Abstract
Using the fruit fly Drosophila melanogaster as model host, we have identified mutants of the bacterium Pseudomonas aeruginosa with reduced virulence. Strikingly, all strains strongly impaired in fly killing also lacked twitching motility; most such strains had a mutation in pilGHIJKL chpABCDE, a gene cluster known to be required for twitching motility and potentially encoding a signal transduction system. The pil chp genes appear to control the expression of additional virulence factors, however, since the wild-type fly-killing phenotype of a subset of mutants isolated on the basis of their compact colony morphology indicated that twitching motility itself was not required for full virulence in the fly.
The extraordinary versatility of bacteria in the genus Pseudomonas is reflected in the diversity of studies using these organisms: studies of biochemical pathway evolution (27), biodegradation of toxic waste (31), and pathogenesis. There is a pressing need to incorporate the results of all these investigations into an improved understanding of the biology of Pseudomonas aeruginosa, a bacterium so versatile that it is not only a major cause of opportunistic human infection but also virulent toward plants (29, 30), insects (4, 17, 30), and the soil-dwelling nematode worm Caenorhabditis elegans (9, 23). Because of this broad spectrum of virulence, a variety of models that allow efficient screening of bacterial mutants have been developed (18). Recently, for instance, P. aeruginosa genes required for killing the nematode C. elegans have been identified (23, 41; L. A. Gallagher and C. Manoil, unpublished data), as has a worm gene required for sensitivity to killing (9).
Innate immunity in the fruit fly Drosophila melanogaster protects it from overwhelming infection in a life rich in interactions with microorganisms (3). The signal transduction cascade underlying this system is under intense investigation, and these studies have revealed striking similarities to the mammalian innate immune response (2, 16). In both insects and mammals, Toll family receptors signal through Rel family transactivators (2, 16), mediating responses that are specific to different classes of pathogens (22). These responses include secretion of antimicrobial peptides (3, 20). The fruit fly thus may be an especially good model organism to reveal the complex interactions between P. aeruginosa virulence factors and host defenses which underlie human disease such as the chronic respiratory infections of cystic fibrosis patients (15, 36).
P. aeruginosa PAO1 kills fruit flies.
Previous studies showed that P. aeruginosa is a particularly virulent pathogen of fruit flies; pricking with a needle dipped into a culture of P. aeruginosa strain α1 is lethal: the bacteria grow exponentially within the fly until and even after the death of the fly (4). This observation suggested that the fruit fly could be used to screen for Pseudomonas mutants with reduced virulence. We therefore tested whether PAO1, the best-characterized strain of P. aeruginosa (38), was also proficient at fly killing.
PAO1 (from the laboratory of B. Iglewski) was grown with 1 mM succinate as the carbon source in M63 minimal medium (35) without the added iron and with 1 mM MgSO4. Cultures were grown for 12 h to stationary phase in a rotary shaker at 37°C. The fruit fly D. melanogaster Canton S was grown with standard cornflour-molasses medium at 25°C. Adult female flies 2 to 4 days old were pricked in the dorsal thorax with a 25-gauge needle dipped directly into the PAO1 culture; the females were then returned to standard fly culture vials with food. Flies generally died 16 to 28 h after infection with wild-type bacteria (Fig. 1) and became noticeably lethargic 1 h before death. Female flies were used since pricking with a sterile needle was consistently harmless while up to 15% of male flies died, presumably because of their smaller size.
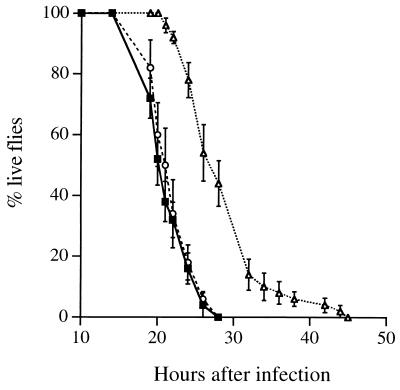
FIG. 1.
