Abstract
Fungal species in the family Mortierellaceae are important for their remarkable capability to synthesize large amounts of polyunsaturated fatty acids, especially arachidonic acid (ARA). Although many genomes have been published, the quality of these data is not satisfactory, resulting in an incomplete understanding of the lipid pathway in Mortierellaceae. We provide herein two novel and high-quality genomes with 55.32% of syntenic gene pairs for Mortierella alpina CGMCC 20262 and M. schmuckeri CGMCC 20261, spanning 28 scaffolds of 40.22 Mb and 25 scaffolds of 49.24 Mb, respectively. The relative smaller genome for the former is due to fewer protein-coding gene models (11,761 vs. 13,051). The former yields 45.57% of ARA in total fatty acids, while the latter 6.95%. The accumulation of ARA is speculated to be associated with delta-5 desaturase (Delta5) and elongation of very long chain fatty acids protein 3 (ELOVL3). A further genomic comparison of 19 strains in 10 species in three genera in the Mortierellaceae reveals three types of fatty acid synthase (FAS), two of which are new to science. The most common type I exists in 16 strains of eight species of three genera, and was discovered previously and consists of a single unit with eight active sites. The newly revealed type II exists only in M. antarctica KOD 1030 where the unit is separated into two subunits α and β comprised of three and five active sites, respectively. Another newly revealed type III exists in M. alpina AD071 and Dissophora globulifera REB-010B, similar to type II but different in having one more acyl carrier protein domain in the α subunit. This study provides novel insights into the enzymes related to the lipid metabolism, especially the ARA-related Delta5, ELOVL3, and FAS, laying a foundation for genetic engineering of Mortierellaceae to modulate yield in polyunsaturated fatty acids.
Keywords: Mucoromyceta, microbial lipids, phylogenomics, polyunsaturated fatty acids, arachidonic acid
1. Introduction
Environmental protection and sustainable development are now giving urgency to identifying alternatives for oil plants and animals. Therefore, oleaginous fungi such as Aspergillus flavus Link [=A. oryzae (Ahlb.) Cohn], Mortierella alpina Peyronel, M. wolfii B.S. Mehrotra & Baijal, Mucor racemosus Fresen., Lichtheimia corymbifera (Cohn) Vuill., Syncephalastrum racemosum Cohn ex J. Schröt., Geotrichum candidum Link, Umbelopsis isabellina (Oudem.) W. Gams (=Mortierella isabellina Oudem.), and Yarrowia lipolytica (Wick., Kurtzman & Herman) Van der Walt & Arx have recently gained attention and played an important role in the studies of biodiesel and polyunsaturated fatty acids (PUFAs) [1,2,3,4,5,6,7,8,9,10,11]. These oleaginous fungi mainly belong to Ascomycota, Mortierellomycota, and Mucoromycota.
Compared with other microorganisms such as microalgae and algae, oleaginous fungi are distinct in possessing a shorter life cycle, assimilating a variety of carbon sources, and adapting to various weather and seasons [1,7,9]. Meanwhile, the species in the Mortierellaceae of Mortierellomycota are characterized by synthesizing a large amount of PUFAs [1,12]. At present, 122 species of seven genera are accommodated in the family Mortierellaceae [13], and almost all tested strains of Mortierellaceae were found to be able to synthesize arachidonic acid (ARA, C20:4) [1,12,14,15,16], which is a vital nutrient for the elderly and infants, possessing multiple functions such as the protection of brain and muscles and potential against tumors and inflammation [16,17,18].
Mortierella alpina synthesizing lipids more than 50% of its dry cell weight and ARA accounting for 30–70% of the total fatty acids has recently been one of the important model organisms for PUFA metabolism [12,16,19,20,21,22]. Based on genetic manipulation in M. alpina, numerous enzymes such as AMP deaminase, omega-3 desaturase, delta-5 desaturase, and fatty acid synthase were proved to play key functions in the biosynthesis of PUFAs [12,16,23,24,25]. For example, overexpression of homologous AMP deaminase in M. alpina resulted in a significant increase of 15.0–34.3% in lipid yield [23].
In 2011, the first draft genome of Mortierella alpina was completed for the strain ATCC 32222, initializing studies on lipid metabolic pathway in the species [11]. Until now, nine genomes of M. alpina have been published in the NCBI database (https://www.ncbi.nlm.nih.gov, accessed on 2 July 2022) [12,26,27,28]. Other genomes of the Mortierellaceae were assembled for Actinomortierella wolfii (B.S. Mehrotra & Baijal) Vandepol & Bonito, Dissophora globulifera (O. Rostr.) Vandepol & Bonito, Mortierella amoeboidea W. Gams, M. antarctica Linnem., M. elongata Linnem., M. epicladia W. Gams & Emden, M. gamsii Milko, and M. verticillata Linnem. [27,29,30,31]. However, comprehensive analyses of these genomes, especially the key genes involved in fatty acid synthesis, are not satisfactory due to a lack of good-quality sequence data.
In the present study, we sequence two strains, Mortierella alpina CGMCC 20262 and M. schmuckeri Linnem. CGMCC 20261, by combining next-generation with PacBio sequencing. A total of 40 strains of basal fungi are used to reconstruct their phylogenomic relationship, and 13 protein-coding genes involved in the lipid metabolism are characterized in 19 strains of Mortierellaceae.
2. Materials and Methods
2.1. Strains, Media, and Fermentation
Mortierella alpina CGMCC 20262 and M. schmuckeri CGMCC 20261 were deposited in the China General Microbiological Culture Collection Center, Beijing, China (CGMCC). Cultures were incubated with potato dextrose agar (PDA: 200 g/L potato, 20 g/L glucose, 20 g/L agar, and 1000 mL distilled water) and ampicillin (100 μg/mL) at 20 °C for ten days. A shake flask fermentation was then carried out following a previous study [1]. In brief, 1 mL of spore suspension (1 × 106) was incubated at 20 °C and 140 rpm for seven days in a 250 mL flask with 100 mL of modified Kendrick media (50 g/L glucose, 2 g/L diammonium tartrate, 7 g/L KH2PO4, 2 g/L Na2HPO4, 1.5 g/L MgSO4·7H2O, 1.5 g/L yeast extract, 0.1 g/L CaCl2·2H2O, 8 mg/L FeCl3·6H2O, 1 mg/L ZnSO4·7H2O, 0.1 mg/L CuSO4·5H2O, 0.1 mg/L CO(NO3)2·6H2O, 0.1 mg/L MnSO4·5H2O, and pH 6.0) [1,23,32].
