Abstract
Extracellular vesicle‐bound DNA (evDNA) is an understudied extracellular vesicle (EV) cargo, particularly in cancer‐unrelated research. Although evDNA has been detected in urine, little is known about its characteristics, localization, and biomarker potential for kidney pathologies. To address this, we enriched EVs from urine of well‐characterized kidney transplant recipients undergoing allograft biopsy, characterized their evDNA and its association to allograft injury. The SEC‐based method enriched pure EVs from urine of kidney transplant recipients, regardless of the allograft injury. Urinary evDNA represented up to 29.2 ± 8% (mean ± SD) of cell‐free DNA (cfDNA) and correlated with cfDNA in several characteristics but was less fragmented (P < 0.001). Importantly, using DNase treatment and immunogold labelling TEM, we demonstrated that evDNA was bound to the surface of urinary EVs. Normalised evDNA yield (P = 0.042) and evDNA copy number (P = 0.027) significantly differed between patients with normal histology, rejection injury and non‐rejection injury, the later groups having significantly larger uEVs (mean diameter, P = 0.045) and more DNA bound per uEV. ddDNA is detectable in uEV samples of kidney allograft recipients, but its quantity is highly variable. In a proof‐of‐principle study, several evDNA characteristics correlated with clinical and histological parameters (P = 0.040), supporting that the potential of evDNA as a biomarker for kidney allograft injury should be further investigated.
Keywords: cell‐free DNA, donor‐derived DNA, extracellular vesicles, kidney allograft injury, kidney allograft rejection, liquid biopsy, urine
1. INTRODUCTION
Liquid biopsy, a minimally invasive test performed on biofluid samples to detect distant pathology, is a promising alternative to the established invasive biopsies used in cancer and organ transplant diagnostics. Most studies focus on circulating cells, circulating RNA or cell‐free DNA (cfDNA), with extracellular vesicles (EVs) emerging as a promising novel analyte (Szilágyi et al., 2020; Tamura et al., 2021). EVs are a heterogeneous group of spherical membrane‐bound particles, which play a prominent role in intercellular communication (Yáñez‐Mó et al., 2015). They can differ in their biogenesis, cell of origin, size, molecular composition (nucleic acids, proteins, lipids, carbohydrates, and metabolites), and biological role (Van Niel et al., 2018). All cells studied so far release EVs, which in vivo accumulate to high concentrations in all body fluids. As stable carriers of (patho)physiological signals that reflect the state of the parental cell, they are being studied as biomarkers in various diseases (González & Falcón‐Pérez, 2015). These studies focused mainly on miRNA, mRNA, and protein EV cargo, but neglected the EV‐bound DNA (evDNA).
In blood, evDNA exists as single‐ or double‐stranded molecule of genomic or mitochondrial origin, with size range from 200 bp in small EVs to up to >2 million bp in large EVs (reviewed in Malkin & Bratman, 2020). In cancer, evDNA was shown to represent the entire genome and mutational status of the cell of origin (Lázaro‐Ibáñez et al., 2019; Vagner et al., 2018). DNA can be attached to EV surface or is present in its lumen (Lázaro‐Ibáñez et al., 2019; Maire et al., 2021; Németh et al., 2017; Shelke et al., 2016; Vagner et al., 2018). Exact mechanism of DNA sorting is not known but might include sequestering of cytosolic DNA during outward plasma membrane budding or intraluminal vesicles formation, or shuttling of collapsed micronuclei to multivesicular bodies (reviewed in Malkin & Bratman, 2020). The biological function of evDNA is poorly understood; however, it may play a role in cellular homeostasis, transfer of genetic material or immune response (Takahashi et al., 2017; Torralba et al., 2018). Despite the growing literature on evDNA in blood, there is a paucity of studies on DNA bound to urinary EVs (uEVs). Study by Lee et al. showed that urinary evDNA reflected tumour mutation status in nine urothelial carcinoma patients (Lee et al., 2018), but little is known about the evDNA characteristics, localization, and biomarker potential.
Kidney transplantation is a perfect model to study the biomarker potential of urinary evDNA. It is the most effective therapy for end‐stage kidney disease, but unrecognised and subsequently untreated injury of the allograft can contribute to the loss of function over time and decreased allograft survival (Nankivell & Kuypers, 2011). Long‐term allograft survival remains a major challenge, mostly due to acute and chronic rejection. Rejection can be broadly categorized as humoral, when mediated by the presence of anti‐donor‐specific antibodies (antibody‐mediated rejection; ABMR), or cellular, when caused by infiltration of the interstitium by T cells and macrophages (T‐cell mediated rejection; TCMR). Although the rate of acute rejection has decreased in the modern era of immunosuppression, recent reported incidence of acute rejections ranges from 11% to 26% (Christakoudi et al., 2019; Sigdel et al., 2018; Suthanthiran et al., 2013). This has been associated with poor long‐term allograft survival (Hariharan et al., 2021). Serum creatinine, estimated glomerular filtration rate (eGFR), and urinary protein excretion are traditional biomarkers used to monitor the kidney allograft, but they lack sensitivity and specificity (Anglicheau et al., 2016). Histopathologic characterization of kidney biopsies based on the Banff classification remains the standard for diagnosis of kidney allograft injury (KDIGO Transplant Work Group, 2009; Solez et al., 1993). However, biopsies are invasive and costly procedures that can be associated with significant morbidity and are inappropriate for continuous monitoring (Hogan et al., 2016). In addition, the biopsy sample may not accurately represent the state of the entire kidney, and histopathologic interpretation is subject to interobserver variability (Mengel et al., 2007). Therefore, novel non‐invasive biomarkers are needed to allow frequent monitoring and earlier detection of kidney allograft injury.
Urinary EVs have a proven role in kidney physiology and are promoters of several kidney diseases (reviewed in Pomatto et al., 2017; Yates et al., 2022a, 2022b). They are released from cells of every nephron segment, bladder, and prostate in males, as well as resident immune cells, and are easily routinely accessible (Erdbrügger et al., 2021; Ranghino et al., 2015). As biomarkers for kidney allograft injury, elevated levels of uEV‐bound tetraspanin‐1, hemopexin (Lim et al., 2018) or specific set of mRNAs (El Fekih et al., 2021) were associated with rejection. Protein composition of uEVs also reflects delayed graft function, reduction in ischaemia‐reperfusion injury, or the inflammatory and stress response of the kidney allograft (Alvarez et al., 2013; Braun et al., 2020; Oshikawa‐Hori et al., 2019; Park et al., 2017). Studies have so far neglected uEV‐bound DNA, although cell‐free DNA (cfDNA) is a promising biomarker for allograft health and function (reviewed in Oellerich et al., 2021; Verhoeven et al., 2018). Organ transplantation is namely also genome transplantation, which allows specific detection of donor‐derived cfDNA (dd‐cfDNA) released from the damaged allograft into the recipient's body fluids (Oellerich et al., 2021). The pivotal DART study has shown that dd‐cfDNA levels >1% in the plasma of recipients indicate kidney allograft rejection (Bloom et al., 2017). This has been confirmed by several larger cohort and multicentre studies, and absolute quantification of dd‐cfDNA has been proposed as an alternative readout (Gielis et al., 2020; Halloran et al., 2022; Huang et al., 2019; Oellerich et al., 2019; Sigdel et al., 2018; Whitlam et al., 2019). In one of the few studies investigating urine samples, Sigdel et al. showed that urinary dd‐cfDNA was significantly higher in patients with acute rejection than in patients with stable kidney allograft (Sigdel et al., 2013). Urinary cfDNA analysis is also part of the multimarker test for early detection of kidney allograft rejection (Watson et al., 2019; Yang et al., 2020). It would be interesting to evaluate the utility of urine evDNA as a biomarker for kidney allograft injury.
The aim of this study was to investigate DNA as a cargo of urinary EVs and to explore its utility as a biomarker for kidney allograft injury. To this end, we isolated EVs, evDNA, and cfDNA from urine of well‐characterized kidney transplant recipients undergoing surveillance or indication biopsy. We characterized uEV concentration and size by transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA); and DNA yield, copy number, integrity index, and donor‐derived DNA (ddDNA) by fluorometry, genotyping and droplet digital PCR (ddPCR). Localization of evDNA was investigated by DNase assay and immunogold labelling for TEM. Finally, the association between evDNA characteristics and clinical characteristics of patients was tested in a proof‐of‐principle study on the utility of evDNA as a biomarker for kidney allograft injury.
2. MATERIALS AND METHODS
2.1. Study design and data collection
Forty‐one adult patients, who underwent a deceased donor kidney transplantation at the Center for Kidney Transplantation, Department of Nephrology, University Medical Center Ljubljana, Slovenia, were enrolled in this prospective observational study at the time of kidney allograft biopsy. From November 2018 to December 2020, 21 enrolled patients underwent surveillance biopsy 1 year after transplantation, whereas 20 patients underwent for‐cause biopsy due to clinical indication (i.e., an increase in serum creatinine >20% from baseline without apparent cause; a new‐onset or increase in proteinuria). Exclusion criteria included multiple‐solid organ transplants. Among the 41 study participants, one patient was later confirmed to have active infection with BK polyomavirus and was excluded from further analysis. The study was approved by the National Medical Ethics Committee of the Republic of Slovenia (approval nr.: 0120–216/2019) and was conducted in accordance with the Declaration of Helsinki and compliant with the Good Clinical Practice Guidelines. An informed consent form was signed by all study participants. All mandatory laboratory health and safety procedures were followed in performing the reported experimental work.
Fully anonymized patient data, collected at the time of biopsy, included patient demographics (age, sex), recipient and donor clinical parameters (expanded criteria donor (ECD; Kauffman et al., 1997), human leukocyte antigen (HLA) mismatch, delayed graft function (DGF), time from transplantation to biopsy and laboratory data (serum creatinine (S‐creatinine), urine creatinine (U‐creatinine), estimated glomerular filtration rate (eGFR), urinary estimated protein excretion rate (ePER), serum C‐reactive protein (S‐CRP) and donor‐specific antibodies (DSA)). Histopathologic examination of blinded biopsy samples was performed at the Institute of Pathology (Faculty of Medicine, University of Ljubljana) by the dedicated pathologist, in accordance with the 2017 Banff Working Groups’ criteria (Haas et al., 2018). TCMR was reported as integrated tubulo‐interstitial (borderline and grade IA/B) or vascular (grade IIA/B). The phenotypes of ABMR were classified as acute or chronic active. The diagnosis of acute ABMR was based on morphologic evidence of acute tissue injury (i.e., peritubular capillaritis and/or glomerulitis) and positive C4d staining. The diagnosis of chronic ABMR was based on the morphologic evidence of antibody‐mediated chronic tissue injury, specifically glomerular double contours compatible with chronic glomerulopathy on light and/or electron microscopy and peritubular basement membrane multilamelation on electron microscopy. Biopsy specimen reports with a diagnosis of mixed ABMR and TCMR were grouped with the ABMR subgroup. All recipients were on an immunosuppressive regimen consisting of a calcineurin inhibitor, mycophenolate mofetil or azathioprine, and steroids. The study patient characteristics are presented in Table 1. Any missing patient data are clearly indicated.
TABLE 1.
Kidney allograft recipients' demographic, clinical and laboratory characteristics
| Variables* | Study cohort N = 40 | Normal histology N = 14 | Rejection injury N = 10 | Non‐rejection injury N = 16 | P‐value | |
|---|---|---|---|---|---|---|
| Patient age | Years | 55 (46–64) | 56 (42–62) | 51 (41–65) | 55 (48–64) | 0.836 a |
| <65 | 31 (78) | 11 (79) | 8 (80) | 12 (75) | 1.000 b | |
| >65 | 9 (23) | 3 (21) | 2 (20) | 4 (25) | ||
| Patient sex | M | 32 (80.0) | 13 (92.9) | 8 (80.0) | 11 (68.8) | 0.315 b |
| F | 8 (20.0) | 1 (7.1) | 2 (20.0) | 5 (31.3) | ||
| ECD | No | 17 (45.9) [3] | 7 (50.0) | 5 (62.5) [2] | 5 (33.3) [1] | 0.407 b |
| Yes | 20 (54.1) | 7 (50.0) | 3 (37.5) | 10 (66.79) | ||
| HLA‐mm | Number | 3 (3–4) | 3 (2–4) | 3 (2.8–3) | 3 (3–4) | 0.329 a |
| DGF | Present | 9 (22.5) | 4 (28.6) | 1 (10.0) | 4 (25.0) | 0.637 |
| Bx | Protocol | 21 (52.5) | 12 (85.7) | 3 (30.0) | 6 (37.5) | 0.008 b |
| For‐cause | 19 (47.5) | 2 (14.3) | 7 (70.0) | 10 (62.5) | ||
| Tx to Bx | Days | 404 (375–1926) | 379 (373–419) | 418 (250–3925) | 924 (379–2993) | 0.170 a |
| S‐creatinine | μmol/L | 140.5 (110.5–165.8) | 132.5 (107–160.3) | 140.5 (110.5–170.3) | 141.5 (112.5–167.3) | 0.754 a |
| U‐creatinine | mmol/L | 7.5 (4.4–9.8) [3] | 8.6 (6.4–10.4) [1] | 5.1 (3.9–8.8) [2] | 5.2 (4.3–11) | 0.318 a |
| eGFR | ml/min/1.73m2 | 46 (39–56) | 51 (42–65) | 45 (40–54) | 46 (32–55) | 0.499 a |
| ePER | g/day/1.73m2 | 0.28 (0.17–0.54) [20] | 0.18 (0.14–0.21) [8] | 0.32 (0.23–0.86) [5] | 0.33 (0.20–2.65) [7] | 0.057 a |
| S‐CRP | mg/L | 1.33 (0.73–3.55) | 1.57 (0.97–4.08) | 0.97 (0.55–3.26) | 1.37 (0.92–2.86) | 0.740 a |
| de‐novo DSA | Present | 6 (15.0) | 1 (7.1) | 4 (40.0) | 1 (6.3) | 0.077 b |
Abbreviations: M, male; F, female; Tx, transplantation; Bx, biopsy; DSA, donor specific antibodies; DGF, delayed graft function; HLA‐mm, human leukocyte antigen mismatch; ECD, expanded criteria donor; Protocol, surveillance biopsy; S‐creatinine, serum creatinine; eGFR, estimated glomerular filtration rate; S‐ CRP, serum C‐reactive protein; U‐creatinine, urinary creatinine; ePER, urinary estimated protein excretion rate.
Kruskal‐Wallis test. Significant P‐values are indicated in bold.
Fisher's exact test.
Data are presented as total numbers (percentages) or medians (25%–75%). [No. of missing data].
Second morning spot urine and whole blood sampling was performed in the morning, prior to biopsy, and processed within 4 h, always following the same protocols. Peripheral whole venous blood (4 ml) of patients was collected into ethylenediaminetetraacetic acid (EDTA) vacutainer tubes (BD Vacutainer, BD). Next, blood was centrifuged at 2500 × g for 10 min at room temperature (RT). After the removal of plasma, residual blood containing blood cells (2 ml) was stored at –20°C for recipients’ genomic DNA extraction. Second morning spot urine (25–50 ml) of study patients was collected into sterile urine cups (Vacuette, Greiner Bio‐one), transferred to conical polypropylene centrifuge tubes (TPP) and centrifuged at 2000 × g for 15 min at RT. The supernatant of each patient's urine sample was transferred to a fresh centrifuge tube, separated into two aliquots (for uEVs enrichment and cfDNA extraction) and stored at −80°C. Transplanted kidney tissue was collected during the surveillance or for‐cause kidney biopsy. Biopsy was performed by a medical professional, using the 16‐ or 18‐gauge biopsy needle. A portion of one core kidney tissue sample (3 mm in length) was immediately stabilized in 500 μl of RNAlater Stabilization Solution (Thermo Fisher Scientific), incubated at RT for approximately 2 h to allow penetration into the tissue, and stored at –80°C for later donor genomic DNA extraction.
