Abstract
Background: Ultraviolet (UV) irradiation can modulate host immune responses and this approach is a novel application for treating endodontic infections and inflammation in root canals. Methods: A dataset of UV-induced molecules was compiled from a literature search. A subset of this dataset was used to calculate expression log2 ratios of endodontic tissue molecules from HEPM cells and gingival fibroblasts after 255, 405, and 255/405 nm UV irradiation. Both datasets were analyzed using ingenuity pathway analysis (IPA, Qiagen, Germantown, MD, USA). Statistical significance was calculated using Fisher’s exact test and z-scores were calculated for IPA comparison analysis. Results: The dataset of 32 UV-induced molecules contained 9 antimicrobial peptides, 10 cytokines, 6 growth factors, 3 enzymes, 2 transmembrane receptors, and 2 transcription regulators. These molecules were in the IPA canonical pathway annotations for the wound healing signaling pathway (9/32, p = 3.22 × 10−11) and communication between immune cells (6/32, p = 8.74 × 10−11). In the IPA disease and function annotations, the 32 molecules were associated with an antimicrobial response, cell-to-cell signaling and interaction, cellular movement, hematological system development and function, immune cell trafficking, and inflammatory response. In IPA comparison analysis of the 13 molecules, the predicted activation or inhibition of pathways depended upon the cell type exposed, the wavelength of the UV irradiation used, and the time after exposure. Conclusions: UV irradiation activates and inhibits cellular pathways and immune functions. These results suggested that UV irradiation can activate innate and adaptive immune responses, which may supplement endodontic procedures to reduce infection, inflammation, and pain and assist tissues to heal.
Keywords: ultraviolet irradiation, UV, UVC, UVB, UVA, antimicrobial peptides, chemokines, cytokines, endodontic, inflammation, pain, tissue healing
1. Introduction
Physical traumas, factures, erosions, and local infections, including caries and periodontal disease, on human teeth are among the conditions that lead to endodontic disease [1]. These conditions often create ‘barrier defects’ that allow entry of opportunistic oral microbiota into the underlying dental pulp tissue. Invading microorganisms can then develop into polymicrobial biofilms containing Archaebacteria, Eubacteria, yeast, and fungal species [2,3,4]. The resulting infections can induce inflammation and pain in the root canal systems. The associated craniofacial pain can be severe and significantly impact the patient’s quality of life and day-to-day comfort.
Current standard-of-care treatment for endodontic emergencies and treatment include opening the tooth; exposing underlying inflammatory tissue or infection; and creating an open instrumented root canal to the tooth apex, removing the inflammatory tissue, canal exudate, necrotic tissue, and tissue debris [3]. The root canal is then irrigated with sodium hypochlorite (NaOCl) to cleanse and dissolve the canal of the remaining tissue and debris, followed by the irrigation of ethylenediaminetetraacetic acid (EDTA) to remove the smear layer and open the dentinal tubules. The root canal is completed by sealing the canal space with endodontic sealer and gutta percha. The access opening is filled with amalgam or a composite material, and in some cases, a crown is advocated to enhance tooth integrity and adequate seal.
Unfortunately, the standard-of-care treatment is unable to completely clean the canal space and residual pulp tissue debris, missed areas of infection, and remnant microorganisms in the dentin tubules along the sides of the root canal can be left; resulting in potential persist reinfections and apical chronic inflammation [3]. Irrigation with NaOCl is the gold standard, but extrusion of this irrigant apically can be extremely detrimental to the apical tissues. The damaged apical tissues by the NaOCl have little chance to regenerate and heal with no opportunities for activation of local innate or adaptive immune responses in the area.
An emerging and novel concept is to treat instrumented root canals with ultraviolet (UV) irradiation during the standard-of-care treatment procedure [5,6]. UVC irradiation kills microorganisms isolated from endodontic infections [6,7,8]. Brief treatment with UV irradiation would also modulate host immune responses [9,10]. UV irradiation induces an influx of cells and the production of antimicrobial peptides (AMPs); chemokines, cytokines, and biomarkers (CCBMs); and other molecules that alter lesion pathogenesis and facilitates local tissue healing [9,10]. UVC induces the secretion of CCBMs in HEPM cells and gingival fibroblasts related to endodontic tissue regeneration [6].
In this study, we were interested in identifying molecules expressed or secreted from cells and tissues after UV irradiation. We first searched the PubMed literature to identify AMPs or CCBMs reported to have up- or down-levels of mRNA expression or secretion in response to UV irradiation treatment. We used a bioinformatics approach with ingenuity pathway analysis (IPA, Qiagen, Germantown, MD, USA) to associate their expression with specific innate and adaptive immune responses. We then used the concentrations of 13 CCBMs in tissue culture media of HEPM cells and gingival fibroblasts after treatment with 255, 405, or 255/405 UV irradiation [6] as (i) wet lab experimental data to validate claims from the bioinformatics data, and (ii) to assess the ability of UV irradiation to activate or inhibit cellular pathways related to innate and adaptive immune responses. We hypothesized that UV irradiation would induce host cells and tissues to express AMPs and CCBMs (Figure 1) and these molecules would be important in future treatments designed to reduce endodontic infection and inflammation, modulate endodontic pain, and assist in endodontic tissue healing.
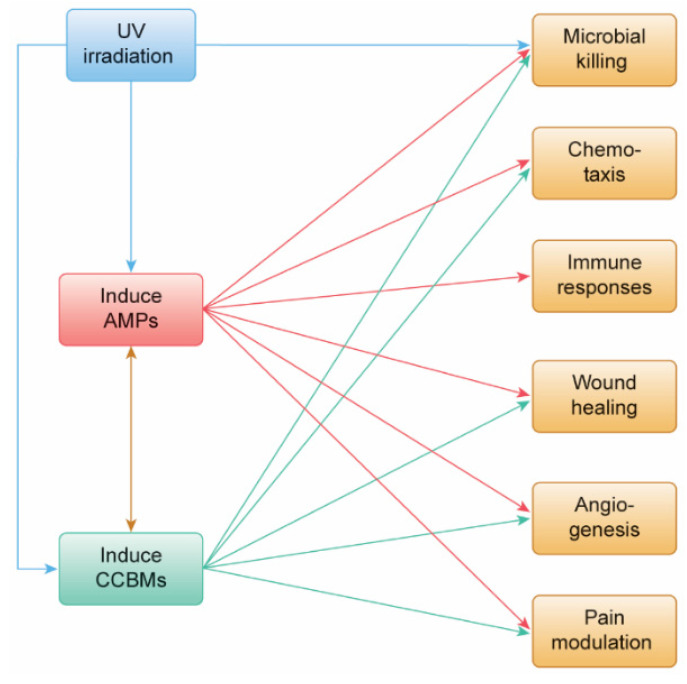
Figure 1.
A schematic diagram of the proposed effects of UV irradiation on endodontic infection and inflammation, pain, and tissue healing. UV irradiation can kill microorganisms directly (blue line). UV irradiation can also induce host cells to express antimicrobial peptides (AMPs) and chemokines, cytokines, and biomarkers (CCBMs) (blue lines). AMPs can kill microorganisms (red line); induce the production of CCBMs (brown line); and induce chemotaxis, modulate immune responses, assist in wound healing, play a role in angiogenesis, and reduce pain (red lines). CCBMs can kill microorganisms (green line); induce the production of AMPs (brown line); and induce chemotaxis, modulate immune responses, assist in wound healing, play a role in angiogenesis, and reduce pain (green lines).
2. Materials and Methods
2.1. Dataset of UV-Induced Molecules
We searched the PubMed literature using UVC, UVB, UVA, chemokines, cytokines, and antimicrobial peptides as search terms linked in various combinations, using Boolean operators to identify AMPs or CCBMs reported to have up- or down-levels of mRNA expression or secretion in response to UV irradiation. AMPs and CCBMs were combined into a single dataset (Table 1).
Table 1.
