Abstract
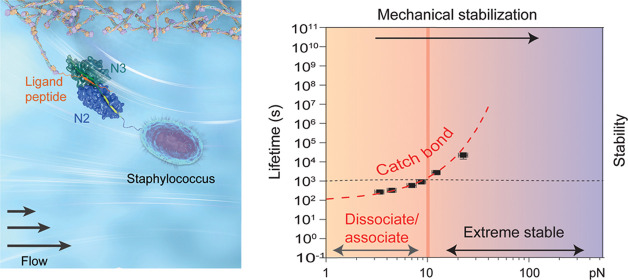
The adhesions between Gram-positive bacteria and their hosts are exposed to varying magnitudes of tensile forces. Here, using an ultrastable magnetic tweezer-based single-molecule approach, we show the catch-bond kinetics of the prototypical adhesion complex of SD-repeat protein G (SdrG) to a peptide from fibrinogen β (Fgβ) over a physiologically important force range from piconewton (pN) to tens of pN, which was not technologically accessible to previous studies. At 37 °C, the lifetime of the complex exponentially increases from seconds at several pN to ∼1000 s as the force reaches 30 pN, leading to mechanical stabilization of the adhesion. The dissociation transition pathway is determined as the unbinding of a critical β-strand peptide (“latch” strand of SdrG that secures the entire adhesion complex) away from its binding cleft, leading to the dissociation of the Fgβ ligand. Similar mechanical stabilization behavior is also observed in several homologous adhesions, suggesting the generality of catch-bond kinetics in such bacterial adhesions. We reason that such mechanical stabilization confers multiple advantages in the pathogenesis and adaptation of bacteria.
Introduction
Gram-positive pathogenic bacteria produce cell wall-anchored (CWA) surface proteins that are important for the bacteria to colonize the host and promote infections.1,2 The most prevalent CWA proteins are the microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), which are critical in bacterial “adhesion, invasion, and immune evasion.”3 During colonization and infection, these MSCRAMM-mediated adhesions are subjected to mechanical forces associated with flow stress induced by the dynamic oscillated blood pressure, airflow, and other hydrostatic pressures.4−7 The tensile forces applied to a single adhered bacterial cell span over a wide range of magnitude, from the nanonewton (nN) range in the urinary tract needed to withstand the high speed of urinary flow down to piconewton (pN) in capillaries according to the reported shear stress.8,9
Many MSCRAMMs promote specific binding to the host peptide ligand through a “dock, lock, and latch” (DLL) mechanism,1,10 where the ligand is bound (dock) and buried (lock) in between the two tandemly arrayed immunoglobulin (Ig)-like folded domains, N2 and N3, in the N-terminal region of MSCRAMMs. The ligand is further secured by a “latch” strand located at the C-terminal of the N3 domain, which binds to N2 by β-strand complementation1,11 (latch, Figure 1A), resulting in a unique adhesin–ligand complex. It has been shown that MSCRAMM-mediated adhesion confers extreme mechanical stability capable of withstanding large shear force.12−16 Recent single-molecule studies using atomic force microscopy (AFM) reported rupturing forces of MSCRAMM adhesion in the order of nN,12,13,15 reaching the mechanical stability of covalent bonds.17,18 In the typical force loading rates of 104–107 pN s–1 in the AFM experiment, the adhesion complex exhibits slip-bond kinetics where the adhesion stability decreases as the force increases in the nN force range.12,15 Steered molecular dynamics (SMD) simulations15 suggested directly pulling out the ligand through the latched binding pocket as the rupturing transition pathway, which requires the breakage of the hydrogen network in a shear force geometry (i.e., the applied force is along the direction of the interaction interface).
Figure 1.
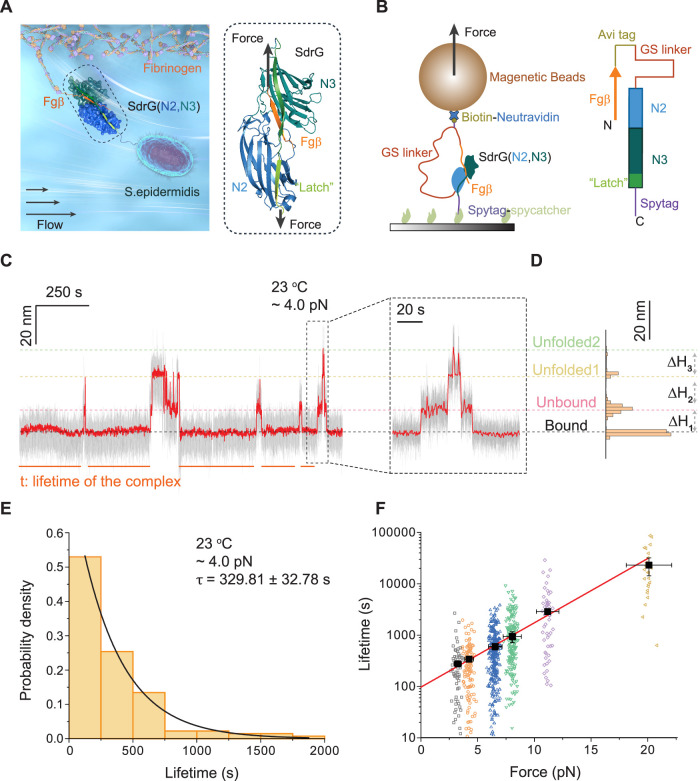
Force-dependent dissociation of the SdrG–Fgβ complex. (A) Illustration of Staphylococcus epidermidis binding to the fibrinogen surface via an adhesion complex formed between the N2 and N3 domains of bacterial SdrG and the Fgβ peptide ligand. The right panel shows the molecular structure and the force geometry of the complex (N2: blue, N3: green, Fgβ: orange). (B) Schematic diagram of the recombinant protein construct tethered between a glass substrate and a superparamagnetic microbead. The right panel shows the domain map of the SdrG–Fgβ protein construct. (C) A representative bead height–time trace at 4.0 ± 0.4 pN. The right panel shows an enlarged time trace. Raw data and smoothed data are indicated by gray and red, respectively. (D) The histogram of the smoothed bead height shows four peaks corresponding to different structural states of the tethered construct. (E) Normalized histogram of the lifetime of the SdrG–Fgβ complex at 4.0 ± 0.4 pN (n = 134). An average lifetime of τ = 329.81 ± 32.78 s was obtained by single exponential function fitting (black curve). (F) Force-dependent lifetimes of the SdrG–Fgβ complex at 3.2 ± 0.3 pN (open black squares, n = 64), 4.0 ± 0.4 pN (open orange circles, n = 134), 6.5 ± 0.7 pN (open blue up triangles, n = 257), 8.0 ± 0.8 pN (open green down triangles, n = 186), 10.5 ± 1.0 pN (open purple diamonds, n = 54), and 20.0 ± 0.2 pN (open yellow left-triangles, n = 24). The average lifetime is indicated by black solid squares. The red solid line is the best fitting to the force-dependent average lifetime using Bell’s model (eq 1). Error bars indicate the mean ± standard error.
The high resilience of MSCRAMM-mediated adhesion to large stresses enables firm anchorage to a host surface, while its mechanical stability under lower forces from pN to tens of pN, also a physiologically important range, is still unexplored. It has been hypothesized that the MSCRAMM-mediated adhesion may have a unique advantageous mechanical stabilization property, a counterintuitive phenomenon in which the stability of the complexes increases as the force increases, which is referred to as catch-bond kinetics.13,15,16,18−20 The catch-bond kinetics has been observed in various biomolecular systems in both humans and bacteria.21−28 However, due to the ultrastable MSCRAMM-mediated adhesions, direct exploration of their mechanical properties at significantly lower forces has been inaccessible to the previous AFM technologies. In addition, there are potential desorption processes during bacteria diffusion and spreading across surfaces, in which detachment of the adhesion is required.4,19,29,30 It is thus imperative to investigate the mechanical responses and the potential modulations of MSCRAMM-mediated adhesion under lower forces.
