Summary
African populations are the most diverse in the world yet are sorely underrepresented in medical genetics research. Here, we examine the structure of African populations using genetic and comprehensive multi-generational ethnolinguistic data from the Neuropsychiatric Genetics of African Populations-Psychosis study (NeuroGAP-Psychosis) consisting of 900 individuals from Ethiopia, Kenya, South Africa, and Uganda. We find that self-reported language classifications meaningfully tag underlying genetic variation that would be missed with consideration of geography alone, highlighting the importance of culture in shaping genetic diversity. Leveraging our uniquely rich multi-generational ethnolinguistic metadata, we track language transmission through the pedigree, observing the disappearance of several languages in our cohort as well as notable shifts in frequency over three generations. We find suggestive evidence for the rate of language transmission in matrilineal groups having been higher than that for patrilineal ones. We highlight both the diversity of variation within Africa as well as how within-Africa variation can be informative for broader variant interpretation; many variants that are rare elsewhere are common in parts of Africa. The work presented here improves the understanding of the spectrum of genetic variation in African populations and highlights the enormous and complex genetic and ethnolinguistic diversity across Africa.
Keywords: diverse populations, genotypes, population genetics, linguistics, population structure, Africa
Graphical abstract
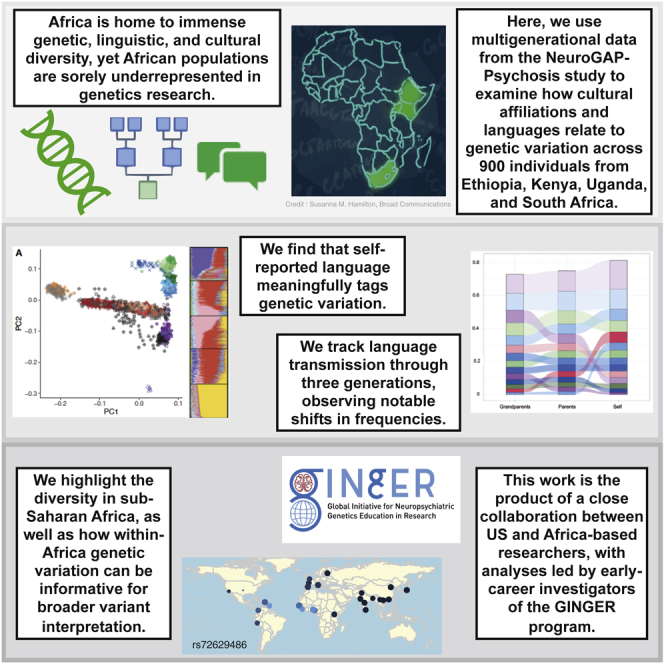
Africa has immense genetic, linguistic, and cultural diversity. Here, we examine how languages relate to genetics across individuals from Ethiopia, Kenya, Uganda, and South Africa. We find self-reported language tags genetic variation, and we observe shifts in language transmission through three generations. African genetic variation is informative for broader variant interpretation.
Introduction
Humans originated in Africa, resulting in more genetic variation on the African continent than anywhere else in the world; the average African genome has nearly a million more genetic variants than the average non-African genome.1 Africa is also immensely culturally and ethno-linguistically diverse; while the rest of the world averages 3.2 to 4.7 ethnic groups per country, African countries have an average of greater than 8 each and account in total for 43% of the world’s ethnic groups.2 Despite this diversity, African ancestry individuals are sorely underrepresented in genomic studies, making up only about 2% of GWAS participants.3,4 Furthermore, the vast majority of African ancestry populations currently represented in genetic studies are African Americans or Afro-Caribbeans (72%–93% in the GWAS catalog and ≥90% in gnomAD) with primarily West African ancestral origins.5 These resources thus currently leave out substantial diversity from regions of Africa that also would be informative for human genetics.
Populations underrepresented in genetic studies contribute disproportionately to our understanding of biomedical phenotypes relative to European ancestry populations. Despite their paltry representation in GWASs, African ancestry populations contribute 7% of genome-wide significant associations.5,6 African population genetic studies are especially informative given their unique evolutionary history, high level of genetic variation, and rapid linkage disequilibrium decay.7 The Eurocentric bias in current genomics studies and resources also makes African descent individuals less likely to benefit from key genomic findings that do not translate fully across populations, contributing to health disparities.8 In this study, we better characterize the immense genetic and ethnolinguistic diversity in four countries in eastern and southern Africa, offering insights into their population history and structure. Data are from 900 genotype samples that are part of the Neuropsychiatric Genetics of African Populations-Psychosis study (NeuroGAP-Psychosis), a major research and capacity building initiative in Ethiopia, Kenya, South Africa, and Uganda.9,10
Genetic variation in Africa has been previously described as following not only isolation-by-distance expectations, but as being influenced by multiple interconnected ecological, historical, environmental, cultural, and linguistic factors.11, 12, 13, 14, 15, 16 These factors capture variation that differs from that tagged by genetics and can be informative for understanding population substructure. Better characterization of the ethnolinguistic composition of these samples is a key initial step towards running well-calibrated statistical genomics analyses including association studies. If ethnolinguistic variation tags additional structure than that captured by geography, explicit incorporation of relevant cultural information into such analyses may be the optimal analytic strategy. In addition to the covariation of culture and genetics,17, 18, 19 individuals’ cultural environments influence how phenotypes are expressed and whether assortative pairing impacts the distribution of traits.20, 21, 22 We measure how ethnolinguistic culture has changed in parallel to and independently of genetics, which provides a foundation for the study of phenotypes of medical interest. In this study, we explore the genetics of the NeuroGAP-Psychosis dataset, which comprises five collection sites across four countries in Africa, and how individuals’ cultural affiliations and languages are related to genetic variation. We also explore ongoing linguistic changes and consider the impact they will have on genetics.
Material and methods
Collection strategy
Proper informed consent was obtained for this study. Ethical clearances to conduct this study have been obtained from all participating sites, including:
-
•
Ethiopia: Addis Ababa University College of Health Sciences (#014/17/Psy) and the Ministry of Science and Technology National Research Ethics Review Committee (#3.10/14/2018)
-
•
Kenya: Moi University School of Medicine Institutional Research and Ethics Committee (#IREC/2016/145, approval number: IREC 1727), Kenya National Council of Science and Technology (#NACOSTI/P/17/56302/19576), KEMRI Centre Scientific Committee (CSC# KEMRI/CGMRC/CSC/070/2016), KEMRI Scientific and Ethics Review Unit (SERU# KEMRI/SERU/CGMR-C/070/3575)
-
•
South Africa: The University of Cape Town Human Research Ethics Committee (#466/2016)
-
•
Uganda: The Makerere University School of Medicine Research and Ethics Committee (SOMREC #REC REF 2016-057) and the Uganda National Council for Science and Technology (UNCST #HS14ES)
-
•
USA: The Harvard T.H. Chan School of Public Health (#IRB17-0822)
As described in more detail in the published protocol,9 NeuroGAP-Psychosis was designed as a case-control study recruiting participants from more than two dozen hospitals and medical clinics in Ethiopia, Kenya, South Africa, and Uganda. Participants were recruited in languages in which they are fluent, including Acholi, Afrikaans, Amharic, English, Kiswahili, Luganda, Lugbara, Oromiffa/Oromigna, Runyankole, and isiXhosa. After consenting to be in the study, participants gave a saliva sample using an Oragene kit (OG-500.005) for DNA extraction. Study staff then asked a range of questions on demographics, mental health, and physical health and took participants' blood pressure, heart rate, height, and weight. The whole study visit lasted approximately 60–90 min. Table S1 contains details about the dataset and country of origin of all populations included in analyses in this manuscript.
Ethnolinguistic phenotypes
Multiple phenotypes related to self-reported ethnolinguistic categorizations have been collected as part of the recruitment process. This includes multi-generational data including each participants’ birth country as well as primary, secondary, and tertiary language and ethnicity. All linguistic data were collected from participants both for themselves as well as for each of their parents and grandparents, giving an unusually rich depth of information. The specific phrasing of questions collected are as follows:
Primary language (lang_self_1): “What primary language do you speak?”
2nd language (lang_self_2): “What 2nd language do you speak?”
3rd language (lang_self_3): “What 3rd language do you speak?”
