Summary
Understanding the genetic basis of human diseases and traits is dependent on the identification and accurate genotyping of genetic variants. Deep whole-genome sequencing (WGS), the gold standard technology for SNP and indel identification and genotyping, remains very expensive for most large studies. Here, we quantify the extent to which array genotyping followed by genotype imputation can approximate WGS in studies of individuals of African, Hispanic/Latino, and European ancestry in the US and of Finnish ancestry in Finland (a population isolate). For each study, we performed genotype imputation by using the genetic variants present on the Illumina Core, OmniExpress, MEGA, and Omni 2.5M arrays with the 1000G, HRC, and TOPMed imputation reference panels. Using the Omni 2.5M array and the TOPMed panel, ≥90% of bi-allelic single-nucleotide variants (SNVs) are well imputed (r2 > 0.8) down to minor-allele frequencies (MAFs) of 0.14% in African, 0.11% in Hispanic/Latino, 0.35% in European, and 0.85% in Finnish ancestries. There was little difference in TOPMed-based imputation quality among the arrays with >700k variants. Individual-level imputation quality varied widely between and within the three US studies. Imputation quality also varied across genomic regions, producing regions where even common (MAF > 5%) variants were consistently not well imputed across ancestries. The extent to which array genotyping and imputation can approximate WGS therefore depends on reference panel, genotype array, sample ancestry, and genomic location. Imputation quality by variant or genomic region can be queried with our new tool, RsqBrowser, now deployed on the Michigan Imputation Server.
Keywords: genotype imputation, whole-genome sequencing, genotyping array
This manuscript presents—across multiple ancestries, genotype arrays, and imputation reference panels—the minor-allele frequency thresholds above which array genotyping and imputation accurately capture genetic variants genotyped with deep whole-genome sequencing.
Introduction
Short-read deep whole-genome sequencing (WGS) accurately captures most single-nucleotide variants (SNVs) and short indels across the genome and minor-allele frequency (MAF) spectrum.1 Advances in sequencing technologies, and corresponding decreases in sequencing cost, have enabled ever larger human sequencing studies.2, 3, 4, 5 Such studies have identified rare alleles that cause Mendelian diseases6, 7, 8 and contribute to risk of common diseases9 and variation in quantitative traits.2,4 However, deep WGS remains prohibitively expensive and computationally intensive for large studies.1,10
In contrast to WGS, genotype arrays assay hundreds of thousands to millions of variants, representing only a small fraction of genetic variation but at a much lower cost. Variants that are not array genotyped can be statistically inferred by comparing sample haplotypes to an external reference panel of sequenced haplotypes via genotype imputation.11 Most common (MAF > 5%) variants are present in recent reference panels and can be imputed with high accuracy from genotype arrays.5,12,13 However, low-frequency (0.5% < MAF ≤ 5%) and rare (MAF ≤ 0.5%) variants appear less often or may be absent from the reference panel, making their imputation less accurate or impossible.14 Therefore, using inexpensive genotype arrays and imputation in place of costly deep WGS can result in lower coverage and less accurate genotyping of rare genetic variation.
Reference panel, genotype array, sample ancestry, and genomic location all influence imputation quality.12, 13, 14, 15 Previous studies have evaluated imputation quality with the multiethnic 1000 Genomes Phase 3 (1000G),12 the predominantly European Haplotype Reference Consortium (HRC),13 and two releases of the multiethnic Trans-Omics for Precision Medicine (TOPMed)5,16 panels, finding that larger, more deeply sequenced panels support more accurate imputation. Closer ancestry matching between the sample and reference panel haplotypes also improves imputation quality, particularly for rare variants, as demonstrated by the use of population-specific reference panels in isolated17, 18, 19 and non-isolated European populations.2,20,21 Likewise, denser genotype arrays are associated with higher imputation quality,13,14,22 although the effect of array size on imputation quality has not been studied with the TOPMed panels. Regional variability in imputation quality with the 1000G panel is associated with genomic features including repeats and GC content,15 but the degree to which imputation quality varies across the genome with the larger HRC or TOPMed panels is unknown. It is also unknown to what extent individual-level imputation quality varies within populations for any reference panel.
Here, we determine the extent to which genotyping with the Illumina Core, OmniExpress, MEGA, and Omni 2.5M arrays followed by imputation with the 1000G, HRC, and TOPMed reference panels can approximate deep WGS in studies with individuals of African, Hispanic/Latino, non-Finnish European, and Finnish ancestries. Depending on the MAF of variants relevant to the research question, study ancestry, and genomic location, we found that array genotyping and imputation can approximate WGS. Our findings, together with our new RsqBrowser tool for querying imputation quality, should help guide investigator decisions between these two technologies.
Material and methods
Genetic data resources
WGS data and processing
We used WGS data from the BioMe,23 InPSYght, METSIM,24,25 and MLOF26 studies. Detailed descriptions of sample collection, sequencing, and data processing for BioMe and MLOF are provided by the TOPMed Informatics Research Center.5 Corresponding information is available for the METSIM study.25 The InPSYght study is a deep whole-genome sequencing US-based case-control study of individuals of admixed African-European or African genetic ancestry. Cases have either bipolar disorder or schizophrenia. The study is composed of samples from the Genomic Psychiatry Cohort (GPC),27,28 Consortium on the Genetics of Schizophrenia (COGS),29 Bipolar Genome Study (BIGS),30 Lithium Treatment Moderate Dose Use Study (LiTMUS),31 and Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD) studies,32 all obtained from the NIMH repository. Whole-genome sequencing of the samples (mean depth 27 + −5.5 X) was performed at the Broad Institute. We excluded individuals with sex mismatches (n = 20), non-XX or XY sex karyotypes (n = 17), duplicates (n = 366), >5% DNA contamination (n = 4), an excess of singletons (n = 39), or <25% global African ancestry as determined by ADMIXTURE33 analysis of array genotype data or for whom <98% of sites were at a sequencing depth of ≥10 (n = 14). We complied with TOPMed data use agreements for the BioMe and MLOF studies. The InPSYght study was approved by the institutional review boards at each site. The METSIM study was approved by the ethics committee at the University of Eastern Finland and the institutional review board at the University of Michigan. All InPSYght and METSIM participants provided informed consent.
Participants from the BioMe biobank self-reported as Hispanic/Latino and were recruited at the Mount Sinai Health System in New York City (n = 4,677; Table 1). Participants in the MLOF study self-reported as non-Hispanic White and were recruited throughout the US (n = 2,987). Participants in the METSIM study were recruited in Kuopio, Finland (n = 3,045). On the basis of recruiting location and self-reported and genetic ancestry, we designated the population groups Hispanic/Latino (BioMe), African (InPSYght), Finnish (METSIM), and European (MLOF) ancestry for the purposes of this study.
Table 1.
Whole-genome sequencing (WGS) datasets
| Study | Ancestry | Mean depth |
Sample size |
Number of variants in 2,429 samples used in analyses |
|||||
|---|---|---|---|---|---|---|---|---|---|
| total | unrelated |
bi-allelic |
multi-allelic |
total | |||||
| SNV | indel | SNV | indel | ||||||
| InPSYght22, 23, 24, 25, 26, 27 | African | 27 | 7,717 | 7,169 | 72.6M | 1.3M | 5.3M | 0.2M | 79.3M |
| BioMe18 | Hispanic/Latino | 37 | 4,677 | 3,141 | 63.2M | 0.9M | 4.5M | 0.1M | 68.8M |
| MLOF21 | European | 39 | 2,987 | 2,429 | 57.3M | 0.8M | 4.1M | 0.1M | 62.2M |
| METSIM19,20 | Finnish | 24 | 3,045 | 2,703 | 20.5M | 0.1M | 1.4M | 10K | 22.0M |
The study name, ancestry, mean sequencing depth, sample size (total and unrelated subset), and number of variants, including single-nucleotide variants (SNVs) and indels, for the four WGS datasets.
In all studies, we removed participants inferred by KING34 to be related at a second degree or closer relationship to any other individual genotyped in TOPMed Freeze 9 (n = 157,675), including all participants in these four studies and all individuals in the TOPMed imputation reference panel. This filtering yielded 3,141 participants in BioMe, 7,169 in InPSYght, 2,703 in METSIM, and 2,429 in MLOF. We then randomly downsampled to 2,429 individuals in each study (Figure S1).