In this work, using an ultrastable magnetic tweezer-based single-molecule approach, we set to address several outstanding questions related to the low-force mechanical responses of MSCRAMM-mediated adhesions: (1) How is the lifetime of the MSCRAMM-mediated adhesions regulated by tensile forces? (2) Does the hypothesized catch-bond kinetics exist at lower forces? (3) What are the molecular and physical mechanisms governing the mechanical stability of the MSCRAMM-mediated adhesions? By direct quantification of the force-dependent lifetimes of a prototypical adhesin protein complex formed between SD-repeat protein G (SdrG) from S. epidermidis and the ligand peptide from the β chain of human fibrinogen (Fgβ), as well as several homologous complexes, we show that increasing force leads to drastically increased lifetimes; thus, the catch-bond hypothesis is directly tested to be true. The transition pathway over this low-force range (from pN to tens of pN) is identified to be the unbinding of the “latch” peptide away from the N2 binding cleft of SdrG, different from that reported at the high-force range (nN range).15 Analysis based on the Arrhenius law of kinetics and the structural elastic property31 of the identified transition pathway reveals that the highly pre-extended “latch” structure of the complex confers the observed catch-bond kinetics. In addition, the highly sensitive temperature dependence of the SdrG–Fgβ complex is revealed, indicated by a drastically decreased lifetime at the human body temperature (37 °C) from that at 23 °C. Collectively, our studies provide insights into the molecular and physical mechanisms underlying the force-dependent stability and the mechanical regulation of such MSCRAMM adhesion complexes, which may provide multiple advantages in the adaptation of the bacterial adhesion to the hosts.
Results
Catch-Bond Behavior in the Dissociation of the SdrG–Fgβ Complex
To reversibly probe the rupturing events of the single-molecule SdrG–Fgβ complex, a recombinant protein construct was designed by linking the Fgβ ligand peptide to the N2 and N3 domains of SdrG through an unstructured peptide linker (Figure 1B). The N1 and B domains of SdrG that span the N2 and N3 domains are not included, since structurally they do not clash with the SdrG–Fgβ binding interface10 and thus should not affect the mechanical stability of the SdrG–Fgβ complex. The Fgβ ligand peptide is located at the N-terminus, followed by biotinylated AviTag, a flexible unstructured peptide linker (glycine-serine, GS linker), the N2 and N3 domains, and finally SpyTag at the C-terminus. The construct was tethered between a Neutravidin-coated superparamagnetic microbead and a SpyCatcher-coated coverslip surface via a specific covalent SpyTag–SpyCatcher bond32 and noncovalent Neutravidin–biotin interactions that confer strong mechanical stability33−35 (Figure 1B). An external force was applied to the microbead using magnetic tweezers, subjecting the SdrG–Fgβ complex to tension along both the C-termini of the ligand and the N3 domain, mimicking the native force geometry.10,15,18 Once the ligand was dissociated from SdrG, the bead height would increase by an amount of ΔH1 (equal to the extension change of the protein construct from the released GS linker; Figure S1), which was tracked at a nanometer resolution in real time.
We found that the SdrG–Fgβ complex could rupture at pN forces over a time scale of hundreds of seconds to hours at room temperature (∼23 °C). A representative bead height–time trace obtained at 4.0 ± 0.4 pN is shown in Figure 1C. The three jumps of the bead height in the time trace correspond to the extension changes from the released GS linker upon the dissociation of SdrG–Fgβ, followed by the successive unfolding of N2 and N3 domains (Figures 1C and S1). Similar representative time traces at different forces from 3.2 ± 0.3 to 20.0 ± 2.0 pN are provided in Figure S2. The histogram of the bead height of the representative time trace in Figure 1C is shown in Figure 1D, from which the step sizes of the complex dissociation and domains’ unfolding were quantified. Single-molecule measurements with force-jumping cycles between 1.5 ± 0.2 pN and different testing forces were implemented to obtain rupturing events from multiple independent tethers (N > 30; Figure S3). Combining with control experiments, the rupturing signal of the complex was confirmed and distinguished from the unfolding signals of the N2 and N3 domains (Figures S4–S6 and Supporting Text 1). We note here that our force calibration approach leads to a force uncertainty of less than 10% (see the Supporting Information).
The lifetimes of the SdrG–Fgβ complex were obtained from the time traces (Figure 1C) and plotted as histograms (Figures 1E and S7). The histograms can be well fitted by a single exponential decay function, from which the average lifetime (τ) of the SdrG–Fgβ complex at ∼4.0 pN was determined to be 329.81 ± 32.78 s (Figure 1E; hereafter written as the means ± standard error of the fitting unless otherwise indicated). The average lifetime increases exponentially from 268.34 ± 22.28 to 22 067.47 ± 8333.96 s when the force is increased from 3.2 ± 0.3 to 20.0 ± 2.0 pN (Figure 1F), respectively. The results show the catch-bond behavior of the SdrG–Fgβ complex, where the adhesion complex is stabilized by increasing forces over the low-force range. Due to the further drastically increased lifetime of the complex (Figure S8 and Supporting Text 2), we could not obtain enough number of rupturing events to estimate the average lifetime at higher forces.
The obtained τ(f) could be well fitted by Bell’s model (Figure 1F)36,37
| 1 |
where τ0 represents the extrapolated zero-force lifetime, Δx‡ is the transition distance, and kBT is the Boltzmann constant times the temperature. The best-fitting values of the parameters are τ0 = 97.24 ± 12.49 s and Δx‡ = −1.20 ± 0.07 nm. τ0 is close to the lifetime measured in the absence of force from previously reported bulk experiments.10 The negative value of Δx‡ suggests a force-dependent increase in the energy barrier (−fΔx‡) that leads to the catch-bond kinetics. The larger the absolute value of Δx‡, the stronger the effect of the catch-bond kinetics of the adhesion complex.
Effect of “Bulgy Plug” on the Stability of the SdrG–Fgβ Complex
Two phenylalanines (Phe10 and Phe11) with benzyl side chains from the Fgβ ligand are crucial for ligand recognition and binding affinity of the complex.10 Buried behind the locking strand, the phenylalanine residue has been described as a “bulgy plug” that intuitively stops the ligand from wiggling through the narrow constriction under applied force10 (Figure 2A,B). Previous results have revealed a dissociation pathway of the SdrG–Fgβ complex in the nN force range, which involves directly pulling out the peptide ligand from the binding pocket.15 The measured force-dependent dissociation only marginally depends on phenylalanines, suggesting that the “bulgy plug” is not a dominant contributor to high-force resilience.15
Figure 2.
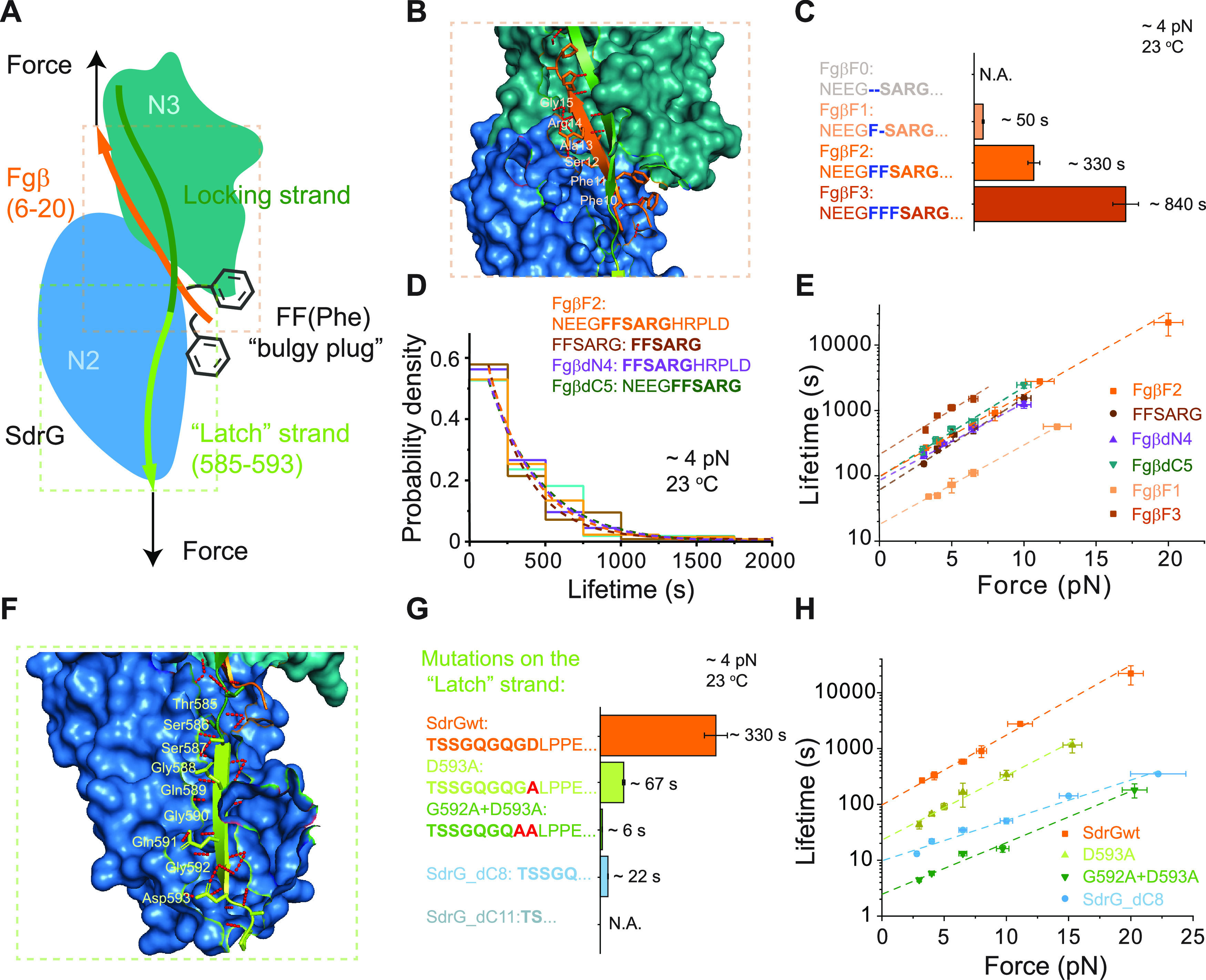
