Abstract
Purpose of review.
Non-coding RNAs (ncRNAs) including microRNAs (miRNAs) and circular RNAs (circRNAs) are pivotal regulators of mRNA and protein expression that critically contribute to cardiovascular pathophysiology. Although little is known about the origin and function of such ncRNAs, they have been suggested as promising biomarkers with powerful therapeutic value in cardiovascular disease (CVD). In this review, we summarize the most recent findings on ncRNAs biology and their implication on cholesterol homeostasis and lipoprotein metabolism that highlight novel therapeutic avenues for treating dyslipidemia and atherosclerosis.
Recent findings.
Clinical and experimental studies have elucidated the underlying effects that specific miRNAs impose both directly and indirectly regulating circulating high-density lipoprotein (HDL), low-density lipoprotein (LDL) and very-low density lipoprotein (VLDL) metabolism and cardiovascular risk. Some of these relevant miRNAs include miR-148a, miR-128-1, miR-483, miR-520d, miR-224 miR30c, miR-122, miR-33, miR-144 and miR-34. circRNAs are known to participate in a variety of physiological and pathological processes due to their abundance in tissues and their stage-specific expression activation. Recent studies have proven that circRNAs may be considered targets of CVD as well. Some of these cirRNAs are circ-0092317, circ_0003546, circ_0028198 and cirFASN that have been suggested to be strongly involved in lipoprotein metabolism however their relevance in CVD is still unknown.
Summary.
MicroRNA and cirRNAs have been proposed as powerful therapeutic targets for treating cardiometabolic disorders including atherosclerosis. Here, we discuss the recent findings in the field of lipid and lipoprotein metabolism underscoring the novel mechanisms by which some of these ncRNAs influences lipoprotein metabolism and CVD.
Keywords: lipoprotein metabolism, miRNAs, reverse cholesterol transport, ABCA1, miR-33
1. INTRODUCCION
Cardiovascular disease (CVD) is the leading cause of death and morbidity in the Western population. Atherosclerosis is the most common underlying cause of CVD manifestations, including heart attack and stroke [1]. Atherosclerosis is regarded as a chronic inflammatory disease that can arise from different factors, such as hyperlipidemia, hypertension, diabetes or smoking, among which, dysregulations in lipid and lipoprotein metabolism are the most important and common risk factors [1]. The progression of the disease involves a number of different cells, from macrophages, which participate in initiation, growth and rupture of the plaque, to endothelial cells (ECs) and vascular smooth muscle cells (VSMCs), the essential cells constituting the artery walls.
Over the last years, several studies have identified numerous non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs), involved in the regulation of the expression of genes associated to lipiid metabolism and CVD [2, 3]. miRNAs, the best characterized class of ncRNAs, are small non-coding RNAs of ~22 nucleotides that regulate gene expression at the post-transcriptional level by direct binding to the 3’UTR of a target mRNA. One important feature of miRNA biology resides in the ability of a single miRNA to target a wide range of mRNAs (hundreds to thousands), while, on the opposite, one mRNA can be targeted by several miRNAs. A number of studies have identified dysregulation of miRNA levels or miRNA-targeted site in CVD [4], highlighting the relevant role of miRNAs in atherosclerosis [5].
Contrary to miRNAs, the relevance of other ncRNAs including circRNAs in CVDs has not been deeply characterized. circRNAs consist of a large class of non-coding RNAs that results from a non-canonical splicing of the RNA (back-splicing), in which a closed circular RNA molecule is produced [6]. Although circRNAs were initially considered irrelevant by-products of transcription, it is now known that circRNAs regulate gene expression by different mechanism. circRNAs can be derived from both exonic and intronic regions of the genome, generating exonic circular RNAs (ecircRNAs), intronic circular RNAs (ciRNAs) and exon-intron circular RNAs (EIciRNAs) [6]. Most known circRNAs are ecircRNAs, therefore being these simply referred to as circRNAs, which are found in the cytoplasm where generally act as endogenous sponges for miRNAs or protein [6]. In the recent years, several studies have shown that a number of circRNAs are important for the pathophysiology of CVDs, mainly through the regulation of cellular processes in ECs, VSMCs and macrophages relevant to the progression of atherosclerosis [7, 8]. However, little has been described so far about their potential role in regulating lipoprotein metabolism and atherosclerosis.
In this review article, we summarize the most relevant and novel findings of the roles of miRNAs and circRNAs in regulating lipoprotein metabolism and atherosclerosis and their potential therapeutic value for treating this disease.
2. miRNAs REGULATION OF CHOLESTEROL METABOLISM
A number of studies from our laboratory and others have elucidated several miRNAs involved in the regulation of cholesterol homeostasis and lipoprotein metabolism [3, 5, 8–13]. These miRNAs control the expression of genes regulating HDL, LDL and VLDL metabolism and include miR-148a, miR-128-1, miR-483, miR-520d, miR-224, miR30c, miR-122, miR-33, miR-144 and miR-34 [10, 14–23] (Figure 1).
Figure 1. miRNAs and circRNAs regulation of lipoprotein metabolism.
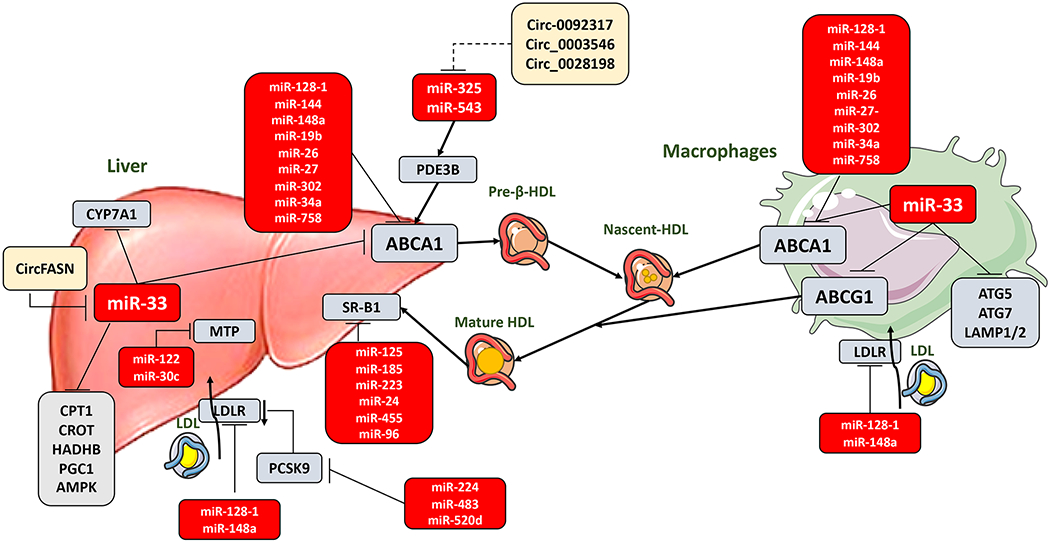
miRNAs have been described to regulate the expression of genes involved in HDL biogenesis and maturation, cholesterol efflux and RCT from peripheral tissues to the liver; as well as genes that regulate triglyceride synthesis for incorporation into VLDLs and genes involved in LDL clearance from the circulation. The major transporter that regulates HDL biogenesis and efflux from the liver, ABCA1, is regulated by miR-33 and several other miRNAs in liver and macrophages. Moreover, miR-33 regulates a broad spectrum of genes that participate in liver and macrophages function, regulating HDL and lipid metabolism, RCT, autophagy and inflammation. Other miRNAs participate in RCT through the regulation of SR-B1 the liver receptor responsible of HDL uptake from the circulation. Finally, VLDL/LDL formation and uptake can be regulated by different miRNAs that regulate MTP expression in the liver and LDLR expression both in liver and macrophages. LDLR can be also indirectly regulated by miRNAS targeting PCSK9, which induces LDLR degradation. Besides miRNAs, circRNAs are arising as potential regulators of lipid metabolism, usually through the regulation of miRNAs. However, not many circRNAs have been proved for a relevant role in lipoprotein metabolism being merely suggested or potentially identified (illustrated as dashed line) based on target prediction. HDL: high-density lipoprotein; RCT: reverse cholesterol transport; VLDL: very low-density lipoprotein; LDL: low-density lipoprotein; ABCA1: ATP-binding cassette A1; ABCG1: ATP-binding cassette subfamily G member 1; SR-B1: scavenger receptor class B type 1; MTP: microsomal triglyceride transfer protein; LDLR: low-density lipoprotein receptor; PCSK9: proprotein convertase subtilisin/kexin type 9; circRNAs: circular RNAs; CYP7A1: cholesterol 7α-hydroxylase; CircFASN: Fatty acid synthase; PDE3B: phosphodiesterase 3B; CPT1: carnitine palmitoyltransferase 1a; CROT: carnitine O-octanoyltransferase; HADHB: hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta; PGC1: peroxisome proliferator-activated receptor gamma coactivator 1-alpha; AMPK: AMP-activated protein kinase; ATG5: autophagy related 5; ATG7: autophagy related 7; LAMP1/2: lysosomal membrane proteins 1 and/or 2. This figure was created using Servier Medical Art illustration resources.