Antimicrobial peptides (AMPs); chemokines, cytokines, and biomarkers (CCBMs); and other mediators were identified from ingenuity pathway analysis (IPA, Qiagen, Germantown, MD, USA), combined into a single dataset below, annotated for their IPA symbol, Entrez Gene name, Entrez Gene ID (human), cellular location, and function type.
| Symbol | Entrez Gene Name | Entrez Gene ID (Human) | Location | Function Type |
|---|---|---|---|---|
| BMP10 | Bone morphogenetic protein 10 | 27302 | Extracellular space | Growth factor |
| CAMP | Cathelicidin antimicrobial peptide | 820 | Cytoplasm | Other |
| CCL2 | C-C motif chemokine ligand 2 | 6347 | Extracellular space | Cytokine |
| CCL20 | C-C motif chemokine ligand 20 | 6364 | Extracellular space | Cytokine |
| CSF2 | Colony stimulating factor 2 | 1437 | Extracellular space | Cytokine |
| CXCL1 | C-X-C motif chemokine ligand 1 | 2919 | Extracellular space | Cytokine |
| CXCL2 | C-X-C motif chemokine ligand 2 | 2920 | Extracellular space | Cytokine |
| CXCL3 | C-X-C motif chemokine ligand 3 | 2921 | Extracellular space | Cytokine |
| CXCL8 | C-X-C motif chemokine ligand 8 | 3576 | Extracellular space | Cytokine |
| DEFB1 | Defensin beta 1 | 1672 | Extracellular space | Other |
| DEFB103B | Defensin beta 103B | 55894 | Extracellular space | Other |
| DEFB4A | Defensin beta 4A | 1673 | Extracellular space | Other |
| FGF1 | Fibroblast growth factor 1 | 2246 | Extracellular space | Growth factor |
| FGF2 | Fibroblast growth factor 2 | 2247 | Extracellular space | Growth factor |
| FN1 | Fibronectin 1 | 2335 | Extracellular space | Enzyme |
| ICAM1 | Intercellular adhesion molecule 1 | 3383 | Plasma membrane | Transmembrane receptor |
| IL6 | Interleukin 6 | 3569 | Extracellular space | Cytokine |
| IL10 | Interleukin 10 | 3586 | Extracellular space | Cytokine |
| PI3 | Peptidase inhibitor 3 | 5266 | Extracellular space | Other |
| PIGF | Phosphatidylinositol glycan anchor biosynthesis class F |
5281 | Cytoplasm | Enzyme |
| RNASE7 | Ribonuclease A family member 7 | 84659 | Extracellular space | Enzyme |
| S100A7 | S100 calcium binding protein A7 | 6278 | Cytoplasm | Other |
| S100A8 | S100 calcium binding protein A8 | 6279 | Cytoplasm | Other |
| S100A9 | S100 calcium binding protein A9 | 6280 | Cytoplasm | Other |
| S100A12 | S100 calcium binding protein A12 | 6283 | Cytoplasm | Other |
| SELE | Selectin E | 6401 | Plasma membrane | Transmembrane receptor |
| SMAD3 | SMAD family member 3 | 4088 | Nucleus | Transcription regulator |
| SMAD4 | SMAD family member 4 | 4089 | Nucleus | Transcription regulator |
| TGFA | Transforming growth factor alpha | 7039 | Extracellular space | Growth factor |
| TGFB1 | Transforming growth factor beta 1 | 7040 | Extracellular space | Growth factor |
| TNF | Tumor necrosis factor | 7124 | Extracellular space | Cytokine |
| VEGFA | Vascular endothelial growth factor A | 7422 | Extracellular space | Growth factor |
2.2. Subset of Endodontic Tissue Molecules
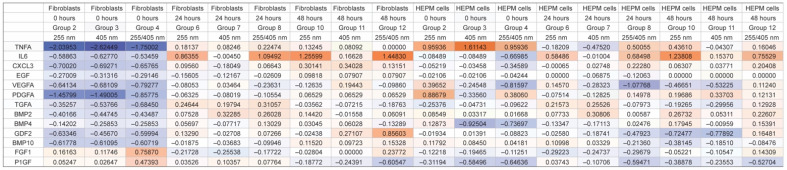
Morio et al. reported the concentrations of 13 CCBMs in tissue culture media of HEPM cells and gingival fibroblasts at 0, 24, and 48 h after treatment with 255, 405, or 255/405 UV irradiation [6]. We used these CCBMs as (i) wet lab experimental data to validate claims from the bioinformatics data, and (ii) to assess the ability of UV irradiation to activate or inhibit cellular pathways related to immune functions. For this, levels of CCBM expression were calculated as the expression log2 ratios of the mean of each treatment after UV irradiation, divided by the mean of the respective control for the same treatment (Figure 2).
Figure 2.
Expression Log2 ratios of 13 chemokine, cytokine, and biomarker (CCBMs) concentrations reported in tissue culture media of HEPM cells and gingival fibroblasts at 0, 24, and 48 h after treatment with 255 nm, 405 nm, or 255/405 nm UV irradiation. Expression was calculated as the log2 ratio of the mean of each treatment after UV irradiation over the mean of the untreated control for that same cell type, UV irradiation wavelength, and time period. Groups are shown as a heatmap, where blue represents inhibition (negative values), white represents midpoint, and orange represents activation (positive values).
2.3. Analysis
We used ingenuity pathway analysis (IPA, Qiagen, Germantown, MD, USA) to assess whether the molecules in this study were related to the activation of innate and immune mechanisms. Two types of analysis were performed.
IPA core analysis was run on the list of 32 molecules from the literature dataset in Table 1 and used to assess whether the IPA canonical pathway and IPA diseases and function annotations were predicted to be associated with relevant diseases, immune pathways, and immune functions. Statistical significance was calculated using Fisher’s exact test and significant p values (p < 0.05) were reported.
IPA comparison analysis was run on the expression log2 ratios of the 13 molecules and used to assess whether the IPA canonical pathway and IPA diseases and function annotations were predicted to be activated or inhibited after UV irradiation. Statistical differences were determined using activation z-scores calculated from the mean of each treatment expression log2 ratio. The activation z-score was determined by IPA as reported by Kramer et al. [11] and makes predictions based on the direction of gene activation or inhibition.
IPA comparison analysis was also used to assess whether the IPA canonical pathway predicted any effects of the activated or inhibited 13 molecules on downstream regulation of gene expression for other innate or adaptive immune functions.
3. Results
3.1. Dataset of 32 UV-Induced Molecules
We identified 32 unique molecules reported to be expressed after UV irradiation of cells and tissues (Table 1). There were 9 antimicrobial peptides (Supplementary Table S1), 10 cytokines, 6 growth factors, 3 enzymes, 2 transmembrane receptors, and 2 transcription regulators (Supplementary Table S2).
IPA analysis predicted that UV irradiation can induce molecules that are involved in innate and adaptive immune responses. IPA canonical pathway annotations of these 32 molecules (Table 1) were predicted to be associated with cellular stress and injury. This category included the wound healing signaling pathway (9/32, p = 3.22 × 10−11). Annotations were also associated with cellular immune responses, and this category included the role of cytokines in mediating communication between immune cells (6/32, p = 8.74 × 10−11), communication between innate and adaptive immune cells (5/32, p = 7.44 × 10−3), the Th1 and Th2 activation pathway (4/32, p = 8.01 × 10−5), the Th1 pathway (3/32, p = 5.81 × 10−4), and the Th2 pathway (3/32, p= 8.13 × 10−4). The top relevant pathway annotations are listed in Table 2 and all the relevant annotations ranked by their −log(p value) are listed in Supplementary Table S3. This list contains relevant annotations within categories on cytokine signaling; growth factor signaling; intracellular and second messenger signaling; cellular growth, proliferation, and development; cellular immune response; and organismal growth and development.
Table 2.
Ingenuity pathway analysis (IPA, Qiagen, Germantown, MD, USA) was used to assess whether the biomarkers in the literature dataset (n = 32) would participate in the activation of innate and immune mechanisms applicable to reducing endodontic infection, reducing inflammation, and assisting in endodontic tissue healing. Representative IPA canonical pathways annotations were associated with cellular stress and injury; cytokine signaling; cellular immune response. Representative IPA diseases or functions annotations were associated with antimicrobial response, cell-to-cell signaling and interaction, cellular movement, hematological system development and function, immune cell trafficking, and inflammatory response.