Primary, 2nd, and 3rd ethnicity (ethnicity_1): “What is your ethnicity or tribe?”
Reports for other relatives followed similar phrasing. The primary language question for each is listed, with “primary” replaced by “2nd” or “3rd” for the second and third reported languages for that family member.
Mother (lang_mat_1): “What was the primary language that your biological mother spoke?”
Father (lang_pat_1): “What was the primary language that your biological father spoke?”
Maternal grandmother (lang_mgm_1): “What primary language did your biological mother’s mother speak?”
Maternal grandfather (lang_mgf_1): “What primary language did your biological mother’s father speak?”
Paternal grandmother (lang_pgm_1): “What primary language did your biological father’s mother speak?
Paternal grandfather (lang_pgf_1): “What primary language did your biological father’s father speak?
Table S2 contains raw data for language transmission counts for all languages reported in the NeuroGAP-Psychosis dataset. Table S3 indicates matrilineal or patrilineal classification of all self-reported ethnicities in the dataset.
Genetic data quality control
Quality control (QC) procedures for NeuroGAP-Psychosis data were done using the Hail python library (see web resources). All of the data was stored on Google Cloud. The QC steps and filters used were adapted from Ricopili23 and Anderson et al.24 The data were genotyped using the Illumina Global Screening Array. For each of the five NeuroGAP-Psychosis sites, a VCF file with genotyping data was stored on Google Cloud. Before QC, each VCF contained 192 samples and 687,537 variants. When looking at the data pre-QC, we discovered elevated deviations in Hardy Weinberg equilibrium. We found that autocall call rate, Illumina’s custom genotype calling algorithm, explained these deviations. The QC filtering steps thus took place after removing individuals with an autocall call rate less than 0.95. Of the original 960 individuals, 937 remained. These 960 individuals were used for the linguistic transmission analyses presented here (with some missing data for specific familial categories), while for genetic analyses further QC on variants was conducted.
The site-specific VCF files were imported as Hail matrix tables and annotated with appropriate data from the metadata file before being merged. The resulting matrix table had 937 samples and 687,537 variants. Prior to QC, the joint dataset was split into autosomes, PAR, and nonPAR regions of the X chromosome. QC filtering was conducted separately for the autosome and X chromosome regions. Pre-QC, the autosomal dataset had 937 samples and 669,346 variants. The following is a list of the QC steps and parameters used for autosomal QC. (1) Removing variants with a call rate less than 95%. After filtering, 638,235 variants remained. (2) Removing individuals with a call rate less than 98%. After filtering, 930 individuals remained. (3) Removing individuals whose reported sex did not match their genotypic sex. After filtering, 923 individuals remained. (4) Removing variants with a minor allele frequency less than 0.5%. After filtering, 360,321 variants remained. This large drop in variants was expected as the GSA array poorly tags common variation in samples with African ancestry.25 (5) Removing variants with a Hardy Weinberg equilibrium p value less than 1 × 10−3. After filtering, 331,667 variants remained. (6) Using PC-Relate with 10 PCs, removing individuals with a kinship coefficient greater than 0.125. After filtering, 900 individuals remained. After autosomal QC, 900 individuals and 331,667 variants remained.
The PAR and nonPAR regions of the X chromosome were subset to the 900 samples which passed autosomal QC before going through variant QC. The same variant thresholds used for autosomal QC were used to conduct QC on the PAR region. Pre-QC, the PAR region had 900 samples and 518 variants. (1) After SNP call rate filtering, 515 variants remained. (2) After MAF filtering, 411 variants remained. (3) After HWE filtering, 402 variants remain. Post-QC, the PAR region had 900 samples and 402 variants. For the nonPAR region, the dataset was split by sex. The female nonPAR dataset had 441 samples and 17,673 variants. Variant QC was carried out on the females using the following metrics. (1) Removing variants with a call rate less than 98%. After filtering, 16,261 variants remained. (2) Removing variants with a minor allele frequency less than 1%. After filtering, 11,113 variants remained. (3) Removing variants with a Hardy Weinberg equilibrium p value less than 1 × 10−6. After filtering, 11,104 variants remained. After nonPAR QC on the females, the male nonPAR dataset was merged with the female QC’d nonPAR dataset. The final nonPAR dataset had 900 samples and 11,104 variants. After QC, the autosomal, PAR, and nonPAR datasets were merged into one matrix table. The final merged post-QC dataset contained 900 samples and 343,173 variants. The counts of variants/individuals per site after autosomal and X QC can be found in Tables S4 and S5.
After QC, the dataset was merged with three reference panel datasets: the 1000 Genomes Project (1kGP), Human Diversity Project (HGP), and the African Genome Variation Project (AGVP).26, 27, 28 Before merging with HGDP and 1kGP, the AGVP and NeuroGAP-Psychosis datasets were merged and lifted over from GRCh37 to GRCh38. Before this initial merge, AGVP had 1,297 samples and 1,778,578 variants while NeuroGAP-Psychosis had 900 samples and 343,173 variants. Prior to merging these two datasets, multi-allelic variants were removed from the NeuroGAP-Psychosis dataset resulting in 343,165 variants. The two datasets were then combined using plink –bmerge. The resulting dataset had 2,197 samples and 1,908,204 variants after intersection. A 5% genotyping rate filter using the --geno plink command was then run on the dataset which gave the final merged dataset counts prior to liftover of 2,197 samples and 206,240 variants. The liftover of the merged AGVP, NeuroGAP-Psychosis dataset was conducted using Hail. After liftover, there were 2,197 samples and 206,156 variants. Next, the AGVP NeuroGAP-Psychosis dataset was merged with a subset of the gnomAD v.3.1 release (see web resources) which consisted of newly sequenced HGDP and 1kGP datasets. This dataset of HGDP and 1kGP had 4,097 samples and 155,648,020 variants prior to merging. After combining AGVP plus NeuroGAP-Psychosis with HGDP plus 1kGP using plink --bmerge, the resulting dataset contained 6,294 samples and 149,518 variants. A 5% --geno filter was then run on the dataset which resulted in the final counts of 6,294 samples and 148,488 variants for the NeuroGAP-Psychosis and reference panels dataset.
Population structure and admixture analyses
Cohort data from the five NeuroGAP-Psychosis plates were merged with African reference populations from the 1000 Genomes Project26,27, Human Genome Diversity Panel,29 and the African Genome Variation Project.28 These populations provide reasonably comprehensive geographic coverage across sub-Saharan African from currently available reference panels and contain populations which are co-located in the same countries as all NeuroGAP-Psychosis samples. We intend for the term “sub-Saharan Africa” to be interpreted exclusively in the geographical sense to be more specific about the geographic scope of the variation we consider in this article. We also note that there are additional datasets of African genetic variation beyond the reference panels incorporated into these analyses, including H3Africa, the Uganda Genome Resource, and many cohorts from specific collection sites.30, 31, 32 PCA was run using flashPCA.33 Detailed examination of admixture was conducted using the program ADMIXTURE34 with 5-fold cross validation error to inform the correct number of clusters. We used the unsupervised mode of ADMIXTURE ten times for each value of k to capture any different modes present in the data. All runs treated data as diploid. Plots from ADMIXTURE output were generated with pong.35 ADMIXTURE was run using a tailored representation of global genetic data consisting of all continental African populations, the CHB population from China to capture East Asian admixture, the GBR from Britain to capture European admixture, and the GIH from India to capture South Asian ancestry. Regions were assigned as according to the UN Statistics division geoscheme (see web resources).
FST estimates across populations were generated using smartPCA.36 FST heatmaps were generated in R using the package corrplot (see web resources). The relationship between ancestry composition on the autosomes vs X chromosome was examined using Wilcoxon rank tests and Mantel tests in R with the package ade4.37
Relationship between genetics and language
To measure linguistic variation, we made use of the PHOIBLE 2.0 phonemic database (see web resources), which contains phoneme inventories and phoneme qualities for languages around the world. For every individual, we identified all languages spoken—excluding English—which were present in the PHOIBLE database (84.5% of languages spoken by the individuals themselves, and 81.1% of languages spoken by their relatives). Using the phoneme inventories (including both primary phonemes and their allophones) from PHOIBLE, we found the mean phoneme presence for each individual's or each relative's spoken languages (if one of two spoken languages contained the sound /g/, the /g/ value for that individual would be 0.5). The resulting matrices (of individuals or their relatives, and mean phoneme presences) were transformed using PCA conducted in R to create three sets of principal components (PCs): from personally spoken languages, from those spoken by matrilineal relatives (mother and maternal grandmother), and from those of patrilineal relatives (father and patrilineal grandfather).
