2E,6E)-FPP removes pyrophosphate ionization, electrophilically attacks the double bond at the other end, and undergoes 1,11 cyclization reaction 11-membered ring carbocation intermediate trans-humulyl cation; Clade IV: After (3R)-NPP is deionized by pyrophosphate, it electrophilically attacks the double bond and forms a 6-membered ring carbocation intermediate (6R)-β-bisabolol cation through a 1,6 cyclization reaction.
Figure 2.
Cyclization patterns of sesquiterpenes in basidiomycetes.
Figure 3.
Sesquiterpene biosynthesis by STS of clades I–IV in basidiomycetes. All mentioned STSs have undergone biochemical verification.
Over the past 20 years, 122 STSs and 2 fusion enzymes STSs have been discovered and identified from 26 species of basidiomycetes (Supplementary File S1), which are responsible for the biosynthesis of hundreds of sesquiterpenes in four ways. The various STSs and their catalytic production of sesquiterpenes are summarized and discussed in this review.
3. STSs in Basidiomycota
3.1. Agaricales
Agaricales is the largest mushroom-forming flora, comprising more than 400 genera and 13,000 species [32]. To date, 10 species have been experimentally identified with 58 different STSs.
3.1.1. Macrolepiota albuminosa
Macrolepiota albuminosa (Termitomyces albuminosus) is a special mushroom in China that belongs to the Agaricaceae family. Bioinformatic analysis of the genome revealed the presence of 22 terpene synthases [33], 3 of which (STC4, STC9, and STC15) were identified as STSs and heterologously expressed by Escherichia coli [34]. Using FPP as a precursor, STC4 synthesized intermedeol (1) via C1,10 cyclization, which in turn enabled germacrene D-4-ol (2) synthesization by STC15. Through C1,6 cyclization, γ-cadinene (3) was synthesized by STC9 with NPP as a substrate.
3.1.2. Coprinopsis cinerea
Coprinopsis cinerea belongs to the Psathyrellaceae family. Six STSs (Cop1–6) have been identified in this fungus [29] and heterologously expressed in Saccharomyces cerevisiae and E. coli. With FPP as the precursor, Cop1 and Cop2 synthesized germacrene A (4) by C1,10 cyclization, and Cop3 synthesized α-muurolene (5). Using NPP as the precursor, Cop4 synthesized δ-cadinene (6) by C1,10 cyclization, and Cop6 synthesized α-cuprenene (7) by C1,6 cyclization [35]. Cop5 cannot be functionally expressed in either system.
3.1.3. Taiwanofungus camphoratus
Taiwanofungus camphoratus (Antrodia cinnamomea) belongs to the mushroom family Fomitopsidaceae and is a rare medicinal fungus found in Taiwan, China. A total of 10 terpene synthases (AcTPS1–7, 9–11) have been identified in it and they are heterologously expressed in E.coli, among which three were identified as STSs (AcTPS4, AcTPS5, and AcTPS9) [31]. T-cadinol (8) was synthesized by C1,10 cyclization of AcTPS5 with FPP as a substrate, and cubebol (9) and zonarene (10) were synthesized from AcTPS9 and AcTPS4, respectively, via C1,10 cyclization with NPP as a substrate.
3.1.4. Clitopilus pseudo-pinsitus
Clitopilus pseudo-pinsitus, belonging to the family Entolomataceae, currently has 18 related STSs (CpSTS1–18) recorded [36]. Apart from the lack of conserved CpSTS10 sequences, the remaining 17 were heterologously expressed by Aspergillus oryzae, and CpSTS15 was found to be inactive. The biosynthesis of the remaining STSs can be summarized as follows: Δ6-protoilludene (11) was synthesized by CpSTS4 using FPP as a precursor and C1,11 cyclization. Sterpurene (12), pentalenene (13), and α-farnesene (14) were synthesized from CpSTS1, CpSTS6, and CpSTS7, respectively. After C1,10 cyclization, δ-cadinene (6) was synthesized from CpSTS2, aristolene (15) was synthesized from CpSTS16, and alloaromadendrene (16) and 9-alloaromadendrene (17) were synthesized from CpSTS8 and CpSTS11, respectively. CpSTS9 and CpSTS12 synthesized virifloridol (18), and CpSTS13 synthesized ledene (19) with NPP as the substrate. CpSTS3 synthesized α-muurolene (5) and δ-cadinol (20). CpSTS5 synthesized α-muurolene (5) with FPP as the substrate. Through C1,6 cyclization, using NPP as a precursor, CpSTS14 synthesized β-elemene (21) and β-farnesene (22), CpSTS17 synthesized β-caryophyllene (23), and CpSTS18 synthesized γ-cadinene (3).
3.1.5. Hypholoma fasciculare
Hypholoma fasciculare is a clustered fungus belonging to the family Strophariaceae. A total of 17 STSs have been previously identified in their genome using bioinformatic methods [37], of which 4 (Hfas94a, Hfas94b, Hfas255, and Hfas344) were heterologously expressed in A. oryzae. Using FPP as a precursor, Hfas94a and Hfas94b mainly synthesized α-humulene (24) through C1,11 cyclization. Hfas255 did not produce any products, and Hfas344 synthesized an oxidized sesquiterpene with spectral data similar to that of β-caryophyllene.
3.1.6. Hypholoma lateritium
Hypholoma lateritium (Hypholoma sublateritium) belongs to the same genus as H. fasciculare and is widely distributed in China. Only one STS (Hypsu1_138665) has been identified in this mushroom [38]. Using E. coli heterologous expression with FPP as the precursor, Hypsu1_138665 was cyclized by C1,11 cyclization to synthesize Δ6-protoilludene (11).
3.1.7. Cyclocybe aegerita
Cyclocybe aegerita (Agrocybe aegerita), also known as pioppino mushroom, is a basidiomycete belonging to the Strophariaceae family. Eleven STSs have been identified, all of which are heterologously expressed in E. coli [38]. Two of these (Agr10 and Agr11) failed to detect the product. Using NPP as the precursor, δ-cadinene (6) was synthesized from Agr1 and Agr4, and viridiflorene (25) was synthesized from Agr2 and Agr5 by C1,10 cyclization. Using FPP as a precursor, Agr3 synthesized α-muurolene (5) by C1,10 cyclization, Agr6 and Agr7 synthesized Δ6-protoilludene (11), and Agr8 synthesized γ-muurolene (26) by C1,11 cyclization. Agr9 synthesized an unknown sesquiterpene alcohol.
3.1.8. Armillaria gallica
Armillaria gallica is a saprophytic or parasitic fungus belonging to the Physalacriaceae family. Using bioinformatics analysis of its genome, 20 STSs were predicted [38], but only 1 (Pro1) was heterologously expressed in E. coli [39]. Using FPP as a precursor, Pro1 synthesized Δ6-protoilludene (11) via C1,11 cyclization.
3.1.9. Galerina marginata
Galerina marginata is a common poisonous mushroom belonging to the Hymenogastraceae family that contains amino peptides. Only one related STS (Galma_104215) has been identified and is heterologously expressed in E. coli [38]. Using NPP as the precursor, Galma_104215 synthesized β-gurjunene (27) by C1,10 cyclization.
3.1.10. Omphalotus olearius
Omphalotus olearius belongs to the family Omphalotaceae and emits green fluorescence. Ten STSs from this fungus have been identified [30]. E. coli was used for heterologous expression. With FPP as the precursor, Omp1 and Omp3 were used to synthesize α-muurolene (5) by C1,10 cyclization, and Omp5a/b was used to synthesize γ-cadinene (3); Omp6 and Omp7 synthesized Δ6-protoilludene (11) by C1,11 cyclization. With NPP as the precursor, Omp4 was used to synthesize δ-cadinene (6) by C1,10 cyclization, Omp9 synthesized α-barbatene (28), and Omp10 mainly guided the synthesis of (E)-dauca-4(11), 8-diene (29) by C1,6 cyclization. Omp8 is a homologue of Omp9/10 that lacks approximately 100 amino acids at its N terminus and was not functional when expressed in E. coli.
3.2. Polyporales
Polyporales contains approximately 1800 species of fungi, representing approximately 1.5% of all known fungal species [40]. At present, 8 species of fungi in this order have been identified to contain 39 different STSs and 1 fusion enzyme.
3.2.1. Lignosus rhinocerus
Lignosus rhinocerus (Lignosus rhinocerotis), also known as tiger milk mushroom, is a macrofungal belonging to the Polyporaceae family. Twelve terpene synthase genes have been found [41], seven of which are actively expressed in the sclerotium. Three STSs (GME3634, GME3638, and GME9210) were heterologously expressed by S. cerevisiae, producing nineteen, eight, and two sesquiterpenes, respectively (29 in total). Using FPP as the precursor, through C1,10 cyclization, GME3634 mainly synthesized α-cadinol (30), and GME3638 mainly synthesized torreyol (31). Using FPP as the precursor, GME9210 mainly synthesized 1,3,4,5,6,7-hexahydro-2,5,5-trimethyl-2H-2,4a-ethanonaphthalene (32) and 1-napthalenol (33).