Time course of infection of wild-type flies with PAO1 (■), a pilD mutant (○), and a chpA mutant (▵). Bacteria were grown as described for PAO1 and introduced into flies by pricking with a syringe needle. The chpA37 mutant (see the legend to Fig. 2) was used for this and subsequent experiments. The values plotted are the averages of five replicate experiments, each with 10 flies, and the standard error of the mean is shown for each point.
Immediately after infection and at various subsequent time points, individual flies were ground with a Teflon pestle in an Eppendorf tube with 100 μl of 10 mM MgSO4 and serial dilutions of the homogenate were spread on Luria-Bertani (LB) agar to determine viable bacterial cell counts. These experiments showed that 400 to 2,000 PAO1 cells were introduced into and onto the fly by pricking and that the flies died when the bacterial titer reached 1 × 106 to 40 × 106 cells. These results are consistent with those of the previous study using a different strain of P. aeruginosa (4).
Screening for P. aeruginosa mutants impaired in fly killing.
P. aeruginosa PAO1 was mutagenized using plasmid pUT carrying transposon ISphoA/hah essentially as described for Escherichia coli (24) but with the following modifications: the chloramphenicol resistance gene in the transposable element had been replaced with a tetracycline resistance gene (Gallagher and Manoil, unpublished), mating recipient PAO1 cells were grown in 5 ml of LB broth for 12 h at 42°C without shaking, the mating duration was 30 min, and PAO1 cells carrying a chromosomal insertion of the transposable element were selected on LB agar containing 10 μg of chloramphenicol per ml (to counterselect E. coli) and 60 μg of tetracycline per ml after incubation for 48 h at 30°C. Single colonies were patched to a selective master plate and also used to inoculate 200-μl cultures of M63-succinate medium in a 96-well tissue culture dish. After growth for 12 h at 37°C without shaking, 100 μl from each well was transferred to the lid of a 48-well tissue culture dish (to avoid the floating film of cells in the unshaken culture) and three flies per PAO1 mutant were pricked with a 25-gauge needle dipped directly into this culture. Each set of three flies was then transferred to a well in a 48-well tissue culture dish capped with cotton and in which 200 μl of apple juice-dextrose agar had solidified as a slant. The dishes were incubated at 25°C. PAO1 mutants that failed to kill more than one of the three flies after 30 to 32 h were scored as potentially fly-killing impaired and retested.
Approximately 1,500 independent PAO1 ISphoA/hah insertion mutants were screened, yielding 54 candidates for strains with reduced virulence. Mutants which grew slowly might appear less virulent in the fly simply because of the growth defect. Of the 54 candidates, therefore, only 33 (2% of the total screened) were analyzed further, i.e., the strains that appeared to have a wild-type growth rate (based on colony size and time of appearance) on plates with M63-succinate medium.
PAO1 twitching-motility mutants are enriched for strains impaired in fly killing.
The 33 mutants were impaired in fly killing to various degrees. Eight mutants had a strong phenotype, with a delay in 50% killing of approximately 6 h and a delay in 100% killing of up to 18 h (typified by the chpA37 mutant in Fig. 1). Strikingly, all eight of these strains displayed the compact, smooth-edged colony morphology characteristic of mutants lacking twitching motility (1), a form of motility used by Pseudomonas to move on solid surfaces and requiring type IV pili (5, 34).
To explore the correlation between colony phenotype and the defect in fly killing, PAO1 twitching-motility mutants in addition to the 8 already assayed were identified by visual inspection of colonies, 11 from the 1,500 strains already generated and 30 from 4,500 newly generated ISphoA/hah PAO1 mutants. Of the total 6,000 mutant strains generated in this study, 49 appeared to lack twitching motility and 24 of the 49 were impaired in fly killing (data not shown). For 45 of these 49 strains, the mutated gene was successfully identified by a semirandom PCR protocol (6) using primers described previously (24) together with the PAO1 genome sequence (Pseudomonas Genome Project). Insertions were identified in each of the chromosomal loci known to contain twitching-motility genes (1, 33), except for the isolated gene pilF (Fig. 2). Only two mutants, independently isolated, had identical insertions (within pilR).