2.2. Biomass, Fatty Acid Measurement, and Profiling
Biomass, fatty acid measurement, and profiling followed the method by Zhao et al. [1]. Fresh biomasses were drained with a vacuum pump, and then freeze-dried for two days to attain a constant dry cell weight (DCW). The freeze-dried biomasses were hydrolyzed with HCl solution (6 mol/L), and then extracted with ethanol, anhydrous ether, and petroleum ether following previous studies [1,33]. The extract was then dried with an oven by slowly warming to 80 °C until a constant total lipid weight (TLW) was attained. Total lipid content (TLC) was calculated as the TLW being divided by the DCW. For fatty acid profiling, the gas chromatography–mass spectrometry (GC/MS, QP2010, Shimadzu Corp., Japan) was performed with a 30 m × 0.25 mm × 0.25 μm column (Rtx-5MS, RESTEK, Bellefonte, PA, USA), and nonanoic acid (C9:0) was selected as internal standard according to Zhao et al. [1].
2.3. Genome Sequencing and Assembly
Mortierella alpina CGMCC 20262 and M. schmuckeri CGMCC 20261 were incubated with PDA at 20 °C for five days. Total cell DNAs were extracted from mycelia using a kit (O-GPLF-400, GeneOnBio Corporation, Changchun, China) according to the manufacturer’s protocol, and then detected by DNA/Protein Analyzer and 1% agarose gel electrophoresis. High-quality DNAs were sequenced at Beijing Novogene Bioinformatics Technology Co., Ltd. (Beijing, China) by using a PacBio Sequel and Illumina NovaSeq 6000 platform with 20 kb and 350 bp library, respectively. Low-quality reads (less than 500 bp) were removed by quality control from the raw data produced with the PacBio Sequel platform, and then the controlled high-quality reads were de novo assembled using SMRT Link v5.1.0 [34]. The assembled genomes were assessed by Quast v5.0.2 [35] and BUSCO v5.2.2 [36].
2.4. Gene Prediction and Functional Annotation
Protein-coding gene models of Mortierella alpina CGMCC 20262 and M. schmuckeri CGMCC 20261 were de novo predicted using Augustus v3.3.3 [37]. Amino acid and DNA sequences were functionally annotated using NR (https://www.ncbi.nlm.nih.gov/protein/, accessed on 2 July 2022), NT (https://www.ncbi.nlm.nih.gov/nucleotide/, accessed on 2 July 2022), Pfam [38], GO [39], KEGG [40], CAZymes [41,42], and Eggnog [43] databases. All sequences were mapped onto these databases using Diamond v2.0.1 [44] with an e-value less than 1 × 10−5. The antiSMASH fungal version [45] was used to annotate the gene clusters of secondary metabolites with default parameters. Repetitive elements were identified using the Extensive de novo TE Annotator (EDTA) pipeline v1.9.5 [46]. RNAmmer v1.2 [47] and tRNAscan-SE v2.0.5 [48] were used to predict rRNAs and tRNAs, respectively.
2.5. Phylogenomic and Phylogenetic Analyses
A total of 40 strains of early diverging fungi, including 38 strains downloaded from online databases and two strains sequenced herein, were used for phylogenomic analyses (Table 1). A total of 192 clusters of orthologous proteins were identified with HMMER v3.3.1 [49] and Trimal v1.4.4 [50], following the methods described by Spatafora et al. [51] and James et al. [52], and then their amino acid sequences were aligned with MAFFT v7 [53]. Phylogenomic analyses were carried out with a Maximum Likelihood (ML) algorithm using RaxML v8.1.12 [54]. Maximum Likelihood analyses adopted the PROTGAMMALGX substitution model with 100 bootstrap replications. Phylogenetic analyses on amino acids of delta-5 desaturase and fatty acid synthase were performed using IQ-TREE v1.0 [55] with an ML algorithm, WAG substitution model, and 1000 bootstrap replications.
Table 1.
The genomic information used for phylogenomic analyses in this study.
| Species | Strains | BioSample | References |
|---|---|---|---|
| Absidia glauca | CBS 101.48 | SAMEA3923633 | [56] |
| Acaulospora morrowiae | CL551 | SAMEA8911292 | |
| Apophysomyces elegans | B7760 | SAMN02351510 | |
| Actinomortierella wolfii * | NRRL 6351 | SAMN05720777 | [27] |
| Choanephora cucurbitarum | KUS-F282377 | SAMN04532838 | |
| Cunninghamella elegans | B9769 | SAMN02351511 | |
| Dissophora globulifera * | REB 010B | SAMN05720531 | [27] |
| Diversispora eburnea | AZ414A | SAMEA8911293 | |
| Endogone sp. | FLAS-F59071 | SAMN09071421 | [57] |
| Gongronella butleri | GbKAU | SAMN15221701 | |
| Lichtheimia corymbifera | FSU 9682 | SAMEA2189700 | [58] |
| L. ramosa | FSU 6197 | SAMN05179542 | [59] |
| Mortierella alpina * | CGMCC 20262 | SAMN29490473 | This study |
| M. alpina * | AD071 | SAMN05720461 | [27] |
| M. alpina * | AD072 | SAMN05720462 | [27] |
| M. alpina * | ATCC 32222 | SAMN02981246 | [12] |
| M. alpina * | B6842 | SAMN02370960 | [26] |
| M. alpina * | CCTCC M-207067 | SAMN03658567 | |
| M. alpina * | CK1249 | SAMN05720518 | [27] |
| M. alpina * | GBA31 | SAMN05720773 | [27] |
| M. alpina * | LL118 | SAMN20056918 | [28] |
| M. alpina * | NRRL 66262 | SAMN10361219 | [27] |
| M. amoeboidea * | CBS 889.72 | SAMN19911466 | [31] |
| M. antarctica * | KOD1030 | SAMN05720520 | [27] |
| M. elongata * | AG-77 | SAMN02745706 | [30] |
| M. epicladia * | AD058 | SAMN05720441 | [27] |
| M. gamsii * | NVP60 | SAMN05720530 | [27] |
| M. verticillata * | NRRL 6337 | SAMN00699802 | [29] |
| M. schmuckeri * | CGMCC 20261 | SAMN29492047 | This study |
| Mucor circinelloides | 1006PhL | SAMN00103456 | [60] |
| M. lusitanicus | CBS 277.49 | SAMN00120579 | [61] |
| Parasitella parasitica | CBS 412.66 | SAMEA278055 | [56] |
| Rhizopus arrhizus | GL8 | SAMN14162349 | [62] |
| R. microsporus | ATCC 52813 | SAMN06821222 | [63] |
| Phycomyces blakesleeanus | NRRL 1555 | SAMN00189023 | [61] |
| Smittium culicis | GSMNP | SAMN04489870 | [64] |
| Syncephalastrum monosporum | B8922 | SAMN02370995 | |
| Thermomucor indicae-seudaticae | HACC 243 | SAMN03070115 | |
| Umbelopsis isabellina | WA0000067209 | SAMN16393839 | [65] |
| U. vinacea | WA0000051536 | SAMN16393840 |
Note: The star mark “*” represents members in the Mortierellaceae. The two genomes newly generated in this study are in bold.