Effects of mutations on the ligand or the “latch” strand. (A) A sketch of the SdrG–Fgβ complex under force. The Fgβ ligand (orange) is buried into the binding cleft between N2 (blue) and N3 (green) domains, where the phenylalanine side chains (gray) acting as a “bulgy plug” are restricted by the locking strand (dark green). The complex is further secured by the “latch” strand (light green). (B) A structure showing the hydrogen networks (red dotted line) between the Fgβ ligand and N2 and N3 domains, where major six residues of the ligand involved in the binding are highlighted. (C) Comparison of the average lifetimes of the complex formed between SdrG and FgβF0 (gray, n = 0), FgβF1 (light orange, n = 163), FgβF2 (orange, n = 134), and FgβF3 (dark orange, n = 52) at 4.0 ± 0.4 pN. (D) Normalized lifetime histograms of the complex formed between SdrG and Fgβ truncations at 4.0 ± 0.4 pN, including FgβF2 (orange, n = 134), FFSARG (brown, n = 126), FgβdN4 (purple, n = 135), and FgβdC5 (green, n = 55). Dotted lines represent the best fitting of each normalized histogram to a single exponential decay function. (E) Force-dependent average lifetimes of the complex formed between SdrG and FgβF2, FFSARG, FgβdN4, FgβdC5, FgβF1, and FgβF3 at 23 °C. The dotted lines are best fittings to eq 1. (F) A structure showing the “latch” strand binds to the N2 domain, where the amino acids (residue 585-593) of the “latch” strand involved in the complementation are labeled. (G) Comparison of the average lifetimes of the complex formed in various protein constructs containing “latch” mutations at 4.0 ± 0.4 pN, including SdrGwt (orange, n = 134), D593A (lemon green, n = 162), G592A + D593A (green, n = 164), SdrG_dC8 (light blue, n = 285), and SdrG_dC11 (gray, n = 0). (H) Force-dependent average lifetimes of the complexes formed in the wild-type construct, D593A, G592A + D593A, and SdrG_dC8. The dotted lines are the best fittings to eq 1. All fitting parameters involved in figure are listed in Table S1. Error bars of the lifetime indicate mean ± standard errors.
We found that the phenylalanines are critical for the low-force mechanical stability of the SdrG–Fgβ complex. Ligands containing 0, 1, or 3 phenylalanines (F), respectively, termed FgβF0, FgβF1, or FgβF3, were adopted into the protein construct by alanine replacement or by addition of an F, to replace the wild-type ligand (FgβF2). It has been shown that the FgβF3 mutant has a higher affinity, while the FgβF1 mutant has a lower affinity for SdrG compared to FgβF2.10 The probability of the complex formation after a certain waiting time at 1.5 pN depended on the phenylalanines, where no binding of FgβF0 to SdrG was observed even after hours of incubation (Figure S9 and Supporting Text 3). The average lifetime of the SdrG–FgβF1 complex decreased by about 7-fold, while that of the SdrG–FgβF3 complex increased by more than 2.5-fold compared to the wild-type complex at each force (Figures 2C,E, S10, and S11). These results suggest that the “bulgy plug” plays a critical role in the mechanical stability of the SdrG–Fgβ complex in the low-force range.
We also prepared a protein construct with the truncated Fgβ ligand keeping the minimal six-residue peptide (FFSARG, Figure 2B) that is necessary for the high-affinity binding and sufficient for the extreme mechanical stability withstanding nN forces.15 The complex formed between truncated Fgβ and SdrG showed similar mechanical stability to that of the wild-type complex (Figures 2D,E and S12). Similar results were obtained for the SdrG–FgβdN4 complex, where the four amino acids NEEG at the N-terminus were truncated, and the SdrG–FgβdC5 complex, where the five amino acids HRPLD at the C-terminus were removed (Figures 2D,E, S13, and S14).
The insensitivity of the transition distances (slope of the fitting lines in Figure 2E) to the perturbations on the Fgβ ligand (Table S1) suggests that the dissociation of the complex in the low-force range is likely via a different transition pathway from directly pulling out the ligand.
Dissociation Pathway via the “Latch” Strand Unbinding
It has been known that the “latch” region secures the ligand in the binding pocket and stabilizes the complex (Figure 2F), whose presence was shown to be crucial for binding affinity10 but not high-force stability.15 We postulate that the low-force rupturing of the complex follows a transition pathway that involves the unbinding of the “latch” strand from N2 (sequential breakage of the short-range interactions between the residues), which leads to disruption of the ligand. To test this hypothesis, we first investigated two mutated constructs by substituting the last one or two residues of the “latch” strand involved in the binding to N2 with alanine, which would gradually weaken the “latch” strand binding from its C-terminus. Hereafter the two protein constructs are referred to as D593A and G592A + D593A, respectively. The lifetimes of mutated complexes formed in D593A and G592A + D593A are about 1/5 and 1/20 of that of the wild-type SdrG–Fgβ complex, respectively (Figures 2G,H, S15, and S16). The magnitudes of the phenomenological transition distances by fitting to Bell’s model become shorter than that of the wild-type complex, Δx‡ = −1.08 ± 0.05 nm for D593A and Δx‡ = −0.87 ± 0.10 nm for G592A + D593A (Figure 2H and Table S1).
We next investigated two other constructs with half or total truncated “latch” region (Figure 2G). Half truncation (SdrG_dC8) on the “latch” region slowed down the complex formation, while the total truncation (SdrG_dC11) resulted in the loss of a stable complex formation (Figure S17, Supporting Text 3). The force-dependent average lifetimes of the SdrG_dC8 construct were about 10-fold less than that of the wild-type complex (Figures 2G,H and S18), with a further decreased magnitude of the transition distance (Δx‡ = −0.68 ± 0.04 nm, Table S1).
Together, these results showed that the perturbations on the “latch” strand change both the force-dependent lifetimes and transition distances, confirming that the sequential unbinding of the “latch” peptide of SdrG away from the N2 domain is the transition pathway over the low-force range (Supporting Text 4).
Temperature-Dependent Mechanical Stability of the Adhesion Complex
As bacterial adhesion is subjected to changes in temperatures across a multitude of environmental and physiological conditions, we next investigated the mechanical stability of the SdrG–Fgβ complex under human body temperature, which is approximately 37 °C. The lifetime of the wild-type SdrG–Fgβ complex at 37 °C is much shorter than that at 23 °C. A representative time trace obtained at 4.0 pN at 37 °C is shown in Figure 3A. The lifetimes from multiple rupturing events are plotted as a normalized histogram in Figure 3B. The average lifetime of the SdrG–Fgβ complex is determined to be 5.72 ± 0.20 s (Figure 3B), about 2 orders of magnitude shorter than that at 23 °C at the same force (Figure 3C). More representative time traces at forces from 3.0 to 30 pN at 37 °C and corresponding lifetime histograms are provided in Figures S19 and S20, respectively. The force-dependent average lifetimes of the complex at 37 °C still show catch-bond behavior (Figure 3D). The extrapolated zero-force lifetime, τ0 = 1.97 ± 0.34 s, and the phenomenological transition distance, Δx‡ = −0.89 ± 0.06 nm, were determined. It should be noted that at several pN, the average lifetimes are in the order of a few seconds, indicating that the SdrG–Fgβ mediated adhesion is unstable at 37 °C. However, the stability of this adhesion rapidly increases with increasing force. At ∼30 pN, the average lifetime already reaches 1000 s. This result suggests that the SdrG–Fgβ complex acts as a highly dynamic mechanosensor, unstable at several pN forces and mechanically stabilized when it senses elevated forces at 37 °C.
Figure 3.