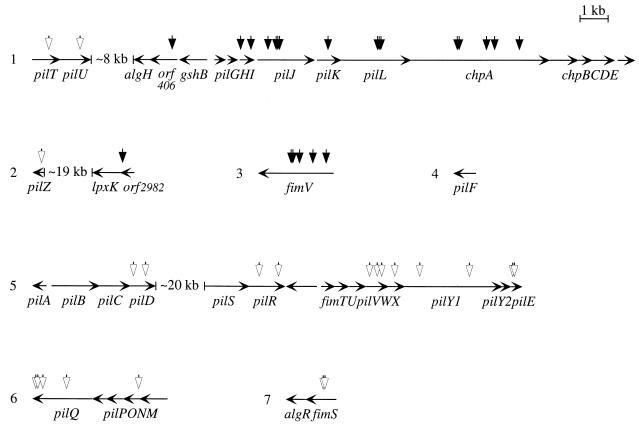
FIG. 2.
Chromosomal genes disrupted in PAO1 twitching-motility mutants. The seven depicted loci are dispersed along the PAO1 chromosome. Horizontal arrows denote the extent of individual genes and their direction of transcription (adapted from Fig. 2 in reference 1); the organization of the pil chp gene cluster is derived from nucleotide sequences with GenBank accession numbers L10831 (pilG), L22036 (pilHIJ), U11382 (pilK), and U79580 (pilL chpABCDE). The sites of insertion of the ISphoA/hah transposable element are indicated by vertical arrows. Black arrowheads denote mutations that impair fly killing (i.e., that delay 50% fly killing by 2 to 24 h relative to infection with wild-type bacteria), and white arrowheads denote mutations in strains that appear to have wild-type virulence in the fly. The strain with the chpA37 mutation, the second insertion from the 5′ end of chpA (at position 458351 in the PAO1 genome), was used for the experiment in Fig. 1 as well as subsequent experiments; the insertion in orf406 is at position 448168, the insertion in orf2982 is at position 3339667, and the insertion in the pilU mutant with a unique colony morphology is at position 438420 in the PAO1 genome. The first insertion from the 5′ end of pilR was identified in two independently generated mutants.
Twitching-motility mutants impaired in fly killing.
The 22 sequenced twitching-motility mutants also impaired in fly killing (Fig. 2) grew on plates with LB or M63-succinate media at a rate indistinguishable from PAO1 (based again on colony size and time of appearance). Of these 22 mutants, 15 had an insertion in pilGHIJKL chpABCDE (1, 10, 12, 38; C. Whitchurch, M. Young, A. Leech, A. Semmler, and J. Mattick, Pseudomonas '99: Biotechnology and Pathogenesis, abstr. S41, 1999), a cluster of genes most similar to the che genes for flagellum-mediated chemotaxis in E. coli and the frz and dif genes involved in type IV pilus-mediated social gliding motility in Myxococcus xanthus (25, 37, 40, 43, 48). The extent to which each of these 15 insertions affects the expression of genes downstream in the cluster is unknown (11). However, this study has identified multiple gene clusters required for full virulence in the fly (Fig. 2), and for the 15 pil chp mutants, the fly-killing phenotype is typified by that of the chpA37 mutant, with 50% fly killing delayed by approximately 6 h (Fig. 1). This strain was therefore used as a representative in subsequent experiments. An equivalent phenotype was observed for five mutants, each with a different insertion in fimV (Fig. 2), a newly discovered gene required for twitching motility (33). The amino acid sequence of FimV and the effect of FimV overexpression on cell shape suggest a role in peptidoglycan remodeling necessary for pilus biogenesis and function (33).
