Abstract
The network structure and the metabolic fluxes in central carbon metabolism were characterized in aerobically grown cells of Saccharomyces cerevisiae. The cells were grown under both high and low glucose concentrations, i.e., either in a chemostat at steady state with a specific growth rate of 0.1 h−1 or in a batch culture with a specific growth rate of 0.37 h−1. Experiments were carried out using [1-13C]glucose as the limiting substrate, and the resulting summed fractional labelings of intracellular metabolites were measured by gas chromatography coupled to mass spectrometry. The data were used as inputs to a flux estimation routine that involved appropriate mathematical modelling of the central carbon metabolism of S. cerevisiae. The results showed that the analysis is very robust, and it was possible to quantify the fluxes in the central carbon metabolism under both growth conditions. In the batch culture, 16.2 of every 100 molecules of glucose consumed by the cells entered the pentose-phosphate pathway, whereas the same relative flux was 44.2 per 100 molecules in the chemostat. The tricarboxylic acid cycle does not operate as a cycle in batch-growing cells, in contrast to the chemostat condition. Quantitative evidence was also found for threonine aldolase and malic enzyme activities, in accordance with published data. Disruption of the MIG1 gene did not cause changes in the metabolic network structure or in the flux pattern.
Since the whole genome of Saccharomyces cerevisiae has been sequenced (15), several kinds of analyses have been applied in order to assign function to orphan genes. In many of these analyses, the aim has been to determine how the different genes (both the ones with known function and those open reading frames that have no assigned function) interact with each other, enabling the cells to take up nutrients, grow, divide, regulate their metabolism, release products to the environment, and respond to different stimuli (31).
The different approaches that have been used for this purpose can be classified as transcriptome, proteome, and metabolome analyses, depending on the type of compounds measured, i.e., transcripts, proteins, or metabolites (31). Besides these approaches, other types of phenotypic investigations have been performed, such as growth on synthetic or rich media, growth on different carbon sources, determination of the ability to consume oxygen, and measurement of enzyme activities in cell extracts. In recent work, Entian and coworkers (9) showed that the type and accuracy of the methods used are very important for the identification of phenotypes in single-deletion mutants.
A very prominent phenotypic investigation method that has been employed for the analysis of cells in different environments, as well as for the analysis of different mutants, is the quantification of metabolic fluxes in cells grown under balanced conditions, such as in a steady-state chemostat or during the exponential phase of a batch culture. This quantification can be carried out by metabolite balancing (30, 40, 41, 42), through which the intracellular fluxes are calculated from a few measured fluxes by using mass balances for the intracellular metabolites. For a more accurate quantification of the fluxes, one may use 13C-labeled substrates followed by measurements of the isotopomers using gas chromatography and mass spectrometry (GC-MS) or nuclear magnetic resonance (38). Thus, balances can be set up for the individual carbon atoms, and this gives a significant redundancy in the measurements, leading to more robust flux estimates. The use of 13C-labeled substrates enables both identification of the metabolic network structure and quantification of the metabolic fluxes and is therefore referred to as metabolic network analysis (5).
This kind of analysis can be applied to investigate the metabolic network of S. cerevisiae, which is known to be under carbon catabolite repression when grown on rapidly fermentable carbon sources (13). As glucose is often the carbon source present in laboratory media as well as in industrial processes, this type of transcriptional control is often termed glucose repression (3, 17). This phenomenon has been known for decades and refers to the repression of the transcription of genes involved in different cellular functions, such as the uptake and catabolism of alternate carbon sources, respiration, tricarboxylic acid (TCA) cycle enzymes, gluconeogenesis, and peroxisomal functions (3). However, the mechanism by which yeast cells sense glucose, and how the signal is transduced from this initial sensing to the actual repression of gene transcription at the end of the cascade, is not yet fully elucidated. There is evidence that glucose concentration, rather than glucose uptake, triggers the glucose-repression cascade (27). There is also evidence that hexokinase PII plays a major role in the early part of the cascade (10, 20), but it is not known how the signal is transduced from glucose to Snf1p, a protein kinase that plays a central role in the cascade (3). Snflp phosphorylates Mig1p (29, 32, 39), a protein that binds to the promoter region of glucose-repressible genes. When glucose levels are high, Snflp is not active and Mig1p is underphosphorylated and located in the nucleus, repressing the transcription of genes. When glucose levels are low, Snflp phosphorylates Mig1p, causing its migration to the cytoplasm, releasing repression (3).
Transcriptional repression via Mig1p is a dual-type regulation, as the protein can bind to structural genes as well as to their repressors or activators. A physiological role for Mig1p has been shown in the repression of genes involved in the catabolism of sugars such as galactose, maltose, and sucrose, as well as of the CAT8 gene, which codes for a derepressor of gluconeogenic genes (19). However, putative binding sites for Mig1p exist in the promoter region of several other genes which are involved in central metabolic functions, such as gluconeogenesis and respiration. Furthermore, TCA cycle genes, such as CIT1, CIT3, KGD1, KGD2, and LPD1, may be indirectly regulated by Mig1p, as it can bind to the HAP4 gene, which codes for the transcriptional activator of these TCA cycle genes (19). Yet, it is not clearly known to what extent glucose repression affects the central metabolism of S. cerevisiae.
In this work, we grew S. cerevisiae cells under different conditions of glucose repression. Different environmental conditions were applied both to a reference strain and to a mig1 disruption mutant. Metabolic network analysis was performed by combining labeling experiments with mathematical modelling (5).
MATERIALS AND METHODS
Strains.
Two strains were used throughout this work: the reference strain CEN.PK113-7D (MATa MAL2-8c SUC2) and the mig1 disruption mutant T468 (MATa mig1Δ::MELI MAL2-8c SUC2), which was constructed by Birgitte Rønnow, Danisco Cultor, as described previously (18).
Cultivations.
A total of four cultivations were carried out in a bioreactor that was specially designed for labeling experiments. The temperature was controlled at 30°C, the pH was controlled at 5.00 ± 0.05 by addition of 0.5 N NaOH, and the dissolved oxygen concentration was kept above 60% saturation by introducing sterile air through a needle and agitating the medium with a magnetic stirrer at 700 min−1. The working volume was 200 ml in the batch cultures and 150 ml in the continuous cultures, and the volume was kept constant by withdrawing liquid through a continuously operating pump.
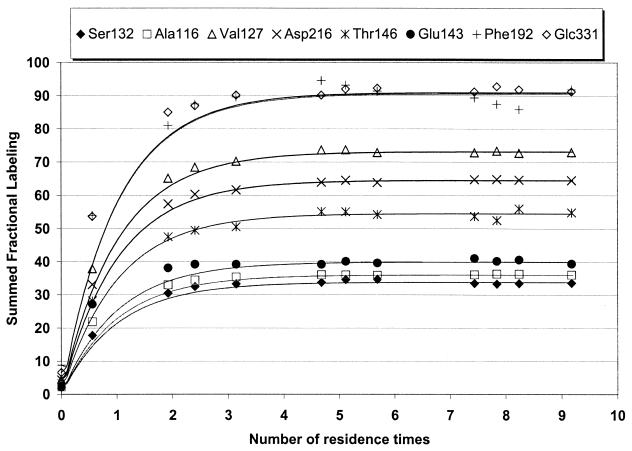
In all cases, cells from yeast-peptone-dextrose (YPD) plates were transferred to 500-ml baffled flasks containing 100 ml of the following medium: glucose, 10 g/liter; (NH4)2SO4, 7.5 g/liter; KH2PO4, 14.4 g/liter; MgSO4 · 7H2O, 0.48 g/liter; trace element solution, 2.0 ml/liter; vitamin solution, 1.0 ml/liter; pH was adjusted to 6.5. After about 24 h on a rotatory shaker at 30°C and 150 min−1, cells from this preculture were used to inoculate the bioreactor to an optical density at 600 nm (OD600) of 0.15 in the following medium (44): (NH4)2SO4, 5.0 g/liter; KH2PO4, 3.0 g/liter; MgSO4 · 7H2O, 0.5 g/liter; trace element solution, 1.0 ml/liter; vitamin solution, 1.0 ml/liter. In all the labeling experiments, 100% 1-13C-labeled glucose was used as the sole carbon source. In the batch cultures, 5 g/liter was used as the initial glucose concentration, and the cultivations were interrupted in the late exponential phase after at least 4 generation times, when cells were still growing at the maximum specific growth rate, μmax. In the continuous cultures, an initial batch with 2 g of glucose/liter was carried out, after which a solution containing 2 g of glucose/liter and the same composition as the batch culture medium was continuously fed to the reactor at a dilution rate of 0.1 h−1. When steady state was achieved (after time enough for replacing four times the reactor content, or in other words after 4 residence times), the feeding medium was switched to a similar solution containing 2 g of [1-13C]glucose/liter, and the cultivations were carried on for at least 5 more residence times. The achievement of steady state was monitored by determining the absorbance of the culture, and it was later confirmed via analyses of excreted metabolites and also via labeling incorporation (see Fig. 1).