To observe the broader linguistic changes taking place, all languages were assigned the highest-level classifications available in Glottolog 4.2.1 (see web resources). These classifications were modified to minimize the number of high-level classifications while maintaining an element of geographic origin. Several classifications were consolidated into Nilo-Saharan (made up of Nilotic, Central Sudanic, Kuliak, and Gamuz classifications) and Khoisan (Khoe-Kwadi, Kxa, and Tuu), and Afro-Asiatic was expanded (with Ta-Ne-Omotic and Dizoid). Indo-European was split to account for the recent history of its speakers: Afrikaans and Oorlams were placed into a unique category, the languages of Europe into another, and those of the Indian subcontinent (Hindi and Urdu) into a third. We excluded languages that were unclassified or identified as speech registers.
Every individual was associated with a survey location, meaning the geographic coordinates where the sample was collected, and we used the spoken languages to assign a different, linguistic location. To do this, using all languages an individual spoke, and these languages' locations from Glottolog, we calculated the mean location of each individual's languages.
To compare linguistic, genetic, and geographic variation, we used a set of Procrustes analyses implemented in R.38 For linguistic and genetic variation, the first three PCs of variation were used. Since Procrustes minimizes the sum of squared euclidean distances, the geographic coordinates of each individual were converted to points on a sphere. To measure the correlation between geographic variation and linguistic or genetic variation, the linguistic and genetic PCs were transformed (via rotation and scaling) to minimize the sum of squared distance between individuals' geographic locations and the transformed genetic or linguistic PCs. The first three PCs of Procrustes-transformed linguistic and genetic variation—representing their similarity to geographic variation—were then plotted onto a map.
We additionally calculated the transmission frequency of languages from sets of family members. Given the discrepancy in number of languages in the matri-vs patri-groupings, patrilineal languages were additionally subsampled to the same overall number as matrilineal and rates were recalculated.
Anthropological variables
To identify relevant anthropological data, we accessed data from the Ethnographic Atlas (EA)39 using D-Place.40 We associated each ethnicity reported in the NeuroGAP-Psychosis survey data to a society in the EA (if possible), and used variable EA076: Inheritance rule for movable property. For ethnicities with data, individuals whose ethnicities were associated with consistent inheritance rules or marital residence patterns were assigned that rule or pattern. Of the 907 NeuroGAP-Psychosis individuals, 779 were assigned an inheritance rule (matrilineal or patrilineal). Additionally, 751 individuals could be assigned a post-marital residence rule (patrilocal, neolocal, or virilocal-like) using EA012: Marital residence with kin, but matrilocality and other forms of residence were not found among the sampled ethnicities, and we did not use these for our analyses. Similarly, other variables such as EA074: Inheritance rule for real property (land) did not vary for available individuals (all available ethnicities traditionally practiced either patrilineal, male-biased, or neutral patterns of real property inheritance). Only a single ethnicity corresponding to eight individuals could be assigned a matrilineal inheritance pattern based on EA043:Descent: major type.
Results
Genetic population structure and admixture
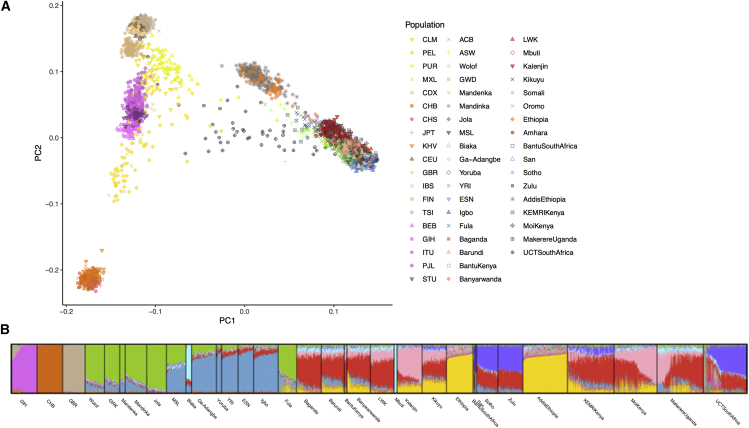
We compared the ancestral composition of our samples relative to global reference data from the 1000 Genomes Project, Human Genome Diversity Panel, and the African Genome Variation Project (AGVP) to see the full breadth of genetic diversity.26,28,29 Most NeuroGAP-Psychosis samples appear genetically similar to their geographically closest reference samples when compared to global datasets (Figures 1 and S1). However, large amounts of admixture is visible within some individuals, particularly among South African individuals (supplemental methods). In South Africa, some individuals cluster wholly within the European reference cluster; this is expected based on the demographic composition of Cape Town, where these samples were collected, which is home to a substantial fraction of people of Dutch ancestry (Afrikaners) and individuals of mixed ancestry.12,15,29,41,42
Figure 1.
Genetic and admixture composition of the NeuroGAP-Psychosis samples against a global reference
(A) First two principal components showing NeuroGAP-Psychosis samples as projected onto global variation of the full 1000 Genomes, HGDP, and AGVP. While most samples fall on a cline of African genetic variation, some South African samples exhibit high amounts of admixture and European genetic ancestry. Color scheme for global PCA plot: Latin American, yellow; East Asian, dark orange; European, tan; South Asian, fuschia; West African, green/blue; East African, red/orange; South African, purple; NeuroGAP-Psychosis collections, gray.
(B) ADMIXTURE plot at best fit k (k = 10) of all African samples as well as three representative non-African populations from the 1000 Genomes Project. The GIH, CHB, and GBR were included to capture South Asian, East Asian, and European admixture, respectively. Individuals are represented as bar charts sorted by population, and ancestry components for each person are visualized with different colors. A key describing the country of origin for all populations can be found in Table S1.
We additionally investigated the degree of admixture within samples and how genetic groups cluster in the data. We ran ADMIXTURE,34 which partitions genetic variation into a given number of distinct genetic clusters. This helps to visualize the groups that are most genetically distinct from one another, as each additional component can be thought of as representing the next most differentiated ancestry component in the data, akin to principal components analysis (PCA). We identified the best fit k value, using 5-fold cross validation, to be 10 using a tailored global reference.
Examining the ancestry composition at the best fit k, we identify several ancestry components unique to areas within Africa (Figures 1B and S1). Notably, several such components, including those unique to Ethiopia (yellow), West Africa (blue), and South Africa (purple), appear at earlier values of k than that separating South Asians from East Asians and Europeans (fuschia from tan and orange). While sample sizes affect the ordering of components identified in ADMIXTURE analyses, this suggests a high level of genetic differentiation between areas of the African continent rivaling that between those out-of-Africa continental ancestries, as has been previously demonstrated.28,32 We also note that Ethiopian participants have evidence of Eurasian admixture, possibly related to historical back-migration into the African continent.30,41,43,44
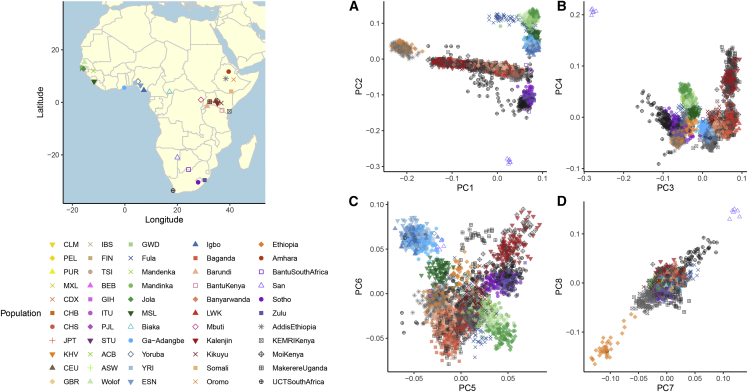
Projecting our samples onto PC space generated from only African reference samples, the top two principal components (PCs) separate geography, and more specifically east-west and north-south patterns of variation within Africa (Figure 2), mirroring isolation by geographic distance in human genetic data. At higher PCs, however, there is fine-scale structure in the data separating different geographically proximal groups within the East African individuals, shown in red. We thus focus our deeper examinations into the East African samples to assess potential drivers of this differentiation (Figures S2–S6). Clear structure is visible in the data to PC8, with higher PCs resolving substructure within geographic regions. For example, two clusters are evident among PCs within participants enrolled in the study from Moi University in Kenya, who tend to speak distinct languages in the Afro-Asiatic and Niger-Congo families (Figure S4). For a detailed discussion of genetic variation within each country, see the supplemental methods. The percent variance explained for PCA plots including the global reference panel as well as the African tailored reference panel can be found in Figure S7.
