Abstract
SigD is translocated into eucaryotic cells by a type III secretion system. In this work, evidence that the putative chaperone SigE directly interacts with SigD is presented. A bacterial two-hybrid system demonstrated that SigE can interact with itself and SigD. In addition, SigD was specifically copurified with SigE-His6 on a nickel column.
Many gram-negative pathogenic bacteria produce type III secretion systems (TTSS) required for virulence in an animal model of infection (for a review, see reference 15). These systems secrete and frequently translocate effector proteins into eucaryotic cells such as epithelial cells and macrophages (3, 4, 8, 10, 11, 17, 24, 29). Once in the eucaryotic cytoplasm, these effectors can stimulate events such as cytoskeletal rearrangements, ion flux, or apoptosis (1, 2, 13, 19, 23, 29, 30). Salmonella enterica serovar Typhimurium has at least two TTSS, one of which is encoded on a large pathogenicity island called SPI1 (22). The SPI1 system is required for invasion of salmonellae into epithelial cells as well as processes leading to fluid secretion in intestinal models of infection (for a review, see reference 6). SigD (also known as SopB in S. enterica serovar Dublin) is secreted by this TTSS and was found to be required for efficient invasion into epithelial cells in vitro (14). SopB was shown to have an inositol phosphatase activity within eucaryotic cells (23) and cause fluid secretion in a calf model of intestinal infection (11). SigD, which is not encoded within SPI1, was identified in a screen for invasion genes (14); however, the original Tn10dTc mutation was actually in the downstream gene sigE. This mutation resulted in the absence of SigD, but not other proteins, in culture supernatants. Because TTSS effectors often require a cognate chaperone, we hypothesized that SigE is a specific chaperone for SigD.
Chaperones have functions ranging from preventing premature association of one effector with another (20) to preventing degradation of the effector within the bacterium prior to secretion (9, 27, 28). Although we hypothesized that SigE is a specific chaperone for the SigD polypeptide, it was also possible that SigE affected the transcription of sigDE. Previous work has shown that another chaperone, SicA, is autoregulated and is required for the transcription of sipBCDA and sigDE (7). Therefore, we wanted to determine if the sigD promoter, which is dependent on SicA for expression (7), was dependent on sigE for transcription. A sigD-lacZYA reporter plasmid (pHD5) was previously constructed (5) and integrated into the chromosome of the wild-type and sigE::Tn10dTc strains. Transduction analysis was used to confirm the linkage of the reporter fusion (conferring chloramphenicol resistance) to the transposon insertion (conferring tetracycline resistance) (18). This reporter did not disrupt secretion of wild-type levels of SigD (7). β-Galactosidase activities from the wild-type and sigE serovar Typhimurium 14028s (American Type Culture Collection) strains were measured and found to be almost identical (57 ± 1 and 60 ± 2 Miller units for wild type and sigE mutant, respectively). Therefore, sigE is not required for transcription from the sigD promoter.
To see if sigE was required for the stability of the SigD polypeptide in the cytoplasm of the bacteria, Western blotting was performed on whole-cell proteins from overnight cultures of wild-type and sigE::Tn10dTc strains. Proteins that were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (7.5% gel) were transferred to polyvinylidene difluoride (Immobilon) membranes and incubated with antibodies raised against the first 192 amino acids of SigD fused to maltose binding protein (5). SigD was undetectable in whole-cell preparations of the sigE mutant (Fig. 1, lane 2) and the sigD mutant (lane 5). SigD could be restored in the sigE strain when transformed with a plasmid encoding sigE (pHH26) (13) (lane 4). In addition, overexpression of sigDE on a medium-copy-number plasmid significantly increased the amount of SigD in whole cells (lane 6). When sigE was disrupted with transposon TnMax2 (12), less SigD was observed (lane 7). This result suggests that SigD can be translated in the absence of SigE and that SigE is probably required for the stability of the SigD protein. Nevertheless, we cannot absolutely rule out the possibility that SigE has a role in translation of the sigDE transcript.
FIG. 1.
Immunoblot analysis using polyclonal antibodies directed against SigD (amino acids 1 to 192) of proteins from 14028s (wild type), SVM167 (sigE::Tn10dTc), SVM167 pWKS30 (vector), SVM167 pHH26 (pWSK29-sigE), SVM255 (sigD), SVM167 pHH10 (sigDE+), and pHH10-23 (sigD+) strains.