The remaining two mutants each had an insertion in a locus not described previously in the context of twitching motility: an open reading frame (ORF) that could encode a protein similar to various putative homologs of E. coli TonB (up to 36% amino acid identity over 277 residues) and an ORF that could encode a protein similar to various putative homologs of E. coli ExbD, an inner membrane protein that complexes with TonB for transport of receptor-bound substrates (up to 64% amino acid identity over 145 residues). The tonB-like ORF (designated orf406) is adjacent to the pil chp cluster and immediately upstream of algH (Fig. 2), encoding a global regulator affecting the expression of alginate as well as several quorum-sensing-controlled virulence factors (32); disruption of orf406 caused a delay of several hours in 50% fly killing (data not shown). The exbD-like ORF (designated orf2982) is immediately upstream of the PAO1 homolog of E. coli lpxK (Fig. 2), encoding lipid A 4′-kinase; disruption of this ORF caused a particularly strong phenotype, with 50% killing delayed by approximately 24 h (data not shown). For this mutant, as well as the chpA37 mutant and strains with a mutation in fimV or orf406, the number of viable bacterial cells used to infect flies (determined with fly homogenates made immediately after infection) varied within the range observed for PAO1 (data not shown).
Twitching-motility mutants not impaired in fly killing.
Genes with diverse roles in twitching motility (1) were disrupted in strains not impaired in fly killing (Fig. 2): pilR and fimS are regulatory genes representing one member of each of the two known two-component sensor-regulator pairs; pilQ, pilY1, and pilN are putative structural genes, as are pilW, pilX, and pilE, which encode pilin subunit (PilA)-like proteins; pilT and pilU encode putative nucleotide binding proteins which could mediate pilus retraction (5, 26, 47) as a basis for motility; and the role of pilZ in twitching motility is unknown. pilD encodes the leader peptidase, which processes components not only of the type IV pilus but also of the type II secretion apparatus, whose substrates include the exoenzyme virulence factors phospholipase C, elastase, and exotoxin A (39); it was therefore unexpected that the pilD mutant would be unimpaired in fly killing (Fig. 1). Also surprising was the observation that the pilU mutant had a unique colony morphology: cells grew slightly slower than did wild-type cells on LB plates and formed colonies with a dry wrinkled surface, distinct from the wet smooth surface of colonies of cells of each of the other mutants in this study. Although pilT and pilU encode homologous proteins, previous studies noted a difference in the virulence (7) and phage sensitivity (45) of pilT and pilU mutants.
A chpA mutant kills flies more slowly than PAO1 even though it grows at the same rate within the fly.
The pil chp genes have recently been proposed (Whitchurch et al., Pseudomonas '99) to function as a signal transduction pathway adding a layer of regulation to PAO1 virulence factor expression. We therefore analyzed the chpA37 mutant in more detail in the fly-killing assay. Individual batches of five flies infected with either PAO1 or the chpA37 mutant were homogenized at various time points before the first flies began to die. An infection with either strain followed an identical course: the number of viable bacterial cells in fly homogenates decreased over the first 4 h after infection and then increased exponentially, doubling approximately every 1.3 h (Fig. 3). The fact that individual data points, essentially representing separate experiments, consistently indicated the same bacterial growth rate within the fly, testifies to the reproducibility of the assay. Furthermore, although the number of viable bacterial cells in single-fly homogenates made immediately after pricking varied between 400 and 2,000, this variability was not sufficient to obscure a delay in fly killing; indeed, for the data shown in Fig. 3, the initial difference between the number of viable cells of the chpA37 mutant and of PAO1 was completely eliminated by 16 h after infection. Based on the colony morphology of cells of the chpA37 mutant recovered from the fly, twitching motility was not regained over the course of the infection.
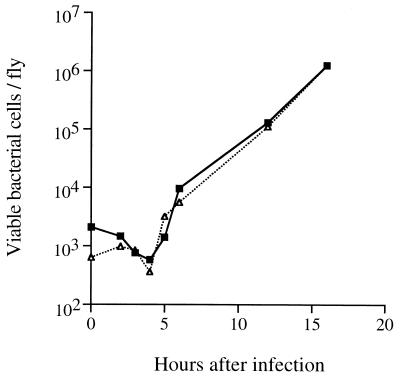
FIG. 3.