Figure 2.
Genetic composition of subcontinental African structure in the NeuroGAP-Psychosis samples
PCA plots for PCs 1–8 with an African reference panel. A map of collection locations is shown to the left of PCA plots. Points are colored by region to assist in interpretation: green, west; blue, west central/central; red, east; orange, Ethiopia; purple, south. See Figures S2–S6 for plots highlighting each cohort individually.
Self-reported population composition
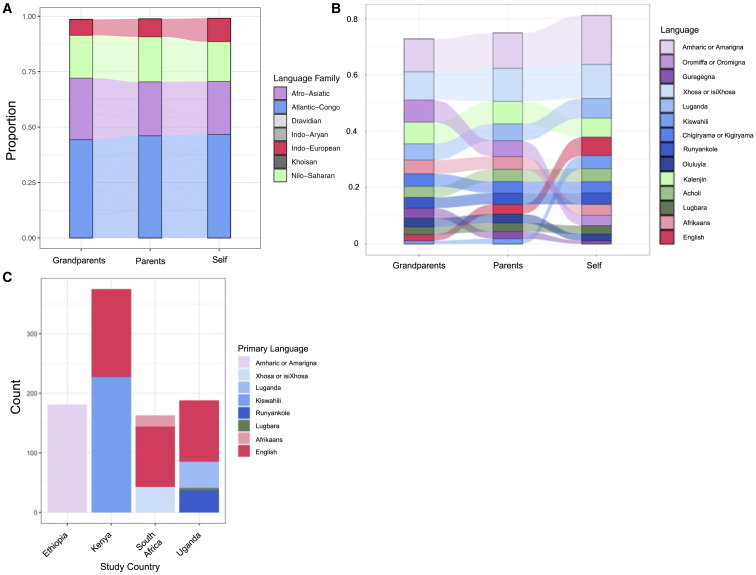
Across samples with self-reported ethnolinguistic information, we observe 62 primary ethnicities and 107 primary languages in the 960 NeuroGAP-Psychosis samples. We also find that languages have shifted in frequency over time, with English increasing in reporting frequency in the current generation, and several grandparental languages disappearing in our dataset (Figures 3 and S8).
Figure 3.
Primary self-reported language shifts over three generations
(A) Individual languages were re-classified into broader language families for comparable granularity. Note that while all languages in the legend are represented in the plot, not all are visible due to being at low frequency in the data.
(B) All languages reported with at least 3% frequency in any generation are shown across the generations. Note the increase in endorsement of English and drop in Oromiffa/Oromigna in the present generation.
(C) Primary language reported by the individuals within each NeuroGAP-Psychosis study country.
Genetic variation partitions with language
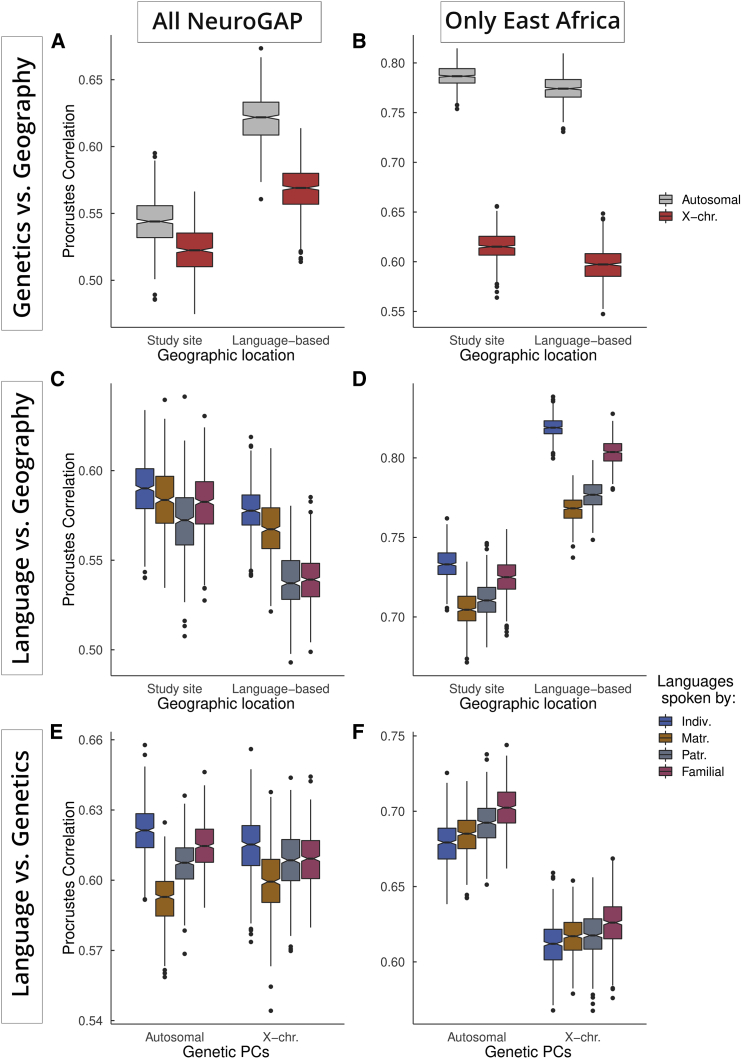
To assess the correlation between the primary self-reported language that an individual reports to be their primary and the genetic partitioning that we observed, we conducted Procrustes analyses to measure the correlation between genetic, linguistic, and geographic variation. Procrustes analysis rotates and scales one set of coordinates to minimize the total distance between it and a second set, providing both a metric of similarity between these sets of coordinates and a visualization of their overlap. We use this to compare each individual’s first three PCs of genetic variation to their geographic locations. We do not have access to the locations where participants live or were born, so we use either the locations at which individuals were sampled (study sites) or the centroids of non-English languages that they reported speaking (language based). By including a database of phonemes (units of sound) found in the self-reported languages of individuals and their families and comparing these to the first three PCs of autosomal and X chromosome variation, we found consistent correlations between genetic, linguistic, and geographic variation throughout Africa (Figure 4 and Table 1). We also plotted the genetic PCs, superimposed onto geography via Procrustes, to visualize the geographic distribution of genetic variation (Figure S9). Because the autosomes and X chromosomes have considerably different numbers of single nucleotide polymorphisms (SNPs), we additionally compared X chromosome variation to chromosome 7, which has a similar length to the X chromosome, and chromosome 22, which is most similar in SNP count to chromosome X (variant counts with/without reference panel intersection: X = 603/1,348, chr22 = 705/1,455; Figure S9). To measure linguistic variation, we queried the PHOIBLE 2.0 phonemic database (see web resources), which contains phoneme inventories and phoneme qualities for many languages around the world. The resulting matrices of mean phoneme presences were used in a PCA to create four sets of linguistic PCs: a score from languages spoken by the participant, a combined score from those spoken by matrilineal relatives (the participant’s mother and maternal grandmother), a combined score from those of patrilineal relatives (father and paternal grandfather), and a composite score that includes all relatives weighted according to relatedness to the individual (1 for languages spoken by each individual, 0.5 for those of parents, and 0.25 for grandparents). Here, “matrilineal” and “patrilineal” refer to the traits associated with direct lines of descent following exclusively mothers and fathers, respectively. We use the languages spoken by these relatives to understand whether sex-biased language transmission may have taken place and whether it parallels sex-biased gene flow. By comparing linguistic variation associated with these relatives to genetic data and spatial positions, we can explore whether norms and traditions have shaped linguistic and genetic variation.
Figure 4.
