Summary
Neisseria meningitidis protects itself from complement-mediated killing by binding complement factor H (FH). Previous studies associated susceptibility to meningococcal disease (MD) with variation in CFH, but the causal variants and underlying mechanism remained unknown. Here we attempted to define the association more accurately by sequencing the CFH-CFHR locus and imputing missing genotypes in previously obtained GWAS datasets of MD-affected individuals of European ancestry and matched controls. We identified a CFHR3 SNP that provides protection from MD (rs75703017, p value = 1.1 × 10−16) by decreasing the concentration of FH in the blood (p value = 1.4 × 10−11). We subsequently used dual-luciferase studies and CRISPR gene editing to establish that deletion of rs75703017 increased FH expression in hepatocyte by preventing promotor inhibition. Our data suggest that reduced concentrations of FH in the blood confer protection from MD; with reduced access to FH, N. meningitidis is less able to shield itself from complement-mediated killing.
Keywords: Complement, factor H, Neisseria meningitidis, Meningococcal disease, Factor H-related protein, CFH, CFHR3, infectious disease, genotyping, in-depth sequencing
Neisseria meningitidis evades complement-mediated clearance by hijacking host complement regulator factor H (FH). Kumar et al. investigate the genetic variations in the CFH locus associating with meningococcal disease. A regulatory region in the adjacent CFHR3 gene controls CFH expression, thereby determining FH plasma amounts and susceptibility towards meningococcal disease.
Introduction
Meningitis and sepsis caused by Neisseria meningitidis remain amongst the most feared bacterial infections world-wide. Although immunization has decreased the incidence of invasive meningococcal disease (MD) in some countries, there are no vaccines effective against all serogroups, and the emergence of new serogroups and strains1,2 poses new challenges to international vaccination strategies. Epidemics and outbreaks continue to occur in many countries, particularly in the meningitis belt of sub-Saharan Africa.3, 4, 5, 6
A remarkable feature of N. meningitidis is that it is a harmless commensal for the majority of the world’s population and is carried in the nasopharynx repeatedly throughout life. Invasive disease occurs in 0.16–20 per 100,000 people in developed countries, but there is wide variation in incidence, and epidemics occur.1,2,7,8
There is good evidence that genetic factors play a role in MD.9, 10, 11 Rare Mendelian defects in complement genes are associated with familial MD.12,13 Our previous genome-wide association study (GWAS) identified an association between MD and a broad genomic region spanning complement factor H (CFH [MIM: 134370]) and the complement factor H-related protein (in genetic order; CFHR3 [MIM: 605336], CFHR1 [MIM: 134371], CFHR4 [MIM: 605337], CFHR2 [MIM: 600889], and CFHR5 [MIM: 608593]) genes.10 Identification of the causal gene and characterization of the functional variant(s) have been difficult because of the complexity of the region; CFH shows sequence similarity to the five adjacent CFHR genes on human chromosome 1.14
Factor H (FH) is a serum glycoprotein that is synthesized mostly in the liver and acts as a negative regulator of the alternative complement activation pathway.15 FH is a crucial factor in preventing host cell damage by uncontrolled complement activation,16 and genetic variation in CFH or the CFHR genes is associated with several diseases, including systemic lupus erythematosus (SLE [MIM: 152700]),17 glomerulonephritis,18 IgA nephropathy,19 atypical hemolytic uremic syndrome (aHUS [MIM: 235400])20 and age-related macular degeneration (AMD [MIM: 603075),21,22 although the mechanistic process leading to disease is unclear for all these diseases.
N. meningitidis expresses several membrane proteins that bind human FH; Neisserial surface protein A (NspA),23 Porin B2 (PorB2),24 Porin B3 (PorB3),25 and FH-binding protein (fHbp)26 and is believed to survive and replicate in human blood by using the surface bound FH in a “Trojan horse” process to inhibit complement-mediated killing. Genetically regulated differences in FH plasma concentrations might thus alter susceptibility to N. meningitidis. Furthermore, inhibition of complement by “hijacking” FH has been adopted as an immune evasion strategy by several pathogens, including fungi, parasites, and viruses next to bacteria (reviewed in27). We aimed to identify the mechanism underlying the association of variants within the CFH-CFHR region with susceptibility and resistance to MD.
Methods
Study sample sets
The design for our study and the composition of clinical cohorts are shown in Figure S1 and Table S1. Clinical details of individuals with MD in UK, Spanish, and other European cohorts have been reported previously, as have the diagnostic criteria, recruitment procedure, and ethical approvals10,28 (supplementary Appendix). 238 individuals with MD and 237 controls from the Central European cohort (CEC) were used for deep sequencing the CFH-CFHR region. Replication of the most significant SNPs was undertaken in 1,522 individuals with MD and 2,672 controls (755 individuals with MD and 1,253 controls from the UK, 279 individuals with MD and 395 controls from Central Europe, and 488 individuals with MD and 1,024 controls from Spain).10,11 Previously genome-wide-genotyped cohorts totaling 1,246 individuals with MD and 7,197 controls (472 individuals with MD and 4,614 controls from the UK; 358 individuals with MD and 1,770 controls from Central Europe;29 and 416 individuals with MD and 813 controls from Spain9,10) were newly imputed, and the data were used for a subsequent meta-analysis. Convalescent serum was available from 367 individuals with MD (308 UK, 59 Dutch) and 124 healthy, unrelated Dutch controls for measurement of FH and FHR-3 concentrations; of the 308 UK individuals with MD, 295 were included in protein quantitative trait loci (pQTL) analysis, together with 56 healthy, unrelated controls from Central Europe.
Sequencing and genotyping of the CFH-CFHR region
To identify functional variants driving the association with MD susceptibility, we devised a capture-targeted sequencing strategy with tiling arrays (designed by Roche NimbleGen) covering more than 85% of the CFH-CFHR region spanning 359 kb on chromosome 1 (chr1: 196,620,000–196,979,000, GRCh37/hg19) and then performed sequencing with Illumina HiSeq 2000 by using 100 bp paired-end reads (stage 1, see supplemental information). The average depth of sequencing was 227× (Figure S2). We validated the most significant SNPs (stage 2, see supplemental information) by using a Sequenom Multiplex MassArray (San Diego, USA).
Genetic association testing was carried out with Fisher’s exact test for rare SNPs (MAF < 1%) and logistic regression analysis for common SNPs and copy-number variants (CNVs) under an additive genetic model. To mitigate the effect of population stratification, we analyzed association of SNPs with MD separately in all three replication cohorts under the additive model and performed meta-analysis for both SNPs and CNVs by combining summary statistics of stage 1 (deep sequencing) and stage 2 (Sequenom validation) by using the Cochran-Mantel-Haenszel (CMH) test. For CNVs, all samples were combined and analyzed under a genotypic and additive model.
Detection of copy-number variation
CNVs were detected in the resequencing dataset (238 individuals with MD and 237 controls from Central Europe) with cnvCapSeq (version 0.1.230) and cross-validated by quantitative PCR in the same cohort (Taqman qPCR, Table S2). In the second-stage validation of the 51 SNPs across the Central Europe, UK, and Spanish cohorts, detection of CNVs was done with the Taqman qPCR assays. For pQTL data, multiplex ligation-dependent probe amplification (MLPA) and Taqman assays were used for identifying CNVs (supplemental information).
Genotype-phenotype correlation of SNPs and CNVs were analyzed for FH and FHR-3 concentrations via linear regression analysis. We used ANCOVA, with sex as a covariate, to estimate the overall difference in the protein concentrations across six genotype groups. Differences in protein concentrations between two genotype groups were evaluated by t test.