2.1. miRNAS regulation of LDL-C and VLDL-C metabolism
Cholesterol and TAGs are water-insoluble molecules and they must be packaged within lipoproteins to be transported effectively [24]. Lipoproteins are large spherical complexes formed by lipids and proteins. Their central core contains cholesterol esters and triglycerides that are surrounded by free cholesterol, phospholipids, and apolipoproteins. There are seven classes of plasma lipoproteins based on size, lipid composition, and apolipoproteins (chylomicrons, chylomicron remnants, very low-density lipoprotein (VLDL), intermediate-density lipoprotein (IDL), LDL, HDL, and lipoprotein a (lp (a)) [11, 24].
Formation of VLDL is the first step of the endogenous lipoprotein pathway that takes place in the liver [24]. In muscle and adipose tissue, triglycerides (TAGs) carried in VLDL are break down by lipoprotein lipase releasing free fatty acids and IDL are formed [24]. IDL are further metabolized to LDL, which are taken up by LDLR primarily in liver, the key site of uptake [11, 24]. In liver, VLDL production and LDL-C clearance via (low-density lipoprotein receptor) LDLR regulate plasma LDL-C levels [3, 11].
Circulating apo-B containing lipoproteins (LDL-C and VLDL) can transcytose across the endothelium and accumulate in the arterial wall, leading to the formation of atherosclerotic plaques [12, 25–27]. Recent studies have identified miRNAs that control plasma LDL-C and VLDL-C levels by regulating hepatic LDL receptor (LDLR) expression and VLDL production [11]. Following we summarize some of the well-known miRNAs that regulates LDL and VLDL metabolism.
2.1.1. miR-148a
Our group and Näär laboratory identified miR-148a as an important regulator of LDL-C and HDL-C metabolism using a high-throughput genome-wide screening assay in vitro and human genome-wide association study (GWAS) respectively [10, 14]. miR-148a is encoded within an intergenic region of human chromosome 7 and its highly expressed in human and mouse hepatocytes, macrophages and adipocytes [10, 14, 28]. miR-148a inhibits the expression of LDLR and ATP-binding cassette A1 (ABCA1), a transporter that regulates cellular cholesterol efflux and HDL biogenesis, thus suppressing cellular cholesterol uptake and efflux respectively [10–12, 14]. Notably, recent studies have identified a single nucleotide polymorphisms (SNP) in the promoter region of miR-148a in human liver associated with altered circulating levels of total cholesterol, LDL-C and TAGs [29–31]. In agreement with these findings, antagonism of miR-148a using antisense oligonucleotides (ASO) reduces circulating LDL-C and increases plasma HLD-C [10, 14]. Taken together these studies suggest that suppression of hepatic miR-148a might be an useful therapeutic approach to treat dyslipidemias and atherosclerotic CVD.
2.1.2. miR-128-1
miR-128-1, an intronic miRNA encoded within the R3HDM1 gene located in chromosome 2 is highly expressed in human liver, spinal cord, brain, adipocytes and testis [32–37]. Similar to mR-148a, miR-128-1 targets the 3′UTR of LDLR and ABCA1, underscoring a fundamental role in cholesterol and lipoprotein metabolism [14]. Similar to miR-148a, miR-128-1 has been stipulated to regulate hepatic LDLR expression [11, 14]. Studies in ApoE−/− mice and human hepatoma cells have revealed alterations of miR-128-1 that notably affected LDL-C clearance and LDL uptake respectively [14]. Related to its ABCA1 target, miR-128-1 has been pointed to regulate cholesterol efflux capacity mediated by ApoA1 in mouse macrophages and in hepatoma cells [11, 12, 14]. Moreover, antisense inhibition of miR-128-1 in Apoe−/− mice decreases circulating TAGs and hepatic steatosis and improves insulin sensitivity and glucose tolerance [14, 38]. In addition to controlling cholesterol and lipid metabolism, miR-128-1 also regulates hepatic insulin signaling and glucose homeostasis by directly modulating the expression of the insulin receptor (INSR), insulin receptor substrate 1 (IRS-1), and Akt signaling [14, 38]. miR-128-1 has been suggested to regulate genes involved in lipogenesis such as fatty acid synthase (FASN), sirtuin 1 (SIRT1), NAD+-dependent energy sensor and lysine deacetylase [14]. Moreover, a very recent study has demonstrated that miR-128 can regulate energy expenditure and this is associated with the fact that miR-128-1 is located at the 2q21.3 locus that harbors the lactase gene which is known to be linked to an increased risk of obesity and type 2 diabetes [15]. This work has proved that the inhibition and genetic ablation of miR-128-1 in mouse models of metabolic diseases caused reductions in body weight gain and fat accumulation and ameliorated high-fat-diet-induced obesity and protected from insulin resistance [15]. All these data suggest that inhibition of miR-128-1 represents an attractive therapeutic strategy against cardiometabolic diseases however new exhaustive studies are needed to reveal the precise function of miR-128-1 in regulating lipoprotein transport, lipogenesis and glucose homeostasis to combat atherosclerosis, type 2 diabetes and obesity.
The relevance of miRNA as potent regulators of LDLR expression in humans have been recently supported with the identification of a rare 2.5 kilobase (kb) deletion (del2.5) on the distal end of the 3‘UTR of LDLR [39]. This deletion results in shortening of the LDLR mRNA 3‘UTR where the target sites for some miRNAs such as miR-128-1 and miR-148a are located, increasing LDLR expression and reducing circulating LDL-C [39].