| IPA Function | p-Value | No. | Identification of Molecules |
|---|---|---|---|
| Canonical Pathway Annotations | |||
| Cellular Stress and Injury | |||
| Wound healing signaling pathway | 3.22 × 10−11 | 9 | CSF2, CXCL8, FGF2, FN1, IL6, TGFA, TGFB1, TNF, VEGFA |
| Cytokine Signaling | |||
| IL17 signaling | 8.43 × 10−20 | 13 | CCL2, CCL20, CSF2, CXCL1, CXCL3, CXCL8, DEFB1, DEFB103A/DEFB103B, DEFB4A/DEFB4B, IL6, TGFB1, TNF, VEGFA |
| IL6 signaling | 2.53 × 10−5 | 4 | CXCL8, IL6, TNF, VEGFA |
| IL10 signaling | 1.23 × 10−4 | 3 | IL10, IL6, TNF |
| IL8 signaling | 1.73 × 10−4 | 4 | CXCL1, CXCL8, ICAM1, VEGFA |
| Cellular Immune Response | |||
| Role of cytokines in mediating communication between immune cells | 8.74 × 10−11 | 6 | CSF2, CXCL8, IL10, IL6, TGFB1, TNF |
| Th1 and Th2 activation pathway | 8.01 × 10−5 | 4 | ICAM1, IL10, IL6, TGFB1 |
| Th1 pathway | 5.81 × 10−4 | 3 | ICAM1, IL10, IL6 |
| Th2 pathway | 8.13 × 10−4 | 3 | ICAM1, IL10, TGFB1 |
| Communication between innate and adaptive immune cells | 7.44 × 10−3 | 5 | CSF2, CXCL8, IL10, IL6, TNF |
| Diseases or Functions Annotations | |||
| Antimicrobial Response, Inflammatory Response | |||
| Antibacterial response | 1.66 × 10−22 | 13 | CAMP, CCL20, DEFB1, DEFB103A/DEFB103B, DEFB4A/DEFB4B, IL10, IL6, RNASE7, S100A12, S100A7, S100A8, S100A9, TNF |
| Antimicrobial Response, Inflammatory Response | |||
| Chemoattraction | 5.09 × 10−23 | 12 | CAMP, CCL2, CCL20, CSF2, CXCL1, CXCL3, CXCL8, DEFB4A/DEFB4B, FN1, TGFB1, TNF, VEGFA |
| Cellular Movement, Hematological System Development and Function, Immune Cell Trafficking, Inflammatory Response | |||
| Chemotaxis | 4.28 × 10−37 | 27 | CAMP, CCL2, CCL20, CSF2, CXCL1, CXCL2, CXCL3, CXCL8, DEFB1, DEFB103A/DEFB103B, DEFB4A/DEFB4B, FGF2, FN1, ICAM1, IL10, IL6, S100A12, S100A7, S100A8, S100A9, SELE, SMAD3, SMAD4, TGFA, TGFB1, TNF, VEGFA |
| Chemotaxis of leukocytes | 1.43 × 10−35 | 24 | CAMP, CCL2, CCL20, CSF2, CXCL1, CXCL2, CXCL3, CXCL8, DEFB1, DEFB103A/DEFB103B, DEFB4A/DEFB4B, FN1, ICAM1, IL10, IL6, S100A12, S100A7, S100A8, S100A9, SELE, SMAD3, TGFB1, TNF, VEGFA |
| Inflammatory Response | |||
| Inflammatory response | 9.34 × 10−29 | 26 | CAMP, CCL2, CCL20, CSF2, CXCL1, CXCL2, CXCL3, CXCL8, DEFB1, DEFB103A/DEFB103B, DEFB4A/DEFB4B, FGF1, FGF2, FN1, ICAM1, IL10, IL6, S100A12, S100A7, S100A8, S100A9, SELE, SMAD3, TGFB1, TNF, VEGFA |
| Proinflammatory response | 4.92 × 10−15 | 7 | CCL2, CXCL3, CXCL8, IL10, IL6, TNF, VEGFA |
| Innate immune response | 5.94 × 10−14 | 10 | CAMP, CXCL1, CXCL8, FN1, IL10, IL6, RNASE7, S100A12, SMAD3, TNF |
| Tissue Development | |||
| Healing of wound | 1.53 × 10−20 | 13 | CSF2, FGF1, FGF2, FN1, ICAM1, IL10, IL6, SMAD3, SMAD4, TGFA, TGFB1, TNF, VEGFA |
| Cell-To-Cell Signaling and Interaction, Cellular Movement, Hematological System Development and Function, Immune Cell Trafficking, Inflammatory Response | |||
| Cell movement of monocytes | 1.37 × 10−31 | 19 | CAMP, CCL2, CCL20, CSF2, CXCL3, CXCL8, DEFB1, DEFB103A/DEFB103B, FN1, ICAM1, IL10, IL6, S100A12, S100A7, SELE, SMAD3, TGFB1, TNF, VEGFA |
| Cell movement of neutrophils | 2.01 × 10−28 | 21 | CAMP, CCL2, CSF2, CXCL1, CXCL2, CXCL3, CXCL8, DEFB1, DEFB103A/DEFB103B, DEFB4A/DEFB4B, FN1, ICAM1, IL10, IL6, S100A12, S100A8, S100A9, SELE, SMAD3, TGFB1, TNF |
| Cell-To-Cell Signaling and Interaction, Cellular Movement, Hematological System Development and Function, Immune Cell Trafficking, Inflammatory Response | |||
| Recruitment of cells | 7.85 × 10−29 | 21 | BMP10, CAMP, CCL2, CCL20, CSF2, CXCL1, CXCL2, CXCL3, CXCL8, DEFB4A/DEFB4B, FGF2, FN1, ICAM1, IL10, IL6, S100A8, SELE, SMAD3, TGFB1, TNF, VEGFA |
| Recruitment of leukocytes | 1.05 × 10−25 | 19 | BMP10, CAMP, CCL2, CCL20, CSF2, CXCL1, CXCL2, CXCL3, CXCL8, DEFB4A/DEFB4B, FN1, ICAM1, IL10, IL6, S100A8, SELE, SMAD3, TGFB1, TNF |
| Cell-To-Cell Signaling and Interaction, Inflammatory Response | |||
| Immune response of cells | 2.70 × 10−18 | 18 | CAMP, CCL2, CCL20, CSF2, CXCL1, CXCL3, CXCL8, FN1, ICAM1, IL10, IL6, S100A12, S100A8, S100A9, SMAD3, TGFB1, TNF, VEGFA |
| Immune response of myeloid cells | 2.85 × 10−17 | 13 | CAMP, CCL2, CSF2, CXCL1, CXCL3, CXCL8, FN1, ICAM1, IL10, IL6, S100A9, TGFB1, TNF |
| Cellular Growth and Proliferation | |||
| Angiogenesis | 1.74 × 10−23 | 24 | BMP10, CAMP, CCL2, CSF2, CXCL1, CXCL2, CXCL8, FGF1, FGF2, FN1, ICAM1, IL10, IL6, PIGF, S100A12, S100A8, S100A9, SELE, SMAD3, SMAD4, TGFA, TGFB1, TNF, VEGFA |
| Proliferation of vascular cells | 3.32 × 10−20 | 16 | CAMP, CCL2, CXCL1, CXCL8, FGF1, FGF2, FN1, IL10, IL6, S100A8, S100A9, SMAD3, SMAD4, TGFB1, TNF, VEGFA |
| Cellular Movement, Hematological System Development and Function, Immune Cell Trafficking | |||
| Cell survival | 1.73 × 10−17 | 24 | CAMP, CCL2, CSF2, CXCL1, CXCL2, CXCL3, CXCL8, DEFB103A/DEFB103B, DEFB4A/DEFB4B, FGF1, FGF2, FN1, ICAM1, IL10, IL6, S100A8, S100A9, SELE, SMAD3, SMAD4, TGFA, TGFB1, TNF, VEGFA |
| Cell viability | 2.74 × 10−15 | 22 | CAMP, CCL2, CSF2, CXCL1, CXCL2, CXCL3, CXCL8, FGF1, FGF2, FN1, ICAM1, IL10, IL6, S100A8, S100A9, SELE, SMAD3, SMAD4, TGFA, TGFB1, TNF, VEGFA |
IPA disease and function annotations of these 32 molecules (Table 1) were predicted to be associated with innate and adaptive immune responses, applicable to reducing infection and inflammation and assisting in tissue healing. These included antimicrobial response, inflammatory response, cell-to-cell signaling and interaction, cellular movement, immune cell trafficking, cell death and survival, cellular growth and proliferation, cellular development, hematological system development and function, hematopoiesis, lymphoid tissue structure and development, tissue development, and tissue morphology. Examples of these functions are listed in Table 2 and all the relevant IPA disease and function annotations are listed in Supplementary Table S4.