Imputation of genome-wide genotyped data
To confirm our resequencing analysis by using current genome assemblies, we re-analyzed our original UK GWAS data,10 including newly genome-wide-genotyped cohorts from Central Europe29 and Spain11 (stage 3, see supplemental information). After pre-processing (supplemental information), we used BEAGLE (version 5.130) to perform haplotype estimation and imputation of missing genotypes by utilizing alternately the Haplotype Reference Consortium (HRC [http://www.haplotype-reference-consortium.org]; HRC release 1.1 [https://ega-archive.org/datasets/EGAD00001002729]) and the 1000 Genomes Project phase 3 (1KGP [http://www.internationalgenome.org]) as reference genomes.
After extraction of the individually calculated allele dose, which is the sum of the two allele probabilities based on a hidden Markov model, we applied a univariate linear mixed-model algorithm (uLMM) using a centered relatedness matrix implemented in GEMMAsoftware (version 0.98.131) to perform genetic association testing for quantitative traits under an additive model. To additionally account for population stratification, we used the first two or four principal components (PCs; Figure S4) as covariates in each individual cohort. Furthermore, the genomic control function implemented in the GWAMA software (version 2.2.232) was used for the subsequent meta-analysis of the single summary statistics, resulting in an overall genomic control lambda (λGC) of 1.002 (95% CI 0.094–1.010) when all variants were used and 1.007 (95% CI 0.971–1.0432) when only genotyped variants were used (Figure S5).
Serum concentrations of FH and FHR-3
FH and FHR-3 concentrations were determined by specific ELISAs as previously described.33 In brief, the antigen was captured with monoclonal antibody anti-FH.16 and anti-FHR-3.1 for FH and FHR-3, respectively (Sanquin Research, Amsterdam, The Netherlands). Bound FH was subsequently detected by the use of polyclonal goat anti-human FH antibodies, and bound FHR-3 was detected with monoclonal anti-FHR-3.4 (Sanquin Research).
Differentiation of human embryonic stem cells
Wild-type and CRISPR/Cas-targeted H1 human embryonic stem (hES) cells were differentiated to hepatocyte-like cells over 18 days as previously described.34
Genome editing of differentiated hepatocytes by CRISPR/Cas9
Guide RNAs flanking the liver-specific regulatory region of interest in CFHR3 were designed, incorporated in plasmids, and transfected via electroporation into H1 hES cells (Table S3). After incubation for two days, Clover+ cells were seeded at 500–1,000 cells per well of a 6-well plate. After 2-3 weeks culture, single colonies were picked and expanded for screening. Deletion of liver specific regulatory region was determined by PCR with primers spanning the targeted region (Table S3). Confirmed deletion clones and wild-type controls were used for detecting RNA expression by RT-PCR (see supplemental information).
Results
Fine mapping of the CFH-CFHR region identifies CFHR3 as the lead association
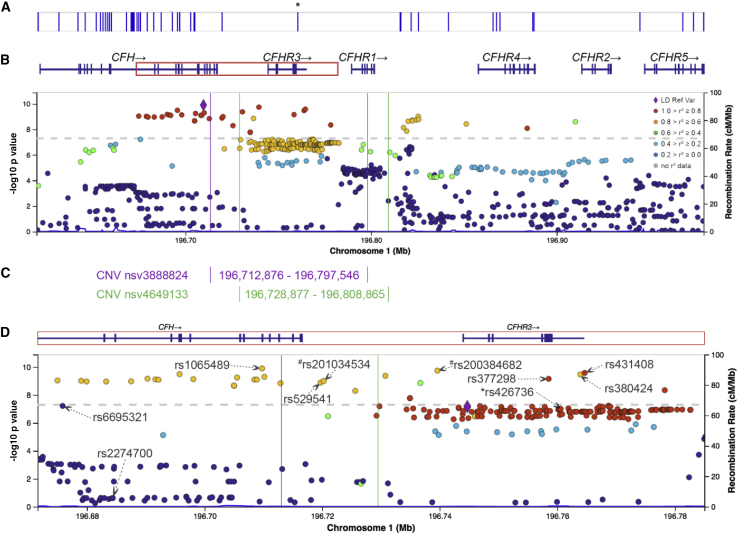
Deep sequencing of the CFH-CFHR region in 238 individuals with MD and 237 healthy controls identified 4,369 SNPs after application of stringent quality-control filters (Table S4). The strongest signal of association was identified on CFHR3 in a region with high linkage disequilibrium (LD, D′ = 0.92) with the previously reported lead variant, rs1065489 in CFH.10,11,35 The 51 SNPs with the strongest association with MD were selected for validation, and 44 SNPs were successfully typed (Table S5) in the UK, Spanish, and Central European cohorts (n = 4,194). 13 SNPs, in a tight LD block within CFHR3, achieved genome-wide significance in the meta-analysis (Figure 1, Table 1), confirming the genetic association with CFHR3. The lead SNP (Table 1), rs75703017 (p value = 1.1 × 10−16), located in intron 1 of CFHR3, showed consistent odds ratios (OR = 0.62) for susceptibility to MD in all cohorts, indicating a protective effect (Figure 2).
Figure 1.
Fine mapping by sequencing of the CFH-CFHR locus
Plot showing association results of all the SNPs (arranged according to their GRCh37/hg19 build chromosomal position on the x axis) from deep sequencing (circle) and from meta-analysis (square) with combined stage 1 and stage 2 cohorts. The top SNP from the analysis is labeled (rs75703017). The color intensity of each symbol reflects the extent of LD with the top GWAS SNP.
Table 1.
Unadjusted p values and ORs of top SNPs from sequencing, genotyping, and the combined analysis
| SNP ID |
Sequence |
Replication |
Combined CMHa |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Central Europe |
Central Europe |
UK |
Spain |
|||||||||||||||
| MAF MD | MAF con | p value | OR | MAF MD | MAF Con | p value | OR | MAF MD | MAF con | p value | OR | MAF MD | MAF Con | p value | OR | p value | OR | |
| rs75703017 | 0.12 | 0.21 | 3.11 × 10−3 | 0.62 | 0.14 | 0.21 | 3.60 × 10−3 | 0.68 | 0.14 | 0.21 | 8.34 × 10−6 | 0.71 | 0.18 | 0.26 | 2.26 × 10−4 | 0.72 | 1.11 × 10−16 | 0.62 |
| rs620015 | 0.13 | 0.21 | 6.18 × 10−3 | 0.65 | 0.14 | 0.22 | 3.62 × 10−3 | 0.68 | 0.15 | 0.21 | 2.27 × 10−5 | 0.73 | 0.20 | 0.26 | 1.01 × 10−3 | 0.75 | 9.55 × 10−15 | 0.64 |
| rs387107 | 0.14 | 0.21 | 9.35 × 10−3 | 0.67 | 0.14 | 0.21 | 4.49 × 10−3 | 0.68 | 0.15 | 0.21 | 4.97 × 10−5 | 0.74 | 0.20 | 0.27 | 3.11 × 10−4 | 0.73 | 1.24 × 10−14 | 0.65 |