A bacterial two-hybrid system using the Bordetella pertussis adenylate cyclase gene (cya) (16) was used to determine if SigE forms dimers as has been suggested for other type III secretion chaperones in vitro (28). In addition, this system was used to test if SigE interacts with SigD. The adenylate cyclase protein (Cya) can be separated into two domains (designated T25 for the N-terminal domain and T18 for the C-terminal domain) which cannot function independently. When fused to interacting proteins, the Cya domains can potentially interact and function, resulting in the production of cyclic AMP (cAMP). cAMP production can be indirectly measured by maltose or lactose metabolism in an Escherichia coli cya mutant (DHP1) (16). The N-terminal region of SigD (SigD′) and the entire SigE protein were each fused to Cya domains T25 and T18 encoded in plasmids pT25 and pT18, respectively. The first 101 codons of SigD were amplified using Pfu polymerase (Stratagene) with primers SigDfKpnI (5′-TTACGGTACCTATGCAAATACAGAGCTTCTATCAC-3′) and SigDr102KpnI (5′- TTGAGGTACCATTGACGTTAGAACCGGGTCTTG-3′) (Life Technologies). The primers used to amplify SigE were SigEfKpnI (5′-TTGAGGTACCTATGGAAAGTCTATTAAATCG-3′) and SigErKpnI (5′-ATTAGGTACCGCATAATGCTCTTTCAATTG-3′). KpnI-digested fragments were cloned into the KpnI sites of pT18 and pT25. Clones were sequenced to check for the correct orientation of the inserts and any mutations that may have been incorporated during the amplification or cloning process. E. coli strain DHP1 was transformed with these plasmids, and β-galactosidase activity from each strain was measured.
When SigE was fused to both domains of Cya, a high level of β-galactosidase activity (21) was measured in liquid overnight cultures grown at 26°C (Table 1). No activity was detected when a SigE fusion was combined with a SicA fusion. When an N-terminal portion (amino acids 1 to 100) of SigD was fused to the T18 domain of Cya (SigD′-T18) and combined with T25-SigE, the level of β-galactosidase activity measured was higher than background activity (Table 1). The same SigD′-T18 construct yielded no activity when combined with T25 fused to the chaperone SicA. Interestingly, when SigD′ was fused to the T25 domain of Cya (T25-SigD′), no activity was measured. It was possible that the SigE binding domain on SigD was occluded by the T25 domain in this construct or that this fusion was not stable, producing no T25-SigD′.
TABLE 1.
Complementation of cya in E. coli DHP1 using sigE and sigD fusions to B. pertussis cya domains
| Test plasmid pair | Mean β-galactosidase activity (Miller units) ± SDa |
|---|---|
| pT18 + pT25 (vectors) | 81 ± 2 |
| pT18 + pT25-SigD′ | 55 ± 2 |
| pSigD′-T18 + pT25 | 61 ± 0 |
| pSigE-T18 + pT25 | 61 ± 5 |
| pSigE-T18 + pT25-SigE | 2,405 ± 220 |
| pSigE-T18 + pT25-SicA | 62 ± 5 |
| pSigD′-T18 + pT25-SigE | 439 ± 43 |
| pSigD′-T18 + pT25-SicA | 68 ± 4 |
| pSigE-T18 + pT25-SigD′ | 93 ± 3 |
E. coli strain DHP1 containing the indicated plasmids was grown overnight (18 h) in Luria-Bertani broth supplemented with chloramphenicol (25 μg/ml) and ampicillin (100 μg/ml) in 13-cm test tubes at 26°C on a roller drum. β-Galactosidase activity was measured as described in the text.
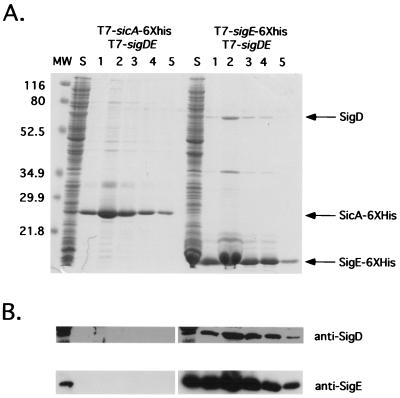
In addition to the two-hybrid system, we used a biochemical approach to determine if SigD could interact with SigE. Hexahistidine (His6) fusions to the C termini of chaperones SicA and SigE were constructed in the expression vector pET24(+) (Novagen) and transformed into the E. coli strain BL21(DE3), which encodes an inducible T7 polymerase gene (25). In addition, these strains were transformed with plasmid pHH22, which contains sigDE downstream of a T7 promoter (14). One-liter cultures of each strain were induced with 100 μM isopropyl-β-d-thiogalactopyranoside during exponential phase (optical density at 600 nm of 0.6), and cell lyates were prepared under native conditions according to the QIAexpressionist manual (Qiagen). When sigDE was coexpressed with SicA-His6, SigD was not coeluted along with SicA-His6 from the nickel column (Fig. 2, lanes 3 to 7). The sicA-His6 contruct should have produced an active protein because it was able to activate transcription of a sicA-lacZYA reporter plasmid when coexpressed with invF in E. coli (data not shown). When sigE-His6 and sigDE were coexpressed, SigD copurified with SigE-His6, as seen in either a 12.5% Coomassie-stained SDS-polyacrylamide gel or Western blot (separated on a 7.5% gel and transferred to 0.2-μm-pore-size Schleicher & Schuell nitrocellulose) using antibodies to SigD (Fig. 2A, lanes 9 to 13, and 2B).