2.6. Comparative Genomic Analyses
The genome sequences of Mortierella alpina CGMCC 20262 and M. schmuckeri CGMCC 20261 were aligned using MCScanX for all protein-coding gene models, and then genomic collinearity was analyzed using a dual synteny plotter package [61]. For comparison of genes in lipid metabolism, especially those encoding fatty acid synthase and delta-5 desaturase, all 19 genomes of Mortierellaceae were reannotated using the KEGG database [41].
3. Results
3.1. Fatty Acid Profiles of Mortierella alpina and M. schmuckeri
Mortierella alpina CGMCC 20262 is much lower than M. schmuckeri CGMCC 20261 in dry cell weight (DCW, 6.6 g/L vs. 11.7 g/L), total lipid weight (TLW, 1.1 g/L vs. 8.0 g/L), and total lipid content (TLC, 17% vs. 70%; Figure 1a). However, M. alpina CGMCC 20262 yields 75.54% of polyunsaturated fatty acids (PUFAs; 15.41% of C18:2, 8.78% of C18:3, 3.40% of C20:3, 45.57% of C20:4, and 2.39% of C20:5) in total fatty acids (Figure 1b), more than the percentage of 23.03% in M. schmuckeri CGMCC 20261 (8.30% C18:2, 4.36% of C18:3, 3.43% of C20:3, 6.95% of C20:4, and no C20:5). Obviously, these two strains are significantly different, C20:4 (45.57%) being the top one in M. alpina CGMCC 20262, and C18:1 (27.54%) and C16:0 (24.68%) being the top two in M. schmuckeri CGMCC 20261 (Figure 1b).
Figure 1.
Lipids of Mortierella alpina CGMCC 20262 and M. schmuckeri CGMCC 20261. (a) Lipid characters: dry cell weight (DCW, g/L), total lipid weight (TLW, g/L), and total lipid content (TLC); (b) fatty acid profiles: others include C10:0, C12:0, C15:0, C20:0, and C20:1.
3.2. Genomic Features of Mortierella alpina and M. schmuckeri
Assembled genomes span 28 scaffolds of 40.22 Mb with GC content of 50.92% in Mortierella alpina CGMCC 20262, and 25 scaffolds of 49.24 Mb with GC content of 47.46% in M. schmuckeri CGMCC 20261 (Table 2). The number of predicted protein-coding gene models of M. schmuckeri CGMCC 20261 is slightly more than that of M. alpina CGMCC 20262 (13,051 vs. 11,761; Table 2). Among these gene models, 66.17%, 33.97%, 47.35%, 42.69%, 73.20%, 37.71%, and 1.70% are mapped onto Pfam, NT, NR, GO, Eggnog, KEGG, and CAZymes databases in M. schmuckeri CGMCC 20261, and 72.33%, 34.23%, 41.70%, 46.01%, 77.17%, 41.31%, and 1.95% in M. alpina CGMCC 20262 (Table 2). Repetitive elements of M. alpina CGMCC 20262 and M. schmuckeri CGMCC 20261 account for 7.29% and 7.45% of the whole genomes (Table 2). Furthermore, 38 rRNAs (11 of 8s rRNA, 12 of 18s rRNA, and 15 of 28s rRNA) and 29 rRNAs (9 of 8s rRNA, 8 of 18s rRNA, and 12 of 28s rRNA) are predicted in M. alpina CGMCC 20262 and M. schmuckeri CGMCC 20261, respectively. More characteristics of genomes are listed in Table 2.
Table 2.
Genomic features of Mortierella sequenced and de novo assembled in this study.
| Species | M. alpina CGMCC 20262 | M. schmuckeri CGMCC 20261 | |
|---|---|---|---|
| Genome size (Mb) | 40.22 | 49.24 | |
| Scaffolds | 28 | 25 | |
| Largest scaffolds (Mb) | 4.35 | 4.78 | |
| GC (%) | 50.92 | 47.46 | |
| N50 (Mb) | 2.53 | 2.71 | |
| L50 | 6 | 8 | |
| Assembly BUSCO coverage (%) | 97.4 | 97.6 | |
| PCG models | 11,761 | 13,051 | |
| Pfam | 8507 | 8636 | |
| NT | 4026 | 4434 | |
| NR | 4904 | 6180 | |
| GO | 5411 | 5571 | |
| Eggnog | 9076 | 9436 | |
| KEGG | 4858 | 4921 | |
| CAZymes | 229 | 222 | |
| Gene clusters of secondary metabolites | |||
| Terpene | 4 | 2 | |
| Fungal-RiPP | 0 | 1 | |
| NRPS | 15 | 0 | |
| NRPS-like | 1 | 2 | |
| Siderophore | 2 | 0 | |
| Repetitive elements (% in genomes) | 7.29 | 7.45 | |
| ncRNA | |||
| rRNA | 38 | 29 | |
| tRNA | 226 | 262 |
3.3. Phylogenomic Placements of Mortierella alpina and M. schmuckeri
The Maximum Likelihood phylogenomic tree (Figure 2) suggests that Mortierella alpina CGMCC 20262 is closely related to other strains of M. alpina (Maximum Likelihood bootstrap values, MLBV = 100%), and M. schmuckeri CGMCC 20261 is a sister to M. gamsii (MLBV = 100%) and closely related to M. elongata (MLBV = 100%).