| rs385390 | 0.14 | 0.22 | 6.04 × 10−3 | 0.65 | 0.15 | 0.22 | 9.74 × 10−3 | 0.71 | 0.15 | 0.21 | 1.50 × 10−5 | 0.72 | 0.20 | 0.27 | 7.11 × 10−4 | 0.75 | 1.35 × 10−14 | 0.65 |
| rs12409571 | 0.13 | 0.21 | 5.04 × 10−3 | 0.64 | 0.14 | 0.21 | 8.20 × 10−3 | 0.70 | 0.15 | 0.20 | 1.10 × 10−4 | 0.75 | 0.19 | 0.26 | 2.46 × 10−4 | 0.73 | 2.38 × 10−14 | 0.65 |
| rs425524 | 0.13 | 0.21 | 7.46 × 10−3 | 0.65 | 0.36 | 0.45 | 4.97 × 10−3 | 0.73 | 0.31 | 0.42 | 2.63 × 10−9 | 0.66 | 0.46 | 0.50 | 9.96 × 10−2 | 0.87 | 8.26 × 10−14 | 0.69 |
| rs401188 | 0.13 | 0.21 | 5.98 × 10−3 | 0.65 | 0.22 | 0.25 | 3.36 × 10−1 | 0.87 | 0.20 | 0.34 | 1.14 × 10−13 | 0.46 | 0.34 | 0.38 | 3.22 × 10−2 | 0.76 | 3.32 × 10−13 | 0.64 |
| rs1738741 | 0.14 | 0.21 | 9.35 × 10−3 | 0.67 | 0.14 | 0.21 | 1.06 × 10−2 | 0.71 | 0.15 | 0.21 | 1.39 × 10−5 | 0.72 | 0.22 | 0.27 | 1.43 × 10−2 | 0.81 | 1.54 × 10−12 | 0.67 |
| rs376841 | 0.13 | 0.21 | 4.42 × 10−3 | 0.64 | 0.10 | 0.16 | 1.55 × 10−3 | 0.56 | 0.10 | 0.15 | 1.29 × 10−5 | 0.63 | 0.14 | 0.18 | 4.74 × 10−4 | 0.65 | 3.06 × 10−11 | 0.66 |
| rs1329423 | 0.19 | 0.27 | 4.10 × 10−3 | 0.63 | 0.21 | 0.26 | 3.43 × 10−2 | 0.75 | 0.21 | 0.25 | 9.72 × 10−4 | 0.78 | 0.23 | 0.30 | 7.31 × 10−5 | 0.70 | 5.07 × 10−10 | 0.73 |
| rs11807997 | 0.13 | 0.21 | 5.12 × 10−3 | 0.64 | 0.19 | 0.18 | 8.97 × 10−1 | 1.02 | 0.15 | 0.21 | 1.24 × 10−4 | 0.74 | 0.19 | 0.25 | 1.12 × 10−3 | 0.75 | 1.34 × 10−9 | 0.71 |
| rs12408446 | 0.13 | 0.20 | 7.96 × 10−3 | 0.65 | 0.20 | 0.18 | 5.19 × 10−1 | 1.08 | 0.15 | 0.21 | 8.96 × 10−5 | 0.74 | 0.19 | 0.25 | 1.36 × 10−3 | 0.76 | 6.86 × 10−9 | 0.72 |
| rs116249058 | 0.14 | 0.21 | 7.53 × 10−3 | 0.66 | 0.12 | 0.14 | 2.54 × 10−1 | 0.83 | 0.12 | 0.15 | 1.74 × 10−2 | 0.81 | 0.14 | 0.21 | 8.85 × 10−5 | 0.67 | 9.59 × 10−9 | 0.70 |
CMH = Cochran-Mantel-Haenszel test; MD = individuals with meningococcal disease; con = healthy controls; MAF = minor-allele frequency; OR = odds ratio.
Figure 2.
Forest plot of the top SNP, rs75703017
Plot showing odds ratios (ORs) and confidence intervals in each of the four cohorts; the summary estimate is shown below. The dotted vertical line indicates no effect. Cohort names with numbers of MD individuals and healthy controls are on the left, ORs and 95% CI are on the right.
Imputation across the CFH-CFHR region confirms a broad region of association
Imputation of genome-wide genotyped data in three different European cohorts including 1,246 individuals with MD and 7,197 controls was complicated by the reported CNVs nsv3888824 (deletion) spanning 84,671 bases and resulting in a hybrid CFH/CFHR1 gene and nsv4649133 (deletion) spanning 79,989 bases and resulting in a complete deletion of CFHR3 and CFHR1 (Figure 3C). Using a subset of the HRC reference panel identified only four of the 13 SNPs found within and adjacent to CFHR3 by our resequencing work, whereas use of the 1KGP reference panel, which identified 11 SNPs with high confidence in a tight LD block closely around the genome-wide significance level of 5 × 10−8 (Table 2 and Figure 3). The usage of updated references and bioinformatic tools for the association mapping (Figure 3) allowed us to impute variants within the complex region in and adjacent to CFHR3, but we were still unable to refine the location of the causative variant within the CFH-CFHR locus as a result of the apparent tight LD. Indeed, we observed discrepancies between the FH associations reported by Sun et al.36 and our findings based on the 1KGP controls (Table S6); we suspect these discrepancies are due to differences in the imputation methods used for estimating the CNVs. Using imputation for the CNVs, we found peak association with MD outside the CNVs; whreas direct measurement of the CNVs shows the peak association to be within them (Figure 1).
Figure 3.
Fine mapping of the CFH-CFHR locus by GWAS
(A) Known variants reported in the NHGRI-EBI catalog of human genome-wide association studies. An asterisk represents the location of the rs426736 SNP within the CNVs and annotated as associated with MD.10
(B) The plot represents the genes located in the captured region (ranging from CFH to CFHR5) of the sequencing approach and shows association results of all variants (SNPs and InDels arranged according to their GRCh37/hg19 build chromosomal position on the x axis) from GWAS meta-analysis with the lead SNP, rs1065489, set as a reference variant (purple diamond). The color intensity of each symbol reflects the extent of LD with the top GWAS SNP.
(C) dbVar (https://www.ncbi.nlm.nih.gov/dbvar/)-annotated common CNVs with partial (nsv3888824 results in a CFH/CFHR1 hybrid gene) or complete (nsv4649133) deletion of CFHR3 and CFHR1.
(D) Plot showing association results of all variants (SNPs and InDels arranged according to their GRCh37/hg19 build chromosomal position on the x axis) from a GWAS meta-analysis with the lead SNP, rs75703017, from stages 1+2 set as a reference variant (purple diamond) mapping to a smaller genetic area focused on the start of the CNVs. Variants, which were either previously reported10,36 or notable findings from this study (stages 1–3) are annotated within the plot. #Annotated variants represent small InDels within the CNVs. Violet vertical and green lines represent the start of the CNVs nsv3888824 and nsv4649133, respectively.
Table 2.