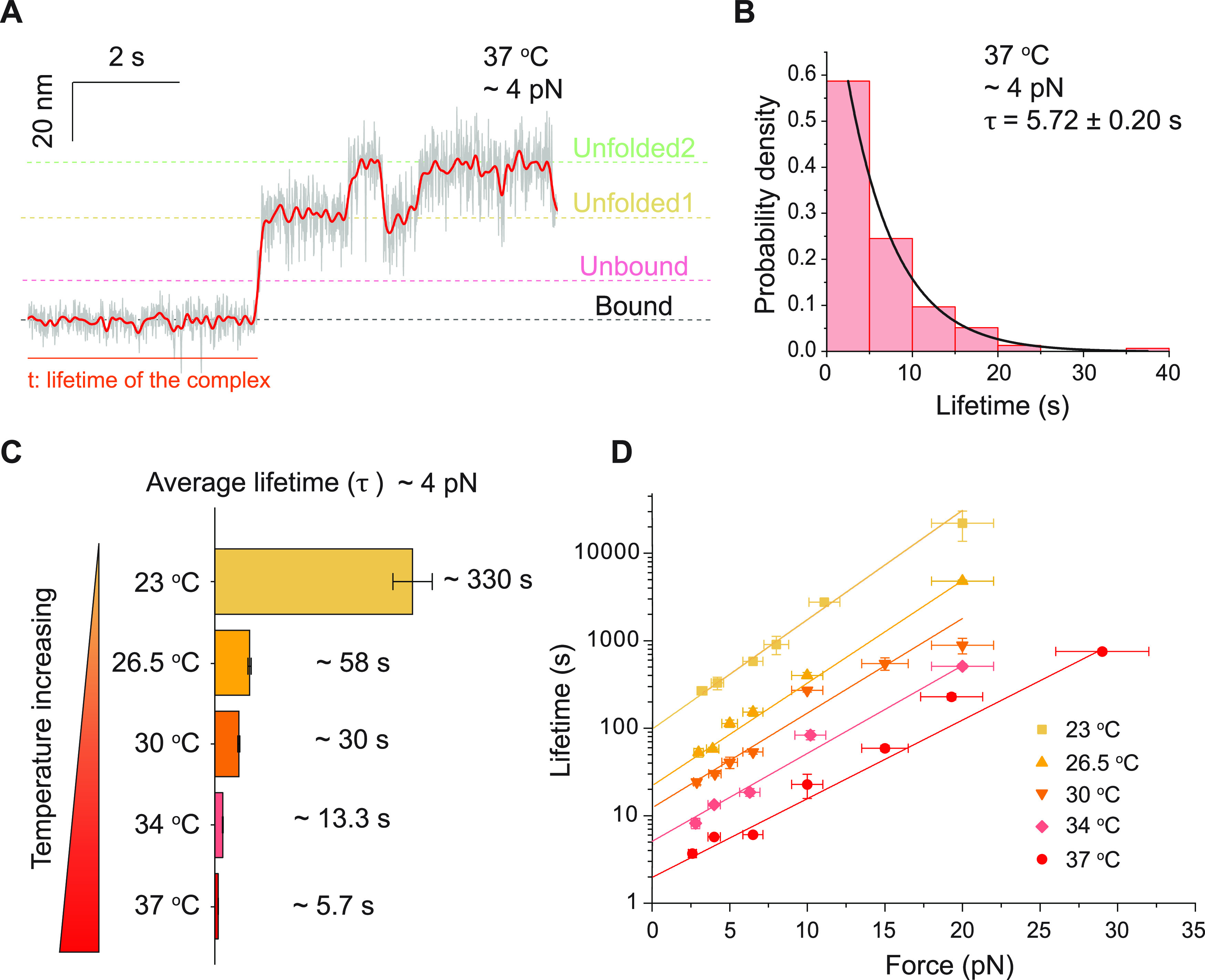
Temperature-dependent catch-bond kinetics. (A) A representative bead height–time trace at 4.0 ± 0.4 pN at 37 °C. (B) Normalized lifetime histogram of the SdrG–Fgβ complex at 4.0 ± 0.4 pN at 37 °C (n = 155, from more than five independent tethers). The solid black curve is the best fitting to the exponential decay function. (C) Comparison of the average lifetimes of the SdrG–Fgβ complex at 4.0 ± 0.4 pN at different temperatures. (D) Force-dependent average lifetimes of the SdrG–Fgβ complex at different temperatures. The colored solid lines are the best fittings to eq 1 at corresponding temperatures. All fitting parameters are listed in Table S1. Error bars of the lifetimes indicate mean ± standard errors.
We also performed similar experiments under three other temperatures of 26.5, 30, and 34 °C (Figures S21–S23). The force-dependent lifetimes monotonically decrease as the temperature increases (Figure 3C,D). The best-fitting values of τ0 and Δx‡ are summarized in Table S1. The results show that both τ0 and the magnitude of Δx‡ gradually drop as the temperature increases (Supporting Text 5).
Generality of the Mechanical Stabilization in Homologous Adhesion Complexes
The DLL mechanism has been reported in many homologous MSCRAMM systems involving the “latch” strand structure.1 To investigate whether the catch-bond kinetics is a general feature among the “latch” strand-containing MSCRAMMs, we quantified the force-dependent average lifetimes of five homologous adhesion complexes that mediate the adhesion between Staphylococcus aureus and its host. The crystal structures of the five homologs of SdrG (SD-repeat protein E (SdrE), bone sialoprotein binding protein (Bbp), clumping factor A (ClfA), clumping factor B (ClfB), and fibronectin-binding protein A (FnbpA)), bound with the corresponding ligands (peptides from the complement factor H (CFH) binding to SdrE, fibrinogen α chain (Fgα) binding to Bbp, dermokine (DK) binding to ClfB, and fibrinogen γ chain (Fgγ) binding to ClfA and FnbpA), have been reported.38−42 Analogous to the SdrG–Fgβ complex, these complexes are also under shear force geometries (Figure 4A). The recombinant protein constructs were prepared according to their respective native shear force geometries and were investigated using magnetic tweezers.
Figure 4.
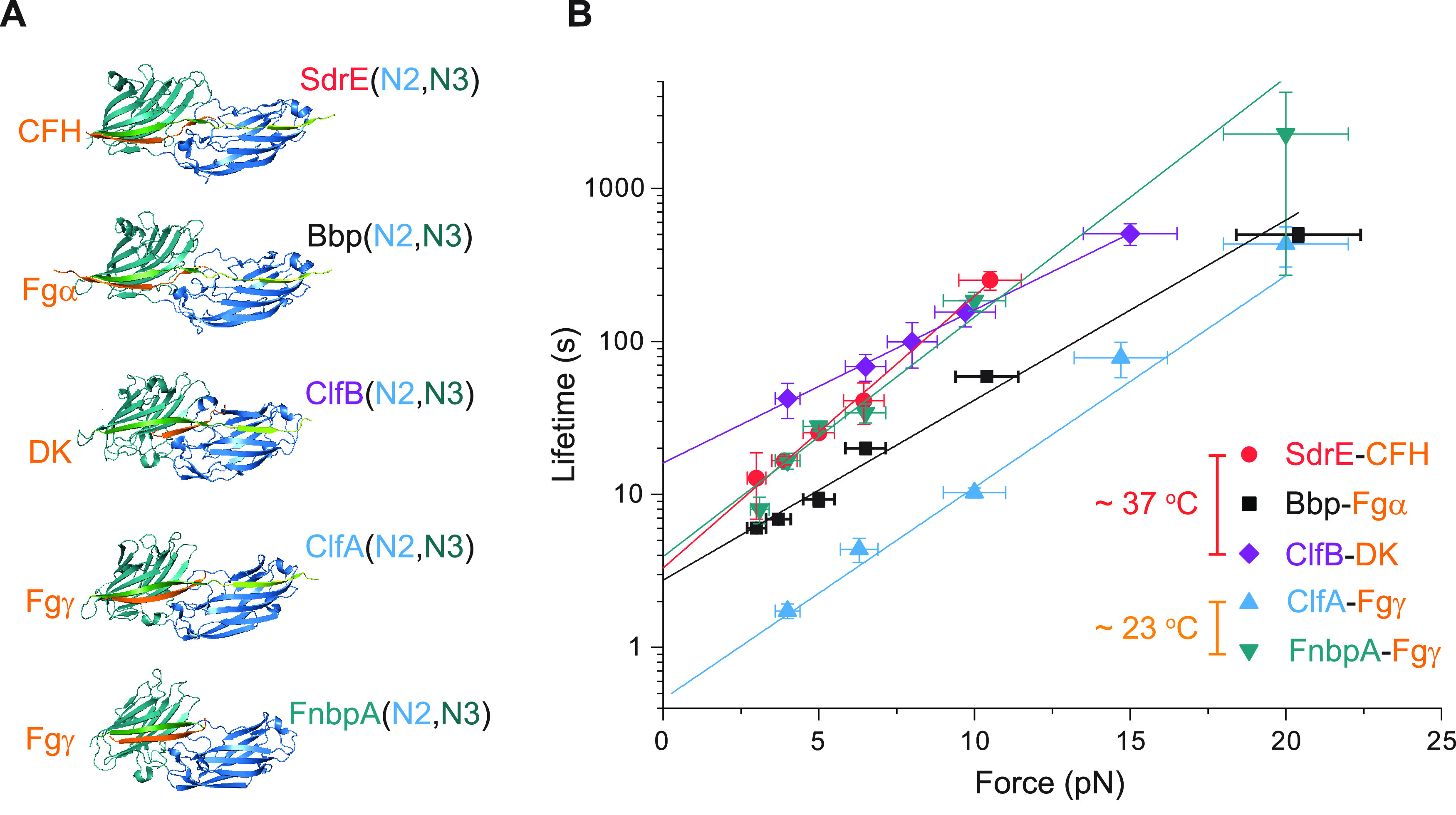
Catch-bond kinetics in homologous adhesion systems. (A) Adhesion complex structures of five homologs of SdrG (SdrE, Bbp, ClfB, ClfA, and FnbpA; N2 domain: blue, N3 domain: green) bound to their target peptide ligands (CFH, Fgα, DK, and Fgγ in orange). (B) The force-dependent average lifetimes of the homolog complexes at 37 °C for SdrE–CFH (red circles), Bbp–Fgα (black squares), and ClfB–DK (purple diamonds) and at 23 °C for ClfA–Fgγ (blue up triangles) and FnbpA–Fgγ (green down triangles). The colored solid lines are the best fittings to eq 1. All fitting parameters are listed in Table S1. Error bars of the lifetime indicate mean ± standard errors.