2.1.3. miR-483
miR-483 is expressed in human and mouse skeletal muscle, adipocytes, breast, whole blood and hepatocytes [16, 40–42]. miR-483 comprises both miR-483-3p and miR-483-5p, that are intronic microRNAs located on human chromosome 11 that are encoded together with their parental gene IGF2 [16, 43]. miR-483-5p affects cholesterol homeostasis by targeting the 3′-UTR of proprotein convertase subtilisin/kexin type 9 (PCSK9), which in turn increased hepatocyte expression of LDLR reducing LDL-C levels and the prevalence of cardiovascular events [16]. Circulatory levels of miR-483-5p were found lower in subjects with elevated LDL-C levels [44] and miR-483 overexpression in hypercholesterolemic mouse models remarkably reduced plasma cholesterol and LDL-C levels [16]. Additionally, miR-483 directly targeting the 3′-UTR of PCSK9 could mitigate the unwanted effect of statins in increasing plasma PCSK9 protein levels. These results demonstrate the potential use of miR-483 in combination with statins for treating hypercholesterolemia. This mechanism of cholesterol lowering via PCSK9 targeting by miR-483 may have therapeutic efficacy. Besides miR-483, other miRNAs such as miR-222, -191, and -224, have been predicted to target PCSK9 [45]. However, the relevance of this miRNAs in regulating hepatic LDLR expression circulating LDL-C in vivo has not been assessed yet.
2.1.4. miR-520d and miR-224
miR-520d and miR-224 located on human chromosomes 19 and X respectively and they are expressed by liver [17]. miR-520d and miR-224 share several targets such as PCSK9 and inducible degrader of the LDLR (IDOL) that are implicated in the degradation of LDLR [17]. In vitro studies using hepatocyte cell lines have shown that miR-224 and miR-520d reduced mRNA and protein levels of PCSK9, IDOL, and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), resulting in increased LDLR expression and LDL binding [17]. Additional experiments will be important to define the relevance of these miRNAs in regulating lipoprotein metabolism and atherosclerosis in vivo.
2.1.5. miR-30c
miR-30c is located at chromosome 6 and resides in intron 5 of the nuclear transcription factor Y subunit C (NFY-C) gene and it is highly expressed in the heart, skeletal muscle and kidney [18]. MiR-30c is a member of the relevant and complex miR-30 family that plays a variety of physiological and pathological roles in vivo including the regulation of lipoprotein metabolism. miR-30c changes have been associated with altered levels of circulating cholesterol and TAGs [18]. This microRNA inhibits the expression of microsomal triglyceride transfer protein (MTP) which is highly expressed in liver, suppressing VLDL biogenesis. Additionally, miR-30c repress hepatic lipid synthesis by targeting lysophosphatidyl glycerol acyltransferase 1 (LPGAT1) [18]. As a result, overexpression of miR-30c using lentivirus or miRNA mimics markedly reduces circulating VLDL-C and LDL-C and attenuates atherogenesis in Apoe−/− mice [18]. By contrary, antagonism of miR-30c increased plasma VLDL-C and LDL-C levels and promotes the progression of atherosclerosis [18]. Given that miR-30c decreases lipid biosynthesis and lipoprotein production, this microRNA might be useful in treating hyperlipidemias and associated disorders.
2.1.6. miR-122
miR-122 coding sequence comprises a single locus on the human chromosome 18 [46]. miR-122 is a liver-specific microRNA that accounts for the 72% and 52% of all cloned miRNAs in the adult liver of both mice and humans respectively [19, 20], being one of the most abundant miRNAs expressed in this tissue [19–21]. Therefore miR-122 is a key regulator of liver physiology by playing a central role in liver development, differentiation and homeostasis [21]. miR-122 also has a crucial role in the hepatic cholesterol homeostasis and FA metabolism. miR-122 antagonism decreases the expression of several genes involved in hepatic lipid metabolism and cholesterol biosynthesis, including acetyl-CoA carboxylase alpha (ACC1), acetyl-CoA carboxylase beta (ACC2), ATP citrate lyase (ACLY), stearoyl-CoA desaturase 1 (SCD1), sterol regulatory element-binding protein 2 (SREBP2), FA synthase (FASN) and phosphomevalonate kinase (PMVK) [47]. 1-Acylglycerol-3-phosphate O-acyltransferase 1 (AGPAT1) that regulates the energy state [48] and CIDEC that controls triglyceride storage and FA oxidation [49] have been also identified as potential targets of miR-122. Experimental and clinical studies have demonstrated that miR-122 is a key factor of fibrogenesis, liver disease and cardiovascular injury [21, 50]. Inhibition of miR-122 decreased plasma cholesterol levels and hepatic FA synthesis rate, improving liver steatosis in a high-fat fed mice [47]. Notably, miR-122 levels were shown to be upregulated in subjects with atherosclerotic lesions, and serum miR-122 levels were positively correlated with atherosclerotic severity [51–53]. Additionally, circulating miR-122 levels were elevated in subjects with metabolic syndrome or type 2 diabetes and correlated with high levels of saturated and monounsaturated fatty acids delineating miR-122 as a marker of hepatic lipid metabolism disorders including dyslipidemias, metabolic syndrome and type 2 diabetes [54]. However, although these results point out miR-122 antagonism as a promising strategy in CVD, raising concerns about the therapeutic value of suppressing miR-122 expression have been postulated as both whole-body and liver-specific deficiency aged mice develop microsteatosis and inflammation which progress to steatohepatitis, fibrosis and hepatocellular carcinoma [55, 56].
2.2. miRNAs regulation of HDL-C metabolism and transport
In mammals, excess cholesterol cannot be degraded and the only way to remove cholesterol is through a tightly orchestrated process known as RCT [57]. In this process, the excess cholesterol is transported from peripheral tissues to the liver, where it must be further metabolized and excreted as bile acids into feces [58]. The levels of plasma HDL and its component apolipoprotein AI (ApoAI) are the major transporters involved in RCT, a process that involve the regulation of several proteins and steps. As first step, ABCA1 expression in the liver is critical for HDL formation [59], while ABCA1 and ABCG1 in macrophages are essential in order to mediate cholesterol efflux to HDL [60]. HDL in circulation needs to be mature return to the liver and finalize RCT, which is regulated by scavenger receptor class B type 1 (SR-B1) [61]. Back in the liver, the enzyme cholesterol 7α-hydroxylase (CYP7A1) is the main responsible of converting cholesterol transported by HDL into bile acids [62], that is finally secreted by a group of ATP-binding cassette transporters: ATP-binding cassette, sub-family B, member 11 (ABCB11 or BSEP); ATP-binding cassette sub-family G, members 5/8 (ABCG5 and ABCG8) and ATP-binding cassette, sub-family B, member 4 (ABCB4) [62]. Several miRNAs have been found to contribute to the regulation of HDL biogenesis, cholesterol efflux, RCT and bile acid metabolism [13, 63–65].
2.2.1. miR-33 regulation of HDL-C metabolism and Reverse cholesterol transport
Among different miRNAs found to be involved in HDL-C metabolism and RCT, miR-33 family members, miR-33a and miR-33b, are the best studied and described miRNAs. miR-33a and miR-33b isoforms are encoded within the intronic regions of SREBP2 and SREBP1 genes, respectively, and their expression is transcriptionally controlled together with their host genes [13, 66]. The transcription of miR-33a together with its host gene SREBP2 involves the regulation of a fine-tuned network aiming to control the cholesterol levels. Recently, it has been identified a novel regulator of miR-33 expression in humans, the primate specific long non-coding RNA CHROME. CHROME regulates the expression of several miRNAs including miR-33, miR-27 and miR-128, thus derepressing the posttranscriptional inhibition of ABCA1. Notably, CHROME is upregulated in serum and plaques of CVD patients [67]. miR-33 controls cholesterol metabolism mostly through the direct repression of ABCA1 and ABCG1 transporters [13, 66, 68]. As a result, overexpression of miR-33 downregulates ABCA1 and ABCG1, decreasing HDL-C levels in plasma, while antagonism miR-33 using antisense oligonucleotides (ASOs) is sufficient to upregulate hepatic ABCA1 and ABCG1, increasing circulating HDL-C [13, 66]. These studies where further confirmed in miR-33 deficient mice and non-human primates treated with miR-33 inhibitors [69–72]. However, global deficiency of miR-33 was found to have detrimental metabolic long-term effects in mice, including obesity, dyslipidemia, insulin resistance and steatosis [73, 74]. Some of these metabolic disruptions associated to miR-33 deficiency have counteracted the benefit in terms atherosclerosis, hampering the development of anti-miR-33 based therapies and urging for a better understanding of miR-33 biology. In this context, our group has recently shown that ABCA1 targeting by miR-33 is the major responsible for the protection against atherosclerosis observed in the miR-33 deficient mice [75]. In this study, we generated a mouse model with specific mutations in the miR-33 binding sites in the 3’UTR of Abca1 using CRISPR/Cas9 genome editing. The disruption of ABCA1-miR-33 targeting was sufficient to improve RCT and atherosclerosis in mice, suggesting that although other target genes may be affected by miR-33, the regulation of ABCA1 is key driver of the processes regulated by miR-33 associated with the progression of atherosclerosis [75].