FIG. 1.
Label incorporation into fragments of selected intracellular metabolites of S. cerevisiae in a continuous cultivation. Time zero corresponds to the medium switch from naturally labeled glucose to [1-13C]glucose as the limiting substrate. The curves shown assume first-order incorporation kinetics, according to the equation SFL = SFLFINAL × (1 − e−t).
The trace metal solution had the following composition (44) (in grams per liter): EDTA, 15; ZnSO4 · 7H2O, 4.5; MnCl2 · 2H2O, 0.84; CoCl2 · 6H2O, 0.30; CuSO4 · 5H2O, 0.30; Na2MoO4 · 2H2O, 0.40; CaCl2 · 2H2O, 4.5; FeSO4 · 7H2O, 3.0; H3BO3, 1.0; KI, 0.10. The composition of the vitamin solution (44) was as follows (in grams per liter): d-biotin, 0.05; calcium pantothenate, 1.0; nicotinic acid, 1.0; myoinositol, 25.0; thiamine chloride hydrochloride, 1.0; pyridoxol hydrochloride, 1.0; p-aminobenzoic acid, 0.20.
With the aim of obtaining precise values for batch cultivation parameters such as the maximum specific growth rate and the cell yield on glucose for both strains employed in this work, two batch cultivations were carried out in 4-liter bioreactors, i.e., one cultivation with each strain. These reactors were equiped with two Rushton turbines, and the cultivations were performed under the same temperature, pH, and medium conditions as specified above for the 200-ml reactor. The only difference between the 200-ml and the 4-liter cultivations was the initial glucose concentration, which was 5 g/liter in the former and 10 g/liter in the latter.
Sample treatment.
In all cases, 1.5-ml samples were taken from the cultivation for the determination of absorbance at 600 nm, which was carried out using a Shimadzu flow spectrophotometer (model CL-720; Kyoto, Japan). Thereafter, filtration of the remaining sample volume was carried out on 0.45-μm-pore-size acetate filters (Osmonics, Minnetonka, Minn.), and the filtrate was frozen at −20°C and later used for sugar and excreted metabolite analyses.
When the batch cultivations were interrupted (late exponential phase, after at least 4 generation times, when the glucose concentration was around 2.7 g/liter), part of the reactor content was used for a dry cell weight determination, whereas the rest was filtered through a 0.45-μm-pore-size nitrocellulose membrane (Gelman Sciences, Ann Arbor, Mich.) and washed twice with distilled water, and the wet biomass obtained was frozen at −80°C and later used for determining the fractional labeling of intracellular metabolites. When the continuous cultivations were interrupted (always after steady state had been achieved), the content of the reactor was treated in the same way as described above. In the continuous cultivation with the reference strain, the incorporation of labeling was followed from the point when the medium containing nonlabeled glucose was switched to the medium containing labeled glucose. This was performed by collecting samples from the reactor outlet in centrifuge tubes in an ice-water bath. For each sample, the liquid was collected during 1 h, which corresponded to a volume of ca. 15 ml. Subsequently, each sample was centrifuged at 4°C, 3,000 × g for 10 min. After this, the cell pellet was resuspended in ca. 20 ml of distilled water and centrifuged again, and the resulting pellet was transferred to Eppendorf tubes using 1.5 ml of distilled water. Centrifugation was then carried out at 4°C, 5,000 × g for 5 min, and the pellet was frozen at −80°C and later used for determining the fractional labeling of intracellular metabolites.
Measurement of sugar and excreted metabolite concentrations.
Glucose, ethanol, glycerol, acetate, succinate, and pyruvate were separated on an Aminex HPX-87H ion-exclusion column (Bio-Rad, Hercules, Calif.) at 65°C, using 5 mM H2SO4 as the mobile phase at a flow rate of 0.6 ml/min. Glucose, ethanol, glycerol, and succinate were detected on a Waters 410 differential refractometer detector (Millipore, Milford, Mass.), whereas acetate and pyruvate were detected on a Waters 486 tunable absorbance detector set at 210 nm. The two detectors were connected in series.
Measurement of fractional labeling of intracellular metabolites.
For the analysis of the fractional labeling of intracellular metabolites, ca. 20 mg of wet biomass at −80°C was transferred to 800 μl of 6 M HCl. The mixture was then separated into two fractions. One fraction, of ca. 700 μl, was used for determining the fractional labeling of amino acid fragments, whereas the other fraction, of ca. 100 μl, was used for determining the fractional labeling of intracellular glucose. The first fraction was hydrolyzed for 12 to 20 h at 105°C. After this, ca. 800 μl of distilled water was added to the sample, which was then centrifuged at 3,000 × g for 5 min for separation of the cell debris. The supernatant was then separated into 200-μl fractions, which were dried at 105°C. After this, the dried samples were separately derivatized by two different agents, ethylchloroformate (ECF) and (N,N)-dimethylformamide dimethyl acetal. In both cases, the protocol described by Christensen and Nielsen (6) was employed. The only difference was in the derivatization with ECF, in which trifluoroacetic anhydride was not added. The second fraction was hydrolyzed for only 20 to 30 min at 105°C, after which 100 μl of distilled water was added. Derivatization of glucose to glucose pentaacetate (GPA) was carried out according to the protocol described previously (7).
In all cases, the prepared samples were injected into the GC-MS (model HP-G1723A, Hewlett-Packard). At least two injections were performed for each derivatization, and the conditions employed were described previously (6). The output of the measurements is a set of clusters, each of which corresponds to the intensities of the mass isotopomers of an amino acid fragment (or of a glucose fragment, in the case of glucose derivatization). These intensities were corrected for the natural labeling in the derivative part, as described elsewhere (22, 23). Finally, the fractional labeling of each fragment, termed summed fractional labeling (SFL), was calculated with the corrected intensities in the following way (7):
0 · m01–3 + 1 · m11–3 + 2 · m21–3 + 3 · m31–3 SFL(Ala1–3) =
m01–3 + m11–3 + m21–3 + m31–3
where the superscripts indicate the carbon atoms present in the fragment and mn indicates the corrected intensity of the mass isotopomer with mn labeled carbon atoms. In the equation, the fragment of alanine containing all three carbon atoms of this amino acid was used as an example. It is important to note that the SFL of a fragment, as calculated from the GC-MS measurements, is also equal to the sum of the fractional labelings of the individual carbon atoms in the fragment. Thus, if SFL(Ala1–3) and SFL(Ala2–3) can be measured, it is possible to calculate the fractional labeling of C-1 of alanine by subtracting the latter from the former.
Mathematical modelling.
The SFLs, as described above, were used as inputs to a mathematical routine that is used for quantifying the fluxes in the central carbon metabolism of S. cerevisiae. The mathematical framework has been described previously (7) and will not be shown in detail here. Briefly, in each iteration of the numerical procedure, a guess is made on the fluxes and is used for calculating the SFLs via the mathematical model. Subsequently, two errors are generated: one by comparing the calculated SFLs with the values measured experimentally (via GC-MS, as described above), and the other by comparing the guessed fluxes with the ones measured experimentally. (Alternatively, literature values for the biomass composition can be used as the measured fluxes). If the sum of these two errors is smaller then the stored error value, the guess on fluxes used in this iteration is considered the best one. Otherwise, this guess is disregarded. A new iteration is started and the process goes on until an acceptable error is achieved.
The model used in this work is shown in the Appendix, indicating both the reactions considered and the corresponding carbon atom transitions. The metabolic functions considered were the central ones: the Embden-Meyerhoff-Parnas (EMP) and pentose-phosphate (PP) pathways, as well as the TCA cycle. Besides these pathways, other reactions have been included based on both specific biochemical knowledge of S. cerevisiae and on the labeling results attained throughout this work (see Results and Discussion for further details). Compartmentation of some metabolites was crucial for fitting the calculated SFLs to the measured values (7), and thus oxaloacetate, acetyl coenzyme A (acetyl-CoA), and pyruvate were separated into two pools each. The anaplerotic reaction catalyzed by pyruvate carboxylase was included in the model as a cytosolic step (16, 45), the reaction catalyzed by threonine aldolase was included as an alternative biosynthetic route for glycine (24, 28), and the reaction catalyzed by malic enzyme was included as mitochondrial step, as there is evidence that it takes place when yeast cells grow on glucose (1). Excretion of succinate and pyruvate were not included in the model, as they represent a very small fraction (<0.1%) of the carbon consumed by the cells as glucose.