2.2. EV enrichment from urine
EVs were enriched from 20 ml of cell‐free urine following our previously optimized protocol (Sedej et al., 2021). Briefly, upon urine thawing at RT, 400 μl of 0.5 M EDTA and 5 ml of 5× phosphate buffered saline (PBS, pH ∼ 7.4) were added for inhibition of uromodulin aggregation and Ca‐oxalate formation, and pH neutralization, respectively (Kobayashi & Fukuoka, 2001). Samples were then concentrated to a volume of 500 μl on 100 kDa centrifugal filter units (Amicon, Merck Millipore), loaded on size‐exclusion chromatography colons (SEC; qEVoriginal, Izon Q) for particle separation and eluted with 1× PBS. Twenty fractions of 500 μl were collected in protein low binding tubes (Eppendorf). Next, soluble protein content was determined by measuring absorbance at 280 nm (Synergy 2, BioTek) for each of the two combined fractions (1 ml). Protein‐free SEC fractions 5–12 were pooled together and concentrated on 100 kDa centrifugal filter units (Amicon, Merck Millipore) to a volume of 70−80 μl, representing patient's population of urine EVs (uEV sample). Samples were stored in DNA low binding tubes (Eppendorf) at −20°C and processed further within 1 month.
2.3. uEV sample characterization
2.3.1. Transmission electron microscopy (TEM) for purity and uEV size
Briefly, upon defrosting, 4 μl of uEV samples were adsorbed for 3 min at RT on freshly glow discharged, formvar coated and carbon stabilized copper grids (SPI), and stained with 1% (w/v) water solution of uranyl‐acetate (SPI). The grids were observed by transmission electron microscope TALOS L120 (Thermo Fisher Scientific), operating at 100 kV. At least 10 grid squares were examined thoroughly, and representative micrographs (camera Ceta 16 M) were taken at different places on the grid. The Velox software (Thermo Fisher Scientific) was used to process images and measure the sizes of uEVs displaying characteristic morphology (N = 100 per sample).
2.3.2. Western blot (WB) analysis for uEV proteins and purity
WB of uEV sample enriched from 20 ml of pooled urine from five kidney transplant recipients was performed to assess typical EV proteins (Hsc70, flotilin‐1, TSG101, GAPDH, CD9) and sample purity (calnexin, uromodulin), as described in detail in Supplementary Material (Section S1).
2.3.3. Nanoparticle Tracking Analysis (NTA) for particle concentration and size
uEV samples were diluted (400–3000×) in particle‐free Dulbecco's PBS (dPBS, Sigma‐Aldrich) to reach an optimum concentration range of 1 × 107–109 particles/ml. Five 60‐s movies per sample, at camera level 15, were recorded on the NanoSight NS300 instrument (488 nm blue laser) connected to the NanoSight Sample Assistant (Malvern Panalytical). Captured videos were manually examined and eliminated in the event of significant abnormalities, but not more than two videos per sample. Measurements were performed in duplicates. Raw data were analysed by the NanoSight NTA 3.3 program at the following settings: water viscosity, temperature of 25°C, detection threshold of 5, minimum track length of 10 and default minimum expected particle size and blur settings. Output data were expressed as uEV concentration, that is, the number of particles per millilitre of input urine, and uEV size, that is, the mean, modal, and median hydrodynamic diameter in nm.
2.4. DNA extraction
2.4.1. Genomic DNA extraction
For genotyping, donor genomic DNA was extracted from kidney tissue and recipient genomic DNA from study participants’ blood cells. For donor genomic DNA extraction, approximately 3 mm fragment of kidney tissue sample, stored in RNAlater solution (Thermo Fisher Scientific), was transferred to a fresh microcentrifuge tube (Eppendorf), and washed with 1× PBS for 15 min to remove recipient's blood. Next, DNA was extracted using the QIAamp DNA Micro Kit (Qiagen), according to manufacturer's instructions. In the last step, 20 μl of the elution buffer was applied to the center of the column membrane and incubated for 5 min at RT to increase the yield of extracted genomic DNA. After centrifugation, the elution step was repeated. DNA concentration was determined by measuring the absorbance at 260 nm (Synergy 2, BioTek) and the samples frozen at –20°C.
For recipient genomic DNA extraction, the residual blood cells (2 ml) were thawed at RT and the recipient's genomic DNA extracted with the EZNA SQ Blood DNA Kit II (Omega Bio‐tek) as per manufacturer's protocol for 1–3 ml Whole Blood. After pelleting the cells (protocol step 4), 10 ml of distilled water was added, and the centrifugation step repeated. After discarding the supernatant, the DNA precipitate was air dried. Genomic DNA was dissolved in 500 μl elution buffer in a water bath heated to 65°C for 2 hours. The concentration of extracted DNA was determined by measuring the absorbance at 260 nm (Synergy 2, BioTek) and the sample frozen at –20°C.
2.4.2. Cell‐free DNA and EV‐bound DNA extraction
cfDNA was extracted from 4 ml of urine with Quick‐DNA Urine Kit (Zymo Research) according to the manufacturer's protocol, and eluted with 15 μl of elution buffer. Quick‐DNA Urine Kit was the best performing kit of five tested commercial kits for cfDNA extraction from urine, as described in more detail in Supplementary Material (Section S3).
evDNA was extracted with QIAamp DNA Micro Kit (Qiagen) following the manufacturer's protocol for extraction of DNA from small volumes of blood. Use of carrier RNA was omitted. Elution of DNA was performed twice, each time adding 20 μl of elution buffer and incubating 5 min at RT before centrifugation.
cfDNA and evDNA concentrations were measured using Qubit dsDNA HS Assay Kit and Qubit 3 Fluorometer (both Invitrogen). Determined DNA concentration was used to calculate DNA yield.
2.5. Genotyping by real‐time PCR
Extracted genomic DNA from blood (recipient) and kidney tissue (donor) was genotyped by real‐time PCR for six single nucleotide polymorphisms (SNPs; rs1707473, rs2691527, rs7687645, rs1420530, rs9289628, rs6070149) loci using Custom TaqMan SNP Genotyping Assays (Thermo Fisher). All primers and probes are described in more detail in Supplementary Material (Section S4). 5 μl reactions with TaqMan Genotyping Master Mix (Applied Biosystems) and 5–10 ng genomic DNA per reaction were run in a 384‐well format on the ViiA 7 Real‐Time PCR System (Applied Biosystems) using the following thermal profile: 60°C for 30 s (DC), 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 1 min 30 s (DC), and finally 60°C for 30 s (DC). The ramp rate was set to 1.6°C/s for all steps. Data were collected in the steps marked with (DC). Positive controls prepared with gBlock Gene Fragments (Integrated DNA Technologies; described in detail in Supplementary Material, section S5) or genomic DNA with known genotype were included for all genotypes. When donor genotype was not clear, due to contamination of donor kidney tissue with recipient's blood, donor genomic DNA was additionally genotyped by ddPCR by comparing copy numbers of the six SNP amplicons and/or the SRY amplicon in allografts that were sex‐mismatched, and subtracting the background copy number of known patient's genotype.
2.6. Preamplification of DNA
Targeted multiplex PCR preamplification of six SNP loci, RPP30 and RPPH1 was performed on cfDNA and evDNA samples included in the study. As previously described (Andersson et al., 2015), 100 μl PCR reactions were prepared with 40 nM final concentration of each of 16 primers using Q5 Hot Start High‐Fidelity 2× Master Mix (New England BioLabs). Denaturation at 98°C for 3 min was followed by 11 or 12 amplification cycles (cfDNA and evDNA, respectively) of 10 s at 98°C, 3 min at 64°C and 20 s at 72°C, and a 2‐min final elongation step at 72°C. When finished, reactions were diluted 10× in Tris EDTA and either analysed with ddPCR in the same day or stored at –20°C until analysis. Preamplification reaction was validated for preamplification efficiency, potential bias and inhibitory effect of possible chemical contaminants as described in Supplementary Material (Section S5).
2.7. Droplet digital PCR
QX100 and QX200 Droplet Digital PCR Systems (Bio‐Rad) were used. Reactions were prepared using ddPCR Supermix for Probes (No dUTP) (Bio‐Rad) and up to 9 μl of preamplified cfDNA or evDNA. In cases of higher yields of DNA, the preamplified DNA was additionally diluted with nuclease‐free water to avoid saturation of the ddPCR reaction. For quantification of RPP30 and RPPH1 amplicons, duplex ddPCR reactions were prepared using final concentration of 900 nM primers and 250 nM probes (all from Integrated DNA Technologies). The same six SNP assays were used for quantification of ddDNA as for genotyping of genomic DNA at 1× concentration. The SRY assay was performed in duplex format along with RPP30, as described for RPP30 and RPPH1 quantification. All reactions were performed in triplicate. Droplets were generated using the QX100, QX200, or AutoDG droplet generator (Bio‐Rad). PCR cycling conditions were 95°C for 10 min, 40 cycles of 94°C for 30 s, and 60°C for 1 min 30 s, with a final incubation step of 98°C for 10 min. The ramp rate was set at 1.5°C/s. Droplets were read using the QX100 or QX200 reader (Bio‐Rad). QuantaSoft software version 1.7.4 (Bio‐Rad) was used to manually determine the thresholds. The raw data were then exported and analysed in Excel (Microsoft).
The initial copy number of an amplicon (before preamplification) was calculated as the amplicon copy number determined by ddPCR, adjusted for the dilution factor, and divided by the theoretical factor of PCR preamplification assuming 100% efficiency (211 in the case of 11 and 212 in the case of 12 cycles, respectively). Samples with initial copy number less than 5 were designated as insufficient DNA copy number and excluded from further analysis. An exception was made for RPP30 quantification of DNase‐treated uEV samples to allow analysis of samples after DNA degradation, but those samples were flagged as samples with less reliable quantification. The DNA copy number refers to the average of initial RPP30 copy numbers calculated from three separate measurements by ddPCR.
To evaluate evDNA fraction of cfDNA, second morning spot urine from seven kidney transplant recipients was collected and pooled. Next, we followed the protocol for enrichment of EVs from 20 ml of urine as described in Materials and Methods (Section 2.2), and collected samples at different steps in the protocol, as described in more detail in Supplementary Material (Section S7, Figure S8). The fraction of evDNA of total cfDNA was calculated as B / (A + B) × 100 [%], or as (C – A) / C × 100 [%] to account for losses during evDNA extraction from uEVs.
The ddDNA fraction in the samples was calculated from the data obtained by the SNP ddPCR reactions. For each sample, at least two informative SNP assays were identified that allowed us to distinguish between recipient and donor DNA. When recipients had a homozygous genotype (AA) and donors had a heterologous homozygous (BB) or heterozygous (AB) genotype, ddDNA fractions were calculated as in Beck et al. (2018), 100 × B / (A + B) for a homozygous donor and 2 × 100 × B / (A + B) for a heterozygous donor. When recipients had a heterozygous genotype (AB) and donors had a homozygous genotype (AA), ddDNA fractions were calculated as 100 × (A – B) / (A + B). The average ddDNA fraction, standard deviation, and coefficient of variation for each sample were calculated from data of at least two SNP assays, each analysed in triplicate. The ddDNA copy number was calculated as the ddDNA fraction multiplied by the DNA copy number.
DNA integrity index was determined by calculating the ratio of RPPH1 (135 bp) to RPP30 (62 bp) copy numbers, representing the ratio of large to short amplicons (Mouliere et al., 2011). DNA yield, DNA copy number and ddDNA copy number were normalised to U‐creatinine, which was measured in mmol/ml.
2.8. Localization of uEV‐bound DNA
2.8.1. DNase treatment of uEV samples
60 or 65 μl uEV samples were treated with 1 U of Ambion DNase I (Thermo Fisher Scientific) in 1× DNase buffer in 70 or 80 μl total reaction volume. Reactions were incubated at 37°C for 30 min. After treatment, DNase was inactivated by incubation at 75°C for 10 min or alternatively, denaturing buffer was added for inactivation and DNA extraction performed immediately.
2.8.2. dsDNA immunogold labelling for TEM
For detection of dsDNA in uEV samples, negative staining method was combined with immunogold labelling at electron microscopy level. 3 μl of defrosted uEV samples were applied on freshly glow discharged copper grids for 3 min, followed by floating the grids on a drop of 1% bovine serum albumin (Sigma) in 0.1 M phosphate buffer to block non‐specific antibody binding, and then incubated on drop of mouse monoclonal antibodies against dsDNA (Abcam, ab 27156). After washing the grids, we incubated them on the drop of goat‐anti‐mouse IgG (Aurion), labelled with gold particles with 6 nm in diameter (Aurion Anionic Gold Tracers). Finally, the grids were stained with 1% (w/v) water solution of uranyl‐acetate. As a negative control, anti‐dsDNA antibodies were omitted from the labelling procedure for each uEV sample. Additionally, all uEV samples were also analysed only by negative staining method, as described above.
2.9. Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics, version 27.0 (IBM Corporation, Armonk, NY, USA). Continuous variables were described as median and interquartile range (25%–75%), whereas categorical variables were described using frequencies. Fisher's exact test was used to compare the distribution of categorical variables between the different groups. Non‐parametric Kruskal‐Wallis and Mann‐Whitney tests with Benjamini‐Hochberg adjustment for pairwise comparisons were used to compare the distribution of continuous variables between the different groups. The Wilcoxon signed‐rank test was used for comparison of related samples. Spearman's rho correlation coefficient (ρ) was used to assess correlations between continuous variables. All statistical tests were two sided and the significance level was set at 0.05.
Figures were prepared in GraphPad Prism, version 9.3.1 (for Mac OS X, GraphPad Software), RStudio, version 1.4.1106 (RStudio Team, 2020), using packages ggplot2, version 3.3.5 (Wickham, 2016), ggpubr, version 0.4.0, finalfit, version 1.0.3, gridExtra, version 2.3 and cowplot, version 1.1.1. In the boxplots, the median, first and third quartiles are presented. Whiskers extend up/down to the largest/smallest value no further than 1.5× interquartile range from the hinges. Data beyond the end of the whiskers were defined as outliers.
2.10. Data availability
We have submitted all relevant experimental data to the EV‐TRACK knowledgebase (EV‐TRACK ID: EV210292) (Van Deun et al., 2017).