3.2.2. Cerrena unicolor
Cerrena unicolor belongs to the Polyporaceae family. A total of 14 STSs have been found. Heterologous expression in E. coli was performed [42]. Four of these (Cun5765, Cun6114, Cun7487, and Cun0802) were not produced, and the loss of Cun6114, Cun7487, and Cun5765 activities might be caused by the difficulty in predicting introns [43]. The failure of Cun0802 gene cloning may be related to its low transcription level [44,45], and the product of Cun9106 could not be identified. The other 9 STSs produced 10 different sesquiterpenes. Using NPP as the precursor, β-cubebene (34) was synthesized by Cun3157, and δ-cadinene (6) was synthesized by Cun3158. Using FPP as a precursor, δ-cadinol (20) was synthesized by Cun7050, α-copaene (35) was synthesized by Cun3574, α-muurolene (5) was synthesized by Cun0759, γ-cadinene (3) was synthesized by Cun3817, and germacrene D (36) was synthesized by Cun0773. Aromadendrene (37) was synthesized by C1,11 cyclization of Cun5155 with FPP as the precursor; δ-cadinol (20) was synthesized by C1,6 cyclization from Cun0716 with NPP as the precursor.
3.2.3. Rhodonia placenta
Rhodonia placenta (Postia placenta), formerly known as brown rot fungus, is a common diseased wood-rot fungus belonging to the Polyporaceae family that can grow a large area of mycelium. It is known that 6 STSs have been isolated from it; they were heterologously expressed by S. cerevisiae. A total of 25 different sesquiterpenoids were synthesized with FPP or NPP precursors [46]. Using FPP as a precursor, through C1,10 cyclization, PpSTS01 successfully synthesized α-muurolene (5), δ-cadinene (6), and β-elemene (21), PpSTS03 synthesized α-cadinene (38) and γ-cadinene (3), and PpSTS06 synthesized α-gurjunene (39); Δ6-protoilludene (11) and pentalenene (13) were synthesized from PpSTS08 and PpSTS14, respectively, by C1,11 cyclization. δ-Cadinene (6) was synthesized from PpSTS10 by C1,10 cyclization.
3.2.4. Fomitopsis pinicola
Fomitopsis pinicola is a brown rot basidiomycete species belonging to the family Fomitopsidaceae, commonly collected from dead conifer trees. One STS (Fompi1) was identified and heterologously expressed in E. coli [30]. α-Cuprenene (7) was synthesized through C1,6 cyclization by Fompi1 with NPP as the precursor.
3.2.5. Ganoderma lucidum
Ganoderma lucidum, which belongs to the Ganodermataceae family, is a well-known medicinal fungus. However, only two STSs, GL26009 [47] and GISTS6 [48], have been isolated and identified from this fungus and expressed heterologously in E. coli. Using FPP as the precursor, GL26009 synthesized γ-muurolene (26) and α-muurolene (5), and GISTS6 synthesized γ-cadinene (3).
3.2.6. Ganoderma sinense
Ganoderma sinense is a medicinal fungus belonging to the same genus as G. lucidum in the Ganodermataceae family. At present, six STSs have been isolated and identified from this fungus and expressed in E. coli. (GS11330, GS14272, GS02363, GsSTS43, GsSTS45a, and GsSTS45b) [48,49,50]. GsSTS45a has no function; GS02363 synthesized α-cadinol (30), δ-cadinene (6), α-muurolene (5), and γ-muurolene (26); GS11330 synthesized α-cuprenene (7); GS14272 synthesized α-muurolene (5); and GsSTS43 and GsSTS45b synthesized γ-cadinene (3).
3.2.7. Phanerodontia chrysosporium
Phanerodontia chrysosporium (Phanerochaete chrysosporium) belongs to the family Phanerochaetaceae. Eleven STSs have been recorded [51], of which seven were heterologously expressed in S. cerevisiae and cultured in SDL medium. PcSTS01 synthesized γ-muurolene (26), α-muurolene (5), and δ-cadinene (6). PcSTS02 and PcSTS04 synthesized β-copaene (40),β-farnesene (22), cadina-1(6),4-diene (41), and δ-cadinene (6). Epicubenol (42) was synthesized from PcSTS03. PcSTS06 synthesized α-barbatene (28) and β-barbatene (43). (E)-α-Bisabolene (44) was synthesized from PcSTS08, and PcSTS11 synthesized α-santalene (45).
3.2.8. Steccherinum ochraceum
Steccherinum ochraceum belongs to the family Meruliaceae. Six STSs were deduced from its genome, of which fusion enzyme A8411 was heterologously expressed in A. oryzae [52]. Hirsutene (46) was synthesized from A8411.
3.3. Russulales
Russulales comprises approximately 1767 species belonging to 80 genera and 12 families [1]. Two fungi of this order have been verified to contain fifteen different STSs and one fusion enzyme.
3.3.1. Stereum hirsutum
Stereum hirsutum belongs to the family Stereaceae. Nearly 50 related sesquiterpenoids have been found [53] as well as 18 STSs (ShSTS1-18, of which ShSTS2, 6, 9, 14 have not been studied due to their high homology to other genes) and 1 fusion protein (HS-HMGS), which were functionally verified by heterologous expression in E. coli or A. oryzae [36,38,54,55]. Their synthetic routes are summarized as follows: using NPP as a substrate, via C1,10 cyclization, ShSTS10 and ShSTS11 can synthesize δ-cadinene (6), ShSTS8 can synthesize 1-epi-cubenol (47), ShSTS12 can synthesize α-cubebene (48), and ShSTS10 can synthesize germacrene D (36). Simultaneously, ShSTS1 synthesized β-barbatene (43), ShSTS3 synthesized α-farnesene (14) and β-farnesene (22), ShSTS4 synthesized hirsutene (46), and ShSTS5 synthesized γ-cadinene (3) by C1,6 cyclization. Using FPP as a substrate, through C1,11 cyclization, ShSTS13 can synthesize β-caryophyllene (23), HS-HMGS can synthesize hirsutene (46), and ShSTS15, ShSTS16, ShSTS17, and ShSTS18 can synthesize Δ6-protoilludene (11); ShSTS7 synthesized δ-cadinene (6) via C1,10 cyclization.
3.3.2. Heterobasidion annosum
Heterobasidion annosum belongs to the Bondarzewiaceae family of the order Russulales. It is a forest pathogen that grows on large, perennial basidiocarps. Only one STS (Hetan2_454193) was identified in this fungus [40] and was heterologously expressed in E. coli. Using FPP as the precursor, Hetan2_454193 synthesized Δ6-protoilludene (11) by C1,11 cyclization.
3.4. Other Basidiomycota
The basidiomycetes in this region cannot be classified by order. There are 6 species of fungi in this part, and 10 different STSs have been verified by experiments.
3.4.1. Boreostereum vibrans
Boreostereum vibrans, originally named Stereum vibrans, is a macrofungus belonging to the Gloeophyllaceae family of the order Gloeophyllales. Many sesquiterpenes have been isolated from it [56]. BvCS is heterologously expression in E. coli [57]. δ-Cadinol (20) was synthesized by C1,10 cyclization of BvCS with FPP as a precursor.
3.4.2. Sphaerobolus stellatus
Sphaerobolus stellatus belongs to the order Geastrales and family Geastraceae. One STS (Sphst_47084) has been identified in this fungus [38] and heterologously expressed in E. coli. Using NPP as the precursor, viridiflorol (49) was synthesized via C1,10 cyclization of Sphst_47084.
3.4.3. Sanghuangporus baumii
Sanghuangporus baumii belongs to the Hymenochaetaceae family of the order Hymenochaetales and is an important medicinal fungus. Only one STS has been isolated from this species and heterologously expressed by E. coli, named SbTps1 [58].
3.4.4. Coniophora puteana
Coniophora puteana belongs to the Coniophoraceae family within the order Boletales. Four STSs have been isolated from it (Copu2, 3, 5 and 9) [59,60]. β-Copaene (40) and cubebol (9) were synthesized by C1,10 cyclization from Copu2 and Copu3 with NPP as the precursor. Using FPP as the precursor, Copu5 and Copu9 synthesized δ-cadinol (20) through C1,10 cyclization.
3.4.5. Serendipita indica
Serendipita indica is an endophytic root-colonizing species belonging to the order Sebacinales and family Serendipitaceae. One STS has been recorded [61], which is hetero-expressed in E. coli. Viridiflorol (49) was synthesized by C1,10 cyclization from SiTPS, using FPP as the precursor.
3.4.6. Dendrodontia bispora
Dendrodontia bispora (Dendrothele bispora) is a basidiomycete species belonging to the Corticiaceae family in Corticiales order. Two STSs (Denbi1_659367 and Denbi1_816208) have been isolated from this fungus [38] and hetero-expressed in E. coli. Δ6-protoilludene (11) was synthesized by C1,11 cyclization of Denbi1_659367. Viridiflorol (49) was synthesized by Denbi1_816208.
