Abstract
Regeneration of lost or injured organs is an intriguing process where numerous cellular events take place to form the new structure. Studies of this process during reconstitution of the intestine have been performed in echinoderms, particularly in holothurians. Many cellular events triggered during regeneration have been described using the sea cucumber Holothuria glaberrima as a research model. More recent experiments have targeted the molecular mechanism behind the process, a task that has been eased by the new sequencing technologies now available. In this review we present the studies involving cellular processes and the genes that have been identified to be associated with the early events of gut regeneration. We also present the ongoing efforts to perform functional studies necessary to establish the role(s) of the identified genes. A synopsis of the studies is given with the course of the regenerative process established so far.
Keywords: gut regeneration, sea cucumber, echinoderms, holothurians
Introduction
Echinoderms (Echinodermata) encompass an important group of organisms that have been used as model systems throughout the years, particularly in the field of developmental biology. Part of their appeal is their embryological development which assigns them to the Deuterostomata clade (García-Arrarás et al., 2019). This classification places echinoderms in the same evolutionary branch as vertebrates along with tunicates, hemichordates and cephalochordates. This close relationship to vertebrates makes echinoderms interesting model organisms, particularly when compared with other invertebrates such as Drosophila melanogaster and Caenorhabditis elegans, that lie in a different evolutionary branch (Protostomata), but have still provided important information concerning cellular and molecular events common to most animals (García-Arrarás and Dolmatov, 2010). Among the less studied properties of the echinoderms are their regenerative capacities. This is a somewhat surprising fact when one takes into account that echinoderms are among the most highly regenerative deuterostomes.
One of the echinoderm classes, Holothuroidea or sea cucumbers, contains species that are able to regenerate complex organs, such as the digestive tract, muscle, tentacles, and nerve cord, among others. In fact, many holothurians have the natural ability to expel their digestive tract under stressful conditions, in a process known as evisceration. This process has been assumed to be a defensive strategy, and can be artificially induced in the laboratory with intracoelomic injections of a KCl solution or distilled water (García-Arrarás and Greenberg, 2001). Evisceration triggers regeneration of the digestive tract along with other associated viscera, providing a “natural” system where regeneration can be studied. This is the topic of this review, specifically, the use of an echinoderm, the sea cucumber Holothuria glaberrima as a model system to study intestinal regeneration (Fig. 1).
Fig 1. The model system portrayed in this review- The sea cucumber Holothuria glaberrima.

An echinoderm commonly found in rocky coastal regions from Florida to Brazil. Side view, anterior (oral) end is to the right.
Digestive Tract of Holothurians
Below, we provide a brief overview of the organization of the sea cucumber digestive system (Fig. 2). The digestive tract begins at the most anterior end of H. glaberrima with a mouth that leads to a pharynx (Fig. 2 A) that lies within an anatomical complex known as the pharyngeal bulb. The pharynx opens into a short esophagus. Behind the esophagus, lies a long looping intestine that terminates in a broad cloaca. The intestine accounts for over 90% of the digestive tract length and is subdivided into three regions: the anterior descending small intestine, an ascending small intestine – the region associated with the hemal system – and a posterior descending large intestine (Fig. 2 A).
Fig 2. Anatomical organization and tissue architecture of the sea cucumber digestive.
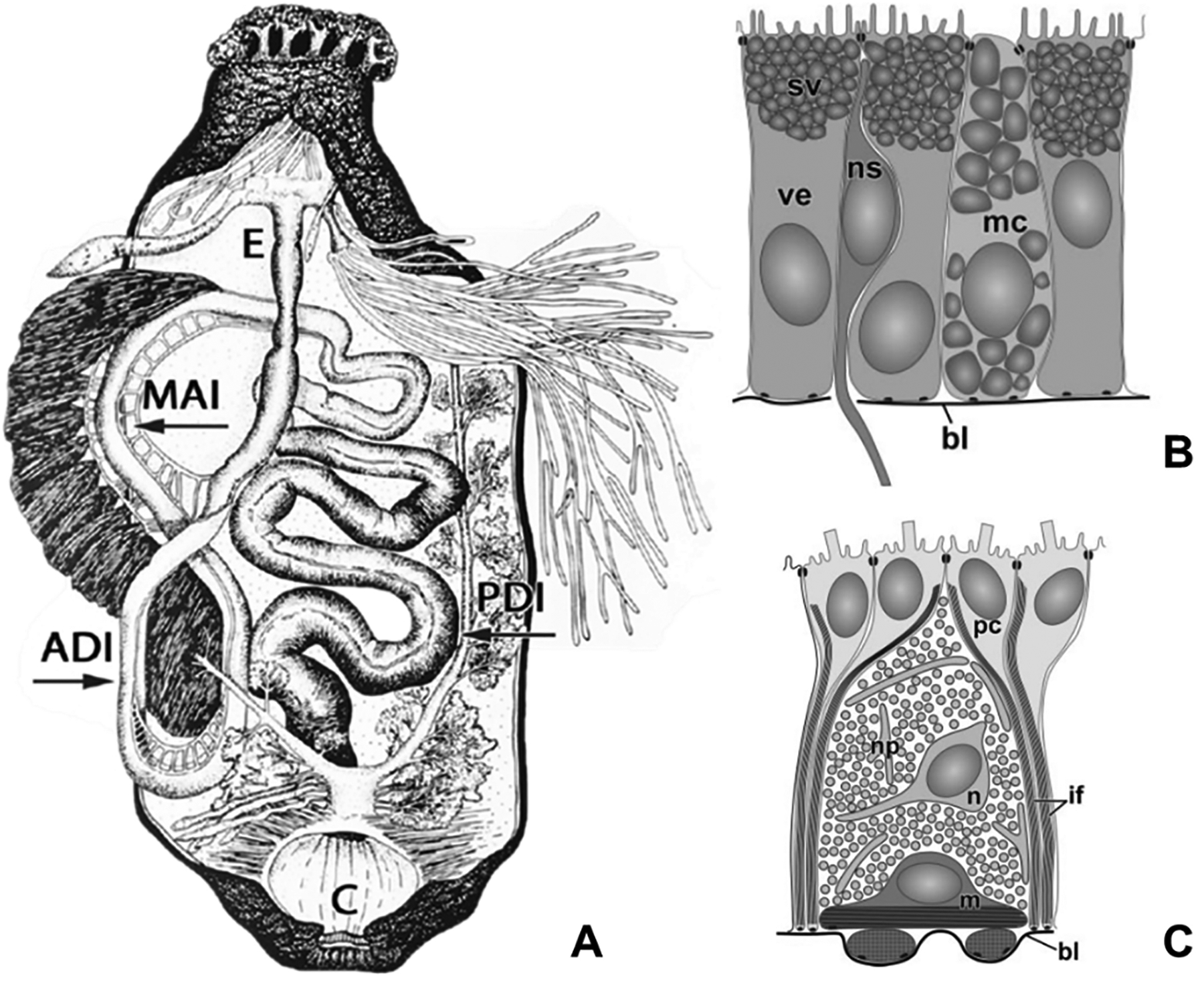
(A) Visceral anatomy of sea cucumber. The intestine is subdivided into the anterior descending small intestine, ascending small intestine, and the posterior descending large intestine. Abbreviations: short esophagus (E); anterior descending intestine (ADI); medial ascending intestine (MAI); posterior descending intestine (PDI). (B) Tissue organization of the luminal epithelium and (C) mesothelium found throughout the intestine of H. glaberrima. Abbreviations: basal lamina (bl); bundles of intermediate filaments in peritoneocytes (if); myoepithelial cell (m); mucocyte (mc); neuron (n); neurosecretory cell (ns); nervous plexus (np); peritoneal cell (pc); secretory vacuoles (sv); vesicular enterocyte (ve). Fig. 2 A adapted from García-Arrarás et al., 2019, Seminars in Cell & Developmental Biology 92, p. 47 and fig 2 B, C from Mashanov & García-Arrarás, 2011, Biological Bulletin, 221, p. 95. Copyright 2019 by Elsevier (2 A) and 2011 by The University of Chicago Press (2 B, C). Adapted with permission.
Throughout its length, the wall of the digestive tube is composed of three tissue layers: the inner luminal epithelium, the outer muscular mesothelium and the connective tissue that is sandwiched between the basal laminae of the two epithelial layers. The inner luminal epithelium is composed mainly of enterocytes, a cell type, which is assumed to play important roles in the absorption and accumulation of nutrients, synthesis and release of digestive enzymes and phagocytosis of food particles. Other cells present in the luminal epithelium include enteroendocrine cells and mucocytes (Fig. 2 B). In the case of the outer layer (mesothelium), its organization is complex with an apical surface composed of monociliated epithelial cells (peritoneocytes) facing the outer side of the intestine and connected to each other by intercellular junctions (García-Arrarás et al., 1998; García-Arrarás and Greenberg, 2001). The basal half of the mesothelium is occupied by myoepithelial cells. Neuronal cell bodies can also be found interspersed among the peritoneocytes and muscle cells (Fig. 2 C). The connective tissue is mostly composed of extracellular matrix (ECM) where a few mesenchymal and neuronal cells are present (Feral & Massin, 1982; Mashanov et al., 2004; Mashanov & García-Arrarás, 2011). A depicted the morphology of the digestive tract can be observed in figure 2, whereas figure 3 portrays the organization of the mentioned tissues during early stages of regeneration with the exception of the luminal epithelium.
Fig 3. Cellular dedifferentiation and proliferation at the early stage of the regenerative process.