Experiments performed at 37 °C for SdrE–CFH, Bbp–Fgα, and ClfB–DK revealed catch-bond kinetics for each of these complexes (Figures 4B and S24–S28). The FnbpA–Fgγ and ClfA–Fgγ complexes are unstable at 37 °C; thus, their lifetime cannot be measured at our sampling rate. Instead, we performed experiments for these two complexes at 23 °C where they have longer lifetimes (Figures S27, S29, and S30). Similar catch-bond kinetics is also observed (Figure 4B). The best-fitting values of τ0 and Δx‡ are summarized in Table S1 in comparison with SdrG–Fgβ. These results show that the catch-bond behavior over the low-force range among the adhesions mediated by MSCRAMMs is a generic feature.
Discussion
In summary, using ultrastable magnetic tweezers, we have investigated and characterized the low-force (pN to tens of pN) mechanical stabilities of the SdrG–Fgβ complex alongside its homologous systems. These adhesion complexes exhibit catch-bond kinetics, where the complexes are stabilized by increasing force. Through mutations on the ligand or “latch” strand of the SdrG–Fgβ complex, the low-force mechanical stability of the complex is found to be dependent on the “bulgy plug” of the ligand; additionally, the dissociation transition pathway in this force range is identified to be the unbinding of the “latch” strand peptide away from the N2 domain. The average lifetime of the SdrG–Fgβ adhesion complex is sensitive to temperature, which decreases by 2 orders of magnitude when the temperature is increased from 23 to 37 °C.
Bell’s model fitting to the force-dependent average lifetimes (τ(f)) leads to negative phenomenological transition distances Δx‡, which provides an understanding of the observed catch-bond behavior. However, it does not reveal the underlying physical essence of the catch-bond kinetics. A recently reported structural elastic theory31 highlights the importance of the entropic extension fluctuation of biomolecules at pN to tens of pN on the mechanical stability of biomolecular complexes. It predicts catch-bond kinetics as a common feature for transitions involving a pathway via unbinding of a pre-extended peptide strand under a shear force geometry43 (Figure S31). As the “latch” strand exhibits a highly pre-extended conformation when it binds to the N2 domain, the observed catch-bond kinetics could be explained by this theory. Indeed, the τ(f) of SdrG–Fgβ could be reasonably fitted by the structural elastic model, from which the most probable transition state structure was identified, where most of the “latch” strand (seven to nine amino acids from the last binding residue of the “latch” strand, 593D) is dissociated away from the N2 domain (Figure S31, Table S2, and Supporting Text 6). Additionally, we have fitted the force-dependent lifetimes of the adhesion complex in D593A and G592A + D593A, (Figure S31), resulting in a gradually reduced number of bound residues between the latch peptide and N2 (Figure S31, Tables S3 and S4, and Supporting Text 6). While the structural elastic model provides a plausible mechanism of the observed catch-bond kinetics, we do not exclude other potential mechanisms such as force-induced additional interactions along the transition pathway.
It has been known that MSCRAMM adhesions confer high resilience to large stress, which plays a crucial role in enabling stable anchorage of the bacterial cells to host sites, such as artery/large vessel, respiratory tract, and urethra, which are subjected to high flow stress. The forces from pN to tens of pN investigated in our study belong to a previously less explored physiologically relevant force range, related to bacterial adhesion to the cell surface of capillaries or organs. The initial anchoring of a bacterium to the blood vessel, which is needed for bacterial spreading and infection to different organs,44,45 is via a single adhesion complex (e.g, the SdrG–Fgβ complex). The initial anchorage subjects the complex to a tensile force from a few pN to tens of pN, which can be estimated based on the fluid shear stress in a range of 5–100 dynes cm–2 in capillaries8,9 or by the Stokes equation considering the blood viscosity (η ≈ 5 × 10–2 Pa), blood flow speed (v ≈ 1 mm s–1), and the dimension of the bacterial cells (L ≈ 1 μm).46 Therefore, the mechanical stability of MMSCARM-mediated adhesions quantified in the study is of physiological importance.
The low-force mechanical stability of SdrG–Fgβ and
its homologous complexes is unusual compared to many other biomolecular
complexes24−28,47−51 that also exhibit catch-bond kinetics over a similar
force range. Most known biomolecular complexes exhibit a catch-to-slip
kinetics switch when the applied force exceeds a certain threshold
value (switch force) in the range of 10–30 pN (Table S5). The catch-bond kinetics of SdrG–Fgβ
and its homologous complexes persists over the tested force range
(3–30 pN) with a constant slope and does not show any drops
at 50 pN, suggesting that the potential switch force far exceeds 50
pN. Indeed, a switch force higher than 300 pN is estimated by 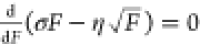 based on the structural elastic force-dependent
lifetime expression.43 Such a surprisingly
high switch force is a direct result from the highly pre-extended
conformation of the “latch” peptide, where the end-to-end
distance of the latch peptide bound on the N2 domain reaches >90%
of the contour length. However, at the predicted high switch force,
potential elastic deformation of the rigid protein domains may affect
the transition kinetics significantly. This effect is not considered
in the simple expression of the force-dependent lifetime; therefore,
we refrain ourselves from further discussion on the exact value of
the switch force.
based on the structural elastic force-dependent
lifetime expression.43 Such a surprisingly
high switch force is a direct result from the highly pre-extended
conformation of the “latch” peptide, where the end-to-end
distance of the latch peptide bound on the N2 domain reaches >90%
of the contour length. However, at the predicted high switch force,
potential elastic deformation of the rigid protein domains may affect
the transition kinetics significantly. This effect is not considered
in the simple expression of the force-dependent lifetime; therefore,
we refrain ourselves from further discussion on the exact value of
the switch force.
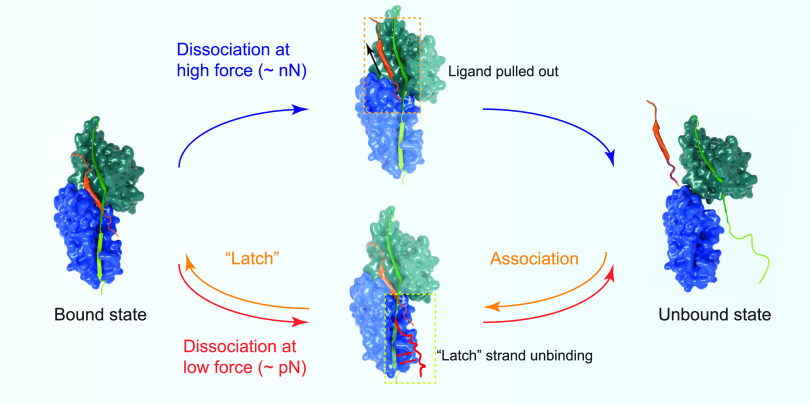
The transition pathway at the low-force range via unbinding of the “latch” strand is distinctly different from that at high forces in the nN range, where the high-force dissociation transition follows a pathway of directly pulling out the ligand from the latched binding pocket.15 The low-force dissociation pathway starts from the “latch” strand detachment, which is a reverse of the association process of SdrG–Fgβ following the DLL mechanism where the “latch” strand binds N2 as the last step (Figure 5). The existence of two pathways, one governing the high-force dissociation kinetics and the other governing the low-force dissociation kinetics, together provide a comprehensive and systematic understanding of the adhesion stability across a wide physiologically relevant force range (Figure 5). Yet, a full understanding requires measuring τ(f) in the force range of 102–103 pN, inaccessible by currently available technologies due to the ultralong lifetime, which warrants future studies.