Besides the targeting of ABCA1 and ABCG1, miR-33 regulates additional genes in the liver involved in RCT. miR-33 directly targets CYP7A1, which mediates cholesterol transformation into bile salts and bile salt exporter ABCB11 and ATP8B1, controlling in this way the final elimination of cholesterol from the body in fecal bile acids [63, 64].
Globally, several studies have highlighted the direct regulation exerted by miR-33 in multiple steps of RCT. Noteworthy, other studies have identified miR-33 as a key regulator of several target genes related to cholesterol, lipid metabolism and macrophage function, processes that can indirectly affect the efficiency of RCT. These other processes will be briefly mentioned in the next section, where other functions of miR-33 in cardiometabolic disease are reviewed.
A number of studies have used novel approaches to better understand the mechanism affected by miR-33 in atherosclerosis. By performing bone marrow (BM) transplants from miR-33 deficient mice into Apoe−/− mice, Horie et al. showed that reduced lipid accumulation in atherosclerotic plaques was produced in mice with whole-body and BM-specific miR-33 deficiency [76]. However, the effects were more dramatic in the mice with the global deficient of miR-33, suggesting a role of miR-33 in macrophages and liver in atherosclerosis [76]. More recent work from Price el at. used double knock-out mice for miR-33 and Ldlr to address the impact of removing miR-33 in this model of atherosclerosis. However, no differences on plaque size or lipid accumulation were observed, probably due to the resulting obesity, dyslipidemia and insulin resistance associated with whole body miR-33 deficiency [77]. In contrast, BM transplant from miR-33 deficient mice into Ldlr deficient mice protects against the progression of atherosclerosis [77]. The study efficiently proved that deficiency of miR-33 in macrophages improves RCT and reduces inflammation, improving atherosclerosis [77]. The specific role of hepatic miR-33 in atherosclerosis has been controversial due numerous factors observed in global miR-33 deficient mice. Recent work from our laboratory demonstrated that genetic specific genetic deletion of miR-33 in the liver increase RCT, improves metabolic homeostasis without developing any adverse effect associated with the global deficiency of miR-33 (e.g. obesity, insulin resistance, steatosis, hypercholesterolemia) [78]. However, miR-33 deficiency in hepatocytes did not have any effect in atherosclerosis despite the increase in HDL and RCT [78]. This lack of effect is attributed to the downregulation of both SREBP2 and miR-33 in the liver under hyperlipidemia conditions [74] , offsetting any effect of miR-33 deletion upon HDL metabolism [78]. Remarkably, this study was performed in mice, where only the miR-33a isoform is expressed, thus, in humans the contribution of miR-33b isoform in the liver could still be important in atherosclerosis, as long as its regulation is subjected to SREBP1, which is not downregulated under hyperlipidemia conditions. Thus, given the shared targets between miR-33a/b isoforms, and the ability of miR-33b to regulate HDL levels [79], it is possible that hepatic miR-33 in humans could to regulate lipid and lipoprotein metabolism impacting hyperlipidemia and atherosclerosis. In fact, studies using human miR-33b isoform knock-in mice, which regulates cholesterol metabolism in mice [79] and promotes atherosclerosis [80], has shown miR-33b expression is increased in response to high-cholesterol diet feeding [74]. Moreover, higher levels of miR-33b in the liver, suggests this is the dominant miR-33 isoform expressed in the liver, especially under atherosclerosis conditions [74]. These results are consistent with a novel study showing that mice expressing only the miR-33b isoform has lower HDL levels and larger atherosclerotic plaques than WT animals expressing only miR-33a [81]. The relevance of hepatic miR-33b in humans still need to be addressed, as novel findings regarding miR-33a/b regulation by lncRNA CHROME¸ which is highly upregulate in the liver in hyperlipidemic conditions, involves a more complex regulation of the pathways regulated by miR-33 [67]. Overall, the study by Price et al. supports the finding that, at least in rodents, the most important atheroprotective effects of miR-33 deficiency are related to miR-33 functions in macrophages. However, although miR-33 deficiency in the liver improves metabolic functions, HDL biogenesis and RCT it is not sufficient to improve atherosclerosis outcome [78].
2.2.1.1. Other regulatory functions of miR-33 in cardiometabolic disease
Progress made in atherosclerosis and CVD treatments have been dampened in the last years by the alarming growth of metabolic diseases that increase the risk of CVD, including obesity, diabetes, insulin resistance and other metabolic syndrome conditions [82, 83]. In addition to its role in regulating ABCA1/ABCG1 and its influence in HDL-C and RCT, miR-33 has been shown target a wide variety of genes and biological processes that could also impact atherosclerosis and other metabolic diseases. miR-33 has been shown to regulate several proteins involved in FA metabolism and mitochondrial function, such as AMPK, carnitine palmitoyltransferase 1a (CPT1a), (PGC1a), NADH-ubiquinone oxidoreductase 1 alpha subcomplex subunit 5 (NDUFA5), pyruvate dehydrogenase kinase isozyme 4 (PDK4) and solute carrier family 25 (SLC25A25) [65, 84]. The regulation of these genes by miR-33 can affect mitochondrial function, energy homeostasis, and ATP availability, which is described to alter the efficiency of ABCA1 and ABCG1 to mediate cholesterol transport and efflux [84].
In other studies, some of the targets described to be regulated by miR-33, and newly described targets involved in lysosomal formation were found to control macrophage autophagy and efferocytosis to impact cholesterol efflux and lipid accumulation in these cells [85]. Finally, miR-33 is also known to regulate macrophage polarization [86], which together with the regulation of energy homeostasis, confers miR-33 deficient macrophages with an anti-inflammatory like phenotype that can improve atherosclerotic plaque burden. Some other functions of miR-33 that have been related with cardiovascular disease are the regulation of Niemann-Pick disease, type C1 (NPC1) [13] and oxysterol binding protein-like 6 (OSBPL6) [85] (important for internal cholesterol trafficking); and with regulation of proliferation and lipid rafts preservation in cardiac fibrosis and vascular smooth muscle cells [87, 88].
Given the number of genes targeted by miR-33 described through the last decade and its ability to regulate HDL-C and RCT, miR-33 has been considered as an interesting target for the treatment of atherosclerosis. Several studies have proved the beneficial effect of targeting miR-33 by genetic deletion or inhibition in reducing atherosclerosis [69–71]. However, long-term detrimental effect associated with miR-33 deficiency have been shown, including development of obesity, dyslipidemia, and steatosis attributed with miR-33 functions in different cells and tissues [9, 73]. These undesired consequences of miR-33 targeting highlighted the promiscuity of miRNA-based therapies, raising flag in their development and the need of a better knowledge on the potential unknown effects of targeting a single miRNA. In this context, the use of new approaches, such as conditional knock-out models and tissue-specific delivery methods of miRNAs, will help to overcome these concerns.