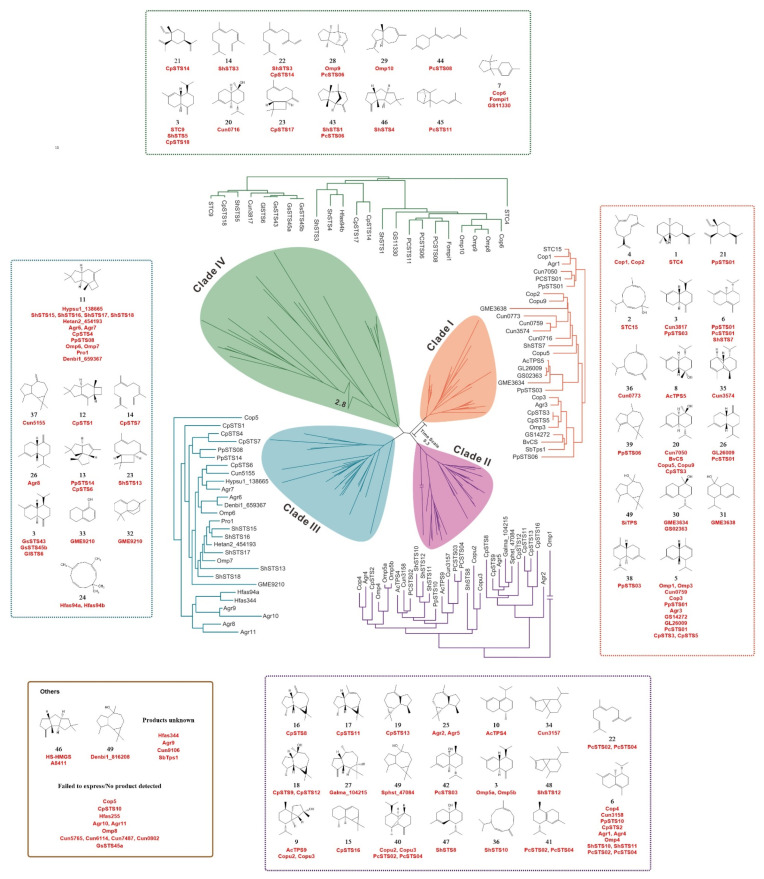
The taxonomic data in the above content come from GBIF (Global Biodiversity Information Facility, https://www.gbif.org/, accessed on 28 June 2022). A summary of information on sesquiterpene biosynthesis in Basidiomycota is presented in Table 1.
Table 1.
Classification of STSs from Basidiomycota.
| Type of Cyclization |
Precursor | Metabolite | Producer | Biochemically Verified Enzyme | |
|---|---|---|---|---|---|
| Clade I | C1,10 | FPP | Intermedeol (1) | Macrolepiota albuminosa | STC4 |
| Germacrene D-4-ol (2) | Macrolepiota albumi-nosa | STC15 | |||
| Germacrene A (4) | Coprinopsis cinerea | Cop1, Cop2 | |||
| T-Cadinol (8) | Taiwanofungus camphoratus | AcTPS5 | |||
| α-Muurolene (5) | Omphalotus olearius | Omp1, Omp3 | |||
| Cerrena unicolor | Cun0759 | ||||
| Coprinopsis cinerea | Cop3 | ||||
| Rhodonia placenta | PpSTS01 | ||||
| Cyclocybe aegerita | Agr3 | ||||
| Ganoderma sinense | GS14272 | ||||
| Ganoderma lucidum | GL26009 | ||||
| Phanerodontia chrysosporium | PcSTS01 | ||||
| Clitopilus pseudo-pinsitus | CpSTS3, CpSTS5 | ||||
| α-Cadinol (30) |
Lignosus rhinoceros
Ganoderma sinense |
GME3634
GS02363 |
|||
| Torreyol (31) | Lignosus rhinocerus | GME3638 | |||
| δ-Cadinol (20) | Cerrena unicolor | Cun7050 | |||
| Boreostereum vibrans | BvCS | ||||
| Coniophora puteana | Copu5, Copu9 | ||||
| Clitopilus pseudo-pinsitus | CpSTS3 | ||||
| α-Copaene (35) | Cerrena unicolor | Cun3574 | |||
| γ-Cadinene (3) | Cerrena unicolor | Cun3817 | |||
| Rhodonia placenta | PpSTS03 | ||||
| Germacrene D (36) | Cerrena unicolor | Cun0773 | |||
| δ-Cadinene (6) | Rhodonia placenta | PpSTS01 | |||
| Phanerodontia chrysosporium | PcSTS01 | ||||
| Stereum hirsutum | ShSTS7 | ||||
| β-Elemene (21) | Rhodonia placenta | PpSTS01 | |||
| α-Cadinene (38) | Rhodonia placenta | PpSTS03 | |||
| α-Gurjunene (39) Viridiflorol (49) γ-Muurolene (26) |
Rhodonia placenta
Serendipita indica Ganoderma lucidum Phanerodontia chrysosporium |
PpSTS06
SiTPS GL26009 PcSTS01 |
|||
| Clade II | C1,10 | NPP | δ-Cadinene (6) | Coprinopsis cinerea | Cop4 |
| Cerrena unicolor | Cun3158 | ||||
| Rhodonia placenta | PpSTS10 | ||||
| Clitopilus pseudo-pinsitus | CpSTS2 | ||||
| Cyclocybe aegerita | Agr1, Agr4 | ||||
| Omphalotus olearius | Omp4 | ||||
|
Stereum hirsutum
Phanerodontia chrysosporium |
ShSTS10, ShSTS11
PcSTS02, PcSTS04 |
||||
| Cubebol (9) |
Taiwanofungus camphoratus
Coniophora puteana |
AcTPS9
Copu2, Copu3 |
|||
| 1-epi-Cubenol (47) | Stereum hirsutum | ShSTS8 | |||
| Zonarene (10) | Taiwanofungus camphoratus | AcTPS4 | |||
| Ledene (19) | Clitopilus pseudo-pinsitus | CpSTS13 | |||
| Virifloridol (18) | Clitopilus pseudo-pinsitus | CpSTS9, CpSTS12 | |||
| Viridiflorol (49) | Sphaerobolus stellatus | Sphst_47084 | |||
| Alloaromadendrene (16) | Clitopilus pseudo-pinsitus | CpSTS8 | |||
| 9-Alloaromadendrene (17) | Clitopilus pseudo-pinsitus | CpSTS11 | |||
| Aristolene (15) | Clitopilus pseudo-pinsitus | CpSTS16 | |||
| Viridiflorene (25) | Cyclocybe aegerita | Agr2, Agr5 | |||
| γ-Cadinene (3) | Omphalotus olearius | Omp5a, Omp5b | |||
| α-Cubebene (48) | Stereum hirsutum | ShSTS12 | |||
| β-Cubebene (34) | Cerrena unicolor | Cun3157 | |||
| Germacrene D (36) | Stereum hirsutum | ShSTS10 | |||
| β-Copaene (40) |
Coniophora puteana
Phanerodontia chrysosporium |
Copu2, Copu3
PcSTS02, PcSTS04 |
|||
| β-Gurjunene (27) Epicubenol (42) β-Farnesene (22) Cadina-1(6),4-diene (41) |
Galerina marginata
Phanerodontia chrysosporium Phanerodontia chrysosporium Phanerodontia chrysosporium |
Galma_104215
PcSTS03 PcSTS02, PcSTS04 PcSTS02, PcSTS04 |
|||
| Clade III | C1,11 | FPP | α-Humulene (24) | Hypholoma fasciculare | Hfas94a, Hfas94b |
| Δ6-Protoilludene (11) | Hypholoma lateritium | Hypsu1_138665 | |||
| Stereum hirsutum | ShSTS15, ShSTS16, ShSTS17, ShSTS18 | ||||
| Heterobasidion annosum | Hetan2_454193 | ||||
| Cyclocybe aegerita | Agr6, Agr7 | ||||
| Clitopilus pseudo-pinsitus | CpSTS4 | ||||
| Rhodonia placenta | PpSTS08 | ||||
| Omphalotus olearius | Omp6, Omp7 | ||||
| Armillaria gallica | Pro1 | ||||
| Dendrodontia bispora | Denbi1_659367 | ||||
| Aromadendrene (37) | Cerrena unicolor | Cun5155 | |||
| β-Caryophyllene (23) | Stereum hirsutum | ShSTS13 | |||
| Pentalenene (13) | Rhodonia placenta | PpSTS14 | |||
| Clitopilus pseudo-pinsitus | CpSTS6 | ||||
| Sterpurene (12) | Clitopilus pseudo-pinsitus | CpSTS1 | |||
| α-Farnesene (14) | Clitopilus pseudo-pinsitus | CpSTS7 | |||
| γ-Muurolene (26) | Cyclocybe aegerita | Agr8 | |||
| 1,3,4,5,6,7-Hexahydro-2,5,5-trimethyl-2H-2,4a-ethanonaphthalene (32) | Lignosus rhinocerus | GME9210 | |||
| 1-Napthalenol (33) | Lignosus rhinocerus | GME9210 | |||
| γ-Cadinene (3) | Ganoderma sinense | GsSTS43, GsSTS45b | |||
| Ganoderma lucidum | GISTS6 | ||||
| Clade IV | C1,6 | NPP | α-Cuprenene (7) |
Coprinopsis cinerea
Ganoderma sinense Fomitopsis pinicola |
Cop6
GS11330 Fompi1 |
| α-Barbatene (28) |
Omphalotus olearius
Phanerodontia chrysosporium |
Omp9
PcSTS06 |
|||
| β-Barbatene (43) | Phanerodontia chrysosporium | PcSTS06 | |||
| Stereum hirsutum | ShSTS1 | ||||
| α-Farnesene (14) | Stereum hirsutum | ShSTS3 | |||
| β-Farnesene (22) |
Stereum hirsutum
Clitopilus pseudo-pinsitus |
ShSTS3
CpSTS14 |
|||
| Hirsutene (46) | Stereum hirsutum | ShSTS4 | |||
| γ-Cadinene (3) |
Termitomyces
albuminosus |
STC9 | |||
| Stereum hirsutum | ShSTS5 | ||||
| Clitopilus pseudo-pinsitus | CpSTS18 | ||||
| β-Elemene (21) | Clitopilus pseudo-pinsitus | CpSTS14 | |||
| β-Caryophyllene (23) | Clitopilus pseudo-pinsitus | CpSTS17 | |||
| (E)-Dauca-4(11),8-diene (29) | Omphalotus olearius | Omp10 | |||
| δ-Cadinol (20) | Cerrena unicolor | Cun0716 | |||
| (E)-α-Bisabolene (44) | Phanerodontia chrysosporium | PcSTS08 | |||
| α-Santalene (45) | Phanerodontia chrysosporium | PcSTS11 | |||
| others | — | Coprinopsis cinerea | Cop5 | ||
| — | Clitopilus pseudo-pinsitus | CpSTS10 | |||
| — | Hypholoma fasciculare | Hfas255 | |||
| Unknown | Hypholoma fasciculare | Hfas344 | |||
| Unknown | Cyclocybe aegerita | Agr9 | |||
| — | Cyclocybe aegerita | Agr10, Agr11 | |||
| — | Omphalotus olearius | Omp8 | |||
| — | Cerrena unicolor | Cun5765, Cun6114, Cun7487, Cun0802 | |||
