Abstract
The genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 comprises many open reading frames (ORFs) which putatively encode eukaryotic-type protein kinase and protein phosphatase. Based on gene disruption analysis, a region of the hypothetical ORF sll1575, which retained a part of the protein kinase motif, was found to be required for normal motility in the original isolate of strain PCC 6803. Sequence determination revealed that in this strain sll1575 was part of a gene (designated spkA) which harbored an entire eukaryotic-type Ser/Thr protein kinase motif. Strain ATCC 27184 and a glucose-tolerant strain derived from the same isolate as the PCC strain had a frameshift mutation dividing spkA into ORFs sll1574 and sll1575. The structural integrity of spkA agreed well with the motility phenotype, determined by colony morphology on agar plates. The spkA gene was expressed in Escherichia coli as a His-tagged protein, which was purified by Ni2+ affinity chromatography. With [γ-32P]ATP, SpkA was autophosphorylated and transferred the phosphate group to casein, myelin basic protein, and histone. SpkA also phosphorylated several proteins in the membrane fraction of Synechocystis cells. These results suggest that SpkA is a eukaryotic-type Ser/Thr protein kinase and regulates cellular motility via phosphorylation of the membrane proteins in Synechocystis.
Protein phosphorylation-dephosphorylation is a mechanism widely used to regulate proteins. In prokaryotes, phosphotransfer of the protein His kinase to the Asp residue in the response regulator is predominant in various signal transduction pathways (15). However, determination of the complete genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 (10) revealed a number of open reading frames (ORFs) that are homologous to the eukaryotic-type protein kinase and protein phosphatase. Recent progress in genome analysis has further shown that bacteria and archaea universally have several types of Ser/Thr protein kinase and protein phosphatase, which were originally believed to be specific to eukaryotes (3, 11, 13, 21). These findings strongly suggested that prokaryotes have signal transduction systems in addition to the well-known sensor His kinase and response regulator systems.
Bacteria lacking flagella such as cyanobacteria can move by gliding motility. Photoresponsive gliding motility is unique to cyanobacteria and has been studied mainly in filamentous organisms for many decades (6, 7). Despite extensive studies, very little has been established with respect to the regulatory mechanism of motility in cyanobacteria. On the other hand, twitching or swimming motility in unicellular cyanobacteria, though not as conspicuous as in filamentous cyanobacteria, has been described (5, 14, 17). The motility of Synechocystis strain sp. PCC 6803, though described as sporadic and very slow (14), seems to be a feasible target for molecular analysis, since the complete genome has been determined (10). It was recently suggested that an alternative sigma factor, SigF, and putative pilin subunit gene, sll1694, are essential for the motility of this cyanobacterium (1).
In an earlier study, we showed that a putative Ser/Thr protein phosphatase gene, slr2031, plays a crucial role in motility of Synechocystis cells (9). To extend these findings, we evaluated the counteracting protein kinase as a regulator of motility by means of targeted disruption of genes with a Ser/Thr protein kinase motif. Here we reported that the protein kinase SpkA is required for the normal motility of Synechocystis cells. The spkA gene was not listed in the original annotation of the Synechocystis genome (10) because of a frameshift mutation in the sequenced strain.
MATERIALS AND METHODS
Strains and culture conditions.
Strains PCC 6803 and ATCC 27184 were obtained as the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 from the Pasteur Culture Collection and American Type Culture Collection, respectively; both were independently deposited by R. Kunisawa as isolates from the same strain (Berkeley strain number 6803) (12, 14). The glucose-tolerant strain, isolated by Williams (18), was a kind gift from W. Vermaas (Arizona State University). Standard strains and mutants were grown in BG11 medium buffered with N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid-KOH (pH 7.8) at 31°C at a light intensity of 20 μE m−2 s−1. Solid medium was supplemented with 0.8% (wt/vol) agar and 0.3% (wt/vol) sodium thiosulfate and used for examination of motility by colony morphology. Kanamycin (20 μg/ml) was added to maintain gene-disrupted mutants; antibiotics were not included for characterization of the mutant phenotype. For cloning and subcloning of plasmids in Escherichia coli, strains XL10 and JM109 were used; BL21 (DE3)pLysS was used for expression with pET28a.