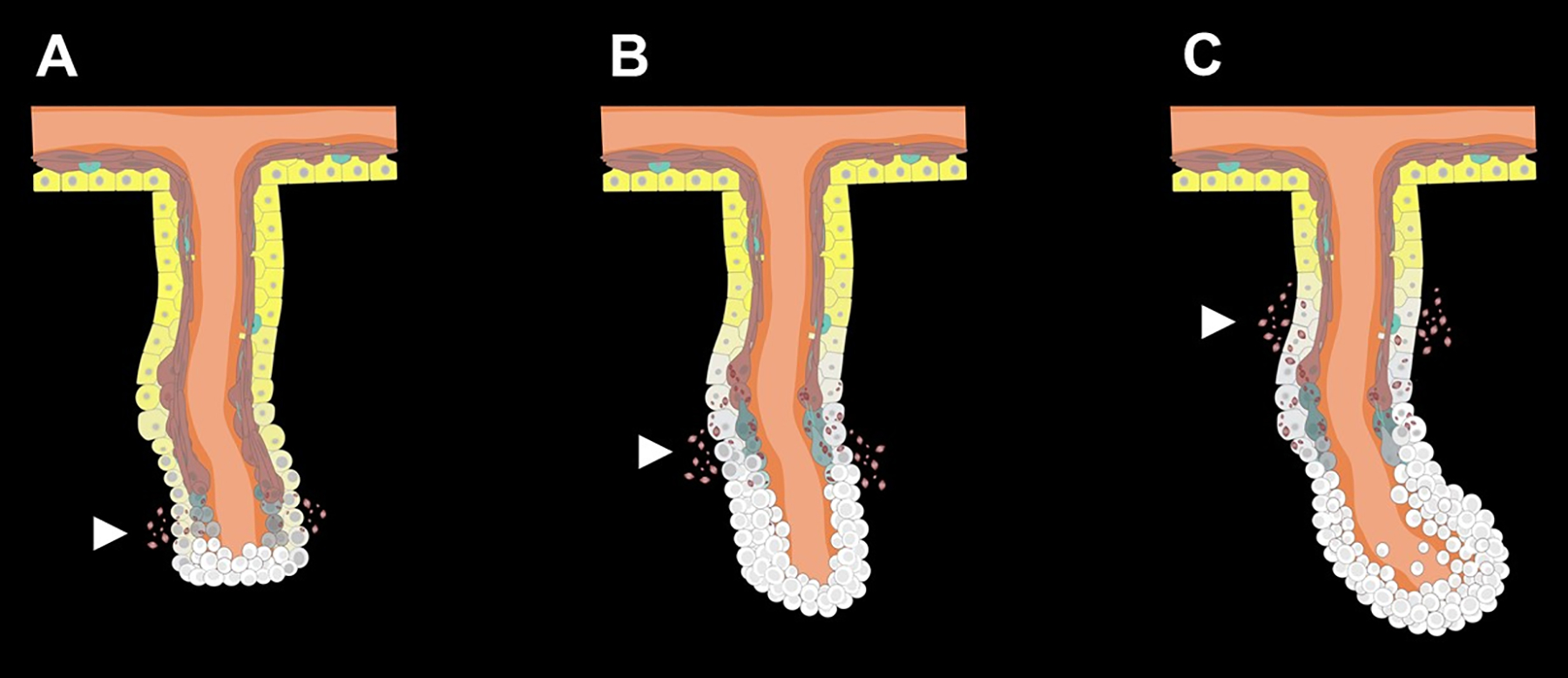
(A) Cells in the free end of the mesentery begin to dedifferentiate soon after evisceration. (B) Cells accumulate at the free edge of mesentery forming the early blastema-like structure. (C) As regeneration proceeds, dedifferentiation spreads along the mesentery and involves regions closer to the body wall, while cell proliferation remains mainly restricted to the blastema-like structure composed of the dedifferentiated cells. White arrows point out SLSs from the dedifferentiation process. White cells represent dedifferentiated cells originated from peritoneocytes (yellow) and muscle cells (reddish-brown) of the mesothelium. Connective tissue (orange) is positioned between the layers of the mesothelium and extends to the body wall (upper part).
Mesentery
The entire length of the intestine is attached to the body wall by a continuous mesentery. The organization of the mesentery along the body wall varies according to the region of the attached intestine. The descending small intestine is attached to the dorsal body wall; the ascending small intestine is attached to the lateral body wall and the large intestine is attached to the ventral body wall. The mesentery is composed of two layers of mesothelium separated by a thin layer of connective tissue. Both the mesothelia and connective tissue of the mesentery are continuous with the respective layers of the intestinal wall at one end and with the body wall mesothelium and dermis at the other. Nervous elements are found within the mesothelial layer associated mostly with the muscle layer, while the connective tissue contains some dispersed nerve cells as well (Nieves-Ríos et al., 2019). This tissue arrangement in the sea cucumber mesentery generally follows the pattern characteristic of other coelomates. However, when compared to more complex organisms, such as mammals, the lymphatic component is mostly absent and the hemal component is much reduced in H. glaberrima.
During evisceration, the intestine detaches from the mesentery and also breaks at its anterior end from the esophagus and at the posterior end from the cloaca. The autotomized digestive tube is then expelled through the anus. This leaves the mesentery attached to the body wall with the free torn edges lying within the body cavity (García-Arrarás et al., 2019). Regeneration begins with the healing of these edges within the first 24 hours after injury. The new intestinal rudiment starts as a swelling that develops at the free edge of the mesentery, initially forming a thick solid rod that connects the esophagus to the cloaca.
In H. glaberrima, cells that form the new intestine originate from two sources. This early regenerate is composed of a connective tissue covered with the mesothelium and initially lacks a lumen. In H. glaberrima, the tissue layers of the new intestine originate from the respective components of the mesentery and esophageal and cloacal stumps. First, the cells of the remaining mesentery dedifferentiate and proliferate to provide most, if not all, the cells for the new mesothelium and connective tissue. The second cell source is the luminal epithelia of the esophagus and cloaca that proliferate and invade the connective tissue swelling at the free edge of the mesentery to form the new lumen and luminal epithelium (García-Arrarás et al., 2011). Nonetheless, it is important to highlight that in other sea cucumber species, the mesentery mesothelium has been proposed to be the source for all tissue layers of the anterior portion of the new intestine (Vladimir S. Mashanov et al., 2005a; Vladimir S. Mashanov et al., 2015b).
Cellular Events Underlying Intestinal Regeneration
Several cellular events, such as cell division, cell death, cell migration, ECM remodeling and cell dedifferentiation, have been found to take place during intestinal regeneration. Our studies suggest that these events occur in a particular order. We have proposed that the early regeneration immediately after evisceration involves reorganization and remodeling of the tissues in the mesentery (García-Arrarás et al., 2019), which both contributes cells to the new intestine and also serves as a source of important signals that guide the regenerative process. Evisceration triggers dedifferentiation in peritoneocytes and muscle cells in the mesothelium of the mesentery. This process is characterized by the reversion to a less differentiated cell phenotype, giving rise to precursor cells that eventually will divide and then re-differentiate into the cellular components of the regenerating organ (García-Arrarás, 2017). Dedifferentiating peritoneocytes undergo fragmentation of their bundles of intermediate filaments (V.S. Mashanov et al., 2004). However, it is the dedifferentiation of the muscle cells that have been best studied (San Miguel-Ruiz and García-Arrarás, 2007). In H. glaberrima, muscle cell dedifferentiation involves elimination of the contractile apparatus through condensation of myofilaments into membrane bound spindle-like structures (SLSs). SLSs are often expelled from the dedifferentiating myocytes into the coelomic cavity or adjacent connective tissue and subsequently degraded by phagocytic amoebocytes (Candelaria et al., 2006). The presence of these structures is an indicator of dedifferentiation that can be conveniently used to track the onset of the intestinal regeneration.
At the organ level, muscle cell dedifferentiation unfolds in a spatio-temporal gradient. During the early stage, cells undergoing dedifferentiation are only localized near the free margin of the mesentery where the intestine was formerly attached, while the rest of the mesentery remains in mostly differentiated state (Fig. 3 A). As regeneration progresses, the middle zone of the mesentery starts to dedifferentiate as well, whereas the region closest to the body wall still has very few dedifferentiating cells (Fig. 3 B). Thus, by determining the presence and localization of SLSs a gradient of dedifferentiation can be observed at about a week following evisceration. At this time, the mesentery can be divided into three regions. The first one comprises the region closest to the forming intestinal rudiment where there are no muscle fibers and little or no SLSs. The second region that lies in the middle of the mesentery, has some muscle fibers and abundant SLSs. This region is undergoing active dedifferentiation. Finally, the third region, at the attachment to the body wall, remains mostly differentiated, has numerous muscle fibers and very few SLSs (García-Arrarás et al., 2019). During the dedifferentiation process, the mesothelial basal lamina disintegrates, but the dedifferentiated cells retain their epithelial organization and remain connected by intercellular junctions (Quiñones et al., 2002; Vladimir S. Mashanov et al., 2005; García-Arrarás et al., 2011).
Cells that have undergone dedifferentiation in the rudiment and adjacent mesenteric regions eventually re-enter the cell cycle to yield the precursors that will form the intestinal mesothelia and connective tissue cells. Cell division is also spatially separated with more extensive proliferation at the distal tip of the mesentery and in the growing rudiment. Few dividing cells are found in the middle region of the mesentery or in the proximal region, next to the body wall. Experiments have shown that cells undergoing apoptosis are also mainly localized to the growing rudiment and to the adjacent region of the mesentery (García-Arrarás et al., 2011).
It has also been shown that following the growth of the new intestinal rudiment, some of the mesothelial cells within the regenerating structure undergo an epithelial to mesenchymal transition (EMT) and ingress into the underlying connective tissue (García-Arrarás et al., 2011). These cells contribute to the mesenchyme of the new intestine connective tissue and may also be involved in the remodeling and/or deposition of the ECM that is observed to take place during regeneration. The increase in the cell number in the distal part of the mesentery together with the possible ECM deposition results in the growth of the intestinal rudiment. Therefore, at about a week after evisceration, the intestinal rudiment appears as a swelling at the free edge of the mesentery and is composed of a large number of mesenchymal cells forming the inner connective tissue surrounded by a dedifferentiated coelomic epithelium (Fig. 3 C).
Few or no neuronal cells can be found within the growing intestinal rudiment at this early stage. This might be due to the fact that the intestinal neurons in this region of the mesentery had been eliminated through apoptosis. In addition to the disappearance of the neuronal cell bodies, neurites originating from more distant neurons undergo a retrograde axonal degeneration which is followed by regrowth and eventual reinnervation of the newly formed tissues including the muscle (Tossas et al., 2014). With the exception of the loss of the nervous plexus from the free end of the mesentery and subsequent regeneration of the neural elements in the new intestine, the nervous component of the rest of the mesentery, however, appears to remain unaltered during regeneration despite the dramatic changes in the surrounding tissues of the mesentery (Nieves-Ríos et al., 2019).
In the next two weeks a lumen will form in the intestinal rudiment and reconstitution of mucosal epithelium will be achieved by the intrusion of tubular projections of the mucosal epithelium that migrate from the esophagus and the cloaca (García-Arrarás et al., 2019). During this period the new intestine will be innervated by fibers originating from the mesentery and new neurons will appear forming a new enteric nervous system similar to the one in the original organ (Tossas et al., 2014). The formation of the luminal epithelium and the lumen, although of great interest, cannot be expanded in this review, which will be limited to the early events that culminate in the formation of the intestinal rudiment consisting of a thickening at the mesenterial tip.