2.2.2. Other miRNAs regulating HDL-C and RCT
In addition to miR-33, other microRNAs have been reported to regulate HDL-C biogenesis and RCT regulating several genes associated to both processes. Some miRNAs directly regulate ABCA1 expression in the liver or macrophages, including miR-144, miR-26, miR-19b, miR-758, miR-27, miR-148a, miR-128-1, miR-302 and, miR10b and miR-34a [10, 14, 22, 23, 89–95]. Notably, miR-144 regulates ABCA1 in hepatocytes and macrophages, affecting HDL levels in circulation RCT, and consequently atherosclerosis, by their inhibition or overexpression in mice [22, 96]. Recently, it has been shown that silencing miR-144 in mice also regulates CYP7B1 affecting HDL remodeling and RCT, only males, which suggest a sex-dependent role of miR-144 [97]. Similarly, miR-128-1 and 148a regulates ABCA1 expression in macrophages and plasma HDL-C levels [10, 14]. However, their relevance during the progression of atherosclerosis needs to be evaluated.
A recent study has uncovered the role of miR-34 in regulating cholesterol metabolism in macrophages and liver [23]. Similar to miR-33, miR-34a inhibits ABCA1/ABCG1 expression in macrophages regulating cholesterol efflux and RCT. Moreover, miR-34a controls macrophage polarization through the repression of liver X receptor (LXR) and Krüppel-like factor 4 (KLF4), attenuating inflammation and atherosclerosis [98, 99]. In this study, the authors show that miR-34a has a role also in the liver, where regulates CYP7A1 and CYP8B1, with the consequent effect on cholesterol transformation and export into bile acids. This novel study globally shows that the deletion or inhibition of miR-34a in macrophages and liver promotes atherosclerosis regression orchestrating several pathways. Considering the aberrant levels of miR-34a in different liver disease conditions, we take into consideration that this work from Xu et al., along with further studies about this miRNA will be of utmost relevance in cardiometabolic disease research.
The last step of RCT is regulated by the hepatic SR-B1 expression, which also can be targeted by miRNAs, including miR-455, miR-125a, miR-185, miR-96, miR-223 and miR-24 [100, 101]. However, the importance of these miRNAs in atherosclerotic murine models and their therapeutic value need to be further evaluated.
Overall, these studies demonstrate the importance of miRNAs in HDL-C regulation and RCT, mainly though the regulation of ABCA1 and ABCG1 in different cells and with particular interest in macrophages and hepatocytes.
3. circRNAs REGULATION OF CHOLESTEROL METABOLISM
Besides miRNAs, other non-coding RNAs with regulatory functions have been described, including circRNAs. circRNAs usually regulate gene expression acting as natural sponges of miRNAs or even proteins. However, circRNAs biological roles are less described than miRNAs, particularly in the field of lipoprotein metabolism [102]. Most functions associated with circRNAs in atherosclerosis have been associated with proliferation and contractility of ECs, VSMCs and macrophages instead of metabolism [8, 103]. One of the most relevant studies in the field recently identified circFASN as mechanism of repression of miR-33 in hepatocytes [104]. Although the study was carried out in patients with dyslipidemia after liver transplantation, it may be of relevance for the field of lipid metabolism and other metabolic disease given the key role of miR-33 in the regulation of metabolic processes.
Some other circRNAs have been suggested as potential regulators of lipoprotein metabolism, but most of them are based on bioinformatics analysis and their relevance in atherosclerotic models still need to be addressed. For example, prediction target studies identified circ_0092317/circ_0003546 sponge miR-326, while circ_0028198/circ_0092317 target miR-543, two miRNAs that are known to inhibit PDE3B, which in turn would result in altered regulation of ABCA1 function [105]. Given the role of miR-122 in hepatic function and its targeting of MTP, this study could be also of relevance in the context of lipoprotein metabolism. Similarly, other described circRNAs have been suggested to be of potential interest based on the role of their targets, but they have not been described neither in macrophages nor in the context of atherosclerosis. In this case, circACC1 emerges among other for their regulatory role of AMPK in hepatocytes, an energy sensor involved in mitochondrial function, LXR signaling and autophagy [105]. The results from this study suggest that circACC1 regulation of AMPK in hepatocytes and macrophages may be important in atherosclerosis. Another novel study described circRNA_002581 sponges miR-122 in hepatocytes, altering autophagy and nonalcoholic steatohepatitis (NASH) progression [106].
Overall, emerging functions regarding circRNAs and metabolism have been recently elucidated. Although most of these studies have focused on metabolic functions of circRNAs in adipogenesis and steatosis, the potential value of these circRNAs in lipoprotein metabolism and atherosclerosis remains to be studied, but they have emerged as a promising field of research [107].
4. CONCLUSIONS
The discovery of miRNAs involved in lipid and lipoprotein metabolism has turned the spotlight on their potential use in the field of atherosclerosis research and cardiometabolic diseases. Despite the great advance in the identification of miRNAs involved in VLDL, LDL and HDL metabolism, the promiscuity of miRNAs derived from their ability to target several mRNAs and pathways in the same or different tissues has evidenced the risk that miRNA-based therapies still face. Thus, there is still an important caveat that requires to be addressed. The most relevant concerns to be addressed regarding miRNA undesired effect are discussed in the review. We also highlighted the most recent findings regarding these concerns, particularly for miR-33, and the potential technical advances that will help to acquire a better understanding for the development of miRNA-based therapies.
Finally, this review also gives insights into the potential role of circRNAs, another class of non-coding RNAs, which still need to be intensely explored in the upcoming years. With the development of databases and predictive resources, we anticipate a fast growth of this field, which will bring a great opportunity for the development of novel therapies.
FUNDING
CF-H and PF-T are supported by grants from National Institutes of Health (R35HL135820 to CF-H), American Heart Association (20TPA35490416 to CF-H) and a Postdoctoral Fellow from The Basque Government to PF-T.