Figure 2.
A Maximum Likelihood phylogenomic tree illustrating the placements of Mortierella alpina CGMCC 20262 and M. schmuckeri CGMCC 20261 based on 192 clusters of orthologous proteins. New genomes obtained in this study are in bold. Maximum Likelihood bootstrap values (MLBV ≥ 50%) are indicated along branches. A scale bar in the upper left indicates substitutions per site.
3.4. Synteny between Mortierella alpina and M. schmuckeri
A total of 13,726 gene models located in all scaffolds are collinear between Mortierella alpina CGMCC 20262 and M. schmuckeri CGMCC 20261, accounting for 55.32% of all the 24,812 gene models in both strains. Figure 3 shows the synteny of the ten largest scaffolds where 16,927 gene models are predicted and 11,777 (69.58%) gene models are syntenic.
Figure 3.
Genomic synteny of Mortierella alpina CGMCC 20262 and M. schmuckeri CGMCC 20261 based on protein-coding gene models of the 10 largest scaffolds.
3.5. Lipid Metabolism in Mortierellaceae
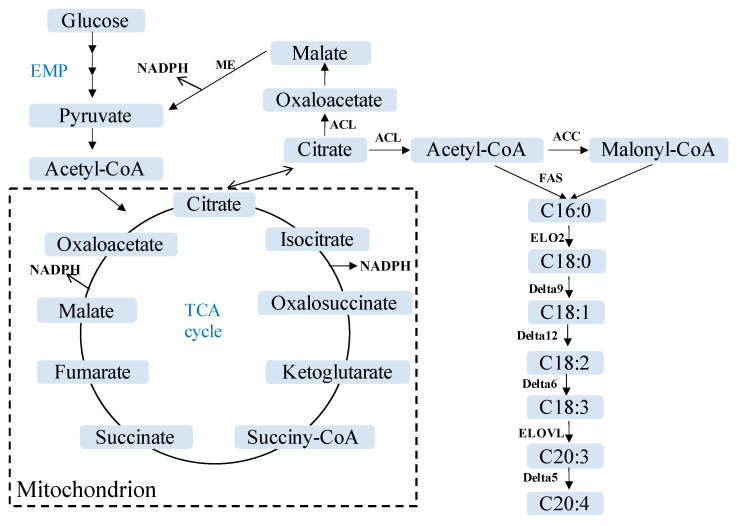
To explore the lipid metabolism in Mortierellaceae, genomes of 19 strains in 10 species are de novo annotated or reannotated. We focus on the 13 genes associated with NADPH, precursors, desaturation, and elongation (Figure 4 and Supplementary Figure S2). The gene Delta5 (encoding delta-5 desaturase) is not found in M. gamsii NVP60, and neither is Delta6 (encoding delta-6 desaturase) in M. alpina LL118. With these two exceptions, each gene possesses between one and six copies. The genes HK (encoding hexokinase, a key enzyme in EMP or the glycolytic pathway) and ME (encoding malate dehydrogenase) are remarkable as both have between three and six copies (Supplementary Figure S2 and Supplementary Table S1).
Figure 4.
The polyunsaturated fatty acid synthesis mechanism in Mortierellaceae. EMP: Glycolytic pathway; TCA cycle: Tricarboxylic acid cycle; ME: Malate dehydrogenase; NADPH: Nicotinamide adenine dinucleotide phosphate; ACC: Acetyl-CoA carboxylase; ACL: ATP citrate (pro-S)-lyase; FAS: Fatty acid synthase; ELO2: Fatty acid elongase 2; Delta5: delta-5 desaturase; Delta6: delta-6 desaturase; Delta9: delta-9 desaturase; Delta12: delta-12 desaturase; ELOVL: Elongation of very long chain fatty acids protein.
Three types of fatty acid synthase are identified in the family Mortierellaceae (Figure 5, Supplementary File S1). Type I is ubiquitous (Actinomortierella wolfii NRRL 6351, Mortierella alpina AD072, M. alpina ATCC 32222, M. alpina B6842, M. alpina CCTCC M-207067, M. alpina CGMCC 20262, M. alpina CK1249, M. alpina GBA31, M. alpina LL118, M. alpina NRRL 66262, M. amoeboidea CBS 889.72, M. elongata AG-77, M. epicladia AD058, M. schmuckeri CGMCC 20261, and M. verticillata NRRL 6337), and unites together eight active sites (Figure 5). Type II is found in M. antarctica KOD 1030 only and includes two subunits, α subunit involving KR, KS, and PPT active sites, and β subunit comprising AT, ER, DH, MPT, and ACP active sites. Type III is identified in Dissophora globulifera REB-010B and Mortierella alpina AD071, similar to type II but with one more ACP active site in α subunit.
Figure 5.
Fatty acid synthases in Mortierellaceae. AT: Acyl transferase domain; ER: Enoyl reductase domain; DH: dehydratase; MPT: Malonyl CoA-acyl carrier protein transacylase; ACP: Acyl carrier protein domain; KR: β-ketoacyl reductase; KS: β-ketoacyl synthase; PPT: Phosphopantetheinyl transferase.
Four desaturases, i.e., Delta5, Delta6, Delta9, and Delta12, are found in the 19 strains of Mortierellaceae (Supplementary Figure S2 and Supplementary Table S1). Among these, the Delta5 catalyzes the synthesis of arachidonic acid (ARA, C20:4; Figure 4), which has diverged into three clades in Mortierellaceae (Figure 6 and Supplementary File S2).
Figure 6.
A Maximum Likelihood phylogenetic tree of the gene encoding delta-5 desaturase in the family Mortierellaceae. A scale bar in the upper left indicates substitutions per site.
The elongation of very long chain fatty acids protein (ELOVL) family consists of four members in Mortierellaceae, i.e., ELOVL2, ELOVL3, ELOVL4, and ELOVL6 (Supplementary Figure S1 and Supplementary File S3). ELOVL2 is shared by all strains of Mortierellaceae, while ELOVL3, ELOVL4, and ELOVL6 are species-specific. In detail, ELOVL3 is identified in M. alpina and M. amoeboidea, and ELOVL4 and ELOVL6 are annotated in M. verticillata and D. globulifera.