Intestinal regeneration shares many similarities with the regeneration process that takes place following nerve cord transection. In the latter, dedifferentiation is also one of the initial events where radial glial cells respond to the injury by losing their radial morphology and degrading their cytoskeletal filaments. Dedifferentiated glial cells proliferate and differentiate into the new neurons and glial cells that form the regenerated section of the radial nerve cord and that are eventually integrated into the central nervous system circuitry (Mashanov et al., 2014).
Molecular basis of intestinal regeneration
As described above, big advances have been made in dissecting out the cellular mechanisms using immunohistochemical and other microscopic techniques. At the same time, we are also beginning to understand the molecular mechanisms driving the various cellular processes. Some efforts have been done to characterize the genes and their products that are involved in the intestinal regeneration in H. glaberrima. The early attempts to identify differentially expressed genes during regeneration employed differential screening (DS) and differential display (DD). The first technique involved radiolabeled probes of cDNA to detect the presence of expressed genes and the second one was based on real time polymerase chain reaction (RT-PCR) (Roig-López et al., 2001). The latter method revealed the presence of mRNA for serum amyloid A (SAA) protein during intestinal regeneration in H. glaberrima. This was the first SAA ortholog found in a non-vertebrate deuterostome. Northern blot revealed SAA overexpression during mid-to-late stages of regeneration (day 15 post-evisceration). This expression coincided with the formation of the lumen and the organization of muscular layers of the digestive tract (Santiago et al., 2000).
Transcripts of other developmental genes were subsequently identified in the regenerating intestinal tissues. PCR amplification from genomic DNA and cDNA libraries yielded several HOX gene homologs. Analysis of the detected nine HOX sequences showed an association with the three HOX groups: anterior (3), medial (1) and posterior (5) (Méndez et al., 2000). A sequence corresponding to ependymin mRNA, a molecule previously associated with regenerative phenomena, was also reported for the first time in an echinoderm and shown to be overexpressed during intestinal regeneration (Suárez-Castillo et al., 2004). Further studies using a more high-throughput approach based on expressed sequence tags (ESTs) from cDNA libraries led to identify 5173 sequences from regenerating intestines, allowing analysis of differentially expressed genes during regeneration (Rojas-Cartagena et al., 2007).
In the last decade different molecular strategies such as microarrays and transcriptome-wide RNA-seq have been used to identify additional genes associated with gut regeneration. In H. glaberrima, gene expression profiles were determined at three time points (days 3, 7 and 14) and compared against those of normal intestine. Numerous individual candidate genes associated with development, extracellular matrix remodeling, cytoskeleton and wound healing processes showed differential expression that were validated with semiquantitative RT-PCR (Ortiz-Pineda et al., 2009). Further RNA-seq experiments have been done during the early stages of regeneration when most of the dedifferentiation process occurs (Quispe et al., unpublished manuscript). In these experiments, a total of 1859 annotated genes were found to be significantly differentially expressed, with more being downregulated than upregulated (Fig. 4A). Gene ontology and pathway enrichment analyses were performed by comparing gene expression at day 3 in regenerating tissues with normal mesentery of non-eviscerated individuals. Differentially expressed genes were significantly enriched with annotation terms associated with cofactor binding, oxidation-reduction and ECM remodeling. Further pathway enrichment analysis resulted in high number of protein processing and metabolic pathways of some nutrients like butanoate (Fig. 4B). The overrepresentation of these pathways during day 3 could be associated with changes in metabolic processes during cell dedifferentiation that are connected to metabolism of available nutrients for subsequent proliferation.
Fig 4. Differential gene expression and pathway enrichment analysis of day 3 regenerating intestine vs normal mesentery.
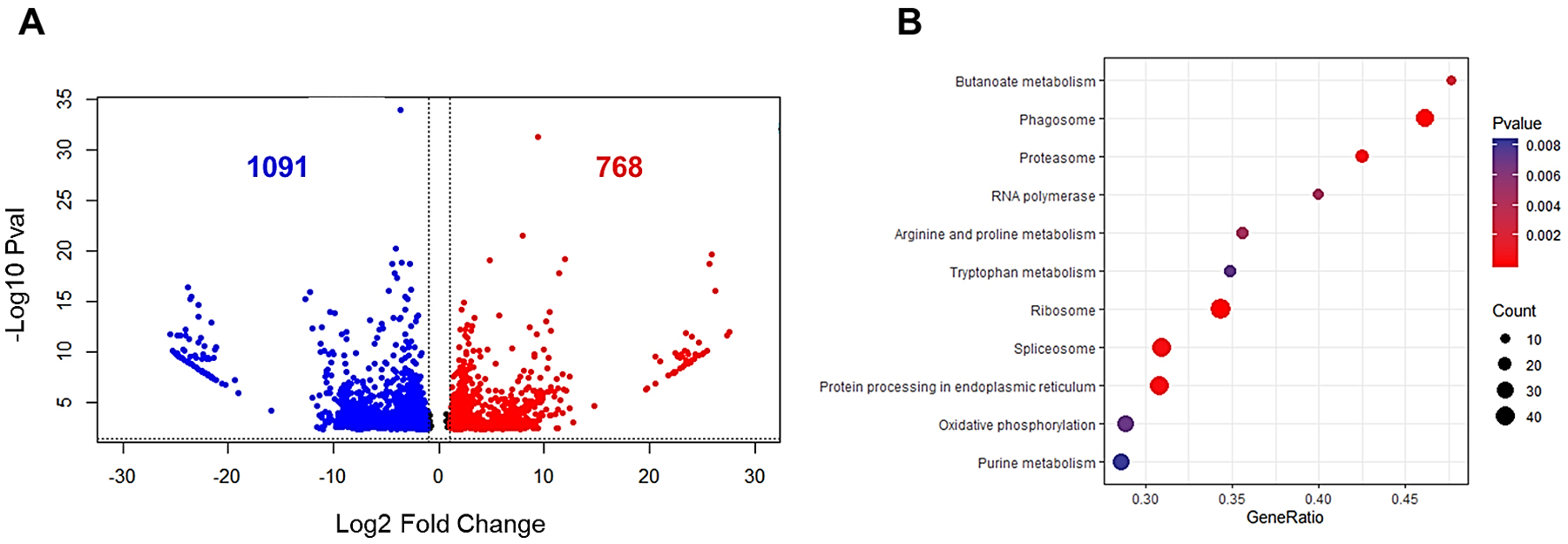
(A) Volcano plot showing downregulated (blue) and upregulated (red) genes. Gene expression shown as log2 fold change is on the x-axis and −log10 adjusted p-values are plotted on the y-axis. Expressed genes were selected with an adjusted p-value lower than 0.05 and a log2 fold change different than 0. (B) Pathway enrichment analysis obtained for differentially expressed genes that had an adjusted p-value <0.05. Gene ratio describes the representation of differentially expressed genes involved in the components of the pathway and count number describes the quantity of those genes in each pathway.
RNA-seq experiments performed in a different sea cucumber species, Apostichopus japonicus, have confirmed our findings and have served to expand the number of genes that are associated with intestinal regeneration (Sun et al., 2013). Certain findings of this study stand out, among them: the quantity of upregulated (~2400) and downregulated (~1000) genes in the early to mid-stages of regeneration (days 3–7) when compared to normal intestine, and the differentially expressed genes involved in processes concerning ECM remodeling, development and cytoskeletal genes. Many of the genes within these groups overlapped with the microarray study done in H. glaberrima supporting the idea of a conserved mechanism in both organisms (Ortiz-Pineda et al., 2009).
Selected Relevant Functional Gene Groups
Identification of genes that are differentially expressed during intestinal regeneration is a first step in determining the underlying molecular mechanisms involved. It is noteworthy that many of these genes are associated with the previously observed cellular events. Thus, genes associated with cell division, apoptosis and ECM remodeling are among those that have been shown to be differentially expressed.
ECM remodeling-
Establishment of new dedifferentiated cells in the intestinal rudiment requires a proper migration process achieved by a remodeling of the connective tissue layers. Moreover, it has been shown that the ECM influences cell differentiation and even the choice of the cell fate (Engler et al., 2006). Thus, it should be expected that to accomplish this new ECM, genes involved in the degradation of fibrillar collagen and remodeling of the ECM are activated (Quiñones et al., 2002; Miao et al., 2017). Microarray analysis of regenerating H. glaberrima intestine detected differentially expressed genes associated with cytoskeletal and ECM remodeling (Ortiz-Pineda et al., 2009). Simultaneous changes in expression of these two gene cohorts suggests coordination between ECM remodeling and cellular dedifferentiation/migration processes. In addition, a decrease in the expression of another ECM molecule, gelsolin, was documented. Gelsolin has been previously linked with inducing scar formation, suggesting an inhibitory activity during wound healing and regenerating processes (Cowin et al., 2003). Furthermore, over expression of actin isoforms was associated with migration of cells into regenerating tissue (Murray and García-Arrarás, 2004). All genes associated with ECM remodeling were found to be upregulated at the early stages of regeneration. These included genes encoding the proteins Echinonectin, Collagen Alpha-1, Laminin alpha, Tenascin-R and three matrix metalloproteases (MMP-15, MMP-11 and MMP-14). The higher activity of these proteins is thought to form a transient ECM that promotes cellular events associated with the regenerative response. Previous functional experiments had established that proper level of MMP activity is required for regeneration (Quiñones et al., 2002; Dolmatov et al., 2019). Therefore, we posit that it is the activation/inactivation of these genes that set the basis for ECM remodeling which then serves as support for the mobilization of the new cells that are going to form the new intestinal rudiment.
Developmental processes-
Another group of genes that are differentially expressed in intestinal regeneration comprise those associated with embryological development. This should be expected, since regeneration has been shown to activate similar processes known to take place during the development of the organism. Among those that could be involved in regeneration are the broad family of HOX genes. HOX are a subset of the homeobox genes that function as transcription factors in regional patterning of metazoan embryos (Zakany and Duboule, 2007). Several HOX genes orthologs have been described in a number of echinoderm groups, including asteroids, ophiuroids and echinoids (Ben Khadra et al., 2014; Martinez et al., 1999). In H. glaberrima HOX9, HOX10 and HOX12 have been shown to be differentially expressed at early-medium stages of regeneration (Méndez et al., 2000; Ortiz-Pineda et al., 2009).