Construction of spkA disruption mutant.
Disruption of spkA was achieved as disruption of sll1575. A part of sll1575 was amplified by PCR using primer 1 (5′-GGGTCAAGTCTACCGAGC-3′), primer 2 (5′-ATCCGACTAGGCATGGGC-3′), and Taq polymerase (Ampli-Taq; PE Applied Biosystems, Foster City, Calif.) and cloned into pT7Blue-T vector (Novagen, Madison, Wis.). sll1575 was interrupted at the MscI site by insertion of the Tn5-derived kanamycin resistance cassette in the same direction as sll1575. Although the cassette allows expression of downstream genes due to lack of transcription termination, the map (Fig. 1A) suggests that insertion of the cassette does not affect expression of the flanking ORFs. Mutants were generated by transformation of Synechocystis cells with this DNA and selected on BG11 plates containing kanamycin (20 μg/ml). Complete segregation was confirmed by PCR with the primers described above (not shown).
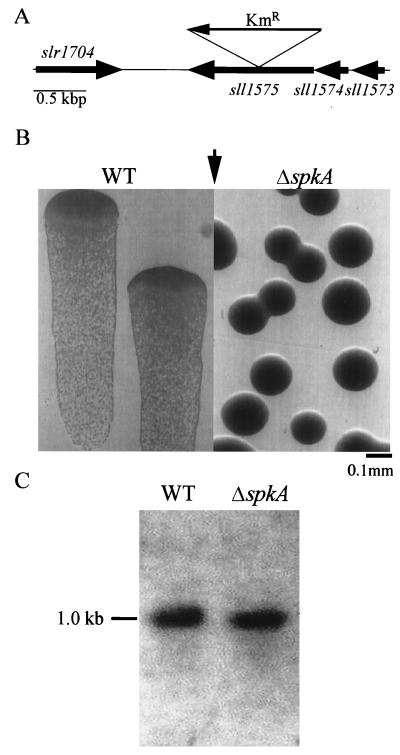
FIG. 1.
Characterization of the sll1575-disrupted mutant. (A) Gene map showing the relative positions of sll1573, sll1574, and sll1575 and insertion of the kanamycin resistance (KmR) cassette. (B) Colony morphology of wild-type (WT) and mutant cells grown under lateral illumination at 20 μE m−2 s−1 (arrow). Cells were grown as single colonies on 0.8% agar–BG11 medium for 5 days at 31°C. (C) Expression of pilA1 (sll1694) as revealed by Northern hybridization.
DNA sequence analysis.
The full-length DNA of spkA of the PCC and glucose-tolerant strains and a region around the border of sll1574 and sll1575 of the ATCC strain were determined by the BigDye terminator fluorescence detection method (PE Applied Biosystems), using a capillary sequencer (ABI PRISM 310 Genetic Analyzer; PE Applied Biosystems).
Cloning of spkA.
The coding region of spkA was amplified from the genomic DNA of the motile PCC strain by PCR with primer 3 (5′-GATGCTAGCGCTATGACCCCTG-3′) and primer 4 (5′-ATGAGCTCACAATCCTGAAACCT-3′), containing NheI and SacI sites, respectively. PCR was performed with Pfu DNA polymerase (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions. Following initial denaturation at 95°C for 1 min, each sample was subjected to 35 cycles consisting of denaturation at 95°C for 30 s, annealing at 57°C for 1.5 min, and elongation at 72°C for 4 min. The PCR product was cloned into pPCR-Script (Stratagene) according to the manufacturer's instructions and propagated in E. coli XL10 (Epicurian Coli XL10-Gold Kan; Stratagene). The cloned spkA was sequenced, excised with NheI and SacI (New England Biolabs, Beverly, Mass.) and then inserted into pET28a (Novagen) as a fusion with the N-terminal His tag.
Isolation of total RNA and Northern blotting.