3. RESULTS
3.1. SEC‐based method enriches pure EVs from the urine of kidney allograft recipients
To investigate DNA as uEV cargo, we first enriched EVs from urine of 40 enrolled patients, following a previously established protocol based on SEC (Figure 1a; Sedej et al., 2021). Because the phenotype of kidney allograft injury could influence the presence of DNA in uEV samples, study patients were divided into three groups according to histological assessment of the allograft. TCMR (N = 6) and acute and chronic active ABMR (N = 4) were integrated to form the rejection group (N = 10), and we distinguished them from samples with no major abnormalities (normal histology group; N = 14), and samples that were classified as having non‐rejection injury (non‐rejection group; N = 16). Non‐rejection injury included specimens with isolated Banff scores (interstitial inflammation (i), tubulitis (t), and interstitial inflammation in the areas of fibrosis (i‐IFTA), arteritis (v), glomerulitis (g), peritubular capillaritis (ptc), and C4d positivity in peritubular capillaries (C4d)) that did not meet rejection criteria, calcineurin inhibitor nephrotoxicity, or recurrence of original kidney disease. The three patient groups did not differ in most of the characteristics studied, except in proportion of for‐cause biopsies (P = 0.008) that was higher in the rejection injury group (Table 1). The median time (25%–75%) between transplantation and biopsy was 1943 (378–3646) days for for‐cause biopsies and 382 (373–412) days for surveillance biopsies (P = 0.005).
FIGURE 1.
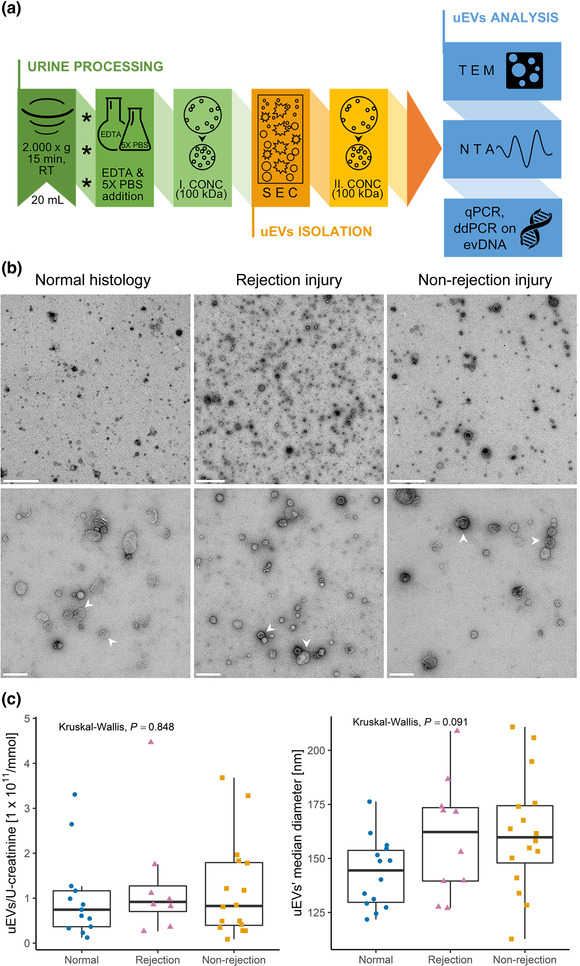
Size exclusion chromatography‐based method enriches pure extracellular vesicles from urine of kidney allograft recipients. (a) Enrichment of EVs from urine (uEV): the processing of second morning spot urine upon collection is presented in green (asterisks indicate sample freezing at −80°C before further processing), the uEV isolation step in orange, and the uEV analyses methods in blue. See Materials and Methods section for details. (b) TEM micrographs of representative negative stained uEV samples from the normal histology (left), rejection injury (middle) and non‐rejection injury (right) patient groups. White arrows indicate two (among many) uEVs. Scale bars: 1 μm (top row), 200 nm (bottom row). (c) NTA analysis of patients’ uEV samples: (left) Distribution of uEV concentrations [particles/ml urine] normalised to U‐creatinine [mmol/ml] in patient groups with normal histology (N = 13, blue circles), rejection injury (N = 8, purple triangles), or non‐rejection injury (N = 16, orange squares). (right) Distribution of median diameters of uEVs in normal histology (N = 14), rejection injury (N = 10) and non‐rejection injury (N = 16). Kruskal‐Wallis test was used to compare the uEV characteristics between all three patient groups. RT, room temperature; EDTA, ethylenediaminetetraacetic acid; PBS, phosphate buffered saline; CONC, concentration; SEC, size exclusion chromatography; TEM, transmission electron microscopy; NTA, nanoparticle tracking analysis; qPCR, quantitative real‐time PCR; ddPCR, digital droplet PCR; evDNA, extracellular vesicle‐bound DNA; U‐creatinine, urinary creatinine
To determine the purity of the uEV samples, representative uEV samples from the three patient groups were analysed by TEM (Figure 1b). Representative micrographs show that the SEC‐based method efficiently enriched EVs with typical morphology from the urine of all patient groups. The uEV samples were in general free of impurities, with some proteins visible in the sample from the patient with rejection injury (Figure 1b middle). To independently confirm the presence of uEVs and absence of protein impurities, representative uEV sample from pooled urine was analysed by WB (Suppl. Figure S1). Typical EV proteins flotilin‐1, TSG101 and CD9 were detected in uEV sample, while endoplasmatic reticulum protein calnexin was absent and uromodulin was detected at low levels. To summarize, using the SEC‐based protocol we were able to enrich pure EVs from the urine of kidney transplant recipients, regardless of the phenotype of allograft injury.
Next, concentration and size of particles in patients’ uEV samples were determined by NTA. As TEM and WB confirmed the presence of EVs and absence of impurities in uEV samples, we are referring to particles as uEVs. To account for interpatient variability and overall urine concentration, uEV concentration values were normalised to U‐creatinine, previously identified as a reliable normalization marker (Blijdorp et al., 2021; Rabant et al., 2015). U‐creatinine data were not available for three of the patients, so these samples were excluded from further analysis. Normalised uEV concentration and median diameter for each included patient are presented in Figure 1c, while uEV size distribution profiles are presented in Suppl. Figure S2. When comparing the uEV size among the patient groups, the uEVs mean (P = 0.045) or median (P = 0.031) diameter were significantly higher in the rejection and non‐rejection injury group compared to the normal histology group (Figure S3, Figure 1c). This was supported by measuring sizes of uEVs displaying characteristic morphology in representative samples under TEM (Figure 1b). uEVs from patients with normal histology namely showed a median diameter (25%–75%) of 68.9 (56.4–103.2) nm, while uEVs from patients with rejection injury or non‐rejection injury were 84.0 (69.6–116.2) nm and 83.6 (68.3–100.0) nm in size, respectively. Overall, uEVs from patients with rejection and non‐rejection allograft injury were larger compared to patients with normal histology.
3.2. Quantity of urine EV‐bound DNA reflects the kidney allograft injury
To investigate if DNA is a cargo of EVs in urine, DNA was extracted from uEV samples enriched from 20 ml of patients’ urine. In parallel, cfDNA (which also includes any DNA bound to uEVs) was extracted directly from 4 ml of the same urine samples for comparison. Evaluation of five commercially available kits for cfDNA extraction from urine is described in Supplementary Material (Section S3, Table S1, Figure S4). Under such conditions, the median (25%–75%) yield of extracted evDNA was 3.4 (2.0–6.6) ng compared to 18.5 (8.4–53.0) ng for cfDNA (Figure 2a). Nevertheless, the yields of evDNA and cfDNA correlated well (ρ = 0.680, P < 0.001, Figure 2b). Importantly, in a separate experiment, described in detail in Supplementary Material (Section S7), we show that in pooled urine from seven kidney transplant recipients, evDNA represents 15.1 ± 8.3% (mean ± SD), or 29.2 ± 8.0% of cfDNA if accounting for losses during evDNA extraction from uEVs (Figure S8).
FIGURE 2.
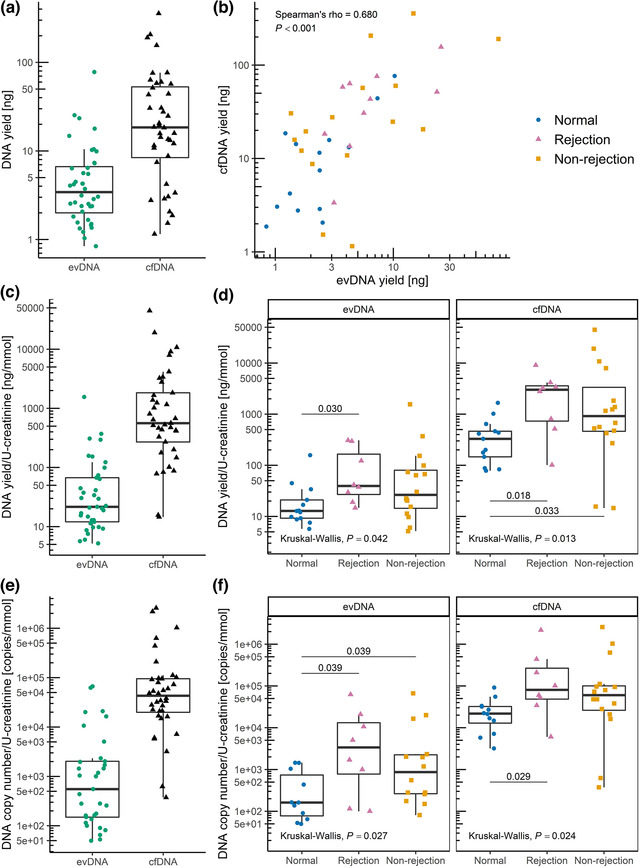
uEV samples of kidney allograft recipients contain DNA. DNA was extracted from uEVs (extracellular vesicle‐bound DNA; evDNA) or directly from urine (cell‐free DNA; cfDNA) and analysed for yield by fluorometry and for DNA copy number by detection of RPP30 copies by ddPCR. (a) Distribution of evDNA and cfDNA extraction yields (N = 40) and (b) their correlation (tested with Spearman correlation coefficient). Distribution of DNA yield [ng/ml urine] normalised to U‐creatinine [mmol/ml] in (c) the patient cohort (N = 37) and (d) in patient groups with normal histology (N = 13), rejection injury (N = 8), and with non‐rejection injury (N = 16). Distribution of DNA copy number normalised to U‐creatinine in (e) the patient cohort (N = 33 for evDNA, N = 36 for cfDNA, respectively) and (f) in patient groups with normal histology (N = 11 for evDNA, N = 12 for cfDNA, respectively), rejection injury (N = 8 for evDNA, N = 8 for cfDNA, respectively), and with non‐rejection injury (N = 14 for evDNA, N = 16 for cfDNA, respectively). Data of patients with normal histology are presented as blue circles, rejection injury as purple triangles and non‐rejection injury as orange squares. Kruskal‐Wallis test was used to compare the characteristics between all three patient groups and Mann‐Whitney test with Benjamini‐Hochberg adjustment for pairwise comparisons between the different groups (P‐values labelled above the lines connecting different groups in (d, f))
DNA yields were normalised to U‐creatinine (Figure 2c), except for three patients for whom the U‐creatinine data were not available. The normalised DNA yields in the three groups were statistically significantly different (P = 0.042 for evDNA and P = 0.013 for cfDNA) (Figure 2d). More urinary evDNA and cfDNA were obtained in patients with rejection injury (P = 0.030 for evDNA and P = 0.018 for cfDNA) and non‐rejection injury (P = 0.033 for cfDNA), than in patients with normal histology. Significance was maintained even when comparing the rejection and non‐rejection injury combined with the normal histology group (P = 0.018 for evDNA and P = 0.004 for cfDNA). When normalizing patients’ evDNA yields to uEV concentrations, patients with rejection and non‐rejection injury had more DNA bound per uEV (median (25%–75%); 10.6 × 10−9 (5.04 × 10−9–23.0 × 10−9) and 10.6 × 10−9 (6.05 × 10−9–18.6 × 10−9), respectively) compared to patients with normal histology (3.90 × 10−9 (1.63 × 10−9–9.54 × 10−9)).
For further analysis of evDNA and cfDNA, the entire volumes of extracted DNA were first preamplified to obtain enough sample for analysis with multiple ddPCR assays and replicates. Importantly, in a series of additional experiments described in more detail in Supplementary Material (Section S5), we showed that preamplified DNA reflects the original quantities of measured DNA fragments (Suppl. Figures S5, S6, S7). The preamplified evDNA and cfDNA for all samples were next analysed by ddPCR.
DNA copy number of evDNA and cfDNA samples was determined by measuring the RPP30 copy number (Figure 2e). In four evDNA and one cfDNA samples, RPP30 copy number was too low for reliable quantification, so they were excluded from further analysis. evDNA copy numbers correlated with evDNA yields (ρ = 0.604, P < 0.001). evDNA copy number, normalised to U‐creatinine, was significantly different between the three patient groups (P = 0.027) (Figure 2f). More evDNA copies were measured in patients with rejection injury (P = 0.039) and non‐rejection injury (P = 0.039), than in patients with normal histology. Significance was maintained even when comparing the rejection and non‐rejection injury combined with the normal histology group (P = 0.007). The cfDNA copy numbers correlated strongly with cfDNA yields (ρ = 0.918, P < 0.001), and also with evDNA copy numbers (ρ = 0.626, P < 0.001). cfDNA copy number, normalised to U‐creatinine, was significantly different between the three patient groups (P = 0.024) (Figure 2f). More cfDNA copies were measured in patients with rejection and non‐rejection injury combined than in patients with normal histology (P = 0.008). Altogether, quantity of urine evDNA and cfDNA, as measured by normalised DNA yield and DNA copy number, reflects the kidney allograft injury.
3.3. EV‐bound DNA is less fragmented than cfDNA in urine of kidney allograft recipients
The relative degree of fragmentation of evDNA and cfDNA samples was determined by calculating the DNA integrity index, defined as the ratio of RPPH1 (135 bp) to RPP30 (62 bp) amplicon copy numbers. In intact human DNA, the copy number of the two amplicons is the same. However, the more the DNA is degraded, fewer amplicon copies are measured, especially for the longer amplicons. The ratio of long to short amplicons is therefore a good estimate of the DNA fragmentation level (Mouliere et al., 2011). The RPPH1 copy number could not be reliably determined for three evDNA samples, so the evDNA integrity index was not calculated for seven samples (including four in which the RPP30 copy number was already too low). The determined integrity indexes ranged from 0.16 to 0.70 for evDNA, and from 0.13 to 0.44 for cfDNA (Figure 3), but there were no significant differences between the patient groups (P = 0.608 for evDNA and P = 0.065 for cfDNA). Interestingly, cfDNA integrity indexes did not correlate with evDNA integrity indexes (ρ = 0.231, P = 0.203), but were significantly lower compared to evDNA in the whole cohort (P < 0.001, Figure 3a) as well as in all individual patient groups (Figure 3b) suggesting that cfDNA is more fragmented than evDNA.
FIGURE 3.