| Unable to identify | Cerrena unicolor | Cun9106 | |||
| — | Ganoderma sinense | GsSTS45a | |||
| Unknown | Sanghuangporus baumii | SbTps1 | |||
| Viridiflorol (49) | Dendrodontia bispora | Denbi1_816208 | |||
| Hirsutene (46) | Stereum hirsutum | HS-HMGS | |||
| Steccherinum ochraceum | A8411 |
4. Research Process and Tools for STSs in Basidiomycetes
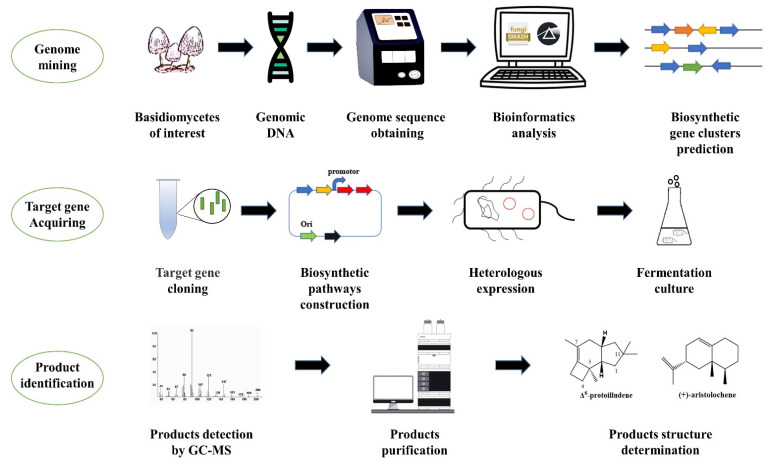
At present, the research process of basidiomycete STSs is mainly divided into three parts (Figure 4), and the latest research tools are developed around the core steps of these three parts (genome sequencing, basidiomycete culture methods, etc.).
Figure 4.
Technical research route of STS in basidiomycetes. The STS genome mining of basidiomycetes can be divided into three steps. The first step of genome mining is mainly based on the whole gene sequence, using bioinformatics tools to mine and predict the biosynthetic gene cluster. The second step is to obtain the target gene from the biosynthetic gene cluster of STS, and then heterologously express the target gene to obtain the product. The final step is to purify and identify the product to determine whether the gene is an STS.
4.1. Long-Read Whole-Genome Sequencing
The average genome size of basidiomycetes and ascomycetes is 46 Mb and 37 Mb, respectively [62], and the genome size of archaea and bacteria is usually within 6 Mb [63]. This means that bacterial genomes can be sequenced using short-read sequencing, but long-read sequencing and proper assembly are required for fungal genomes. Inexpensive nanopore sequencing [64] has been applied to whole-genome sequencing of basidiomycetes. For example, nanopore sequencing technology was used for whole-genome sequencing of the basidiomycete Clathrus columnatus and Inonotus obliquus, and genome assembly was completed [65,66].
Although nanopore technology enables long-read sequencing and is inexpensive, the accuracy of base calling is 85–94% depending on the sequencing method [67]. However, if nanopore long-read sequencing is used in combination with short-read sequencing, it may be possible to assemble a higher quality genome, if the software that assembles the genome has this capability. This functionality is currently available for bacterial and fungi genomes, and the Pilon software can combine Illumina and Nanopore sequence data to polish assemblies [68].
4.2. Basidiomycetes Cultures
Many basidiomycetes have high requirements for their growth environment; they can only grow under specific conditions, and most of them cannot be cultivated artificially, which greatly limits isolation and identification of sesquiterpenoids in basidiomycetes. Fungal growth can be simply divided into two stages: (i) germination of fungal spores, and (ii) subsequent filamentous growth, forming a network of hyphae called mycelium [69].
The conditions that induce or inhibit the germination of fungal spores have always puzzled researchers. So far, the main research directions are growth factor induction, activator induction, co-culture, volatile organic compound induction, and physical factors [70]. Taking Agaricomycetes ectomycorrhizal (EcM) mushrooms as an example, the M factor (growth-promoting metabolites in addition to b vitamins and amino acids are essential for the growth of tree mycorrhizal fungi), as a growth factor, promotes its growth [71]. Placing EcM mushrooms together with specific tree seedlings on lipid or gel medium can also promote spore germination, although this approach often fails [71,72]. There is evidence that EcM mushroom spore germination can also be promoted when co-cultured with bacteria [73].
The growth factors that induce the growth of fungal hyphae are mostly root exudates. Studies on EcM fungi have shown that in addition to M factor, palmitic acid, stearic acid, and cytokinins, such as kinetin, zeatin, and isopentenyl aminopurine, are also growth-promoting factors. Root exudates can induce their growth [74].
At the same time, fungal gene expression is very complex and is affected by RNAi silencing [75] and trans-acting elements of genome structure [76]. Therefore, many biosynthetic gene clusters are silent. However, by adjusting the culture conditions (changing the physical conditions of the culture, adding compounds, growth factors, etc.) [70], utilizing co-culture [77], and chromatin-based transcriptional regulation [78], silenced biosynthetic gene clusters can be activated. Studies on these operations are still in preliminary stages. However, the metabolites produced by basidiomycete fungi in different growth cycles are different [79], and many genetic regulators that control fungal development also control the production of secondary metabolites [80,81]. Studies on the basidiomycetes Coprinopsis cinerea [82] and Lentinula edodes [83] have shown that gene expression differs at the developmental stages of fruiting bodies, limiting the mining of active ingredients.
Cultivation technology for basidiomycetes has always been inadequate, but in recent years, the development of new laboratory-level cultivation techniques has brought new opportunities for artificial cultivation. For example, basidiomycetes are cultivated using microfluidic culture technology [84].
4.3. Exogenous Expression Platforms and Bioinformatics Tools for Basidiomycetes STSs
Due to the complexity of basidiomycetes genes, traditional heterologous expression platforms cannot meet the functional identification of basidiomycetes STS. Although E. coli can express STS genes, it lacks a post-translational modification system to express complex proteins and entire biosynthetic pathways, and the eukaryotic expression system in yeast cannot remove introns of fungal genes [85]. At present, A. oryzae is a relatively successful heterologous expression platform, which can more accurately splice the intron of the basidiomycetes terpenoid synthase gene [37] and correctly express the entire gene cluster [86]. Ustilago maydis has also been developed as a heterologous expression platform for the production of terpenoids [87]. It offers the advantage of metabolic compatibility and potential tolerance of substances toxic to other microorganisms.