Total RNA was isolated by using an RNeasy Midi kit (QiaGEN, Hilden, Germany). The standard protocol for breakage of cells was modified as follows. Synechocystis cells collected from a 100-ml culture (A730 = 0.6 to 1.0) were disrupted with a Mini-Bead Beater (Biospec, Bartlesville, Okla.) and zircon beads (100 μm in diameter; Biospec) for three pulses of 50 s at 4°C in 0.9 ml of buffer provided in the kit. After removal of the beads by brief centrifugation, the volume of cell lysate and ethanol concentration were adjusted and subjected to a spin column chromatography according to the manufacturer's instructions. Total RNA (10 μg) thus isolated was fractionated in a 1.2% denaturing agarose gel and blotted onto a Hybond-N+ membrane (Amersham Pharmacia, Uppsala, Sweden). As a probe, pilA1 (sll1694) was amplified with primer 5 (5′-CACATATGGCTAGTAATTTTAAATTC-3′) and primer 6 (5′-GGCACGTGTTTAATTACTTCAGCACC-3′). Labeling of the probe and detection were done by using ECL (enhanced chemiluminecence) direct nucleic acid labeling and detection systems (Amersham Pharmacia) according to the manufacturer's instructions.
Expression and purification of SpkA.
pET28a carrying spkA was introduced into E. coli BL21(DE3)pLysS. Cells were grown at 37°C in 250 ml of Luria broth medium containing kanamycin (20 μg/ml) and chloramphenicol (37 μg/ml) to an A600 of about 0.5. Then isopropyl-β-d-thiogalactoside (IPTG) was added to a final concentration of 0.5 mM, and the cultures were incubated for 2 h at 25°C. The cells were harvested by centrifugation, washed with 50 mM Tris-HCl (pH 7.5) containing 100 mM NaCl and 1 mM phenylmethylsulfonyl fluoride, and resuspended in 25 ml of the same medium plus 10% (wt/vol) glycerol. The cell suspension was once frozen at −85°C for 15 min, thawed on ice, and then sonicated at 4°C for 9 min (three cycles of 3-min bursts with a cooling period) in a sonicator (model 200M; Kubota Co., Tokyo, Japan). The cell extract was centrifuged at 16,000 × g for 30 min, and the supernatant was subjected to Ni2+ affinity chromatography.
A Hi-Trap chelating column (Amersham Pharmacia) charged with Ni2+ was equilibrated with 50 mM Tris-HCl (pH 7.5) containing 100 mM NaCl, 10% (wt/vol) glycerol, and 5 mM imidazole (buffer A). The column was loaded with the cell extract and washed with buffer A; then His-tagged SpkA protein was eluted using linear gradient from 5 to 500 mM imidazole. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by staining with Coomassie brilliant blue R-250. Alternatively, proteins resolved in the SDS-gel were blotted to a polyvinylidene difluoride membrane (Immobilon; Millipore, Bedford, Mass.) and the His tag portion was visualized with His-Probe (Pierce, Rockford, Ill.) as instructed by the manufacturer.
Assay of protein kinase activity.
Autophosphorylation of SpkA and phosphorylation of myelin basic protein (MBP), histone, or casein were assayed in vitro with [γ-32P]ATP. About 0.5 μg of the purified SpkA protein was added to 10 μl of phosphorylation buffer containing 20 mM 2-morpholinoethanesulfonic acid (pH 6.5), 10 mM MgCl2, and 0.1 mM [γ-32P]ATP (3,000 Ci mol−1) with or without 2.5 μg of bovine MBP (Sigma, St. Louis, Mo), 2.5 μg of histone (type IIIS, calf thymus; Sigma), or 6.25 μg of bovine casein (partially dephosphorylated; Sigma) and incubated for 15 min at 30°C. Control phosphorylation experiments were done with a crude extract from E. coli before induction. SDS (final concentration, 1%) and dithiothreitol (final concentration, 60 mM) were added to stop the reaction. After boiling for 5 min, proteins were resolved by SDS-PAGE. The gels were stained with Coomassie brilliant blue R-250, dried, and then subjected to autoradiography with X-ray film (X-Omat Blue XB-1; Eastman Kodak, Rochester, N.Y.).
In vitro phosphorylation of cell extracts.