p values and ORs of the top 20 variants from the GWAS meta-analysis and of the 11 SNPs from the sequencing and genotyping
| Variant ID | BP | Variant | Alt | Ref | MAF | OR | pmeta | I2 | DR2 |
|---|---|---|---|---|---|---|---|---|---|
| rs1065489a | 196,709,774 | missense | T | G | 0.17 | 0.69 | 1.25 × 10−10 | 0.74 | gt |
| rs200384682 | 196,739,608b | indel | CA | C | 0.18 | 0.69 | 1.81 × 10−10 | 0.72 | 0.90 |
| rs431408 | 196,764,663b | intron | G | T | 0.20 | 0.69 | 2.62 × 10−10 | 0.61 | 0.91 |
| rs3753396 | 196,695,742 | synonymous | G | A | 0.17 | 0.69 | 3.21 × 10−10 | 0.75 | gt |
| rs380424 | 196,763,939b | downstream | C | T | 0.18 | 0.69 | 3.38 × 10−10 | 0.70 | 0.97 |
| rs72482676 | 196,730,755b | intergenic | C | T | 0.16 | 0.68 | 4.21 × 10−10 | 0.73 | 0.96 |
| rs11582939a | 196,710,157 | intron | T | C | 0.17 | 0.70 | 4.99 × 10−10 | 0.72 | gt |
| rs742855a | 196,705,520 | intron | C | T | 0.17 | 0.70 | 5.66 × 10−10 | 0.76 | gt |
| rs141408533 | 196,690,281 | intron | T | TA | 0.17 | 0.70 | 6.77 × 10−10 | 0.75 | 1.00 |
| rs377298 | 196,758,541b | 3′ UTR | C | A | 0.19 | 0.70 | 6.98 × 10−10 | 0.61 | 0.93 |
| rs77302817 | 196,698,082 | indel | C | CTCTG | 0.17 | 0.70 | 7.32 × 10−10 | 0.76 | 1.00 |
| rs12402808 | 196,691,625 | intron | A | C | 0.17 | 0.70 | 7.65 × 10−10 | 0.76 | 1.00 |
| rs11799380 | 196,708,455 | intron | G | A | 0.17 | 0.70 | 7.91 × 10−10 | 0.73 | 1.00 |
| rs2336221 | 196,708,891 | intron | T | G | 0.17 | 0.70 | 7.91 × 10−10 | 0.73 | 1.00 |
| rs11801630 | 196,692,148 | intron | T | C | 0.17 | 0.70 | 8.03 × 10−10 | 0.75 | gt |
| rs1048663 | 196,674,982 | intron | A | G | 0.17 | 0.70 | 9.14 × 10−10 | 0.75 | 1.00 |
| rs74861068 | 196,825,380 | intron | A | G | 0.13 | 0.65 | 9.60 × 10−10 | 0.36 | 0.99 |
| rs74213209 | 196,679,010 | intron | G | A | 0.17 | 0.70 | 9.65 × 10−10 | 0.75 | 1.00 |
| rs201034534 | 196,720,267b | indel | A | AAAAC | 0.17 | 0.70 | 1.00 × 10−9 | 0.74 | 0.99 |
| rs10489456a | 196,687,515 | intron | A | G | 0.17 | 0.70 | 1.05 × 10−9 | 0.73 | 1.00 |
| rs12409571 | 196,768,726b | intergenic | G | A | 0.20 | 0.73 | 5.80 × 10−8 | 0.57 | 0.87 |
| rs116249058 | 196,767,218b | downstream | G | A | 0.20 | 0.74 | 6.37 × 10−8 | 0.53 | 0.87 |
| rs75703017 | 196,744,699b | intron | A | C | 0.20 | 0.74 | 6.80 × 10−8 | 0.56 | 0.88 |
| rs387107 | 196,757,881b | missense | T | G | 0.21 | 0.74 | 7.19 × 10−8 | 0.51 | 0.87 |
| rs11807997 | 196,743,213b | upstream | G | A | 0.20 | 0.74 | 8.68 × 10−8 | 0.56 | 0.87 |
| rs401188 | 196,757,083b | intron | T | C | 0.21 | 0.74 | 1.30 × 10−7 | 0.50 | 0.87 |
| rs12408446 | 196,741,197b | upstream | A | G | 0.21 | 0.74 | 1.38 × 10−7 | 0.53 | 0.88 |
| rs620015 | 196,748,676b | intron | G | A | 0.21 | 0.74 | 1.47 × 10−7 | 0.51 | 0.87 |
| rs376841 | 196,746,600b | intron | C | T | 0.21 | 0.75 | 1.53 × 10−7 | 0.51 | 0.87 |
| rs385390 | 196,743,927b | 5′ UTR | C | A | 0.21 | 0.75 | 2.36 × 10−7 | 0.50 | 0.87 |
| rs1329423 | 196,646,387 | exon | C | T | 0.26 | 0.79 | 4.09 × 10−7 | 0.71 | 0.99 |
BP = base position (GRCh37/hg19); MAF = minor-allele frequency; OR = odds ratio (estimated from LMM beta effects according to https://shiny.cnsgenomics.com/LMOR/); DR2 = mean dosage R-squared from the three single cohorts. gt = genotyped, no DR2 score. SNPs from the sequencing and genotyping are indicated in italics.
Previously reported as associated with MD susceptibility.
Within a CNV (nsv3888824, nsv4649133).
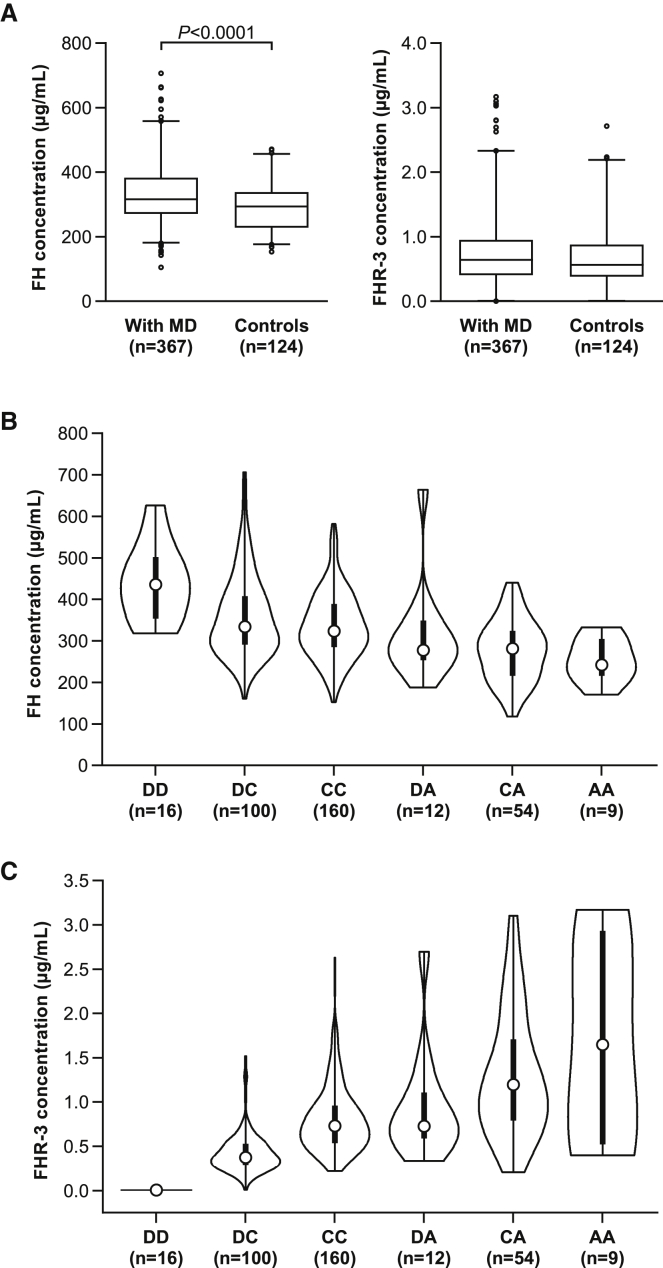
Serum concentrations of FH, but not FHR-3, are lower in controls than in individuals who survived MD
To explore the relationship between serum concentrations of FH, FHR-3, and MD, we measured concentrations in serum from individuals who survived invasive MD, at least six months after the acute illness. Serum concentrations of FH were significantly lower in healthy controls than in those with MD (Figure 4A). In contrast, FHR-3 serum concentrations were not significantly different between MD survivors and controls (Figure 4A).
Figure 4.
FH and FHR-3 concentrations in MD survivors
(A) Box plots with 95% range of FH serum concentrations (left panel), determined by ELISA. FH serum concentrations are increased (p < 0.0001, Mann-Whitney test) in MD survivors (n = 367) compared to healthy controls (n = 124), whereas FHR-3 serum concentrations are not significantly different between the two groups.
(B) Violin plotsof FH serum concentrations delineated by genotype for the SNP (rs75703017) most associated with FH concentrations; p = 1.41 × 10−11.
(C) Violin plot of FHR-3 serum concentrations delineated by genotype for the SNP (rs75703017), p value < 2 × 10−16.
For both (B) and (C), the x and y axes indicate genotypes and protein concentration (µg/mL), repsectively, with D = CFHR3/CFHR1 deletion, A = minor allele, and C = major allele for rs75703017. The white dots indicate median concentrations, the thick black bars indicate the interquartile range, and the thin black bars represent total range. p values were estimated by ANCOVA.
Low serum concentrations of FH are associated with both SNPs and CNVs in CFHR3
To investigate the effect of the top associated SNPs on the serum concentrations of FH and FHR-3, we undertook pQTL analysis, relating protein concentrations and genotype. The minor allele (A) of the lead SNP (rs75703017), shown to confer protection against MD (OR = 0.63), also showed the most significant association with lower FH serum concentrations (p = 1.4 × 10−11, Figure 4B), as confirmed in a pairwise comparison between genotypes carrying the minor allele (A) of rs75703017 (Figures 4B and 4C and Tables S7 and S11).