4. Discussion
Among numerous oleaginous fungi, Mortierella alpina is now attracting more and more attention because of its ability to accumulate large amounts of arachidonic acid (ARA) [12,16,22]. Previous studies suggested that M. alpina synthesized lipids up to 50% of dry cell weight (DCW) and ARA 30–70% of total lipid content (TLC) [12]. Herein, M. alpina CGMCC 20262 falls in these ranges, yielding lipids 17% of DCW and ARA 45.57% of TCL (Figure 1). In addition, we find a novel oleaginous fungus, M. schmuckeri CGMCC 20261, which accumulates lipids 70% of DCW (Figure 1), much more than M. alpina (17%).
The draft genome of M. alpina was published in 2011, and consequently lipid synthesis pathway was predicted [12]. Since then, more genomes of members in the family Mortierellaceae have shed light on their lipid synthesis mechanism. For example, Mortierella sp. BCC40632 has an n-6 series fatty acid synthesis pathway due to a lack of delta-15 or omega-3 desaturase [25,26,27,30,31]. In this paper, the genomes of two more strains, M. alpina CGMCC 20262 and M. schmuckeri CGMCC 20261, are sequenced to provide more insights into lipid metabolism (Table 2).
A large number of genes hitherto have been found to affect lipid synthesis in Mortierella alpina, such as acetyl-CoA carboxylase, AMP deaminase, malate dehydrogenase, delta-5 desaturase, delta-6 desaturase, and elongase 2 [12,22,23,66,67,68,69]. In this study, we compare 13 genes of 19 strains in Mortierellaceae (Supplementary Figure S2), finding differences in genes encoding hexokinase, delta-5 desaturase, and elongation of very long chain fatty acids protein. Compared with M. schmuckeri CGMCC 20261, the M. alpina CGMCC 20262 possesses one more copy of the HK gene, more copies of Delta5 and ELOVL genes but fewer copies of the Delta9 gene (Supplementary Figure S2 and Supplementary Table S1). Delta-5 desaturase and elongation of very long chain fatty acids protein play an essential role in the synthesis of polyunsaturated fatty acids (PUFAs, Figure 4), which might be one of the reasons for M. alpina CGMCC 20262 yielding more PUFAs than M. schmuckeri CGMCC 20261.
PUFA biosynthesis requires an activity of elongation of very long chain fatty acids proteins, as well as fatty acid desaturases such as delta-5, delta-6, delta-12, and omega-3 desaturase [12,16]. For example, the M. alpina 1S-4 mutant in delta-5 desaturase resulted in a huge accumulation of dihomo-γ-linolenic acid (DGLA), up to 43.3% of total fatty acids [70]. In the present study, the delta-5 desaturase of M. alpina is significantly different from other species in the family Mortierellaceae based on the phylogenetic analyses (Figure 6 and Supplementary File S2), which is probably one of the reasons for synthesizing high content of ARA [12,71,72]. Elongation of very long chain fatty acids proteins (ELOVLs) are involved in the long-chain polyunsaturated fatty acid synthetic pathway, and four gene types, ELOVL2, ELOVL3, ELOVL4, and ELOVL6, are identified herein in 19 strains of Mortierellaceae (Supplementary Figure S1 and Supplementary File S3).
The fatty acid synthase plays an important role in lipid synthesis, catalyzing the synthesis of saturated fatty acids from acetyl-CoA and malonyl-CoA. A variety of fatty acid synthases were found in fungi, such as Aspergillus oryzae, Mortierella alpina, Saccharomyces cerevisiae, and Yarrowia lipolytica [6,12,73,74]. In this study, three types of fatty acid synthases are found in Mortierellaceae (Supplementary Figure S2 and Supplementary File S1). Type I is the most common and similar to that reported in Mortierella alpina ATCC 32222, a single subunit consisting of eight catalytic domains/active sites [17]. Types II and III are new to science. Type II presents in M. antarctica KOD 1030 only, and type III in Dissophora globulifera REB-010B and Mortierella alpina AD071 (Supplementary Figure S2 and Supplementary File S1).
Overall, the rapid increase in genomic data is providing more materials for tackling fatty acid biosynthesis, discovering more potential genes, and finalizing the understanding of the genetic foundation of lipid metabolism.
5. Conclusions
In this paper, two new genomes, Mortierella alpina CGMCC 20262 and M. schmuckeri CGMCC 20261, were sequenced using the PacBio Sequel and Illumina NovaSeq 6000 platform. To explore the lipid metabolism in Mortierellaceae, a total of 19 genomes were reannotated. The results suggest that delta-5 desaturase and elongation of very long chain fatty acids protein 3 probably promoted the accumulation of polyunsaturated fatty acids, especially arachidonic acid. Besides, three types of fatty acid synthase, including two novel ones, were identified. Consequently, with the increase in public genomic data, a comprehensive analysis on lipid metabolism will discover more genes or protein-encoding models, providing more information for subsequent genetic engineering of oleaginous fungi.
Acknowledgments
We thank Beijing Novogene Bioinformatics Technology Co., Ltd. (Beijing, China).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8090891/s1. Supplementary Table S1. The gene number of lipid metabolism in the 19 strains of Mortierellaceae; Supplementary Figure S1. A Maximum Likelihood tree of elongation of very long chain fatty acids protein in the family Mortierellaceae; Supplementary Figure S2. The heatmap of gene numbers associated with fatty acid synthesis annotated in 19 strains of Mortierellaceae; Supplementary File S1. The amino acid sequences of fatty acid synthase in Mortierellaceae; Supplementary File S2. The aligned dataset of amino acid sequences of delta-5 desaturase; Supplementary File S3. The amino acid sequences of elongation of very long chain fatty acids protein in Mortierellaceae.
Author Contributions
H.Z.; validation, writing—original draft preparation, and writing—review, Y.N.; formal analysis, Y.J.; and S.W.; investigation, T.-Y.Z.; data curation, X.-Y.L.; funding acquisition, projection administration, and writing—review. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The newly identified genomics sequences have been deposited in GenBank (Table 2).
Conflicts of Interest
All authors declare no conflict of interest.