WGS variant calling for all four studies was performed jointly with TOPMed Freeze 9 by the TOPMed Informatics Research Center (IRC) with the TOPMed Variant Calling/GotCloud pipeline.5,35 We analyzed bi-allelic SNVs, multi-allelic SNVs, bi-allelic indels, and multi-allelic indels separately with n-allele variants recoded and analyzed as n − 1 bi-allelic variants at the same position.
Array genotyping in METSIM
METSIM participants were genotyped with the Illumina Human OmniExpress array. Variants with poor mapping of probes to GRCh37, call rate < 95%, or deviations from Hardy-Weinberg equilibrium (p < 10−6) were removed.36
Genotype imputation
For each study, we subsetted WGS variants to those present on the Illumina Infinium Core (0.3M markers), Illumina Omni Express (0.7M), Infinium Omni 2.5M (2.4M), and Multi-Ethnic Genotyping (MEGA; 1.8M) arrays (Table S1). We refer to these WGS variant subsets as WGS-based arrays. For each study, we phased the selected variants with Eagle 2.4.1 and imputed genotypes by using Minimac4 on the Michigan Imputation Server (pipeline version 1.2.4)37 with the (1) 1000 Genomes Phase 3 (n = 2,504), (2) Haplotype Reference Consortium (n = 32,470), and (3) modified TOPMed (n = 88,804) reference panels. MLOF and BioMe are included in the full, publicly available TOPMed R2 (n = 97,256) panel. To avoid overlap of participants and the presence of close relatives in the reference panel, we removed 4,694 BioMe (4,668 Hispanic/Latino and 26 missing ethnicity) and 3,758 MLOF (2,977 non-Hispanic White and 781 missing race/ethnicity) individuals from the full TOPMed R2 panel to create our modified TOPMed panel.
Evaluation of imputation quality
Observed imputation r2
For each variant, we calculated the observed imputation r2 as the squared Pearson correlation coefficient between the imputed genotype dosages and the sequence-based genotypes. We assigned r2 = 0 for any variant present in the sequenced individuals but absent from the reference panels and so not imputed. We also assigned r2 = 0 for any variant with undefined correlation due to being monomorphic in the imputed data. For each variant category (bi-allelic SNVs, bi-allelic indels, multi-allelic SNVs, and multi-allelic indels) and each WGS-based array, we calculated the proportion of variants that were well-imputed (observed imputation r2 > 0.8) within study-specific MAF bins of size 0.00025 for MAF between 0.0002 and 0.002 and of size 0.001 for MAF > 0.002. We also calculated the mean r2 for each MAF bin. Each minor allele for multi-allelic variants was analyzed independently of the other minor alleles of the same variant so that multi-allelic variants had the same number of r2 measurements as minor alleles.
Genotype concordance
Separately for common, low-frequency, and rare bi-allelic SNVs, we calculated the heterozygous concordance rate between the imputed best-guess genotypes and sequenced-based genotypes as the proportion of heterozygous variants in WGS that were present in the reference panel that were also heterozygous in the imputed data by using bed-diff.38 We excluded bi-allelic SNVs that were absent from the reference panels in these calculations.
Predicted variant consequences
In each study, we used VEP39 to predict the functional consequences of bi-allelic SNVs. We partitioned variants into four classes based on the predicted impact on protein coding: high, moderate, low, and modifier. While variants in the high and moderate classes are likely to change protein behavior, variants in the low impact class are unlikely to do so. Modifier variants are mostly non-coding with no evidence of impacting protein coding.
Fine-scale ancestry estimation
For InPSYght, we estimated the proportion of African ancestry present in each individual by using RFMix40 with two reference groups representing African and European ancestry from 1000G. For BioMe, participants had previously been grouped by continental origin and into identity-by-descent (IBD) communities representing groups with shared recent genetic ancestry.23 We labeled BioMe participants as from a Caribbean population if their continental origin was Caribbean or if they were members of the Puerto Rican or Dominican IBD communities. We labeled all other BioMe participants with non-missing continental origin as non-Caribbean. Participants not from Puerto Rican or Dominican IBD communities and with missing continental origin information were not included in comparisons between Caribbean and non-Caribbean populations.
We performed principal-component analysis (PCA) to obtain fine-scale ancestry information for all four studies. We used genotypes subset to those present on the Omni 2.5M array to project participants from each of the four studies onto the 938 reference samples from the Human Genome Diversity Project41 by using the LASER server.42
Effect of regional genomic features on imputation quality
Genomic features datasets
We downloaded GC content over 5 bp intervals, the genomic positions of segmental duplications, the genomic positions of structural variants annotated with the Database of Genomic Variants, and the genomic positions of repeats identified with RepeatMasker from the UCSC Genome Browser database.43 Recombination rate was calculated with the HapMap GrCh38 genetic map44 as centimorgans per megabase.
Relationships between genomic features and TOPMed imputation quality
In each study, we performed LD pruning to obtain a set of near-independent bi-allelic SNVs on chromosome 20, retaining variants with pairwise r2 < 0.2 within a sliding 50 kb window with a five variant step size with PLINK v2.0.45, 46, 47 For each retained variant, we defined five aggregate measures of genomic features over 10 kb windows centered at the variant: mean GC content, number of repeats, number of structural variants, presence of ≥1 segmental duplication, and mean recombination rate. We defined the linear distance of the variant from the nearest array-genotyped variant. For each of the six genomic features, we performed a logistic regression to test the association between dichotomous imputation quality (observed imputation r2 > 0.8 versus ≤0.8) and the feature, adjusting for variant MAF as a categorical variable with nine bins and breaks at 0, 0.0003, 0.0006, 0.0009, 0.001, 0.0032, 0.01, 0.032, 0.1, and 0.5. We also performed zero-one inflated beta regression to test the association between the continuous observed imputation r2 and each feature with the same MAF adjustment. Zero-one inflated beta regression models the association of the genomic features with the observed imputation r2 in the open interval 0 < r2 < 1 (mean and variance ) and the probabilities of observed imputation r2 = 0 () and observed imputation r2 = 1 () in a piecewise manner.48 In both regression models, we centered and scaled continuous and count predictors for comparability.
Effect of real versus WGS-based array genotypes on evaluation of imputation quality
To determine whether the WGS-based imputation results were consistent with the genotype array-based results, we imputed the real OmniExpress arrays with each of the three reference panels in METSIM. For each reference panel, we compared the observed imputation r2 and genotype concordance metrics for the real array-based genotype imputation to WGS-based array genotype imputation results (from above).
Effect of variant caller on evaluation of imputation quality
Variants in TOPMed and the four study datasets were called with the TOPMed Variant Calling/GotCloud pipeline.5,35 To assess the impact of variant calling tool, we used an additional set of WGS variants called with Haplotype Caller from GATK version 3.549 in the METSIM study.50 Variants that deviated from Hardy-Weinberg equilibrium (p < 10−6), had >2% missingness, or had allelic imbalance <0.3 or >0.7 were excluded from this callset. Variants in regions of low complexity, centromeres, segmental duplications, or satellite regions were also excluded.50 After filtering, 21.8M variants remained in the subset of 2,429 individuals used for imputation analysis in METSIM. We then created each of the four WGS-based arrays by using both METSIM callsets and evaluated imputation performance by comparing the imputed variants to the respective sequenced variants.
Imputability tool for the Michigan Imputation Server
We developed RsqBrowser, a tool that allows researchers to query for the observed imputation r2 for variants or regions of interest. Users specify the genomic position in build GRCh38 and select the genotype array, imputation reference panel, and sample ancestry. RsqBrowser returns a table with the position and observed imputation r2 for all variants in the specified regions or genes. We have deployed this tool on the Michigan Imputation Server.
Results
Whole-genome sequencing studies of four ancestries
We used WGS data in four studies as gold standard genotypes. These four represent three major US populations, African American/African Diaspora (African), Hispanic/Latino, and European American (European) ancestry, and a population isolate, Finnish ancestry from Finland (Table 1). We observed that our primary metric of imputation quality, the observed imputation r2, was upwardly biased in small samples for low-frequency (0.5% < MAF ≤ 5%) and rare (MAF ≤ 0.5%) variants (Figure S1). To avoid any biases comparing across datasets of different sample sizes, we randomly downsampled the African, Hispanic/Latino, and Finnish ancestry datasets to 2,429 individuals to match the smaller European ancestry dataset. After all sample- and variant-level filtering, we included in our analysis 79.3M, 68.8M, 62.2M, and 22.0M variants in the African, Hispanic/Latino, European, and Finnish ancestry studies, respectively (Table 1). In each study, >91% of these variants were bi-allelic SNVs. The others were multi-allelic SNVs (0.3%–1.6%), and bi-allelic (6.5%–6.6%) and multi-allelic indels (0.005%–0.2%).