3.2. Subset of 13 Endodontic Tissue Molecules
IPA analysis also predicted that different wavelengths of UV irradiation might selectively regulate gene expression for innate or adaptive immune functions. IPA comparison analysis of the log2 ratios of the concentrations of 13 CCBMs in tissue culture media of HEPM cells and gingival fibroblasts at 0, 24, and 48 h after treatment with 255, 405, or 255/405 UV irradiation identified differences in expression across 18 observations, representing 2 cell lines, 3 UV treatments, and 3 time periods (Figure 3). The expression log2 ratios of the 13 CCBMs varied from −2.6245 to 1.6114. Relevant IPA canonical pathways in Figure 3 were related to cellular stress and injury (wound healing signaling pathway, the CLEAR signaling pathway, HIF1α signaling pathway, and autophagy), cytokine signaling (IL17 signaling, IL6 signaling, and NF-κB signaling), growth factor signaling (regulation of the epithelial mesenchymal transition by growth factors pathway), and organismal growth and development (ID1 signaling pathway). IPA comparison analysis predicted that UV irradiation activated or inhibited pathways depending upon the cell type, wavelength of treatment, and time after treatment. Numerous IPA canonical pathway annotations were inhibited shortly after irradiation (0 h) but activated at 24 and 48 h. Fibroblasts and HEPM cells both were strongly activated by 405 nm and 255/405 nm UV irradiation treatments (Figure 3).
Figure 3.
IPA comparison analysis of 18 observations from fibroblasts (observations 1–9) and HEPM cells (observations 10–18) at 0 h (observations 1–3, 10–12), 24 h (observations 4–6, 13–15), and 48 h (observations 7–9, 16–18) after treatment with 255 nm (observations 1, 4, 7, 10, 13, and 16), 405 nm (observations 2, 5, 8, 11, 14, 17), or 255/405 nm (observations 3, 6, 9, 12, 15, and 18) irradiation. Groups are shown as a heatmap, where blue represents inhibition (negative values), white represents midpoint, and orange represents activation (positive values). Numerous IPA canonical pathways were inhibited shortly after irradiation (0 h) but activated at 24 and 48 h. Fibroblasts and HEPM cells both were strongly activated by 405 nm and 255/405 nm UV irradiation treatments.
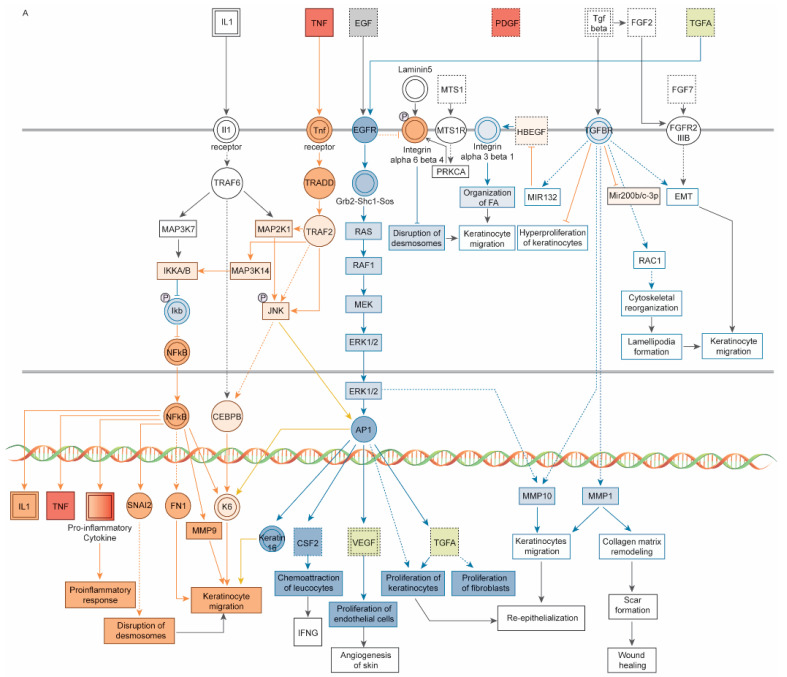
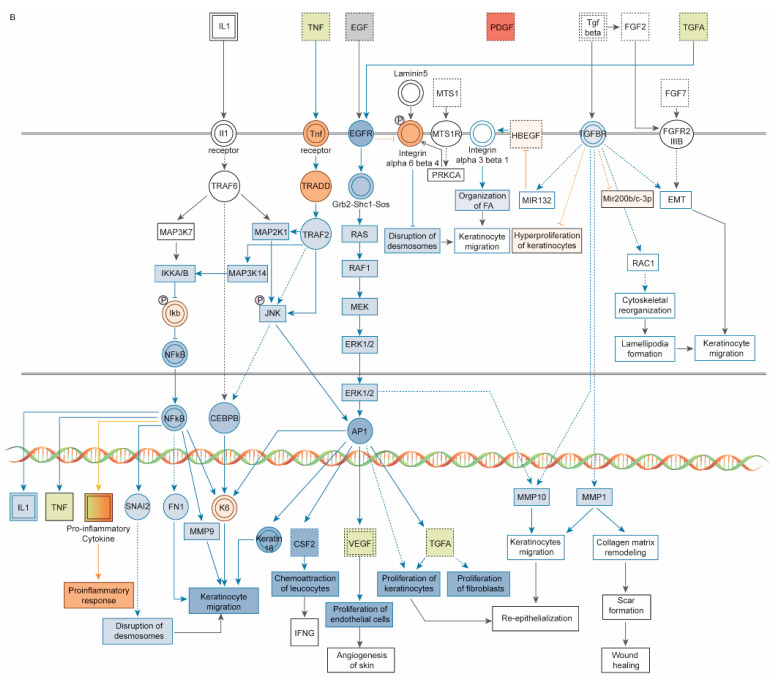
Within the wound healing signaling pathway, for example, after 0 h of 255, 405, and 255/405 nm irradiation, fibroblasts and HEPM cells (observations 1–3, 10–12) had negative z-scores, predicting inhibition of pathway signaling, whereas after 24 and 48 h after 255, 405, and 255/405 nm irradiation, fibroblasts (observations 4–9) and after 48 h of 255, 405, and 255/405 nm irradiation, HEPM cells (observations 16–18) had positive z-scores, predicting activation of pathway signaling. There were also conditions predicted to activate some pathways yet inhibit others (observations 3, 9, 12, 15, and 18). Here, treatment with cells with 255/405 nm irradiation contained both inhibited and activated pathway signaling. After 48 h of 255/405 nm irradiation, HEPM cells (observation 18) had a positive z-score but contained both inhibited and activated pathway signaling (Figure 4A–C). Signaling was predicted to occur through binding of molecules to the TNF receptor, EGFR, and TGFBR and signaling through JNK and ERK1/2 to transcription factors NF-κB, CEBPB, and AP-1. This was predicted to activate additional CCBMs, leading to proinflammatory responses, disruption of desmosomes, chemoattraction of leukocytes, migration and proliferation of fibroblasts and cells, collagen matrix remodeling, and wound healing pathways.
Figure 4.
Schematic diagrams of the wound healing signaling pathway, prepared using Ingenuity Pathway Analysis software (IPA, Qiagen, Germantown, MD), showing both inhibited and activated pathway signaling in HEPM cells 48 h after (A) 255 nm irradiation, (B) 405 nm irradiation, and (C) 255/405 nm irradiation. Signaling starts via TNF binding to the TNF receptor; EGF and TGFA binding to the EGFR; and TGFB binding to the TGFBR. These pathways signal through TRADD and TRAF2 to JNK and through RAS, RAF, and MEK to ERK1/2. Signaling continues to NF-κB, CEBPB, and AP-1 to activate additional CCBMs, leading to proinflammatory responses, disruption of desmosomes, chemoattraction of leukocytes, migration and proliferation of fibroblasts and cells, collagen matrix remodeling, and wound healing pathways. Pathway molecules in red indicate activation and molecules in green indicated inhibition. Signaling connections in orange indicate pathway activation and signaling connections in blue indicate pathway inhibition.
3.3. Modulating Endodontic Pain
While UV irradiation is not known to be directly anti-nociceptive, it does act on ERK, p38 MAPK, JNK, and NF-κB signaling pathways (Supplementary Figure S1) that produce AMPs and CCBMs that are involved in pain reduction (Supplementary Figure S2). Many of the molecules induced by UV irradiation (Table 1) can be pro-nociceptive, nociceptive, and anti-nociceptive stimuli. AMPs can be anti-nociceptive stimuli or pro-nociceptive stimuli, chemokines can be pro-nociceptive or nociceptive stimuli, and growth factors can be anti-nociceptive stimuli.