Procrustes correlations between genetics, geography, and language
Procrustes correlations (all p < 5E−5) are shown between geography and genetics (A and B), geography and language (C and D), and genetics and language (E and F). The left column includes results for the entire NeuroGAP-Psychosis collection. The right column contains results subset to the four cohorts in East Africa. For linguistic analyses, linguistic variation is measured by the first three PCs of phoneme inventories from languages reported by individuals as spoken by themselves and their relatives. Matrilineal relatives include the mother and maternal grandmother. Patrilineal relatives include the father and paternal grandfather. Familial refers to a weighted average of all reported family members. Note that Y-axis labels vary between plots.
Table 1.
Procrustes correlation between genetics, geography, and language
| Genetic | Geography | Self | Maternal languages | Paternal languages | |
|---|---|---|---|---|---|
| All individuals | autosomal | 0.5426 | 0.6223 | 0.5935 | 0.6078 |
| All individuals | X chrom. | 0.5231 | 0.6078 | 0.5988 | 0.6082 |
| East African cohorts | autosomal | 0.7868 | 0.6815 | 0.6856 | 0.6924 |
| East African cohorts | X chrom. | 0.6170 | 0.6103 | 0.6178 | 0.6200 |
All p < 5E−5. The first three PCs of autosomal and X chromosome variation were used for comparisons. Linguistic variation was calculated as a function of mean phoneme presence across all languages reported by the individual across their pedigree. Maternal language contains results from the languages spoken by the participants’ mother and maternal grandmother; paternal contains results from their father and paternal grandfather.
The first three PCs of both autosomal and X chromosome variation are less correlated to geography (ρ = 0.543 and 0.523; p < 5E−5) than are the first three PCs of linguistic variation (ρ = 0.589; p < 5E−5). Both autosomal and X chromosomal variation are similarly correlated to linguistic variation when considering all individuals together. Looking within East African cohorts alone, X chromosome variation is less correlated to geography (Table 1, Figure 4). Similarly, linguistic variation associated with individuals’ matrilineal relatives is less correlated to geography and to genetics than that of patrilineal relatives (Figures 4D and 4F).
Language transmission through families
Using detailed multi-generational ethnolinguistic information (see Ethnolinguistic phenotypes in Material and methods), we computed overall transmission rates of language families over three generations. We initially examined the raw self-reported information of the participant with respect to the primary, second, and third languages spoken. We assessed the frequency with which the primary language reported by the participant matched that of each of their older relatives (i.e., maternal and paternal grandparents, mother, and father) as well as the frequency with which the participants’ primary reported language matched any (primary, secondary, or tertiary) of the languages reported for their relatives (Table 2). We find that transmission rates are similar between family members of the same generation when looking at primary language matching any language, regardless of whether including or excluding English.
Table 2.
Language transmission rates from relatives
| Family member | Overall | Patrilineal | Patrilineal (downsampled) | Matrilineal |
|---|---|---|---|---|
| Father | 0.810 | 0.837 | 0.901 | 0.871 |
| Mother | 0.802 | 0.811 | 0.837 | 0.800 |
| Paternal grandfathers | 0.778 | 0.726 | 0.775 | 0.926 |
| Paternal grandmothers | 0.773 | 0.738 | 0.779 | 0.939 |
| Maternal grandfathers | 0.762 | 0.708 | 0.736 | 0.903 |
| Maternal grandmothers | 0.758 | 0.726 | 0.750 | 0.812 |
Frequency of a participant’s reported primary language matching one of the top three reported languages spoken by relatives. Rates were calculated excluding English. Given that all but one of the NeuroGAP-Psychosis populations with linguistic data were collected in East Africa, we conducted an additional suite of analyses zooming into this region to examine transmission in this part of the continent. In East Africa, individuals were thus additionally partitioned by their affiliation with ethnic groups with either a matrilineal or patrilineal transmission of movable property. Patrilineal languages were run in their entirety as well as downsampled to 105 to match the sample size available for matrilineal languages.
To take a closer look at language transmission across the pedigree, we calculated the rate of transmission between various relatives in our family tree. We note that we have information only for the four countries present in the NeuroGAP-Psychosis dataset, so our results do not capture the full breadth of ethnolinguistic diversity across the continent. In these calculations, we ran tests excluding English to get a better sense of the transmission of languages that have been present in the continent for a longer period of time. We additionally identified whether individuals came from ethnic groups in which the transmission of movable property was historically matrilineal or patrilineal according to the Ethnographic Atlas (EA)39 and recalculated the transmission rates within those two classifications. Here, we were interested in measuring whether the inheritance of property through the male (patrilineal) or female (matrilineal) lines parallels the transmission of languages. Partitioning east African individuals by the presence of matrilineal vs patrilineneal transmission of movable property in their traditional societies (from Murdock's Ethnographic Atlas, code EA07639,40), we see a significantly higher transmission rate from individuals assigned to a matrilineal classification (p = 0.028). As our sample size for matrilineal groups is quite small (n = 105 and 674 for matrilineal and patrilineal transmission, respectively), we subsampled the patrilineal groupings to the same size as matrilineal and re-ran our analyses. Considered altogether, the trend disappears (p = 0.097). However, when looking at familial relationships at just the parental and grandparental levels, we do detect a significantly higher language transmission rate for individuals assigned to matrilineal groups (p = 0.012).
The results from our various tests of language transmission and those from Procrustes analysis support the anthropological data that classified the peoples in the regions of Africa that we studied as patrilocal,39,45 but how language was transmitted is uncertain. The lower geography-X chromosome correlation in East African cohorts suggests that the cultural norms (a predominance of patrilocality) have had effects on genetic variation. We cannot conclusively determine whether language transmission here was historically sex biased, as recent changes to cultures (including movement to urban areas, colonial histories, access to markets, etc.), has affected the associations between groups of people and languages. There is no evidence of recent sex-biased language transmission in this region from our linguistic data alone. However, the decreased association between X-chromosome variation and language (Figure 4B) suggests that east Africa has a history of patrilineal language transmission, which parallels the region’s historically predominant patrilocal social structure.39
Interestingly, we also find that 12 languages reported for earlier generations were not spoken by the participants, indicating that they have disappeared from our dataset. Khoekhoe, Somali, and Urdu disappeared in the parental generation, and Amba, Afar, Argobba, Gumuz, Harar, Hindi, Soddo, Soo, and Tamil were no longer reported languages in the participants’ generation. We caution, however, that many of these languages were observed at very low rates overall.
Testing for evidence of sex-biased demography
To examine whether there was evidence for sex-biased gene flow in our samples, we assessed the similarity of ancestry proportions on the X chromosome versus autosomes. Ancestry fractions were highly correlated across these genomic regions, indicating no evidence for sex-biased demography at this scale, although care should be taken in interpretation given the difference in effective sample size for the X chromosome as compared to the autosomes (Figures S11 and S12). Wilcoxon signed rank tests comparing the fractions of ancestry on X versus autosomes from ADMIXTURE at k = 4 did not find a significant difference in the means, nor for PC1 vs PC2 (p = 0.3754). Similarly, the Mantel tests indicated an observed correlation of 0.987 (simulated p value = 0.001) for the X chromosome compared to the autosomal FST values. We additionally ran Procrustes analyses comparing genetic and linguistic variation on the X chromosome as compared to the autosomes in their entirety (chromosome 7 was most similar in length to chrX and chromosome 22 was most similar in SNP count to chrX) (Figure S10). Similarly, the Procrustes tests showed significant correlation between the first three PCs of X and autosomal variation (ρ = 0.850 for sub-Saharan Africa and ρ = 0.753 for East African cohorts alone). Compared to chromosomes 7 and 22, results were similar (ρ = 0.826 and 0.780 for sub-Saharan Africa, and ρ = 0.728 and 0.691 for East African cohorts).