Impact of reference panel on genotype imputation quality
For each WGS study participant, we subsetted WGS genotypes to those present on the Illumina Core (0.3M markers), OmniExpress (0.7M), Multi-Ethnic Genotyping (MEGA) (1.8M), and Omni 2.5M (2.4M) arrays. We then carried out genotype imputation on these genotype array subsets by using the 1000G and HRC imputation reference panels, as well as a modified TOPMed panel. Because the Hispanic/Latino and European WGS datasets were included in the TOPMed panel, we restricted the TOPMed panel to a subset of 88,804 reference samples that did not overlap our WGS datasets for all analyses. To measure the imputation quality of each sequenced variant in the study, we calculated the squared Pearson correlation between sequenced genotypes and imputed genotype dosages (observed imputation r2). We considered two aggregate measures of imputation quality: the mean observed imputation r2 and the proportion of well-imputed variants (observed imputation r2 > 0.8) by MAF bin. We selected a stringent threshold (>90% of variants well imputed) to define sets of variants for which array genotyping followed by imputation can approximate WGS. For variants satisfying this threshold, the two technologies are expected to provide very similar information.51
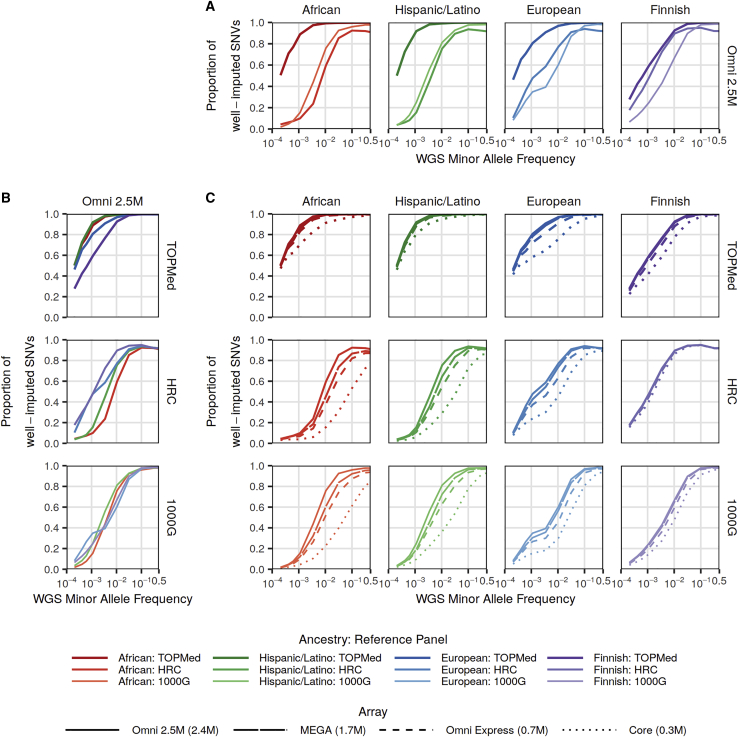
As expected, across all combinations of reference panels, ancestries, and MAF, the densest genotype array (Omni 2.5M) had both the highest mean observed imputation r2 and highest number and proportion of well-imputed variants (Figure 1, Figure S2, Tables S2 and S3). For the Omni 2.5M array and in all ancestries, TOPMed-based imputation approximated WGS for variants of lower MAF compared to the HRC or 1000G panels. TOPMED-based panel imputation approximated WGS at lower MAF thresholds in African and Hispanic/Latino ancestry (0.14% and 0.11%) than in European or Finnish ancestry (0.35% and 0.85%) (Figures 1A and 1B, Table S4). The ordering of imputation quality by reference panel, genotyping array, and ancestry did not change when using mean observed imputation r2 as a metric of average imputation quality (Figure S2). The ordering of results for both metrics also did not change by either the use of subsets of WGS genotypes instead of actual genotyping arrays (Figure S3) or the choice of variant caller (Figure S4).
Figure 1.
Proportion of well-imputed (r2 > 0.8) bi-allelic SNVs by reference panel, study ancestry, and genotyping array
The proportion of sequenced variants that are well-imputed (r2 > 0.8) with the TOPMed, HRC, and 1000G imputation reference panels.
(A) Comparison across the reference panels using the Illumina Omni 2.5M array.
(B) Comparison across the four studies using the Illumina Omni 2.5M array.
(C) Comparison across four Illumina genotyping arrays: Omni 2.5M, MEGA, Omni Express, and Core by ancestry (columns) and imputation reference panels (rows). In all plots, the x axes show minor-allele frequency (MAF) calculated separately by study. Sequenced bi-allelic SNVs not present in reference panels were assigned r2 = 0. Bi-allelic SNVs were then aggregated by MAF bins of width 0.00025 MAF for MAF between 0.0002 and 0.002 and of size 0.001 MAF for MAF > 0.002; those plotted here correspond to singletons, doubletons, and tripletons in each study, as well as those with mean MAF closest to the values 0.001, 0.0032, 0.01, 0.032, 0.1, 0.32, and 0.5.
Less influence of genotype array size with TOPMed compared to 1000G- and HRC-based imputation
For all four ancestries and all three imputation reference panels, imputation quality increased with larger array size (Figure 1C). However, the difference in TOPMed imputation quality among the Omni 2.5M, MEGA, and OmniExpress arrays was minimal. For example, in the African ancestry study, TOPMed imputation approximated WGS for variants with MAF ≥ 0.14% with the Omni 2.5M array, ≥0.16% with the MEGA array, and ≥0.24% with the OmniExpress array (Table S4). This threshold was higher with the smaller Core array (≥0.84%). In contrast, genotype array size had a larger effect on imputation quality with the HRC and 1000G panels in African and Hispanic/Latino ancestry studies. For African ancestry, 1000G imputation approximated WGS at a much lower MAF with the Omni 2.5M array (≥2.5%) compared to the OmniExpress array (≥14.0%). With the HRC panel, imputation with the OmniExpress array could not approximate WGS at any MAF in African ancestry.
Individual-level imputation accuracy varies with finer-scale ancestry
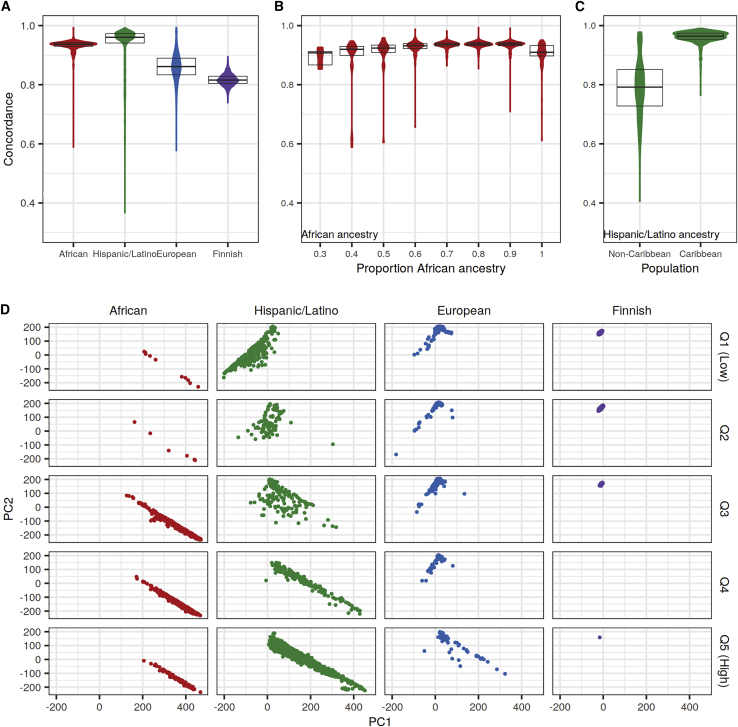
Because imputation quality depends on the shared ancestry between reference panel and sample haplotypes,11 we hypothesized that imputation quality within the four WGS studies would vary with finer-scale ancestry. To measure individual-level imputation quality, we calculated concordance rates between heterozygous sequenced and imputed genotypes separately for study-specific rare, low-frequency, and common bi-allelic SNVs in each individual. As expected, concordance rates varied across individuals more for rare variants than for low-frequency and common variants (TOPMed: Figures 2, S5, and S6; all panels: Table S5). With the TOPMed panel, mean heterozygous concordance rates for rare variants were higher in individuals of African and Hispanic/Latino ancestry (0.93 in both) compared to individuals of European and Finnish ancestry (0.86 and 0.82) (Figure 2A). Concordance rates varied most within Hispanic/Latino individuals (10th–90th percentile: 0.80–0.98).