ABBREVIATIONS
- ABCA1
ATP-binding cassette A1
- ABCG1
ATP-binding cassette subfamily G member 1
- ABCB11
ATP-binding cassette, sub-family B, member 11
- ABCB4
ATP-binding cassette, sub-family B, member 4
- ABCG5
ATP-binding cassette sub-family G, members 5
- ABCG8
ATP-binding cassette sub-family G, members 8
- ACC1
acetyl-CoaA carboxylase alpha
- ACC2
acetyl-CoaA carboxylase beta
- ACLY
ATP citrate lyase
- AFs
adventitial fibroblasts
- AGPAT1
1-acylglycerol-3-phosphate O-acyltransferase 1
- AMPK
AMP-activated protein kinase
- ApoAl
component apolipoprotein AI
- ApoE
apolipoprotein E
- ASOs
antisense oligonucleotides
- BM
bone marrow
- CFs
cardiac fibroblasts
- CIDEC
cell death Inducing DFFA like effector C
- ciRNAs
intronic circular RNAs
- CircRNAs
circular RNAs
- CMs
cardiomyocytes
- CPT1a
carnitine palmitoyltransferase 1a
- CVD
cardiovascular disease
- CYP7A1
cholesterol 7α-hydroxylase
- EcircRNAs
exonic circular RNAs
- ECs
endothelial cells
- ElciRNAs
exon-intron circular RNAs
- FASN
fatty acid synthase
- GWAS
genome-wide association study
- HDL-C
high-density lipoprotein cholesterol
- HMGCR
3-hydroxy-3-methylglutaryl-CoA reductase
- IDL
intermediate-density lipoprotein
- IDOL
inducible degrader of the LDLR or E3 ubiquitin ligase-Inducible degrader of the low density
- INSIG1
insulin-induced gene 1
- INSR
insulin receptor
- IRS-1
insulin receptor substrate 1
- KLF4
Krüppel-like factor 4
- LDL-C
low-density lipoprotein cholesterol
- LDLR
low-density lipoprotein receptor
- LncRNAs
long non-coding RNAs
- Lp (a)
lipoprotein (a)
- LPGAT1
lysophosphatidyl glycerol acyltransferase 1
- LXR
liver X receptor
- MiRNAs
microRNAs
- MTP
microsomal triglyceride transfer protein
- NASH
nonalcoholic steatohepatitis
- NcRNAs
non-coding RNAs
- NDUFA5
NADH-ubiquinone oxidoreductase 1 alpha subcomplex subunit 5
- NPC1
Niemann-Pick disease, type C1
- OSBPL6
oxysterol binding protein like 6
- ox-LDL
oxidized low-density lipoprotein
- PAS
Per-Arnt-Sim
- PCSK9
proprotein convertase subtilisin/kexin type 9
- PDE3B
phosphodiesterase 3B
- PDK4
pyruvate dehydrogenase kinase isozyme 4
- PGC1a
peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PMVK
phosphomevalonate kinase
- RCT
reverse cholesterol transport
- SCD1
stearoyl-coenzyme A desaturase 1
- SIK1
salt inducible kinase 1
- SIRT1
sirtuin 1
- SNPs
single nucleotide polymorphisms
- SLC25A25
solute carrier family 25
- SR-B1
scavenger receptor class B type 1
- SREBP1
sterol regulatory element binding protein 1
- SREBP2
sterol regulatory element-binding protein 2
- SREBPc1
sterol regulatory element binding protein 1C
- TAGs
triglycerides
- TC
total cholesterol
- UTR
untranslated region
- VLDL-C
very low-density lipoprotein cholesterol
- VSMCs
vascular smooth muscle cells
Footnotes
Conflict of Interest
CF-H has a patent on the use of miR-33 inhibitors to treat inflammation. The other authors report no conflicts.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
REFERENCES
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Glass CK, Witztum JL: Atherosclerosis. the road ahead. Cell 2001, 104(4):503–516. [DOI] [PubMed] [Google Scholar]
- 2.Olson EN: MicroRNAs as therapeutic targets and biomarkers of cardiovascular disease. Sci Transl Med 2014, 6(239):239ps233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Price NL, Fernandez-Hernando C: Non-coding RNAs in lipid metabolism. Vascul Pharmacol 2019, 114:93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambros V: The functions of animal microRNAs. Nature 2004, 431(7006):350–355. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Hernando C, Suarez Y, Rayner KJ, Moore KJ: MicroRNAs in lipid metabolism. Curr Opin Lipidol 2011, 22(2):86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J: The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019, 20(11):675–691. [DOI] [PubMed] [Google Scholar]
- 7.Cao Q, Guo Z, Du S, Ling H, Song C: Circular RNAs in the pathogenesis of atherosclerosis. Life Sci 2020, 255:117837. [DOI] [PubMed] [Google Scholar]
- 8.Zhang S, Wang W, Wu X, Zhou X: Regulatory Roles of Circular RNAs in Coronary Artery Disease. Mol Ther Nucleic Acids 2020, 21:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goedeke L, Salerno A, Ramirez CM, et al. : Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice. EMBO Mol Med 2014, 6(9):1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goedeke L, Rotllan N, Canfran-Duque A, et al. : MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat Med 2015, 21(11):1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goedeke L, Wagschal A, Fernandez-Hernando C, Naar AM: miRNA regulation of LDL-cholesterol metabolism. Biochim Biophys Acta 2016, 1861(12 Pt B):2047–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aryal B, Singh AK, Rotllan N, Price N, Fernandez-Hernando C: MicroRNAs and lipid metabolism. Curr Opin Lipidol 2017, 28(3):273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rayner KJ, Suarez Y, Davalos A, et al. : MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328(5985):1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagschal A, Najafi-Shoushtari SH, Wang L, et al. : Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat Med 2015, 21(11):1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Sinnott-Armstrong N, Wagschal A, et al. : A MicroRNA Linking Human Positive Selection and Metabolic Disorders. Cell 2020, 183(3):684–701 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong J, He M, Li J, et al. : microRNA-483 ameliorates hypercholesterolemia by inhibiting PCSK9 production. JCI Insight 2020, 5(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salerno AG, van Solingen C, Scotti E, et al. : LDL Receptor Pathway Regulation by miR-224 and miR-520d. Front Cardiovasc Med 2020, 7:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM: MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med 2013, 19(7):892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T: Identification of tissue-specific microRNAs from mouse. Curr Biol 2002, 12(9):735–739. [DOI] [PubMed] [Google Scholar]
- 20.Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A: miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol 2008, 48(4):648–656. [DOI] [PubMed] [Google Scholar]
- 21.Bandiera S, Pfeffer S, Baumert TF, Zeisel MB: miR-122--a key factor and therapeutic target in liver disease. J Hepatol 2015, 62(2):448–457. [DOI] [PubMed] [Google Scholar]
- 22.Ramirez CM, Rotllan N, Vlassov AV, et al. : Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ Res 2013, 112(12):1592–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Xu Y, Zhu Y, et al. : Macrophage miR-34a Is a Key Regulator of Cholesterol Efflux and Atherosclerosis. Mol Ther 2020, 28(1):202–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feingold KR, Grunfeld C: Introduction to Lipids and Lipoproteins. In: Endotext. Edited by Feingold KR, Anawalt B, Boyce A, et al. South Dartmouth (MA); 2000. [Google Scholar]
- 25.Merkel M, Velez-Carrasco W, Hudgins LC, Breslow JL: Compared with saturated fatty acids, dietary monounsaturated fatty acids and carbohydrates increase atherosclerosis and VLDL cholesterol levels in LDL receptor-deficient, but not apolipoprotein E-deficient, mice. Proc Natl Acad Sci U S A 2001, 98(23):13294–13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez-Hernando C, Yu J, Suarez Y, et al. : Genetic evidence supporting a critical role of endothelial caveolin-1 during the progression of atherosclerosis. Cell Metab 2009, 10(1):48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez CM, Zhang X, Bandyopadhyay C, et al. : Caveolin-1 Regulates Atherogenesis by Attenuating Low-Density Lipoprotein Transcytosis and Vascular Inflammation Independently of Endothelial Nitric Oxide Synthase Activation. Circulation 2019, 140(3):225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi C, Zhang M, Tong M, et al. : miR-148a is Associated with Obesity and Modulates Adipocyte Differentiation of Mesenchymal Stem Cells through Wnt Signaling. Sci Rep 2015, 5:9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willer CJ, Schmidt EM, Sengupta S, et al. : Discovery and refinement of loci associated with lipid levels. Nat Genet 2013, 45(11):1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Do R, Willer CJ, Schmidt EM, et al. : Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet 2013, 45(11):1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huan T, Rong J, Liu C, et al. : Genome-wide identification of microRNA expression quantitative trait loci. Nat Commun 2015, 6:6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen J, Ambrosone CB, Zhao H: Novel genetic variants in microRNA genes and familial breast cancer. Int J Cancer 2009, 124(5):1178–1182. [DOI] [PubMed] [Google Scholar]