Figure 5.
Illustrations of the high-force dissociation pathway15 via directly pulling out the ligand and the low-force dissociation pathway via unbinding of the “latch” strand from N2.
As important opportunistic pathogens, S. epidermidis is the leading cause of medical device and implant-related nosocomial infections,2 while S. aureus causes both superficial and invasive, potentially life-threatening infections.3 During the adhesion and invasion onto the surface of host cells or devices that are immobilized with protein ligands, they are subjected to a wide range of hydrodynamic flow stress. Besides surface-immobilized ligands, there is a large fraction of soluble protein ligands (fibrinogen, etc.). An important question is how bacterial cells could avoid the binding by the soluble protein ligands. In other words, what is the mechanism behind the specific binding to the surface-immobilized ligands? The seconds of lifetime of the SdrG–Fgβ complex at 37 °C at near-zero forces suggests that adhesion complexes formed with these soluble ligands, which are not under tensile force, can dissociate quickly. In sharp contrast, once attached to the surface-immobilized ligands, the adhesion complex becomes stabilized under flow-induced stress through the catch-bond kinetics (Figure S32A). We propose that this mechanism provides the specificity of the stable complexes formed on surface-immobilized ligands and confers an elevated efficiency of the MSCRAMM-mediated adhesion to the surface.
Through the catch-bond kinetics, these adhesion complexes can resist the shear force in the range of tens pN to nN, which is crucial for pathogen adhesion and infection (Figure S32B).15 Meanwhile, due to the seconds lifetime of the complex at pN forces at the human body temperature, we propose that the pathogens can dissociate from the ligand on the surface and associate to a different site under low flow stress conditions (Figure S32C), which would benefit the spreading and the evasion of the pathogens.
The low-force mechanical stability of the bacterial adhesion complexes investigated in this study requires mechanical manipulation of a single tethered complex over a long-time scale (hours to days) without losing the spatial resolution. This is achieved by an in-house-made ultrastable magnetic tweezer setup.52,53 As piconewton forces are not only involved in bacterial adhesion systems but also prevalent in force-bearing mechanosensing cytoskeleton proteins of mammalian cells,54−56 the ultrastable magnetic tweezer technology can be broadly applied to studies of the mechanical stabilities and force-dependent interactions of biomolecules involved in various biological processes.
Experimental Section
Reagents and Materials
(3-Aminopropyl)triethoxysilane (APTES), glutaraldehyde solution (70% in H2O), dithiothreitol (DTT), bovine serum albumin (BSA), l-ascorbic acid, ampicillin, isopropyl β-d-1-thiogalactopyranoside (IPTG), phenylmethylsulfonyl fluoride (PMSF), biotin, mineral oil, PBS, and Tris buffer were purchased from Sigma-Aldrich. Neutravidin biotin-binding protein and Dynabeads M-270 Epoxy beads were purchased from Thermo Scientific.
Gene Construction
The N2 and N3 domain genes of SdrG from S. epidermidis (residue 274-597, Uniprot Q9KI13, PDB 1R17) and the N2 and N3 domain genes from homologs: SdrE (residue 263-601, Uniprot Q2FJ77, PDB 5WTB), Bbp (residue 271-601, Uniprot Q14U76, PDB 5CFA), ClfA (residue 225-548, Uniprot: Q2G015, PDB 2VR3), ClfB (residue 212-541, Uniprot Q6GDH2, PDB 4F20), and FnbpA (residue 187-514, Uniprot P14738, PDB 4B60) from S. aureus, as well as the short peptide ligand genes: Fgα (SKQFTSSTSYNRGDS, from human fibrinogen α-chain, Uniprot P02671), Fgβ (NEEGFFSARGHRPLD, residue 6-20 from the N-terminal region of human fibrinogen β-chain, Uniprot P02675), Fgγ (GEGQQHHLGGAKQAGDV, from the C-terminal of human fibrinogen γ-chain, Uniprot: P02679-2), CFH from human complement factor H (RLSSRSHTLRTTCWDGKLEYP, Uniprot P08603), and DK from human dermokine 10 (QSGSSGSGSNGD, Uniprot Q6E0U4) were codon-optimized and synthesized for expression in Escherichia coli as gBlocks gene fragments (Integrated DNA Technologies (IDT)) with suitable overhangs. The genes of the flexible GS linker were synthesized as gBlocks gene fragments. A flexible FH1 linker gene from human formin was obtained from our previous work.57 Genes were cloned into pET151 vectors (ampicillin resistance) with 6 × His-tag for purification, AviTag for biotinylation, and SpyTag using the NEB HiFi assembly strategy (New England Biolabs, MA). The construction of SpyTag-binding protein SpyCatcher was referred to in previous works.58 The mutations, truncations, or insertions on the ligands or the “latch” strand of SdrG were introduced by the Site-Directed Mutagenesis kit (New England Biolabs, MA). The complete sequences of all recombinant protein constructs used are listed in the Supporting Information (the “Protein Construction and Sequence” section).
Protein Expression and Purification
All protein constructs were expressed in E. coli DE3 with biotin protein ligase (BirA) and purified using a His-tag affinity column following the previous protocol.58 Basically, the colonies transfected with each of the corresponding plasmids were precultured in 10 mL of LB medium containing 100 μg mL–1 ampicillin at 37 °C overnight. The precultures were then inoculated into 1 L of ampicillin-containing LB medium and grown at 37 °C for around 4–6 h until the optical density (OD600) reached ∼0.6. Then, 0.4 mM IPTG and 50 μM biotin were then added to the cultures, and the resulting mixture was grown at 20 °C overnight. The biotin could be catalyzed by BirA to conjugate to the AviTag of proteins in the cell. Bacteria were harvested by centrifugation at 6000g, and pellets were stored frozen at −80 °C until further purification.
In the purification steps, bacterial pellets were resuspended with lysis buffer (50 mM Tris, 300 mM NaCl, pH = 7.4) including 1 mM PMSF, and cells were mechanically lysed using a French press, followed by centrifugation at 40 000g for 30 min. The resultant supernatants were allowed to bind to the Co2+-NTA column (Thermo Scientific, DE) for 1 h. After repeated washes (wash buffer: 50 mM Tris, 300 mM NaCl, 10 mM imidazole, pH = 7.4), the proteins were eluted into elution buffer (50 mM Tris, 300 mM NaCl, 200 mM imidazole, pH = 7.4). These proteins were further purified with gel filtration chromatography (Superdex 200, Äkta Pure system, GE Healthcare, MA). The protein-containing fractions were verified using sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis, dialyzed into PBS buffer, and frozen in aliquots with 15% (v/v) glycerol by liquid nitrogen to be stored at −80 °C for use. Protein concentration was measured by spectrophotometry at 280 nm (NanoDrop 1000, Thermo Scientific, DE).
Magnetic Tweezer-Based Single-Molecule Force Approach
Single-molecule manipulation and measurement of the proteins were performed on a homemade magnetic tweezer (MT) setup as previously described.59 A novel antidrift procedure based on focal plane adjustment by piezo was coupled into the home-written Labview program, providing a steady applying force to the single-molecule measurement for days without any drift. The protein constructs were tethered between the Neutravidin-coated magnetic beads and SpyCatcher-coated cover glass in the laminar flow channels. Experiments were performed with a standardized solution (1 × PBS, 1% (m/m) BSA, 1 mM DTT, 10 mM L-ascorbate acid, pH = 7.4) unless otherwise mentioned. The temperature of the experiment was held at 23, 26.5, 30, 34, or 37 °C controlled by an objective heating system (Bioptechs) and calibrated by a thermometer. The detailed protocols of channel and microbead preparation, sample preparation, magnetic tweezer setup, single-protein force-jumping experiment, and force calibration were published in our previous studies58,59 and are briefly shown in the Supporting Information.
Data Analysis
Raw time trace data were recorded at a sampling rate of 200 Hz. In all of the figures of the main text and the Supporting Information, the time traces were smoothed using the fast Fourier transform (FFT) smooth function of OriginPro 9.0 unless otherwise mentioned. The lifetimes (dwell times) of the dissociation, the step sizes of the rupture of the ligand from the N2 and N3 domains, and the step sizes of the unfolding of the N2 and N3 domains were measured from the raw time trace data. Due to the sampling rate, the time resolution of our experiment is around 5 μs.