Figure 2.
Heterozygous genotype concordance rates for rare variants by ancestry with TOPMed panel imputation
Heterozygous concordance rates were calculated between sequenced and TOPMed-imputed genotypes for rare (MAF < 0.5%, calculated separately in each study) bi-allelic SNVs with the Omni 2.5M array.
(A) Distribution of concordance rates in each of the four studies. Boxplots correspond to 25th, 50th, and 75th percentiles.
(B) Distribution of concordance rates by bins of estimated proportion of African ancestry in the admixed African study.
(C) Distribution of concordance rates in Caribbean and non-Caribbean populations in the Hispanic/Latino study.
(D) Principal-component analysis (PCA) by genotype concordance quintile and ancestry. PCA was performed by projecting onto the Human Genome Diversity Project reference samples. Genotype concordance quintiles were calculated across all four studies and correspond to concordance rates of 0.37–0.82 (Q1), 0.82–0.86 (Q2), 0.86–0.93 (Q3), 0.93–0.95 (Q4), and 0.95–0.99 (Q5). Points are colored by ancestry.
We next stratified African and Hispanic/Latino study participants by finer-scale measures of ancestry. The African American population in the United States is primarily of African and European ancestries.52 We therefore estimated the proportion of African ancestry for each individual in the African ancestry study assuming two populations, which ranged from 0.26 to 1.00 (mean 0.82). For the 2,307 individuals with an estimated African ancestry < 0.95, individuals with higher proportions of African ancestry had higher genotype concordance rates with the TOPMed panel (Figure 2B). For instance, concordance rates for those with an estimated proportion between 0.86 and 0.95 were higher (mean 0.93) than for those between 0.26 and 0.35 (mean 0.89). In contrast, concordance rates for the 122 individuals with estimated proportion of African ancestry > 0.95 were lower (mean 0.91) than for individuals with smaller estimated proportions of African ancestry.
Hispanic/Latino populations in the United States are admixed with primarily European, Native American, and African ancestry, and individuals of Caribbean origin usually have more African ancestry.53 The concordance rates for individuals from Caribbean populations were higher (mean 0.96) compared to those from non-Caribbean (mean 0.79) populations with the TOPMed panel (Figure 2C).
We also estimated finer-scale ancestry in all four studies with principal-component analysis (PCA), projecting the study individuals onto 938 reference samples from the Human Genome Diversity Project (HGDP).41 The first two principal components (PCs) reflect clines of European (high PC2), African (high PC1 and low PC2), and Native American (low PC1 and low PC2) ancestries (Figure S7). To see how TOPMed imputation quality varied with fine-scale ancestry, we divided individuals into concordance rate quintiles calculated jointly across all four studies. In all studies, individuals clustering closer to HGDP individuals of African ancestry were more likely to be in higher concordance rate quintiles, while those clustering closer to Native American or European populations were more likely to be in lower concordance rate quintiles for rare (Figure 2D) and low-frequency variants (Figure S5D). As expected, there was little variability in common variant imputation quality (Figure S6D).
Taken together, these results demonstrate that TOPMed imputation quality varies across individuals with finer-scale ancestry. Among the populations studied here, population subsets with large proportions of African ancestry, including Hispanic/Latino ancestry individuals of Caribbean origin, were on average the most accurately imputed for rare variants. However, individuals with the greatest proportions of African ancestry in the African study were not the most accurately imputed. The heterozygote concordance rates from HRC and 1000G imputation also varied with finer-scale ancestry for rare variants (Table S5).
Imputation quality varies across the genome
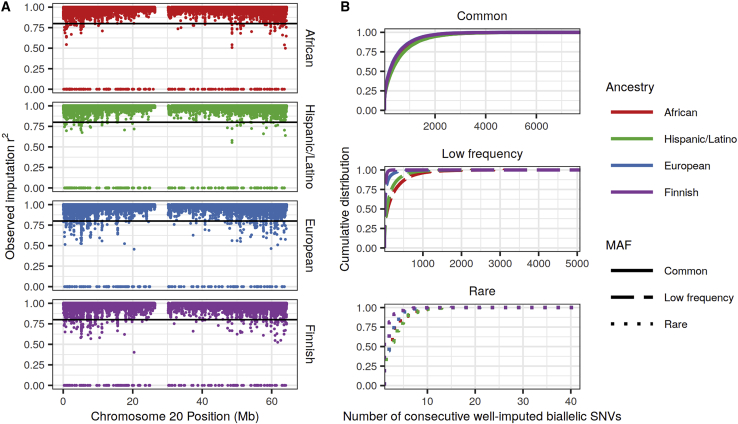
Sequence quality and genotype array density are not uniform across the genome. Because these factors influence imputation, we sought to quantify the regional variability in imputation quality. We first visualized the observed imputation r2 for common variants (MAF > 5%) across the chromosomes. Although the vast majority (>99.6%) of common variants are well imputed (observed imputation r2 > 0.8) in all four ancestries with the Omni 2.5M array and TOPMed reference panel (Table S3), we identified clusters of common variants that were not well imputed at the same genomic positions across ancestries and genotype arrays (Figures 3A and S8). There are likewise regions with better-than-average imputation quality, including the HLA region on chromosome 6 (Figure S8), which is characterized by high LD and dense genotype array coverage.22,54
Figure 3.
Regional variability in TOPMed reference panel imputation quality
(A) Observed imputation r2 by genomic position (Mb) of common (MAF > 0.05) bi-allelic SNVs on chromosome 20. Sequenced bi-allelic SNVs not present in reference panels were assigned r2 = 0. The horizontal line at r2 = 0.8 corresponds to the threshold used to determine well-imputed variants.
(B) Cumulative distribution of the number of consecutively well-imputed (r2 > 0.8) bi-allelic SNVs in each MAF category: common (MAF ≥ 0.05), low frequency (0.005 ≤ MAF < 0.05), and rare (MAF < 0.005), as calculated separately in each study. For common variants, European and Finnish curves appear to overlap and African and Hispanic/Latino curves appear to overlap.
To assess regional variability in imputation quality, we calculated the lengths of runs of consecutively well-imputed variants separately for rare, low-frequency, and common bi-allelic SNVs across the genome (Figure 3B, Tables S6 and S7). We identified a large variability in the number of consecutively well-imputed common and low-frequency variants with the TOPMed panel (e.g., interquartile range [IQR] in African ancestry is 41–750 common variants [10.4–253.2 kb] and 9–287 low-frequency variants [2.3–84.3 kb] with the Omni 2.5M array). As expected, the lengths of consecutive well-imputed rare variants were much shorter, having a maximum length of 34–45 variants depending on ancestry.
Local genomic features explain little variability in imputation quality
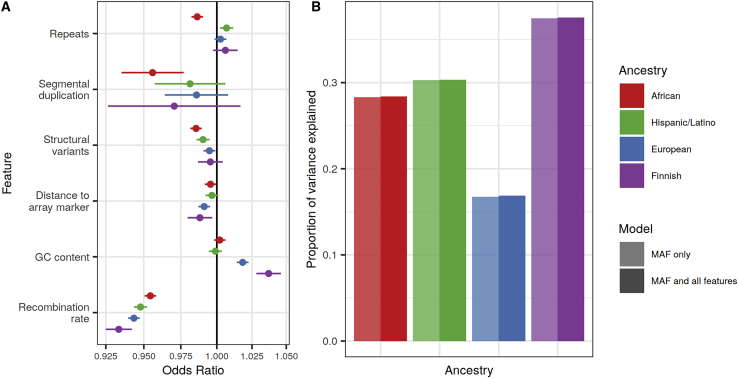
Genomic features including high GC content and the presence of large duplications or repeats have been associated with regions of poor imputation quality in Europeans when using the 1000G panel.15 To test the effects of genomic features on imputation quality with the TOPMed panel, we performed logistic regressions in each of our four studies with the imputed quality status (observed imputation r2 > 0.8) as the dichotomous outcome for independent variants on chromosome 20. In separate models, we tested the associations of distance to the nearest genotyped variant and the following features aggregated over a 10 kb window centered at the variant: mean GC content, mean recombination rate, number of repeats, number of structural variants, and presence of ≥1 segmental duplication, adjusting for bins of MAF. We found that higher recombination rate, lower GC content, greater distance to genotype array variants, more structural variants, and the presence of segmental duplications were all associated with lower imputation quality (Figure 4A, Table S8). The effect of nearby repeats was not consistent across ancestries or repeat class, although nearby simple repeats were associated with worse imputation quality in all ancestries (Figure S9, Table S9). However, none of the tested genomic features meaningfully impacted the proportion of variability in imputation quality beyond variant MAF (Figure 4B). Results were similar when modeling imputation quality as a continuous variable (Figure S10, Table S10) and were consistent across reference panels (Table S8).