Successful characterization of the biosynthesis of basidiomycetes products requires not only genetic engineering and heterologous expression, but also metabolic analysis [88]. Bioactivity-guided methods for isolating metabolites have been gradually replaced by more sensitive methods, such as tandem mass spectrometry (MS/MS), for untargeted metabolomics data analysis, resulting in data that can be compared with known spectral databases. Researchers can also identify unknown metabolites and intermediates through the Global Natural Products Social Molecular Networking (GNPS) [88,89] and infer biosynthetic pathways. New technologies and tools are also being developed to assist in the identification of STSs in basidiomycetes. Bioinformatics techniques can be used to establish a general prediction framework for STS and to improve the accuracy of genome-based tools for predicting biosynthetic gene clusters [58], such as antiSMASH [90] and PRISM [91]. Although it can predict the monomer sequences assembled into PKS and NRPS biosynthetic lines based on module specificity, the accuracy and specificity need to be further improved, which is also the key to identifying STS [91].
5. Discussion
Sesquiterpenes play a very important role in basidiomycetes. They can attract insects for pollination [92], defend against other organisms or parasites [93], and play an important role in basidiomycete’s physiological effect. Moreover, when basidiomycetes form a symbiotic relationship with plants, these sesquiterpenoids produced by basidiomycetes can act as phytohormones [13,14]. Therefore, basidiomycetes produce many kinds of sesquiterpenes to help them better adapt to the living environment. The influence of the ambient environment on the production of sesquiterpenes by basidiomycetes includes external physical factors (light, temperature, etc.) and chemical factors (exogenous chemical substances, etc.). The changes of sesquiterpenes during basidiomycete development and their biological roles are still unclear. At present, there are studies on the changes of sesquiterpenes during the development of the fruiting bodies of the Cyclocybe aegerita AAE-3 strain. In particular, the development of the fruiting body changes resulted in greater changes during the sporulation process. In the early stage of sporulation, mainly alcohols and ketones appeared, while in the later stage of sporulation, sesquiterpenes such as Δ6-protoilludene (11), α-cubebene (48) and δ-cadinene (6) appeared. After sporulation, sesquiterpenoids decreased and other compounds appeared, mainly octan-3-one [94].
Site-specific mutations are tools to study enzyme structure, function, and catalytic mechanism, and they include single and combinatorial mutations [95]. In the study of sesquiterpene synthases of basidiomycetes, the point mutations at residues near the conserved region are mostly used. The current point mutation experiments for sesquiterpene synthase of basidiomycetes are concentrated near the conserved regions of the RY Pair and NSE Triad (Figure 5).
Figure 5.
(a) Comparison of the conserved regions of H-α1 loop and NSE/DTE motif. (b) Comparison of conserved regions of the RY Pair to the Thirteen Positions Upstream of the RY Pair. The MUSCLE algorithm was used to compare the following protein sequences: Cop3 (XP_001832925), Cop4 (XP_001836356), Cop6 (XP_001832549) in Coprinopsis cinerea; STC4 (KAH0582448), STC9 (KAH0583476), STC15 (KAG5341349) in Macrolepiota albuminosa.
Cop3, Cop4, and Cop6 were experimentally point mutated at the sites of their H-α1 loops, respectively. K233 in Cop4 and K251 in Cop3 did not play a major role in the side chain and ligand interaction network formed during active-site closure; the mutations in Cop4 (K233, H235, T236, N238, and N239) showed that the mutations in the H-α1 loop region site significantly altered the type of product, with the mutation of N239L having the greatest effect on the product; the mutation of Cop6 (C236, E237, and N240) showed that the mutation of the H-α1 loop region site did not alter the product of Cop6. Structural modeling of the Cop enzyme pointed to a potential interaction between the H-α1 loop and the conserved residues of the two metal-binding motifs (DDXXDD and NSE/DTE). Potential interactions between the conserved Asp/Glu and the Arg, Asn and Lys sites in some sesquiterpene synthases in several fungi and plants may stabilize the closed enzyme conformation by closing the H-α1 loop [96]. STC4 was transformed into germacrene A (4) synthase after the single site W335F mutation; various variants of the W314 point mutation in STC15 were unable to obtain expressed protein, and enzyme activity was reduced after the mutation of the putative C311 active site in STC9 [34]. A triple mutant Cop2(17H2) was obtained by error-prone PCR. Cop2(17H2) contains three mutations in L59H, T65A and S310Y, and the three mutations tend to make Cop2(17H2) products be specific. Moreover, compared to the original Cop2, Cop2(17H2) is more inclined to produce Germacrene D-4-ol (2) [97].
6. Conclusions
A comparison of the reported genome sequences revealed that each basidiomycete contained, on average, more than 12 STSs. Although the reasons for the existence of many STSs are unclear, it is speculated that they are closely related to their biological activities. The development of molecular tools for basidiomycetes research will allow researchers to further explore these microbial taxa. These efforts have definitely resulted in a global push for the discovery and characterization of fungal STSs, and they provide hope for the future of fungal sesquiterpenoid discovery.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8090913/s1, Table S1: Sesquiterpene skeleton and production source of basidiomycetes from different species; Table S2: Basidiomycetes sesquiterpenoid skeleton type and the number of compounds; File S1: Gene name and amino acid sequence of sesquiterpene synthase from basidiomycetes.
Author Contributions
Data curation, J.W., X.Y., Y.D., J.Q. and C.L.; writing original draft, J.W. and X.Y.; discussion of the contents, P.W. and J.Q.; writing—review and editing, J.-M.G. and C.L.; proofreading and approval of the manuscript, C.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was funded by the Innovation and Development Joint Fund of the Natural Science Foundation of Shandong Province (Project No. ZR2021LSW022), the Shaanxi Key Laboratory of Natural Products & Chemical Biology Open Foundation (Project No. SXBPCB 2021001), and the Fundamental Research Funds for the Central Universities (Project No. 2572022BD03).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Blackwell M. The Fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 2.Halbwachs H., Harper C.J., Krings M. Encyclopedia of Mycology. Elsevier; Amsterdam, The Netherlands: 2021. Fossil Ascomycota and Basidiomycota, with Notes on Fossil Lichens and Nematophytes; pp. 378–395. [Google Scholar]
- 3.Yang Y.-L., Yu W.-W., Li Z.-H., Liu J.-K., Feng T. Antrodiellins A–C, triquinane sesquiterpenoids from fungus Antrodiella albocinnamomea with their antibacterial activity. Phytochem. Lett. 2021;47:24–27. doi: 10.1016/j.phytol.2021.11.001. [DOI] [Google Scholar]
- 4.Zhu J., Xiong P., Li Z., Li J., Lin L., Fu X., Huang Y., Xiong Y., Li C. Antifungal sesquiterpenes with post-harvest anthracnose control effect on bananas from the fungus Fusarium lateritium. Nat. Prod. Res. 2021;36:1245–1252. doi: 10.1080/14786419.2021.1872569. [DOI] [PubMed] [Google Scholar]
- 5.Bunbamrung N., Intaraudom C., Dramae A., Boonyuen N., Veeranondha S., Rachtawee P., Pittayakhajonwut P. Antimicrobial activity of illudalane and alliacane sesquiterpenes from the mushroom Gloeostereum incarnatum BCC41461. Phytochem. Lett. 2017;20:274–281. doi: 10.1016/j.phytol.2017.05.017. [DOI] [Google Scholar]