Reference panels miss meaningful allele frequency resolution within Africa
We visualized allele frequencies for functionally important variants across our five collection sites as compared to reference data from the 1000 Genomes Project. One example variant, rs2071348 (GenBank: NC_000011.9; g.5264146T>A), key in beta-thalassemia, dramatically varies in frequency depending on the precise location in Africa considered. The NeuroGAP-Psychosis allele frequencies observed across the five sites were 12.26% in Ethiopia, 11.35% in Kenya (KEMRI), 9.71% in Kenya (Moi), 13.11% in Uganda, and 5.7% in South Africa. As this variant has direct consequences on human health, consideration of the difference in frequency across the continent is meaningful. For another example, rs72629486 (GenBank: NC_000001.10; g.2938924T>G [p.Leu225Trp], Figure S13), a missense coding single nucleotide variant in the gene ACTRT2, ranges in minor allele frequency (MAF) in NeuroGAP-Psychosis from 5% in Ethiopia down to 1.3% in Uganda (other frequencies were 2.6% in Kenya [KEMRI], 2.6% in Kenya [Moi], and 2.3% in South Africa). This is nearly the full range of the frequency distribution for all global populations in the gnomAD database,46 which lists the variant in the AFR as 5.5%, missing finer subcontinental resolution. rs72629486 is predicted to be deleterious and probably damaging by SIFT and PolyPhen, respectively, and has a combined annotation dependent depletion score of 22.9, highlighting that this variant is likely to be highly functionally important.47, 48, 49
Discussion
Africa is a highly diverse continent, home to immense genetic, linguistic, and cultural diversity. This ethnolinguistic variation is extremely complex and is meaningful to disentangle prior to statistical genetics analyses. Here, we measured the correlation between genetic, linguistic, and geographic variation focusing on four African countries for which we have collected data as part of the NeuroGAP-Psychosis study. We find that genetic and linguistic variation are closely correlated to each other as well as to geography. This is consistent with previous work examining global patterns of diversity as well as the expansion of Bantu speaking people, one of the largest demographic events in African history.11,12,16,50, 51, 52, 53, 54, 55 We find that across the regions of Africa that we surveyed, language is closely correlated to both genetics and geography, a phenomenon that has been noted in Europe and Ethiopia previously.44,56, 57, 58 The patterns of linguistic and genetic variation in this dataset suggest a history of patrilocal residence, in accord with previous studies of the region.45,59,60 We find that individuals collected from the same geographic location show significant genetic differentiation by language family, particularly in east Africa, where there is especially immense linguistic diversity. Previous studies have examined this covariation between culture and genetics produced by a history of migrations and population expansions,44,61, 62, 63 and we explore how this affects genetic datasets by examining both genetics and ongoing changes to culture. These data should be synthesized to influence how population substructure is controlled for in genetic tests, and we suggest that anthropological data should be incorporated into a nuanced treatment of genetic clusters if these affiliations explain associations with the phenotype that are not captured by genetics. For example, future work exploring the direct incorporation of ethnolinguistic affiliations into linear mixed models would be useful, e.g., in the context of including it as a random effect covariance matrix to better control for stratification.64
As there is such immense genetic variation across Africa,28,31,32,29,65 we highlight cases where such variability may be particularly informative. Africa is not simply one monolithic location, as it is sometimes treated in genomics resources that include primarily or exclusively individuals of the African diaspora as representatives of genetic variation for the entire African continent.46,66 Rather, there is an extraordinary amount of genetic variability within it. These example loci highlight both the diversity of variation within the African continent, as well as the fact that within-Africa variation can be informative for broader variant interpretation; many variants appearing rare elsewhere are common in parts of Africa.
As part of the NeuroGAP-Psychosis study’s recruitment process, multi-generational self-reported ethnolinguistic data were collected from participants, including individual ethnicity and at least primary, second, and third language from participants for themselves, as well as for each of their parents and grandparents. This provides us with an unusually rich depth of multigenerational demographic information from participants, a unique strength of our dataset that affords us the opportunity to investigate language transmission through the pedigree and shifts in language frequencies over time. First, we examined the overall change in self-reported language frequencies over three generations. Perhaps most striking is the increase in the reporting frequency of English by participants as their primary language as compared to their reports for older generations of their family. Twelve languages reported for earlier generations were no longer reported as spoken by the participants, suggesting their loss from this cohort. Interestingly, these languages represent a mix of both historically spoken and imported languages for the countries that enrolled participants in the NeuroGAP-Psychosis study. These results emphasize that the analysis of phenotypes should consider not only how they relate to genetics, but how phenotypes may be affected by a rapidly changing cultural environment. While these results are intriguing, we stress that our participants are not necessarily representative of the local populations from which they come and do not begin to cover the full breadth of variation across Africa. A further consideration is a potential upwards bias towards reporting of English and Amharic as a primary language due to a preference toward reporting the language of consent as primary, as well as toward languages taught in local educational systems. This additionally highlights the importance of careful consideration of items on self-report forms to ensure accurate and representative phenotype collection.
In summary, better understanding the composition of samples is a key first step to calibrating subsequent statistical genetics analyses. Cultural factors such as language can dramatically impact the structure of cohort data; we find that self-reported language classifications meaningfully track underlying genetic variation that varies independently from geography. The work presented here improves the understanding of the immense spectrum of genetic and ethnolinguistic variation found across multiple African populations and sheds light on the shifts in reported language over the past three generations in five collection sites.
Consortia
The members of the NeuroGAP-Psychosis Study Team are Tamrat Abebe, Dickens Akena, Melkam Alemayehu, Amantia A. Ametaj, Fred K. Ashaba, Elizabeth G. Atkinson, Lukoye Atwoli, Mark Baker, Sinéad B. Chapman, Lori B. Chibnik, Shareefa Dalvie, Abebaw Fekadu, Katelyn Flowers, Bizu Gelaye, Stella Gichuru, Wilfred E. Injera, Roxanne James, Allan Kalungi, Symon M. Kariuki, Gabriel Kigen, Hannah H Kim, Sam Kinyanjui, Nastassja Koen, Karestan C. Koenen, Zan Koenig, Kristina J. Korte, Edith Kwobah, Joseph Kyebuzibwa, Lerato Majara, Alicia R. Martin, Supriya Misra, Henry Musinguzi, Rehema M. Mwema, Benjamin M. Neale, Carter P. Newman, Charles R. J. C. Newton, Fredrick Ochieng, Ana Maria Olivares, Sam Pollock, Kristianna Post, Adele Pretorius, Raj Ramesar, Welelta Shiferaw, Ilina Singh, Dan J. Stein, Anne Stevenson, Rocky E. Stroud II, Solomon Teferra, and Zukiswa Zingela.
Acknowledgments
We thank all participants for their willingness to contribute their data to this effort. John Mugane and Ana Maria Olivares provided advice and assistance related to this project. This work was supported by funding from the NIH/National Institute of Mental Health (K01MH121659 and T32MH017119 to E.G.A.; K99/R00MH117229 to A.R.M.; L.B.C., K.C.K., B.G., D.J.S., S.T., and D.A. are supported in part by R01MH120642).
Author contributions
Conceptualization, E.G.A., A.R.M., K.C.K.; Formal Analysis, E.G.A., S.D., Y.P., A.K., L.M.; Data Curation, M.B., Z.K., C.P.N., A.S., R.E.S.; Investigation: S.G., J.K., R.J., R.M.M., M.A., W.E.I., G.K., W.S., F.K.A., H.M., L.M.; Resources, T.A., D.A., M.A., F.K.A., L.A., A.F., S.G., W.E.I., R.J., S.M.K., G.K., E.K., J.K., H.M., R.M.M., C.R.J.N., R.R., W.S., D.J.S., S.T., Z.Z.; Writing - Original Draft, E.G.A., A.R.M.; Writing - Review & Editing, all authors; Visualization, E.G.A., S.D., Y.P., A.K., L.M., Z.K.; Supervision, N.C., M.J.D., B.M.N., K.C.K., S.R., A.S., T.A., D.A., L.A., A.F., W.E.I., S.M.K., G.K., E.K., C.P.N., C.R.J.N., R.R., D.J.S., R.E.S., S.T., Z.Z., L.B.C., B.G.; Project Administration, A.S., R.E.S., M.A., S.G., R.J., J.K., L.O., R.M.M., C.P.N.; Funding Acquisition, M.J.D., B.M.N., K.C.K.
Declaration of interests
A.R.M. has consulted for 23andMe and Illumina and received speaker fees from Genentech, Pfizer, and Illumina. B.M.N. is a member of the Deep Genomics Scientific Advisory Board. He also serves as a consultant for the Camp4 Therapeutics Corporation, Takeda Pharmaceutical, and Biogen. M.J.D. is a founder of Maze Therapeutics. The remaining authors declare no competing interests.
Published: September 1, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.07.013.