The rupturing lifetimes (dwell times) for each adhesion complex under specific force were collected and plotted as histograms. The y-axis of the histogram was the count of the events observed in this dwell time range (Supporting Figures). The corresponding histograms presented in the maintext are converted to relative probability densities. The number of the rupturing events (n) and the number of the tethers (N) for each histogram were indicated in the figures and legends.
The average lifetimes (τ) of the dissociation of all of the adhesion complexes under forces were fitted from the rupturing lifetime (t) histograms using a single exponential decay equation, y(t) = B* exp(−t/τ), where y is the count or relative probability density of the histogram and B is the amplitude of the histogram. The error bars indicate the mean ± standard error of the fitting.
Acknowledgments
The authors thank Prof. Yi Cao for the helpful advice and proofreading of the manuscript. The authors thank the High-throughput Molecular Genetics core of the Mechanobiology Institute for the help with protein expression.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c03961.
Additional experimental details and protein construct sequences; additional texts including description about the three stepwise jumps, ultra-long lifetime of the complex at high force, complex reformation, temperature-dependent low-force transition pathway, and physical mechanism of the catch-bond kinetics; additional figures including detailed illustrations, representative time traces, control experiments, mechanical responses and force-dependent lifetime distributions of all the proteins, the structural-elastic model, and the proposed mechanisms; tables including summary of the best-fitting parameters using Bell's model, best-fitting parameters using structural-elastic model, and comparison of the catch-bond kinetics (PDF)
This work was supported by the Singapore Ministry of Education Academic Research Fund Tier 2 (MOE-T2EP50220-0015 and MOE2019-T2-1-099), the Ministry of Education under the Research Centres of Excellence Programme, and the Ministry of Education under the Research Scholarship Block.
The authors declare no competing financial interest.
Supplementary Material
References
- Foster T. J. The MSCRAMM family of cell-wall-anchored surface proteins of gram-positive cocci. Trends Microbiol. 2019, 27, 927–941. 10.1016/j.tim.2019.06.007. [DOI] [PubMed] [Google Scholar]
- Otto M. Staphylococcus epidermidis—the’accidental’pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J.; Geoghegan J. A.; Ganesh V. K.; Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persat A.; Nadell C. D.; Kim M. K.; Ingremeau F.; Siryaporn A.; Drescher K.; Wingreen N. S.; Bassler B. L.; Gitai Z.; Stone H. A. The Mechanical World of Bacteria. Cell 2015, 161, 988–997. 10.1016/j.cell.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi R.; Guasto J. S.; Stocker R. Bacterial transport suppressed by fluid shear. Nat. Phys. 2014, 10, 212–217. 10.1038/nphys2883. [DOI] [Google Scholar]
- Guasto J. S.; Rusconi R.; Stocker R. Fluid mechanics of planktonic microorganisms. Annu. Rev. Fluid Mech. 2012, 44, 373–400. 10.1146/annurev-fluid-120710-101156. [DOI] [Google Scholar]
- Genova L. A.; Roberts M. F.; Wong Y.-C.; Harper C. E.; Santiago A. G.; Fu B.; Srivastava A.; Jung W.; Wang L. M.; Krzemiński Ł.; et al. Mechanical stress compromises multicomponent efflux complexes in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 25462–25467. 10.1073/pnas.1909562116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pries A. R.; Secomb T.; Gessner T.; Sperandio M.; Gross J.; Gaehtgens P. Resistance to blood flow in microvessels in vivo. Circ. Res. 1994, 75, 904–915. 10.1161/01.RES.75.5.904. [DOI] [PubMed] [Google Scholar]
- Mikaty G.; Soyer M.; Mairey E.; Henry N.; Dyer D.; Forest K. T.; Morand P.; Guadagnini S.; Prévost M. C.; Nassif X.; Duménil G. Extracellular Bacterial Pathogen Induces Host Cell Surface Reorganization to Resist Shear Stress. PLoS Pathog. 2009, 5, e1000314 10.1371/journal.ppat.1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnuraj K.; Bowden M. G.; Davis S.; Gurusiddappa S.; Moore D.; Choe D.; Xu Y.; Hook M.; Narayana S. V. L. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 2003, 115, 217–228. 10.1016/S0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- Bowden M. G.; Heuck A. P.; Ponnuraj K.; Kolosova E.; Choe D.; Gurusiddappa S.; Narayana S. V. L.; Johnson A. E.; Hook M. Evidence for the “dock, lock, and latch” ligand binding mechanism of the staphylococcal microbial surface component recognizing adhesive matrix molecules (MSCRAMM) SdrG. J. Biol. Chem. 2008, 283, 638–647. 10.1074/jbc.M706252200. [DOI] [PubMed] [Google Scholar]
- Herman P.; El-Kirat-Chatel S.; Beaussart A.; Geoghegan J. A.; Foster T. J.; Dufrêne Y. F. The binding force of the staphylococcal adhesin SdrG is remarkably strong. Mol. Microbiol. 2014, 93, 356–368. 10.1111/mmi.12663. [DOI] [PubMed] [Google Scholar]
- Herman-Bausier P.; Labate C.; Towell A. M.; Derclaye S.; Geoghegan J. A.; Dufrêne Y. F. Staphylococcus aureus clumping factor A is a force-sensitive molecular switch that activates bacterial adhesion. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 5564–5569. 10.1073/pnas.1718104115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzieleghem T.; Herman-Bausier P.; Dufrene Y. F.; Mahillon J. Staphylococcus epidermidis affinity for fibrinogen-coated surfaces correlates with the abundance of the SdrG adhesin on the cell surface. Langmuir 2015, 31, 4713–4721. 10.1021/acs.langmuir.5b00360. [DOI] [PubMed] [Google Scholar]
- Milles L. F.; Schulten K.; Gaub H. E.; Bernardi R. C. Molecular mechanism of extreme mechanostability in a pathogen adhesin. Science 2018, 359, 1527–1533. 10.1126/science.aar2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitry P.; Valotteau C.; Feuillie C.; Bernard S.; Alsteens D.; Geoghegan J. A.; Dufrêne Y. F. Force-induced strengthening of the interaction between Staphylococcus aureus clumping factor B and loricrin. mBio 2017, 8, e01748-17 10.1128/mBio.01748-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbois M.; Beyer M.; Rief M.; Clausen-Schaumann H.; Gaub H. E. How strong is a covalent bond?. Science 1999, 283, 1727–1730. 10.1126/science.283.5408.1727. [DOI] [PubMed] [Google Scholar]
- Milles L. F.; Gaub H. E. Extreme mechanical stability in protein complexes. Curr. Opin. Struct. Biol. 2020, 60, 124–130. 10.1016/j.sbi.2019.11.012. [DOI] [PubMed] [Google Scholar]
- Dufrêne Y. F.; Persat A. Mechanomicrobiology: how bacteria sense and respond to forces. Nat. Rev. Microbiol. 2020, 18, 227–240. 10.1038/s41579-019-0314-2. [DOI] [PubMed] [Google Scholar]
- Mathelié-Guinlet M.; Viela F.; Pietrocola G.; Speziale P.; Alsteens D.; Dufrêne Y. F. Force-clamp spectroscopy identifies a catch bond mechanism in a Gram-positive pathogen. Nat. Commun. 2020, 11, 5431 10.1038/s41467-020-19216-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W. E.; Vogel V.; Sokurenko E. Biophysics of catch bonds. Annu. Rev. Biophys. 2008, 37, 399–416. 10.1146/annurev.biophys.37.032807.125804. [DOI] [PubMed] [Google Scholar]
- Liu B.; Kolawole E. M.; Evavold B. D. Mechanobiology of T Cell Activation: To Catch a Bond. Annu. Rev. Cell Dev. Biol. 2021, 37, 65–87. 10.1146/annurev-cellbio-120219-055100. [DOI] [PubMed] [Google Scholar]
- Thomas W. Catch bonds in adhesion. Annu. Rev. Biomed. Eng. 2008, 10, 39–57. 10.1146/annurev.bioeng.10.061807.160427. [DOI] [PubMed] [Google Scholar]