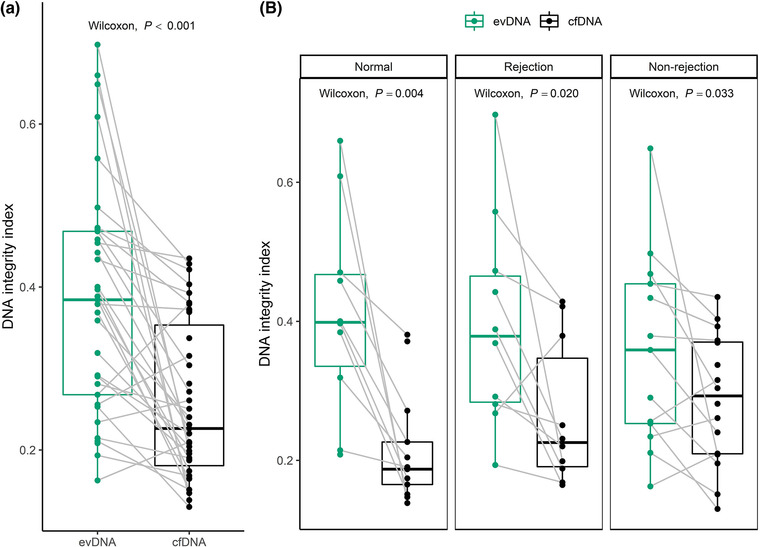
Extracellular vesicle‐bound DNA (evDNA) is less fragmented than cell‐free DNA (cfDNA) in urine of kidney allograft recipients. DNA integrity index is defined as the RPPH1/RPP30 (long/short amplicon) ratio, as determined by ddPCR. Distribution of DNA integrity indexes of evDNA (green) and cfDNA (black) in (a) the patient cohort (N = 33 for evDNA, N = 39 for cfDNA, respectively) and in (b) patient groups with normal histology (N = 10 for evDNA, N = 13 for cfDNA, respectively), rejection injury (N = 10 for evDNA, N = 10 for cfDNA, respectively), or with non‐rejection injury (N = 13 for evDNA, N = 16 for cfDNA, respectively). Gray lines connect paired evDNA and cfDNA samples. The Wilcoxon signed‐rank test was used for comparison of DNA integrity indexes of related evDNA and cfDNA
3.4. Donor‐derived DNA is detectable in uEV samples of kidney allograft recipients
For analysis of ddDNA content, recipient and donor genomic DNA were genotyped at six SNP loci by real‐time PCR and, in ambiguous cases, additionally by ddPCR. For all patients, the recipient and donor genotypes differed in at least two SNP loci, allowing measurement of ddDNA content. Nevertheless, in seven evDNA samples, the number of SNP allelic copies of the donor was too low to allow ddDNA quantification. The ddDNA fraction of evDNA and cfDNA was highly variable, ranging from 2.7% to 99.7% for evDNA, and from 1.8% to 98.6% for cfDNA (Figure 4a). Interestingly, the ddDNA fractions of evDNA and cfDNA correlated strongly (ρ = 0.907, P < 0.001), with no significant differences between the patient groups (P = 0.303 for evDNA and P = 0.113 for cfDNA) (Figure 4b). However, dd‐cfDNA fractions were significantly higher in patients with rejection and non‐rejection injury combined than in patients with normal histology (P = 0.039). When comparing ddDNA copies normalised to U‐creatinine, slightly higher ddDNA content was observed in patients with rejection injury, although the differences between patient groups were not statistically significant (P = 0.123 for evDNA and P = 0.218 for cfDNA) (Figure 4c). When subgroups of histologically confirmed rejection cases were compared, significantly more normalised dd‐evDNA copies were detected in ABMR than in TCMR (P = 0.036) (Figure 4c, Table S4).
FIGURE 4.
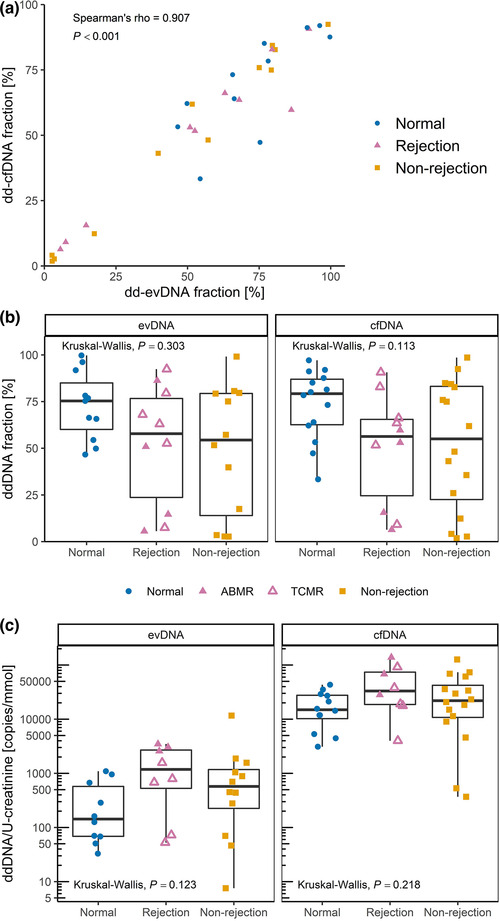
Donor‐derived DNA (ddDNA) is detectable in uEV samples of kidney allograft recipients. ddDNA fraction of extracellular vesicle‐bound DNA (evDNA) and cell‐free DNA (cfDNA) was calculated from ddPCR quantification of at least two single nucleotide polymorphisms (SNPs) that differed between the kidney allograft donor and recipient. The ddDNA copy number was calculated by multiplying ddDNA fraction with DNA copy number. (a) Fractions of dd‐evDNA and dd‐cfDNA correlate strongly (N = 33; tested with Spearman correlation coefficient). (b) Distribution of ddDNA fractions determined for evDNA (left) and cfDNA (right) in patient groups with normal histology (N = 11 for evDNA, N = 14 for cfDNA, respectively), rejection injury (N = 10 for evDNA, N = 10 for cfDNA, respectively), or with non‐rejection injury (N = 12 for evDNA, N = 16 for cfDNA, respectively). (c) distribution of dd‐evDNA (left) and dd‐cfDNA (right) absolute copy numbers normalised to U‐creatinine [mmol/ml] in patient groups with normal histology (N = 10 for evDNA, N = 12 for cfDNA, respectively), rejection injury (N = 8 for evDNA, N = 8 for cfDNA, respectively), or with non‐rejection injury (N = 12 for evDNA, N = 16 for cfDNA, respectively). Data of patients with normal histology are presented as blue circles, rejection injury as purple triangles and non‐rejection injury as orange squares. In the rejection group, full triangles represent antibody mediated rejection (ABMR), while empty triangles represent T‐cell mediated rejection (TCMR). Patients with diagnosis of mixed ABMR and TCMR were grouped within the ABMR subgroup. Kruskal‐Wallis test was used to compare the characteristics between all three patient groups
3.5. DNA is bound to the surface of EVs enriched from urine
The above data indicate that urinary evDNA is better protected from degradation than cfDNA. To test whether evDNA is packaged inside or bound to the surface of uEVs, we first treated uEVs of nine study patients (three from each patient group) with DNase I. After treatment, the reaction mixtures were heat inactivated, before proceeding with evDNA extraction, preamplification and characterization. A large reduction in evDNA copy number was observed in DNase‐treated compared to untreated uEV samples from the same patient (Figure 5a), as only 2%–38% evDNA copies (mean 12%) remained undigested in the analysed patients’ uEV samples (Figure 5b). In a separate experiment, described in the Supplementary Material (Section S6), we have shown that DNase I retains most of its activity even after heat treatment, which may contribute to the observed evDNA degradation. To further evaluate this, we performed additional experiments on three uEV samples, in which DNase was inactivated by addition of denaturing buffer immediately after DNase treatment at 37°C. Similar to heat inactivation, on average only 14% of the evDNA copies remained undigested after DNase treatment of the uEV samples (Figure 5b). We confirmed that DNase I did not retain activity after inactivation by denaturing buffer (Supplementary Material, Section S6).
FIGURE 5.
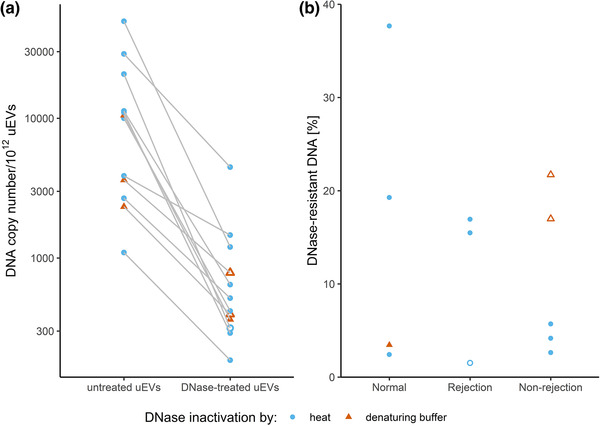
DNase treatment degrades most of the DNA in uEV samples of kidney allograft recipients. DNA copy number in uEV samples, isolated from selected patients with normal histology, rejection injury or non‐rejection injury, without or with DNAse treatment was determined by measuring the RPP30 copies by ddPCR. DNase was inactivated by heat (light blue circles) or by immediate DNA extraction (red triangles). Empty circles and triangles represent less reliable measurements of DNA copies in EV samples, as concentration of DNA after DNase treatment was very low. (a) DNA copy numbers normalised to 1012 uEVs in untreated and DNase‐treated uEV samples. (b) DNase‐resistant fraction of DNA in treated uEV samples
This suggests that most of evDNA is sensitive to degradation by DNase I, but as shown here, evDNA is also less fragmented compared to cfDNA in urine. To test whether evDNA is protected from degradation in urine by binding to the surface of EVs, we labelled DNA in representative uEV samples from patients with normal histology, rejection injury, or non‐rejection injury and analysed them using TEM (Figure 6). We used antibodies against dsDNA, which in turn were recognised by IgG antibodies conjugated to gold particles. Representative micrographs show that at least some of the uEVs in analysed samples were labelled with aggregated gold particles, indicating binding of dsDNA to the surface of uEVs (Figure 6b). On the other hand, gold particles were not present in representative uEV samples where incubation with antibodies against dsDNA was omitted (Figure 6a). Interestingly, despite the absence of dsDNA‐specific antibodies, the uromodulin filament was labelled with gold particles in one of the samples (Figure S9), as previously noted (Rhodes et al., 1993). In summary, DNA is bound to the surface of EVs enriched from urine, independent of the patients’ allograft injury status.
FIGURE 6.
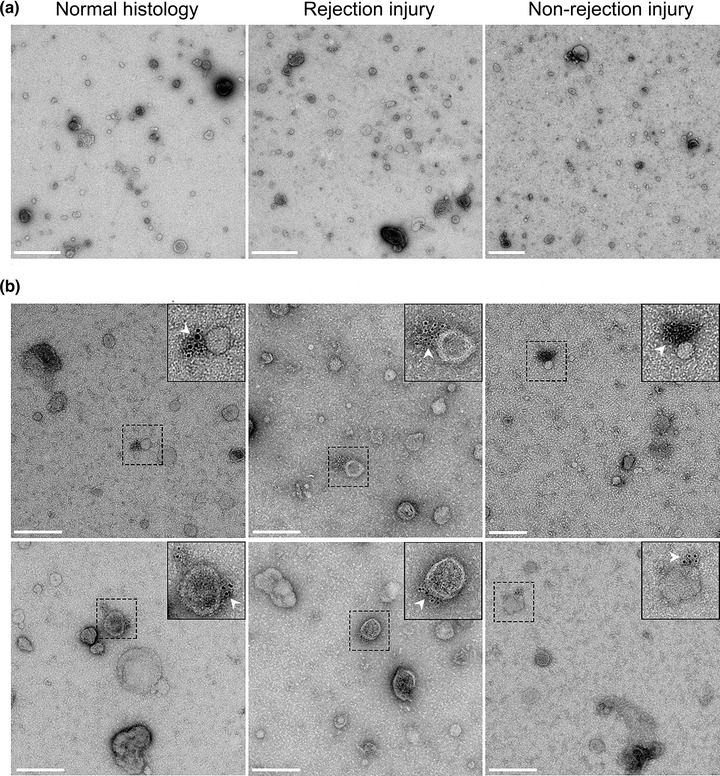
DNA in uEV samples of kidney allograft recipients is bound to the surface of extracellular vesicles. (a) Transmission electron micrographs of representative negative‐stained uEV samples from the normal histology (left), rejection injury (middle) and non‐rejection injury (right) patient groups. Scale bar: 500 nm. (b) Micrographs of two representative fields of vision of uEV samples of the three patients groups, labelled with antibodies against dsDNA and IgG conjugated with gold particles. White arrows mark colloidal gold particles (6 nm), indicating the location of the DNA. Scale bar: 200 nm.
3.6. Proof‐of‐principle study to assess potential of urinary evDNA as a biomarker for kidney allograft injury
Finally, to evaluate the potential of urinary evDNA as a biomarker for kidney allograft injury, the association of evDNA characteristics with patients’ clinical characteristics was assessed in a proof‐of‐principle study. Associations with categorical variables are presented in Table 2 and Table S3, and with continuous variables in Table S5. Among the clinical characteristics assessed, normalised evDNA copy numbers were significantly increased (P = 0.025) in patients with for‐cause biopsy compared to those with surveillance biopsy. Normalised evDNA yield, evDNA copy number, and dd‐evDNA copy number were significantly higher in patients with an interstitial inflammation score > 0 (inflammatory lesions graded with scores 1, 2, or 3), compared to those with a score 0 (absence or evidence of less than 10% inflammation) (all P < 0.050, Table 2). We divided the Banff scores into two categories because the patient cohort was small and associations with specific score values cannot be tested. Trend of higher interstitial inflammation scores were detected in patients with allograft rejection injury, particularly in those with TCMR (Figure 7a). For the combined parameters of glomerulitis and peritubular capillaritis, normalised evDNA yield, evDNA copy number and dd‐evDNA copy number were also significantly higher in patients with Banff lesion scores > 0 compared to those with score 0 (all P < 0.050). Trend of higher combined scores for glomerulitis and peritubular capillaritis were observed in patients with ABMR (Figure 7b). For inflammation in areas of fibrosis, normalised evDNA copy numbers were higher in patients with Banff lesion scores > 0 (P = 0.040). Trend of higher inflammation in areas of fibrosis score was observed in patients with rejection or non‐rejection injury, compared to patients with normal histology (Figure 7c). Interestingly, several evDNA characteristics (dd‐evDNA fraction, evDNA integrity index, normalised evDNA yield, size) significantly differed with respect to patients’ sex (Table 2, Table S3).
TABLE 2.