The Wnt/β-catenin pathway is known to be important for development, as well as essential for regulation of cellular differentiation and proliferation (Katoh and Katoh, 2007). It has recently been proposed, based on its positive selection in regenerative echinoderms, that the Wnt signaling pathway is a key signaling pathway (Yuan et al., 2019). In H. glaberrima several orthologs of genes coding for Wnt ligands have been identified and also found to be differentially expressed. For example, WNT9 was upregulated during intestinal regeneration between days 3 and 14 (Ortiz-Pineda et al., 2009). This expression has also been observed by the detection of WNT transcripts in the mesothelium of the intestine using in situ hybridization (Mashanov et al., 2012). Studies in A. japonicus have also focused on WNT6 and HOX6 due to their upregulation during intestinal regeneration (Sun et al., 2013).
Other developmental genes include the bone morphogenetic proteins (BMPs). These are members of the transforming growth factor β superfamily (TGF-β) and serve as signaling ligands that mediate a multitude of developmental events including dorsal-ventral patterning and imaginal disk patterning in organisms like Drosophila (Martindale, 2005). In the context of regeneration, BMP has been found to induce a regenerative response in the digit of neonatal mice and to play an essential role in regeneration of bilaterally symmetric fragments and dorsoventral signaling in planarians (Reddien et al., 2007; Yu et al., 2010). In H. glaberrima, expression of BMP1 has been observed using microarrays of day 7 regenerating intestines (Ortiz-Pineda et al., 2009). This expression was also detected by in situ hybridization in the luminal epithelium and mesothelium during days 2–14 of regeneration. This timeframe overlaps with the timing of drastic changes in the luminal epithelium caused by cell dedifferentiation (Mashanov et al., 2012). The period of expression of BMP genes is similar to that of WNT9 and HOX genes mentioned earlier.
Finally, the Notch signaling pathway is known to be implicated in regulation of several cellular processes including proliferation, differentiation, stem cell maintenance, fate decision and apoptosis (Kopan and Ilagan, 2009). In the regeneration context, its importance has been shown in studies with varied model organisms such as Hydra (Münder et al., 2013), newts (Nakamura and Chiba, 2007), planarians (Zhao et al., 2014) and zebrafish (Wenemoser et al., 2012). In echinoderms, the genes encoding the major components of the pathway have been identified in Strongylocentrotus purpuratus (Walton et al., 2006) and their differential expression has been studied in the regeneration of nerve cord in H. glaberrima (Mashanov et al., 2014). Furthermore, pharmacological inhibition of the pathway in the brittle star Ophioderma brevispina showed involvement of Notch signaling in regulation of ECM composition/remodeling, innate immune response, apoptosis, cell proliferation, cell migration and even activity of transposable elements, highlighting its involvement in many mechanisms associated with regeneration (Mashanov et al., 2019).
Cell proliferation-
Genes involved in proliferation of the cells that will eventually form the blastema-like structure are common to many processes including development and tissue renewal in the gastrointestinal tract. Previously mentioned genes like WNT, BMP and HOX have been associated with modulation of intestinal stem cell proliferation (Yen and Wright, 2006) and their expression during gut regeneration has been confirmed in H. glaberrima and A. japonicus (Ortiz-Pineda et al., 2009; L Sun et al., 2013). Downstream components of these pathways have a regulatory role as effectors for gene activation. Among these downstream components, β-catenin and Myc have been identified as important elements in gut regeneration. The former is a transcription factor that has an essential role as signal transducer in the canonical Wnt signaling pathway. The activity of β-catenin is guided by its phosphorylation state along with TCF to form a complex (β-catenin/TCF) that increases transcriptional activity. Experiments with antibodies to label phosphorylation in the Y489 residue of β-catenin have suggested the importance of this process in the activity of Wnt pathway during intestinal regeneration (Bello et al., 2019). Likewise, Myc is another transcription factor, one of the Yamanaka factors that were used to induce pluripotency in mammalian cells (Takahashi and Yamanaka, 2006). In H. glaberrima, upregulation of MYC has been found at early stages of both visceral and neural regeneration (Vladimir S. Mashanov et al., 2015). Both β-catenin and Myc have been shown to play important roles in the regulation and activation of cell proliferation in Wnt, BMP and Notch pathways in other non-echinoderm models (Huang et al., 2014; Toofan & Wheadon, 2016).
Apoptosis-
Regeneration requires a regulated interaction between proliferation and cell death (Bergmann and Steller, 2010). Previously, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay in H. glaberrima have identified a peak in the number of apoptotic cells during the first week of gut regeneration (Mashanov et al., 2010). In situ hybridization coupled with TUNEL showed that programmed cell death is regionally regulated through expression of two anti-apoptotic genes survivin (Fig. 5A) and mortalin (Fig. 5B) (Mashanov et al., 2010). Furthermore, Myc has also been studied for its effect in the regulation of programmed cell death in H. glaberrima, it appears to have a variable effect by either triggering apoptosis or protecting cells from apoptosis in neural regeneration (Vladimir S Mashanov et al., 2015). This role seems to be dependent on the context leading to its expression and its effect has yet to be studied during intestinal regeneration.
Fig 5. Correlation of survivin and mortalin expression with apoptosis in the regenerating intestine on day 7.
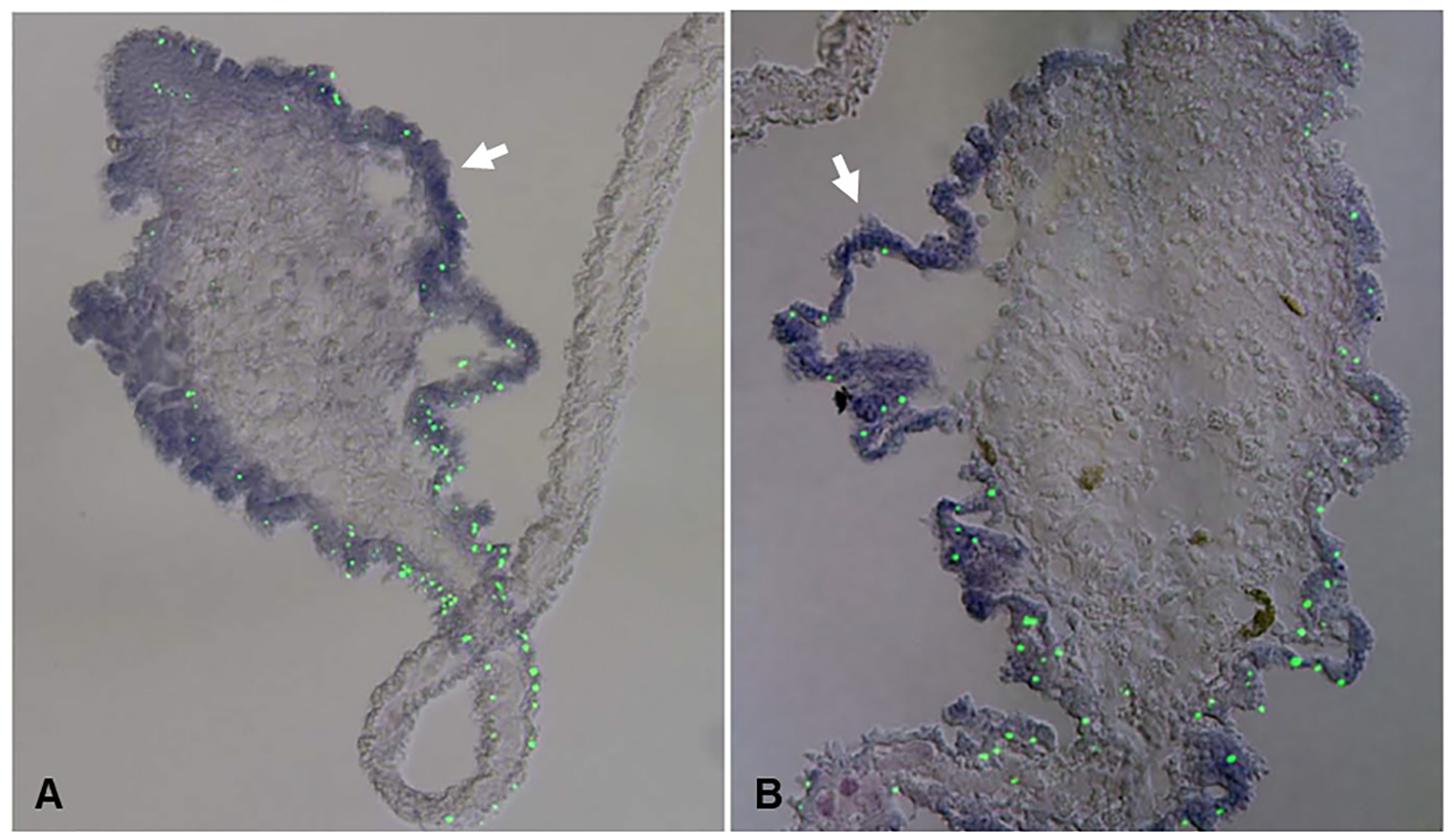
(A) Double labeling with in situ hybridization with a survivin riboprobe and TUNEL assay. (B) Double labeling with a mortalin riboprobe and TUNEL assay. Survivin and mortalin transcripts are showed in violet and TUNEL labeled cells are showed in green. Lower density of TUNEL-positive cells can be observed when higher signal of survivin in situ hybridization is present (white arrows). Scale bar = 100 μm. Adapted from Mashanov et al. 2010, BMC Developmental Biology, 10:117. Copyright ©2010 Mashanov et al; licensee BioMed Central Ltd.
Novel genes-
High throughput sequencing methods have increased our tally of genes that could be important for the regeneration process. Such is the case of the orpin gene identified in H. glaberrima. Over expression of orpin was found during early stages of intestinal regeneration and the use of antibodies showed labeling in luminal epithelium as well as the radial nerve cord (Pineda, 2010). However, orpin’s possible role remains unknown.