Growth of PAO1 and the chpA37 mutant in wild-type flies. Homogenates of batches of five flies were made at various time points after infection and before flies began dying. Homogenates were plated on LB agar to determine viable bacterial cell counts. The bacterial cultures used in this experiment were also used for one of the replicates in the experiment in Fig. 1. Three subsequent experiments gave equivalent results. Symbols: ■, PAO1; ▵, chpA37 mutant.
To compare the end points of infection with PAO1 and with the chpA37 mutant, 10 single flies infected with each strain were homogenized when their limb motion was no longer detectable with the unaided eye (which normally preceded death by less than 30 min). Although most of the first flies to die contained fewer bacterial cells than the last flies to die, the observed range for PAO1 (1 × 106 to 40 × 106 cells) was distinct from that of the chpA37 mutant (10 × 106 to 90 × 106 cells). The generally higher titer of the mutant is consistent with the delay in fly killing seen with this strain (Fig. 1) and suggests that although it grew within the fly as fast as PAO1 (Fig. 3), the chpA37 mutant was less virulent.
A chpA mutant rapidly kills flies that have defects in innate immunity.
Flies have a sophisticated innate immune system evolutionarily related to that of mammals (16). Since it seemed possible that the chpA37 mutant was compromised in evading the fly immune response, we assayed this strain along with PAO1 for its ability to kill Bc,imd mutant flies. Bc is a mutation blocking the phenoloxidase cascade, by which foreign bodies are sequestered by melanization in the fly circulatory system, and imd is a mutation that reduces the expression of antibacterial peptides and renders flies more susceptible specifically to infection with bacteria as opposed to fungi (21, 22). Flies with either mutation are more readily killed by E. coli (21, 22). The distinction between the killing kinetics of PAO1 and the chpA37 mutant was reduced in infections of Bc,imd mutant flies; in particular, the average time at which the last mutant fly died was the same for an infection with either bacterial strain (Fig. 4). When the experiment was repeated using bacterial cultures diluted 10-fold in 10 mM MgSO4 before infection, a similar effect was observed: the average delay in 50% fly killing for flies infected with the chpA37 mutant with respect to PAO1 decreased from 3.7 h for wild-type flies (standard error of the mean, 0.62) to 0.8 h for Bc,imd flies (standard error of the mean, 0.73). Although the observed reductions are somewhat subtle, they are nevertheless consistent with ChpA being important for overcoming fly innate immunity.
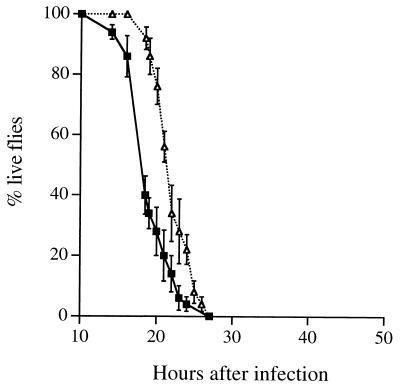
FIG. 4.
Time course of infection of Bc,imd/Bc,imd mutant flies with PAO1 and the chpA37 mutant. Bacteria were grown and introduced into flies as in the experiment in Fig. 1. The values plotted are the averages of five replicate experiments, each with 10 flies, and the standard error of the mean is shown for each point. Symbols: ■, PAO1; ▵, chpA37.
Parallels between the biology of P. aeruginosa and that of M. xanthus.