A common deletion spanning CFHR3 and CFHR1 (including rs75703017) has been shown to influence susceptibility to several inflammatory diseases.17,19,21,37 To establish whether this deletion was also associated with susceptibility to MD, we determined CNVs in the sequenced individuals with MD by using cnvcapSeq,38 which permits detection of CNVs in long-range targeted sequencing data. We then validated the findings by MLPA or qPCR analysis in a subset of samples (1,302 individuals with MD, 1,463 controls) from three European cohorts. Meta-analysis of CNV data revealed an overall lack of association (p = 0.76) between the CFHR3/CFHR1 deletion and susceptibility to MD (Table S8), as previously reported.10,35 Considering that the pQTL data indicated a dominant effect of the minor allele (A) of rs75703017, we performed a second comparison consisting of minor-allele carriers (A) vs. CFHR3/CFHR1 deletion allele carriers. In contrast to the initial overall lack of association, this second comparison revealed that deletion of CFHR3/CFHR1 was in fact associated with higher genetic risk of MD (p = 0.0081, Table S9) and increased FH serum concentrations. This positive genetic association with the CFHR3/CFHR1 deletion was only detected when the combination of three alleles (wild type allele C, minor allele A, and deletion D) in rs75703017 were taken into account. Meta-analysis of the quantitative-trait association removing all samples with the CFHR3/CFHR1 deletion did not modify the results (Figure S3), indicating that the association of rs75703017 persists regardless of CFHR3/CFHR1 deletion status (Table S11).
CFHR3 controls CFH expression through epigenetic long-range interaction
Having established the correlation between “protective genotypes” and lower serum concentrations of FH and between “risk genotypes” and higher concentrations of FH, we next investigated the epigenetic histone marks in various cell lines to provide information on the putative regulatory role of the potential functional SNP in CFHR3. Histone marks (H3K4me3 and H3K9ac) from the Roadmap epigenomics database indicated that all investigated hepatic cell lines have an active regulatory site within CFHR3. Furthermore, no other cell types (non-hepatic) tested showed any indication of regulatory regions, suggesting that this functional site might be specifically active in liver cells,39,40 which is concordant with the liver’s being the main FH-producing organ.14 In line with our hypothesis that there is a regulatory interaction between CFHR3 and CFH, we examined whether the homozygous deletion of CFHR3/CFHR1, carried by 3% of the European population, affected FH protein concentrations. Indeed, the deletion of CFHR3/CFHR1, identified by the lack of FHR-3 in serum, was associated with significantly higher FH protein concentrations (Figure 4B).
Dual-luciferase assays confirm liver-specific activity
To confirm the role of the rs75703017 minor allele identified in our fine mapping in regulating CFH activation, we compared luciferase activity of a liver cell line (HepG2) and of a line originating from embryonic kidney (HEK293T). We compared cells containing an empty vector (pGL3-empty) and three constructs containing the following: rs75703017 major allele C (pGL3-C); rs75703017 minor allele A (pGL3-A); and rs75703017 minor allele A together with minor alleles of two SNPs in close proximity (A of rs446868 and C of rs385390, pGL3-AAC) (Figure S7A, Table S12). Differential expression of a test reporter was detected in HepG2 (pGL3-A vs. pGL3-empty; p < 0.0001, Figure 5A) whereas HEK293T showed no significant change in expression (Figure S7B), supporting the liver-specific activity of the regulatory region.
Figure 5.
Functional validation of the top associated variant
(A) Effect of 2.8 kb regulatory sequence and the lead SNP, rs75703017, on reporter (firefly luciferase) activity in the HepG2 human liver cell line. pGL3-empty is an empty construct only containing a promoter, whereas pGL3-C contains the 2.8 kb regulatory sequence with major allele C at rs75703017, pGL3-A contains minor allele A at rs75703017, and pGL3-AAC contains minor allele A at rs75703017 and minor allele C at rs446868 and rs385390. Firefly luciferase concentrations were normalized to renilla luciferase activity for each sample, and all values were plotted relative to the pGL3-empty construct. The graph is representative of six independent experiments, and error bars represent means with standard deviation. Level of significance, calculated by t test, is indicated.
(B) Schematic depiction of CRISPR/Cas9 targeting of the CFHR3 liver-specific regulatory region comprising 2,844 base pairs (chr1: 196,743,825–196,746,668). Excision sites of each guide RNA, located around the SNP of interest (rs75703017), are indicated by scissors. The position of screening primers designed for selection of positive clones with excision of the targeted region are indicated by red arrows.
(C) n-fold change, relative to the wild type, of CFH transcript expression levels of CRISPR-edited CFHR3 (CRISPR-edited H1; genotype DD) carrying one copy of CFHR3 with allele C (WT-H1; genotype DC) in liver-differentiated H1 human embryonic stem cells. Expression was measured by qRT-PCR. The graph represents three independent experiments with two biological replicates (different sets of gRNA were used for targeting; KO1 WT-Cas9 gRNA 1 and 3 and KO2 nickase-Cas9 gRNA 1, 2, 3, and 4; see Table S2) and one technical replicate of KO1. Error bars represent means with standard deviation. Level of significance, calculated by t test, is indicated. (D = CFHR3/CFHR1 deletion, and C = major allele rs75703017).
Genome editing of the CFHR3 region via CRISPR/Cas9 confirms its regulatory role in FH expression
To confirm that the identified CFHR3 region regulates FH expression, we undertook genome editing by using CRISPR/Cas technology (Figures 5B and 5C, and supplemental Information). This required a liver cell line that constitutively expressed FH and carried at least one copy of CFHR3. Because none of the tested cell lines complied with both requirements, we differentiated human embryonic stem cells (H1 cell line) to hepatocytes.41 H1 cells do not express FH or FHR-3 and carry only one copy of CFHR3. Upon differentiation to hepatocytes (Figure S8), we detected FH expression (Figure 5C, Table S13) supporting the liver-specific expression reported previously.40 Deletion of a 2.8 kb region (chr1: 196,743,825–196,746,668) within CFHR3 containing rs75703017 via CRISPR/Cas9 in H1 cells (Figure 5B), followed by differentiation to hepatocytes, revealed enhanced FH expression, confirming the regulatory function of this region (Figure 5C). This finding is consistent with our t-test analysis (Table S7) of rs75703017 genotypes showing significant differential FH expression between deletion/major allele C (DC) and homozygous deletion (DD) genotypes (p = 6.8 × 10−3) and is further supported by Hi-C sequencing data, a strategy by which one can study three-dimensional architecture of the genome by coupling proximity-based ligation with massive parallel sequencing and that allows identification of long-range genomic interactions.42 In two cell lines a long-range interaction could be observed between CFH and the association interval in CFHR3 (Figure S9). Moreover, these results were concordant with in vivo data of individuals who were homozygous for the CFHR3/CFHR1 deletion and who showed increased FH concentrations (Figure 4B).
Discussion
Genetic variants within CFH and the CFHR genes have been associated with genetic susceptibility to a range of human diseases.17, 18, 19, 20, 21,43 Concordant with our work, deletion of CFHR3-CFHR1 has been reported to alter FH concentrations in serum and modify genetic susceptibility to disease,17,43,44 suggesting that a regulatory region controlling FH concentrations might exist at this locus.17,43 Identification of the causal variants underlying these associations has been difficult because of the complexity of the region; CNVs and sequence homology hamper genotyping and sequencing efforts. Thus, previous reports relied on surrogate markers to identify the deletion. Our strategy here allowed us to type the CNV and polymorphisms in the CFH-CFHR region, to narrow the regulatory element to a short sequence in intron 1 of CFHR3, and to identify the complex interplay of six possible genotypes at one SNP locus, including the lead SNP and copy-number variant, with FH serum concentrations. Recent development of specific monoclonal antibodies for FH and FH-related proteins33 allowed for an accurate detection of serum concentrations of FH and FHR-3.