- 33.Zhang PF, Wang F, Wu J, et al. : LncRNA SNHG3 induces EMT and sorafenib resistance by modulating the miR-128/CD151 pathway in hepatocellular carcinoma. J Cell Physiol 2019, 234(3):2788–2794. [DOI] [PubMed] [Google Scholar]
- 34.Jin J, Guo T, Guo Y, Liu J, Qu F, He Y: Methylationassociated silencing of miR128 promotes the development of esophageal cancer by targeting COX2 in areas with a high incidence of esophageal cancer. Int J Oncol 2019, 54(2):644–654. [DOI] [PubMed] [Google Scholar]
- 35.Lu Q, Meng Q, Qi M, Li F, Liu B: Shear-Sensitive lncRNA AF131217.1 Inhibits Inflammation in HUVECs via Regulation of KLF4. Hypertension 2019, 73(5):e25–e34. [DOI] [PubMed] [Google Scholar]
- 36.He F, Song Z, Chen H, et al. : Long noncoding RNA PVT1-214 promotes proliferation and invasion of colorectal cancer by stabilizing Lin28 and interacting with miR-128. Oncogene 2019, 38(2):164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barad O, Meiri E, Avniel A, et al. : MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res 2004, 14(12):2486–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Motohashi N, Alexander MS, Shimizu-Motohashi Y, Myers JA, Kawahara G, Kunkel LM: Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J Cell Sci 2013, 126(Pt 12):2678–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bjornsson E, Gunnarsdottir K, Halldorsson GH, et al. : Lifelong Reduction in LDL Cholesterol Due to a Gain-of-Function Mutation in LDLR. Circ Genom Precis Med 2020. [DOI] [PubMed] [Google Scholar]
- 40.Bostjancic E, Zidar N, Glavac D: MicroRNAs and cardiac sarcoplasmic reticulum calcium ATPase-2 in human myocardial infarction: expression and bioinformatic analysis. BMC Genomics 2012, 13:552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferland-McCollough D, Fernandez-Twinn DS, Cannell IG, et al. : Programming of adipose tissue miR-483-3p and GDF-3 expression by maternal diet in type 2 diabetes. Cell Death Differ 2012, 19(6):1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W, Zhao LJ, Yang Y, et al. : Retinoic acid induced 16 enhances tumorigenesis and serves as a novel tumor marker for hepatocellular carcinoma. Carcinogenesis 2012, 33(12):2578–2585. [DOI] [PubMed] [Google Scholar]
- 43.Ma N, Wang X, Qiao Y, et al. : Coexpression of an intronic microRNA and its host gene reveals a potential role for miR-483-5p as an IGF2 partner. Mol Cell Endocrinol 2011, 333(1):96–101. [DOI] [PubMed] [Google Scholar]
- 44.Expert Panel on Detection E, Treatment of High Blood Cholesterol in A: Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). JAMA 2001, 285(19):2486–2497. [DOI] [PubMed] [Google Scholar]
- 45.Naeli P, Mirzadeh Azad F, Malakootian M, Seidah NG, Mowla SJ: Post-transcriptional Regulation of PCSK9 by miR-191, miR-222, and miR-224. Front Genet 2017, 8:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jopling C: Liver-specific microRNA-122: Biogenesis and function. RNA Biol 2012, 9(2):137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esau C, Davis S, Murray SF, et al. : miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006, 3(2):87–98. [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi K, Reue K: Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am J Physiol Endocrinol Metab 2009, 296(6):E1195–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vila-Brau A, De Sousa-Coelho AL, Goncalves JF, Haro D, Marrero PF: Fsp27/CIDEC is a CREB target gene induced during early fasting in liver and regulated by FA oxidation rate. J Lipid Res 2013, 54(3):592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Song JW, Lin JY, Miao R, Zhong JC: Roles of MicroRNA-122 in Cardiovascular Fibrosis and Related Diseases. Cardiovasc Toxicol 2020, 20(5):463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez-Micaelo N, Beltran-Debon R, Baiges I, Faiges M, Alegret JM: Specific circulating microRNA signature of bicuspid aortic valve disease. J Transl Med 2017, 15(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang YL, Yu W: Association of circulating microRNA-122 with presence and severity of atherosclerotic lesions. PeerJ 2018, 6:e5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Yang N, Dong B, Yang J, Kou L, Qin Q: MicroRNA-122 promotes endothelial cell apoptosis by targeting XIAP: Therapeutic implication for atherosclerosis. Life Sci 2019, 232:116590. [DOI] [PubMed] [Google Scholar]
- 54.Willeit P, Skroblin P, Moschen AR, et al. : Circulating MicroRNA-122 Is Associated With the Risk of New-Onset Metabolic Syndrome and Type 2 Diabetes. Diabetes 2017, 66(2):347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai WC, Hsu SD, Hsu CS, et al. : MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest 2012, 122(8):2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu SH, Wang B, Kota J, et al. : Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest 2012, 122(8):2871–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tall AR, Yvan-Charvet L, Terasaka N, Pagler T, Wang N: HDL, ABC transporters, and cholesterol efflux: implications for the treatment of atherosclerosis. Cell Metab 2008, 7(5):365–375. [DOI] [PubMed] [Google Scholar]
- 58.Ouimet M, Barrett TJ, Fisher EA: HDL and Reverse Cholesterol Transport. Circ Res 2019, 124(10):1505–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bodzioch M, Orso E, Klucken J, et al. : The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat Genet 1999, 22(4):347–351. [DOI] [PubMed] [Google Scholar]
- 60.Kennedy MA, Barrera GC, Nakamura K, et al. : ABCG1 has a critical role in mediating cholesterol efflux to HDL and preventing cellular lipid accumulation. Cell Metab 2005, 1(2):121–131. [DOI] [PubMed] [Google Scholar]
- 61.Ji Y, Jian B, Wang N, et al. : Scavenger receptor BI promotes high density lipoprotein-mediated cellular cholesterol efflux. J Biol Chem 1997, 272(34):20982–20985. [DOI] [PubMed] [Google Scholar]
- 62.Chiang JY: Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J Hepatol 2004, 40(3):539–551. [DOI] [PubMed] [Google Scholar]
- 63.Li T, Francl JM, Boehme S, Chiang JY: Regulation of cholesterol and bile acid homeostasis by the cholesterol 7alpha-hydroxylase/steroid response element-binding protein 2/microRNA-33a axis in mice. Hepatology 2013, 58(3):1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allen RM, Marquart TJ, Albert CJ, et al. : miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med 2012, 4(9):882–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davalos A, Goedeke L, Smibert P, et al. : miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A 2011, 108(22):9232–9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Najafi-Shoushtari SH, Kristo F, Li Y, et al. : MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010, 328(5985):1566–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.••.Hennessy EJ, van Solingen C, Scacalossi KR, et al. : The long noncoding RNA CHROME regulates cholesterol homeostasis in primate. Nat Metab 2019, 1(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]; The findings from the study uncovers the long non-coding RNA CHROME as a central regulator of a set of microRNAs involved in cholesterol metabolism in hepatocytes and macrophages.
- 68.Marquart TJ, Allen RM, Ory DS, Baldan A: miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A 2010, 107(27):12228–12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horie T, Ono K, Horiguchi M, et al. : MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A 2010, 107(40):17321–17326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rotllan N, Ramirez CM, Aryal B, Esau CC, Fernandez-Hernando C: Therapeutic silencing of microRNA-33 inhibits the progression of atherosclerosis in Ldlr−/− mice--brief report. Arterioscler Thromb Vasc Biol 2013, 33(8):1973–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rayner KJ, Sheedy FJ, Esau CC, et al. : Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest 2011, 121(7):2921–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rottiers V, Obad S, Petri A, et al. : Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci Transl Med 2013, 5(212):212ra162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Horie T, Nishino T, Baba O, et al. : MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nat Commun 2013, 4:2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nishino T, Horie T, Baba O, et al. : SREBF1/MicroRNA-33b Axis Exhibits Potent Effect on Unstable Atherosclerotic Plaque Formation In Vivo. Arterioscler Thromb Vasc Biol 2018, 38(10):2460–2473. [DOI] [PubMed] [Google Scholar]
- 75.•.Price NL, Rotllan N, Zhang X, et al. : Specific Disruption of Abca1 Targeting Largely Mimics the Effects of miR-33 Knockout on Macrophage Cholesterol Efflux and Atherosclerotic Plaque Development. Circ Res 2019, 124(6):874–880. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses innovative techniques to show the specific effect of Abca1 targeting by miR-33 in cholesterol metabolism and atherosclerosis.