Funding Statement
This research was supported by the National Natural Science Foundation of China, Grant Nos. 31970009 and 32170012.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhao H., Lv M.L., Liu Z., Zhang M.Z., Wang Y.N., Ju X., Song Z., Ren L.Y., Jia B.S., Qiao M., et al. High-yield oleaginous fungi and high-value microbial lipid resources from Mucoromycota. BioEnergy Res. 2021;14:1196–1206. doi: 10.1007/s12155-020-10219-3. [DOI] [Google Scholar]
- 2.Reis C.E.R., Valle G.F., Bento H.B., Carvalho A.K., Alves T.M., de Castro H.F. Sugarcane by-products within the biodiesel production chain: Vinasse and molasses as feedstock for oleaginous fungi and conversion to ethyl esters. Fuel. 2020;277:118064. doi: 10.1016/j.fuel.2020.118064. [DOI] [Google Scholar]
- 3.Papanikolaou S., Aggelis G. Sources of microbial oils with emphasis to Mortierella (Umbelopsis) isabellina fungus. World J. Microb. Biot. 2019;35:63. doi: 10.1007/s11274-019-2631-z. [DOI] [PubMed] [Google Scholar]
- 4.Kothri M., Mavrommati M., Elazzazy A.M., Baeshen M.N., Moussa T.A., Aggelis G. Microbial sources of polyunsaturated fatty acids (PUFAs) and the prospect of organic residues and wastes as growth media for PUFA-producing microorganisms. FEMS Microbiol. Lett. 2020;367:fnaa028. doi: 10.1093/femsle/fnaa028. [DOI] [PubMed] [Google Scholar]
- 5.Blazeck J., Hill A., Liu L., Knight R., Miller J., Pan A., Otoupal P., Alper H.S. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 2014;5:3131. doi: 10.1038/ncomms4131. [DOI] [PubMed] [Google Scholar]
- 6.Thammarongtham C., Nookaew I., Vorapreeda T., Srisuk T., Land M.L., Jeennor S., Laoteng K. Genome characterization of oleaginous Aspergillus oryzae BCC7051: A potential fungal-based platform for lipid production. Curr. Microbiol. 2018;75:57–70. doi: 10.1007/s00284-017-1350-7. [DOI] [PubMed] [Google Scholar]
- 7.Hashem A.H., Suleiman W.B., Abu-Elrish G.M., Ei-Sheikh H.H. Consolidated bioprocessing of sugarcane bagasse to microbial oil by newly isolated oleaginous fungus: Mortierella wolfii. Arab. J. Sci. Eng. 2021;46:199–211. doi: 10.1007/s13369-020-05076-3. [DOI] [Google Scholar]
- 8.Hashem A.H., Abu-Elreesh G., El-Sheikh H.H., Suleiman W.B. Isolation, identification, and statistical optimization of a psychrotolerant Mucor racemosus for sustainable lipid production. Biomass Conv. Biorefinery. 2022:1–12. doi: 10.1007/s13399-022-02390-8. [DOI] [Google Scholar]
- 9.Hashem A.H., Hasanin M.S., Khalil A.M.A., Suleiman W.B. Eco-green conversion of watermelon peels to single cell oils using a unique oleaginous fungus: Lichtheimia corymbifera AH13. Waste Biomass Valor. 2022;11:5721–5732. doi: 10.1007/s12649-019-00850-3. [DOI] [Google Scholar]
- 10.Hashem A.H., Suleiman W.B., Abu-elreesh G., Shehabeldine A.M., Khalil A.M.A. Sustainable lipid production from oleaginous fungus Syncephalastrum racemosum using synthetic and watermelon peel waste media. Bioresour. Technol. Rep. 2020;12:100569. doi: 10.1016/j.biteb.2020.100569. [DOI] [Google Scholar]
- 11.Gad A.M., Suleiman W.B., El-Sheikh H.H., Eimezayen H.A., Beltagy A.M. Characterization of cellulase from Geotrichum candidum strain Gad1 approaching bioethanol production. Arab. J. Sci. Eng. 2022;47:6837–6850. doi: 10.1007/s13369-021-06391-z. [DOI] [Google Scholar]
- 12.Wang L., Chen W., Feng Y., Ren Y., Gu Z., Chen H., Wang H., Thomas M.J., Zhang B., Berquin I.M. Genome characterization of the oleaginous fungus Mortierella alpina. PLoS ONE. 2011;6:e28319. doi: 10.1371/journal.pone.0028319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bánki O., Roskov Y., Döring M., Ower G., Vandepitte L., Hobern D., Remsen D., Schalk P., DeWalt R., Keping M., et al. Catalogue of Life Checklist, Version 2022-03-21; Catalogue of Life. 2022. [(accessed on 2 July 2022)]. Available online: https://www.catalogueoflife.org/
- 14.Eroshin V., Dedyukhina E., Chistyakova T., Zhelifonova V., Kurtzman C., Bothast R. Arachidonic-acid production by species of Mortierella. World J. Microbiol. Biotechnol. 1996;12:91–96. doi: 10.1007/BF00327809. [DOI] [PubMed] [Google Scholar]
- 15.Botha A., Paul I., Roux C., Kock J.L., Coetzee D.J., Strauss T., Maree C. An isolation procedure for arachidonic acid producing Mortierella species. Antonie. Leeuw. 1999;75:253–256. doi: 10.1023/A:1001848709005. [DOI] [PubMed] [Google Scholar]
- 16.Kikukawa H., Sakuradani E., Ando A., Shimizu S., Ogawa J. Arachidonic acid production by the oleaginous fungus Mortierella alpina 1S-4: A review. J. Adv. Res. 2018;11:15–22. doi: 10.1016/j.jare.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikura Y., Ikeda G., Akimoto K., Hata M., Kusumoto A., Kidokoro A., Kontani M., Kawashima H., Kiso Y., Koga Y. Arachidonic acid supplementation decreases P300 latency and increases P300 amplitude of event-related potentials in healthy elderly men. Neuropsychobiology. 2009;60:73–79. doi: 10.1159/000236447. [DOI] [PubMed] [Google Scholar]
- 18.Tallima H., El Ridi R. Arachidonic acid: Physiological roles and potential health benefits–a review. J. Adv. Res. 2018;11:33–41. doi: 10.1016/j.jare.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratledge C., Wynn J.P. Advances in Applied Microbiology. Volume 51. Academic Press; San Diego, CA, USA: 2002. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms; pp. 1–51. [DOI] [PubMed] [Google Scholar]
- 20.Ho S.-Y., Jiang Y., Chen F. Polyunsaturated fatty acids (PUFAs) content of the fungus Mortierella alpina isolated from soil. J. Agric. Food Chem. 2007;55:3960–3966. doi: 10.1021/jf0700071. [DOI] [PubMed] [Google Scholar]
- 21.Tedersoo L., Sánchez-Ramírez S., Kõljalg U., Bahram M., Döring M., Schigel D., May T., Ryberg M., Abarenkov K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 2018;90:135–159. doi: 10.1007/s13225-018-0401-0. [DOI] [Google Scholar]