Figure 4.
Genomic features associated with TOPMed imputation quality of bi-allelic SNVs by ancestry
(A) The odds ratios and corresponding unadjusted 95% confidence intervals from logistic regression models. Estimates are from separate models testing the associations between characteristics of regional genomic features and whether or not a variant is well imputed (observed imputation r2 > 0.8), adjusting for variant MAF.
(B) The proportion of variance explained (Nagelkerke R2) for each logistic regression models with MAF only or with MAF and all six tested genomic features in one joint model.
Impact of variant predicted function and type on imputation quality
Protein-coding variants are often of high clinical significance and easier to interpret compared to non-coding variants; they are also more likely to be rare and more difficult to impute.5,55 To determine the extent to which variants that impact protein coding are well imputed, we classified sequenced bi-allelic SNVs by predicted impact on protein coding. With the TOPMed panel and Omni 2.5M array, we found that 45.6%–64.1% of variants predicted to have high impact and 51.0%–66.8% of variants predicted to have moderate impact on protein coding were well imputed (Table S11). We found no meaningful difference in imputation quality between the protein-coding classes when controlling for MAF (Figure S11).
All results presented above are for bi-allelic SNVs. Multi-allelic SNVs and all indels have been shown to have lower imputation quality than bi-allelic SNVs with the 1000G panel,12,15 and indels are absent from the HRC panel.13 To quantify the effect of variant type on imputation quality, we calculated the proportion of well-imputed indels, multi-allelic SNVs, and multi-allelic indels by using the TOPMed panel and all four genotype arrays. We observed very similar MAF thresholds for which imputation could approximate WGS among bi-allelic SNVs, bi-allelic indels, and multi-allelic SNVs (Figure S12). Multi-allelic indels were less well imputed. For example, in African ancestry, TOPMed imputation with the Omni 2.5M array approximated WGS at similar MAF thresholds for bi-allelic SNVs and indels and multi-allelic SNVs (0.14%, 0.24%, and 0.16% respectively) compared to 0.55% for multi-allelic indels (Table S12).
Discussion
Here, we used deep WGS from studies of African, Hispanic/Latino, European, and Finnish ancestries to quantify the extent to which array genotyping followed by genotype imputation can approximate WGS. We performed imputation by using genotypes present on the Illumina Core, OmniExpress, MEGA, and Omni 2.5M arrays with the 1000G, HRC, and TOPMed reference panels. We found that with the largest array (Omni 2.5M) and largest reference panel (TOPMed), array genotyping followed by imputation can approximate WGS at a population level for variants with MAF ≥ 0.14% in African ancestry, ≥0.11% in Hispanic/Latino ancestry, ≥0.35% in European ancestry, and ≥0.84% in Finnish ancestry. Particularly for the African and Hispanic/Latino ancestry studies, TOPMed imputation approximated WGS at much lower MAF than HRC or 1000G imputation, which is consistent with previous analyses showing improvements in these populations even with a smaller version of the TOPMed panel.16 For analyses primarily investigating the genetic effects of common and low-frequency variants, such as single-variant genome-wide association studies (GWASs), in any of the four populations, array genotyping and imputation is sufficient to accurately capture genetic variants and given differences in cost allows for much larger sample sizes than WGS. Large proportions (∼44%–60%) of rare variants with MAF even lower than the reported thresholds were also well-imputed, highlighting the potential for well-powered rare-variant studies without WGS, although not all rare variants can be reliably imputed. Because we restricted the TOPMed panel to reference samples that did not overlap the WGS datasets, we expect that imputation quality with the full TOPMed R2 panel to be even better than reported here. In particular, we would expect higher imputation quality for Hispanic/Latino studies, as a large proportion of the Hispanic/Latino individuals in the TOPMed R2 panel were excluded here.
As previously reported with the 1000G and HRC panels,12,13 imputation quality with the TOPMed panel was higher when using larger arrays. However, the effect of genotype array choice on TOPMed imputation was much smaller than on HRC or 1000G imputation. The difference between the Omni 2.5M, MEGA, and OmniExpress arrays was minimal, suggesting that researchers imputing with the TOPMed panel in these populations may opt for the less expensive OmniExpress array with little loss of information. However, we did find lower imputation quality when using the smaller Core array (∼307k variants) and might expect even lower quality for arrays with fewer markers.
WGS is also used for clinical purposes, including diagnosis, screening, and identifying therapeutic targets.6 Variants predicted to alter protein function are often of high clinical significance.55 In the populations studied here, we found that only 50.7%–66.7% of bi-allelic SNVs with moderate or high predicted impact on protein coding were well imputed, as might be expected given the generally low MAF of these variants. To quantify individual imputation quality in contrast to population-level imputation quality, we calculated the heterozygous concordance rates between sequenced bi-allelic SNVs and imputed best-guess genotypes. For all three reference panels, we found that the concordance rates for rare and low-frequency variants varied widely among individuals in the African, Hispanic/Latino, and European ancestry studies and were associated with finer-scale ancestry. Because of this variability and the large proportion of rare variants that are not accurately imputed with the available imputation reference panels, we believe that WGS cannot currently be reliably approximated in clinical settings with array genotyping and imputation with the reference panels studied here. Recent work in cystic fibrosis-affected individuals has shown that disease-specific WGS panels can provide additional information for imputation of disease-causing variants and may play a role in future clinical use.56
Despite large numbers of individuals with African and Hispanic/Latino ancestry in the TOPMed reference panel, more than half of the individuals in the panel are estimated to be of >50% European ancestry. Still, we found that TOPMed imputation quality was highest for the African and Hispanic/Latino ancestry studies and for individuals with large proportions of African ancestry among the populations studied here. Higher imputation quality of rare variants in African populations compared to European populations was previously reported with the 1000G panel.12 A first possible explanation for the high relative imputation quality in African populations is that there are proportionally more rare variants in non-African populations that have undergone recent bottlenecks and subsequent population growth, as is true in the three US populations studied here (Figure S13). In these populations, it can be more difficult to identify the haplotype background of the rare variation.12,57 A second possible explanation is that individuals with large proportions of African ancestry in these studies match more closely by chance with a subset of TOPMed haplotypes than do the individuals with large proportions of Native American or European ancestry. However, relatively higher imputation quality with the TOPMed panel in samples of African and Hispanic/Latino ancestry compared to European ancestry was previously reported in a separate set of samples.5 Third, admixture could impact the accuracy of haplotype phasing of the sample or reference haplotypes. Taken together, these results emphasize the importance of ancestrally diverse reference panels such as TOPMed and suggest that reference panel composition is not the only factor explaining ancestry differences in imputation quality.
While nearly all common and low-frequency variants are well imputed with the TOPMed panel in the populations studied, there was substantial variability by genomic region in imputation across the MAF spectrum. Some regions, such as the densely genotyped HLA locus, had higher imputation quality than what would be expected on the basis of variant MAF alone. We found that lower recombination rate and higher GC content around a variant were associated with higher imputation quality in all four studies but that none of these features except MAF explained a substantial proportion of variability in imputation quality. For all tested features except for GC content and repeats, the direction of effect was consistent with previous work examining regions of poor imputation quality via the 1000G panel in Europeans.15 The inconsistency in GC content direction could be explained by our focus on well-imputed variants instead of poorly imputed regions and/or by continued improvements in sequencing quality. Given the difficulty of predicting hard-to-impute regions/variants, we developed RsqBrowser, a tool that allows researchers to query empirical imputation quality for specific variants or genomic regions of interest by ancestry, which is available on the Michigan Imputation Server.