Web resources
corrplot, https://cran.r-project.org/web/packages/corrplot/index.html
Glottolog, https://glottolog.org/
Hail 0.2, https://hail.is/
NeuroGAP-Psychosis analytic code, https://github.com/atgu/NeuroGAP
Phoible, https://phoible.org/
United Nations Statistics Division Methodology, https://unstats.un.org/unsd/methodology/m49
Supplemental information
Data and code availability
The genetic data generated during this study for NeuroGAP-Psychosis samples are available on dbGAP. The accession number for the genotype array data reported in this paper is dbGAP: phs2528.v1. Code used to process and analyze data is freely available on Github at: https://github.com/atgu/NeuroGAP.
References
- 1.The 1000 Genomes Project Consortium An integrated map of genetic variation from 1, 092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fearon J.D. Ethnic and Cultural Diversity by Country. J. Econ. Growth. 2003;8:195–222. [Google Scholar]
- 3.Sirugo G., Williams S.M., Tishkoff S.A. The missing diversity in human genetic studies. Cell. 2019;177:1080. doi: 10.1016/j.cell.2019.04.032. [DOI] [PubMed] [Google Scholar]
- 4.Popejoy A.B., Fullerton S.M. Genomics is failing on diversity. Nature. 2016;538:161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin A.R., Teferra S., Möller M., Hoal E.G., Daly M.J. The critical needs and challenges for genetic architecture studies in Africa. Curr. Opin. Genet. Dev. 2018;53:113–120. doi: 10.1016/j.gde.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morales J., Welter D., Bowler E.H., Cerezo M., Harris L.W., McMahon A.C., Hall P., Junkins H.A., Milano A., Hastings E., et al. A standardized framework for representation of ancestry data in genomics studies, with application to the NHGRI-EBI GWAS Catalog. Genome Biol. 2018;19:21. doi: 10.1186/s13059-018-1396-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tishkoff S.A., Verrelli B.C. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu. Rev. Genomics Hum. Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- 8.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevenson A., Akena D., Stroud R.E., Atwoli L., Campbell M.M., Chibnik L.B., Kwobah E., Kariuki S.M., Martin A.R., de Menil V., et al. Neuropsychiatric Genetics of African Populations-Psychosis (NeuroGAP-Psychosis): a case-control study protocol and GWAS in Ethiopia, Kenya, South Africa and Uganda. BMJ Open. 2019;9:e025469. doi: 10.1136/bmjopen-2018-025469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Merwe C., Mwesiga E.K., McGregor N.W., Ejigu A., Tilahun A.W., Kalungi A., Akimana B., Dubale B.W., Omari F., Mmochi J., et al. Advancing neuropsychiatric genetics training and collaboration in Africa. Lancet. Glob. Health. 2018;6:e246–e247. doi: 10.1016/S2214-109X(18)30042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker J.L., Rotimi C.N., Shriner D. Human ancestry correlates with language and reveals that race is not an objective genomic classifier. Sci. Rep. 2017;7:1572. doi: 10.1038/s41598-017-01837-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uren C., Kim M., Martin A.R., Bobo D., Gignoux C.R., van Helden P.D., Möller M., Hoal E.G., Henn B.M. Fine-scale human population structure in Southern Africa reflects ecogeographic boundaries. Genetics. 2016;204:303–314. doi: 10.1534/genetics.116.187369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henn B.M., Botigué L.R., Gravel S., Wang W., Brisbin A., Byrnes J.K., Fadhlaoui-Zid K., Zalloua P.A., Moreno-Estrada A., Bertranpetit J., et al. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 2012;8:e1002397. doi: 10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henn B.M., Botigué L.R., Peischl S., Dupanloup I., Lipatov M., Maples B.K., Martin A.R., Musharoff S., Cann H., Snyder M.P., et al. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc. Natl. Acad. Sci. USA. 2016;113:E440–E449. doi: 10.1073/pnas.1510805112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sikora M., Laayouni H., Calafell F., Comas D., Bertranpetit J. A genomic analysis identifies a novel component in the genetic structure of sub-Saharan African populations. Eur. J. Hum. Genet. 2011;19:84–88. doi: 10.1038/ejhg.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Creanza N., Ruhlen M., Pemberton T.J., Rosenberg N.A., Feldman M.W., Ramachandran S. A comparison of worldwide phonemic and genetic variation in human populations. Proc. Natl. Acad. Sci. USA. 2015;112:1265–1272. doi: 10.1073/pnas.1424033112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Filippo C., Barbieri C., Whitten M., Mpoloka S.W., Gunnarsdóttir E.D., Bostoen K., Nyambe T., Beyer K., Schreiber H., de Knijff P., et al. Y-chromosomal variation in sub-Saharan Africa: insights into the history of Niger-Congo groups. Mol. Biol. Evol. 2011;28:1255–1269. doi: 10.1093/molbev/msq312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karafet T.M., Bulayeva K.B., Nichols J., Bulayev O.A., Gurgenova F., Omarova J., Yepiskoposyan L., Savina O.V., Rodrigue B.H., Hammer M.F. Coevolution of genes and languages and high levels of population structure among the highland populations of Daghestan. J. Hum. Genet. 2016;61:181–191. doi: 10.1038/jhg.2015.132. [DOI] [PubMed] [Google Scholar]
- 19.Barbieri C., Butthof A., Bostoen K., Pakendorf B. Genetic perspectives on the origin of clicks in Bantu languages from southwestern Zambia. Eur. J. Hum. Genet. 2013;21:430–436. doi: 10.1038/ejhg.2012.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coelho M., Sequeira F., Luiselli D., Beleza S., Rocha J. On the edge of Bantu expansions: mtDNA, Y chromosome and lactase persistence genetic variation in southwestern Angola. BMC Evol. Biol. 2009;9:80. doi: 10.1186/1471-2148-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchiyama R., Spicer R., Muthukrishna M. Cultural evolution of genetic heritability. Behav. Brain Sci. 2021:1–147. doi: 10.1017/S0140525X21000893. [DOI] [PubMed] [Google Scholar]
- 22.Creanza N., Kolodny O., Feldman M.W. Cultural evolutionary theory: How culture evolves and why it matters. Proc. Natl. Acad. Sci. USA. 2017;114:7782–7789. doi: 10.1073/pnas.1620732114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam M., Awasthi S., Watson H.J., Goldstein J., Panagiotaropoulou G., Trubetskoy V., Karlsson R., Frei O., Fan C.-C., De Witte W., et al. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics. 2020;36:930–933. doi: 10.1093/bioinformatics/btz633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson C.A., Pettersson F.H., Clarke G.M., Cardon L.R., Morris A.P., Zondervan K.T. Data quality control in genetic case-control association studies. Nat. Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin A.R., Atkinson E.G., Chapman S.B., Stevenson A., Stroud R.E., Abebe T., Akena D., Alemayehu M., Ashaba F.K., Atwoli L., et al. Low-coverage sequencing cost-effectively detects known and novel variation in underrepresented populations. Am. J. Hum. Genet. 2021;108:656–668. doi: 10.1016/j.ajhg.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auton A., Salcedo T. In: Assessing Rare Variation in Complex Traits. Zeggini E., Morris A., editors. Springer; 2015. The 1000 Genomes Project; pp. 71–85. [Google Scholar]
- 27.Byrska-Bishop M., Evani U.S., Zhao X., Basile A.O., Abel H.J., Regier A.A., Corvelo A., Clarke W.E., Musunuri R., Nagulapalli K., et al. High Coverage Whole Genome Sequencing of the Expanded 1000 Genomes Project Cohort Including 602 Trios. bioRxiv. 2021 doi: 10.1101/2021.02.06.430068. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gurdasani D., Carstensen T., Tekola-Ayele F., Pagani L., Tachmazidou I., Hatzikotoulas K., Karthikeyan S., Iles L., Pollard M.O., Choudhury A., et al. The African genome variation project shapes medical genetics in Africa. Nature. 2015;517:327–332. doi: 10.1038/nature13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergström A., McCarthy S.A., Hui R., Almarri M.A., Ayub Q., Danecek P., Chen Y., Felkel S., Hallast P., Kamm J., et al. Insights into human genetic variation and population history from 929 diverse genomes. Science. 2020;367:eaay5012. doi: 10.1126/science.aay5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pagani L., Schiffels S., Gurdasani D., Danecek P., Scally A., Chen Y., Xue Y., Haber M., Ekong R., Oljira T., et al. Tracing the route of modern humans out of Africa by using 225 human genome sequences from Ethiopians and Egyptians. Am. J. Hum. Genet. 2015;96:986–991. doi: 10.1016/j.ajhg.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurdasani D., Carstensen T., Fatumo S., Chen G., Franklin C.S., Prado-Martinez J., Bouman H., Abascal F., Haber M., Tachmazidou I., et al. Uganda genome resource enables insights into population history and genomic discovery in Africa. Cell. 2019;179:984–1002.e36. doi: 10.1016/j.cell.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choudhury A., Aron S., Botigué L.R., Sengupta D., Botha G., Bensellak T., Wells G., Kumuthini J., Shriner D., Fakim Y.J., et al. High-depth African genomes inform human migration and health. Nature. 2020;586:741–748. doi: 10.1038/s41586-020-2859-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abraham G., Qiu Y., Inouye M. FlashPCA2: principal component analysis of Biobank-scale genotype datasets. Bioinformatics. 2017;33:2776–2778. doi: 10.1093/bioinformatics/btx299. [DOI] [PubMed] [Google Scholar]