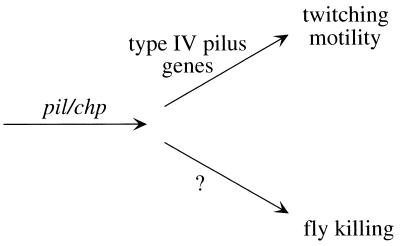
This study identified P. aeruginosa mutants with reduced virulence toward the fruit fly. All strains strongly impaired in fly killing also lacked twitching motility; most such strains had a mutation in pilGHIJKL chpABCDE, a gene cluster which could encode a signal transduction pathway. An additional set of twitching-motility mutants generated in this study, however, was not impaired in fly killing (these strains were identified by their compact colony morphology, indicating a lack of surface spreading). Therefore, twitching motility itself is not required for full virulence in the fly, in turn suggesting that the pil chp genes control the expression of as yet unknown virulence factors which are the true determinants of fly killing (Fig. 5).
FIG. 5.
Model for the role of the pil chp gene cluster in both twitching motility and virulence in the fruit fly. The P. aeruginosa pilGHIJKL chpABCDE gene cluster could encode a signal transduction system and is required for twitching motility mediated by type IV pili (1, 12) and possibly other adaptations for surface growth. Such adaptations appear to include the expression of as yet undetermined virulence factors, since pil chp mutants are impaired in fly killing even though twitching motility is not required for full virulence in the fly.
The first description of the pil chp genes in PAO1 (10) emphasized their similarity to genes involved in gliding motility in M. xanthus, a nonflagellated soil bacterium that glides on surfaces as it hunts the bacteria that are its food. Subsequent studies have greatly expanded the parallels between these two organisms: twitching motility in P. aeruginosa has been shown to be genetically and morphologically equivalent to social gliding motility in M. xanthus (34); genes coordinately regulated with each motility include those for biosynthesis of exopolysaccharides, which could facilitate surface spreading (37), alginate for P. aeruginosa (46); cell-to-cell signaling is crucial for each motility in groups of bacterial cells (14, 37), quorum sensing for P. aeruginosa (14); and under stressful environmental conditions, each motility is required to form a structured community of resistant cells (28, 42), a biofilm for P. aeruginosa (8, 13, 28, 44). Incorporating models proposed for M. xanthus (40, 43, 48), the chemotaxis-like pil chp genes in PAO1 could encode a signal transduction system that controls adaptations for surface growth, adaptations that include twitching motility as well as expression of factors required for full virulence in the fruit fly (Fig. 5).
The pil chp signal transduction system is likely to be important for virulence not only in the fruit fly but also in mammals. This conclusion is supported by a study of P. aeruginosa genes mediating epithelial cell injury (19): part of one such gene was identified as being similar to E. coli cheA, and, using DNA sequences not available at the time of that study, the only exact match to this sequence in the PAO1 genome is chpA. The fruit fly thus may be a particularly good model host to both reveal and characterize new components of P. aeruginosa pathogenicity: this study has shown that bacterial mutants can be efficiently screened in wild-type flies and that individual strains then can be tested in mutant flies with specific defects in an innate immune system evolutionarily related to that of mammals.
Acknowledgments
We thank S. Jackson, M. Terayama, J. Dorman, R. French, K. James, and D. Tran for sharing their expertise with fruit flies and S. Lory for sharing his expertise with Pseudomonas. All PAO1 gene analysis was greatly facilitated by the online Pseudomonas aeruginosa Genome Database at the Centre for Molecular and Cellular Biology, University of Queensland.
This work was supported by National Science Foundation grants MCB-9905048 and IBN-9983207 to C.M. and C.B., respectively, and a Cystic Fibrosis Research Development Program postdoctoral fellowship to D.D.