Synechocystis cells were harvested from a 50-ml culture (A730 = 0.5 to 0.8) by centrifugation at 8,000 × g for 5 min, washed with 20 mM Tris-HCl (pH 7.5) containing 100 mM NaCl, and resuspended in 0.9 ml of the same buffer. The cells were disrupted with the zircon beads in a Mini-Bead Beater for three pulses of 50 s at 4°C. After removal of the beads by brief centrifugation, cell extracts were fractionated into soluble and membrane fractions by centrifugation at 541,000 × g for 30 min at 4°C. The membranes were resuspended in the original volume of the buffer. Soluble and membrane fractions, both derived from 1.25 μg of chlorophyll, were incubated with [γ-32P]ATP in 20 μl of the phosphorylation buffer described above. When stated, 1 μg of the purified SpkA protein was included.
RESULTS
Novel gene is required for motility.
The Synechocystis genome contains genes encoding seven putative Ser/Thr protein kinases which show similarity to typical eukaryotic-type protein kinases (10). Among them, sll1575 is unique; it encodes only the C-terminal part of the conserved protein kinase motif, while the upstream gene sll1574, originally annotated as a hypothetical gene, encodes the remainder. To determine whether sll1575 is functional, we constructed a gene disruption mutant (Fig. 1A) from the motile wild type, which was obtained as PCC strain 6803 (denoted the PCC strain). After complete segregation, it was found that the sll1575-disrupted mutant formed domed, round-shaped colonies on agar plates, while the parent strain formed flat sheet-like colonies, indicative of loss of motility in the mutant. Figure 1B shows typical nonmotile colonies of the spkA mutant in contrast with active movement of the wild type toward the light source (positive phototaxis). This also indicates that the sequence of sll1575 is functional in the PCC strain.
To confirm that the split gene comprising sll1574 and sll1575 encodes a functional protein kinase, we cloned it from the PCC strain and determined the nucleotide sequence. Unexpectedly, the gene was no longer split but consisted of a single ORF due to frameshifting as a result of lack of a single A in the last codon for Asn in sll1574 (Fig. 2). The new ORF encodes a protein of 521 amino acid residues. We designated this novel ORF spkA (for Synechocystis protein kinase). The deduced protein of spkA has the entire motif (subdomains I to XI) common to a number of eukaryotic-type Ser/Thr protein kinases (8). We also determined the nucleotide sequence of spkA in derivatives of this Synechocystis strain and found that ATCC 27184 and the widely used glucose-tolerant strain (12, 18) carried the same frameshift mutation as the sequenced strain (not shown). In accordance with this finding, these substrains were nonmotile on agar plates.
FIG. 2.
Gene and protein sequences of spkA. (A) Sequence variation in spkA. Part of the nucleotide and deduced amino acid sequences of spkA in the PCC, ATCC, and GT strains. The putative initiation codon (GTG) of sll1575 is underlined. One base pair insertion together with the frameshifted codons in the ATCC and glucose-tolerant (GT) strains are shown in reverse type. (B) Amino acid sequence alignment of Synechocystis SpkA (S.6803 spkA) with ORF520 from Anabaena sp. strain PCC 7120 (A.7120 ORF520), ORF517 from Nostoc punctiforme ATCC 29133 (N.p. ORF517), and Pkn2 from Myxococcus xanthus (M.x. pkn2). Residues conserved with those in SpkA are shown in reverse type. Subdomains I to XI typical to eukaryotic-type protein kinases are shown above the alignment, and highly conserved residues are indicated with asterisks (see text for details). Note that only the N-terminal kinase domain of Pkn2 is presented.
Recently it was shown that motility of Synechocystis sp. strain PCC 6803 requires a type IV-like pilus structure, which is supported by a number of subunits and biogenesis factors (1, 2, 19). To determine whether the biogenesis of pili was regulated by the protein kinase SpkA, we examined the mRNA level of pilA1 (sll1694), which encodes a major pilin subunit of the pili. Northern blot analysis revealed no significant difference in pilA1 mRNA levels of the wild-type and the spkA mutant strains (Fig. 1C). We observed cell surface architecture by electron microscopy and found that both the wild type and the spkA mutant had the two types of pili, thick and thin (S. Yoshihara, A. Kamei, and M. Ikeuchi, unpublished results), the latter of which is known to be essential for motility (2, 19). These observations suggest that spkA is not an essential factor but regulates motility via an unidentified signal transduction pathway.