4. Discussion
We identified a list of 32 molecules expressed or secreted from cells and tissues after UV irradiation and used a bioinformatics approach to show that they were related to wound healing and innate and adaptive immune functions, including chemotaxis, movement, growth, and proliferation of cells. We then used a subset of 13 osteoinductive, angiogenic, proliferative, and proinflammatory molecules to show that HEPM cells and gingival fibroblasts treated with 255, 405, and 255/405 nm UV irradiation had different expression profiles. These results suggested that UV irradiation can activate innate and adaptive immune responses, which may supplement endodontic procedures to reduce infection, inflammation, and pain and assist tissues to heal.
Wound healing is a complex process, whereby secreted molecules from infected, injured, or damaged cells and tissues attract a variety of inflammatory cells to the injured site (inflammatory phase). Once present, inflammatory cells release additional CCBMs. These include growth factors that attract and transform fibroblasts and molecules that stimulate attracted cells to proliferate and stimulate other cells to begin forming new capillaries and blood vessels (proliferative phase). Tissue development, angiogenesis, and vasculogenesis occurs (maturation and remodeling phase). IPA identified 13/32 molecules (p = 1.53 × 10−20) in the IPA canonical pathway annotations associated with these phases in the wound healing signaling pathway (Table 2, Supplementary Table S3). Of the 13 molecules, TNFA, a proinflammatory cytokine, is secreted by activated monocytes, macrophages, B-cells, T-cells, and fibroblasts; IL6 regulates immune and inflammatory responses, including B-cell differentiation and antibody production; and IL10 inhibits the expression of pro-inflammatory cytokines, but enhances humoral immune responses and attenuates cell mediated immune reactions. CSF2 stimulates the development of neutrophils and macrophages. Growth factors including FGF1, FGF2, TGFA, TGFB1, and VEGFA are involved in cell motility; cell proliferation; cell growth and differentiation; redistribution of tissue; angiogenesis and vascular permeability of endothelial cells; and synthesis and deposition of the extracellular matrix. FN1 and collagen from fibroblasts allow tissues to contract [12], SMAD3 and SMAD4 play roles in the signaling of TGFB1 [13], and ICAM1 provides adhesion between endothelial cells and leukocytes after stress or injury.
Innate immunity is a type of nonspecific host resistance without memory, involving soluble molecules and cells [14]. Stimulation of receptors activates several cellular pathways, resulting in the production of AMPs and inflammatory cytokines. Stimulation also leads to changes in cellular metabolism, upregulation of numerous genes involved in cell defense and pathogen restriction, and the induction of regulated cell death [14].
AMPs are a large component of innate immune responses and UV irradiation induces their transcription and secretion (Supplementary Table S1). Their ability to modulate both innate immune responses, cellular immunity, and angiogenesis are very well known [15,16,17,18]. IPA identified 14/32 molecules involved in antimicrobial and antibacterial response annotations (Supplementary Table S4). These included CAMP (LL37), the defensin family (DEFB1, DEFB103A/DEFB103B, and DEFB4A/DEFB4B), the S100 family of calcium binding proteins (S100A7, S100A8, S100A9, and S100A12), and RNASE7. It also included the chemokine CCL20, which has antimicrobial activity [19] and cytokines IL6, IL10, TNFA, and TGFB1, which contribute to the antibacterial response. So far, AMP expression and secretion has been reported to occur in a narrow range from 280 to 400 nm (Supplementary Table S1). AMP expression is increased after 280–313 nm irradiation [20,21], but not after 340 to 400 nm irradiation [22,23]. Thus, UV irradiation of an infected root canal would not have to kill 100% of microorganisms, but simply reduce the microbial infection burden to a level that could be managed by UV-induced innate immune mechanisms.
CCBMs are large components of innate immune responses and UV irradiation induces their transcription and secretion (Supplementary Table S2), CCBM expression and secretion has been reported to occur in a wider range from 254 to 404 nm (Supplementary Table S2). In irradiated cells, CCBM expression is increased for many CCBMs after 200 to 320 nm irradiation but decreased for others, such as BMP10 and FGF1 in HEPM cells and VEGF in fibroblasts [6]. At 340 to 405 nm irradiation, cells had increased levels of IL6, CXCL8, and CSF mRNA expression and secretion in keratinocytes [24,25] but decreased levels of secreted FGF1 in fibroblasts [6]. In irradiated human and murine skin at 2–3 MED (minimal erythemal dose), there were increased levels of immunostaining for CCL2, CCL20, CXCL1, CXCL8, ICAM1, IL1, IL10, SELE, SMAD3, SMAD4, TGFA, TGFB, TNFA, and VEGF 24–48 h after exposure [26,27,28].
UV-induced AMPs and CCBMs would have a variety of common functions (Figure 1). In addition to their potent antimicrobial activity mentioned above, these UV-induced molecules can chemoattract a variety of cells important to both immune protection and wound healing. Defensins attract keratinocytes, dendritic cells, and T-cells [29,30] and CAMP (LL37) attracts fibroblasts, microvascular endothelial cells, and human umbilical vein endothelial cells [31]. AMPs can regulate proinflammatory CCBM production [32]. At lower concentrations, defensins do not induce TNFA or IL1B expression in monocytes or macrophages [33,34]. However, at higher concentrations, defensins induce CCBM production in epithelial cells, keratinocytes, monocytes, and macrophages [30,35,36] and CAMP (LL37) induces CXCL8 in epithelial cells and macrophages [37]. Finally, AMPs and CCBMs play a direct role in wound healing, angiogenesis, and vasculogenesis [18]. DEFB4A (HBD2) increases keratinocyte proliferation [30,38] and CAMP (LL37) increases fibroblast proliferation, induces human microvascular endothelial cell and human umbilical vein endothelial cell proliferation and stimulates re-epithelialization [31,39,40].
UV irradiation also suppresses cellular immunity and acts primarily on T-cell-mediated immune reactions [41,42,43]. This application has been used to treat several T-cell-mediated diseases, including graft-versus-host disease and systemic scleroderma [44]. UV irradiation alters antigen specificity, alters antigen-presenting cell function, acts on effector and regulatory T-cells [41] and induces the production of CCBMs [42]. For example, UV irradiated dendritic cells do not present antigens effectively, and thus induce regulatory T-cells (CD4+CD25+), but not effector T-cells [41]. UV irradiation can lead to T-cell tolerance and prevents the priming of antigen-specific CD8+ T-cells (in models of contact hypersensitivity) independent of conventional CD4+ regulatory T-cells [44]. Tolerant CD8+ T-cells prevented migration of dendritic cells and prevented priming of other CD8+ T-cells. TGFB and immunosuppressive IL10 are regulatory T-cell-associated cytokines [10,45].
There are differences based on the specific wavelength. UVB induces the infiltration of immature inflammatory myeloid CD11c+ bDCA1- dendritic cells, which may have a suppressive function [10]. UVA1 does not induce IL10, but does suppress the production of TNFA and IL12, and contributes to cis-UCA isomerization [10,46]. Immune suppression may be dependent upon the extent of UV irradiation-induced damage to DNA [41]. However, to what extent the secondary immunostimulatory effects of UV-induced AMPs and CCBMs offset the immunosuppressive effects of immune cells is not yet known.
Many oral related infections, inflammation, and tissue injury/peripheral nerve injury can be stimuli that activate receptors on the surface of cells, initiating signal transduction through MAPK and NF-κB signaling pathways (Supplementary Figure S2) [47]. The MAPK pathway regulates proinflammatory and pronociceptive molecules involved in inflammation and pain [48]. Nociceptive activity or nerve injury stimuli signal through raf and MEK1/2 to ERK in the cytoplasm and transcription factor CREB in the nucleus [48]. Chemokines (FKN), cytokines (TNFA), and nerve injury stimuli signal through TAK1 and MKK3,6 to p38 MAPK in the cytoplasm and on to transcription factor ATF-2 in the nucleus [48]. Cytokines (TNFA), growth factors BFGF (FGF2), and nerve injury stimuli signal through MLK3/MEKK1 and MKK4,7 to JNK in the cytoplasm and on to transcription factor c-Jun (AP-1) in the nucleus [48].
UV irradiation also alters the expression of ERK, p38 MAPK, JNK, and NF-κB signaling pathways [49] (Supplementary Figure S1) and it is possible that treatment of endodontic infections, inflammation, and tissue injury/peripheral nerve injury can modulate downstream production of CCBMs and be a potential intervention at these nodes to mitigate acute or chronic pain. Phosphorylation of ERK in nerve injury is induced early, is long lasting, and is involved in the induction of pain (Supplementary Figure S2). Suppressing this step in ERK is thought to be a promising strategy for treatment of neuropathic pain [50]. Likewise, targeting the p38 MAPK pathway and its signaling is also thought to be a potential therapeutic strategy for pain management [51].