- Marshall B. T.; Long M.; Piper J. W.; Yago T.; McEver R. P.; Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature 2003, 423, 190–193. 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- Buckley C. D.; Tan J.; Anderson K. L.; Hanein D.; Volkmann N.; Weis W. I.; Nelson W. J.; Dunn A. R. The minimal cadherin-catenin complex binds to actin filaments under force. Science 2014, 346, 1254211 10.1126/science.1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. L.; Bax N. A.; Buckley C. D.; Weis W. I.; Dunn A. R. Vinculin forms a directionally asymmetric catch bond with F-actin. Science 2017, 357, 703–706. 10.1126/science.aan2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F.; García A. J.; Mould A. P.; Humphries M. J.; Zhu C. Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 2009, 185, 1275–1284. 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.; Mallis R. J.; Lang M. J.; Reinherz E. L. The αβ TCR mechanosensor exploits dynamic ectodomain allostery to optimize its ligand recognition site. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 21336–21345. 10.1073/pnas.2005899117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Caballero A.; Echelman D. J.; Tapia-Rojo R.; Haldar S.; Eckels E. C.; Fernandez J. M. Protein folding modulates the chemical reactivity of a Gram-positive adhesin. Nat. Chem. 2021, 13, 172–181. 10.1038/s41557-020-00586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt E. J. G.; Diggle S. P. Defining motility in the Staphylococci. Cell. Mol. Life Sci. 2017, 74, 2943–2958. 10.1007/s00018-017-2507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S.; Tang Q.; Yao M.; You H.; Le S.; Chen H.; Yan J. Structural–elastic determination of the force-dependent transition rate of biomolecules. Chem. Sci. 2018, 9, 5871–5882. 10.1039/C8SC01319E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakeri B.; Fierer J. O.; Celik E.; Chittock E. C.; Schwarz-Linek U.; Moy V. T.; Howarth M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, E690–E697. 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S.; Löf A.; Sedlak S. M.; Benoit M.; Gaub H. E.; Lipfert J. Designed anchoring geometries determine lifetimes of biotin–streptavidin bonds under constant load and enable ultra-stable coupling. Nanoscale 2020, 12, 21131–21137. 10.1039/D0NR03665J. [DOI] [PubMed] [Google Scholar]
- Sedlak S. M.; Schendel L. C.; Melo M. C. R.; Pippig D. A.; Luthey-Schulten Z.; Gaub H. E.; Bernardi R. C. Direction matters: Monovalent streptavidin/biotin complex under load. Nano Lett. 2019, 19, 3415–3421. 10.1021/acs.nanolett.8b04045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlak S. M.; Schendel L. C.; Gaub H. E.; Bernardi R. C. Streptavidin/biotin: tethering geometry defines unbinding mechanics. Sci. Adv. 2020, 6, eaay5999 10.1126/sciadv.aay5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. I. Models for the specific adhesion of cells to cells. Science 1978, 200, 618–627. 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Prezhdo O. V.; Pereverzev Y. V. Theoretical aspects of the biological catch bond. Acc. Chem. Res. 2009, 42, 693–703. 10.1021/ar800202z. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Wu M.; Hang T.; Wang C.; Yang Y.; Pan W.; Zang J.; Zhang M.; Zhang X. Staphylococcus aureus SdrE captures complement factor H’s C-terminus via a novel ’close, dock, lock and latch’ mechanism for complement evasion. Biochem. J. 2017, 474, 1619–1631. 10.1042/BCJ20170085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Wu M.; Zhuo W.; Gu J.; Zhang S.; Ge J.; Yang M. Crystal structures of Bbp from Staphylococcus aureus reveal the ligand binding mechanism with Fibrinogen α. Protein Cell 2015, 6, 757–766. 10.1007/s13238-015-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh V. K.; Rivera J. J.; Smeds E.; Ko Y.-P.; Bowden M. G.; Wann E. R.; Gurusiddappa S.; Fitzgerald J. R.; Höök M. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog. 2008, 4, e1000226 10.1371/journal.ppat.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang H.; Feng Y.; Wang J.; Liu B.; Chen Y.; Liu L.; Deng X.; Yang M. Crystal structures reveal the multi-ligand binding mechanism of Staphylococcus aureus ClfB. PLoS Pathog. 2012, 8, e1002751 10.1371/journal.ppat.1002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemberk V.; Jones R. P. O.; Moroz O.; Atkin K. E.; Edwards A. M.; Turkenburg J. P.; Leech A. P.; Massey R. C.; Potts J. R. Evidence for steric regulation of fibrinogen binding to Staphylococcus aureus fibronectin-binding protein A (FnBPA). J. Biol. Chem. 2014, 289, 12842–12851. 10.1074/jbc.M113.543546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S.; Efremov A. K.; Yan J. Understanding the catch-bond kinetics of biomolecules on a one-dimensional energy landscape. Commun. Chem. 2019, 2, 30 10.1038/s42004-019-0131-6. [DOI] [Google Scholar]
- Kwiecinski J. M.; Horswill A. R. Staphylococcus aureus bloodstream infections: pathogenesis and regulatory mechanisms. Curr. Opin. Microbiol. 2020, 53, 51–60. 10.1016/j.mib.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. M.; Massey R. C. How does Staphylococcus aureus escape the bloodstream?. Trends Microbiol. 2011, 19, 184–190. 10.1016/j.tim.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Ivanov K. P.; Kalinina M. K.; Levkovich Y. I. Blood flow velocity in capillaries of brain and muscles and its physiological significance. Microvasc. Res. 1981, 22, 143–155. 10.1016/0026-2862(81)90084-4. [DOI] [PubMed] [Google Scholar]
- Le Trong I.; Aprikian P.; Kidd B. A.; Forero-Shelton M.; Tchesnokova V.; Rajagopal P.; Rodriguez V.; Interlandi G.; Klevit R.; Vogel V.; Stenkamp R. E.; Sokurenko E. V.; Thomas W. E. Structural Basis for Mechanical Force Regulation of the Adhesin FimH via Finger Trap-like β Sheet Twisting. Cell 2010, 141, 645–655. 10.1016/j.cell.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W. E.; Trintchina E.; Forero M.; Vogel V.; Sokurenko E. V. Bacterial Adhesion to Target Cells Enhanced by Shear Force. Cell 2002, 109, 913–923. 10.1016/S0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- Rakshit S.; Zhang Y.; Manibog K.; Shafraz O.; Sivasankar S. Ideal, catch, and slip bonds in cadherin adhesion. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 18815–18820. 10.1073/pnas.1208349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.; Chen W.; Evavold B. D.; Zhu C. Accumulation of Dynamic Catch Bonds between TCR and Agonist Peptide-MHC Triggers T Cell Signaling. Cell 2014, 157, 357–368. 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D. K.; Feng Y.; Mallis R. J.; Li X.; Keskin D. B.; Hussey R. E.; Brady S. K.; Wang J.-H.; Wagner G.; Reinherz E. L.; Lang M. J. Force-dependent transition in the T-cell receptor β-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 1517–1522. 10.1073/pnas.1424829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Fu H.; Zhu X.; Cong P.; Nakamura F.; Yan J. Improved high-force magnetic tweezers for stretching and refolding of proteins and short DNA. Biophys. J. 2011, 100, 517–523. 10.1016/j.bpj.2010.12.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.; Zeng X.; Lu C.; Yan J. Studying the mechanical responses of proteins using magnetic tweezers. Nanotechnology 2017, 28, 414002 10.1088/1361-6528/aa837e. [DOI] [PubMed] [Google Scholar]
- del Rio A.; Perez-Jimenez R.; Liu R.; Roca-Cusachs P.; Fernandez J. M.; Sheetz M. P. Stretching Single Talin Rod Molecules Activates Vinculin Binding. Science 2009, 323, 638–641. 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M.; Goult B. T.; Klapholz B.; Hu X.; Toseland C. P.; Guo Y.; Cong P.; Sheetz M. P.; Yan J. The mechanical response of talin. Nat. Commun. 2016, 7, 11966 10.1038/ncomms11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Yao M.; Baker K. B.; Gough R. E.; Le S.; Goult B. T.; Yan J. Force-Dependent Interactions between Talin and Full-Length Vinculin. J. Am. Chem. Soc. 2021, 143, 14726–14737. 10.1021/jacs.1c06223. [DOI] [PubMed] [Google Scholar]
- Yu M.; Le S.; Efremov A. K.; Zeng X.; Bershadsky A.; Yan J. Effects of Mechanical Stimuli on Profilin- and Formin-Mediated Actin Polymerization. Nano Lett. 2018, 18, 5239–5247. 10.1021/acs.nanolett.8b02211. [DOI] [PubMed] [Google Scholar]
- Yu M.; Zhao Z.; Chen Z.; Le S.; Yan J. Modulating mechanical stability of heterodimerization between engineered orthogonal helical domains. Nat. Commun. 2020, 11, 4476 10.1038/s41467-020-18323-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Yuan G.; Winardhi R. S.; Yao M.; Popa I.; Fernandez J. M.; Yan J. Dynamics of Equilibrium Folding and Unfolding Transitions of Titin Immunoglobulin Domain under Constant Forces. J. Am. Chem. Soc. 2015, 137, 3540–3546. 10.1021/ja5119368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.