FIG. 2.
SigD copurifies with SigE-His6. (A) Coomassie brilliant blue-stained 12.5% polyacrylamide gel of fractions eluted from a nickel-agarose column. Positions of molecular mass standards (MW) are indicated on the left in kilodaltons. Lane S, soluble fraction after sonication of cell pellets and removal of insoluble debris by centrifugation; lanes 1 to 5, imidazole-eluted fractions. (B) Immunoblot analysis of the same fractions using antibodies to SigD or SigE. The major band labeled SicA-6XHis in panel A was confirmed to be SicA by antibodies specific to SicA (data not shown).
Many putative type III secretion chaperones have been identified in other gram-negative bacteria but have yet to be shown to multimerize or specifically interact with effector molecules (15). Type III chaperones share several characteristics: small size (around 12 to 20 kDa), acidic isoelectric point (∼4), and the presence of an amphipathic alpha helix near the C terminus. The results of this work show that SigE appears to be a chaperone for SigD, a type III secreted effector molecule. The inability of SigD to interact with SicA suggests the interaction with SigD is specific for SigE rather than a general interaction with type III chaperones. In addition, SigE appears to form dimers or other higher-order multimers. SigE is not required for transcription of the sigDE operon. In one case in Salmonella, a mutation in the sicA chaperone results in reduced expression of genes encoding several effector molecules (7). It is notable that a mutation in sicA is likely to also have posttranslational effects on two effectors, SipB and SipC (7, 26). The sigDE operon, however, is more similar to other known chaperone-effector gene pairs, like sicP-sptP (9) from Salmonella and ipgC-ipaBC from Shigella (20), that do not appear to be dependent on a chaperone for transcription activation. Rather than affect transcription, SigE is likely to affect either the translation or stability of SigD. The transcriptional organization of sigDE suggests that sigD and sigE are cotranslated. Moreover, overexpression of sigD in a sigE mutant results in the production, albeit in reduced amounts, of SigD. Together, these results suggest that SigE enhances the stability of SigD in the cytoplasm, but we cannot rule out the possibility that SigE also has a role in sigD translation. Previous studies on other chaperone-effector pairs have also shown that chaperones can directly interact with their cognate effector molecules (9, 20, 26). In this work, both genetic (bacterial two-hybrid) and biochemical (affinity purification) approaches indicate that SigD and SigE can specifically interact with each other. In addition, similar to other chaperone-effector pairs, the N-terminal region of SigD contains at least a part of the SigE chaperone binding domain.
Acknowledgments
We thank Paula Revell for critically reviewing the manuscript and Daniel Ladant for the two-hybrid plasmids and E. coli strain DHP1.
