Abstract
2-Hydroxy-6-oxo-6-phenylhexa-2,4-dienoate (HOPDA) hydrolase (BphD) is a key determinant in the aerobic transformation of polychlorinated biphenyls (PCBs) by Burkholderia sp. strain LB400 (S. Y. K. Seah, G. Labbé, S. Nerdinger, M. Johnson, V. Snieckus, and L. D. Eltis, J. Biol. Chem. 275:15701–15708, 2000). To determine whether this is also true in divergent biphenyl degraders, the homologous hydrolase of Rhodococcus globerulus P6, BphDP6, was hyperexpressed, purified to apparent homogeneity, and studied by steady-state kinetics. BphDP6 hydrolyzed HOPDA with a kcat/Km of 1.62 (± 0.03) × 107 M−1 s−1 (100 mM phosphate [pH 7.5], 25°C), which is within 70% of that of BphDLB400. BphDP6 was also similar to BphDLB400 in that it catalyzed the hydrolysis of HOPDAs bearing chloro substituents on the phenyl moiety at least 25 times more specifically than those bearing chloro substituents on the dienoate moiety. However, the rhodococcal enzyme was significantly more specific for 9-Cl and 10-Cl HOPDAs, catalyzing the hydrolysis of 9-Cl, 10-Cl, and 9,10-diCl HOPDAs two- to threefold respectively, more specifically than HOPDA. Moreover, 4-Cl HOPDA competitively inhibited BphDP6 more effectively than 3-Cl HOPDA, which is the inverse of what was observed in BphDLB400. These results demonstrate that BphD is a key determinant in the aerobic transformation of PCBs by divergent biphenyl degraders, but that there exists significant diversity in the specificity of these biphenyl hydrolases.
Bioremediation is a technology in which biological systems are harnessed to clean up environmental pollutants. This technology has the potential to be less polluting, less expensive, and less invasive than others for destroying pollutants. Among the biological systems with potential for bioremediation, microorganisms are especially promising due to their catabolic diversity and their plasticity. One factor that can limit the efficacy of bioremediation strategies is the inability of existing microbial catabolic activities to degrade the target pollutant (34). Failure to degrade a target pollutant can arise from the inability of catabolic enzymes to transform the pollutant or its metabolites, as well as from the inhibition of these enzymes by metabolites. Such catabolic blocks are particularly problematic for structurally diverse pollutants, such as polychlorinated biphenyls (PCBs), whose occurrence in the biosphere has only recently become widespread.
PCBs were produced over a 50-year period for a wide variety of industrial uses. Although their production has been banned in the industrial world for over 20 years, the destruction of PCBs is of continued relevance due to their pervasiveness and persistence, their impact on fragile ecosystems, and human health concerns (24). A number of microorganisms are able to transform some of the congeners found in commercial formulations of PCBs. This transformation is effected aerobically by a pathway encoded by the bph gene cluster, the first four enzymatic activities of which transform biphenyl via a catecholic intermediate to benzoate and a pentanoate derivative (14). Among PCB-degrading strains, Rhodococcus globerulus P6 (variously identified as Acinetobacter sp. strain P6, Corynebacterium sp. strain MB1, and Arthrobacter sp. strain M5 [3]) and Burkholderia sp. strain LB400 (originally identified as Pseudomonas sp. strain LB400 [6]) transform a particularly large number of congeners. However, even these strains do not effectively mineralize lightly chlorinated congeners and transform but a few highly chlorinated congeners (5, 15, 31). The development of strains to effectively mineralize mixtures of PCBs requires the identification of recalcitrant or inhibitory catabolites and the development of strategies to degrade them.
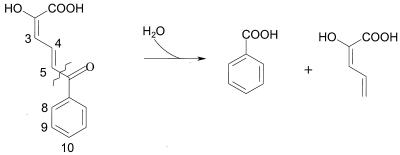
2-Hydroxy-6-oxo-6-phenylhexa-2,4-dienoate (HOPDA) hydrolase BphD (EC 3.7.1.8) has been identified as a key determinant in the aerobic degradation of PCBs in Burkholderia sp. strain LB400 (29). This α/β-fold serine hydrolase catalyzes the fourth step of the bph pathway, hydrolyzing HOPDA at a carbon-carbon bond to yield 2-hydroxypenta-2,4-dienoate and benzoate (Fig. 1) (27). Recent studies (29) with monochlorinated HOPDAs have established that 3-Cl and 4-Cl HOPDAs competitively inhibit BphD of strain LB400 (BphDLB400). Moreover, HOPDAs undergo a spontaneous transformation to acetophenones, although 3-Cl HOPDA is much more stable than 4-Cl HOPDA. Consistent with these observations, HOPDAs and chloroacetophenones accumulate in the culture medium when Burkholderia sp. strain LB400 is incubated with congeners that are predicted to give rise to 3-Cl and 4-Cl HOPDAs, respectively (6, 13, 27).
FIG. 1.
Hydrolysis of HOPDA catalyzed by BphD.
It is unclear whether the specificity of BphDLB400 represents that of other BphDs. For example, R. globerulus P6 transforms 4,4′-dichlorobiphenyl (4,4′-diClB) to an unidentified monochlorinated benzoate (15). As the biotransformation of 4,4′-diClB is expected to proceed via the formation of 3,10-diCl HOPDA, this observation suggests that BphD from R. globerulus P6 (BphDP6) hydrolyzes 3-Cl HOPDAs more specifically than BphDLB400 does. Interestingly, BphDP6 shares only 42% sequence identity with BphDLB400 (22). This is significantly less than the sequence identity between the other respective homologous enzymes of the bph pathway in these strains, which is between 52 and 61% (2). The study of evolutionary divergent BphDs is critical to establish structure-function relationships in this family of hydrolases and may also identify enzymes useful for the effective microbial degradation of PCBs.