In terms of metabolites drained for biosynthesis, two different cell compositions were considered in terms of macromolecules, one for each cultivation condition (Table 1). However, the composition of each type of macromolecule was assumed to be independent of the cultivation condition. Thus, the amino acid composition of a protein was assumed to be the same under both conditions and was taken from Oura (34). Similarly, it was assumed that the lipid composition was the same under both growth conditions and was taken from Bruinenberg et al. (2) and Oura (34). The carbohydrate fraction was assumed to be a polymer of glucose, and the metabolite drained in this case is exclusively glucose-6-phosphate. Nucleic acids were considered to be RNA, as the DNA fraction corresponds to only 0.3% of the dry cell weight (34). Finally, it was assumed that the cell composition was the same for both the reference strain and the mig1 mutant, when compared under the same cultivation conditions. All these approximations are reasonable, as it has been shown that perturbations in the biomass composition do not significantly alter the final flux patterns (8). Combining all these considerations with the biochemical knowledge of biosynthetic pathways for amino acid synthesis (46), it was possible to calculate the drain of precursors to biomass under both cultivation conditions (Table 2). These values were used as the measured fluxes in the mathematical routine, as described previously (7).
TABLE 1.
Cell composition under the cultivation conditions investigated
| Component | D = μ = 0.1 h−1 (chemostat) | μmax = 0.37 h−1 (batch) | Reference(s) |
|---|---|---|---|
| Protein | 42 | 51 | 11, 43 |
| Lipid | 7 | 7 | 30, 32, 41 |
| Carbohydrate | 40 | 27 | 21 |
| RNA | 7 | 11 | 33, 43 |
| Ashes | 4 | 4 | 30, 33 |
TABLE 2.
Precursors drained for biomass formationa
| Precursorb | D = μ = 0.1 h−1 (chemostat) | μmax = 0.37 h−1 (batch) |
|---|---|---|
| ACCOAcyt | 2.44 | 2.50 |
| ACCOAmit | 0.311 | 0.378 |
| AKG | 1.07 | 1.30 |
| C−1 | 0.327 | 0.397 |
| E4P | 0.277 | 0.337 |
| GLY | 0.407 | 0.494 |
| SER | 0.202 | 0.245 |
| G3P (lipids) | 0.091 | 0.091 |
| G6P | 2.47 | 1.67 |
| THR | 0.403 | 0.490 |
| OAA (others) | 0.589 | 0.715 |
| P5P | 0.318 | 0.464 |
| PEP | 0.555 | 0.674 |
| PYR | 1.83 | 2.22 |
Values are given in millimoles of precursor per gram (dry weight).
Abbreviations: ACCOAcyt, acetyl-CoA in the cytosol; ACCOAmit, acetyl-CoA in the mitochondria; AKG, α-ketoglutarate; E4P, erythrose-4-phosphate; G3P, glyceraldehyde-3-phosphate; G6P, glucose-6-phosphate; OAA, oxaloacetate; P5P, pentose-5-phosphate; PEP, phosphoenol pyruvate; PYR, pyruvate.
RESULTS AND DISCUSSION
Method validation.
In order to check the reproducibility of the methodology developed and validated previously for the measurement of SFLs of intracellular metabolite fragments of a filamentous fungus (6), which was here applied to S. cerevisiae, a so-called incorporation experiment was performed. This experiment consisted of a continuous cultivation in which samples were periodically taken from the reactor outlet during a period of 9 residence times after the feeding solution was switched to the medium containing labeled glucose. The whole content of the reactor, as described in Materials and Methods, was the final sample. The incorporation of label into some of the analyzed metabolite fragments is shown in Fig. 1 (only selected fragments are shown, for the sake of clarity). It can be seen that 4 or 5 residence times are enough for the complete incorporation of label into all fragments, which means that the isotopic steady state is achieved at this point. First-order kinetics can be used as an approximation for the incorporation phenomenon, as illustrated in Fig. 1 for all fragments. With first-order kinetics, the SFL is 98.2% of the isotopic steady-state value after 4 residence times. The results shown in Fig. 1 also demonstrate the reproducibility of the measurements, as can be observed from the standard deviations in Table 3, which were calculated based on independent measurements performed on at least six different samples obtained after the isotopic steady state had been achieved.
TABLE 3.
Measured SFLs
| Metabolite | MW of unlabeled isotopomera | C atoms in measured fragment | Reported precursorb | Reported C-atom correspondenceb | SFL of the fragment (%) per condition and strain
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Batch
|
Chemostat
|
SDc | |||||||
| RSd | migl | RS | migl | ||||||
| Glucose | 331§ | 1, 2, 3, 4, 5, 6 | G6P | 1, 2, 3, 4, 5, 6 | 98.2 | 99.9 | 91.0 | 92.2 | 1.7 |
| Ser | 175# | 1, 2 | G3P | 1, 2 | 2.3 | 3.6 | 3.1 | 3.0 | 0.6 |
| Ser | 132# | 2, 3 | G3P | 2, 3 | 44.3 | 43.7 | 33.8 | 33.2 | 0.5 |
| Gly | 175# | 1, 2 | G3P | 1, 2 | 2.2 | 3.6 | 5.9 | 8.0 | 0.3 |
| Gly | 144* | 1, 2 | G3P | 1, 2 | 2.4 | 4.1 | 6.2 | 7.5 | 0.3 |
| Gly | 85* | 2 | G3P | 2 | 1.3 | 1.9 | 3.7 | 4.3 | 0.1 |
| Ala | 116# | 2, 3 | PYR | 2, 3 | 45.2 | 44.4 | 36.0 | 35.2 | 0.3 |
| Ala | 99* | 2, 3 | PYR | 2, 3 | 45.1 | 44.1 | 36.0 | 35.9 | 0.2 |
| Ala | 158* | 1, 2, 3 | PYR | 1, 2, 3 | 46.8 | 45.8 | 38.5 | 39.1 | 0.5 |
| Val | 144# | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 90.6 | NDe | 73.3 | 72.9 | 0.3 |
| Val | 143* | 1, 2 | PYR | 1, 2 | 6.9 | 6.2 | 6.9 | 7.0 | 0.3 |
| Val | 127* | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 90.2 | 89.2 | 73.1 | 73.9 | 0.4 |
| Val | 186* | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 93.0 | 91.8 | 74.9 | 73.2 | 0.8 |
| Leu | 158# | 2, 3, 4, 5, 6 | PYR + AcCoA | 2, 2, 3, 3 + 2 | 130.1 | 126.9 | 106.1 | 105.1 | 0.7 |
| Asp | 188# | 2, 3, 4 | OAA | 2, 3, 4 | 49.9 | 50.3 | 57.1 | 57.8 | 0.3 |
| Asp | 115* | 2 | OAA | 2 | 2.0 | 2.0 | 12.0 | 12.7 | 0.3 |
| Asp | 216* | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 51.7 | 52.3 | 64.4 | 66.5 | 0.4 |
| Thr | 175# | 1, 2 | OAA | 1, 2 | 2.9 | 4.6 | 17.8 | 17.5 | 0.8 |
| Thr | 146# | 2, 3, 4 | OAA | 2, 3, 4 | 47.6 | 52.3 | 54.5 | 57.4 | 1.0 |
| Ile | 158# | 2, 3, 4, 5, 6 | OAA + PYR | 2, 3, 4 + 2, 3 | 95.1 | 94.9 | 93.7 | 95.1 | 0.4 |
| Glu | 143* | 1, 2 | AKG | 1, 2 | 43.9 | 43.0 | 39.9 | 42.6 | 0.6 |
| Glu | 230* | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 95.2 | 95.4 | 100.1 | 99.5 | 0.8 |
| Pro | 142# | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 87.1 | 86.3 | 85.5 | 86.6 | 0.4 |
| Lys | 156# | 2, 3, 4, 5, 6 | AKG + AcCoA | 2, 3, 4, 5 + 2 | 125.7 | 122.6 | 117.2 | 117.2 | 0.3 |
| Phe | 143* | 1, 2 | PEP | 1, 2 | 2.5 | 2.2 | 3.0 | 2.6 | 0.1 |
| Phe | 192# | 2, 3, 4, 5, 6, 7, 8, 9 | E4P + PEP | 1, 2, 3, 4 + 2, 2, 3, 3 | 111.9 | 110.9 | 90.6 | 89.1 | 2.9 |
Superscripts indicate the type of derivatization used: §, GPA; #, ECF; *, DMFDMA. See Materials and Methods for details. MW, molecular weight; GPA, glucose pentaacetate; DMFDMA, (N,N)-dimethylformamide dimethyl acetal.