REFERENCES
- 1.Andersson K, Carballeira N, Magnusson K-E, Persson C, Stendahl O, Wold-Watz H, Fällman M. YopH of Yersinia pseudotuberculosis interrupts early phosphotyrosine signalling associated with phagocytosis. Mol Microbiol. 1996;20:1057–1069. doi: 10.1111/j.1365-2958.1996.tb02546.x. [DOI] [PubMed] [Google Scholar]
- 2.Chen L M, Kaniga K, Galán J E. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 3.Collazo C M, Galán J E. The invasion-associated type III system of Salmonella typhimurium directs the translocation of the Sip proteins into the host cell. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 4.Cornelis G R, Wolf-Watz H. MicroReview: The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 5.Darwin K H, Miller V L. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J Bacteriol. 1999;181:4949–4954. doi: 10.1128/jb.181.16.4949-4954.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darwin K H, Miller V L. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin Microbiol Rev. 1999;12:405–428. doi: 10.1128/cmr.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darwin K H, Miller V L. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol Microbiol. 2000;35:949–959. doi: 10.1046/j.1365-2958.2000.01772.x. [DOI] [PubMed] [Google Scholar]
- 8.Francis M S, Wolf-Watz H. YopD of Yersinia pseudotuberculosis is translocated into the cytosol of HeLa epithelial cells: evidence of a structural domain necessary for translocation. Mol Microbiol. 1998;29:799–813. doi: 10.1046/j.1365-2958.1998.00973.x. [DOI] [PubMed] [Google Scholar]
- 9.Fu Y, Galán J E. Identification of a specific chaperone for SptP, a substrate of the centisome 63 type III secretion system of Salmonella typhimurium. J Bacteriol. 1998;180:3393–3399. doi: 10.1128/jb.180.13.3393-3399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, Galán J E. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol Microbiol. 1998;27:359–368. doi: 10.1046/j.1365-2958.1998.00684.x. [DOI] [PubMed] [Google Scholar]
- 11.Galyov E E, Wood M W, Rosqvust R, Mullan P B, Watson P R, Hedges S, Wallis T S. A secreted effector protein of Salmonella dublin is translocated into eucaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 12.Haas R, Kahrs A F, Facius D, Allmeier H, Schmitt R, Meyer T F. TnMax—a versatile mini-transposon for the analysis of cloned genes and shuttle mutagenesis. Gene. 1993;130:23–31. doi: 10.1016/0378-1119(93)90342-z. [DOI] [PubMed] [Google Scholar]
- 13.Hardt W-D, Chen L-M, Schuebel K E, Bustelo X R, Galán J E. S. typhimurium encodes an activator of rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 14.Hong K H, Miller V L. Identification of a novel Salmonella invasion locus homologous to Shigella ipgDE. J Bacteriol. 1998;180:1793–1802. doi: 10.1128/jb.180.7.1793-1802.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hueck C J. Type III protein secretion in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee V T, Anderson D M, Schneewind O. Targetting of Yersinia Yop proteins into the cytosol of HeLa cells: one-step translocation of YopE across bacterial and eucaryotic membranes is dependent on SycE chaperone. Mol Microbiol. 1998;28:593–601. doi: 10.1046/j.1365-2958.1998.00822.x. [DOI] [PubMed] [Google Scholar]
- 18.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 19.Ménard R, Prévost M-C, Gounon P, Sansonetti P J, Dehio C. The secreted Ipa complex of Shigella flexneri promotes entry into mammalian cells. Proc Natl Acad Sci USA. 1996;93:1254–1258. doi: 10.1073/pnas.93.3.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ménard R, Sansonetti P, Parsot C, Vasselon T. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell. 1994;79:515–525. doi: 10.1016/0092-8674(94)90260-7. [DOI] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 22.Mills D M, Bajaj V, Lee C A. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 23.Norris F A, Wilson M P, Wallis T S, Galyov E E, Majerus P W. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc Natl Acad Sci USA. 1998;95:14057–14059. doi: 10.1073/pnas.95.24.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosqvist R, Forsberg A, Wolf-Watz H. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect Immun. 1991;59:4562–4569. doi: 10.1128/iai.59.12.4562-4569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Studier F, Rosenberg A, Dunn J, Dubendorf J. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 26.Tucker S C, Galán J E. Complex function for SicA, a Salmonella enterica serovar Typhimurium type III secretion-associated chaperone. J Bacteriol. 2000;182:2262–2268. doi: 10.1128/jb.182.8.2262-2268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wattiau P, Bernier B, Deslée P, Michiels T, Cornelis G R. Individual chaperones required for Yop secretion by Yersinia. Proc Natl Acad Sci USA. 1994;91:10493–10497. doi: 10.1073/pnas.91.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wattiau P, Cornelis G R. SycE, a chaperone-like protein of Yersinia enterocolitica involved in the secretion of YopE. Mol Microbiol. 1993;8:123–131. doi: 10.1111/j.1365-2958.1993.tb01209.x. [DOI] [PubMed] [Google Scholar]
- 29.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 30.Zychlinsky A, Kenny B, Ménard R, Prevost M C, Holland I B, Sansonetti P J. IpaB mediates macrophage apoptosis induced by Shigella flexneri. Mol Microbiol. 1994;11:619–627. doi: 10.1111/j.1365-2958.1994.tb00341.x. [DOI] [PubMed] [Google Scholar]