Association of evDNA characteristics with kidney allograft recipients’ clinical and histological parameters (categorical variables)
| U‐creatinine normalised | ||||||
|---|---|---|---|---|---|---|
| Variables | evDNA integrity index median (25%–75%) | dd‐evDNA fraction [%] median (25%–75%) | evDNA yield (ng) median (25%–75%) | evDNA copy number [copies/mmol) median (25%–75%) | dd‐evDNA copy number [copies/mmol] median (25%–75%) | |
| Sex | Male | 0.32 (0.24–0.45) [7] | 75.19 (52.34–82.02) [6] | 19.6 (10.93–42.13) [2] | 494.1 (111.03–1766.45) [6] | 554.15 (70.68–1079.75) [8] |
| Female | 0.47 (0.39–0.54) | 5.58 (2.74–39.76) [1] | 122.5 (30.4–294.7) [1] | 16410.5 (257–20881.1) [1] | 574.05 (54.15–3152.55) [2] | |
| P‐value | 0.025 | <0.001 | 0.026 | 0.060 | 0.820 | |
| ECD | No | 0.41 (0.28–0.51) [3] | 63.03 (7.45–79.52) [2] | 26.4 (13.35–133.05) [1] | 358.3 (111.03–4403.13) [3] | 223.5 (50.85–988.43) [3] |
| Yes | 0.37 (0.25–0.47) [4] | 71.55 (47.41–79.54) [4] | 16.8 (9.8–67.1) [1] | 1000.8 (149.9–1995.7) [3] | 680.9 (125.3–1567.5) [5] | |
| P‐value | 0.423 | 0.495 | 0.367 | 0.710 | 0.252 | |
| DGF | No | 0.37 (0.25–0.47) [5] | 66.02 (44.9–82.02) [5] | 20.65 (11.48–66.28) [3] | 1000.8 (138.05–2174.3) [6] | 670.1 (70.1–1044.8) [8] |
| Yes | 0.45 (0.32–0.47) [2] | 57.19 (17.46–76.82) [2] | 22.4 (10.15–226.1) | 357.95 (139.9–47118.4) [1] | 438.2 (68.1–3477.3) [2] | |
| P‐value | 0.330 | 0.503 | 0.589 | 1.000 | 0.774 | |
| Bx | Protocol | 0.38 (0.28–0.5) [6] | 68.04 (54.43–91.78) [6] | 17.85 (11.48–41.95) [1] | 210.05 (92.2–1029.9) [5] | 219.05 (65.68–762.33) [7] |
| For‐cause | 0.39 (0.26–0.46) [1] | 51.25 (12.8–79.55) [1] | 37.5 (12.55–110.75) [2] | 1699.1 (228.3–7847.85) [2] | 839.5 (161.83–1798.5) [3] | |
| P‐value | 0.817 | 0.126 | 0.297 | 0.025 | 0.166 | |
| t | Score 0 | 0.39 (0.26–0.47) [7] | 64.39 (34.19–79.36) [7] | 20.65 (10.18–72.13) [1] | 358.3 (139.9–2009.3) [5] | 438.2 (69.9–1068.1) [8] |
| Score > 0 | 0.29 (0.27–0.44) | 68.04 (50.86–86.25) | 37.5 (22.15–174.6) [2] | 1699.1 (550.25–7847.85) [2] | 792.1 (366.65–2071.6) [2] | |
| P‐value | 0.590 | 0.620 | 0.328 | 0.419 | 0.416 | |
| i | Score 0 | 0.39 (0.27–0.48) [7] | 64.39 (44.9–78.45) [7] | 19.6 (10.18–59) [1] | 280.55 (120.28–1446.03) [5] | 286.2 (68.9–920.5) [8] |
| Score > 0 | 0.29 (0.25–0.44) | 79.52 (7.45–86.25) | 99 (39.4–301.3) [2] | 5060.3 (1833.8–36488.5) [2] | 1569.6 (1179.8–3025.45) [2] | |
| P‐value | 0.352 | 0.682 | 0.016 | 0.011 | 0.007 | |
| t i | Score 0 | 0.39 (0.3–0.53) [4] | 66.02 (23.39–77.84) [4] | 19.6 (12.2–41.95) | 221.5 (108.53–1592.33) [2] | 219.05 (68.6–839.78) [4] |
| Score > 0 | 0.34 (0.25–0.47) [2] | 57.19 (43.18–80.12) [1] | 37.5 (9.7–157.5) [3] | 1568.4 (458.45–6454.08) [4] | 873.1 (346.08–2049.5) [4] | |
| P‐value | 0.616 | 0.873 | 0.314 | 0.071 | 0.058 | |
| *g+ptc | Score 0 | 0.37 (0.25–0.46) [7] | 67.18 (51.19–79.88) [7] | 19.6 (10.18–59) [1] | 357.65 (120.28–1636.53) [5] | 438.2 (69.9–920.5) [8] |
| Score > 0 | 0.47 (0.29–0.56) | 17.46 (5.58–79.52) | 122.5 (33.95–925.35) [2] | 20881.1 (2670.75–64493.9) [2] | 3044.3 (1290.55–7557.55) [2] | |
| P‐value | 0.131 | 0.109 | 0.016 | 0.005 | 0.037 | |
| i‐IFTA | Score 0 | 0.38 (0.21–0.61) [1] | 75.94 (65.88–91.66) | 16.1 (10.55–30.75) | 162.2 (61.88–995.05) | 143.05 (55.48–736.73) |
| Score > 0 | 0.40 (0.26–0.47) [4] | 52.58 (14.58–79.63) [4] | 29.6 (10.5–137.65) [2] | 1238.65 (168.93–12079.18) [5] | 680.9 (71.05–1722.2) [6] | |
| P‐value | 0.848 | 0.060 | 0.290 | 0.040 | 0.153 | |
Note: [No. of missing data]; score > 0: combined Banff Lesion Scores 1, 2, and 3. Significant P‐values are indicated in bold, and in Italics when observed only for cfDNA or evDNA.
Abbreviations: evDNA, extracellular vesicle bound DNA; dd‐evDNA, donor‐derived extracellular vesicle bound DNA; DGF, delayed graft function; ECD, expanded criteria donor; Protocol, surveillance biopsy; Bx, biopsy; t, tubulitis; i, interstitial inflammation; ti, total inflammation;
g+ptc: glomerulitis (g) and peritubular capillaritis (ptc) combined, i‐IFTA: inflammation in areas of fibrosis.
FIGURE 7.
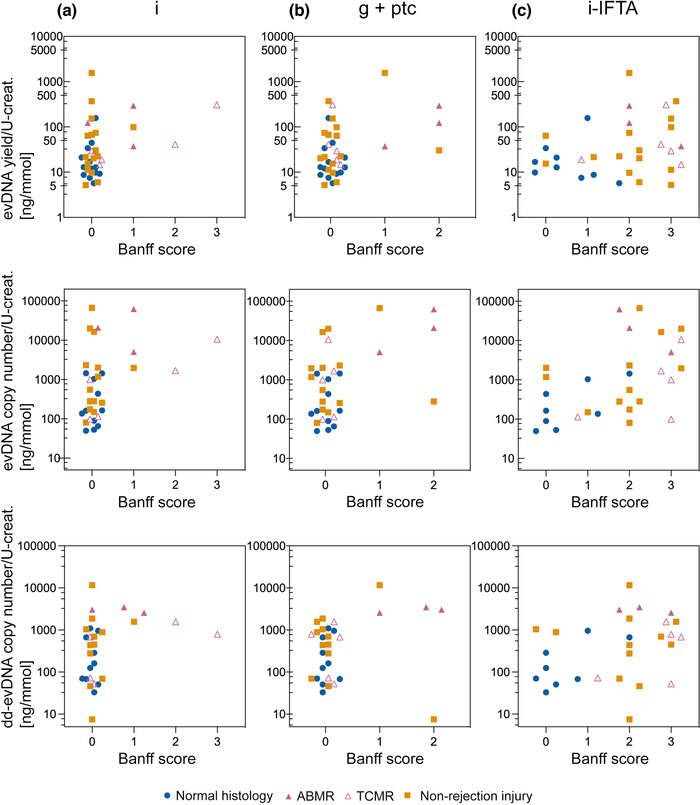
Distribution of normalised evDNA yield, evDNA copy number and dd‐evDNA copy number for each value of the selected Banff parameters. Each data point represents individual patient, with blue circles, purple triangles and orange squares denoting data of patients with normal histology, rejection injury (antibody mediated rejection; ABMR ‐ full, T‐cell mediated rejection; TCMR ‐ empty) or non‐rejection injury, respectfully. Banff score 0 is indicative of absence or detection of less than 10% inflammation in the specific region of the biopsy sample, while inflammatory lesions are graded with scores 1, 2 or 3 depending on the severity. Because of the smaller patient cohort, it was not possible to test associations with specific Banff score values; therefore, in the statistical analysis, patients with Banff score > 0 were compared to those with a score of 0 (Tables 2, 3). i, interstitial inflammation; g+ptc, patients with glomerulitis (g) and peritubular capillaritis (ptc) combined; i‐IFTA, inflammation in areas of fibrosis; evDNA, extracellular vesicle bound DNA; dd‐evDNA, donor‐derived extracellular vesicle bound DNA
To evaluate the relevance of evDNA compared to urine cfDNA as a biomarker for kidney allograft injury, association of cfDNA characteristics with patients’ clinical characteristics was performed (Table 3, Table S6). Similar to evDNA, urine cfDNA measured characteristics were significantly associated with sex, the type of biopsy, and the level of interstitial inflammation, combined glomerulitis and peritubular capillaritis, and inflammation in areas of fibrosis (Table 3). Despite observed similarities, additional cfDNA or evDNA characteristics correlated with patients’ U‐protein/creatinine ratio, biopsy type, combined parameters of glomerulitis and peritubular capillaritis or inflammation in areas of fibrosis (Tables 2 and 3, Table S5; P‐values indicated in italics). The proof‐of‐principle study showed that urinary evDNA is a potential biomarker for kidney allograft injury, but can also be interchanged with urinary cfDNA.
TABLE 3.
Association of cfDNA characteristics with kidney allograft recipients’ clinical and histological parameters (categorical variables)
| U‐creatinine normalised values | ||||||
|---|---|---|---|---|---|---|
| Variables | cfDNA Integrity index median (25%–75%) | dd‐cfDNA fraction [%] median (25%–75%) | cfDNA yield [ng] median (25%–75%) | cfDNA copy number [copies/mmol] median (25%–75%) | dd‐cfDNA copy number [copies/mmol] median (25%–75%) | |
| Sex | Male | 0.2 (0.17–0.28) | 75.41 (54.85–85.08) | 468.85 (169.48–1179.9) [2] | 33304.3 (15511.3–66309.4) [3] | 21148.7 (11874.95–35616.75) [3] |
| Female | 0.38 (0.26–0.41) | 10.95 (3.04–31.5) | 7916.7 (1380–10750) [1] | 438517.7 (79746.6–2175604.9) [1] | 20706.3 (9000.9–69041.1) [1] | |
| P‐value | 0.003 | <0.001 | 0.001 | 0.003 | 0.696 | |
| Bx | Protocol | 0.2 (0.17–0.32) | 78.39 (49.46–89.37) | 372.2 (112.48–966.93) [1] | 26979.1 (6043.2–59993.3) [1] | 17716.1 (5307.5–35290.4) [1] |
| For‐cause | 0.26 (0.2–0.38) | 53.24 (15.53–75.87) | 1237.5 (501.25–3314) [2] | 72625.5 (33027.4–160386.9) [2] | 28165 (11634–64682.4) [2] | |
| P‐value | 0.134 | 0.039 | 0.024 | 0.018 | 0.219 | |
| i | Score 0 | 0.23 (0.18–0.36) | 63.96 (39.35–83.82) | 494.25 (182.98–1325.18) [1] | 33304.3 (16208.9–91592.8) [2] | 18337 (9000.9–35290.4) [2] |
| Score 6 > 0 | 0.22 (0.17–0.38) | 59.69 (9.1–84.38) | 3233.3 (2008.2–6232.25) [2] | 101242.3 (62890.45–1192390.75) [2] | 61281.8 (23598–115222.3) [2] | |
| P‐value | 0.788 | 0.835 | 0.014 | 0.028 | 0.032 | |
| *g+ptc | Score 0 | 0.22 (0.17–0.31) [1] | 73.19 (47.73–85.01) | 494.25 (182.98–1218.3) [1] | 33304.3 (16208.9–79746.6) [2] | 19031 (11379.6–35290.4) [2] |
| Score > 0 | 0.38 (0.21–0.42) | 15.53 (6.37–59.69) | 4153.8 (2079.45–14059.9) [2] | 438517.7 (82376.3–1601223.6) [2] | 68083 (16371.2–132609.6) [2] | |
| P‐value | 0.058 | 0.028 | 0.004 | 0.004 | 0.091 | |
| i‐IFTA areas of fibrosis | Score 0 | 0.15 (0.15–0.17) | 77.13 (66.27–86.98) | 396.25 (159.8–905.88) | 26979.1 (16839.8–55176.9) [1] | 21148.7 (14337.5–35290.4) [1] |
| Score > 0 | 0.27 (0.2–0.38) | 53.24 (15.53–82.75) | 674.3 (419.85–3314) [2] | 47955 (21611.65–160386.9) [2] | 23161.8 (10190.25–49687.9) [2] | |
| P‐value | <0.001 | 0.046 | 0.162 | 0.302 | 0.964 | |
Note: [No. of missing data]; score > 0: combined Banff Lesion Scores 1, 2 and 3. Significant P‐values are indicated in bold, and in Italics when observed only for cfDNA or evDNA.
Only cfDNA parameters that are significantly associated with clinical variables are presented.
Abbreviations: cfDNA: cell‐free DNA; dd‐cfDNA: donor‐derived cell‐free DNA; Protocol: surveillance biopsy; Bx: biopsy; i: interstitial inflammation
g+ptc: glomerulitis (g) and peritubular capillaritis (ptc) combined; i‐IFTA: inflammation in areas of fibrosis.
4. DISCUSSION
The purpose of this study was to investigate DNA as urinary EV cargo, using kidney transplantation as a model. To this end, we enriched EVs from urine of well‐characterized kidney transplant recipients and explored the uEV‐bound DNA, further evaluating its biomarker potential in a proof‐of‐principle study. The SEC‐based method enriched pure EVs from urine of kidney transplant recipients, regardless of the allograft injury. Urinary evDNA represents up to 29.2% ± 8.0% of cfDNA, but is less fragmented, while correlating with cfDNA in several other characteristics. Importantly, using DNase treatment and immunogold labelling TEM, we showed for the first time that evDNA is mostly bound to the surface of EVs in urine. Normalised evDNA yield and evDNA copy number differed significantly between patients with normal histology, rejection injury and non‐rejection injury, with the latter patient groups having significantly larger uEVs and more DNA bound per uEV. ddDNA is detectable in uEV samples of kidney allograft recipients, but its quantity is highly variable. The proof‐of‐principle study showed that urinary evDNA is a potential biomarker for kidney allograft injury, as several evDNA characteristics correlated with clinical and histological parameters, but could also be interchanged with urinary cfDNA.
An important finding of our study is the first experimental evidence that dsDNA is bound to the surface of EVs in urine. evDNA was mostly unprotected from DNase I degradation, but localized to the uEV surface in immunogold DNA‐labelling TEM. This may explain the previous observation that DNA was co‐isolated with microvesicles from human and rat urine, which was at the time of discovery designated as contaminant (Miranda et al., 2010). Nevertheless, up to 38% (mean 12%) of evDNA remained undigested in our study, suggesting that evDNA might also be incorporated inside uEVs to small extent or protected from degradation as part of exomeres (Jeppesen et al., 2019), so far not yet detected in urine (Zhang et al., 2018). Studies on EVs from blood plasma or serum (Kahlert et al., 2014; Lázaro‐Ibáñez et al., 2014; Vagner et al., 2018) and cell cultures (Lázaro‐Ibáñez et al., 2019; Németh et al., 2017) similarly showed that DNA was mostly bound to the surface of small EVs, whereas it was incorporated inside larger EVs. In those studies, DNA EV‐localization was largely determined by DNase degradation assay. One study demonstrated colocalization of DNA dye with EV‐typical proteins CD63 and CD81 using super‐resolution imaging of single EVs (Maire et al., 2021), whereas another study demonstrated localization of DNA dye with EVs labelled with annexin V, anti‐CD63 using flow cytometry (Németh et al., 2017). Moreover, our observations on evDNA localization in the context of kidney transplantation are valuable, because most of what is known about evDNA has so far been studied in the context of cancer or cancer cell lines, with characteristic genome instability not seen in most other pathologies (Lázaro‐Ibáñez et al., 2019; Lee et al., 2018; Vagner et al., 2018).