In the same organism, four genes encoding melanotransferrin (MTf) were found to be overexpressed during early stages of gut regeneration (Hernández-Pasos et al., 2017). This finding was specially compelling due to the fact that only one MTf gene has been reported in all other organisms that have been sequenced to date. MTf expression can be triggered by activation of the immune system using lipopolysaccharide (LPS) (Ramírez-Gómez et al., 2009). The presence and activation of Mtfs provides support for the conjectures that propose that the immune system plays a role in regenerative events (Mescher and Neff, 2005). Furthermore, the recent sequencing of the genome of the sea cucumber A. japonicus has provided new data for comparative genomic and phylogenetic analysis. Two gene families encoding fibrinogen-related proteins and the prostatic secretory protein (PSP94) were found to be overrepresented in the animal’s genome and overexpressed during regeneration (Zhang et al., 2017). The former was previously studied as it was found that gene expression of fibrinogen-like protein A had a gradual increase during intestinal regeneration (Wu et al., 2014). Moreover, a recent study in the sea cucumber Eupentacta fraudatrix has identified 11 genes encoding transcription factors that could be associated with transdifferentiation in holothurian intestinal regeneration, although the evidence for this is strictly circumstantial. These factors are involved in distinct cellular process in other organisms and their expression increases during days 5–7 of regeneration when the transformation of coelomic epithelial cells into enterocytes occurs (Boyko et al., 2020).
Functional Studies
Identification of gene candidates, although an important initial step in determining the molecular basis of the intestinal regenerative process, are only based on correlative evidence with a particular event. Further functional studies showing the necessity of the required gene activity for the process are required. In order to probe further the function of particular genes in intestinal regeneration, researchers have relied on two experimental procedures. The first is the use of pharmacological agents that modulate the activity of particular proteins. This method, although widely used in functional studies, requires caution due to unspecific targets that can trigger a broad spectrum of effects in other cellular processes. Nonetheless, if performed adequately, it can provide important functional information with the use of multiple drugs.
Previous experiments in H. glaberrima have been applied to antagonize the function of MMPs with different compounds. Results from this pioneering study showed disruption of ECM remodeling followed by a reversible inhibition of intestinal regeneration (Quiñones et al., 2002). A similar approach also targeting MMPs was performed in E. fraudatrix, in which a reversible inhibitory effect was confirmed (Lamash and Dolmatov, 2013).
Recent studies using compounds to alter Wnt pathway have contributed to discerning its modulating role in cellular events. Experiments in vivo with iCRT14, an inhibitor of the Wnt/β-catenin pathway, led to a significant reduction in cell proliferation and gut rudiment size compared with control DMSO (Fig. 6A, B). On the other hand, the use of the pathway activator LiCl produced an increase in gut rudiment size when compared with control NaCl (Fig. 6C, D). iCRT14 also produced a delay in muscle differentiation which was measured by the changes in the ratio of SLSs and cell nuclei. Further experiments in vitro using gut explants also revealed a reduction in cell dedifferentiation when iCRT14 was applied, while LiCl had the opposite effect. In the case of cell proliferation, both drugs reduced this process in the gut explants. Although the effects of the drugs significantly affected dedifferentiation and proliferation in both treatments, no significant effect was observed in cell apoptosis. Overall, the results showed a dissociation between proliferation and dedifferentiation when using the LiCl in gut explants in vitro (Bello et al., 2019). More importantly it led the authors to propose that while cell proliferation is mediated via the canonical Wnt pathway effect involving β-catenin and Myc, cell dedifferentiation is modulated by a Wnt-independent, GSK-3 dependent pathway. In A. japonicus pharmacological inhibition using Wnt targeting compound salinomycin, also reduced intestine length in regenerating organisms (Yuan et al., 2019).
Fig 6. Effect of pharmacological modulation on the size of the regenerating gut in vivo.
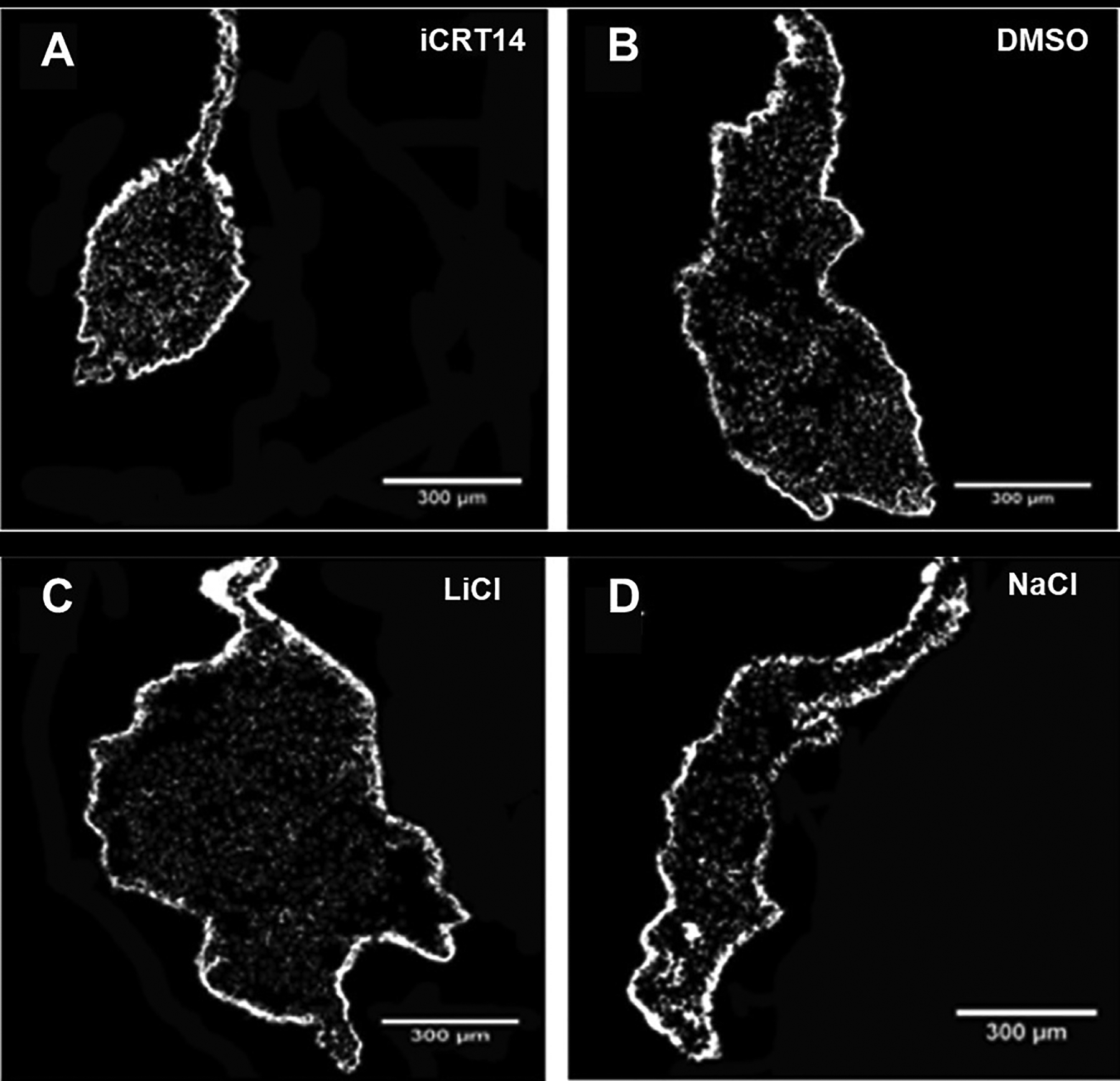
Histological cross sections of the intestine with the attached mesentery show the effect of a Wnt pathway inhibitor iCRT14 (A) and a Wnt-pathway activator LiCl (C) treatment on the gut rudiment size compared with controls DMSO (B) and NaCl (D), respectively. iCRT14 treatment reduces the size of the gut rudiment compared with control DMSO, whereas treatment with LiCl increases the size compared with NaCl. Scale bars = 300 μm. Adapted from Bello et al., 2020, Developmental Biology, 458, p. 15. Copyright 2020 by Elsevier. Adapted with permission.
The second method for functional studies requires the use of RNA interference (RNAi) to inhibit translation of a specific mRNA, thus decreasing the amount of the protein product. In holothurians this technique has only recently been put to work and there is space for much improvements and applications. Nevertheless, its use in H. glaberrima has been established both in vivo and in vitro. The former approach was used to study the role of Myc in neural regeneration. Results suggested that MYC knockdown inhibited dedifferentiation of radial glial cells and apoptosis following neural injury (Vladimir S Mashanov et al., 2015). In vitro inhibition by electroporation with RNAi, allowed the inhibition of Myc and β-catenin transcripts on intestinal explants during regeneration (Delgado, 2019). Reduction in cell proliferation was observed in treated explants with RNAi targeting MYC (Fig. 7 B) when compared with control treatments of RNAi targeting green fluorescent protein (GFP) (Fig. 7 A). Proliferation was also reduced when β-catenin transcripts were targeted with RNAi, while dedifferentiation remained unaffected. Interestingly, a differential effect on programmed cell death was also observed with a reduction in apoptosis in explants treated with RNAi targeting MYC but not β-catenin (Delgado, 2019). These results suggest a prominent implication of the Wnt pathway in cell proliferation, while it confirms the involvement of Myc in cell death control during early stages of gut regeneration. A similar technique was used in vivo to disrupt the Wnt pathway in A. japonicus. Targeting of WNT7 and DVL with RNAi significantly reduced the length of the newly regenerated intestine (Yuan et al., 2019).
Fig 7. Effect RNAi inhibition on cell proliferation of regenerating gut explants in vitro.