The results presented here are limited by the use of high quality but imperfect WGS as a gold standard. We did not consider any variants that were imputed from the reference panels but not detected in the WGS. Comparisons of imputation quality by reference panel or genotyping array did not change when using real array data compared to WGS-based arrays or when using a different variant caller, although data were only available to carry out these sensitivity analyses in the study of Finnish ancestry. We also note that the results presented here cannot necessarily be extended to other populations or population isolates, particularly those such as East and South Asian populations, that are not represented or represented in smaller numbers in the TOPMed panel. Furthermore, we only used WGS from one study for each population that we analyzed. For some ancestries, particularly population isolates, other population-specific reference panels may perform better than the three commonly used imputation panels analyzed here.
While array genotyping and imputation cannot fully replace deep WGS, we found that it can approximate WGS for variants down to specific MAF thresholds depending on genotype array and reference panel choices as well as sample ancestry. Researchers’ decision to invest in one technology over another will depend on these criteria, genomic location, and the MAF of variants relevant to their research questions.
Consortia
The members of the NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium are Namiko Abe, Goncalo Abecasis, Francois Aguet, Christine Albert, Laura Almasy, Alvaro Alonso, Seth Ament, Peter Anderson, Pramod Anugu, Deborah Applebaum-Bowden, Kristin Ardlie, Dan Arking, Donna K. Arnett, Allison Ashley-Koch, Stella Aslibekyan, Tim Assimes, Paul Auer, Dimitrios Avramopoulos, Najib Ayas, Adithya Balasubramanian, John Barnard, Kathleen Barnes, R. Graham Barr, Emily Barron-Casella, Lucas Barwick, Terri Beaty, Gerald Beck, Diane Becker, Lewis Becker, Rebecca Beer, Amber Beitelshees, Emelia Benjamin, Takis Benos, Marcos Bezerra, Larry Bielak, Joshua Bis, Thomas Blackwell, John Blangero, Eric Boerwinkle, Donald W. Bowden, Russell Bowler, Jennifer Brody, Ulrich Broeckel, Jai Broome, Deborah Brown, Karen Bunting, Esteban Burchard, Carlos Bustamante, Erin Buth, Brian Cade, Jonathan Cardwell, Vincent Carey, Julie Carrier, Cara Carty, Richard Casaburi, Juan P. Casas Romero, James Casella, Peter Castaldi, Mark Chaffin, Christy Chang, Yi-Cheng Chang, Daniel Chasman, Sameer Chavan, Bo-Juen Chen, Wei-Min Chen, Yii-Der Ida Chen, Michael Cho, Seung Hoan Choi, Lee-Ming Chuang, Mina Chung, Ren-Hua Chung, Clary Clish, Suzy Comhair, Matthew Conomos, Elaine Cornell, Adolfo Correa, Carolyn Crandall, James Crapo, L. Adrienne Cupples, Joanne Curran, Jeffrey Curtis, Brian Custer, Coleen Damcott, Dawood Darbar, Sean David, Colleen Davis, Michelle Daya, Mariza de Andrade, Lisa de las Fuentes, Paul de Vries, Michael DeBaun, Ranjan Deka, Dawn DeMeo, Scott Devine, Huyen Dinh, Harsha Doddapaneni, Qing Duan, Shannon Dugan-Perez, Ravi Duggirala, Jon Peter Durda, Susan K. Dutcher, Charles Eaton, Lynette Ekunwe, Adel El Boueiz, Patrick Ellinor, Leslie Emery, Serpil Erzurum, Charles Farber, Jesse Farek, Tasha Fingerlin, Matthew Flickinger, Myriam Fornage, Nora Franceschini, Chris Frazar, Mao Fu, Stephanie M. Fullerton, Lucinda Fulton, Stacey Gabriel, Weiniu Gan, Shanshan Gao, Yan Gao, Margery Gass, Heather Geiger, Bruce Gelb, Mark Geraci, Soren Germer, Robert Gerszten, Auyon Ghosh, Richard Gibbs, Chris Gignoux, Mark Gladwin, David Glahn, Stephanie Gogarten, Da-Wei Gong, Harald Goring, Sharon Graw, Kathryn J. Gray, Daniel Grine, Colin Gross, C. Charles Gu, Yue Guan, Xiuqing Guo, Namrata Gupta, David M. Haas, Jeff Haessler, Michael Hall, Yi Han, Patrick Hanly, Daniel Harris, Nicola L. Hawley, Jiang He, Ben Heavner, Susan Heckbert, Ryan Hernandez, David Herrington, Craig Hersh, Bertha Hidalgo, James Hixson, Brian Hobbs, John Hokanson, Elliott Hong, Karin Hoth, Chao (Agnes) Hsiung, Jianhong Hu, Yi-Jen Hung, Haley Huston, Chii Min Hwu, Marguerite Ryan Irvin, Rebecca Jackson, Deepti Jain, Cashell Jaquish, Jill Johnsen, Andrew Johnson, Craig Johnson, Rich Johnston, Kimberly Jones, Hyun Min Kang, Robert Kaplan, Sharon Kardia, Shannon Kelly, Eimear Kenny, Michael Kessler, Alyna Khan, Ziad Khan, Wonji Kim, John Kimoff, Greg Kinney, Barbara Konkle, Charles Kooperberg, Holly Kramer, Christoph Lange, Ethan Lange, Leslie Lange, Cathy Laurie, Cecelia Laurie, Meryl LeBoff, Jiwon Lee, Sandra Lee, Wen-Jane Lee, Jonathon LeFaive, David Levine, Dan Levy, Joshua Lewis, Xiaohui Li, Yun Li, Henry Lin, Honghuang Lin, Xihong Lin, Simin Liu, Yongmei Liu, Yu Liu, Ruth J.F. Loos, Steven Lubitz, Kathryn Lunetta, James Luo, Ulysses Magalang, Michael Mahaney, Barry Make, Ani Manichaikul, Alisa Manning, JoAnn Manson, Lisa Martin, Melissa Marton, Susan Mathai, Rasika Mathias, Susanne May, Patrick McArdle, Merry-Lynn McDonald, Sean McFarland, Stephen McGarvey, Daniel McGoldrick, Caitlin McHugh, Becky McNeil, Hao Mei, James Meigs, Vipin Menon, Luisa Mestroni, Ginger Metcalf, Deborah A. Meyers, Emmanuel Mignot, Julie Mikulla, Nancy Min, Mollie Minear, Ryan L. Minster, Braxton D. Mitchell, Matt Moll, Zeineen Momin, May E. Montasser, Courtney Montgomery, Donna Muzny, Josyf C. Mychaleckyj, Girish Nadkarni, Rakhi Naik, Take Naseri, Pradeep Natarajan, Sergei Nekhai, Sarah C. Nelson, Bonnie Neltner, Caitlin Nessner, Deborah Nickerson, Osuji Nkechinyere, Kari North, Jeff O'Connell, Tim O'Connor, Heather Ochs-Balcom, Geoffrey Okwuonu, Allan Pack, David T. Paik, Nicholette Palmer, James Pankow, George Papanicolaou, Cora Parker, Gina Peloso, Juan Manuel Peralta, Marco Perez, James Perry, Ulrike Peters, Patricia Peyser, Lawrence S. Phillips, Jacob Pleiness, Toni Pollin, Wendy Post, Julia Powers Becker, Meher Preethi Boorgula, Michael Preuss, Bruce Psaty, Pankaj Qasba, Dandi Qiao, Zhaohui Qin, Nicholas Rafaels, Laura Raffield, Mahitha Rajendran, Vasan S. Ramachandran, D.C. Rao, Laura Rasmussen-Torvik, Aakrosh Ratan, Susan Redline, Robert Reed, Catherine Reeves, Elizabeth Regan, Alex Reiner, Muagututi‘a Sefuiva Reupena, Ken Rice, Stephen Rich, Rebecca Robillard, Nicolas Robine, Dan Roden, Carolina Roselli, Jerome Rotter, Ingo Ruczinski, Alexi Runnels, Pamela Russell, Sarah Ruuska, Kathleen Ryan, Ester Cerdeira Sabino, Danish Saleheen, Shabnam Salimi, Sejal Salvi, Steven Salzberg, Kevin Sandow, Vijay G. Sankaran, Jireh Santibanez, Karen Schwander, David Schwartz, Frank Sciurba, Christine Seidman, Jonathan Seidman, Frederic Sériès, Vivien Sheehan, Stephanie L. Sherman, Amol Shetty, Aniket Shetty, Wayne Hui-Heng Sheu, M. Benjamin Shoemaker, Brian Silver, Edwin Silverman, Robert Skomro, Albert Vernon Smith, Jennifer Smith, Josh Smith, Nicholas Smith, Tanja Smith, Sylvia Smoller, Beverly Snively, Michael Snyder, Tamar Sofer, Nona Sotoodehnia, Adrienne M. Stilp, Garrett Storm, Elizabeth Streeten, Jessica Lasky Su, Yun Ju Sung, Jody Sylvia, Adam Szpiro, Daniel Taliun, Hua Tang, Margaret Taub, Kent D. Taylor, Matthew Taylor, Simeon Taylor, Marilyn Telen, Timothy A. Thornton, Machiko Threlkeld, Lesley Tinker, David Tirschwell, Sarah Tishkoff, Hemant Tiwari, Catherine Tong, Russell Tracy, Michael Tsai, Dhananjay Vaidya, David Van Den Berg, Peter VandeHaar, Scott Vrieze, Tarik Walker, Robert Wallace, Avram Walts, Fei Fei Wang, Heming Wang, Jiongming Wang, Karol Watson, Jennifer Watt, Daniel E. Weeks, Bruce Weir, Scott T. Weiss, Lu-Chen Weng, Jennifer Wessel, Cristen Willer, Kayleen Williams, L. Keoki Williams, Carla Wilson, James Wilson, Lara Winterkorn, Quenna Wong, Joseph Wu, Huichun Xu, Lisa Yanek, Ivana Yang, Ketian Yu, Seyedeh Maryam Zekavat, Yingze Zhang, Snow Xueyan Zhao, Wei Zhao, Xiaofeng Zhu, Michael Zody, and Sebastian Zoellner.