Protein kinase activity of SpkA.
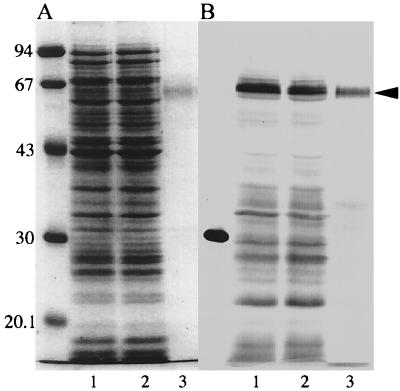
We tried to express the functional spkA gene with an N-terminal His tag in E. coli under control of the T7 promoter. However, the dye-stained profile was not changed after induction, even though expression was induced at 25°C (Fig. 3). We could detect the His-tagged SpkA protein only by Western blotting with His-Probe. Not only the major 63-kDa band but also many bands in the low-molecular-mass region were visualized with His-Probe, indicative of rapid degradation in cells. Under the conditions used, very little SpkA was recovered in the inclusion body (not shown). The soluble fraction was subjected to Ni2+ affinity column chromatography. Although a large part of the 63-kDa protein and many degraded proteins were not adsorbed, a small part of the His-tagged SpkA was purified to homogeneity (Fig. 3, lane 3). This may suggest that improperly folded SpkA protein is not stable and is rapidly degraded in the cytoplasm of E. coli.
FIG. 3.
Expression and purification of SpkA. Proteins resolved on 12% polyacrylamide gels were visualized by staining with Coomassie brilliant blue (A) and Western blotting with His-Probe (B). Lane 1, cell extract of E. coli after induction with IPTG; lanes 2 and 3, flowthrough fraction and His-tagged SpkA-enriched fraction in Ni2+ affinity chromatography, respectively. Positions of molecular size markers are shown in kilodaltons at the left (phosphorylase b, 94 kDa; bovine serum albumin, 67 kDab; ovalbumin, 43 kDa; carbonic anhydrase, 30 kDa; trypsin inhibitor, 20.1 kDa). The arrow head shows the 63-kDa SpkA band.
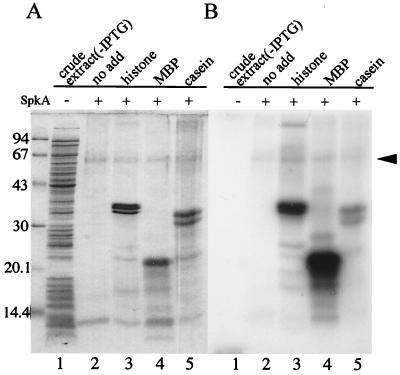
We examined protein kinase activity of SpkA with [γ-32P]ATP (Fig. 4). We could detect weak but significant autophosphorylation activity of SpkA and strong phosphorylation of histone, MBP, and casein, which are general substrates of typical Ser/Thr protein kinases. Among these, MBP was phosphorylated to the greatest extent. A very similar phosphorylation pattern was obtained with the crude soluble extract before chromatography (not shown). As a negative control, we detected no phosphorylation in the crude extract of E. coli before induction (Fig. 4, lane 1). Thus, we conclude that SpkA of Synechocystis is a Ser/Thr protein kinase which belongs to the large family of protein kinases in eukaryotes.
FIG. 4.
Detection of protein kinase activity. Phosphorylated proteins were resolved on 15% polyacrylamide gels and visualized by staining with Coomassie brilliant blue (A) and autoradiography (B). Lane 1, cell extract of E. coli before induction with IPTG; lanes 2 to 5, Ni2+ affinity-purified SpkA protein without (lane 2) or with histone, MBP, and casein, as indicated. Positions of molecular size markers are shown in Kilodaltons at the left; the arrow head shows autophosphorylation of the 63-kDa SpkA band.