At the molecular level, studies on DNA binding to the cellular membrane surface show that binding can be promoted through electrostatic interactions (Laktionov et al., 2004), direct binding of DNA by specific membrane receptors (Rykova et al., 2012), or indirect binding through histones (Kubota et al., 1990). As EVs share characteristics with parental cells, similar mechanism could exist. This is supported by the following observations: (i) EV adhesive capabilities are influenced by their structure (Kawamura et al., 2017), (ii) the cell‐surface DNA‐binding receptors MAC‐1, Ku80, and Ku70 have been identified in uEV proteomic studies (Silvers et al., 2017; Wang et al., 2012), (iii) histones have been detected on EVs (Németh et al., 2017) or DNA‐bound to EVs (Lázaro‐Ibáñez et al., 2019; Takahashi et al., 2017). Regardless, based on observation that DNA weakly linked to cellular surface can be easily detached by 5 mM EDTA (Laktionov et al., 2004), DNA association to the uEVs was stronger in our study, because 8 mM final concentration of EDTA was added to the urine during the processing step to prevent polymerization of uromodulin (Kobayashi & Fukuoka, 2001).
The EV biogenesis process involved in DNA binding to the uEV surface, that is, is DNA bound to EV surface during endosomal biogenesis (Jeppesen et al., 2019) or is it a consequence of DNA binding to cell surface in the process of plasma membrane invagination (Tamkovich & Laktionov, 2019), is not known. Regardless, urinary evDNA and cfDNA correlated in DNA yield, DNA copy number, and ddDNA fraction, implying the common source of DNA. Based on evaluation of the patients’ biopsy samples for histomorphological characteristic of apoptosis and inflammation‐related cell damage associated with necrosis, we speculate that cfDNA is released in urine (at least in part) by cell necrosis. In urine, released necrotic DNA is likely degraded by very active DNA nucleases (Bryzgunova & Laktionov, 2015), resulting in typical cfDNA fragment lengths, as also observed here. Future in‐depth characterization of evDNA and cfDNA sequences, epigenetic modifications and DNA‐bound proteins may help answer these questions.
To our knowledge, our study is the first to correlate urinary evDNA with kidney allograft injury, as previous studies mostly focused on uEV proteins (Alvarez et al., 2013; Dimuccio et al., 2014; Park et al., 2017; Sonoda et al., 2009), RNAs (El Fekih et al., 2021), or size (Dimuccio et al., 2020; Freeman et al., 2018). Normalised evDNA yield and evDNA copy number differed significantly between patient groups, with more urinary evDNA obtained in patients with rejection or non‐rejection allograft injury compared to normal histology. The uEVs from groups with allograft injury were significantly larger and had more DNA bound per uEV compared to normal histology. Monitoring kidney function using urinary evDNA is particularly useful in allogenic kidney transplants, as we can use the differences in genotype to specifically track the DNA released from the allograft (ddDNA) due to apoptosis, necrosis, or active release (Oellerich et al., 2021; Sigdel et al., 2013; Zhang et al., 1999). Here, we have successfully measured dd‐evDNA fraction and dd‐evDNA copy number in the urine of our kidney transplant patient cohort, using our recently developed ddPCR method for determining dd‐cfDNA in plasma (Jerič Kokelj et al., 2021). The ddDNA fraction of evDNA was highly variable, ranging from 2.7% to 99.7%, but no significant differences were detected among the three patient groups. When absolute ddDNA levels were evaluated, a trend toward more U‐creatinine normalised dd‐evDNA copies was observed in patients with rejection injury. Significantly more normalised dd‐evDNA copies were detected in ABMR than in TCMR, but this preliminary observation should be validated in a larger patient cohort. Overall, it appears that absolute quantification of dd‐evDNA in urine is more informative for allograft rejection compared to the dd‐evDNA fraction, similar to the study by Oellerich et al. (2019). This could be due to fluctuations in recipient DNA levels due to infection, exercise, or psychological stress, which can damage the recipient's own cells, resulting in increased levels of recipient extracellular DNA that affect dd‐cfDNA fraction calculations (Oellerich et al., 2021).
Several large, multicentre cohort studies support the use of plasma dd‐cfDNA levels to monitor kidney allograft rejection, performing most robustly for discriminating ABMR (Bloom et al., 2017; Gielis et al., 2020; Halloran et al., 2022; Huang et al., 2019; Oellerich et al., 2019; Sigdel et al., 2018; Whitlam et al., 2019). To date, three commercial blood‐based dd‐cfDNA assays have become available for clinical use (Filippone & Farber, 2021; Paul et al., 2021). Our study on urinary cfDNA is one of the few studies investigating the association between urinary dd‐cfDNA and kidney injury and rejection. Sigdel et al. had previously shown that urinary dd‐cfDNA is significantly higher in patients with acute rejection than in patients with stable kidney allograft (Sigdel et al., 2013), whereas urinary cfDNA analysis is part of the multimarker test for early detection of kidney allograft rejection (Watson et al., 2019; Yang et al., 2020). Here, we showed that the ddDNA fraction of cfDNA varied widely, similar to evDNA. The combined group of patients with rejection or non‐rejection injury had significantly higher dd‐cfDNA fractions compared to patients with normal histology.
The proof‐of‐principle study showed that urinary evDNA and cfDNA characteristics were significantly associated with the type of biopsy and the extent of interstitial inflammation, combined glomerulitis and peritubular capillaritis, and inflammation in areas of fibrosis, supporting evDNA and cfDNA as potential biomarkers for kidney allograft injury. This observation should be interpreted in the context of several limitations of the study. First, the limited group size used in this study should be significantly expanded, both in terms of number of patients and injury phenotypes. Such expansion will lend more power to the data obtained and will allow further analysis of the association between urinary evDNA and kidney allograft injury phenotype. Second, our study has not included an independent validation cohort. Third, although the study provides a valuable contribution to understanding the relationship between urinary evDNA and phenotypes of allograft injury, it is limited in its contribution to understanding how urinary evDNA can be used in either detecting allograft injury or in monitoring transplanted patients. There are also several advantages of our study. It is a single‐centre study in which patients were treated according to the same and well‐established treatment protocol. Importantly, clinical samples were collected according to the same protocol and at fixed time points, which may have helped reduce the impact of preanalytical variables on the measured characteristics. In addition, histologic assessment of allograft injury was performed by a dedicated histopathologist, reducing the possibility of interobserver variability in interpretation of results.
In our study, evDNA and cfDNA are interchangeable as putative biomarkers for kidney allograft injury. cfDNA is more accessible than evDNA, which requires extraction from larger volumes of urine, following a more complex protocol. Urinary cfDNA also showed much higher yields compared to evDNA, still there was enough evDNA to conduct ddPCR analyses for most patients. Considering this, urinary cfDNA seems to perform better compared to evDNA. Nevertheless, evDNA is less fragmented, which may be helpful in developing ddPCR assays with respect to amplicon length. Unlike cfDNA, EVs also carry a variety of biological signals, which allows multiplexing and offers the possibility of identifying the origin of cellular cargo, which in turn could improve detection specificity (Chennakrishnaiah et al., 2019). Furthermore, EVs protect their molecular cargo from degradation, allowing relevant signals to be recovered even from archival samples (Chennakrishnaiah et al., 2019). In biomarker studies of pancreatic ductal adenocarcinoma, a higher rate of KRAS mutations was detected in evDNA than in cfDNA (Allenson et al., 2017), and only evDNA levels correlated with patient outcomes (Bernard et al., 2019). Thus, evDNA and cfDNA perform similarly as potential biomarkers for kidney allograft injury, while evDNA was shown to outperform cfDNA in cancer, especially when detecting specific mutations or other nucleotide changes.
In conclusion, we successfully extracted and characterized evDNA from urine. We were the first to show that evDNA in urine is mostly bound to the surface of EVs and that evDNA quantity reflects kidney allograft injury. Our data on urinary cfDNA also contribute to the limited knowledge on its association with kidney allograft injury. The proof‐of‐principle study supports urinary evDNA and cfDNA as potential biomarkers for kidney allograft injury, but this should be validated in a larger study.
AUTHOR CONTRIBUTIONS
Ivana Sedej: Formal analysis; Investigation; Methodology; Resources; Visualization; Writing – original draft. Maja Štalekar: Formal analysis; Investigation; Methodology; Visualization; Writing – original draft. Magda Tušek Žnidarič: Investigation; Methodology; Visualization; Writing – original draft. Katja Goričar: Formal analysis; Investigation; Methodology; Writing – original draft. Nika Kojc: Resources; Writing – original draft. Polona Kogovšek: Funding acquisition; Resources; Writing – review & editing. Vita Dolžan: Conceptualization; Funding acquisition; Resources; Writing – review & editing. Miha Arnol: Conceptualization; Funding acquisition; Project administration; Resources; Supervision; Writing – review & editing. Metka Lenassi: Conceptualization; Funding acquisition; Project administration; Supervision; Writing – review & editing.
GEOLOCATION INFORMATION
14.518433998551663
CONFLICT OF INTEREST
The authors report no conflict of interest.
Supporting information
Supporting Information.
ACKNOWLEDGEMENTS
We thank Tina Demšar, MSc for soybean gDNA and Le1 assay for its quantification, and Tjaša Jakomin for helping with additional ddPCR experiments. This work was supported by the Slovenian Research Agency (ARRS), Ljubljana, Slovenia under Grants P3‐0323, P1‐0170, P4‐0165 and P4‐0407. I. Sedej was supported by a young researcher scholarship of ARRS.
Sedej, I. , Štalekar, M. , Tušek Žnidarič, M. , Goričar, K. , Kojc, N. , Kogovšek, P. , Dolžan, V. , Arnol, M. , & Lenassi, M. (2022). Extracellular vesicle‐bound DNA in urine is indicative of kidney allograft injury. Journal of Extracellular Vesicles, 11, e12268. 10.1002/jev2.12268
Ivana Sedej, Maja Štalekar, Miha Arnol, and Metka Lenassi contributed equally to this study.
Contributor Information
Miha Arnol, Email: miha.arnol@kclj.si.
Metka Lenassi, Email: metka.lenassi@mf.uni-lj.si.
REFERENCES
- Allenson, K. , Castillo, J. , San Lucas, F. A. , Scelo, G. , Kim, D. U. , Bernard, V. , Davis, G. , Kumar, T. , Katz, M. , Overman, M. J. , Foretova, L. , Fabianova, E. , Holcatova, I. , Janout, V. , Meric‐Bernstam, F. , Gascoyne, P. , Wistuba, I. , Varadhachary, G. , Brennan, P. , …, Alvarez, H. (2017). High prevalence of mutant KRAS in circulating exosome‐derived DNA from early‐stage pancreatic cancer patients. Annals of Oncology: Official Journal of the European Society for Medical Oncology, 28(4), 741–747. 10.1093/annonc/mdx004[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, S. , Suazo, C. , Boltansky, A. , Ursu, M. , Carvajal, D. , Innocenti, G. , Vukusich, A. , Hurtado, M. , Villanueva, S. , Carreño, J. E. , Rogelio, A. , & Irarrazabal, C. E. (2013). Urinary exosomes as a source of kidney dysfunction biomarker in renal transplantation. Transplantation Proceedings, 45(10), 3719–3723. 10.1016/j.transproceed.2013.08.079[CrossRef] [DOI] [PubMed] [Google Scholar]
- Andersson, D. , Akrap, N. , Svec, D. , Godfrey, T. E. , Kubista, M. , Landberg, G. , & Ståhlberg, A. (2015). Properties of targeted preamplification in DNA and cDNA quantification. Expert Review of Molecular Diagnostics, 15(8), 1085–1100. 10.1586/14737159.2015.1057124[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglicheau, D. , Naesens, M. , Essig, M. , Gwinner, W. , & Marquet, P. (2016). Establishing biomarkers in transplant medicine: A critical review of current approaches. Transplantation, 100(10), 2024–2038. 10.1097/TP.0000000000001321[CrossRef] [DOI] [PubMed] [Google Scholar]
- Beck, J. , Oellerich, M. , & Schütz, E. (2018). A universal droplet digital PCR approach for monitoring of graft health after transplantation using a preselected SNP set. Methods in Molecular Biology (Clifton, N.J.), 1768, 335–348. 10.1007/978-1-4939-7778-9_19[CrossRef] [DOI] [PubMed] [Google Scholar]
- Bernard, V. , Kim, D. U. , San Lucas, F. A. , Castillo, J. , Allenson, K. , Mulu, F. C. , Stephens, B. M. , Huang, J. , Semaan, A. , Guerrero, P. A. , Kamyabi, N. , Zhao, J. , Hurd, M. W. , Koay, E. J. , Taniguchi, C. M. , Herman, J. M. , Javle, M. , Wolff, R. , Katz, M. , …, Alvarez, H. A. (2019). Circulating nucleic acids are associated with outcomes of patients with pancreatic cancer. Gastroenterology, 156(1), 108–118.e4. 10.1053/j.gastro.2018.09.022[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blijdorp, C. J. , Tutakhel, O. A. Z. , Hartjes, T. A. , Van Den Bosch, T. P. P. , Van Heugten, M. H. , Rigalli, J. P. , Willemsen, R. , Musterd‐Bhaggoe, U. M. , Barros, E. R. , Carles‐Fontana, R. , Carvajal, C. A. , Arntz, O. J. , Van De Loo, F. A. J. , Jenster, G. , Clahsen‐Van Groningen, M. C. , Cuevas, C. A. , Severs, D. , Fenton, R. A. , Van Royen, M. E. , …, Hoorn, E. J. (2021). Comparing approaches to normalize, quantify, and characterize urinary extracellular vesicles. Journal of the American Society of Nephrology : JASN, 32(5), 1210–1226. 10.1681/ASN.2020081142[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom, R. D. , Bromberg, J. S. , Poggio, E. D. , Bunnapradist, S. , Langone, A. J. , Sood, P. , Matas, A. J. , Mehta, S. , Mannon, R. B. , Sharfuddin, A. , Fischbach, B. , Narayanan, M. , Jordan, S. C. , Cohen, D. , Weir, M. R. , Hiller, D. , Prasad, P. , Woodward, R. N. , Grskovic, M. , & Brennan, D. C. , Circulating Donor‐Derived Cell‐Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study Investigators . (2017). Cell‐free DNA and active rejection in kidney allografts. Journal of the American Society of Nephrology: JASN, 28(7), 2221–2232. 10.1681/ASN.2016091034[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, F. , Rinschen, M. , Buchner, D. , Bohl, K. , Plagmann, I. , Bachurski, D. , Richard Späth, M. , Antczak, P. , Göbel, H. , Klein, C. , Lackmann, J. W. , Kretz, O. , Puelles, V. G. , Wahba, R. , Hallek, M. , Schermer, B. , Benzing, T. , Huber, T. B. , Beyer, A. , …, Müller, R. U. (2020). The proteomic landscape of small urinary extracellular vesicles during kidney transplantation. Journal of Extracellular Vesicles, 10(1), e12026. 10.1002/jev2.12026[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryzgunova, O. E. , & Laktionov, P. P. (2015). Extracellular nucleic acids in urine: Sources, structure, diagnostic potential. Acta naturae, 7(3), 48–54.[CrossRef]. [PMC free article] [PubMed] [Google Scholar]