Acknowledgments
Full acknowledgments are provided in the supplemental information.
Declaration of interests
E.E.K. has received speaker honorarium from Regeneron, 23&Me, and Illumina and sits on the advisory board for Encompass Bio and Galateo Bio.
Published: August 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.07.012.
Web resources
RsqBrowser, https://imputationserver.sph.umich.edu/rsq-browser
Supplemental information
Data and code availability
Genotypes can be accessed through dbGaP for the BioMe (dbGaP: phs001644) and MLOF (dbGaP: phs001515) studies. METSIM and InPSYght genotype data are not currently available because of privacy restrictions. The variant-level r2 statistics generated during this study are available to download from RsqBrowser (https://imputationserver.sph.umich.edu/rsq-browser/downloads). Analysis code and scripts are available at https://github.com/schanks/imputation_rsqbrowser.
References
- 1.Sazonovs A., Barrett J.C. Rare-Variant Studies to Complement Genome-Wide Association Studies. Annu. Rev. Genomics Hum. Genet. 2018;19:97–112. doi: 10.1146/annurev-genom-083117-021641. [DOI] [PubMed] [Google Scholar]
- 2.Walter K., Min J.L., Huang J., Crooks L., Memari Y., McCarthy S., Perry J.R.B., Xu C., Futema M., Lawson D., et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flannick J., Mercader J.M., Fuchsberger C., Udler M.S., Mahajan A., Wessel J., Teslovich T.M., Caulkins L., Koesterer R., Barajas-Olmos F., et al. Exome sequencing of 20, 791 cases of type 2 diabetes and 24, 440 controls. Nature. 2019;570:71–76. doi: 10.1038/s41586-019-1231-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Hout C.V., Tachmazidou I., Backman J.D., Hoffman J.D., Liu D., Pandey A.K., Gonzaga-Jauregui C., Khalid S., Ye B., Banerjee N., et al. Exome sequencing and characterization of 49, 960 individuals in the UK Biobank. Nature. 2020;586:749–756. doi: 10.1038/s41586-020-2853-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. Sequencing of 53, 831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez-Marmiesse A., Gouveia S., Couce M.L. NGS Technologies as a Turning Point in Rare Disease Research , Diagnosis and Treatment. Curr. Med. Chem. 2018;25:404–432. doi: 10.2174/0929867324666170718101946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishiguchi K.M., Tearle R.G., Liu Y.P., Oh E.C., Miyake N., Benaglio P., Harper S., Koskiniemi-Kuendig H., Venturini G., Sharon D., et al. Whole genome sequencing in patients with retinitis pigmentosa reveals pathogenic DNA structural changes and NEK2 as a new disease gene. Proc. Natl. Acad. Sci. USA. 2013;110:16139–16144. doi: 10.1073/pnas.1308243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boycott K.M., Vanstone M.R., Bulman D.E., MacKenzie A.E. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat. Rev. Genet. 2013;14:681–691. doi: 10.1038/nrg3555. [DOI] [PubMed] [Google Scholar]
- 9.Cade B.E., Lee J., Sofer T., Wang H., Zhang M., Chen H., Gharib S.A., Gottlieb D.J., Guo X., Lane J.M., et al. Whole-genome association analyses of sleep-disordered breathing phenotypes in the NHLBI TOPMed program. Genome Med. 2021;13:136. doi: 10.1186/s13073-021-00917-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tam V., Patel N., Turcotte M., Bossé Y., Paré G., Meyre D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 11.Li Y., Willer C., Sanna S., Abecasis G. Genotype Imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auton A., Abecasis G.R., Altshuler D.M., Durbin R.M., Abecasis G.R., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E., et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., et al. A reference panel of 64, 976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das S., Abecasis G.R., Browning B.L. Genotype Imputation from Large Reference Panels. Annu. Rev. Genomics Hum. Genet. 2018;19:73–96. doi: 10.1146/annurev-genom-083117-021602. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q., Cirulli E.T., Han Y., Yao S., Liu S., Zhu Q. Systematic assessment of imputation performance using the 1000 Genomes reference panels. Brief. Bioinform. 2015;16:549–562. doi: 10.1093/bib/bbu035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kowalski M.H., Qian H., Hou Z., Rosen J.D., Tapia A.L., Shan Y., Jain D., Argos M., Arnett D.K., Avery C., et al. Use of >100, 000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet. 2019;15:e1008500. doi: 10.1371/journal.pgen.1008500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudbjartsson D.F., Helgason H., Gudjonsson S.A., Zink F., Oddson A., Gylfason A., Besenbacher S., Magnusson G., Halldorsson B.V., Hjartarson E., et al. Large-scale whole-genome sequencing of the Icelandic population. Nat. Genet. 2015;47:435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 18.Sidore C., Busonero F., Maschio A., Porcu E., Naitza S., Zoledziewska M., Mulas A., Pistis G., Steri M., Danjou F., et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat. Genet. 2015;47:1272–1281. doi: 10.1038/ng.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pistis G., Porcu E., Vrieze S.I., Sidore C., Steri M., Danjou F., Busonero F., Mulas A., Zoledziewska M., Maschio A., et al. Rare variant genotype imputation with thousands of study-specific whole-genome sequences: implications for cost-effective study designs. Eur. J. Hum. Genet. 2015;23:975–983. doi: 10.1038/ejhg.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitt M., Kals M., Pärn K., Gabriel S.B., Lander E.S., Palotie A., Ripatti S., Morris A.P., Metspalu A., Esko T., et al. Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur. J. Hum. Genet. 2017;25:869–876. doi: 10.1038/ejhg.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deelen P., Menelaou A., van Leeuwen E.M., Kanterakis A., van Dijk F., Medina-Gomez C., Francioli L.C., Hottenga J.J., Karssen L.C., Estrada K., et al. Improved imputation quality of low-frequency and rare variants in European samples using the “Genome of The Netherlands. Eur. J. Hum. Genet. 2014;22:1321–1326. doi: 10.1038/ejhg.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verlouw J.A.M., Clemens E., de Vries J.H., Zolk O., Verkerk A.J.M.H., am Zehnhoff-Dinnesen A., Medina-Gomez C., Lanvers-Kaminsky C., Rivadeneira F., Langer T., et al. A comparison of genotyping arrays. Eur. J. Hum. Genet. 2021;29:1611–1624. doi: 10.1038/s41431-021-00917-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belbin G.M., Cullina S., Wenric S., Soper E.R., Glicksberg B.S., Torre D., Moscati A., Wojcik G.L., Shemirani R., Beckmann N.D., et al. Toward a fine-scale population health monitoring system. Cell. 2021;184:2068–2083.e11. doi: 10.1016/j.cell.2021.03.034. [DOI] [PubMed] [Google Scholar]
- 24.Laakso M., Kuusisto J., Stančáková A., Kuulasmaa T., Pajukanta P., Lusis A.J., Collins F.S., Mohlke K.L., Boehnke M. The Metabolic Syndrome in Men study: a resource for studies of metabolic and cardiovascular diseases. J. Lipid Res. 2017;58:481–493. doi: 10.1194/jlr.O072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin X., Chan L.S., Bose D., Jackson A.U., VandeHaar P., Locke A.E., Fuchsberger C., Stringham H.M., Yu K., Silva L.F., et al. Genome-wide association study of 1, 391 plasma metabolites in 6, 136 Finnish men identifies 303 novel signals and provides biological insights into human diseases. medRxiv. 2021 doi: 10.1101/2021.10.19.21265094. Preprint at. [DOI] [Google Scholar]