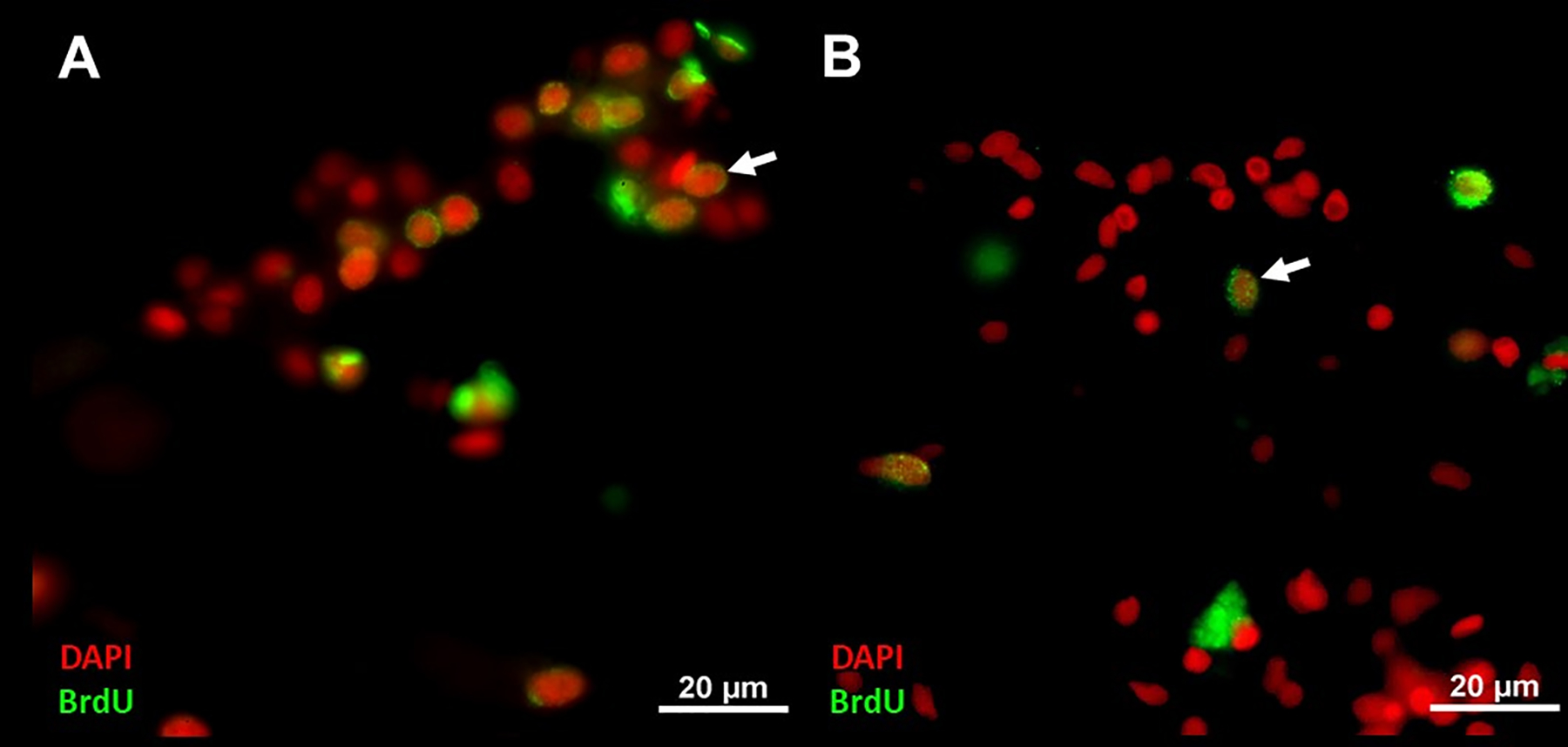
Sections of regenerating gut in vitro treated with control RNAi targeting GFP (A) and MYC (B). BrdU labeled cells are reduced in treatment with RNAi targeting MYC compared with control targeting GFP. Cell nuclei are shown in red (DAPI) and BrdU labeled cell nuclei (white arrows) in green. Scale bar = 20 μm.
As the cellular processes and the genes that participate in the regeneration process are identified, the next step is to provide a roadmap that shows the localization and timing of gene expression, as well as the gene and cell interactions that ensue. Such a roadmap must involve many molecular mechanisms that are tightly regulated in order to modulate the cellular events. A schematic illustrating depicting a timeline of the cellular events and genes with potential roles in the regulation of these processes (suggested by expression and/or functional data) is shown in Fig. 8. In this scenario, cell dedifferentiation and migration are the important initial events in regeneration and are associated with the modulation of developmental genes. These processes mark the starting point for the reconstitution of the lost intestine. Regions of dedifferentiation constitute the origin for new cells that are going to relocate to the edge of the torn mesentery to form the blastema-like structure. The dedifferentiation and movement of cells can be associated with the activation of cytoskeletal genes (Alpha-tubulin, actin) found to be upregulated during the first days of regeneration (Cabrera-Serrano & García-Arrarás, 2004; García-Arrarás et al., 2006) and involved in the modulation of cell protrusion, contraction and retraction (Tang and Gerlach, 2017). This process is followed by ECM remodeling which might be essential for the migration of cells towards the rudiment (Quiñones et al., 2002; Miao et al., 2017). ECM associated genes (MMPs, collagen alpha, tenascin-R, laminin-alpha, echinonectin) involved in the degradation of collagen and generation of a suitable environment for the cell framework have been shown to be upregulated during early to mid-stages after evisceration (Ortiz-Pineda et al., 2009). In all these processes, cell proliferation is triggered in the dedifferentiated cells via mechanisms and signaling pathways that involve previous developmental genes (WNT9, BMP1, HOX, mortalin, survivin). These genes might be involved in the modulation of either the migration and/or proliferative response of cells that invade the free edge to form the gut rudiment. It is interesting to note that there is a spatial separation of the dedifferentiation and the proliferation events. While cellular dedifferentiation mainly takes place in the mesentery, cellular proliferation is highest in the blastema-like structure. Moreover, in situ experiments have shown that many of the over-expressed genes are observed in cells of the coelomic epithelia or in cells within the connective tissue layer of the blastema-like structure. These spatial differences of dedifferentiation and proliferation events suggest that at least two distinct gene activation processes must be occurring simultaneously. One of them is taking place in the mesentery and consists of active dedifferentiation with concomitant ECM remodeling. The second is mainly taking place in the blastema-like structure and includes cell proliferation, apoptosis and a dramatic change in the gene expression profile of the epithelial coelomic cells. It remains to be determined if there is a link between them and whether one of them (dedifferentiation) leads to the other (proliferation) as we have suggested. At the same time, in other regenerating organisms, it has been proposed that the generation of new cells might be linked with apoptosis (Galliot and Chera, 2010). In H. glaberrima, this last event is associated with the increased expression of cancer-related genes such as survivin, which appears to be involved in both proliferation and cell death (Mashanov et al., 2010). MYC gene expression has also been confirmed as important for cell death from previous functional studies (Delgado, 2019). In situ and immunohistochemical studies have also suggested that there is a spatial separation of apoptosis and proliferation, but in this case within the blastema-like structure itself (Mashanov et al., 2010).
Fig 8. Schematic representation of cellular and molecular mechanisms during the first week of regeneration.
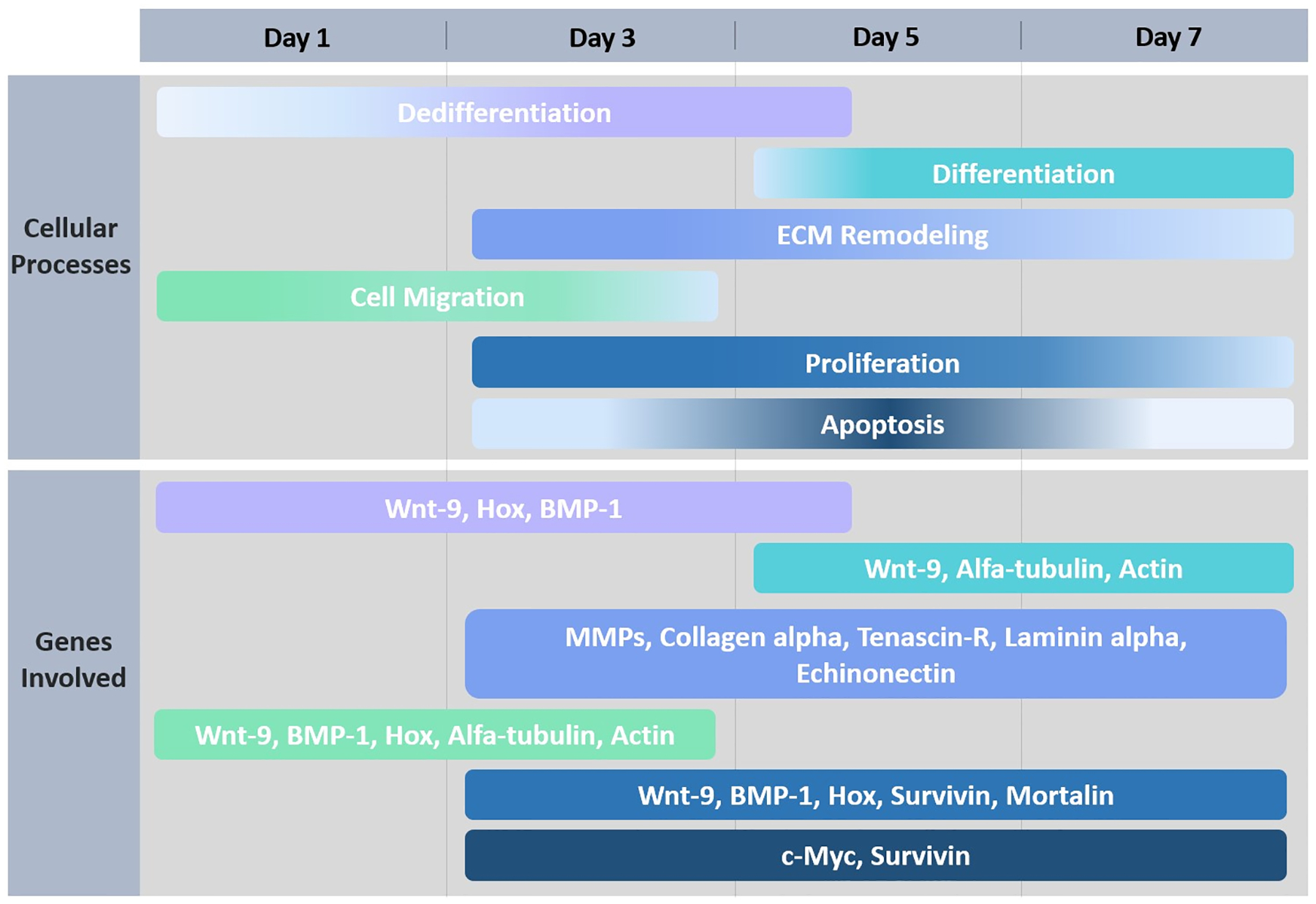
Timeline is represented by days of regeneration after de evisceration process occurs. Colors for each cellular process correspond to the same color for the genes that are associated with that process.
Finally, the transition of early to mid-stages (days 5–7 after evisceration) (Murray and García-Arrarás, 2004), coincides with a period of reduced dedifferentiation and an increase in the differentiation within the intestinal rudiment. In this period, cells from the coelomic epithelium give rise to the new muscle cells, while cells from the luminal epithelium of the esophagus and cloaca induce the formation of the new luminal epithelium in the intestinal rudiment. The genes in charge of triggering differentiation appear to be involved with previous processes including cytoskeletal and developmental genes of Wnt pathway (Yen and Wright, 2006).