- Chennakrishnaiah, S. , Tsering, T. , Aprikian, S. , & Rak, J. (2019). Leukobiopsy – A possible new liquid biopsy platform for detecting oncogenic mutations. Frontiers in Pharmacology, 10, 1608. 10.3389/fphar.2019.01608[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christakoudi, S. , Runglall, M. , Mobillo, P. , Tsui, T. L. , Duff, C. , Domingo‐Vila, C. , Kamra, Y. , Delaney, F. , Montero, R. , Spiridou, A. , Kassimatis, T. , Phin‐Kon, S. , Tucker, B. , Farmer, C. , Strom, T. B. , Lord, G. M. , Rebollo‐Mesa, I. , Stahl, D. , Sacks, S. , …, Chowdhury, P. (2019). Development of a multivariable gene‐expression signature targeting T‐cell‐mediated rejection in peripheral blood of kidney transplant recipients validated in cross‐sectional and longitudinal samples. EBioMedicine, 41, 571–583. 10.1016/j.ebiom.2019.01.060[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimuccio, V. , Peruzzi, L. , Brizzi, M. F. , Cocchi, E. , Fop, F. , Boido, A. , Gili, M. , Gallo, S. , Biancone, L. , Camussi, G. , & Bussolati, B. (2020). Acute and chronic glomerular damage is associated with reduced CD133 expression in urinary extracellular vesicles. American Journal of Physiology. Renal Physiology, 318(2), F486–F495. 10.1152/ajprenal.00404.2019[CrossRef] [DOI] [PubMed] [Google Scholar]
- Dimuccio, V. , Ranghino, A. , Praticò Barbato, L. , Fop, F. , Biancone, L. , Camussi, G. , & Bussolati, B. (2014). Urinary CD133+ extracellular vesicles are decreased in kidney transplanted patients with slow graft function and vascular damage. PloS One, 9(8), e104490. 10.1371/journal.pone.0104490[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Fekih, R. , Hurley, J. , Tadigotla, V. , Alghamdi, A. , Srivastava, A. , Coticchia, C. , Choi, J. , Allos, H. , Yatim, K. , Alhaddad, J. , Eskandari, S. , Chu, P. , Mihali, A. B. , Lape, I. T. , Lima Filho, M. P. , Aoyama, B. T. , Chandraker, A. , Safa, K. , Markmann, J. F. , …, Azzi, J. R. (2021). Discovery and validation of a urinary exosome mRNA signature for the diagnosis of human kidney transplant rejection. Journal of the American Society of Nephrology: JASN, 32(4), 994–1004. 10.1681/ASN.2020060850[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdbrügger, U. , Blijdorp, C. J. , Bijnsdorp, I. V. , Borràs, F. E. , Burger, D. , Bussolati, B. , Byrd, J. B. , Clayton, A. , Dear, J. W. , Falcón‐Pérez, J. M. , Grange, C. , Hill, A. F. , Holthöfer, H. , Hoorn, E. J. , Jenster, G. , Jimenez, C. R. , Junker, K. , Klein, J. , Knepper, M. A. , …, Martens‐Uzunova, E. S. (2021). Urinary extracellular vesicles: A position paper by the Urine Task Force of the International Society for Extracellular Vesicles. Journal of Extracellular Vesicles, 10(7), e12093. 10.1002/jev2.12093[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippone, E. J. , & Farber, J. L. (2021). The monitoring of donor‐derived cell‐free DNA in kidney transplantation. Transplantation, 105(3), 509–516. 10.1097/TP.0000000000003393[CrossRef] [DOI] [PubMed] [Google Scholar]
- Freeman, D. W. , Noren Hooten, N. , Eitan, E. , Green, J. , Mode, N. A. , Bodogai, M. , Zhang, Y. , Lehrmann, E. , Zonderman, A. B. , Biragyn, A. , Egan, J. , Becker, K. G. , Mattson, M. P. , Ejiogu, N. , & Evans, M. K. (2018). Altered extracellular vesicle concentration, cargo, and function in diabetes. Diabetes, 67(11), 2377–2388. 10.2337/db17-1308[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielis, E. M. , Ledeganck, K. J. , Dendooven, A. , Meysman, P. , Beirnaert, C. , Laukens, K. , De Schrijver, J. , Van Laecke, S. , Van Biesen, W. , Emonds, M. P. , De Winter, B. Y. , Bosmans, J. L. , Del Favero, J. , & Abramowicz, D. (2020). The use of plasma donor‐derived, cell‐free DNA to monitor acute rejection after kidney transplantation. Nephrology, Dialysis, Transplantation: Official Publication of the European Dialysis and Transplant Association ‐ European Renal Association, 35(4), 714–721. 10.1093/ndt/gfz091[CrossRef] [DOI] [PubMed] [Google Scholar]
- González, E. , & Falcón‐Pérez, J. M. (2015). Cell‐derived extracellular vesicles as a platform to identify low‐invasive disease biomarkers. Expert Review of Molecular Diagnostics, 15(7), 907–923. 10.1586/14737159.2015.1043272[CrossRef] [DOI] [PubMed] [Google Scholar]
- Haas, M. , Loupy, A. , Lefaucheur, C. , Roufosse, C. , Glotz, D. , Seron, D. , Nankivell, B. J. , Halloran, P. F. , Colvin, R. B. , Akalin, E. , Alachkar, N. , Bagnasco, S. , Bouatou, Y. , Becker, J. U. , Cornell, L. D. , Duong Van Huyen, J. P. , Gibson, I. W. , Kraus, E. S. , Mannon, R. B. , …, Mengel, M. (2018). The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell–mediated rejection, antibody‐mediated rejection, and prospects for integrative endpoints for next‐generation clinical trials. American Journal of Transplantation, 18(2), 293–307. 10.1111/ajt.14625[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halloran, P. F. , Reeve, J. , Madill‐Thomsen, K. S. , Demko, Z. , Prewett, A. , Billings, P. , & Trifecta Investigators . (2022). The Trifecta Study: Comparing plasma levels of donor‐derived cell‐free DNA with the molecular phenotype of kidney transplant biopsies. Journal of the American Society of Nephrology : JASN, 33(2), 387–400. 10.1681/ASN.2021091191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan, S. , Israni, A. K. , & Danovitch, G. (2021). Long‐term survival after kidney transplantation. The New England Journal of Medicine, 385(8), 729–743. 10.1056/NEJMra2014530[CrossRef] [DOI] [PubMed] [Google Scholar]
- Hogan, J. J. , Mocanu, M. , & Berns, J. S. (2016). The native kidney biopsy: Update and evidence for best practice. Clinical Journal of the American Society of Nephrology : CJASN, 11(2), 354–362. 10.2215/CJN.05750515[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, E. , Sethi, S. , Peng, A. , Najjar, R. , Mirocha, J. , Haas, M. , Vo, A. , & Jordan, S. C. (2019). Early clinical experience using donor‐derived cell‐free DNA to detect rejection in kidney transplant recipients. American Journal of Transplantation : Official Journal of the American Society of Transplantation and the American Society of Transplant Surgeons, 19(6), 1663–1670. 10.1111/ajt.15289[CrossRef] [DOI] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Fenix, A. M. , Franklin, J. L. , Higginbotham, J. N. , Zhang, Q. , Zimmerman, L. J. , Liebler, D. C. , Ping, J. , Liu, Q. , Evans, R. , Fissell, W. H. , Patton, J. G. , Rome, L. H. , Burnette, D. T. , & Coffey, R. J. (2019). Reassessment of exosome composition Cell, 177(2), 428–445.e18. 10.1016/j.cell.2019.02.029[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerič Kokelj, B. , Štalekar, M. , Vencken, S. , Dobnik, D. , Kogovšek, P. , Stanonik, M. , Arnol, M. , & Ravnikar, M. (2021). Feasibility of droplet digital PCR analysis of plasma cell‐free DNA from kidney transplant patients. Frontiers in Medicine, 8, 748668. 10.3389/fmed.2021.748668[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert, C. , Melo, S. A. , Protopopov, A. , Tang, J. , Seth, S. , Koch, M. , Zhang, J. , Weitz, J. , Chin, L. , Futreal, A. , & Kalluri, R. (2014). Identification of double‐stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. The Journal of Biological Chemistry, 289(7), 3869–3875. 10.1074/jbc.C113.532267[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman, H. M. , Bennett, L. E. , Mcbride, M. A. , & Ellison, M. D. (1997). The expanded donor. Transplantation Reviews, 11(4), 165–190. 10.1016/S0955-470X(97)80037-7[CrossRef] [DOI] [Google Scholar]
- Kawamura, Y. , Yamamoto, Y. , Sato, T. A. , & Ochiya, T. (2017). Extracellular vesicles as trans‐genomic agents: Emerging roles in disease and evolution. Cancer Science, 108(5), 824–830. 10.1111/cas.13222[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidney Disease: Improving Global Outcomes (KDIGO) Transplant Work Group . (2009). KDIGO clinical practice guideline for the care of kidney transplant recipients. American Journal of Transplantation: Official Journal of the American Society of Transplantation and the American Society of Transplant Surgeons, 9(3), S1–S155. 10.1111/j.1600-6143.2009.02834.x[CrossRef] [DOI] [PubMed] [Google Scholar]
- Kobayashi, K. , & Fukuoka, S. (2001). Conditions for solubilization of Tamm‐Horsfall protein/uromodulin in human urine and establishment of a sensitive and accurate enzyme‐linked immunosorbent assay (ELISA) method. Archives of Biochemistry and Biophysics, 388(1), 113–120. 10.1006/abbi.2000.2265[CrossRef] [DOI] [PubMed] [Google Scholar]
- Kubota, T. , Kanai, Y. , & Miyasaka, N. (1990). Interpretation of the cross‐reactivity of anti‐DNA antibodies with cell surface proteins: The role of cell surface histones. Immunology Letters, 23(3), 187–193. 10.1016/0165-2478(90)90190-2[CrossRef] [DOI] [PubMed] [Google Scholar]
- Laktionov, P. P. , Tamkovich, S. N. , Rykova, E. Y. , Bryzgunova, O. E. , Starikov, A. V. , Kuznetsova, N. P. , & Vlassov, V. V. (2004). Cell‐surface‐bound nucleic acids: Free and cell‐surface‐bound nucleic acids in blood of healthy donors and breast cancer patients. Annals of the New York Academy of Sciences, 1022, 221–227. 10.1196/annals.1318.034[CrossRef] [DOI] [PubMed] [Google Scholar]
- Lázaro‐Ibáñez, E. , Lässer, C. , Shelke, G. V. , Crescitelli, R. , Jang, S. C. , Cvjetkovic, A. , García‐Rodríguez, A. , & Lötvall, J. (2019). DNA analysis of low‐ and high‐density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. Journal of Extracellular Vesicles, 8(1), 1656993. 10.1080/20013078.2019.1656993[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro‐Ibáñez, E. , Sanz‐Garcia, A. , Visakorpi, T. , Escobedo‐Lucea, C. , Siljander, P. , Escobedo‐Lucea, Ã. , & Yliperttula, M. (2014). Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: Apoptotic bodies, microvesicles, and exosomes. The Prostate, 74(14), 1379–1390. 10.1002/pros.22853[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D. H. , Yoon, H. , Park, S. , Kim, J. S. , Ahn, Y. H. , Kwon, K. , Lee, D. , & Kim, K. H. (2018). Urinary exosomal and cell‐free DNA detects somatic mutation and copy number alteration in urothelial carcinoma of bladder. Scientific Reports, 8(1), 14707. 10.1038/s41598-018-32900-6[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J. H. , Lee, C. H. , Kim, K. Y. , Jung, H. Y. , Choi, J. Y. , Cho, J. H. , Park, S. H. , Kim, Y. L. , Baek, M. C. , Park, J. B. , Kim, Y. H. , Chung, B. H. , Lee, S. H. , & Kim, C. D. (2018). Novel urinary exosomal biomarkers of acute T cell‐mediated rejection in kidney transplant recipients: A cross‐sectional study. PloS One, 13(9), e0204204. 10.1371/journal.pone.0204204[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maire, C. L. , Fuh, M. M. , Kaulich, K. , Fita, K. D. , Stevic, I. , Heiland, D. H. , Welsh, J. A. , Jones, J. C. , Görgens, A. , Ricklefs, T. , Dührsen, L. , Sauvigny, T. , Joosse, S. A. , Reifenberger, G. , Pantel, K. , Glatzel, M. , Miklosi, A. G. , Felce, J. H. , Caselli, M. , …, Ricklefs, F. L. (2021). Genome‐wide methylation profiling of glioblastoma cell‐derived extracellular vesicle DNA allows tumor classification. Neuro‐Oncology, 23(7), 1087–1099. 10.1093/neuonc/noab012[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin, E. Z. , & Bratman, S. V. (2020). Bioactive DNA from extracellular vesicles and particles. Cell Death & Disease, 11(7), 584. 10.1038/s41419-020-02803-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengel, M. , Chapman, J. R. , Cosio, F. G. , Cavaillé‐Coll, M. W. , Haller, H. , Halloran, P. F. , Kirk, A. D. , Mihatsch, M. J. , Nankivell, B. J. , Racusen, L. C. , Roberts, I. S. , Rush, D. N. , Schwarz, A. , Serón, D. , Stegall, M. D. , & Colvin, R. B. (2007). Protocol biopsies in renal transplantation: Insights into patient management and pathogenesis. American Journal of Transplantation: Official Journal of the American Society of Transplantation and the American Society of Transplant Surgeons, 7(3), 512–517. 10.1111/j.1600-6143.2006.01677.x[CrossRef] [DOI] [PubMed] [Google Scholar]
- Miranda, K. C. , Bond, D. T. , Mckee, M. , Skog, J. , Păunescu, T. G. , Da Silva, N. , Brown, D. , & Russo, L. M. (2010). Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney International, 78(2), 191–199. 10.1038/ki.2010.106[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouliere, F. , Robert, B. , Arnau Peyrotte, E. , Del Rio, M. , Ychou, M. , Molina, F. , Gongora, C. , & Thierry, A. R. (2011). High fragmentation characterizes tumour‐derived circulating DNA. PloS One, 6(9), e23418. 10.1371/journal.pone.0023418[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nankivell, B. J. , & Kuypers, D. R. (2011). Diagnosis and prevention of chronic kidney allograft loss. Lancet (London, England), 378(9800), 1428–1437. 10.1016/S0140-6736(11)60699-5[CrossRef] [DOI] [PubMed] [Google Scholar]