Our analysis and results support those of Ou and Peterson [9] and Vieyra-Garcia et al. [10] and also suggest that UV irradiation can induce the production of AMPs and CCBMs. Our results also suggest that the production of these molecules can induce the innate and adaptive immune responses involved in attenuating infection, inflammation, and pain and enhancing healing and regeneration of tissue. However, these results are based on bioinformatics analysis of molecules induced by UV irradiation reported in the literature and produced in culture from cells treated with UV irradiation. These concepts and results form a strong hypothesis for future studies and should be examined in detail.
5. Conclusions
In summary, UV irradiation has the ability to kill microorganisms, but could also be used to activate innate and adaptive immune mechanisms in endodontic root canals directly or through UV-induced molecules. UV irradiation-induced effects appear to be wavelength specific and could supplement procedures to reduce infection, to reduce inflammation, and to facilitate local tissue healing.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pharmaceutics14091979/s1, Supplementary Figure S1: A schematic diagram of UV-induced MAPK signaling; Supplementary Figure S2: A schematic diagram of neuropathic pain signaling; Supplementary Table S1: Antimicrobial peptides (AMPs) expressed or secreted in cells, tissues, and tissue explants after irradiation with UVC, UVB, or UVA; Supplementary Table S2: Chemokines, cytokines, and biomarkers (CCBMs) expressed or secreted in cells, tissues, and tissue explants after irradiation with UVC, UVB, or UVA; Supplementary Table S3: Antimicrobial peptides and biomarkers in the literature dataset (n = 32) participating in the activation of innate and immune mechanisms, as determined using IPA canonical pathways annotations; and Supplementary Table S4: Antimicrobial peptides and biomarkers in the literature dataset (n = 32) participating in the activation of innate and immune mechanisms as determined using IPA diseases and function annotations. Refs. [52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71] are cited in Supplementary Materials.
Author Contributions
The authors contributed to the study in the following aspects: Conceptualization, K.A.M., R.H.S. and K.A.B.; Methodology, K.A.M., E.Z. and K.A.B.; Validation, E.Z.; Formal Analysis, K.A.M., E.Z. and K.A.B.; Data Curation, K.A.M. and K.A.B.; Writing—Original Draft Preparation, K.A.M., R.H.S. and K.A.B.; Writing—Review and Editing, K.A.M., R.H.S., E.Z. and K.A.B.; and Project Administration, K.A.M. and K.A.B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Morio et al. reported the concentrations of 13 CCBMs in tissue culture media of HEPM cells and gingival fibroblasts at 0, 24, and 48 h after treatment with 255, 405, or 255/405 UV irradiation [6]. In the current study, we used these CCBM values as a subset of data to assess the ability of UV irradiation to activate or inhibit cellular pathways related to immune functions.
Conflicts of Interest
The authors declare no conflict of interest. We have no financial affiliation (e.g., employment, direct payment, stock holdings, retainers, consultantships, patent licensing arrangements, or honoraria), or involvement with any commercial organization with direct financial interest in the subject or materials discussed in this manuscript, nor have any such arrangements existed in the past 3 years. Kimberly A. Morio is an Endodontist at Apex Endodontics and an Adjunct Instructor at the University of Iowa; Robert H. Sternowski is the President of Softronics, Ltd.; Erliang Zeng is a Biostatistician and a Computational Biologist at the University of Iowa; and Kim A. Brogden is an Emeritus Professor at the University of Iowa. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results. The company had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fouad A.F. Microbiological Aspects of Traumatic Injuries. J. Endod. 2019;45:S39–S48. doi: 10.1016/j.joen.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Delikan E., Caliskan S., Cankilic M.Y., Aksu S., Kesim B., Ulger S.T. Microbiota of endodontically infected primary and permanent teeth. Pediatr. Dent. 2021;43:102–110. [PubMed] [Google Scholar]
- 3.Wong J., Manoil D., Näsman P., Belibasakis G.N., Neelakantan P. Microbiological aspects of root canal infections and disinfection strategies: An update review on the current knowledge and challenges. Front. Oral Health. 2021;2:672887. doi: 10.3389/froh.2021.672887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siqueira J.F., Jr., Rocas I.N. Present status and future directions: Microbiology of endodontic infections. Int. Endod. J. 2022;55:512–530. doi: 10.1111/iej.13677. [DOI] [PubMed] [Google Scholar]
- 5.Metzger Z., Better H., Abramovitz I. Immediate root canal disinfection with ultraviolet light: An ex vivo feasibility study. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2007;104:425–433. doi: 10.1016/j.tripleo.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 6.Morio K.A., Thayer E.L., Bates A.M., Brogden K.A. 255-nm light-emitting diode kills Enterococcus faecalis and induces the production of cellular biomarkers in human embryonic palatal mesenchyme cells and gingival fibroblasts. J. Endod. 2019;45:774–783.e6. doi: 10.1016/j.joen.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metzger Z., Dotan M., Better H., Abramovitz I. Sensitivity of oral bacteria to 254 nm ultraviolet light. Int. Endod. J. 2007;40:120–127. doi: 10.1111/j.1365-2591.2006.01191.x. [DOI] [PubMed] [Google Scholar]
- 8.Morio K.A., Sternowski R.H., Brogden K.A. Dataset of endodontic microorganisms killed at 265 nm wavelength by an ultraviolet C light emitting diode in root canals of extracted, instrumented teeth. Data Brief. 2022;40:107750. doi: 10.1016/j.dib.2021.107750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ou Y., Petersen P.M. Application of ultraviolet light sources for in vivo disinfection. Jpn. J. Appl. Phys. 2021;60:100501. doi: 10.35848/1347-4065/ac1f47. [DOI] [Google Scholar]
- 10.Vieyra-Garcia P.A., Wolf P. A deep dive into UV-based phototherapy: Mechanisms of action and emerging molecular targets in inflammation and cancer. Pharmacol. Ther. 2021;222:107784. doi: 10.1016/j.pharmthera.2020.107784. [DOI] [PubMed] [Google Scholar]
- 11.Kramer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morykwas M.J., Mark M.W. Effects of ultraviolet light on fibroblast fibronectin production and lattice contraction. Wounds. 1998;10:111–117. [Google Scholar]
- 13.Dennler S., Itoh S., Vivien D., Dijke P., Huet S., Gauthier J.M. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagai T., Chen X., Miragaia R.J., Rostom R., Gomes T., Kunowska N., Henriksson J., Park J.E., Proserpio V., Donati G., et al. Gene expression variability across cells and species shapes innate immunity. Nature. 2018;563:197–202. doi: 10.1038/s41586-018-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai Y., Gallo R.L. AMPed up immunity: How antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brogden K.A., Bates A.M., Fischer C.L. Antimicrobial peptides in host defense: Functions beyond antimicrobial activity. In: Harder J., Schroeder J.M., Kaufmann S.H., Mercer A.A., Weber B., editors. Antimicrobial Peptides—Role in Human Health and Disease. Springer International Publishing; Cham, Switzerland: 2016. pp. 129–146. Birkhauser Advances in Infectious Diseases. [DOI] [Google Scholar]