The overlapping events depicted (Fig. 8) provide a description of possible molecular networks and cell interaction that can be tested. It is in this realm that the recent experiments from our lab gain importance (Delgado, 2019; Bello et al., 2020). In these experiments, using regenerating intestinal explants, the dedifferentiation process is uncoupled from the cellular proliferation using pharmacological and RNAi tools. The results support our hypothesis that muscle cells undergo dedifferentiation prior to entering the cell proliferation cycle. This experiments also propose the signaling pathways that might be involved: GSK-3 for dedifferentiation and Wnt for cell proliferation. The cells where these pathways are activated remain undetermined, although recent immunohistochemical studies with an antibody against phosphorylated β-catenin suggest that they are the cells of the rudiment mesothelium (Bello et al., 2020). The combination of the different molecular mechanisms involved and the subsequent cellular events triggered, opens more possibilities for studies unraveling the key elements of gut regeneration.
In summary, great advances have been made to decipher the mechanisms by which the holothurian intestine regenerates. The cellular events have been described and multiple genes proposed as candidates for the molecular events underlying the cellular interactions. Various laboratories are now developing the necessary tools to advance toward functional studies that will allow to clearly identify the triggering factors and interactions among cells and the underlying network of gene activity. We are hopeful that the next decade will bring new discoveries in the quest to understand the process of regeneration not only in the holothurian intestine, but to understand the common aspects of regenerative processes in the metazoan.
Acknowledgements
This project was funded by NIH (Grant R15NS01686 and R21AG057974) and the University of Puerto Rico. We want to thank Drs. Vladimir Mashanov and David Forsthoefel for their critical comments on the manuscript. Finally, we express our gratitude to Joshua Medina for his help with the preparation of Figure 3.
Abbreviations used in this paper:
- SLSs
spindle like structures
- BMP
bone morphogenetic proteins
- DMSO
dimethyl sulfoxide
- RNA-seq
RNA sequencing
- PCR
polymerase chain reaction
- RT-PCR
real time polymerase chain reaction
- ECM
extracellular matrix
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- MTf
melanotransferrin
- MMP
matrix metalloproteases
- RNAi
RNA interference
- GFP
green fluorescent protein
References
- BELLO SA, TORRES-GUTIÉRREZ V, RODRÍGUEZ-FLORES EJ, TOLEDO-ROMÁN EJ, RODRÍGUEZ N, DÍAZ-DÍAZ LM, VÁZQUEZ-FIGUEROA LD, CUESTA JM, GRILLO-ALVARADO V, AMADOR A, REYES J, GARCÍA-ARRARÁS JE (2019). Insights into intestinal regeneration signaling mechanisms. Developmental Biology. Available at: https://linkinghub.elsevier.com/retrieve/pii/S0012160618305037 [Accessed January 22, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELLO SA, TORRES-GUTIÉRREZ V, RODRÍGUEZ-FLORES EJ, TOLEDO-ROMÁN EJ, RODRÍGUEZ N, DÍAZ-DÍAZ LM, VÁZQUEZ-FIGUEROA LD, CUESTA JM, GRILLO-ALVARADO V, AMADOR A, REYES J, GARCÍA-ARRARÁS JE (2020). Insights into intestinal regeneration signaling mechanisms. Developmental Biology 458: 12–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEN KHADRA Y, SAID K, THORNDYKE M, MARTINEZ P (2014). Homeobox Genes Expressed During Echinoderm Arm Regeneration. Biochem Genet 52: 166–180. [DOI] [PubMed] [Google Scholar]
- BERGMANN A, STELLER H (2010). Apoptosis, Stem Cells, and Tissue Regeneration. Sci Signal 3: re8–re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOYKO AV, GIRICH AS, TKACHEVA ES, DOLMATOV IYu (2020). The Eupentacta fraudatrix transcriptome provides insights into regulation of cell transdifferentiation. Scientific Reports 10. Available at: http://www.nature.com/articles/s41598-020-58470-0 [Accessed February 4, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CABRERA-SERRANO A, GARCÍA-ARRARÁS JE (2004). RGD-containing peptides inhibit intestinal regeneration in the sea cucumber Holothuria glaberrima. Dev Dyn 231: 171–178. [DOI] [PubMed] [Google Scholar]
- CANDELARIA AG, MURRAY G, FILE SK, GARCÍA-ARRARÁS JE (2006). Contribution of mesenterial muscle dedifferentiation to intestine regeneration in the sea cucumber Holothuria glaberrima. Cell Tissue Res 325: 55–65. [DOI] [PubMed] [Google Scholar]
- COWIN AJ, HATZIRODOS N, TEUSNER JT, BELFORD DA (2003). Differential effect of wounding on actin and its associated proteins, paxillin and gelsolin, in fetal skin explants. J Invest Dermatol 120: 1118–1129. [DOI] [PubMed] [Google Scholar]
- DELGADO MA (2019). Functional Role of B-catenin and Myc as Active Players in the Canonical Wnt Signaling Pathway During Intestinal Regeneration of the Sea Cucumber Holothuria Glaberrima. Universidad de Puerto Rico.
- DOLMATOV IY, SHULGA AP, GINANOVA TT, ELISEIKINA MG, LAMASH NE (2019). Metalloproteinase inhibitor GM6001 delays regeneration in holothurians. Tissue Cell 59: 1–9. [DOI] [PubMed] [Google Scholar]
- ENGLER AJ, SEN S, SWEENEY HL, DISCHER DE (2006). Matrix elasticity directs stem cell lineage specification. Cell 126: 677–689. [DOI] [PubMed] [Google Scholar]
- FERAL J, MASSIN C (1982). Digestive system: Holothuroidea. Echinoderm Nutrition, M Jangoux and J Lawrence: 192–212. [Google Scholar]
- GALLIOT B, CHERA S (2010). The Hydra model: disclosing an apoptosis-driven generator of Wnt-based regeneration. Trends in Cell Biology 20: 514–523. [DOI] [PubMed] [Google Scholar]
- GARCÍA-ARRARÁS JE (2017). Dedifferentiation as a cell source for organ regeneration. In Regenerative Engineering and Developmental Biology (Ed. Gardiner DM). CRC Press, pp. 373–394. Available at: https://www.taylorfrancis.com/books/9781498723329/chapters/10.1201/9781315120188-20 [Accessed January 18, 2020]. [Google Scholar]
- GARCÍA-ARRARÁS JE, BELLO SA, MALAVEZ S (2019). The mesentery as the epicenter for intestinal regeneration. Seminars in Cell & Developmental Biology 92: 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARCÍA-ARRARÁS JE, DOLMATOV IYu (2010). Echinoderms; potential model systems for studies on muscle regeneration. Curr Pharm Des 16: 942–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARCÍA-ARRARÁS JE, ESTRADA-RODGERS L, SANTIAGO R, TORRES II, DÍAZ-MIRANDA L, TORRES-AVILLÁN I (1998). Cellular mechanisms of intestine regeneration in the sea cucumber, Holothuria glaberrima Selenka (Holothuroidea:Echinodermata). Journal of Experimental Zoology 281: 288–304. [DOI] [PubMed] [Google Scholar]
- GARCÍA-ARRARÁS JE, GREENBERG MJ (2001). Visceral regeneration in holothurians: Holothurian Regeneration. Microscopy Research and Technique 55: 438–451. [DOI] [PubMed] [Google Scholar]
- GARCÍA-ARRARÁS JE, SCHENK C, RODRÍGUES-RAMÍREZ R, TORRES II, VALENTÍN G, CANDELARIA AG (2006). Spherulocytes in the echinoderm Holothuria glaberrima and their involvement in intestinal regeneration. Dev Dyn 235: 3259–3267. [DOI] [PubMed] [Google Scholar]
- GARCÍA-ARRARÁS JE, VALENTÍN-TIRADO G, FLORES JE, ROSA RJ, RIVERA-CRUZ A, SAN MIGUEL-RUIZ JE, TOSSAS K (2011). Cell dedifferentiation and epithelial to mesenchymal transitions during intestinal regeneration in H. glaberrima. BMC Dev Biol 11: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERNÁNDEZ-PASOS J, VALENTÍN-TIRADO G, GARCÍA-ARRARÁS JE (2017). Melanotransferrin: New Homolog Genes and Their Differential Expression during Intestinal Regeneration in the Sea Cucumber Holothuria glaberrima. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 328: 259–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG H, WENG H, ZHOU H, QU L (2014). Attacking c-Myc: Targeted and Combined Therapies for Cancer. Current Pharmaceutical Design 20: 6543–6554. [DOI] [PubMed] [Google Scholar]
- KATOH Masuko, KATOH Masaru (2007). WNT Signaling Pathway and Stem Cell Signaling Network. Clin Cancer Res 13: 4042–4045. [DOI] [PubMed] [Google Scholar]
- KOPAN R, ILAGAN MaXG (2009). The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell 137: 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAMASH NE, DOLMATOV IY (2013). Proteases from the Regenerating Gut of the Holothurian Eupentacta fraudatrix. PLoS One 8. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3591370/ [Accessed February 11, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTINDALE MQ (2005). The evolution of metazoan axial properties. Nat Rev Genet 6: 917–927. [DOI] [PubMed] [Google Scholar]
- MARTINEZ P, RAST JP, ARENAS-MENA C, DAVIDSON EH (1999). Organization of an echinoderm Hox gene cluster. Proc Natl Acad Sci U S A 96: 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASHANOV V, AKIONA J, KHOURY M, FERRIER J, REID R, MACHADO DJ, ZUEVA O, JANIES D (2019). Active Notch Signaling is Required for Arm Regeneration in a Brittle Star. Developmental Biology. Available at: http://biorxiv.org/lookup/doi/10.1101/2019.12.13.875401 [Accessed January 23, 2020]. [DOI] [PMC free article] [PubMed]
- MASHANOV VS, DOLMATOV IYu, HEINZELLER T (2004). Gut formation during development and regeneration in Eupentacta fraudatrix. Taylor & Francis. [Google Scholar]
- MASHANOV VS, DOLMATOV IYu, HEINZELLER T (2005). Transdifferentiation in Holothurian Gut Regeneration. The Biological Bulletin 209: 184–193. [DOI] [PubMed] [Google Scholar]