We have fine mapped the complex CFH-CFHR region in individuals of European ancestry with MD and found that susceptibility and resistance to the disease is associated with a single SNP locus within intron 1 of CFHR3. This locus is affected by a well-known copy-number variant. Furthermore, by accounting for the protective effect of the minor allele (A) and the risk effect of the wild-type allele (C), we now demonstrate that the CFHR3 deletion does associate, although to a lesser extent than the identified SNP, with increased susceptibility for MD. Previous studies10,35 have missed this effect because their deletion analysis has combined the protective and risk alleles. Interestingly, the intronic lead SNP in CFHR3, rs75703017 (p = 1.1 × 10−16, OR = 0.63, 95% CI 0.55–0.71) lies in a liver-specific regulatory region that has been shown to loop and interact with CFH at the genomic level. This interaction seems to regulate CFH transcription activity. Protective homozygous rs75703017 A allele CFHR3 genotypes were associated with low FH serum concentrations (p = 1.41 × 10−11), the homozygous rs75703017C allele genotype had higher FH serum concentrations. In our analyses, deletion of this region through genome editing in human embryonic stem cells differentiated to hepatocytes also showed a substantial increase (p value < 0.05) of CFH transcript concentrations and expression of FH protein.
We showed that individuals surviving MD had higher serum concentrations of FH than controls and that low concentrations of FH were protective for MD. This is concordant with our previous report showing that addition of excess FH to blood increases the survival of N. meningitidis.45 Our data demonstrate that FH is a critical complement regulatory protein associated with MD susceptibility and that its serum concentrations are controlled through a cis-regulatory element in intron 1 of CFHR3, independent of FHR-3 concentrations. Whereas previous studies have suggested that competition between FHR-3 and FH for the fHbp on the surface of N. meningitidis could be the mechanism controlling susceptibility to MD,46 we suggest that serum concentrations of FHR-3 are too low to affect binding of the (on average) 132-fold more abundant FH to fHbp.33 Our genetic analysis confirms this assumption. In fact, our data indicate that the effect on MD susceptibility is predominantly defined by regulation of FH concentrations in serum by genetic variation in CFHR3, irrespective of serum FHR-3 concentrations. A schematic explanation of the inhibition of meningococcal bactericidal activity of complement in human blood by FH and its regulation by genetic variation in CFHR3 is shown in Figure 6. Importantly, our strongest genetic association is between low concentrations of serum FH and protection from disease, whereas high protein concentrations were less strongly associated with susceptibility. This suggests that N. meningitidis is able to harvest sufficient FH to prevent complement activity (thus ensuring serum survival) in most individuals and that high serum concentrations of FH only offer marginal additional bacterial protection as compared to average concentrations.
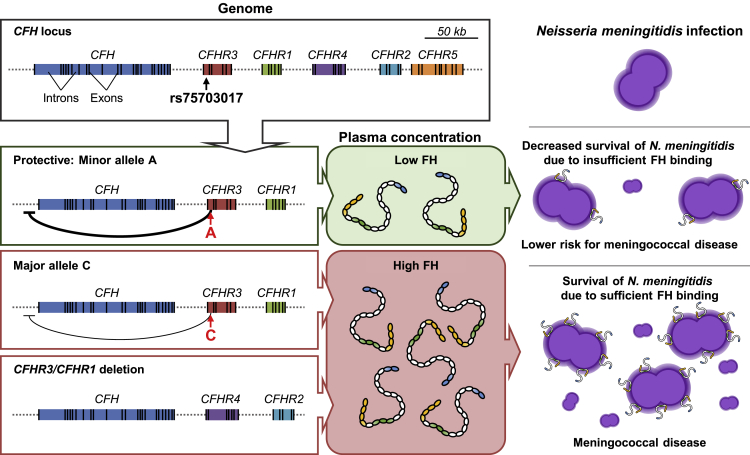
Figure 6.
Schematic model of the effect of CFHR3/CFHR1 deletion and SNP (rs75703017) on FH concentrations, interactions with N. meningitidis, and susceptibility to MD
The schematic diagram shows the structure of the gene region containing CFH and five CFHR genes. Carriers of the minor allele, A, on rs75703017 show the lowest FH concentrations. Increased concentrations of FH are found with the major allele, C, on rs7570317, whereas deletion of CFHR3/CFHR1 is associated with the highest concentrations of FH. Susceptibility to MD is driven by FH serum availability, which increases binding to the meningococcal surface protein fHbp. This binding results in FH’s impairing complement-mediated killing and allowing survival and growth of N. meningitidis in blood.
Our findings show that serum concentrations of FH are genetically regulated by a locus within CFHR3. Complement activation is an important immune protection mechanism against infections, but uncontrolled or excessive complement activation is potentially damaging to host cells and tissues. FH is a major regulator of complement-mediated damage to host cells47 as highlighted by the severe diseases associated with inadequate concentration or function of FH; such diseases include TTP/aHUS,20,37 glomerulonephritis,18 other inflammatory diseases,19 and AMD.21 Next to N. meningitidis, many other pathogens (see also Moore et al.27), including Streptococcus pneumoniae,48 group A streptococcus,49 Borrelia burgdorferi,50 and Plasmodium falciparum51 possess FH-binding proteins and might use FH to evade complement-mediated killing. The genomic regulation of serum FH concentration that we have identified through genetic variation in CFHR3 may thus be relevant to many other infectious and inflammatory diseases.
Acknowledgments
We would like to thank all individuals who participated in this study. All samples have been collected under country-specific institutional review boards (UK EC3263; Netherlands: 37986.091.11/RvB12.51320; Austria: 24-116 ex 11/12; Spain: 2011/298; Swiss: Cantonal Ethics Committee, Inselspital, University of Bern, no. KEK- 029/11). This work has been partially supported by the European Seventh Framework Programme for Research and Technological Development (FP7) under EUCLIDS Grant Agreement no. 279185, and by funding from the Agency for Science, Technology, and Research of Singapore (A∗STAR). Additional funding supporting the establishment of the MD cohorts used in this study are kindly acknowledged and listed in the supplemental information.
Declaration of interests
R.B.P., M.C.B., D.W., and T.W.K. are co-inventors of patents or patent applications describing FH potentiating antibodies and uses thereof. A.J.P. is chair of the UK Department of Health and Social Care’s Joint Committee on Vaccination and Immunisation. F.M.-T. has received honoraria from GSK group of companies, Pfizer Inc, Sanofi Pasteur, MSD, Seqirus, Biofabri, and Janssen for taking part in advisory boards and expert meetings and for acting as a speaker in congresses outside the scope of the submitted work. F.M.-T. has also acted as principal investigator in randomized controlled trials of the above-mentioned companies as well as Ablynx, Gilead, Regeneron, Roche, Abbott, Novavax, and MedImmune, with honoraria paid to his institution. All other authors declare no relevant competing interest related to the contents of this manuscript.
Published: September 1, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.08.001.
Contributor Information
Taco W. Kuijpers, Email: t.w.kuijpers@amsterdamumc.nl.
Sonia Davila, Email: gmssmdd@nus.edu.sg.
Supplemental information
‘C’ and ‘A’ refer to wild-type and mutant alleles, respectively, and 'D' refers to CFHR3/CFHR1 deletion.
Data and code availability
Summary statistics of the genotyped analysis generated during this study are available at LocusZoom (https://my.locuszoom.org/gwas/552110/) and FUMA (https://fuma.ctglab.nl/browse/469). Other datasets supporting the current study have not been deposited in a public repository but are available from the corresponding authors upon reasonable request.