- 26.Johnsen J.M., Fletcher S.N., Dove A., McCracken H., Martin B.K., Kircher M., Josephson N.C., Shendure J., Ruuska S., Valentino L.A., et al. Results of Genetic Analysis of 11, 341 Participants Enrolled in the My Life, Our Future (MLOF) Hemophilia Genotyping Initiative. Blood. 2020;136:19. doi: 10.1111/jth.15805. [DOI] [PubMed] [Google Scholar]
- 27.Pato M.T., Sobell J.L., Medeiros H., Abbott C., Sklar B.M., Buckley P.F., Bromet E.J., Escamilla M.A., Fanous A.H., Lehrer D.S., et al. The genomic psychiatry cohort: partners in discovery. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2013;162B:306–312. doi: 10.1002/ajmg.b.32160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bigdeli T.B., Genovese G., Georgakopoulos P., Meyers J.L., Peterson R.E., Iyegbe C.O., Medeiros H., Valderrama J., Achtyes E.D., Kotov R., et al. Contributions of common genetic variants to risk of schizophrenia among individuals of African and Latino ancestry. Mol. Psychiatry. 2020;25:2455–2467. doi: 10.1038/s41380-019-0517-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swerdlow N.R., Gur R.E., Braff D.L. Consortium on the Genetics of Schizophrenia (COGS) assessment of endophenotypes for schizophrenia: an introduction to this Special Issue of Schizophrenia Research. Schizophr. Res. 2015;163:9–16. doi: 10.1016/j.schres.2014.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith E.N., Bloss C.S., Badner J.A., Barrett T., Belmonte P.L., Berrettini W., Byerley W., Coryell W., Craig D., Edenberg H.J., et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol. Psychiatry. 2009;14:755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nierenberg A.A., Friedman E.S., Bowden C.L., Sylvia L.G., Thase M.E., Ketter T., Ostacher M.J., Leon A.C., Reilly-Harrington N., Iosifescu D.V., et al. Lithium treatment moderate-dose use study (LiTMUS) for bipolar disorder: a randomized comparative effectiveness trial of optimized personalized treatment with and without lithium. Am. J. Psychiatry. 2013;170:102–110. doi: 10.1176/appi.ajp.2012.12060751. [DOI] [PubMed] [Google Scholar]
- 32.Sklar P., Smoller J.W., Fan J., Ferreira M.a.R., Perlis R.H., Chambert K., Nimgaonkar V.L., McQueen M.B., Faraone S.V., Kirby A., et al. Whole-genome association study of bipolar disorder. Mol. Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jun G., Wing M.K., Abecasis G.R., Kang H.M. An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. 2015;25:918–925. doi: 10.1101/gr.176552.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teslovich T.M., Kim D.S., Yin X., Stancáková A., Jackson A.U., Wielscher M., Naj A., Perry J.R.B., Huyghe J.R., Stringham H.M., et al. Identification of seven novel loci associated with amino acid levels using single-variant and gene-based tests in 8545 Finnish men from the METSIM study. Hum. Mol. Genet. 2018;27:1664–1674. doi: 10.1093/hmg/ddy067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M., et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang H.M. 2020. APIGenome - Big Data Genomics Analysis Libraries & Tools. https://github.com/hyunminkang/apigenome. [Google Scholar]
- 39.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maples B.K., Gravel S., Kenny E.E., Bustamante C.D. RFMix: A Discriminative Modeling Approach for Rapid and Robust Local-Ancestry Inference. Am. J. Hum. Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Myers R.M. Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 42.Taliun D., Chothani S.P., Schönherr S., Forer L., Boehnke M., Abecasis G.R., Wang C. LASER server: ancestry tracing with genotypes or sequence reads. Bioinformatics. 2017;33:2056–2058. doi: 10.1093/bioinformatics/btx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Browning B.L. 2021. Beagle 5.3. https://faculty.washington.edu/browning/beagle/b5_3.html. [Google Scholar]
- 45.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7–015. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purcell S.M., Chang C.C. 2022. PLINK 2.0. https://www.cog-genomics.org/plink/2.0/ [Google Scholar]
- 48.Stasinopoulos D.M., Rigby R.A. Generalized Additive Models for Location Scale and Shape (GAMLSS) in R. J. Stat. Softw. 2008;23:1–46. [Google Scholar]
- 49.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ganel L., Chen L., Christ R., Vangipurapu J., Young E., Das I., Kanchi K., Larson D., Regier A., Abel H., et al. Mitochondrial genome copy number measured by DNA sequencing in human blood is strongly associated with metabolic traits via cell-type composition differences. Hum. Genomics. 2021;15:34. doi: 10.1186/s40246-021-00335-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Auer P.L., Johnsen J.M., Johnson A.D., Logsdon B.A., Lange L.A., Nalls M.A., Zhang G., Franceschini N., Fox K., Lange E.M., et al. Imputation of Exome Sequence Variants into Population- Based Samples and Blood-Cell-Trait-Associated Loci in African Americans: NHLBI GO Exome Sequencing Project. Am. J. Hum. Genet. 2012;91:794–808. doi: 10.1016/j.ajhg.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parra E.J., Marcini A., Akey J., Martinson J., Batzer M.A., Cooper R., Forrester T., Allison D.B., Deka R., Ferrell R.E., Shriver M.D. Estimating African American Admixture Proportions by Use of Population-Specific Alleles. Am. J. Hum. Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bryc K., Velez C., Karafet T., Moreno-Estrada A., Reynolds A., Auton A., Hammer M., Bustamante C.D., Ostrer H. Genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc. Natl. Acad. Sci. USA. 2010;107:8954–8961. doi: 10.1073/pnas.0914618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Bakker P.I.W., Mcvean G., Sabeti P.C., Miretti M.M., Green T., Marchini J., Ke X., Monsuur A.J., Whittaker P., Delgado M., et al. A high resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tennessen J.A., Bigham A.W., O’Connor T.D., Fu W., Kenny E.E., Gravel S., McGee S., Do R., Liu X., Jun G., et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun Q., Liu W., Rosen J.D., Huang L., Pace R.G., Dang H., Gallins P.J., Blue E.E., Ling H., Corvol H., et al. Leveraging TOPMed imputation server and constructing a cohort-specific imputation reference panel to enhance genotype imputation among cystic fibrosis patients. HGG Adv. 2022;3:100090. doi: 10.1016/j.xhgg.2022.100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wojcik G.L., Fuchsberger C., Taliun D., Welch R., Martin A.R., Shringarpure S., Carlson C.S., Abecasis G., Kang H.M., Boehnke M., et al. Imputation-Aware Tag SNP Selection To Improve Power for Large-Scale, Multi-ethnic Association Studies. G3. 2018;8:3255–3267. doi: 10.1534/g3.118.200502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotypes can be accessed through dbGaP for the BioMe (dbGaP: phs001644) and MLOF (dbGaP: phs001515) studies. METSIM and InPSYght genotype data are not currently available because of privacy restrictions. The variant-level r2 statistics generated during this study are available to download from RsqBrowser (https://imputationserver.sph.umich.edu/rsq-browser/downloads). Analysis code and scripts are available at https://github.com/schanks/imputation_rsqbrowser.