REFERENCES
- 1.Alm R A, Mattick J S. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. doi: 10.1016/s0378-1119(96)00805-0. [DOI] [PubMed] [Google Scholar]
- 2.Anderson K V. Toll signaling pathways in the innate immune response. Curr Opin Immunol. 2000;12:13–19. doi: 10.1016/s0952-7915(99)00045-x. [DOI] [PubMed] [Google Scholar]
- 3.Boman H G. Antibacterial peptides: key components needed in immunity. Cell. 1991;65:205–207. doi: 10.1016/0092-8674(91)90154-q. [DOI] [PubMed] [Google Scholar]
- 4.Boman H G, Nilsson I, Rasmuson B. Inducible antibacterial defence system in Drosophila. Nature. 1972;237:232–235. doi: 10.1038/237232a0. [DOI] [PubMed] [Google Scholar]
- 5.Bradley D E. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol. 1980;26:146–160. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 6.Chun K T, Edenberg H J, Kelley M R, Goebl M G. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast. 1997;13:233–240. doi: 10.1002/(SICI)1097-0061(19970315)13:3<233::AID-YEA88>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 7.Comolli J C, Hauser A R, Waite L, Whitchurch C B, Mattick J S, Engel J N. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect Immun. 1999;67:3625–3630. doi: 10.1128/iai.67.7.3625-3630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 9.Darby C, Cosma C L, Thomas J H, Manoil C. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:15202–15207. doi: 10.1073/pnas.96.26.15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darzins A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1994;11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 11.Darzins A. The Pseudomonas aeruginosa pilK gene encodes a chemotactic methyltransferase (CheR) homologue that is translationally regulated. Mol Microbiol. 1995;15:703–717. doi: 10.1111/j.1365-2958.1995.tb02379.x. [DOI] [PubMed] [Google Scholar]
- 12.Darzins A, Russell M A. Molecular genetic analysis of type-4 pilus biogenesis and twitching motility using Pseudomonas aeruginosa as a model system—a review. Gene. 1997;192:109–115. doi: 10.1016/s0378-1119(97)00037-1. [DOI] [PubMed] [Google Scholar]
- 13.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 14.Glessner A, Smith R S, Iglewski B H, Robinson J B. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J Bacteriol, 1999;181:1623–1629. doi: 10.1128/jb.181.5.1623-1629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann J A, Kafatos F C, Janeway Jr C A, Ezekowitz R A B. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 17.Jander G, Rahme L G, Ausubel F M. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol. 2000;182:3843–3845. doi: 10.1128/jb.182.13.3843-3845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson C D, Liu L X. Novel antimicrobial targets from combined pathogen and host genetics. Proc Natl Acad Sci USA. 2000;97:958–959. doi: 10.1073/pnas.97.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang P J, Hauser A R, Apodaca G, Fleiszig S M J, Wiener-Kronish J, Mostov K, Engel J N. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol Microbiol. 1997;24:1249–1262. doi: 10.1046/j.1365-2958.1997.4311793.x. [DOI] [PubMed] [Google Scholar]
- 20.Lehrer R I, Ganz T, Selsted M E. Defensins: endogenous antibiotic peptides of animal cells. Cell. 1991;64:229–230. doi: 10.1016/0092-8674(91)90632-9. [DOI] [PubMed] [Google Scholar]
- 21.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart J-M, Hoffmann J A. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci USA. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemaitre B, Nicolas E, Michaut L, Reichhart J-M, Hoffmann J A. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 23.Mahajan-Miklos S, Tan M-W, Rahme L G, Ausubel F M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 24.Manoil C. Tagging exported proteins using Escherichia coli alkaline phosphatase gene fusions. Methods Enzymol. 2000;326:35–47. doi: 10.1016/s0076-6879(00)26045-x. [DOI] [PubMed] [Google Scholar]
- 25.McBride M J, Weinberg R A, Zusman D R. “Frizzy” aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc Natl Acad Sci USA. 1989;86:424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merz A J, So M, Sheetz M P. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 27.Ornston L N, Parke D. The evolution of induction mechanisms in bacteria: insights derived from the study of the β-ketoadipate pathway. Curr Top Cell Regul. 1977;12:209–262. doi: 10.1016/b978-0-12-152812-6.50011-1. [DOI] [PubMed] [Google Scholar]