- 6.Niu S., Yang L., Zhang G., Chen T., Hong B., Pei S., Shao Z. Phenolic bisabolane and cuparene sesquiterpenoids with anti-inflammatory activities from the deep-sea-derived Aspergillus sydowii MCCC 3A00324 fungus. Bioorg. Chem. 2020;105:104420. doi: 10.1016/j.bioorg.2020.104420. [DOI] [PubMed] [Google Scholar]
- 7.Lin L.-B., Xiao J., Gao Y.-Q., Zhang Q., Han R., Qi J.-Z., Han W.-B., Xu B., Gao J.-M. Trinor- and tetranor-eremophilane sesquiterpenoids with anti-neuroinflammatory activity from cultures of the fungus Septoria rudbeckiae. Phytochemistry. 2021;183:112642. doi: 10.1016/j.phytochem.2020.112642. [DOI] [PubMed] [Google Scholar]
- 8.Hegazy M.-E.F., El-Beih A.A., Hamed A.R., El Aty A.A.A., Mohamed N.S., Paré P.W. 3-Oxo-γ-costic acid fungal-transformation generates eudesmane sesquiterpenes with in vitro tumor-inhibitory activity. Bioorg. Med. Chem. Lett. 2017;27:3825–3828. doi: 10.1016/j.bmcl.2017.06.057. [DOI] [PubMed] [Google Scholar]
- 9.Yin R.-H., Zhao Z.-Z., Chen H.-P., Yin X., Ji X., Dong Z.-J., Li Z.-H., Feng T., Liu J.-K. Tremulane sesquiterpenes from cultures of the fungus Phellinus igniarius and their vascular-relaxing activities. Phytochem. Lett. 2014;10:300–303. doi: 10.1016/j.phytol.2014.10.019. [DOI] [Google Scholar]
- 10.Qi Q.-Y., Ren J.-W., Sun L.-W., He L.-W., Bao L., Yue W., Sun Q.-M., Yao Y.-J., Yin W.-B., Liu H.-W. Stucturally Diverse Sesquiterpenes Produced by a Chinese Tibet Fungus Stereum hirsutum and Their Cytotoxic and Immunosuppressant Activities. Org. Lett. 2015;17:3098–3101. doi: 10.1021/acs.orglett.5b01356. [DOI] [PubMed] [Google Scholar]
- 11.Abraham W.-R. Bioactive Sesquiterpenes Produced by Fungi are they Useful for Humans as Well. Curr. Med. Chem. 2001;8:583–606. doi: 10.2174/0929867013373147. [DOI] [PubMed] [Google Scholar]
- 12.Rohlfs M., Churchill A.C.L. Fungal secondary metabolites as modulators of interactions with insects and other arthropods. Fungal Genet. Biol. 2011;48:23–34. doi: 10.1016/j.fgb.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Plett J.M., Martin F. Blurred boundaries: Lifestyle lessons from ectomycorrhizal fungal genomes. Trends Genet. 2011;27:14–22. doi: 10.1016/j.tig.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Wu J., Kawagishi H. Plant growth regulators from mushrooms. J. Antibiot. 2020;73:657–665. doi: 10.1038/s41429-020-0352-z. [DOI] [PubMed] [Google Scholar]
- 15.Kashiwabara M., Kamo T., Makabe H., Shibata H., Hirota M. Repraesentins D, E and F, New Plant Growth Promoters from Lactarius repraesentaneus. Biosci. Biotechnol. Biochem. 2006;70:1502–1505. doi: 10.1271/bbb.50639. [DOI] [PubMed] [Google Scholar]
- 16.Kamo T., Matsue M., Kashiwabara M., Hirota M. 1,2-Dehydrolactarolide A, a New Plant Growth Regulatory Lactarane Sesquiterpene from Lactarius vellereus. Biosci. Biotechnol. Biochem. 2006;70:2307–2309. doi: 10.1271/bbb.60148. [DOI] [PubMed] [Google Scholar]
- 17.Hirota M., Shimizu Y., Kamo T., Makabe H., Shibata H. New Plant Growth Promoters, Repraesentins A, B and C, from Lactarius repraesentaneus. Biosci. Biotechnol. Biochem. 2003;67:1597–1600. doi: 10.1271/bbb.67.1597. [DOI] [PubMed] [Google Scholar]
- 18.Del Val A.G., Platas G., Arenal F., Orihuela J.C., Garcia M., Hernandez P., Royo I., De Pedro N., Silver L.L., Young K., et al. Novel illudins from Coprinopsis episcopalis (syn. Coprinus episcopalis), and the distribution of illudin-like compounds among filamentous fungi. Mycol. Res. 2003;107:1201–1209. doi: 10.1017/S0953756203008487. [DOI] [PubMed] [Google Scholar]
- 19.Alexandre J., Raymond E., Kaci M.O., Brain E.C., Lokiec F., Kahatt C., Faivre S., Yovine A., Goldwasser F., Smith S.L., et al. Phase I and Pharmacokinetic Study of Irofulven Administered Weekly or Biweekly in Advanced Solid Tumor Patients. Clin. Cancer Res. 2004;10:3377–3385. doi: 10.1158/1078-0432.CCR-03-0349. [DOI] [PubMed] [Google Scholar]
- 20.Tanasova M., Sturla S.J. Chemistry and Biology of Acylfulvenes: Sesquiterpene-Derived Antitumor Agents. Chem. Rev. 2012;112:3578–3610. doi: 10.1021/cr2001367. [DOI] [PubMed] [Google Scholar]
- 21.Wu P.-F., Ding R., Tan R., Liu J., Hu E.-M., Li C.-Y., Liang G.-Y., Yi P. Sesquiterpenes from cultures of the fungus Phellinus igniarius and their Cytotoxicities. Fitoterapia. 2020;140:104415. doi: 10.1016/j.fitote.2019.104415. [DOI] [PubMed] [Google Scholar]
- 22.Chen H.-P., Liu J.-K. Secondary Metabolites from Higher Fungi. In: Kinghorn A.D., Falk H., Gibbons S., Kobayashi J., editors. Progress in the Chemistry of Organic Natural Products 106. Volume 106. Springer International Publishing; Cham, Switzerland: 2017. pp. 1–201. [DOI] [PubMed] [Google Scholar]
- 23.Dai Q., Zhang F.-L., Feng T. Sesquiterpenoids Specially Produced by Fungi: Structures, Biological Activities, Chemical and Biosynthesis (2015–2020) J. Fungi. 2021;7:1026. doi: 10.3390/jof7121026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt-Dannert C. Biosynthesis of Terpenoid Natural Products in Fungi. In: Schrader J., Bohlmann J., editors. Biotechnology of Isoprenoids. Volume 148. Springer International Publishing; Cham, Switzerland: 2014. pp. 19–61. Advances in Biochemical Engineering/Biotechnology. [Google Scholar]
- 25.Shao Y.-Z., Li Y.-T., Gong T., Zhu P., Yu S.-S. Research advances in methods of cyclezation mechanism of sesquiterpenes. China J. Chin. Mater. Med. 2021;46:3797–3805. doi: 10.19540/j.cnki.cjcmm.20210416.601. [DOI] [PubMed] [Google Scholar]
- 26.Christianson D.W. Structural and Chemical Biology of Terpenoid Cyclases. Chem. Rev. 2017;117:11570–11648. doi: 10.1021/acs.chemrev.7b00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starks C.M., Back K.W., Chappell J., Noel J.P. Structural Basis for Cyclic Terpene Biosynthesis by Tobacco 5-Epi-Aristolochene Synthase. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 28.Cane D.E., Xue Q., Fitzsimons B.C. Trichodiene Synthase. Probing the Role of the Highly Conserved Aspartate-Rich Region by Site-Directed Mutagenesis. Biochemistry. 1996;35:12369–12376. doi: 10.1021/bi961344y. [DOI] [PubMed] [Google Scholar]
- 29.Agger S., Lopez-Gallego F., Schmidt-Dannert C. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol. Microbiol. 2009;72:1307–1308. doi: 10.1111/j.1365-2958.2009.06743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wawrzyn G.T., Quin M.B., Choudhary S., López-Gallego F., Schmidt-Dannert C. Draft Genome of Omphalotus olearius Provides a Predictive Framework for Sesquiterpenoid Natural Product Biosynthesis in Basidiomycota. Chem. Biol. 2012;19:772–783. doi: 10.1016/j.chembiol.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Y.-L., Ma L.-T., Lee Y.-R., Shaw J.-F., Wang S.-Y., Chu F.-H. Differential Gene Expression Network in Terpenoid Synthesis of Antrodia cinnamomea in Mycelia and Fruiting Bodies. J. Agric. Food Chem. 2017;65:1874–1886. doi: 10.1021/acs.jafc.6b05386. [DOI] [PubMed] [Google Scholar]
- 32.Kirk P.M., Cannon P.F., Minter D.W., Stalpers J.A., editors. Ainswoth & Bisby’s Dictionary of the Fungi. 10th ed. CABI Europe; Wallingford, UK: 2008. p. 445. [Google Scholar]
- 33.Kreuzenbeck N.B., Seibel E., Schwitalla J.W., Fricke J., Conlon B.H., Schmidt S., Hammerbacher A., Köllner T.G., Poulsen M., Hoffmeister D., et al. Comparative Genomic and Metabolomic Analysis of Termitomyces Species Provides Insights into the Terpenome of the Fungal Cultivar and the Characteristic Odor of the Fungus Garden of Macrotermes natalensis Termites. mSystems. 2022;7:e01214-21. doi: 10.1128/msystems.01214-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burkhardt I., Kreuzenbeck N.B., Beemelmanns C., Dickschat J.S. Mechanistic characterization of three sesquiterpene synthases from the termite-associated fungus Termitomyces. Org. Biomol. Chem. 2019;17:3348–3355. doi: 10.1039/C8OB02744G. [DOI] [PubMed] [Google Scholar]