- MASHANOV VS, FROLOVA LT, DOLMATOV IYu (2004). Structure of the Digestive Tube in the Holothurian Eupentacta fraudatrix (Holothuroidea: Dendrochirota). Russian Journal of Marine Biology 30: 314–322. [Google Scholar]
- MASHANOV VS, GARCÍA-ARRARÁS JE (2011). Gut regeneration in holothurians: a snapshot of recent developments. Biol Bull 221: 93–109. [DOI] [PubMed] [Google Scholar]
- MASHANOV Vladimir S., ZUEVA OR, GARCÍA-ARRARÁS JE (2015). Expression of pluripotency factors in echinoderm regeneration. Cell Tissue Res 359: 521–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASHANOV VS, ZUEVA OR, GARCIA-ARRARAS JE (2012). Expression of Wnt9, TCTP, and Bmp1/Tll in sea cucumber visceral regeneration. Gene Expr Patterns 12: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASHANOV Vladimir S, ZUEVA OR, GARCÍA-ARRARÁS JE (2015). Myc regulates programmed cell death and radial glia dedifferentiation after neural injury in an echinoderm. BMC Dev Biol 15. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4448152/ [Accessed February 3, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASHANOV VS, ZUEVA OR, GARCÍA-ARRARÁS JE (2014). Transcriptomic changes during regeneration of the central nervous system in an echinoderm. BMC Genomics 15: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASHANOV VS, ZUEVA OR, ROJAS-CATAGENA C, GARCIA-ARRARAS JE (2010). Visceral regeneration in a sea cucumber involves extensive expression of survivin and mortalin homologs in the mesothelium. BMC Developmental Biology 10: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MÉNDEZ AT, ROIG-LÓPEZ JL, SANTIAGO P, SANTIAGO C, GARCÍA-ARRARÁS JE (2000). Identification of Hox Gene Sequences in the Sea Cucumber Holothuria glaberrima Selenka (Holothuroidea: Echinodermata). Mar Biotechnol 2: 231–240. [DOI] [PubMed] [Google Scholar]
- MESCHER AL, NEFF AW (2005). Regenerative Capacity and the Developing Immune System. In Regenerative Medicine I: Theories, Models and Methods (Ed. Yannas IV). Springer, Berlin, Heidelberg, pp. 39–66. Available at: 10.1007/b99966 [Accessed March 11, 2020]. [DOI] [PubMed] [Google Scholar]
- MIAO T, WAN Z, SUN L, LI X, XING L, BAI Y, WANG F, YANG H (2017). Extracellular matrix remodeling and matrix metalloproteinases (ajMMP-2 like and ajMMP-16 like) characterization during intestine regeneration of sea cucumber Apostichopus japonicus. Comp Biochem Physiol B, Biochem Mol Biol 212: 12–23. [DOI] [PubMed] [Google Scholar]
- MÜNDER S, TISCHER S, GRUNDHUBER M, BÜCHELS N, BRUCKMEIER N, ECKERT S, SEEFELDT CA, PREXL A, KÄSBAUER T, BÖTTGER A (2013). Notch-signalling is required for head regeneration and tentacle patterning in Hydra. Dev Biol 383: 146–157. [DOI] [PubMed] [Google Scholar]
- MURRAY G, GARCÍA-ARRARÁS JE (2004). Myogenesis during holothurian intestinal regeneration. Cell and Tissue Research 318: 515–524. [DOI] [PubMed] [Google Scholar]
- NAKAMURA K, CHIBA C (2007). Evidence for Notch signaling involvement in retinal regeneration of adult newt. Brain Res 1136: 28–42. [DOI] [PubMed] [Google Scholar]
- NIEVES-RÍOS C, ALVAREZ-FALCÓN S, MALAVEZ S, RODRIGUEZ-OTERO J, GARCÍA-ARRARÁS JE (2019). The nervous system component of the mesentery of the sea cucumber Holothuria glaberrima in normal and regenerating animals. Cell Tissue Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ORTIZ-PINEDA PA, RAMÍREZ-GÓMEZ F, PÉREZ-ORTIZ J, GONZÁLEZ-DÍAZ S, SANTIAGO-DE JESÚS F, HERNÁNDEZ-PASOS J, DEL VALLE-AVILA C, ROJAS-CARTAGENA C, SUÁREZ-CASTILLO EC, TOSSAS K, MÉNDEZ-MERCED AT, ROIG-LÓPEZ JL, ORTIZ-ZUAZAGA H, GARCÍA-ARRARÁS JE (2009). Gene expression profiling of intestinal regeneration in the sea cucumber. BMC Genomics 10: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PINEDA PAO (2010). Analysis and Characterization of Genes Associated with Intestinal Regeneration in the Sea Cucumber (Echinodermata: Holothuroidea). Universidad de Puerto Rico.
- QUIÑONES JL, ROSA R, RUIZ DL, GARCÍA-ARRARÁS JE (2002). Extracellular matrix remodeling and metalloproteinase involvement during intestine regeneration in the sea cucumber Holothuria glaberrima. Dev Biol 250: 181–197. [DOI] [PubMed] [Google Scholar]
- QUISPE D, MEDINA J, CRUZ-GONZÁLEZ S, ORTIZ-ZUAZAGA H, GARCÍA-ARRARÁS JE Transcriptomic Analysis of Early Stages of Regeneration in Holothuria glaberrima. Unpublished manuscript. [DOI] [PMC free article] [PubMed]
- RAMÍREZ-GÓMEZ F, ORTIZ-PINEDA PA, RIVERA-CARDONA G, GARCÍA-ARRARÁS JE (2009). LPS-Induced Genes in Intestinal Tissue of the Sea Cucumber Holothuria glaberrima. PLoS One 4. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2702171/ [Accessed February 4, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REDDIEN PW, BERMANGE AL, KICZA AM, ALVARADO AS (2007). BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development 134: 4043–4051. [DOI] [PubMed] [Google Scholar]
- ROIG-LÓPEZ J, SANTIAGO P, GARCÍA-ARRARÁS J (2001). Strategies to identify differentially expressed genes during regeneration. Echinoderms. [Google Scholar]
- ROJAS-CARTAGENA C, ORTÍZ-PINEDA P, RAMÍREZ-GÓMEZ F, SUÁREZ-CASTILLO EC, MATOS-CRUZ V, RODRÍGUEZ C, ORTÍZ-ZUAZAGA H, GARCÍA-ARRARÁS JE (2007). Distinct profiles of expressed sequence tags during intestinal regeneration in the sea cucumber Holothuria glaberrima. Physiological Genomics 31: 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAN MIGUEL-RUIZ JE, GARCÍA-ARRARÁS JE (2007). Common cellular events occur during wound healing and organ regeneration in the sea cucumber Holothuria glaberrima. BMC Developmental Biology 7: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANTIAGO P, ROIG-LÓPEZ J, SANTIAGO C, GARCÍA-ARRARÁS J (2000). Serum Amyloid A Protein in an Echinoderm: Its Primary Structure and Expression During Intestinal Regeneration in the Sea Cucumber Holothuria glaberrima. Journal of Experimental Zoology Part B, Molecular and Developmental Evolution: 335–344. [DOI] [PubMed] [Google Scholar]
- SUÁREZ-CASTILLO EC, MEDINA-ORTÍZ WE, ROIG-LÓPEZ JL, GARCÍA-ARRARÁS JE (2004). Ependymin, a gene involved in regeneration and neuroplasticity in vertebrates, is overexpressed during regeneration in the echinoderm Holothuria glaberrima. Gene 334: 133–143. [DOI] [PubMed] [Google Scholar]
- SUN L, YANG H, CHEN M, MA D, LIN C (2013). RNA-Seq Reveals Dynamic Changes of Gene Expression in Key Stages of Intestine Regeneration in the Sea Cucumber Apostichopus japonicas Ed. T Unver. PLoS ONE 8: e69441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKAHASHI K, YAMANAKA S (2006). Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126: 663–676. [DOI] [PubMed] [Google Scholar]
- TANG DD, GERLACH BD (2017). The roles and regulation of the actin cytoskeleton, intermediate filaments and microtubules in smooth muscle cell migration. Respir Res 18. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5385055/ [Accessed February 13, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOOFAN P, WHEADON H (2016). Role of the bone morphogenic protein pathway in developmental haemopoiesis and leukaemogenesis. Biochemical Society Transactions 44: 1455–1463. [DOI] [PubMed] [Google Scholar]
- TOSSAS K, QI-HUANG S, CUYAR E, GARCÍA-ARRARÁS JE (2014). Temporal and spatial analysis of enteric nervous system regeneration in the sea cucumber Holothuria glaberrima. Regeneration (Oxf) 1: 10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WALTON KD, CROCE JC, GLENN TD, WU S-Y, MCCLAY DR (2006). Genomics and expression profiles of the Hedgehog and Notch signaling pathways in sea urchin development. Developmental Biology 300: 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WENEMOSER D, LAPAN SW, WILKINSON AW, BELL GW, REDDIEN PW (2012). A molecular wound response program associated with regeneration initiation in planarians. Genes Dev 26: 988–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU Y, YAO F, MEI Y, CHU B, CHENG C, LIU Y, LI X, ZOU X, HOU L (2014). Cloning and expression analysis of the gene encoding fibrinogen-like protein A, a novel regeneration-related protein from Apostichopus japonicus. Mol Biol Rep 41: 2617–2627. [DOI] [PubMed] [Google Scholar]
- YEN T-H, WRIGHT NA (2006). The gastrointestinal tract stem cell niche. Stem Cell Reviews 2: 203–212. [DOI] [PubMed] [Google Scholar]
- YU L, HAN M, YAN M, LEE E-C, LEE J, MUNEOKA K (2010). BMP signaling induces digit regeneration in neonatal mice. Development 137: 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YUAN J, GAO Y, SUN L, JIN S, ZHANG X, LIU C, LI F, XIANG J (2019). Wnt Signaling Pathway Linked to Intestinal Regeneration via Evolutionary Patterns and Gene Expression in the Sea Cucumber Apostichopus japonicus. Front Genet 10. Available at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00112/full [Accessed November 19, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZAKANY J, DUBOULE D (2007). The role of Hox genes during vertebrate limb development. Current Opinion in Genetics & Development 17: 359–366. [DOI] [PubMed] [Google Scholar]
- ZHANG X, SUN L, YUAN J, SUN Y, GAO Y, ZHANG L, LI S, DAI H, HAMEL J-F, LIU C, et al. (2017). The sea cucumber genome provides insights into morphological evolution and visceral regeneration Ed. C Tyler-Smith. PLOS Biology 15: e2003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO L, BORIKOVA AL, BEN-YAIR R, GUNER-ATAMAN B, MACRAE CA, LEE RT, BURNS CG, BURNS CE (2014). Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. PNAS 111: 1403–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]