- 22.Kikukawa H., Sakuradani E., Ando A., Okuda T., Shimizu S., Ogawa J. Microbial production of dihomo-γ-linolenic acid by Δ5-desaturase gene-disruptants of Mortierella alpina 1S-4. J. Biosci. Bioeng. 2016;122:22–26. doi: 10.1016/j.jbiosc.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Chang L., Tang X., Lu H., Zhang H., Chen Y.Q., Chen H., Chen W. Role of adenosine monophosphate deaminase during fatty acid accumulation in oleaginous fungus Mortierella alpina. J. Agric. Food Chem. 2019;67:9551–9559. doi: 10.1021/acs.jafc.9b03603. [DOI] [PubMed] [Google Scholar]
- 24.Rong C., Chen H., Tang X., Gu Z., Zhao J., Zhang H., Chen Y., Chen W. Structural determinants of substrate specificity of omega-3 desaturases from Mortierella alpina and Rhizophagus irregularis by domain-swapping and molecular docking. Int. J. Mol. Sci. 2019;20:1603. doi: 10.3390/ijms20071603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vorapreeda T., Thammarongtham C., Palasak T., Srisuk T., Jenjaroenpun P., Wongsurawat T., Nookaew I., Laoteng K. Systematic genome analysis of a novel arachidonic acid-producing strain uncovered unique metabolic traits in the production of acetyl-CoA-derived products in Mortierella fungi. Gene. 2020;741:144559. doi: 10.1016/j.gene.2020.144559. [DOI] [PubMed] [Google Scholar]
- 26.Etienne K.A., Chibucos M.C., Su Q., Orvis J., Daugherty S., Ott S., Sengamalay N.A., Fraser C.M., Lockhart S.R., Bruno V.M. Draft genome sequence of Mortierella alpina isolate CDC-B6842. Genome Announc. 2014;2:e01180-13. doi: 10.1128/genomeA.01180-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandepol N., Liber J., Desirò A., Na H., Kennedy M., Barry K., Grigoriev I.V., Miller A.N., O’Donnell K., Stajich J.E. Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Divers. 2020;104:267–289. doi: 10.1007/s13225-020-00455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang S., Vinatzer B.A. Draft Genome sequence of Mortierella alpina Strain LL118, isolated from an Aspen (Populus tremuloides) leaf litter sample. Microbiol. Resour. Announc. 2021;10:e00864-21. doi: 10.1128/MRA.00864-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seif E., Leigh J., Liu Y., Roewer I., Forget L., Lang B.F. Comparative mitochondrial genomics in zygomycetes: Bacteria-like RNase P RNAs, mobile elements and a close source of the group I intron invasion in angiosperms. Nucleic Acids Res. 2005;33:734–744. doi: 10.1093/nar/gki199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uehling J., Gryganskyi A., Hameed K., Tschaplinski T., Misztal P., Wu S., Desirò A., Vande Pol N., Du Z., Zienkiewicz A. Comparative genomics of Mortierella elongata and its bacterial endosymbiont Mycoavidus cysteinexigens. Environ. Microbiol. 2017;19:2964–2983. doi: 10.1111/1462-2920.13669. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y., Liu X.Y., Huang B. The complete mitochondrial genome of Linnemannia amoeboidea (W. Gams) Vandepol & Bonito (Mortierellales: Mortierellaceae) Mitochondrial DNA B. 2022;7:374–376. doi: 10.1080/23802359.2022.2039080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kendrick A., Ratledge C. Desaturation of polyunsaturated fatty acids in Mucor circinelloides and the involvement of a novel membrane-bound malic enzyme. Eur. J. Biochem. 1992;209:667–673. doi: 10.1111/j.1432-1033.1992.tb17334.x. [DOI] [PubMed] [Google Scholar]
- 33.Betina V., Koman V. Changes in the lipid composition during the photo-induced conidiation of Trichoderma viride. Folia Microbiol. 1980;25:295. doi: 10.1007/BF02876608. [DOI] [PubMed] [Google Scholar]
- 34.Chin C.S., Alexander D.H., Marks P., Klammer A.A., Drake J., Heiner C., Clum A., Copeland A., Huddleston J., Eichler E.E. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 35.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 37.Stanke M., Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19:ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 38.El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H., Yohe T., Entwustke S., Wu P.Z., Yang Z.L., Busk P.K., Xu Y., Yin Y.B. dbCAN2: A meta server for automated carbohydrate-activate enzyme annotation. Nucleic Acids Res. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S.K., Cook H., Mende D.R., Letunic I., Rattei T., Jensen L.J. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 45.Medema M.H., Blin K., Cimermancic P., De Jager V., Zakrzewski P., Fischbach M.A., Weber T., Takano E., Breitling R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ou S., Su W., Liao Y., Chougule K., Agda J.R., Hellinga A.J., Lugo C.S.B., Elliott T.A., Ware D., Peterson T. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 2019;20:275. doi: 10.1186/s13059-019-1905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lagesen K., Hallin P., Rødland E.A., Stærfeldt H.H., Rognes T., Ussery D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lowe T.M., Eddy S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finn R.D., Clements J., Eddy S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sánchez R., Serra F., Tárraga J., Medina I., Carbonell J., Pulido L., de María A., Capella-Gutíerrez S., Huerta-Cepas J., Gabaldón T. Phylemon 2.0: A suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Res. 2011;39:W470–W474. doi: 10.1093/nar/gkr408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spatafora J.W., Chang Y., Benny G.L., Lazarus K., Smith M.E., Berbee M.L., Bonito G., Corradi N., Grigoriev I., Gryganskyi A. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia. 2016;108:1028–1046. doi: 10.3852/16-042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.James T.Y., Pelin A., Bonen L., Ahrendt S., Sain D., Corradi N., Stajich J.E. Shared signatures of parasitism and phylogenomics unite Cryptomycota and microsporidia. Curr. Biol. 2013;23:1548–1553. doi: 10.1016/j.cub.2013.06.057. [DOI] [PubMed] [Google Scholar]