- 35.Lopez-Gallego F., Agger S.A., Abate-Pella D., Distefano M.D., Schmidt-Dannert C. Sesquiterpene Synthases Cop4 and Cop6 from Coprinus cinereus: Catalytic Promiscuity and Cyclization of Farnesyl Pyrophosphate Geometric Isomers. ChemBioChem. 2010;11:1093–1106. doi: 10.1002/cbic.200900671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagamine S., Liu C., Nishishita J., Kozaki T., Sogahata K., Sato Y., Minami A., Ozaki T., Schmidt-Dannert C., Maruyama J.-I., et al. Ascomycete Aspergillus oryzae Is an Efficient Expression Host for Production of Basidiomycete Terpenes by Using Genomic DNA Sequences. Appl. Environ. Microbiol. 2019;85:e00409-19. doi: 10.1128/AEM.00409-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Salihi S.A.A., Dao T.T., Williams K., Bailey A.M., Foster G.D. The Biogenetic Origin of the Biologically Active Naematolin of Hypholoma Species Involves an Unusual Sesquiterpene Synthase. Mol. Biotechnol. 2019;61:754–762. doi: 10.1007/s12033-019-00199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang C., Chen X., Orban A., Shukal S., Birk F., Too H.-P., Ruehl M. Agrocybe aegerita Serves as a Gateway for Identifying Sesquiterpene Biosynthetic Enzymes in Higher Fungi. ACS Chem. Biol. 2020;15:1268–1277. doi: 10.1021/acschembio.0c00155. [DOI] [PubMed] [Google Scholar]
- 39.Engels B., Heinig U., Grothe T., Stadler M., Jennewein S. Cloning and Characterization of an Armillaria gallica cDNA Encoding Protoilludene Synthase, Which Catalyzes the First Committed Step in the Synthesis of Antimicrobial Melleolides. J. Biol. Chem. 2011;286:6871–6878. doi: 10.1074/jbc.M110.165845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Justo A., Miettinen O., Floudas D., Ortiz-Santana B., Sjökvist E., Lindner D., Nakasone K., Niemelä T., Larsson K.-H., Ryvarden L., et al. A revised family-level classification of the Polyporales (Basidiomycota) Fungal Biol. 2017;121:798–824. doi: 10.1016/j.funbio.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 41.Yap H.-Y.Y., Muria-Gonzalez M.J., Kong B.-H., Stubbs K.A., Tan C.-S., Ng S.-T., Tan N.-H., Solomon P.S., Fung S.-Y., Chooi Y.-H. Heterologous expression of cytotoxic sesquiterpenoids from the medicinal mushroom Lignosus rhinocerotis in yeast. Microb. Cell Factories. 2017;16:103. doi: 10.1186/s12934-017-0713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Püth N., Ersoy F., Krings U., Berger R.G. Sesquiterpene Cyclases from the Basidiomycete Cerrena unicolor. Catalysts. 2021;11:1361. doi: 10.3390/catal11111361. [DOI] [Google Scholar]
- 43.Misiek M., Hoffmeister D. Processing sites involved in intron splicing of Armillaria natural product genes. Mycol. Res. 2008;112:216–224. doi: 10.1016/j.mycres.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 44.Bayram Ö., Braus G.H. Coordination of secondarymetabolism and development in fungi: The velvet familyof regulatory proteins. FEMS Microbiol. Rev. 2012;36:1–24. doi: 10.1111/j.1574-6976.2011.00285.x. [DOI] [PubMed] [Google Scholar]
- 45.Merhej J., Richard-Forget F., Barreau C. The pH regulatory factor Pac1 regulates Tri gene expression and trichothecene production in Fusarium graminearum. Fungal Genet. Biol. 2011;48:275–284. doi: 10.1016/j.fgb.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 46.Ichinose H., Kitaoka T. Insight into metabolic diversity of the brown-rot basidiomycete Postia placenta responsible for sesquiterpene biosynthesis: Semi-comprehensive screening of cytochrome P450 monooxygenase involved in protoilludene metabolism. Microb. Biotechnol. 2018;11:952–965. doi: 10.1111/1751-7915.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L.-Z., Pu X.-D., Tan S.-Q., Sun S.-J., Bi Y.-T., Sun C., Chen S.-L., Wang H.-Y. Cloning and expression of the sesquiterpene synthase gene from Ganoderma lucidum. J. Agric. Univ. Hebei. 2017;40:67–72. doi: 10.13320/j.cnki.jauh.2017.0036. [DOI] [Google Scholar]
- 48.Cao R., Wu X., Wang Q., Qi P., Zhang Y., Wang L., Sun C. Characterization of γ-Cadinene Enzymes in Ganoderma lucidum and Ganoderma sinensis from Basidiomycetes Provides Insight into the Identification of Terpenoid Synthases. ACS Omega. 2022;7:7229–7239. doi: 10.1021/acsomega.1c06792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei J., Pu X., Wang L., Sun S., Sun C., Wang H. Cloning and characterization of sesquiterpene synthase genes from the Ganoderma sinense genome. Sci. Sin. Vitae. 2018;48:447–454. doi: 10.1360/N052017-00190. [DOI] [Google Scholar]
- 50.Chu L.-H., Wang L.-Z., Chen S.-L., Zeng X.-Y., Xu J., Li Y., Sun C. Functional Identification of a Multi-product Sesquiterpene Synthase from Ganoderma sinense. Chin. J. Exp. Tradit. Med. Formulae. 2019:151–157. doi: 10.13422/j.cnki.syfjx.20191216. [DOI] [Google Scholar]
- 51.Ichinose H., Ukeba S., Kitaoka T. Latent potentials of the white-rot basidiomycete Phanerochaete chrysosporium responsible for sesquiterpene metabolism: CYP5158A1 and CYP5144C8 decorate (E)-α-bisabolene. Enzym. Microb. Technol. 2022;158:110037. doi: 10.1016/j.enzmictec.2022.110037. [DOI] [PubMed] [Google Scholar]
- 52.Wang Q., Liu J.-K., Zhao Q., He Q.-L. Mechanistic investigations of hirsutene biosynthesis catalyzed by a chimeric sesquiterpene synthase from Steccherinum ochraceum. Fungal Genet. Biol. 2022;161:103700. doi: 10.1016/j.fgb.2022.103700. [DOI] [PubMed] [Google Scholar]
- 53.Lei C.-Z., Han H.-Y., Liu C.-W., Zhao M. A review of research on the secondary metabolites of Stereum hirsutum. Mycosystema. 2021;40:19181937. doi: 10.13346/j.mycosystema.210136. [DOI] [Google Scholar]
- 54.Quin M.B., Flynn C.M., Wawrzyn G.T., Choudhary S., Schmidt-Dannert C. Mushroom Hunting by Using Bioinformatics: Application of a Predictive Framework Facilitates the Selective Identification of Sesquiterpene Synthases in Basidiomycota. ChemBioChem. 2013;14:2480–2491. doi: 10.1002/cbic.201300349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flynn C.M., Schmidt-Dannert C. Sesquiterpene Synthase–3-Hydroxy-3-Methylglutaryl Coenzyme A Synthase Fusion Protein Responsible for Hirsutene Biosynthesis in Stereum hirsutum. Appl. Environ. Microbiol. 2018;84:e00036-18. doi: 10.1128/AEM.00036-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding J.-H., Feng T., Li Z.-H., Li L., Liu J.-K. Twelve new compounds from the basidiomycete Boreostereum vibrans. Nat. Prod. Bioprospect. 2012;2:200–205. doi: 10.1007/s13659-012-0060-x. [DOI] [Google Scholar]
- 57.Zhou H., Yang Y.-L., Zeng J., Zhang L., Ding Z.-H., Zeng Y. Identification and Characterization of a δ-Cadinol Synthase Potentially Involved in the Formation of Boreovibrins in Boreostereum vibrans of Basidiomycota. Nat. Prod. Bioprospect. 2016;6:167–171. doi: 10.1007/s13659-016-0096-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wumuti B., Tang Y.-Q., Wang S.-T., Li Y.-W., Zou L. Cloning and Function Identification of the Sesquiterpenes Synthase Gene SbTps1 in Sanghuangporus baumii. For. Eng. 2021;37:33–39. doi: 10.16270/j.cnki.slgc.2021.04.009. [DOI] [Google Scholar]
- 59.Mischko W., Hirte M., Fuchs M., Mehlmer N., Brück T.B. Identification of sesquiterpene synthases from the Basidiomycota Coniophora puteana for the efficient and highly selective β-copaene and cubebol production in E. coli. Microb. Cell Factories. 2018;17:164. doi: 10.1186/s12934-018-1010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ringel M., Dimos N., Himpich S., Haack M., Huber C., Eisenreich W., Schenk G., Loll B., Brück T. Biotechnological potential and initial characterization of two novel sesquiterpene synthases from Basidiomycota Coniophora puteana for heterologous production of δ-cadinol. Microb. Cell Factories. 2022;21:64. doi: 10.1186/s12934-022-01791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ntana F., Bhat W., Johnson S., Jørgensen H., Collinge D., Jensen B., Hamberger B. A Sesquiterpene Synthase from the Endophytic Fungus Serendipita indica Catalyzes Formation of Viridiflorol. Biomolecules. 2021;11:898. doi: 10.3390/biom11060898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mohanta T.K., Bae H. The diversity of fungal genome. Biol. Proced. Online. 2015;17:8. doi: 10.1186/s12575-015-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kellner S., Spang A., Offre P., Szöllősi G.J., Petitjean C., Williams T.A. Genome size evolution in the Archaea. Emerg. Top. Life Sci. 2018;2:595–605. doi: 10.1042/etls20180021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deamer D., Akeson M., Branton D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016;34:518–524. doi: 10.1038/nbt.