In vitro phosphorylation of cyanobacterial proteins.
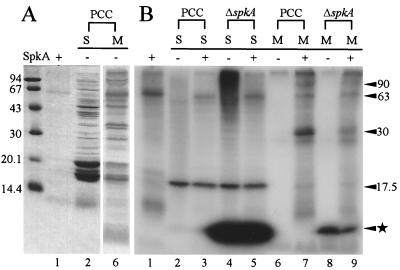
To determine the intrinsic substrate of SpkA, we performed in vitro phosphorylation in crude extracts from wild-type and spkA mutant cells in the presence or absence of His-tagged SpkA (Fig. 5). We detected SpkA-dependent phosphorylation in a 90-kDa band and a doublet band around 30 kDa in the membrane fractions in addition to autophosphorylation of the exogenous SpkA (Fig. 5, lanes 7 and 9). These bands were not detected in the soluble fractions or did not correspond to the dye-stained bands in the membrane fractions. These findings suggest that minor membrane-associated proteins of about 90 and 30 kDa are the intrinsic substrates of SpkA. On the other hand, there were heavily labeled bands around 17.5 kDa and in the low-molecular-mass region of the soluble fractions even in the absence of SpkA (Fig. 5, lane 4). The 17.5-kDa band was located just below the major stained band of phycocyanin. The prominent label in the low-molecular-mass region was not due to the unreacted [γ-32P]ATP. Curiously, labeling of the latter was very weak in the wild type. Disruption of spkA may have changed the expression of other protein kinases. Anyway phosphorylation of these bands was independent of SpkA, suggestive of an intrinsic protein kinase which is yet to be identified.
FIG. 5.
In vitro phosphorylation of Synechocystis proteins with SpkA. Proteins of soluble (S) and membrane fractions (M) from Synechocystis strain PCC and ΔspkA cells were incubated with (+) or without (−) purified SpkA and resolved on an SDS–15% polyacrylamide gel. (A) Dye-stained gel; (B) autoradiogram of the same gel. Lane 1, Ni2+ affinity-purified SpkA without cell extracts. Purified SpkA protein was extraneously added in lanes 3, 5, 7, and 9. Positions of molecular size markers are shown in Kilodaltons at the left; positions of the major phosphorylated polypeptides are indicated with the arrowhead and asterisk on the right.
DISCUSSION
In this study, we demonstrated experimentally that a novel gene, spkA, encodes a Ser/Thr-type protein kinase resembling typical eukaryotic enzymes. Clearly, SpkA phosphorylated the general substrate proteins, intrinsic membrane proteins, and itself. Before our work, it was assumed that a protein kinase was encoded by a split gene comprising sll1574 and sll1575 (10, 11, 21). We created sll1575-disrupted mutants from both the nonmotile glucose-tolerant strain and the motile PCC strain. As a result, the mutant from the PCC strain lost motility, while that from the glucose-tolerant strain showed no defect. This led us to further confirm the gene sequence in both PCC and glucose-tolerant strains. In fact, the gene in the PCC strain was uninterrupted and expressed an active product in E. coli. On the other hand, the gene in the glucose-tolerant strain was split as in the sequenced Kazusa strain. The glucose-tolerant strain has been widely used as standard in many laboratories because of its application to genetic engineering of photosynthetic apparatus (18). Thus, especially in gene analysis we must use a wild type as much as possible, although it is rather difficult to ensure this beforehand. Spontaneous inactivation and its domination in a culture stock such as ATCC may reflect the physiological significance of motility-related phenomena of this cyanobacterium in photosynthetic propagation.