Central metabolic pathways considered here are the EMP, the PP pathway, and the TCA cycle. The information was taken from a standard biochemistry textbook (46). Abbreviations: G6P, glucose-6-phosphate; G3P, glyceraldehyde-3-phosphate; PYR, pyruvate; AcCoA, acetyl-CoA; OAA, oxaloacetate; AKG, α-ketoglutarate; PEP, phosphoenolpyruvate; E4P, erythrose-4-phosphate.
Standard deviations (SD) were determined from independent measurements performed on at least six different samples.
RS, reference strain.
ND, not determined.
Several of the results from Table 3 demonstrate the accuracy of the measurements. It is known that alanine and valine are both derived from pyruvate. According to the SFL values and the C-atom correspondence shown in Table 3, it can be seen that the SFLs obtained for all fragments of these two amino acids are in accordance with each other. For instance, the SFL of the fragment Ala116 (or Ala99) corresponds to half the value of the fragment Val144 (or Val127) in all cultivations. Similar observations can be made for the SFLs of threonine and aspartate fragments, which are both derived from oxaloacetate.
In the case of batch cultivations, it has been also observed that the measurement of labeling of intracellular metabolite fragments is the same when cells are harvested in the middle of the exponential phase (OD600, 1.1) or in the late exponential phase (OD600, 1.3) (data not shown). A similar finding was also reported for Escherichia coli (37).
Yield coefficients.
In Table 4, the yield coefficients are presented for biomass formation on glucose, as well as for the main metabolites excreted during aerobic cultivation of S. cerevisiae. These coefficients provide an idea of the overall metabolic fluxes in the cell. It can be seen that cells cultivated in the chemostat present pure respiratory metabolism, as there is no excretion of metabolites to the medium. In contrast, cells grown in the batch cultures present respiro-fermentative metabolism, leading to excretion of metabolites to the medium, which results in a lower biomass yield on glucose. It can also be seen that there is an apparently smaller excretion of metabolites coupled with a slightly higher biomass yield on glucose in the mig1 mutant compared to that in the reference strain in the batch cultivation. However, in the chemostat it is difficult to draw any quantitative conclusions when the two strains are compared with each other in terms of metabolite yields, as the concentration range of metabolites for detection after high-performance liquid chromatography separation and the available sample volume for cell concentration measurements in terms of dry weight from the 200-ml reactors are both very low. Thus, it is not possible to affirm that dry weights of 0.52 and 0.46 g are necessarily different from each other.
TABLE 4.
Specific growth rates and yield coefficientsa
| Parameter | Condition and strain
|
|||
|---|---|---|---|---|
| Batchb
|
Chemostat
|
|||
| RSd | mig1 | RS | mig1 | |
| μmax/D (h−1) | 0.37 | 0.35 | 0.11 | 0.098 |
| Ysx (g [dry wt]/g) | 0.105 | 0.125 | 0.52 | 0.46 |
| Ys(acetate) (mol/mol) | 0.065 | 0.045 | ∼0 | ∼0 |
| Ys(glycerol) (mol/mol) | 0.055 | 0.037 | ∼0 | NDc |
| Ys(ethanol) (mol/mol) | 1.56 | 1.38 | ∼0 | ∼0 |
Pyruvate and succinate yields on glucose were always below 0.01 mol/mol.
Parameters μmax and Ysx were determined in the 4-liter reactors. The maximum specific growth rates, as calculated from absorbance measurements, were the same in the 4-liter and 200-ml reactors. Metabolite yield values on glucose were also similar in both reactor types.
ND, not determined.
RS, reference strain.
Influence of Mig1p on network structure.
From an analysis of the results presented in Table 3, the first remarkable observation is that almost no difference in the SFLs is found when the two strains are compared under the same cultivation condition. The observation that the data are the same in both chemostats might be explained by the fact that the cells are not under glucose-repressing conditions and thus there is no binding of Mig1p to the promoter of repressible genes. In this case, a different phenotype in terms of fluxes would not be expected in cells that do not express the MIG1 gene compared with the reference strain. On the other hand, it is known that Mig1p binds to glucose-repressible genes, at least in the case of SUC2, when cells are grown under glucose-repressing conditions (5, 12, 15, 19, 21), which is the case for the batch cultivations performed in this work (the high specific growth rate and the residual glucose concentration in the reactor should be enough for provoking repression). With the measurements performed in this work, it is not possible to affirm that Mig1p is binding to the central metabolic genes of S. cerevisiae, which present putative binding sites for this protein. However, the results from the batch cultivation with the mig1 mutant do indicate that the central metabolism is tightly regulated through pathways that operate in parallel to the Mig1p pathway, either by avoiding its binding to the genes or by eliminating the effect that this binding would have on the metabolic fluxes. In the case of gluconeogenic functions, it is known that the activation of gene expression, mediated by Sip4p and Cat8p, plays a more important role than that of repression by Mig1p (5, 35). Thus, gluconeogenic fluxes do not occur in mig1 cells. However, a relief in the repression of respiratory and TCA-cycle genes could be expected in the batch cultivation with the mig1 mutant, and this would have yielded a different SFL pattern for the internal metabolites than that of the reference strain.
The fact that the results are very similar to each other when both strains are compared under the same cultivation condition also points to the robustness and reproducibility of the methodology employed, besides the issues presented above.
Network identification: qualitative inspection of SFL data.
It can be seen from Table 3 that the SFLs of a considerable number of amino acid fragments, as well as of glucose, could be measured by the GC-MS technique employed (6). With these measurements, it was possible to assess the SFLs of some key metabolites in the central metabolic pathways of S. cerevisiae, namely the EMP and PP pathways and the TCA cycle. Inspection of the SFLs allows identification of the network structure (in terms of the activity of different pathways), which is the first step in metabolic network analysis. In the discussion below, no separate analysis will be made with respect to the mig1 mutant strain, as the SFL data obtained for this strain were not significantly different from the data obtained for the reference strain.
(i) Glycine biosynthesis.
In the case of glycine, it is supposed that its precursor is serine, which in turn is exclusively generated from 3-phosphoglycerate (46). However, the results from the chemostats show that the SFL of the fragment Gly175 (or Gly144) is higher than the value obtained for the fragment Ser175. This indicates that there is another route for the formation of glycine, which causes an increase in the SFL of this amino acid. Three genes from S. cerevisiae were cloned and characterized, the inactivation of which are required to generate auxotrophy for glycine (25). Two of these genes, denominated SHM1 and SHM2, encode the mitochondrial and cytosolic serine hydroxymethyltransferases (SHMs), respectively. The third gene, denominated GLY1, was assigned no function. Later, two independent investigations (24, 28) reported that the gene GLY1 of S. cerevisiae encodes a threonine aldolase that catalyzes the cleavage of threonine into glycine and acetaldehyde. Thus, when grown on glucose, two pathways exist for the biosynthesis of glycine: one starting at 3-phosphoglycerate via serine and another starting at oxaloacetate via threonine. When cells are grown on nonfermentable carbon sources, e.g., ethanol and acetate, a third pathway is the major source of glycine. In this case, glyoxylate is converted into glycine via the reaction catalyzed by the enzyme alanine-glyoxylate aminotransferase (28). However, the gene coding for this enzyme has not yet been found. As this pathway is not operative under growth on glucose, the analysis performed here points towards the activity of threonine aldolase, and therefore this reaction has been included in the mathematical model for flux calculations (see Appendix).
The metabolism of the folate coenzymes is another point to be discussed. When the flux calculations were first made for the chemostat, the reaction corresponding to the decarboxylation of glycine was included in the model, but the estimated flux was zero (results not shown). This is in accordance with published results (26), which indicate that the generation of C-1 units is mainly carried out by SHM and that the glycine cleavage system only takes place when the former enzyme is inactive. Thus, it was decided to not include this reaction in further calculations, and the corresponding reaction is not shown in the Appendix.
(ii) Compartmentation of pyruvate.
It is known that valine is derived from mitochondrial pyruvate, as the first step in its biosynthetic route, catalyzed by acetolactate synthase, is mitochondrial (12). However, it is not yet known in which compartment alanine biosynthesis takes place, as there are putative alanine aminotransferases both in the cytosol and in the mitochondrion. Thus, it was decided to separate the pyruvate pool into two compartments in the model (see Appendix), although as already mentioned above, the SFLs of alanine and valine were not different from each other.