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ogiso-Tanaka E., Itagaki H., Ohmae M., Hosoya T., Hosaka K. De Novo Genome Assembly of Stinkhorn Mushroom Clathrus columnatus (Basidiomycota, Fungi) Using Illumina and Nanopore Sequencing Data. Microbiol. Resour. Announc. 2022;11:e01026-21. doi: 10.1128/mra.01026-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duan Y., Han H., Qi J., Gao J.-M., Xu Z., Wang P., Zhang J., Liu C. Genome sequencing of Inonotus obliquus reveals insights into candidate genes involved in secondary metabolite biosynthesis. BMC Genom. 2022;23:314. doi: 10.1186/s12864-022-08511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jain M., Tyson J.R., Loose M., Ip C.L., Eccles D.A., O’Grady J., Malla S., Leggett R.M., Wallerman O., Jansen H.J., et al. MinION Analysis and Reference Consortium: Phase 2 data release and analysis of R9.0 chemistry. F1000Research. 2017;6:760. doi: 10.12688/f1000research.11354.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goldstein S., Beka L., Graf J., Klassen J.L. Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing. BMC Genom. 2019;20:23. doi: 10.1186/s12864-018-5381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shaw B.D., Hoch H.C. Ions Regulate Spore Attachment, Germination, and Fungal Growth. In: Howard R.J., Gow N.A.R., editors. Biology of the Fungal Cell. Volume 8. Springer; Berlin/Heidelberg, Germany: 2007. pp. 219–236. The Mycota. [Google Scholar]
- 70.Rämä T., Quandt C.A. Improving Fungal Cultivability for Natural Products Discovery. Front. Microbiol. 2021;12:706044. doi: 10.3389/fmicb.2021.706044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Melin E. Tree Growth. 1962. Physiological Aspects of Mycorrhizae of Forest Trees; pp. 247–263. [Google Scholar]
- 72.Fries N. Spore Germination in Ectomycorrhizal Fungi; Proceedings of the 6th North American Conference on Mycorrhizae; Bend, OR, USA. 25–29 June 1984; Corvallis, OR, USA: Oregon State University, Forest Research Laboratory; 1985. [Google Scholar]
- 73.Borwn T., Merrill W. Germination of Basidiospores of Fomes applanatus. Phytopathology. 1973;63:547. doi: 10.1094/Phyto-63-547. [DOI] [Google Scholar]
- 74.Sun Y.-P., Fries N. The effect of tree-root exudates on the growth rate of ectomycorrhizal and saprotrophic fungi. Mycorrhiza. 1992;1:63–69. doi: 10.1007/BF00206138. [DOI] [Google Scholar]
- 75.Nakayashiki H. RNA silencing in fungi: Mechanisms and applications. FEBS Lett. 2005;579:5950–5957. doi: 10.1016/j.febslet.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 76.Noble L.M., Andrianopoulos A. Fungal Genes in Context: Genome Architecture Reflects Regulatory Complexity and Function. Genome Biol. Evol. 2013;5:1336–1352. doi: 10.1093/gbe/evt077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu G., Sun Y., Han H., Yan X., Wang Y., Ge X., Qiao B., Tan L. Coculture, An Efficient Biotechnology for Mining the Biosynthesis Potential of Macrofungi via Interspecies Interactions. Front. Microbiol. 2021;12:663924. doi: 10.3389/fmicb.2021.663924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Collemare J., Seidl M.F. Chromatin-dependent regulation of secondary metabolite biosynthesis in fungi: Is the picture complete? FEMS Microbiol. Rev. 2019;43:591–607. doi: 10.1093/femsre/fuz018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calvo A.M., Wilson R.A., Bok J.W., Keller N.P. Relationship between Secondary Metabolism and Fungal Development. Microbiol. Mol. Biol. Rev. 2002;66:447–459. doi: 10.1128/MMBR.66.3.447-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Calvo A.M., Cary J.W. Association of fungal secondary metabolism and sclerotial biology. Front. Microbiol. 2015;6:62. doi: 10.3389/fmicb.2015.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chang P.-K., Bennett J.W., Cotty P.J. Association of aflatoxin biosynthesis and sclerotial development in Aspergillus parasiticus. Mycopathologia. 2002;153:41–48. doi: 10.1023/A:1015211915310. [DOI] [PubMed] [Google Scholar]
- 82.Cheng C.K., Au C.H., Wilke S.K., Stajich J.E., Zolan M.E., Pukkila P.J., Kwan H.S. 5′-Serial Analysis of Gene Expression studies reveal a transcriptomic switch during fruiting body development in Coprinopsis cinerea. BMC Genom. 2013;14:195. doi: 10.1186/1471-2164-14-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miyazaki Y., Nakamura M., Babasaki K. Molecular cloning of developmentally specific genes by representational difference analysis during the fruiting body formation in the basidiomycete Lentinula edodes. Fungal Genet. Biol. 2005;42:493–505. doi: 10.1016/j.fgb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 84.Aleklett K., Ohlsson P., Bengtsson M., Hammer E.C. Fungal foraging behaviour and hyphal space exploration in micro-structured Soil Chips. ISME J. 2021;15:1782–1793. doi: 10.1038/s41396-020-00886-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Harvey C.J.B., Tang M., Schlecht U., Horecka J., Fischer C.R., Lin H.-C., Li J., Naughton B., Cherry J., Miranda M., et al. HEx: A heterologous expression platform for the discovery of fungal natural products. Sci. Adv. 2018;4:eaar5459. doi: 10.1126/sciadv.aar5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu C., Minami A., Ozaki T., Wu J., Kawagishi H., Maruyama J.-I., Oikawa H. Efficient Reconstitution of Basidiomycota Diterpene Erinacine Gene Cluster in Ascomycota Host Aspergillus oryzae Based on Genomic DNA Sequences. J. Am. Chem. Soc. 2019;141:15519–15523. doi: 10.1021/jacs.9b08935. [DOI] [PubMed] [Google Scholar]
- 87.Lee J., Hilgers F., Loeschke A., Jaeger K.-E., Feldbrügge M. Ustilago maydis Serves as a Novel Production Host for the Synthesis of Plant and Fungal Sesquiterpenoids. Front. Microbiol. 2020;11:1655. doi: 10.3389/fmicb.2020.01655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van der Hooft J.J.J., Mohimani H., Bauermeister A., Dorrestein P.C., Duncan K.R., Medema M.H. Linking genomics and metabolomics to chart specialized metabolic diversity. Chem. Soc. Rev. 2020;49:3297–3314. doi: 10.1039/D0CS00162G. [DOI] [PubMed] [Google Scholar]
- 89.Wang M., Carver J.J., Phelan V.V., Sanchez L.M., Garg N., Peng Y., Nguyen D.D., Watrous J., Kapono C.A., Luzzatto-Knaan T., et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S.Y., Medema M.H., Weber T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Skinnider M.A., Merwin N.J., Johnston C.W., Magarvey N.A. PRISM 3: Expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 2017;45:W49–W54. doi: 10.1093/nar/gkx320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Bruyne M., Baker T.C. Odor Detection in Insects: Volatile Codes. J. Chem. Ecol. 2008;34:882–897. doi: 10.1007/s10886-008-9485-4. [DOI] [PubMed] [Google Scholar]
- 93.Qu J., Truhan J.J., Dai S., Luo H., Blau P.J. Ionic liquids with ammonium cations as lubricants or additives. Tribol. Lett. 2006;22:207–214. doi: 10.1007/s11249-006-9081-0. [DOI] [Google Scholar]
- 94.Orban A., Hennicke F., Rühl M. Volatilomes of Cyclocybe aegerita during different stages of monokaryotic and dikaryotic fruiting. Biol. Chem. 2020;401:995–1004. doi: 10.1515/hsz-2019-0392. [DOI] [PubMed] [Google Scholar]
- 95.Yang H., Li J., Du G., Liu L. Biotechnology of Microbial Enzymes. Elsevier; Amsterdam, The Netherlands: 2017. Microbial Production and Molecular Engineering of Industrial Enzymes; pp. 151–165. [Google Scholar]
- 96.Gallego F.L., Wawrzyn G., Schmidt-Dannert C. Selectivity of Fungal Sesquiterpene Synthases: Role of the Active Site’s H-1α Loop in Catalysis. Appl. Environ. Microbiol. 2010;76:7723–7733. doi: 10.1128/AEM.01811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lauchli R., Pitzer J., Kitto R.Z., Kalbarczyk K.Z., Rabe K.S. Improved selectivity of an engineered multi-product terpene synthase. Org. Biomol. Chem. 2014;12:4013–4020. doi: 10.1039/C4OB00479E. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.