References
- 1.Borrow R., Alarcón P., Carlos J., Caugant D.A., Christensen H., Debbag R., De Wals P., Echániz-Aviles G., Findlow J., Head C., et al. The Global Meningococcal Initiative: global epidemiology, the impact of vaccines on meningococcal disease and the importance of herd protection. Expert Rev. Vaccines. 2016;16:313–328. doi: 10.1080/14760584.2017.1258308. [DOI] [PubMed] [Google Scholar]
- 2.Sridhar S., Greenwood B., Head C., Plotkin S.A., Sáfadi M.A., Saha S., Taha M.-K., Tomori O., Gessner B.D. Global incidence of serogroup B invasive meningococcal disease: a systematic review. Lancet Infect. Dis. 2015;15:1334–1346. doi: 10.1016/S1473-3099(15)00217-0. [DOI] [PubMed] [Google Scholar]
- 3.Mustapha M.M., Marsh J.W., Harrison L.H. Global epidemiology of capsular group W meningococcal disease (1970-2015): Multifocal emergence and persistence of hypervirulent sequence type (ST)-11 clonal complex. Vaccine. 2016;34:1515–1523. doi: 10.1016/j.vaccine.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Xie O., Pollard A.J., Mueller J.E., Norheim G. Emergence of serogroup X meningococcal disease in Africa: need for a vaccine. Vaccine. 2013;31:2852–2861. doi: 10.1016/j.vaccine.2013.04.036. [DOI] [PubMed] [Google Scholar]
- 5.Chow J., Uadiale K., Bestman A., Kamau C., Caugant D.A., Shehu A., Greig J. Invasive meningococcal meningitis serogroup C outbreak in Northwest Nigeria, 2015 – Third consecutive outbreak of a new strain. PLoS Curr. 2016;8 doi: 10.1371/currents.outbreaks.06d10b6b4e690917d8b0a04268906143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell H., Parikh S.R., Borrow R., Kaczmarski E., Ramsay M.E., Ladhani S.N. Presentation with gastrointestinal symptoms and high case fatality associated with group W meningococcal disease (MenW) in teenagers, England, July 2015 to January 2016. Euro Surveill. 2016;21 doi: 10.2807/1560-7917.ES.2016.21.12.30175. [DOI] [PubMed] [Google Scholar]
- 7.Bårnes G.K., Kristiansen P.A., Beyene D., Workalemahu B., Fissiha P., Merdekios B., Bohlin J., Préziosi M.P., Aseffa A., Caugant D.A. Prevalence and epidemiology of meningococcal carriage in Southern Ethiopia prior to implementation of MenAfriVac, a conjugate vaccine. BMC Infect. Dis. 2016;16:639. doi: 10.1186/s12879-016-1975-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mbaeyi S., Pondo T., Blain A., Yankey D., Potts C., Cohn A., Hariri S., Shang N., MacNeil J.R. Incidence of meningococcal disease before and after implementation of quadrivalent meningococcal conjugate vaccine in the United States. JAMA Pediatr. 2020;174:843–851. doi: 10.1001/jamapediatrics.2020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haralambous E., Weiss H.A., Radalowicz A., Hibberd M.L., Booy R., Levin M. Sibling familial risk ratio of meningococcal disease in UK Caucasians. Epidemiol. Infect. 2003;130:413–418. [PMC free article] [PubMed] [Google Scholar]
- 10.Davila S., Wright V.J., Khor C.C., Sim K.S., Binder A., Breunis W.B., Inwald D., Nadel S., Betts H., Carrol E.D., et al. Genome-wide association study identifies variants in the CFH region associated with host susceptibility to meningococcal disease. Nat. Genet. 2010;42:772–776. doi: 10.1038/ng.640. [DOI] [PubMed] [Google Scholar]
- 11.Martinón-Torres F., Png E., Khor C.C., Davila S., Wright V.J., Sim K.S., Vega A., Fachal L., Inwald D., Nadel S., et al. Natural resistance to meningococcal disease related to CFH loci: Meta-analysis of genome-wide association studies. Sci. Rep. 2016;6:35842. doi: 10.1038/srep35842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sjöholm A.G., Jönsson G., Braconier J.H., Sturfelt G., Truedsson L. Complement deficiency and disease: an update. Mol. Immunol. 2006;43:78–85. doi: 10.1016/j.molimm.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 13.Fijen C.A., Kuijper E.J., te Bulte M.T., Daha M.R., Dankert J. Assessment of complement deficiency in patients with meningococcal disease in the Netherlands. Clin. Infect. Dis. 1999;28:98–105. doi: 10.1086/515075. [DOI] [PubMed] [Google Scholar]
- 14.Józsi M., Zipfel P.F. Factor H family proteins and human diseases. Trends Immunol. 2008;29:380–387. doi: 10.1016/j.it.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Blaum B.S. The lectin self of complement factor H. Curr. Opin. Struct. Biol. 2017;44:111–118. doi: 10.1016/j.sbi.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Parente R., Clark S.J., Inforzato A., Day A.J. Complement factor H in host defense and immune evasion. Cell. Mol. Life Sci. 2017;74:1605–1624. doi: 10.1007/s00018-016-2418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J., Wu H., Khosravi M., Cui H., Qian X., Kelly J.A., Kaufman K.M., Langefeld C.D., Williams A.H., Comeau M.E., et al. Association of genetic variants in complement factor H and factor H-related genes with systemic lupus erythematosus susceptibility. PLoS Genet. 2011;7:e1002079. doi: 10.1371/journal.pgen.1002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abrera-Abeleda M.A., Nishimura C., Smith J.L.H., Sethi S., McRae J.L., Murphy B.F., Silvestri G., Skerka C., Józsi M., Zipfel P.F., et al. Variations in the complement regulatory genes factor H (CFH) and factor H related 5 (CFHR5) are associated with membranoproliferative glomerulonephritis type II (dense deposit disease) J. Med. Genet. 2006;43:582–589. doi: 10.1136/jmg.2005.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiryluk K., Li Y., Scolari F., Sanna-Cherchi S., Choi M., Verbitsky M., Fasel D., Lata S., Prakash S., Shapiro S., et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat. Genet. 2014;46:1187–1196. doi: 10.1038/ng.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickering M.C., de Jorge E.G., Martinez-Barricarte R., Recalde S., Garcia-Layana A., Rose K.L., Moss J., Walport M.J., Cook H.T., de Córdoba S.R., Botto M. Spontaneous hemolytic uremic syndrome triggered by complement factor H lacking surface recognition domains. J. Exp. Med. 2007;204:1249–1256. doi: 10.1084/jem.20070301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes A.E., Orr N., Esfandiary H., Diaz-Torres M., Goodship T., Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat. Genet. 2006;38:1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- 22.Cheng C.Y., Yamashiro K., Jia Chen L., Ahn J., Huang L., Huang L., Cheung C.M.G., Miyake M., Cackett P.D., Yeo I.Y., et al. New loci and coding variants confer risk for age-related macular degeneration in East Asians. Nat. Commun. 2015;6:6817. doi: 10.1038/ncomms7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis L.A., Ngampasutadol J., Wallace R., Reid J.E.A., Vogel U., Ram S. The meningococcal vaccine candidate neisserial surface protein a (NspA) binds to factor H and enhances meningococcal resistance to complement. PLoS Pathog. 2010;6:e1001027. doi: 10.1371/journal.ppat.1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis L.A., Vu D.M., Vasudhev S., Shaughnessy J., Granoff D.M., Ram S. Factor h-dependent alternative pathway inhibition mediated by porin B contributes to virulence of Neisseria meningitidis. mBio. 2013;4:1–9. doi: 10.1128/mBio.00339-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giuntini S., Pajon R., Ram S., Granoff D.M. Binding of Complement Factor H to PorB3 and NspA Enhances Resistance of Neisseria meningitidis to Anti-Factor H Binding Protein Bactericidal Activity. Infect. Immun. 2015;83:1536–1545. doi: 10.1128/IAI.02984-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider M.C., Prosser B.E., Caesar J.J.E., Kugelberg E., Li S., Zhang Q., Quoraishi S., Lovett J.E., Deane J.E., Sim R.B., et al. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature. 2009;458:890–893. doi: 10.1038/nature07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore S.R., Menon S.S., Cortes C., Ferreira V.P. Hijacking Factor H for Complement Immune Evasion. Front. Immunol. 2021;12:602277–602323. doi: 10.3389/fimmu.2021.602277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agyeman P.K.A., Schlapbach L.J., Giannoni E., Stocker M., Posfay-Barbe K.M., Heininger U., Schindler M., Korten I., Konetzny G., Niederer-Loher A., et al. Epidemiology of blood culture-proven bacterial sepsis in children in Switzerland: a population-based cohort study. Lancet. Child Adolesc. Health. 2017;1:124–133. doi: 10.1016/S2352-4642(17)30010-X. [DOI] [PubMed] [Google Scholar]