- 34.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behr A.A., Liu K.Z., Liu-Fang G., Nakka P., Ramachandran S. pong: fast analysis and visualization of latent clusters in population genetic data. Bioinformatics. 2016;32:2817–2823. doi: 10.1093/bioinformatics/btw327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 38.Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927–930. [Google Scholar]
- 39.Murdock G.P., Textor R., Barry H., III, White D.R., Gray J.P., Divale W. Ethnographic atlas. World Cultures. 1999;10:24–136. [Google Scholar]
- 40.Kirby K.R., Gray R.D., Greenhill S.J., Jordan F.M., Gomes-Ng S., Bibiko H.-J., Blasi D.E., Botero C.A., Bowern C., Ember C.R., et al. D-PLACE: A global database of cultural, linguistic and environmental diversity. PLoS One. 2016;11:e0158391. doi: 10.1371/journal.pone.0158391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pickrell J.K., Patterson N., Loh P.-R., Lipson M., Berger B., Stoneking M., Pakendorf B., Reich D. Ancient west Eurasian ancestry in southern and eastern Africa. Proc. Natl. Acad. Sci. USA. 2014;111:2632–2637. doi: 10.1073/pnas.1313787111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chimusa E.R., Meintjies A., Tchanga M., Mulder N., Seoighe C., Seioghe C., Soodyall H., Ramesar R. A genomic portrait of haplotype diversity and signatures of selection in indigenous southern African populations. PLoS Genet. 2015;11:e1005052. doi: 10.1371/journal.pgen.1005052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.López S., Tarekegn A., Band G., van Dorp L., Bird N. The genetic landscape of Ethiopia: diversity, intermixing and the association with culture. bioRxiv. 2021 doi: 10.1101/756536. Preprint at. [DOI] [Google Scholar]
- 44.Pagani L., Kivisild T., Tarekegn A., Ekong R., Plaster C., Gallego Romero I., Ayub Q., Mehdi S.Q., Thomas M.G., Luiselli D., et al. Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am. J. Hum. Genet. 2012;91:83–96. doi: 10.1016/j.ajhg.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vansina J. Kinshasa: Université Lovanium; 1966. Introduction à l’ethnographie du Congo. [Google Scholar]
- 46.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141, 456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;Chapter 7:Unit7.20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Filippo C., Bostoen K., Stoneking M., Pakendorf B. Bringing together linguistic and genetic evidence to test the Bantu expansion. Proc. Biol. Sci. 2012;279:3256–3263. doi: 10.1098/rspb.2012.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beleza S., Gusmão L., Amorim A., Carracedo A., Salas A. The genetic legacy of western Bantu migrations. Hum. Genet. 2005;117:366–375. doi: 10.1007/s00439-005-1290-3. [DOI] [PubMed] [Google Scholar]
- 52.Li S., Schlebusch C., Jakobsson M. Genetic variation reveals large-scale population expansion and migration during the expansion of Bantu-speaking peoples. Proc. Biol. Sci. 2014;281:20141448. doi: 10.1098/rspb.2014.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patin E., Lopez M., Grollemund R., Verdu P., Harmant C., Quach H., Laval G., Perry G.H., Barreiro L.B., Froment A., et al. Dispersals and genetic adaptation of Bantu-speaking populations in Africa and North America. Science. 2017;356:543–546. doi: 10.1126/science.aal1988. [DOI] [PubMed] [Google Scholar]
- 54.Semo A., Gayà-Vidal M., Fortes-Lima C., Alard B., Oliveira S., Almeida J., Prista A., Damasceno A., Fehn A.-M., Schlebusch C., Rocha J. Along the indian ocean coast: genomic variation in mozambique provides new insights into the bantu expansion. Mol. Biol. Evol. 2020;37:406–416. doi: 10.1093/molbev/msz224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seidensticker D., Hubau W., Verschuren D., Fortes-Lima C., de Maret P., Schlebusch C.M., Bostoen K. Population collapse in Congo rainforest from 400 CE urges reassessment of the Bantu Expansion. Sci. Adv. 2021;7:eabd8352. doi: 10.1126/sciadv.abd8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Longobardi G., Ghirotto S., Guardiano C., Tassi F., Benazzo A., Ceolin A., Barbujani G. Across language families: Genome diversity mirrors linguistic variation within Europe: Genome Diversity Across Language Families. Am. J. Phys. Anthropol. 2015;157:630–640. doi: 10.1002/ajpa.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piazza A., Rendine S., Minch E., Menozzi P., Mountain J., Cavalli-Sforza L.L. Genetics and the origin of European languages. Proc. Natl. Acad. Sci. USA. 1995;92:5836–5840. doi: 10.1073/pnas.92.13.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.López S., Tarekegn A., Band G., van Dorp L., Bird N., Morris S., Oljira T., Mekonnen E., Bekele E., Blench R., et al. Evidence of the interplay of genetics and culture in Ethiopia. Nat. Commun. 2021;12:3581. doi: 10.1038/s41467-021-23712-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wood E.T., Stover D.A., Ehret C., Destro-Bisol G., Spedini G., McLeod H., Louie L., Bamshad M., Strassmann B.I., Soodyall H., Hammer M.F. Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes. Eur. J. Hum. Genet. 2005;13:867–876. doi: 10.1038/sj.ejhg.5201408. [DOI] [PubMed] [Google Scholar]
- 60.Destro-Bisol G., Donati F., Coia V., Boschi I., Verginelli F., Caglià A., Tofanelli S., Spedini G., Capelli C. Variation of female and male lineages in sub-Saharan populations: the importance of sociocultural factors. Mol. Biol. Evol. 2004;21:1673–1682. doi: 10.1093/molbev/msh186. [DOI] [PubMed] [Google Scholar]
- 61.Tishkoff S.A., Gonder M.K., Henn B.M., Mortensen H., Knight A., Gignoux C., Fernandopulle N., Lema G., Nyambo T.B., Ramakrishnan U., et al. History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol. Biol. Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- 62.Hollfelder N., Schlebusch C.M., Günther T., Babiker H., Hassan H.Y., Jakobsson M. Northeast African genomic variation shaped by the continuity of indigenous groups and Eurasian migrations. PLoS Genet. 2017;13:e1006976. doi: 10.1371/journal.pgen.1006976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gomes V., Pala M., Salas A., Álvarez-Iglesias V., Amorim A., Gómez-Carballa A., Carracedo Á., Clarke D.J., Hill C., Mormina M., et al. Mosaic maternal ancestry in the Great Lakes region of East Africa. Hum. Genet. 2015;134:1013–1027. doi: 10.1007/s00439-015-1583-0. [DOI] [PubMed] [Google Scholar]
- 64.Heckerman D., Gurdasani D., Kadie C., Pomilla C., Carstensen T., Martin H., Ekoru K., Nsubuga R.N., Ssenyomo G., Kamali A., et al. Linear mixed model for heritability estimation that explicitly addresses environmental variation. Proc. Natl. Acad. Sci. USA. 2016;113:7377–7382. doi: 10.1073/pnas.1510497113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.1000 Genomes Project Consortium. Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. Sequencing of 53, 831 diverse genomes from the NHLBI TOPMed program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genetic data generated during this study for NeuroGAP-Psychosis samples are available on dbGAP. The accession number for the genotype array data reported in this paper is dbGAP: phs2528.v1. Code used to process and analyze data is freely available on Github at: https://github.com/atgu/NeuroGAP.