- 53.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen L.-T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ellenberger S., Burmester A., Wöstemeyer J. Complete mitochondrial DNA sequence of the mucoralean fungus Absidia glauca, a model for studying host-parasite interactions. Genome Announc. 2016;4:e00153-16. doi: 10.1128/genomeA.00153-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang Y., Desirò A., Na H., Sandor L., Lipzen A., Clum A., Barry K., Grigoriev I.V., Martin F.M., Stajich J.E. Phylogenomics of Endogonaceae and evolution of mycorrhizas within Mucoromycota. New Phytol. 2019;222:511–525. doi: 10.1111/nph.15613. [DOI] [PubMed] [Google Scholar]
- 58.Schwartze V.U., Winter S., Shelest E., Marcet-Houben M., Horn F., Wehner S., Linde J., Valiante V., Sammeth M., Riege K. Gene expansion shapes genome architecture in the human pathogen Lichtheimia corymbifera: An evolutionary genomics analysis in the ancient terrestrial Mucorales (Mucoromycotina) PLoS Genet. 2014;10:e1004496. doi: 10.1371/journal.pgen.1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Linde J., Schwartze V., Binder U., Lass-Flörl C., Voigt K., Horn F. De novo whole-genome sequence and genome annotation of Lichtheimia ramosa. Genome Announc. 2014;2:e00888-14. doi: 10.1128/genomeA.00888-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee S.C., Billmyre R.B., Li A., Carson S., Sykes S.M., Huh E.Y., Mieczkowski P., Ko D.C., Cuomo C.A., Heitman J. Analysis of a food-borne fungal pathogen outbreak: Virulence and genome of a Mucor circinelloides isolate from yogurt. MBio. 2014;5:e01390-14. doi: 10.1128/mBio.01390-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corrochano L.M., Kuo A., Marcet-Houben M., Polaino S., Salamov A., Villalobos-Escobedo J.M., Grimwood J., Álvarez M.I., Avalos J., Bauer D. Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr. Biol. 2016;26:1577–1584. doi: 10.1016/j.cub.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen M.H., Kaul D., Muto C., Cheng S.J., Richter R.A., Bruno V.M., Liu G., Beyhan S., Sundermann A.J., Mounaud S. Genetic diversity of clinical and environmental Mucorales isolates obtained from an investigation of mucormycosis cases among solid organ transplant recipients. Microb. Genom. 2020;6:mgen000473. doi: 10.1099/mgen.0.000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horn F., Üzüm Z., Möbius N., Guthke R., Linde J., Hertweck C. Draft genome sequences of symbiotic and nonsymbiotic Rhizopus microsporus strains CBS 344.29 and ATCC 62417. Genome Announc. 2015;3:e01370-14. doi: 10.1128/genomeA.01370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y., White M.M., Kvist S., Moncalvo J.-M. Genome-wide survey of gut fungi (Harpellales) reveals the first horizontally transferred ubiquitin gene from a mosquito host. Mol. Biol. Evol. 2016;33:2544–2554. doi: 10.1093/molbev/msw126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takeda I., Tamano K., Yamane N., Ishii T., Miura A., Umemura M., Terai G., Baker S.E., Koike H., Machida M. Genome sequence of the Mucoromycotina fungus Umbelopsis isabellina, an effective producer of lipids. Genome Announc. 2014;2:e00071-14. doi: 10.1128/genomeA.00071-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dulermo T., Nicaud J.-M. Involvement of the G3P shuttle and β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab. Eng. 2011;13:482–491. doi: 10.1016/j.ymben.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 68.Beopoulos A., Cescut J., Haddouche R., Uribelarrea J.-L., Molina-Jouve C., Nicaud J.M. Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 2009;48:375–387. doi: 10.1016/j.plipres.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 69.Rodríguez-Frómeta R.A., Gutiérrez A., Torres-Martínez S., Garre V. Malic enzyme activity is not the only bottleneck for lipid accumulation in the oleaginous fungus Mucor circinelloides. Appl. Microbiol. Biot. 2013;97:3063–3072. doi: 10.1007/s00253-012-4432-2. [DOI] [PubMed] [Google Scholar]
- 70.Jareonkitmongkol S., Sakuradani E., Shimizu S. A novel Δ5-desaturase-defective mutant of Mortierella alpina 1S-4 and its dihomo-γ-linolenic acid productivity. Appl. Environ. Microb. 1993;59:4300–4304. doi: 10.1128/aem.59.12.4300-4304.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parker-Barnes J.M., Das T., Bobik E., Leonard A.E., Thurmond J.M., Chaung L.T., Huang Y.S., Mukerji P. Identification and characterization of an enzyme involved in the elongation of n-6 and n-3 polyunsaturated fatty acids. Proc. Natl. Acad. Sci. USA. 2000;97:8284–8289. doi: 10.1073/pnas.97.15.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sakuradani E., Nojiri M., Suzuki H., Shimizu S. Identification of a novel fatty acid elongase with a wide substrate specificity from arachidonic acid-producing fungus Mortierella alpina 1S-4. Appl. Microbiol. Biot. 2009;84:709–716. doi: 10.1007/s00253-009-1999-3. [DOI] [PubMed] [Google Scholar]
- 73.Shin G.H., Veen M., Stahl U., Lang C. Overexpression of genes of the fatty acid biosynthetic pathway leads to accumulation of sterols in Saccharomyces cerevisiae. Yeast. 2012;29:371–383. doi: 10.1002/yea.2916. [DOI] [PubMed] [Google Scholar]
- 74.Tamano K., Bruno K.S., Karagiosis S.A., Culley D.E., Deng S., Collett J.R., Umemura M., Koike H., Baker S.E., Machida M. Increased production of fatty acids and triglycerides in Aspergillus oryzae by enhancing expressions of fatty acid synthesis-related genes. Appl. Microbiol. Biot. 2013;97:269–281. doi: 10.1007/s00253-012-4193-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The newly identified genomics sequences have been deposited in GenBank (Table 2).