(iii) Compartmentation of acetyl-CoA.
The SFLs of amino acids derived from more than one precursor, i.e., leucine, isoleucine, lysine, and phenylalanine, can be used to gain an insight into metabolism. Subtracting the SFL of the fragment Val127 from that of the fragment Leu158, it is possible to assess the SFL of the C-2 atom of acetyl-CoA (39.9% in the batch cultivation with the reference strain, 37.7% in the batch cultivation with the mig1 mutant, 33.0% in the chemostat with the reference strain, and 31.2% in the chemostat with the mig1 mutant). This probably represents the SFL of mitochondrial acetyl-CoA, as the biosynthetic step of leucine formation catalyzed by the enzyme α-isopropylmalate synthase is located in the mitochondrion (36). Subtracting the SFL of the fragment Pro142 from that of the fragment Lys156, it is also possible to assess the SFL of the C-2 atom of acetyl-CoA (38.6% in the batch cultivation with the reference strain, 36.3% in the batch cultivation with the mig1 mutant, 31.7% in the chemostat with the reference strain, and 30.6% in the chemostat with the mig1 mutant). This probably represents the SFL of acetyl-CoA in the nucleus, as the biosynthetic step of lysine formation catalyzed by the two homocitrate synthase isoenzymes (Lys20p and Lys21p) occurs in the nucleus (4). In the model for flux calculations, this acetyl-CoA was included as cytosolic, as a means of differentiating it from the mitochondrial acetyl-CoA. In spite of the fact that the SFL values for mitochondrial and nonmitochondrial acetyl-CoA were not significantly different from each other, compartmentation of this metabolite proved to be important for minimizing the error in the calculations.
(iv) Malic enzyme.
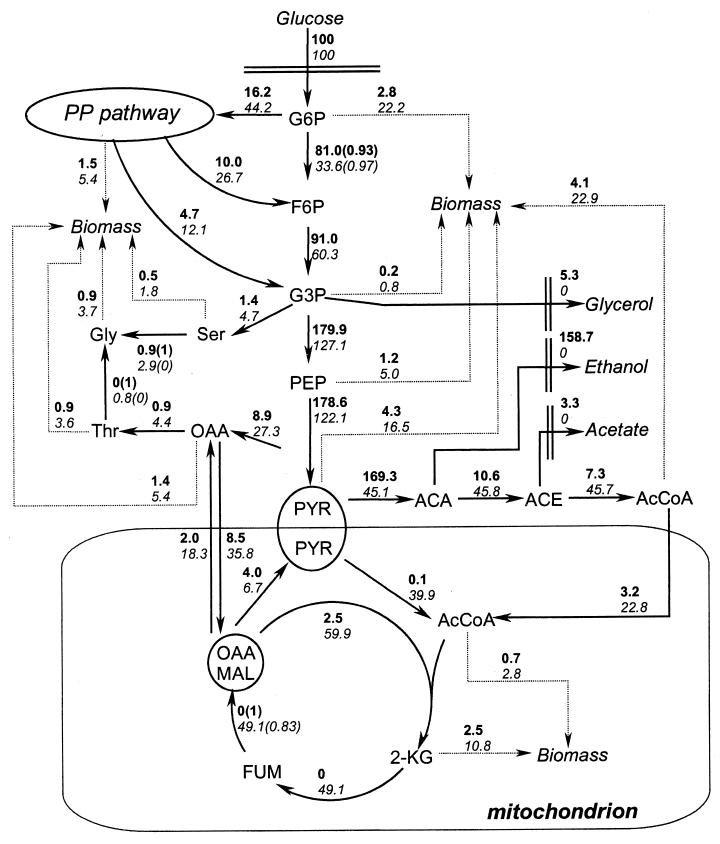
Concerning the SFL of phenylalanine, which is known to be synthesized from erythrose-4-phosphate and phosphoenolpyruvate, it is possible to see from the fragment Phe143 that the phosphoenolpyruvate C-1 and C-2 atoms have a lower SFL when compared to the SFL of pyruvate C-1 and C-2 atoms, as assessed from the fragment Val143. This observation points towards the activity of another pathway for pyruvate formation besides the pathway catalyzed by pyruvate kinase. Boles and coworkers (1) have shown that malic enzyme (Mae1p), the role of which still remains quite intriguing, is active when S. cerevisiae grows on glucose. This enzyme catalyzes the mitochondrial conversion of malate to pyruvate and may therefore explain the differences between the SFLs of pyruvate and phosphoenolpyruvate, as mentioned above. The reaction catalyzed by malic enzyme was also included in the model (see Appendix and Fig. 2).
FIG. 2.
Fluxes in the central metabolism of S. cerevisiae (reference strain). Values in bold correspond to respiro-fermentative metabolism (batch cultivation). Values in italics correspond to respiratory metabolism (chemostat). All the fluxes shown are net fluxes. For reversible reactions, the degree of reversibility is indicated in parentheses by the normalized exchange flux, calculated according to the formula vexch[0,1] = (vexch)/(vexch + vnet). All fluxes are relative to a glucose uptake of 100 (arbitrary units). Abbreviations: G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; G3P, glyceraldehyde-3-phosphate; PEP, phosphoenolpyruvate; PYR, pyruvate; ACA, acetaldehyde; ACE, acetate; AcCoA, acetyl-CoA; OAA, oxaloacetate; MAL, malate; FUM, fumarate; 2-KG, 2-ketoglutarate.
(v) TCA cycle.
When the batch cultivation is compared to the chemostat, significant differences are observed. In the former case, it becomes clear that respiro-fermentative metabolism occurs and that the TCA cycle is not operating as a cycle, as such, but rather as two branches. This can be deduced from the SFL of the C-2 atom of aspartate, which is equal to 2.0%, indicating that this carbon atom cannot originate from α-ketoglutarate but is rather coming from pyruvate via the anaplerotic reaction catalyzed by pyruvate carboxylase, which is cytosolic (16, 45). The C-2 atom of aspartate (which reflects that of oxaloacetate) can only originate from the C-2 atom of pyruvate or from the C-3 and C-4 atoms of α-ketoglutarate, considering the symmetric intermediates of the TCA cycle. As the SFLs of the proline and glutamate fragments were much higher than that of aspartate, it can be concluded that C-2 of oxaloacetate can only arise from pyruvate, which is reflected in the valine fragment and which in turn presents low labeling in the C-1 and C-2 atoms. On the other hand, in the chemostats cells grow with purely respiratory metabolism. In this case, the C-2 atom of aspartate has an SFL of 12.0%, indicating that it is at least in part formed from α-ketoglutarate (based on the high label in the proline and glutamate fragments and the low label in the valine fragments).
(vi) PP pathway.
Another observation is that the amino acid fragments originating from precursor metabolites in the EMP pathway, such as 3-phosphoglycerate and pyruvate, present lower SFLs in the chemostat than in the batch cultivation. This is due to the higher PP pathway flux in the former. A high PP pathway flux is required when the biomass yield is high, because a relatively larger amount of glucose has to be used for generation of NADPH. A high PP pathway flux gives lower SFLs for the lower glycolytic intermediates, since decarboxylation of glucose-6-phosphate carried out in the first steps of the PP pathway removes exactly the C-1 atom of this molecule, which is the one that receives the label from [1-13C]glucose.
The higher SFL observed for the glucose-6-phosphate fragment in the batch culture, when compared to that in the chemostat, reflects the lower PP pathway flux in the former case, as less fructose-6-phosphate is being converted back to glucose-6-phosphate by glucose-6-phosphate isomerase; this in turn causes a smaller “dilution” of the label in the C-1 atom of glucose-6-phosphate (Fig. 2).
Metabolic fluxes.
The analysis above, concerning the identification of the network structure, was made by inspecting the measured SFLs in a qualitative or semiquantitative manner. By combining these measurements with a suitable mathematical model, it was possible to quantify the fluxes in the central metabolism of S. cerevisiae. In this way, it was possible to compare cells growing in a chemostat at steady state, with a specific growth rate of 0.1 h−1, with cells growing in a batch cultivation, with μmax = 0.37 h−1, in a quantitative fashion. The process of estimating fluxes is characterized by the process of finding the most suitable model that describes the metabolism under investigation, in a sort of trial-and-error process, which is based on biochemical knowledge and appropriate assumptions (with the aim of both fixing nondefined biochemical details and avoiding numerical problems in the calculations). The most suitable model for the experiments performed in this work is the one shown in the Appendix. The same model was suitable for both cultivation conditions investigated.