Homology search of the protein database with the deduced SpkA protein revealed that it is a member of Pkn2 family in bacteria, which belongs to the eukaryotic Ser/Thr protein kinase superfamily (13). Although there was no obvious homolog of SpkA in the database, we could identify homologs in ongoing genome projects, namely, ORF520 in Anabaena sp. strain PCC 7120 (http://www.kazusa.or.jp/cyano/anabaena/) and ORF517 in Nostoc punctiforme ATCC 29133 (http://www.jgi.doe.gov/JGI_microbial/html/nostoc_homepage.html). On the other hand, we could not detect the homolog in the genome of the marine cyanobacterium Prochlorococcus marinus MED4 (http://www.jgi.doe.gov/JGI_microbial/html/prochlorococcus_homepage.html). Since the P. marinus genome contain no genes involved in motility such as the pilM cluster (19), they may not retain the ability to regulate the motility. Sequence alignment of the N-terminal halves of the three SpkA homologs and the typical Pkn2 from Myxococcus xanthus (16) revealed the following common features (Fig. 2): GXGXXGXV motif in subdomain I for ATP binding; Lys residue in subdomain II, necessary for phosphotransfer (4); Glu residue in subdomain III; DXKPXN motif in subdomain VI as a Ser/Thr-specific feature; triplet DFG in subdomain VII; Asp residue in subdomain IX; and Arg residue in subdomain XI (8). Recent genome analysis revealed that the Pkn2 family has many components in cyanobacteria (7 genes in Synechocystis sp. strain PCC 6803) and mycobacteria (11 genes in Mycobacterium tuberculosis) but not many in other bacteria (11). We expressed in E. coli the other six proteins of the Pkn2 family in Synechocystis and detected phosphorylation activity in most of them (A. Kamei, and M. Ikeuchi, unpublished data). Thus, it is now clear that SpkA as well as other proteins have Ser/Thr protein kinase activities in Synechocystis.
The C-terminal half of SpkA was also conserved in Anabaena ORF520 and Nostoc ORF517 (Fig. 2). Notably, there is a variable region between the N-terminal kinase motif and the C-terminal conserved domain. This region may simply connect the two domains. Homology search of the database with the C-terminal conserved domain revealed no homology to known proteins or any motif. At the moment, we assume that the C-terminal part of SpkA is important for determination of the substrate specificity or regulation of the kinase activity. In vitro experiments suggested that SpkA regulates motility via phosphorylation of 90- and 30-kDa proteins in the membrane fraction in situ, although their identities are not known (Fig. 5).
The nonmotile phenotype of the spkA-disrupted mutant strongly suggests that protein phosphorylation regulates motility by a molecular mechanism that remains to be clarified. We also recently identified many genes which are essential for the motility of Synechocystis, such as the pilM cluster (19). In the pilM disruptant, motility and transformation competency were abolished, along with loss of the thick pili on the cell surface. By contrast, the spkA mutant was nearly nonmotile on agar plates (Fig. 1B), although it retained both thick and thin pili. The mutant also expressed mRNA of the major pilin gene pilA1 (sll1694) at a level comparable to the wild-type level (Fig. 1C). On the other hand, the mutant showed slight motility on soft agar, as judged by occasional formation of a small fringe of a single-cell layer surrounding the domed colonies (not shown in Fig. 1B). Movement of the mutant cells on soft agar was very weak, and it was difficult to determine whether phototactic properties were also affected. Anyway, these findings suggest that spkA is not essential for motility or biogenesis of the thick pili but stimulates motility by phosphorylation of some yet unidentified component(s) of the motility apparatus or signal transduction pathway to regulate it. In this context, it is of note that the putative protein phosphatase gene slr2031 was also a regulatory factor for motility (9). In theory, it is not impossible that both SpkA kinase and Slr2031 phosphatase attack the same target protein, which is involved in motility. Understanding of the complicated processes of motility in cyanobacteria requires determination of the target protein(s) for these enzymes.
ACKNOWLEDGMENTS
This work was supported by a Research Fellowship for Young Scientists from the Japan Society of the Promotion of Science (to A.K.), Grants-in-Aid for Scientific Research on Priority Areas C “Genome Biology” (12206002) (to M.I.) and for Scientific Research C (08836002) and B (11554035, 09NP1501) (to M.I.) from the Ministry of Education, Science and Culture, Japan, and a grant for Scientific Research from the Human Frontier Science program (to M.I.).