- 17.Drayton M., Deisinger J.P., Ludwig K.C., Raheem N., Muller A., Schneider T., Straus S.K. Host defense peptides: Dual antimicrobial and immunomodulatory action. Int. J. Mol. Sci. 2021;22:11172. doi: 10.3390/ijms222011172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi M., Umehara Y., Yue H., Trujillo-Paez J.V., Peng G., Nguyen H.L.T., Ikutama R., Okumura K., Ogawa H., Ikeda S., et al. The antimicrobial peptide human beta-defensin-3 accelerates wound healing by promoting angiogenesis, cell migration, and proliferation through the FGFR/JAK2/STAT3 signaling pathway. Front. Immunol. 2021;12:712781. doi: 10.3389/fimmu.2021.712781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G. Human antimicrobial peptides and proteins. Pharmaceuticals. 2014;7:545–594. doi: 10.3390/ph7050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallbris L., Edstrom D.W., Sundblad L., Granath F., Stahle M. UVB upregulates the antimicrobial protein hCAP18 mRNA in human skin. J. Investig. Dermatol. 2005;125:1072–1074. doi: 10.1111/j.0022-202X.2005.23872.x. [DOI] [PubMed] [Google Scholar]
- 21.Glaser R., Navid F., Schuller W., Jantschitsch C., Harder J., Schroder J.M., Schwarz A., Schwarz T. UV-B radiation induces the expression of antimicrobial peptides in human keratinocytes in vitro and in vivo. J. Allergy Clin. Immunol. 2009;123:1117–1123. doi: 10.1016/j.jaci.2009.01.043. [DOI] [PubMed] [Google Scholar]
- 22.Kreuter A., Hyun J., Skrygan M., Sommer A., Bastian A., Altmeyer P., Gambichler T. Ultraviolet A1-induced downregulation of human beta-defensins and interleukin-6 and interleukin-8 correlates with clinical improvement in localized scleroderma. Br. J. Dermatol. 2006;155:600–607. doi: 10.1111/j.1365-2133.2006.07391.x. [DOI] [PubMed] [Google Scholar]
- 23.Gambichler T., Skrygan M., Tomi N.S., Altmeyer P., Kreuter A. Changes of antimicrobial peptide mRNA expression in atopic eczema following phototherapy. Br. J. Dermatol. 2006;155:1275–1278. doi: 10.1111/j.1365-2133.2006.07481.x. [DOI] [PubMed] [Google Scholar]
- 24.Park K.C., Jung H.C., Hwang J.H., Youn S.W., Ahn J.S., Park S.B., Kim K.H., Chung J.H., Youn J.I. GM-CSF production by epithelial cell line: Upregulation by ultraviolet A. Photodermatol. Photoimmunol. Photomed. 1997;13:133–138. doi: 10.1111/j.1600-0781.1997.tb00216.x. [DOI] [PubMed] [Google Scholar]
- 25.Imokawa G., Yada Y., Kimura M., Morisaki N. Granulocyte/macrophage colony-stimulating factor is an intrinsic keratinocyte-derived growth factor for human melanocytes in UVA-induced melanosis. Biochem. J. 1996;313:625–631. doi: 10.1042/bj3130625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaudschun R., Sunderkotter C., Brenneisen P., Hinrichs R., Peters T., Schneider L., Razi-Wolf Z., Hunzelmann N., Scharffetter-Kochanek K. Vascular endothelial growth factor causally contributes to the angiogenic response upon ultraviolet B irradiation in vivo. Br. J. Dermatol. 2002;146:581–587. doi: 10.1046/j.1365-2133.2002.04669.x. [DOI] [PubMed] [Google Scholar]
- 27.Oxholm A., Oxholm P., Staberg B., Bendtzen K. Immunohistological detection of interleukin I-like molecules and tumour necrosis factor in human epidermis before and after UVB-irradiation in vivo. Br. J. Dermatol. 1988;118:369–376. doi: 10.1111/j.1365-2133.1988.tb02430.x. [DOI] [PubMed] [Google Scholar]
- 28.Crispin M.K., Fuentes-Duculan J., Gulati N., Johnson-Huang L.M., Lentini T., Sullivan-Whalen M., Gilleaudeau P., Cueto I., Suárez-Fariñas M., Lowes M.A., et al. Gene profiling of narrowband UVB-induced skin injury defines cellular and molecular innate immune responses. J. Investig. Dermatol. 2013;133:692–701. doi: 10.1038/jid.2012.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang D., Chertov O., Bykovskaia S.N., Chen Q., Buffo M.J., Shogan J., Anderson M., Schroder J.M., Wang J.M., Howard O.M., et al. B-defensins: Linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 30.Niyonsaba F., Ushio H., Nakano N., Ng W., Sayama K., Hashimoto K., Nagaoka I., Okumura K., Ogawa H. Antimicrobial peptides human beta-defensins stimulate epidermal keratinocyte migration, proliferation and production of proinflammatory cytokines and chemokines. J. Investig. Dermatol. 2007;127:594–604. doi: 10.1038/sj.jid.5700599. [DOI] [PubMed] [Google Scholar]
- 31.Ramos R., Silva J.P., Rodrigues A.C., Costa R., Guardao L., Schmitt F., Soares R., Vilanova M., Domingues L., Gama M. Wound healing activity of the human antimicrobial peptide LL37. Peptides. 2011;32:1469–1476. doi: 10.1016/j.peptides.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Yang D., Biragyn A., Kwak L.W., Oppenheim J.J. Mammalian defensins in immunity: More than just microbicidal. Trends Immunol. 2002;23:291–296. doi: 10.1016/S1471-4906(02)02246-9. [DOI] [PubMed] [Google Scholar]
- 33.Chaly Y.V., Paleolog E.M., Kolesnikova T.S., Tikhonov I.I., Petratchenko E.V., Voitenok N.N. Neutrophil alpha-defensin human neutrophil peptide modulates cytokine production in human monocytes and adhesion molecule expression in endothelial cells. Eur. Cytokine Netw. 2000;11:257–266. [PubMed] [Google Scholar]
- 34.Barabas N., Rohrl J., Holler E., Hehlgans T. Beta-defensins activate macrophages and synergize in pro-inflammatory cytokine expression induced by TLR ligands. Immunobiology. 2013;218:1005–1011. doi: 10.1016/j.imbio.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Van Wetering S., MannesseLazeroms S.P.G., Dijkman J.H., Hiemstra P.S. Effect of neutrophil serine proteinases and defensins on lung epithelial cells: Modulation of cytotoxicity and IL-8 production. J. Leukoc. Biol. 1997;62:217–226. doi: 10.1002/jlb.62.2.217. [DOI] [PubMed] [Google Scholar]
- 36.Petrov V., Funderburg N., Weinberg A., Sieg S. Human beta defensin-3 induces chemokines from monocytes and macrophages: Diminished activity in cells from HIV-infected persons. Immunology. 2013;140:413–420. doi: 10.1111/imm.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott M.G., Davidson D.J., Gold M.R., Bowdish D., Hancock R.E. The human antimicrobial peptide LL-37 is a multifunctional modulator of innate immune responses. J. Immunol. 2002;169:3883–3891. doi: 10.4049/jimmunol.169.7.3883. [DOI] [PubMed] [Google Scholar]
- 38.Warnke P.H., Voss E., Russo P.A., Stephens S., Kleine M., Terheyden H., Liu Q. Antimicrobial peptide coating of dental implants: Biocompatibility assessment of recombinant human beta defensin-2 for human cells. Int. J. Oral Maxillofac. Implant. 2013;28:982–988. doi: 10.11607/jomi.2594. [DOI] [PubMed] [Google Scholar]
- 39.Heilborn J.D., Nilsson M.F., Kratz G., Weber G., Sorensen O., Borregaard N., Stahle-Backdahl M. The cathelicidin anti-microbial peptide LL-37 is involved in re-epithelialization of human skin wounds and is lacking in chronic ulcer epithelium. J. Investig. Dermatol. 2003;120:379–389. doi: 10.1046/j.1523-1747.2003.12069.x. [DOI] [PubMed] [Google Scholar]
- 40.Nakatsuji T., Gallo R.L. Antimicrobial peptides: Old molecules with new ideas. J. Investig. Dermatol. 2012;132:887–895. doi: 10.1038/jid.2011.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwarz T., Beissert S. Milestones in photoimmunology. J. Investig. Dermatol. 2013;133:E7–E10. doi: 10.1038/skinbio.2013.177. [DOI] [PubMed] [Google Scholar]
- 42.Matos T.R., Sheth V. The symbiosis of phototherapy and photoimmunology. Clin. Dermatol. 2016;34:538–547. doi: 10.1016/j.clindermatol.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 43.Bruhs A., Schwarz T. Ultraviolet radiation-induced immunosuppression: Induction of regulatory T cells. Methods Mol. Biol. 2017;1559:63–73. doi: 10.1007/978-1-4939-6786-5_5. [DOI] [PubMed] [Google Scholar]