The estimated fluxes for both cultivation conditions investigated are shown in Fig. 2. In Table 5, the values of the calculated and the measured SFLs are shown and give an indication of the accuracy of the calculations performed. From these data, it can be seen that almost all SFLs calculated using the mathematical model presented in the Appendix are similar to the measured values if a deviation of one absolute unit for the SFLs is accepted. Reversibility is shown in Fig. 2 for the key reversible reactions in terms of the normalized exchange fluxes. The reversibility in the transaldolases and transketolases in the PP pathway are not shown because the number of measurements of SFLs of the compounds in this pathway does not allow a precise estimation of the reversibility of these reactions. However, the net flux values for these reactions did not change significantly when different calculations were performed.
TABLE 5.
Calculated and measured SFLs in the chemostat and batch cultivations with the reference strain
| Fragmenta | SFL (%)
|
|||
|---|---|---|---|---|
| Chemostat
|
Batch
|
|||
| Calculated | Measured | Calculated | Measured | |
| Glc331 | 91.1 | 91.0 | 98.5 | 98.2 |
| Gly175 | 6.2 | 5.9 | 2.9 | 2.2 |
| Gly144 | 6.2 | 6.2 | 2.9 | 2.4 |
| Gly85 | 3.2 | 3.7 | 1.4 | 1.3 |
| Ser175 | 3.5 | 3.1 | 2.9 | 2.3 |
| Ser132 | 34.4 | 33.8 | 44.3 | 44.3 |
| Ala116 | 34.4 | 36.0 | 43.9 | 45.2 |
| Ala99 | 34.4 | 36.0 | 43.9 | 45.1 |
| Ala158 | 36.6 | 38.5 | 46.2 | 46.8 |
| Leu158 | 105.1 | 106.1 | 128.4 | 130.1 |
| Val144 | 72.7 | 73.3 | 87.9 | 90.6 |
| Val143 | 7.7 | 6.9 | 5.6 | 6.9 |
| Val127 | 72.7 | 73.1 | 87.9 | 90.2 |
| Val186 | 76.2 | 74.9 | 90.2 | 93.0 |
| Asp188 | 58.9 | 57.1 | 53.2 | 49.9 |
| Asp115 | 10.3 | 12.0 | 3.3 | 2.0 |
| Asp216 | 65.4 | 64.4 | 55.4 | 51.7 |
| Ile158 | 95.2 | 93.7 | 97.1 | 95.1 |
| Glu143 | 40.2 | 39.9 | 42.7 | 43.9 |
| Glu230 | 99.6 | 100.1 | 96.0 | 95.2 |
| Pro142 | 85.2 | 85.5 | 87.9 | 87.1 |
| Lys156 | 118.1 | 117.2 | 128.4 | 125.7 |
| Phe192 | 91.8 | 90.6 | 110.3 | 111.9 |
| Phe143 | 3.5 | 3.0 | 3.0 | 2.5 |
For details on the fragments, see footnotes of Table 3. The threonine fragments have not been included in these calculations.
A first observation that can be made is on the PP pathway flux. It can be seen that the value of 44.2 for the chemostat is much higher than 16.2 obtained for the batch cultivation (both values are relative to a glucose uptake of 100 arbitrary units). This is in accordance with the higher need of NADPH for anabolic purposes in the former case (Ysx = 0.5 g/g). In the latter case, although the protein content in the cells is higher (Table 1), ethanol was the main product arising from glucose catabolism, and the cell yield was about 0.1 g/g. If the specific uptake rate of glucose, which was 15.9 mmol/g (dry weight)/h in the batch culture and 1.17 mmol/g (dry weight)/h in the chemostat, is multiplied by the calculated relative PP pathway flux, it is possible to observe that the absolute flux through the PP pathway is actually higher in the batch culture (2.57 mmol/g [dry weight]/h) than in the chemostat (0.51 mmol/g [dry weight]/h), reflecting the higher general glycolytic flux in respiro-fermenting cells compared to respiratory cells. Different values for the PP pathway flux have been reported. Gancedo and Lagunas (14) reported that the PP pathway accounts for only 2.5% of the total metabolism of glucose when cells are grown on glucose and ammonia. This value was calculated from measurements on the radioactivity of the CO2 produced by cells growing in shake flask cultures. Due to the reversibility of the glucose-6-phosphate isomerase-catalyzed reaction, this method is likely to give an underestimation of the PP pathway flux. The amount of NADPH required for biomass formation has been estimated to be 831 or 931 mmol per 100 g (dry weight) (2, 34). Using these values it is possible to make an estimation of the PP pathway flux if we consider that this pathway is the sole route for NADPH formation. With a biomass yield of 0.5 g (dry weight)/g glucose, for each 100 mmol of glucose consumed 9 g (dry weight) are formed, and 74.8 or 83.8 mmol of NADPH are required. As every glucose-6-phosphate molecule that enters the PP pathway generates two molecules of NADPH, a flux of 37.4 or 41.9 mmol per 100 mmol of glucose taken up is obtained, which is not far from the value estimated here. Applying the same reasoning for fermenting cells with a Ysx of 0.1 g (dry weight)/g, values of 7.5 or 8.4 mmol can be calculated for the PP pathway flux. In the case of fermenting cells, the value estimated in our work is higher (16.2 mmol). It is important to note that our analysis is based on two main issues. First, we were able to assess the SFL of glucose-6-phosphate with a high degree of reliability (Table 3), and second, the reversibility of the reaction catalyzed by glucose-6-phosphate isomerase has been included in the calculations (Appendix and Fig. 2).
Concerning the TCA cycle, it can be seen that it operates in a cyclic manner in cells grown in the chemostat, whereas it operates as two branches in the cells grown in the batch cultivation, one of which is oxidative, leading to α-ketoglutarate, and one of which is reductive, leading to fumarate. It can be seen in Fig. 2 that the flux leading from α-ketoglutarate to fumarate is zero in respiro-fermenting cells, and the net flux leading from fumarate to oxaloacetate is also zero, although there is an exchange flux in this case. To our knowledge this is the first report of this in vivo observation. Succinate and succinyl-CoA are not included in the model, and it is therefore not possible to identify exactly where the cycle is interrupted. The compartmentation of oxaloacetate was a key point in fitting the calculated SFLs to the measured values, as the anaplerotic reaction catalyzed by pyruvate carboxylase and the reaction catalyzed by malic enzyme take place in different compartments.
The inclusion of malic enzyme in the model was the only way of accounting for the differences found in the SFLs of pyruvate and phosphoenolpyruvate. Although enzyme activity measurements and Northern blot analyses indicate that the malic enzyme-catalyzed reaction presents a higher flux in a batch culture at maximum specific growth rate than in a chemostat with D = 0.1 h−1 (1), our results indicate similar flux values under both cultivation conditions (Fig. 2). The physiological reasons for this observation and for the general role of malic enzyme remain to be elucidated.
The formation of glycine by two different routes, namely via SHM- and threonine aldolase-catalyzed reactions, has been included in the model. Our results show a higher contribution to glycine formation from the SHM-catalyzed reaction compared to the threonine aldolase-catalyzed reaction (Fig. 2). However, some published results (25) show that growth is more affected when the GLY1 gene (coding for threonine aldolase) is deleted compared to growth after deletion of both the SHM1 and SHM2 genes.
Conclusions.
Metabolic network analysis, as applied in the present work to cells of S. cerevisiae, is a tool that can be utilized for the quantitative inspection of metabolism. The first step in this analysis is the identification of the network structure, which is basically achieved by combining the inspection of the SFLs of intracellular metabolites, as measured by GC-MS, with biochemical knowledge. This step may bring insight into which enzymes are active and which ones are not active and if there is an unknown reaction taking place under the cultivation condition under investigation or in specific mutant cells. The second step in this analysis is a consequence of the first one. A mathematical model describing the metabolic network, as identified in the first step, combined with the measured SFLs with the aim of estimating the metabolic fluxes. These data can be used for the quantitative comparison of cells grown under different environmental conditions or for the comparison of different mutants. Besides yielding important physiological information for well-characterized species, such as the activities of the PP pathway, TCA cycle, threonine aldolase, and malic enzyme, as shown in this work with S. cerevisiae, metabolic network analysis is a promising tool for investigating other poorly characterized species with potential biotechnological applications.
ACKNOWLEDGMENTS
Andreas Karoly Gombert gratefully acknowledges financial support by CAPES (Brasília, Brazil), grant number BEX1098/98-5. Margarida Moreira dos Santos acknowledges Fundação para a Ciência e Tecnologia (Portugal) for financial support.