REFERENCES
- 1.Bhaya D, Watanabe N, Ogawa T, Grossman A R. The role of an alternative sigma factor in motility and pilus formation in the cyanobacterium Synechocystis sp. strain PCC6803. Proc Natl Acad Sci USA. 1999;96:3188–3193. doi: 10.1073/pnas.96.6.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaya D, Bianco N R, Bryant D A, Grossman A R. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol Microbiol. 2000;37:941–951. doi: 10.1046/j.1365-2958.2000.02068.x. [DOI] [PubMed] [Google Scholar]
- 3.Bork P, Brown N P, Hegyi H, Schultz J. The protein phosphatase 2C (PP2C) superfamily: detection of bacterial homologues. Protein Sci. 1996;5:1421–1425. doi: 10.1002/pro.5560050720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrera A C, Alexandrov K, Roberts T M. The conserved lysine of the catalytic domain of protein kinases is actively involved in the phosphotransfer reaction and not required for anchoring ATP. Proc Natl Acad Sci USA. 1993;90:442–446. doi: 10.1073/pnas.90.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castenholz R W. Movements. In: Carr N G, Whitton B A, editors. The biology of blue-green algae. Oxford, United Kingdom: Blackwell Scientific Publications Ltd.; 1973. pp. 320–339. [Google Scholar]
- 6.Diehn B, Feinleib M E, Haupt W, Hildebrand E, Lenci F, Nultchh W. Terminology of behavioral responses of motile microorganisms. Photochem Photobiol. 1979;26:559–560. [Google Scholar]
- 7.Häder D-P. Photosensory behavior in procaryotes. Microbiol Rev. 1987;51:1–21. doi: 10.1128/mr.51.1.1-21.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanks S K, Quinn A M, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 9.Kamei A, Ogawa T, Ikeuchi M. Identification of a novel gene (slr2031) involved in high-light resistance in the cyanobacterium Synechocystis sp. PCC 6803. In: Garab G, editor. Photosynthesis: mechanism and effects. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 2901–2905. [Google Scholar]
- 10.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 11.Leonard C J, Aravind L, Koonin E V. Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res. 1998;8:1038–1047. doi: 10.1101/gr.8.10.1038. [DOI] [PubMed] [Google Scholar]
- 12.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 13.Shi L, Potts M, Kennelly P J. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms. A family portrait. FEMS Microbiol Rev. 1998;22:229–253. doi: 10.1111/j.1574-6976.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 14.Stanier R Y, Kunisawa R, Mandel M, Cohen-Bazire G. Purification and properties of unicellular blue-green algae (order Chroococcales) Bacteriol Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stock J B, Stock A M, Motten J M. Signal transduction in bacteria. Nature. 1990;344:395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- 16.Udo H, Munoz-Dorado J, Inouye M, Inouye S. Myxococcus xanthus, a Gram-negative bacterium, contains a transmembrane protein serine/threonine kinase that blocks the secretion of β-lactamase by phosphorylation. Genes Dev. 1995;9:972–983. doi: 10.1101/gad.9.8.972. [DOI] [PubMed] [Google Scholar]
- 17.Waterbury J B, Willey J M, Franks D G, Valois F W, Watson S W. A cyanobacterium capable of swimming motility. Science. 1985;230:74–76. doi: 10.1126/science.230.4721.74. [DOI] [PubMed] [Google Scholar]
- 18.Williams J G K. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis sp. PCC 6803. Methods Enzymol. 1988;167:766–778. [Google Scholar]
- 19.Yoshihara, S., X. X. Geng, S. Okamoto, K. Yura, T. Murata, M. Go, M. Ohmori, and M. Ikeuchi. Mutational analysis of genes involved in pilus structure, motility and transformation competency in the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol., in press. [DOI] [PubMed]
- 20.Yoshihara S, Suzuki F, Fujita H, Geng X X, Ikeuchi M. Novel putative photoreceptor and regulatory genes required for the positive phototactic movement of the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2000;41:1299–1304. doi: 10.1093/pcp/pce010. [DOI] [PubMed] [Google Scholar]
- 21.Zhang C C, Gonzalez L, Phalip V. Survey, analysis and genetic organization of genes encoding eukaryotic-like signaling proteins on a cyanobacterial genome. Nucleic Acids Res. 1998;26:3619–3625. doi: 10.1093/nar/26.16.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]