- Németh, A. , Orgovan, N. , Sódar, B. W. , Osteikoetxea, X. , Pálóczi, K. , Szabó‐Taylor, K. E. , Vukman, K. V. , Kittel, Ã. , Turiák, L. , Wiener, Z. , Tóth, S. , Drahos, L. , Vékey, K. , Horvath, R. , & Buzás, E. I. (2017). Antibiotic‐induced release of small extracellular vesicles (exosomes) with surface‐associated DNA. Scientific Reports, 7(1), 8202. 10.1038/s41598-017-08392-1[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oellerich, M. , Sherwood, K. , Keown, P. , Schütz, E. , Beck, J. , Stegbauer, J. , Rump, L. C. , & Walson, P. D. (2021). Liquid biopsies: Donor‐derived cell‐free DNA for the detection of kidney allograft injury. Nature Reviews. Nephrology, 17(9), 591–603. 10.1038/s41581-021-00428-0[CrossRef] [DOI] [PubMed] [Google Scholar]
- Oellerich, M. , Shipkova, M. , Asendorf, T. , Walson, P. D. , Schauerte, V. , Mettenmeyer, N. , Kabakchiev, M. , Hasche, G. , Gröne, H. J. , Friede, T. , Wieland, E. , Schwenger, V. , Schütz, E. , & Beck, J. (2019). Absolute quantification of donor‐derived cell‐free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study. American Journal of Transplantation: Official Journal of the American Society of Transplantation and the American Society of Transplant Surgeons, 19(11), 3087–3099. 10.1111/ajt.15416[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshikawa‐Hori, S. , Yokota‐Ikeda, N. , Sonoda, H. , & Ikeda, M. (2019). Urinary extracellular vesicular release of aquaporins in patients with renal transplantation. BMC Nephrology, 20(1), 216. 10.1186/s12882-019-1398-7[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. , Lin, H. Y. , Assaker, J. P. , Jeong, S. , Huang, C. H. , Kurdi, A. , Lee, K. , Fraser, K. , Min, C. , Eskandari, S. , Routray, S. , Tannous, B. , Abdi, R. , Riella, L. , Chandraker, A. , Castro, C. M. , Weissleder, R. , Lee, H. , & Azzi, J. R. (2017). Integrated kidney exosome analysis for the detection of kidney transplant rejection. ACS Nano, 11(11), 11041–11046. 10.1021/acsnano.7b05083[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, R. S. , Almokayad, I. , Collins, A. , Raj, D. , & Jagadeesan, M. (2021). Donor‐derived cell‐free DNA: Advancing a novel assay to new heights in renal transplantation. Transplantation Direct, 7(3), e664. 10.1097/TXD.0000000000001098[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomatto, M. A. C. , Gai, C. , Bussolati, B. , & Camussi, G. (2017). Extracellular vesicles in renal pathophysiology. Frontiers in Molecular Biosciences, 4, 37. 10.3389/fmolb.2017.00037[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabant, M. , Amrouche, L. , Lebreton, X. , Aulagnon, F. , Benon, A. , Sauvaget, V. , Bonifay, R. , Morin, L. , Scemla, A. , Delville, M. , Martinez, F. , Timsit, M. O. , Duong Van Huyen, J. P. , Legendre, C. , Terzi, F. , & Anglicheau, D. (2015). Urinary C‐X‐C motif chemokine 10 independently improves the noninvasive diagnosis of antibody‐mediated kidney allograft rejection. Journal of the American Society of Nephrology: JASN, 26(11), 2840–2851. 10.1681/ASN.2014080797[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranghino, A. , Dimuccio, V. , Papadimitriou, E. , & Bussolati, B. (2015). Extracellular vesicles in the urine: Markers and mediators of tissue damage and regeneration. Clinical Kidney Journal, 8(1), 23–30. 10.1093/ckj/sfu136[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes, D. C. J. , Hinsman, E. J. , & Rhodes, J. A. (1993). Tamm‐Horsfall glycoprotein binds IgG with high affinity. Kidney International, 44(5), 1014–1021. 10.1038/ki.1993.343[CrossRef] [DOI] [PubMed] [Google Scholar]
- RStudio Team . (2020). (n.d.). RStudio: Integrated Development for R. http://www.rstudio.com/
- Rykova, E. Y. , Morozkin, E. S. , Ponomaryova, A. A. , Loseva, E. M. , Zaporozhchenko, I. A. , Cherdyntseva, N. V. , Vlassov, V. V. , & Laktionov, P. P. (2012). Cell‐free and cell‐bound circulating nucleic acid complexes: Mechanisms of generation, concentration and content. Expert Opinion on Biological Therapy, 12(1), S141–S153. 10.1517/14712598.2012.673577[CrossRef] [DOI] [PubMed] [Google Scholar]
- Sedej, I. , Tušek Žnidarič, M. , Dolžan, V. , Lenassi, M. , & Arnol, M. (2021). Optimization of isolation protocol and characterization of urinary extracellular vesicles as biomarkers of kidney allograft injury. Clinical Nephrology, 96(1), 107–113. 10.5414/CNP96S19[CrossRef] [DOI] [PubMed] [Google Scholar]
- Shelke, G. , Jang, S. C. , Yin, Y. , Lässer, C. , & Lötvall, J. (2016). Human mast cells release extracellular vesicle‐associated DNA. Matters (Zürich), 10.19185/matters.201602000034[CrossRef] [DOI] [Google Scholar]
- Sigdel, T. , Archila, F. , Constantin, T. , Prins, S. , Liberto, J. , Damm, I. , Towfighi, P. , Navarro, S. , Kirkizlar, E. , Demko, Z. , Ryan, A. , Sigurjonsson, S. , Sarwal, R. , Hseish, S. C. , Chan‐On, C. , Zimmermann, B. , Billings, P. , Moshkevich, S. , & Sarwal, M. (2018). Optimizing detection of kidney transplant injury by assessment of donor‐derived cell‐free DNA via massively multiplex PCR. Journal of Clinical Medicine, 8(1), 19. 10.3390/jcm8010019[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigdel, T. K. , Vitalone, M. J. , Tran, T. Q. , Dai, H. , Hsieh, S. C. , Salvatierra, O. , & Sarwal, M. M. (2013). A rapid noninvasive assay for the detection of renal transplant injury. Transplantation, 96(1), 97–101. 10.1097/TP.0b013e318295ee5a[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvers, C. R. , Miyamoto, H. , Messing, E. M. , Netto, G. J. , & Lee, Y. F. (2017). Characterization of urinary extracellular vesicle proteins in muscle‐invasive bladder cancer. Oncotarget, 8(53), 91199–91208. 10.18632/oncotarget.20043[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solez, K. , Axelsen, R. A. , Benediktsson, H. , Burdick, J. F. , Cohen, A. H. , Colvin, R. B. , Croker, B. P. , Droz, D. , Dunnill, M. S. , Halloran, P. F. , Häyry, P. , Jennette, J. C. , Keown, P. A. , Marcussen, N. , Mihatsch, M. J. , Morozumi, K. , Myers, B. D. , Nast, C. C. , Olsen, S. , …, Yamaguchi, Y. (1993). International standardization of criteria for the histologic diagnosis of renal allograft rejection: The Banff working classification of kidney transplant pathology. Kidney International, 44(2), 411–422. 10.1038/ki.1993.259[CrossRef] [DOI] [PubMed] [Google Scholar]
- Sonoda, H. , Yokota‐Ikeda, N. , Oshikawa, S. , Kanno, Y. , Yoshinaga, K. , Uchida, K. , Ueda, Y. , Kimiya, K. , Uezono, S. , Ueda, A. , Ito, K. , & Ikeda, M. (2009). Decreased abundance of urinary exosomal aquaporin‐1 in renal ischemia‐reperfusion injury. American Journal of Physiology. Renal Physiology, 297(4), F1006–F1016. 10.1152/ajprenal.00200.2009[CrossRef] [DOI] [PubMed] [Google Scholar]
- Suthanthiran, M. , Schwartz, J. E. , Ding, R. , Abecassis, M. , Dadhania, D. , Samstein, B. , Knechtle, S. J. , Friedewald, J. , Becker, Y. T. , Sharma, V. K. , Williams, N. M. , Chang, C. S. , Hoang, C. , Muthukumar, T. , August, P. , Keslar, K. S. , Fairchild, R. L. , Hricik, D. E. , Heeger, P. S. , & Shaked, A. , Clinical Trials in Organ Transplantation 04 (CTOT‐04) Study Investigators . (2013). Urinary‐cell mRNA profile and acute cellular rejection in kidney allografts. The New England Journal of Medicine, 369(1), 20–31. 10.1056/NEJMoa1215555[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilágyi, M. , Pös, O. , Márton, É. , Buglyó, G. , Soltész, B. , Keserű, J. , Penyige, A. , Szemes, T. , & Nagy, B. (2020). Circulating Cell‐Free Nucleic Acids: Main Characteristics and Clinical Application. International Journal of Molecular Sciences, 21(18), 6827. 10.3390/ijms21186827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A. , Okada, R. , Nagao, K. , Kawamata, Y. , Hanyu, A. , Yoshimoto, S. , Takasugi, M. , Watanabe, S. , Kanemaki, M. T. , Obuse, C. , & Hara, E. (2017). Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nature Communications, 8, 15287. 10.1038/ncomms15287[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkovich, S. , & Laktionov, P. (2019). Cell‐surface‐bound circulating DNA in the blood: Biology and clinical application. IUBMB Life, 71(9), 1201–1210. 10.1002/iub.2070[CrossRef] [DOI] [PubMed] [Google Scholar]
- Tamura, T. , Yoshioka, Y. , Sakamoto, S. , Ichikawa, T. , & Ochiya, T. (2021). Extracellular vesicles as a promising biomarker resource in liquid biopsy for cancer. Extracellular Vesicles and Circulating Nucleic Acids, 10.20517/evcna.2021.06[CrossRef] [DOI] [Google Scholar]
- Torralba, D. , Baixauli, F. , Villarroya‐Beltri, C. , Fernández‐Delgado, I. , Latorre‐Pellicer, A. , Acín‐Pérez, R. , Martín‐Cófreces, N. B. , Jaso‐Tamame, Ã. L. , Iborra, S. , Jorge, I. , González‐Aseguinolaza, G. , Garaude, J. , Vicente‐Manzanares, M. , Enríquez, J. A. , Mittelbrunn, M. A. , & Sánchez‐Madrid, F. (2018). Priming of dendritic cells by DNA‐containing extracellular vesicles from activated T cells through antigen‐driven contacts. Nature Communications, 9(1), 2658. 10.1038/s41467-018-05077-9[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner, T. , Spinelli, C. , Minciacchi, V. R. , Balaj, L. , Zandian, M. , Conley, A. , Zijlstra, A. , Freeman, M. R. , Demichelis, F. , De, S. , Posadas, E. M. , Tanaka, H. , & Di Vizio, D. (2018). Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. Journal of Extracellular Vesicles, 7(1), 1505403. 10.1080/20013078.2018.1505403[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deun, J. , Mestdagh, P. , Agostinis, P. , Akay, Ö. , Anand, S. , Anckaert, J. , Martinez, Z. A. , Baetens, T. , Beghein, E. , Bertier, L. , Berx, G. , Boere, J. , Boukouris, S. , Bremer, M. , Buschmann, D. , Byrd, J. B. , Casert, C. , Cheng, L. , Cmoch, A. , …, Hendrix, A. (2017). EV‐TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nature Methods, 14(3), 228–232. 10.1038/nmeth.4185[CrossRef] [DOI] [PubMed] [Google Scholar]
- Van Niel, G. , D'angelo, G. , & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews. Molecular Cell Biology, 19(4), 213–228. 10.1038/nrm.2017.125[CrossRef] [DOI] [PubMed] [Google Scholar]
- Verhoeven, J. G. H. P. , Boer, K. , Van Schaik, R. H. N. , Manintveld, O. C. , Huibers, M. M. H. , Baan, C. C. , & Hesselink, D. A. (2018). Liquid biopsies to monitor solid organ transplant function: A review of new biomarkers. Therapeutic Drug Monitoring, 40(5), 515–525. 10.1097/FTD.0000000000000549[CrossRef] [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Hill, S. , Luther, J. M. , Hachey, D. L. , & Schey, K. L. (2012). Proteomic analysis of urine exosomes by multidimensional protein identification technology (MudPIT). Proteomics, 12(2), 329–338. 10.1002/pmic.201100477[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, D. , Yang, J. Y. C. , Sarwal, R. D. , Sigdel, T. K. , Liberto, J. M. , Damm, I. , Louie, V. , Sigdel, S. , Livingstone, D. , Soh, K. , Chakraborty, A. , Liang, M. , Lin, P. C. , & Sarwal, M. M. (2019). A novel multi‐biomarker assay for non‐invasive quantitative monitoring of kidney injury. Journal of Clinical Medicine, 8(4), 499. 10.3390/jcm8040499[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlam, J. B. , Ling, L. , Skene, A. , Kanellis, J. , Ierino, F. L. , Slater, H. R. , Bruno, D. L. , & Power, D. A. (2019). Diagnostic application of kidney allograft‐derived absolute cell‐free DNA levels during transplant dysfunction. American Journal of Transplantation : Official Journal of the American Society of Transplantation and the American Society of Transplant Surgeons, 19(4), 1037–1049. 10.1111/ajt.15142[CrossRef] [DOI] [PubMed] [Google Scholar]
- Wickham, H. (2016). ggplot2: Elegant graphics for data analysis (2nd ed.). Springer‐Verlag New York. 10.1007/978-3-319-24277-4 [DOI] [Google Scholar]
- Yáñez‐Mó, M. , Siljander, P. R. , Andreu, Z. , Bedina Zavec, A. , Borràs, F. E. , Buzas, E. I. , Buzas, K. , Casal, E. , Cappello, F. , Carvalho, J. , Colás, E. , Cordeiro‐Da Silva, A. , Fais, S. , Falcon‐Perez, J. M. , Ghobrial, I. M. , Giebel, B. , Gimona, M. , Graner, M. , Gursel, I. , …, De Wever, O. (2015). Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles, 4, 27066. 10.3402/jev.v4.27066[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. Y. C. , Sarwal, R. D. , Sigdel, T. K. , Damm, I. , Rosenbaum, B. , Liberto, J. M. , Chan‐On, C. , Arreola‐Guerra, J. M. , Alberu, J. , Vincenti, F. , & Sarwal, M. M. (2020). A urine score for noninvasive accurate diagnosis and prediction of kidney transplant rejection. Science Translational Medicine, 12(535), eaba2501. 10.1126/scitranslmed.aba2501[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates, A. G. , Pink, R. C. , Erdbrügger, U. , Siljander, P. R. , Dellar, E. R. , Pantazi, P. , Akbar, N. , Cooke, W. R. , Vatish, M. , Dias‐Neto, E. , Anthony, D. C. , & Couch, Y. (2022a). In sickness and in health: The functional role of extracellular vesicles in physiology and pathology in vivo: Part I: Health and Normal Physiology: Part I: Health and Normal Physiology. Journal of Extracellular Vesicles, 11(1), e12151. 10.1002/jev2.12151[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates, A. G. , Pink, R. C. , Erdbrügger, U. , Siljander, P. R. , Dellar, E. R. , Pantazi, P. , Akbar, N. , Cooke, W. R. , Vatish, M. , Dias‐Neto, E. , Anthony, D. C. , & Couch, Y. (2022b). In sickness and in health: The functional role of extracellular vesicles in physiology and pathology in vivo: Part II: Pathology: Part II: Pathology. Journal of Extracellular Vesicles, 11(1), e12190. 10.1002/jev2.12190[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Freitas, D. , Kim, H. S. , Fabijanic, K. , Li, Z. , Chen, H. , Mark, M. T. , Molina, H. , Martin, A. B. , Bojmar, L. , Fang, J. , Rampersaud, S. , Hoshino, A. , Matei, I. , Kenific, C. M. , Nakajima, M. , Mutvei, A. P. , Sansone, P. , Buehring, W. , …, Lyden, D. (2018). Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field‐flow fractionation. Nature Cell Biology, 20(3), 332–343. 10.1038/s41556-018-0040-4[CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Tong, K. L. , Li, P. K. , Chan, A. Y. , Yeung, C. K. , Pang, C. C. , Wong, T. Y. , Lee, K. C. , & Lo, Y. M. (1999). Presence of donor‐ and recipient‐derived DNA in cell‐free urine samples of renal transplantation recipients: Urinary DNA chimerism. Clinical Chemistry, 45(10), 1741–1746.[CrossRef]. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.
Data Availability Statement
We have submitted all relevant experimental data to the EV‐TRACK knowledgebase (EV‐TRACK ID: EV210292) (Van Deun et al., 2017).