- 76.Horie T, Baba O, Kuwabara Y, et al. : MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in ApoE−/− mice. J Am Heart Assoc 2012, 1(6):e003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Price NL, Rotllan N, Canfran-Duque A, et al. : Genetic Dissection of the Impact of miR-33a and miR-33b during the Progression of Atherosclerosis. Cell Rep 2017, 21(5):1317–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.•.Price NL, Zhang X, Fernandez-Tussy P, et al. : Loss of hepatic miR-33 improves metabolic homeostasis and liver function without altering body weight or atherosclerosis. Proc Natl Acad Sci U S A 2021, 118(5). [DOI] [PMC free article] [PubMed] [Google Scholar]; The findings from this work suggests hepatic miR-33 in mice does not influences atherosclerosis and obesity but may be important for metabolic function.
- 79.Horie T, Nishino T, Baba O, et al. : MicroRNA-33b knock-in mice for an intron of sterol regulatory element-binding factor 1 (Srebf1) exhibit reduced HDL-C in vivo. Sci Rep 2014, 4:5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hussain MM, Goldberg IJ: Human MicroRNA-33b Promotes Atherosclerosis in Apoe(−/−) Mice. Arterioscler Thromb Vasc Biol 2018, 38(10):2272–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koyama S, Horie T, Nishino T, et al. : Identification of Differential Roles of MicroRNA-33a and -33b During Atherosclerosis Progression With Genetically Modified Mice. J Am Heart Assoc 2019, 8(13):e012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hayward RA, Reaven PD, Wiitala WL, et al. : Follow-up of glycemic control and cardiovascular outcomes in type 2 diabetes. N Engl J Med 2015, 372(23):2197–2206. [DOI] [PubMed] [Google Scholar]
- 83.Gore MO, McGuire DK, Lingvay I, Rosenstock J: Predicting cardiovascular risk in type 2 diabetes: the heterogeneity challenges. Curr Cardiol Rep 2015, 17(7):607. [DOI] [PubMed] [Google Scholar]
- 84.Karunakaran D, Thrush AB, Nguyen MA, et al. : Macrophage Mitochondrial Energy Status Regulates Cholesterol Efflux and Is Enhanced by Anti-miR33 in Atherosclerosis. Circ Res 2015, 117(3):266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ouimet M, Hennessy EJ, van Solingen C, et al. : miRNA Targeting of Oxysterol-Binding Protein-Like 6 Regulates Cholesterol Trafficking and Efflux. Arterioscler Thromb Vasc Biol 2016, 36(5):942–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ouimet M, Ediriweera HN, Gundra UM, et al. : MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J Clin Invest 2015, 125(12):4334–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nishiga M, Horie T, Kuwabara Y, et al. : MicroRNA-33 Controls Adaptive Fibrotic Response in the Remodeling Heart by Preserving Lipid Raft Cholesterol. Circ Res 2017, 120(5):835–847. [DOI] [PubMed] [Google Scholar]
- 88.Huang K, Bao H, Yan ZQ, et al. : MicroRNA-33 protects against neointimal hyperplasia induced by arterial mechanical stretch in the grafted vein. Cardiovasc Res 2017, 113(5):488–497. [DOI] [PubMed] [Google Scholar]
- 89.Ramirez CM, Davalos A, Goedeke L, et al. : MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler Thromb Vasc Biol 2011, 31(11):2707–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun D, Zhang J, Xie J, Wei W, Chen M, Zhao X: MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS Lett 2012, 586(10):1472–1479. [DOI] [PubMed] [Google Scholar]
- 91.Zhang M, Wu JF, Chen WJ, et al. : MicroRNA-27a/b regulates cellular cholesterol efflux, influx and esterification/hydrolysis in THP-1 macrophages. Atherosclerosis 2014, 234(1):54–64. [DOI] [PubMed] [Google Scholar]
- 92.Meiler S, Baumer Y, Toulmin E, Seng K, Boisvert WA: MicroRNA 302a is a novel modulator of cholesterol homeostasis and atherosclerosis. Arterioscler Thromb Vasc Biol 2015, 35(2):323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lv YC, Tang YY, Peng J, et al. : MicroRNA-19b promotes macrophage cholesterol accumulation and aortic atherosclerosis by targeting ATP-binding cassette transporter A1. Atherosclerosis 2014, 236(1):215–226. [DOI] [PubMed] [Google Scholar]
- 94.Hazen SL, Smith JD: An antiatherosclerotic signaling cascade involving intestinal microbiota, microRNA-10b, and ABCA1/ABCG1-mediated reverse cholesterol transport. Circ Res 2012, 111(8):948–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sala F, Aranda JF, Rotllan N, et al. : MiR-143/145 deficiency attenuates the progression of atherosclerosis in Ldlr−/−mice. Thromb Haemost 2014, 112(4):796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Aguiar Vallim TQ, Tarling EJ, Kim T, et al. : MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ Res 2013, 112(12):1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng J, Cheng A, Clifford BL, et al. : MicroRNA-144 Silencing Protects Against Atherosclerosis in Male, but Not Female Mice. Arterioscler Thromb Vasc Biol 2020, 40(2):412–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P: Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med 2003, 9(2):213–219. [DOI] [PubMed] [Google Scholar]
- 99.Liao X, Sharma N, Kapadia F, et al. : Kruppel-like factor 4 regulates macrophage polarization. J Clin Invest 2011, 121(7):2736–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hu Z, Shen WJ, Kraemer FB, Azhar S: MicroRNAs 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type I in steroidogenic cells. Mol Cell Biol 2012, 32(24):5035–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang L, Jia XJ, Jiang HJ, et al. : MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol Cell Biol 2013, 33(10):1956–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang F, Zhang R, Zhang X, et al. : Comprehensive analysis of circRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of atherosclerosis in rabbits. Aging (Albany NY) 2018, 10(9):2266–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gong X, Wu G, Zeng C: Role of circular RNAs in cardiovascular diseases. Exp Biol Med (Maywood) 2019, 244(2):73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang C, Chen K, Wei R, et al. : The circFASN/miR-33a pathway participates in tacrolimus-induced dysregulation of hepatic triglyceride homeostasis. Signal Transduct Target Ther 2020, 5(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang L, Zheng Z, Feng X, et al. : circRNA/lncRNA-miRNA-mRNA Network in Oxidized, Low-Density, Lipoprotein-Induced Foam Cells. DNA Cell Biol 2019, 38(12):1499–1511. [DOI] [PubMed] [Google Scholar]
- 106.Jin X, Gao J, Zheng R, et al. : Antagonizing circRNA_002581-miR-122-CPEB1 axis alleviates NASH through restoring PTEN-AMPK-mTOR pathway regulated autophagy. Cell Death Dis 2020, 11(2):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen C, Zhang X, Deng Y, et al. : Regulatory roles of circRNAs in adipogenesis and lipid metabolism: emerging insights into lipid-related diseases. FEBS J 2020. [DOI] [PubMed] [Google Scholar]