Appendix
The model used for flux calculations is given in Table A1.
TABLE A1.
Model used for flux calculations; the C atom transitions, which are used by the flux estimation routine, are also indicated
| Measure and substrate | Product(s) |
|---|---|
| % Glucose uptake | |
| GLC | G6P |
| abcdef | abcdef |
| % EMP pathway | |
| G6P | F6P |
| abcdef | abcdef |
| F6P + G6P | G6P + F6P |
| bcdef ghijkl | abcdef ghijkl |
| 6P | G3P + G3P |
| bcdef | cba def |
| 3P | PEP |
| abc | abc |
| PEP | PYR |
| abc | abc |
| % PP-pathway | |
| G6P | CO + P5P |
| abcdef | a bcdef |
| P5P + P5P | S7P + G3P |
| abcde fghij | fgabcde hij |
| S7P + G3P + P5P + P5P | P5P + P5P + S7P + G3P |
| fgabcde hij klmno pqrst | abcde fghij klpqrst mno |
| S7P + G3P | F6P + E4P |
| abcdefg hij | abchij defg |
| F6P + E4P + S7P + G3P | S7P + G3P + F6P + E4P |
| abchij defg klmnopq rst | abcdefg hij klmrst nopq |
| P5P + E4P | F6P + G3P |
| abcde fghi | abfghi cde |
| F6P + G3P + P5P + E4P | P5P + E4P + F6P + G3P |
| abfghi cde mklmn opqr | abcde fghi jkopqr lmn |
| % Ethanol, acetate, and glycerol formation | |
| PYR | ACA + CO |
| abc | bc a |
| ACA | ETH |
| ab | ab |
| ACA | ACE |
| ab | ab |
| G3P | GLYC |
| abc | abc |
| % Formation of AcCoA in the cytosol | |
| ACE | ACCOACYT |
| ab | ab |
| % Anaplerotic reaction (cytosolic) | |
| PYR + CO | OAACYT |
| abc d | abcd |
| % TCA cycle (considering scrambling around FUM) | |
| PYR | ACCOAMIT + CO |
| abc | bc a |
| OAAMIT + ACCOAMIT | ICIT |
| abcd ef | dcbafe |
| ICIT | AKG + CO |
| abcdef | abcef d |
| AKG | FUM + CO |
| abcde | bcde a |
| FUM + FUM | OAAMIT + OAAMIT |
| abcd efgh | abcd hgfe |
| OAAMIT | FUM |
| abcd | abcd |
| % Transports | |
| OAAMIT | OAACYT |
| abcd | abcd |
| OAACYT | OAAMIT |
| abcd | abcd |
| ACCOACYT | ACCOAMIT |
| ab | ab |
| % Threonine, serine, and glycine metabolism (all enzymes assumed to be cytoplasmic) | |
| G3P | SER |
| abc | abc |
| SER | GLY + C-1 |
| abc | ab c |
| SER + Gly + C-1 | SER + GLY + C-1 |
| abc de f | def ab c |
| OAACYT | THR |
| abcd | abcd |
| THR | GLY + ACA |
| abcd | ab cd |
| GLY + ACA + THR | THR + GLY + ACA |
| ab cd efgh | abcd ef gh |
| % Malic enzyme (malate decarboxylation, mitochondrial) | |
| OAAMIT | PYR + CO |
| abcd | abc d |
| % Pyruvate pool | |
| PYRCYT | PYR |
| abc | abc |
| PYRMIT | PYR |
| abc | abc |
| % Drain of intermediates to macromolecules | |
| G6P | G6POUT |
| abcdef | abcdef |
| P5P | P5POUT |
| abcde | abcde |
| E4P | E4POUT |
| abcd | abcd |
| G3P | G3POUT |
| abc | abc |
| PEP | PEPOUT |
| abc | abc |
| PYR | PYROUT |
| abc | abc |
| OAACYT | OAACYTOUT |
| abcd | abcd |
| AKG | AKGOUT |
| abcde | abcde |
| ACCOACYT | ACCOACYTOUT |
| ab | ab |
| ACCOAMIT | ACCOAMITOUT |
| ab | ab |
| SER | SEROUT |
| abc | abc |
| GLY | GLYOUT |
| ab | ab |
| C1 | C1OUT |
| a | a |
| GLYC | GLYCOUT |
| abc | abc |
| THR | THROUT |
| abcd | abcd |
| % Excreted products | |
| ETH | ETHOUT |
| ab | ab |
| ACE | ACEOUT |
| ab | ab |
| % CO2 evolution | |
| CO | COOUT |
| a | a |
Abbreviations: ACA, acetaldehyde; ACE, acetate; ACEOUT, acetate excreted to the medium; ACCOACYT, acetyl-CoA in the cytosol; ACCOACYTOUT, cytosolic acetyl-CoA used for biomass formation; ACCOAMIT, acetyl-CoA in the mitochondria; ACCOAMITOUT, mitochondrial acetyl-CoA used for biomass formation; AKG, α-ketoglutarate; AKGOUT, 2-ketoglutarate used for biomass formation; CO, carbon dioxide; COOUT, carbon dioxide excreted to the medium; E4P, erythrose-4-phosphate; E4POUT, erythrose-4-phosphate used for biomass formation; ETH, ethanol; ETHOUT, ethanol excreted to the medium; F6P, fructose-6-phosphate; FUM, fumarate; GLC, glucose; GLYC, glycerol; GLYOUT, glycine used for biomass formation; G3P, glyceraldehyde-3-phosphate; G3POUT, glyceraldehyde-3-phosphate used for biomass formation; G6P, glucose-6-phosphate; G6POUT, glucose-6-phosphate used for biomass formation; ICIT, isocitrate; OAACYT, oxaloacetate in the cytosol; OAACYTOUT, cytosolic oxaloacetate used for biomass formation; OAAMIT, oxaloacetate in the mitochondria; PEP, phosphoenolpyruvate; PEPOUT, phosphoenolpyruvate used for biomass formation; P5P, pentose-5-phosphate; P5POUT, pentose-5-phosphate used for biomass formation; PYR, pyruvate; PYRCYT, pyruvate in the cytosol; PYRMIT, pyruvate in the mitochondria; PYROUT, pyruvate used for biomass formation; SEROUT, serine used for biomass formation; S7P, sedoheptulose-7-phosphate; THROUT, threonine used for biomass formation. (Standard three-letter abbreviations are used for the amino acids.)