- 28.O'Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 29.Rahme L G, Stevens E J, Wolfort S F, Shao J, Tompkins R G, Ausubel F M. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 30.Rahme L G, Ausubel F M, Cao H, Drenkard E, Goumnerov B C, Lau G W, Mahajan-Miklos S, Plotnikova J, Tan M-W, Tsongalis J, Walendziewicz C L, Tompkins R G. Plants and animals share functionally common bacterial virulence factors. Proc Natl Acad Sci USA. 2000;97:8815–8821. doi: 10.1073/pnas.97.16.8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramos J L, Duque E, Godoy P, Segura A. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J Bacteriol. 1998;180:3323–3329. doi: 10.1128/jb.180.13.3323-3329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlictman D, Kubo M, Shankar S, Chakrabarty A M. Regulation of nucleoside diphosphate kinase and secretable virulence factors in Pseudomonas aeruginosa: roles of algR2 and algH. J Bacteriol. 1995;177:2469–2474. doi: 10.1128/jb.177.9.2469-2474.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semmler A B T, Whitchurch C B, Leech A J, Mattick J S. Identification of a novel gene, fimV, involved in twitching motility in Pseudomonas aeruginosa. Microbiology. 2000;146:1321–1332. doi: 10.1099/00221287-146-6-1321. [DOI] [PubMed] [Google Scholar]
- 34.Semmler A B T, Whitchurch C B, Mattick J S. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology. 1999;145:2863–2873. doi: 10.1099/00221287-145-10-2863. [DOI] [PubMed] [Google Scholar]
- 35.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. p. 219. [Google Scholar]
- 36.Smith J J, Travis S M, Greenberg E P, Welsh M J. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell. 1996;85:229–236. doi: 10.1016/s0092-8674(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 37.Spormann A M. Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol Mol Biol Rev. 1999;63:621–641. doi: 10.1128/mmbr.63.3.621-641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S L, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K-S, Wu Z, Paulsen I T, Reizer J, Saier M H, Hancock R E W, Lory S, Olson M V. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 39.Strom M S, Nunn D, Lory S. Multiple roles of the pilus biogenesis protein PilD: involvement of PilD in excretion of enzymes from Pseudomonas aeruginosa. J Bacteriol. 1991;173:1175–1180. doi: 10.1128/jb.173.3.1175-1180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun H, Zusman D R, Shi W. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr Biol. 2000;10:1143–1146. doi: 10.1016/s0960-9822(00)00705-3. [DOI] [PubMed] [Google Scholar]
- 41.Tan M-W, Ausubel F M. Caenorhabditis elegans: a model genetic host to study Pseudomonas aeruginosa pathogenesis. Curr Opin Microbiol. 2000;3:29–34. doi: 10.1016/s1369-5274(99)00047-8. [DOI] [PubMed] [Google Scholar]
- 42.Wall D, Kolenbrander P E, Kaiser D. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J Bacteriol. 1999;181:24–33. doi: 10.1128/jb.181.1.24-33.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward M J, Zusman D R. Regulation of directed motility in Myxococcus xanthus. Mol Microbiol. 1997;24:885–893. doi: 10.1046/j.1365-2958.1997.4261783.x. [DOI] [PubMed] [Google Scholar]
- 44.Watnick P, Kolter R. Biofilm, city of microbes. J Bacteriol. 2000;182:2675–2679. doi: 10.1128/jb.182.10.2675-2679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitchurch C B, Mattick J S. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol Microbiol. 1994;13:1079–1091. doi: 10.1111/j.1365-2958.1994.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 46.Whitchurch C B, Alm R A, Mattick J S. The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1996;93:9839–9843. doi: 10.1073/pnas.93.18.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfgang M, Park H-S, Hayes S F, van Putten J P M, Koomey M. Suppression of an absolute defect in Type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1998;95:14973–14978. doi: 10.1073/pnas.95.25.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z, Ma X, Tong L, Kaplan H B, Shimkets L J, Shi W. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J Bacteriol. 2000;182:5793–5798. doi: 10.1128/jb.182.20.5793-5798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]