- 44.Hequet O., Nosbaum A., Guironnet-Paquet A., Blasco E., Nicolas-Virelizier E., Griffith T.S., Rigal D., Cognasse F., Nicolas J.F., Vocanson M. CD8(+) T cells mediate ultraviolet A-induced immunomodulation in a model of extracorporeal photochemotherapy. Eur. J. Immunol. 2020;50:725–735. doi: 10.1002/eji.201948318. [DOI] [PubMed] [Google Scholar]
- 45.Schwarz T. Regulatory T cells induced by ultraviolet radiation. Int. Arch. Allergy Immunol. 2005;137:187–193. doi: 10.1159/000086330. [DOI] [PubMed] [Google Scholar]
- 46.Bernard J.J., Gallo R.L., Krutmann J. Photoimmunology: How ultraviolet radiation affects the immune system. Nat. Rev. Immunol. 2019;19:688–701. doi: 10.1038/s41577-019-0185-9. [DOI] [PubMed] [Google Scholar]
- 47.Patil C.S., Kirkwood K.L. p38 MAPK signaling in oral-related diseases. J. Dent. Res. 2007;86:812–825. doi: 10.1177/154405910708600903. [DOI] [PubMed] [Google Scholar]
- 48.Ji R.R., Gereau R.W., Malcangio M., Strichartz G.R. MAP kinase and pain. Brain Res. Rev. 2009;60:135–148. doi: 10.1016/j.brainresrev.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muthusamy V., Piva T.J. A comparative study of UV-induced cell signalling pathways in human keratinocyte-derived cell lines. Arch. Dermatol. Res. 2013;305:817–833. doi: 10.1007/s00403-013-1412-z. [DOI] [PubMed] [Google Scholar]
- 50.Ma W., Quirion R. The ERK/MAPK pathway, as a target for the treatment of neuropathic pain. Expert Opin. Ther. Targets. 2005;9:699–713. doi: 10.1517/14728222.9.4.699. [DOI] [PubMed] [Google Scholar]
- 51.Lin X., Wang M., Zhang J., Xu R. p38 MAPK: A potential target of chronic pain. Curr. Med. Chem. 2014;21:4405–4418. doi: 10.2174/0929867321666140915143040. [DOI] [PubMed] [Google Scholar]
- 52.Bode A.M., Dong Z. Mitogen-activated protein kinase activation in UV-induced signal transduction. Sci. STKE. 2003;2003:167. doi: 10.1126/stke.2003.167.re2. [DOI] [PubMed] [Google Scholar]
- 53.Bender K., Blattner C., Knebel A., Iordanov M., Herrlich P., Rahmsdorf H.J. UV-induced signal transduction. J. Photochem. Photobiol. B. 1997;37:1–17. doi: 10.1016/S1011-1344(96)07459-3. [DOI] [PubMed] [Google Scholar]
- 54.Adachi M., Gazel A., Pintucci G., Shuck A., Shifteh S., Ginsburg D., Rao L.S., Kaneko T., Freedberg I.M., Tamaki K., et al. Specificity in stress response: Epidermal keratinocytes exhibit specialized UV-responsive signal transduction pathways. DNA Cell Biol. 2003;22:665–677. doi: 10.1089/104454903770238148. [DOI] [PubMed] [Google Scholar]
- 55.Seo S.J., Ahn S.W., Hong C.K., Ro B.I. Expressions of beta-defensins in human keratinocyte cell lines. J. Dermatol. Sci. 2001;27:183–191. doi: 10.1016/S0923-1811(01)00135-9. [DOI] [PubMed] [Google Scholar]
- 56.Suo W., Guo H., Wang X., Wang D. Effect of ultraviolet C light on the expression of basic fibroblast growth factor in rat wounds. Chinese J. Phys. Med. Rehabil. 2003;25:651–654. [Google Scholar]
- 57.Suo W., Wang X., Wang D. Effect of ultraviolet C irradiation on expression of transforming growth factor-β in wound. Chin. J. Rehabil. Theory Pract. 2002;8:5–7. [Google Scholar]
- 58.Gallo R.L., Staszewski R., Sauder D.N., Knisely T.L., Granstein R.D. Regulation of GM-CSF and IL-3 production from the murine keratinocyte cell line PAM 212 following exposure to ultraviolet radiation. J. Investig. Dermatol. 1991;97:203–209. doi: 10.1111/1523-1747.ep12479676. [DOI] [PubMed] [Google Scholar]
- 59.Kirnbauer R., Koch A., Kurtmann J., Schwarz T., Urbanski A., Luger T.A. Different effects of UVA and UVB irradiation on epidermal cell-IL6 expression and release; Proceedings of the ESDR-JSID-SID Tricontinental Meeting; Washington, DC, USA. 26 April 1989; pp. 393–548. [Google Scholar]
- 60.Gallo R.L., Brownstein E., Granstein R.D. Secretion of interleukin 3 activity from a transformed murine keratinocyte line after exposure to ultraviolet radiation: Role of membrane signal transduction mechanisms; Proceedings of the ESDR-JSID-SID Tricontinental Meeting; Washington, DC, USA. 26 April 1989; pp. 393–548. [Google Scholar]
- 61.Clingen P.H., Berneburg M., Petit-Frere C., Woollons A., Lowe J.E., Arlett C.F., Green M.H. Contrasting effects of an ultraviolet B and an ultraviolet A tanning lamp on interleukin-6, tumour necrosis factor-alpha and intercellular adhesion molecule-1 expression. Br. J. Dermatol. 2001;145:54–62. doi: 10.1046/j.1365-2133.2001.04281.x. [DOI] [PubMed] [Google Scholar]
- 62.Chung K.Y., Chang N.S., Park Y.K., Lee K.H. Effect of ultraviolet light on the expression of adhesion molecules and T lymphocyte adhesion to human dermal microvascular endothelial cells. Yonsei Med. J. 2002;43:165–174. doi: 10.3349/ymj.2002.43.2.165. [DOI] [PubMed] [Google Scholar]
- 63.Ansel J.C., Luger T.A., Green I. The effect of in vitro and in vivo UV irradiation on the production of ETAF activity by human and murine keratinocytes. J. Investig. Dermatol. 1983;81:519–523. doi: 10.1111/1523-1747.ep12522862. [DOI] [PubMed] [Google Scholar]
- 64.Kupper T.S., Chua A.O., Flood P., McGuire J., Gubler U. Interleukin 1 gene expression in cultured human keratinocytes is augmented by ultraviolet irradiation. J. Clin. Investig. 1987;80:430–436. doi: 10.1172/JCI113090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwarz T., Luger T.A. New trends in photobiology: Effect of UV irradiation on epidermal cell cytokine production. J. Photochem. Photobiol. B: Biol. 1989;4:1–13. doi: 10.1016/1011-1344(89)80097-1. [DOI] [PubMed] [Google Scholar]
- 66.Rhodes L.E., Joyce M., West D.C., Strickland I., Friedmann P.S. Comparison of changes in endothelial adhesion molecule expression following UVB irradiation of skin and a human dermal microvascular cell line (HMEC-1) Photodermatol. Photoimmunol. Photomed. 1996;12:114–121. doi: 10.1111/j.1600-0781.1996.tb00187.x. [DOI] [PubMed] [Google Scholar]
- 67.Kennedy-Crispin M., Billick E., Mitsui H., Gulati N., Fujita H., Gilleaudeau P., Sullivan-Whalen M., Johnson-Huang L.M., Suarez-Farinas M., Krueger J.G. Human keratinocytes’ response to injury upregulates CCL20 and other genes linking innate and adaptive immunity. J. Investig. Dermatol. 2012;132:105–113. doi: 10.1038/jid.2011.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kirnbauer R., Kock A., Neuner P., Forster E., Krutmann J., Urbanski A., Schauer E., Ansel J.C., Schwarz T., Luger T.A. Regulation of epidermal cell interleukin-6 production by UV light and corticosteroids. J. Investig. Dermatol. 1991;96:484–489. doi: 10.1111/1523-1747.ep12470181. [DOI] [PubMed] [Google Scholar]
- 69.Yarosh D., Both D., Kibitel J., Anderson C., Elmets C., Brash D., Brown D. Regulation of TNFalpha production and release in human and mouse keratinocytes and mouse skin after UV-B irradiation. Photodermatol. Photoimmunol. Photomed. 2000;16:263–270. doi: 10.1034/j.1600-0781.2000.160606.x. [DOI] [PubMed] [Google Scholar]
- 70.Gambichler T., Tomi N.S., Skrygan M., Altmeyer P., Kreuter A. Alterations of TGF-beta/Smad mRNA expression in atopic dermatitis following narrow-band ultraviolet B phototherapy: Results of a pilot study. J. Dermatol. Sci. 2006;44:56–58. doi: 10.1016/j.jdermsci.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 71.James L.C., Moore A.M., Wheeler L.A., Murphy G.M., Dowd P.M., Greaves M.W. Transforming growth factor alpha: In vivo release by normal human skin following UV irradiation and abrasion. Skin Pharmacol. 1991;4:61–64. doi: 10.1159/000210925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Morio et al. reported the concentrations of 13 CCBMs in tissue culture media of HEPM cells and gingival fibroblasts at 0, 24, and 48 h after treatment with 255, 405, or 255/405 UV irradiation [6]. In the current study, we used these CCBM values as a subset of data to assess the ability of UV irradiation to activate or inhibit cellular pathways related to immune functions.