REFERENCES
- 1.Boles E, de Jong-Gubbels P, Pronk J T. Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J Bacteriol. 1998;180:2875–2882. doi: 10.1128/jb.180.11.2875-2882.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruinenberg P M, van Dijken J P, Scheffers A. A theoretical analysis of NADPH production and consumption in yeasts. J Gen Microbiol. 1983;129:953–964. doi: 10.1099/00221287-129-4-965. [DOI] [PubMed] [Google Scholar]
- 3.Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 4.Chen S, Brockenbrough J S, Dove J E, Aris J P. Homocitrate synthase is located in the nucleus in the yeast Saccharomyces cerevisiae. J Biol Chem. 1997;16:10839–10846. doi: 10.1074/jbc.272.16.10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen B, Nielsen J. Metabolic network analysis: a powerful tool in metabolic engineering. Adv Biochem Eng Biotechnol. 1999;66:209–231. [PubMed] [Google Scholar]
- 6.Christensen B, Nielsen J. Isotopomer analysis unsing GC-MS. Metab Eng. 1999;1:282–290. doi: 10.1006/mben.1999.0117. [DOI] [PubMed] [Google Scholar]
- 7.Christensen B, Nielsen J. Metabolic network analysis on Penicillium chrysogenum using 13C-labeled glucose. Biotechnol Bioeng. 2000;68:652–659. [PubMed] [Google Scholar]
- 8.Daae E B, Ison A P. Classification and sensitivity analysis of a proposed primary metabolic reaction network for Streptomyces lividans. Metabol Eng. 1999;1:153–165. doi: 10.1006/mben.1998.0112. [DOI] [PubMed] [Google Scholar]
- 9.Entian K-D, et al. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol Gen Genet. 1999;262:683–702. doi: 10.1007/pl00013817. [DOI] [PubMed] [Google Scholar]
- 10.Entian K-D, Schüller H-J. Glucose repression (carbon catabolite repression) in yeast. In: Zimmermann F K, Entian K-D, editors. Yeast sugar metabolism. Basel, Switzerland: Technomic; 1997. pp. 409–434. [Google Scholar]
- 11.Ertugay N, Hamamci H. Continuous cultivation of baker's yeast: change in cell composition at different dilution rates and effect of heat stress on trehalose level. Folia Microbiol. 1997;42:463–467. doi: 10.1007/BF02826554. [DOI] [PubMed] [Google Scholar]
- 12.Falco S C, Dumas K S, Livak K J. Nucleotide sequence of the yeast ILV2 gene which encodes acetolactate synthase. Nucleic Acids Res. 1985;13:4011–4027. doi: 10.1093/nar/13.11.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gancedo J M. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gancedo J M, Lagunas R. Contribution of the pentose-phosphate pathway to glucose metabolism in Saccharomyces cerevisiae: a critical analysis of the use of labelled glucose. Plant Sci Lett. 1973;1:193–200. [Google Scholar]
- 15.Goffeau A. The yeast genome directory. Nature. 1997;387:5–6. [PubMed] [Google Scholar]
- 16.Haarasilta S, Taskinen L. Location of three key enzymes of gluconeogenesis in baker's yeast. Arch Microbiol. 1977;113:159–161. doi: 10.1007/BF00428597. [DOI] [PubMed] [Google Scholar]
- 17.Johnston M. Feasting, fasting and fermenting. Trends Genet. 1999;15:29–33. doi: 10.1016/s0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- 18.Klein C J L, Rasmussen J J, Rønnow B, Olsson L, Nielsen J. Investigation of the impact of MIG1 and MIG2 on the physiology of Saccharomyces cerevisiae. J Biotechnol. 1999;68:197–212. doi: 10.1016/s0168-1656(98)00205-3. [DOI] [PubMed] [Google Scholar]
- 19.Klein C J L, Olsson L, Nielsen J. Glucose control in Saccharomyces cerevisiae: the role of MIG1 in metabolic functions. Microbiology. 1998;144:13–24. doi: 10.1099/00221287-144-1-13. [DOI] [PubMed] [Google Scholar]
- 20.Kraakman L S, Winderickx J, Thevelein J M, de Winde J H. Structure-function analysis of yeast hexokinase: structural requirements for triggering signalling and catabolite repression. Biochem J. 1999;343:159–168. [PMC free article] [PubMed] [Google Scholar]
- 21.Küenzi M T, Fiechter A. Regulation of carbohydrate composition of Saccharomyces cerevisiae under growth limitation. Arch Mikrobiol. 1972;84:254–265. doi: 10.1007/BF00425203. [DOI] [PubMed] [Google Scholar]
- 22.Lee W-N P, Bergner E A, Guo Z K. Mass isotopomer pattern and precursor-product relationship. Biol Mass Spectrom. 1992;21:114–122. doi: 10.1002/bms.1200210210. [DOI] [PubMed] [Google Scholar]
- 23.Lee W-N P, Byerley L O, Bergner E A, Edmond J. Mass isotopomer analysis: theoretical and practical considerations. Biol Mass Spectrom. 1991;20:451–458. doi: 10.1002/bms.1200200804. [DOI] [PubMed] [Google Scholar]
- 24.Liu J-Q, Nagata S, Dairi T, Misono H, Shimizu S, Yamada H. The GLY1 gene of Saccharomyces cerevisiae encodes a low-specific l-threonine aldolase that catalyzes cleavage of l-allo-threonine and l-threonine to glycine. Expression of the gene in Escherichia coli and purification and characterization of the enzyme. Eur J Biochem. 1997;245:289–293. doi: 10.1111/j.1432-1033.1997.00289.x. [DOI] [PubMed] [Google Scholar]
- 25.McNeil J B, McIntosh E M, Taylor B V, Zhang F, Tangt S, Bognar A L. Cloning and molecular characterization of three genes, including two genes encoding serine hydroxymethyltransferases, whose inactivation is required to render yeast auxotrophic for glycine. J Biol Chem. 1994;269:9155–9165. [PubMed] [Google Scholar]
- 26.McNeil J B, Bognar A L, Pearlman R E. In vivo analysis of folate coenzymes and their compartmentation in Saccharomyces cerevisiae. Genetics. 1996;142:371–381. doi: 10.1093/genetics/142.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meijer M M C, Boonstra J, Verkleij A J, Verrips C T. Glucose repression in Saccharomyces cerevisiae is related to the glucose concentration rather than the glucose flux. J Biol Chem. 1998;273:24102–24107. doi: 10.1074/jbc.273.37.24102. [DOI] [PubMed] [Google Scholar]
- 28.Monschau N, Stahmann K-P, Sahm H, McNeil J B, Bognar A L. Identification of Saccharomyces cerevisiae GLY1 as a threonine aldolase: a key enzyme in glycine biosynthesis. FEMS Microbiol Lett. 1997;150:55–60. doi: 10.1111/j.1574-6968.1997.tb10349.x. [DOI] [PubMed] [Google Scholar]
- 29.Nehlin J O, Ronne H. Yeast Mig1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nissen T, Schulze U, Nielsen J, Villadsen J. Flux distributions in anaerobic, glucose-limited continuous cultures of Saccharomyces cerevisiae. Microbiology. 1997;143:203–218. doi: 10.1099/00221287-143-1-203. [DOI] [PubMed] [Google Scholar]
- 31.Oliver S G, Winson M K, Kell D B, Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16:373–378. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 32.Östling J, Ronne H. Negative control of the Mig1p repressor by Snf1-dependent phosphorylation in the absence of glucose. Eur J Biochem. 1998;252:162–168. doi: 10.1046/j.1432-1327.1998.2520162.x. [DOI] [PubMed] [Google Scholar]
- 33.Oura E. The effect of aeration on the growth energetics and biochemical composition of baker's yeast, with an appendix: reactions leading to the formation of yeast cell material from glucose and ethanol. Ph.D. thesis. Helsinki, Finland: Helsinki University; 1972. [Google Scholar]
- 34.Oura E. Biomass from carbohydrates. In: Rehm H-J, Reed G, editors. Biotechnology. Weinheim, Germany: Verlag Chemie; 1983. [Google Scholar]
- 35.Rahner A, Hiesinger M, Schüller H-J. Deregulation of gluconeogenic structural genes by variants of the transcriptional activator Cat8p of the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;34:146–156. doi: 10.1046/j.1365-2958.1999.01588.x. [DOI] [PubMed] [Google Scholar]
- 36.Ryan E D, Tracy J W, Kohlhaw G B. Subcellular localization of the leucine biosynthetic enzymes in yeast. J Bacteriol. 1973;116:222–225. doi: 10.1128/jb.116.1.222-225.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauer U, Lasko D R, Fiaux J, Hochuli M, Glaser R, Szyperski T, Wüttrich K, Bailey J E. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J Bacteriol. 1999;181:6679–6688. doi: 10.1128/jb.181.21.6679-6688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szyperski T. 13C-NMR, MS and metabolic flux balancing in biotechnology research. Q Rev Biophys. 1998;31:41–106. doi: 10.1017/s0033583598003412. [DOI] [PubMed] [Google Scholar]
- 39.Treitel M A, Kuchin S, Carison M. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6273–6280. doi: 10.1128/mcb.18.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vallino J J, Stephanopoulos G. Flux determination in cellular bioreaction networks: applications to lysine fermentations. In: Sikdar S K, Bier M, Todd O, editors. Frontiers in bioprocessing. Boca Raton, Fla: CRC Press; 1990. pp. 205–219. [Google Scholar]
- 41.van Gulik W M, Heijnen J J. A metabolic network stoichiometry analysis of microbial growth and product formation. Biotechnol Bioeng. 1995;48:681–698. doi: 10.1002/bit.260480617. [DOI] [PubMed] [Google Scholar]
- 42.Varma A, Palsson B O. Metabolic flux balancing: basic concepts, scientific and practical use. Bio/Technology. 1994;12:994–998. [Google Scholar]
- 43.Verduyn C. Physiology of yeasts in relation to biomass yields. Antonie Leeuwenhoek. 1991;60:325–353. doi: 10.1007/BF00430373. [DOI] [PubMed] [Google Scholar]
- 44.Verduyn C, Postma E, Scheffers W A, van Dijken J P. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 45.Walker M E, Val D L, Rohde M, Devenish R J, Wallace J C. Yeast pyruvate carboxylase: identification of two genes encoding isoenzymes. Biochem Biophys Res Commun. 1991;176:1210–1217. doi: 10.1016/0006-291x(91)90414-3. [DOI] [PubMed] [Google Scholar]
- 46.Zubay G. Biochemistry. 2nd ed. New York, N.Y: Macmillan; 1988. [Google Scholar]