- 29.Geishofer G., Binder A., Müller M., Zöhrer B., Resch B., Müller W., Faber J., Finn A., Endler G., Mannhalter C., et al. 4G/5G promoter polymorphism in the plasminogen-activator-inhibitor-1 gene in children with systemic meningococcaemia. Eur. J. Pediatr. 2005;164:486–490. doi: 10.1007/s00431-005-1673-4. [DOI] [PubMed] [Google Scholar]
- 30.Browning B.L., Zhou Y., Browning S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018;103:338–348. doi: 10.1016/j.ajhg.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mägi R., Morris A.P. GWAMA: software for genome-wide association meta-analysis. BMC Bioinf. 2010;11:1–6. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pouw R.B., Brouwer M.C., Geissler J., van Herpen L.V., Zeerleder S.S., Wuillemin W.A., Wouters D., Kuijpers T.W. Complement factor H-related protein 3 serum levels are low compared to factor H and mainly determined by gene copy number variation in CFHR3. PLoS One. 2016;11:e0152164. doi: 10.1371/journal.pone.0152164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ang L.T., Tan A.K.Y., Autio M.I., Goh S.H., Choo S.H., Lee K.L., Tan J., Pan B., Lee J.J.H., Lum J.J., et al. A roadmap for human liver differentiation from pluripotent stem cells. Cell Rep. 2018;22:2190–2205. doi: 10.1016/j.celrep.2018.01.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradley D.T., Bourke T.W., Fairley D.J., Borrow R., Shields M.D., Zipfel P.F., Hughes A.E. Susceptibility to invasive meningococcal disease: Polymorphism of complement system genes and Neisseria meningitidis factor H binding protein. PLoS One. 2015;10:e0120757. doi: 10.1371/journal.pone.0120757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun B.B., Maranville J.C., Peters J.E., Stacey D., Staley J.R., Blackshaw J., Burgess S., Jiang T., Paige E., Surendran P., et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zipfel P.F., Edey M., Heinen S., Józsi M., Richter H., Misselwitz J., Hoppe B., Routledge D., Strain L., Hughes A.E., et al. Deletion of complement factor H-related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet. 2007;3:e41. doi: 10.1371/journal.pgen.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bellos E., Kumar V., Lin C., Maggi J., Phua Z.Y., Cheng C.Y., Cheung C.M.G., Hibberd M.L., Wong T.Y., Coin L.J.M., Davila S. cnvCapSeq: detecting copy number variation in long-range targeted resequencing data. Nucleic Acids Res. 2014;42:e158. doi: 10.1093/nar/gku849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward L.D., Kellis M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loh K.M., Ang L.T., Zhang J., Kumar V., Ang J., Auyeong J.Q., Lee K.L., Choo S.H., Lim C.Y.Y., Nichane M., et al. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell. 2014;14:237–252. doi: 10.1016/j.stem.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Werken H.J.G., Landan G., Holwerda S.J.B., Hoichman M., Klous P., Chachik R., Splinter E., Valdes-Quezada C., Öz Y., Bouwman B.A.M., et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat. Methods. 2012;9:969–972. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- 43.Zhu L., Zhai Y.-L., Wang F.-M., Hou P., Lv J.-C., Xu D.-M., Shi S.-F., Liu L.-J., Yu F., Zhao M.-H., et al. Variants in complement factor H and complement factor H-related protein genes, CFHR3 and CFHR1, affect complement activation in IgA nephropathy. J. Am. Soc. Nephrol. 2014;26:1195–1204. doi: 10.1681/ASN.2014010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ansari M., Mckeigue P.M., Skerka C., Hayward C., Rudan I., Vitart V., Polasek O., Armbrecht A.-M., Yates J.R.W., Vatavuk Z., et al. Genetic influences on plasma CFH and CFHR1 concentrations and their role in susceptibility to age-related macular degeneration. Hum. Mol. Genet. 2013;22:4857–4869. doi: 10.1093/hmg/ddt336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haralambous E., Dolly S.O., Hibberd M.L., Litt D.J., Udalova I.A., O’dwyer C., Langford P.R., Simon Kroll J., Levin M. Factor H, a regulator of complement activity, is a major determinant of meningococcal disease susceptibility in UK Caucasian patients. Scand. J. Infect. Dis. 2006;38:764–771. doi: 10.1080/00365540600643203. [DOI] [PubMed] [Google Scholar]
- 46.Caesar J.J., Lavender H., Ward P.N., Exley R.M., Eaton J., Chittock E., Malik T.H., Goiecoechea De Jorge E., Pickering M.C., Tang C.M., Lea S.M. Competition between antagonistic complement factors for a single protein on N. meningitidis rules disease susceptibility. Elife. 2014;3:e04008. doi: 10.7554/eLife.04008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferluga J., Kouser L., Murugaiah V., Sim R.B., Kishore U. Potential influences of complement factor H in autoimmune inflammatory and thrombotic disorders. Mol. Immunol. 2017;84:84–106. doi: 10.1016/j.molimm.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 48.Janulczyk R., Iannelli F., Sjöholm A.G., Pozzi G., Björck L. Hic, a novel surface protein of Streptococcus pneumoniae that interferes with complement function. J. Biol. Chem. 2000;275:37257–37263. doi: 10.1074/jbc.M004572200. [DOI] [PubMed] [Google Scholar]
- 49.Haapasalo K., Jarva H., Siljander T., Tewodros W., Vuopio-Varkila J., Jokiranta T.S. Complement factor H allotype 402H is associated with increased C3b opsonization and phagocytosis of Streptococcus pyogenes. Mol. Microbiol. 2008;70:583–594. doi: 10.1111/j.1365-2958.2008.06347.x. [DOI] [PubMed] [Google Scholar]
- 50.Kraiczy P., Skerka C., Kirschfink M., Brade V., Zipfel P.F. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and factor H. Eur. J. Immunol. 2001;31:1674–1684. doi: 10.1002/1521-4141(200106)31:6<1674::aid-immu1674>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 51.Kennedy A.T., Schmidt C.Q., Thompson J.K., Weiss G.E., Taechalertpaisarn T., Gilson P.R., Barlow P.N., Crabb B.S., Cowman A.F., Tham W.-H. Recruitment of factor H as a novel complement evasion strategy for blood-stage Plasmodium falciparum infection. J. Immunol. 2016;196:1239–1248. doi: 10.4049/jimmunol.1501581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
‘C’ and ‘A’ refer to wild-type and mutant alleles, respectively, and 'D' refers to CFHR3/CFHR1 deletion.
Data Availability Statement
Summary statistics of the genotyped analysis generated during this study are available at LocusZoom (https://my.locuszoom.org/gwas/552110/) and FUMA (https://fuma.ctglab.nl/browse/469). Other datasets supporting the current study have not been deposited in a public repository but are available from the corresponding authors upon reasonable request.