We report here the heterologous expression, purification, and characterization of BphDP6. The specificity of the enzyme for monochlorinated HOPDAs was established by steady-state kinetic studies and was compared to that of BphDLB400. The specificity of both enzymes for two dichlorinated (diCl) HOPDAs was also determined. The specificities of the respective hydrolases are discussed in terms of their implications for the microbial degradation of PCBs.
MATERIALS AND METHODS
Chemicals.
4-Chloro-2,3-dihydroxybiphenyl (4-Cl DHB) was synthesized as described previously (26). All other chlorinated DHBs were synthesized and isolated as described elsewhere (20). Restriction enzymes and PfuI polymerase were from Amersham Pharmacia Biotech (Baie d'Urfe, Quebec, Canada) and Stratagene, respectively. All other chemicals were of analytical grade.
Bacterial strains and plasmids.
Strains used for protein expression or DNA propagation included Escherichia coli DH5α (17), BL21 (DE3) (33), and LE392 (7) and Pseudomonas putida KT2442 (18). Plasmids used in this work were pT7-7 (32, 33), pVLT31 (12), and pEMBL18 (13). E. coli strains were grown at 37°C, while P. putida was grown at 30°C. In experiments designed to test the expression of BphDP6, bacterial strains were grown on Luria-Bertani broth (LB) supplemented with carbenicillin (15 μg/ml; ICN Biomedicals Inc.) or tetracycline (12.5 μg/ml; ICN Biomedicals Inc.). Bacterial strains used for other purposes were grown on LB supplemented with the appropriate antibiotics.
DNA manipulation and amplification.
DNA was purified, digested, and ligated using standard protocols (28). The bphD gene of strain P6 was amplified by PCR using two synthetic oligonucleotides designed to be complementary to the 5′ and 3′ ends of the published bpdF sequence from Rhodococcus sp. strain M5, which encodes a HOPDA hydrolase (22). The respective sequences of these oligonucleotides were 5′-CGGGCATATGATCCAAAAAATTG-3′ and 5′-GGGGCTGCAGGTCAACTTAGATCAA-3′. These primers introduced NdeI and PstI restriction sites at the 5′ and 3′ ends, respectively, of the amplified product, facilitating subsequent cloning of the gene. The PCR mixture contained approximately 0.5 μg of BamH1-digested genomic DNA of R. globerulus P6, 0.75 U of Pfu DNA polymerase (Stratagene), 20 nmol of each deoxynucleoside triphosphate, and 100 pmol of each primer in a final volume of 100 μl. Twenty amplification cycles were performed as follows: 95°C for 1 min, 48°C for 1 min, and 72°C for 2 min. The PCR product was purified using a QIAquick PCR purification kit (Qiagen). DNA was purified from agarose gels using a Qiagen QIAEX-II gel extraction kit. Plasmid DNA was sequenced using an ABI model 373 Stretch DNA sequencer at the Nucleic Acid Analysis Unit, Université Laval. Sequencing reactions were performed according to the ABI dye-deoxy terminator protocol.
Purification of BphDP6.
Buffers containing 20 mM sodium HEPES (pH 7.5) were used throughout the purification. Chromatography was performed using an ÄKTA Explorer with resins and columns from Amersham Pharmacia Biotech. A crude extract prepared from 27 g of cells was loaded onto a Source 15Q anion-exchange column (2 by 9 cm) and eluted with a linear gradient of 0.05 to 0.25 M NaCl in 15 column volumes. Activity-containing fractions (0.11 M NaCl) were pooled and concentrated to 6 ml by ultrafiltration using an Amicon stirred cell equipped with a YM10 regenerated cellulose membrane. This preparation was brought to 5% saturation of ammonium sulfate and loaded onto a phenyl-Sepharose column (1 by 9 cm). The enzyme eluted around 1% saturated ammonium sulfate in a decreasing concentration gradient (5 to 0% in 5 column volumes). Activity-containing fractions from two runs were pooled, concentrated to 2.5 ml, and loaded onto a HiLoad 26/60 Superdex 200 gel filtration column. Fractions containing pure enzyme were pooled, concentrated by ultrafiltration as described above, and rediluted with fresh buffer. This was repeated several times to remove NaCl from the solution. Aliquots of concentrated enzyme were stored at −80°C.
Purification of other enzymes.
Recombinant BphDLB400 was purified as described previously (30). 2,3-Dihydroxybiphenyl dioxygenase (DHBD) of Burkholderia sp. strain LB400 was purified anaerobically as previously described (35).
Determination of protein concentration, purity, and molecular mass.
Protein concentrations were determined using a bicinchoninic acid protein assay reagent kit (Pierce Chemical Co.) and bovine serum albumin as the standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using a Bio-Rad Miniprotean II apparatus, and gels were stained with Coomassie blue according to established procedures (21). Molecular weight markers of the range 14,000 to 200,000 were used (Bio-Rad). The molecular mass of BphD was determined using high-pressure liquid chromatography-electrospray mass spectrometry (HPLC-ESMS). The instrumentation consisted of a microbore HPLC (Michrom UMA) connected on-line to a Perkin Elmer-Sciex model API III ESMS. Samples contained 5 mg of purified protein/ml. The molecular mass of the protein was determined using the Sciex deconvolution software.
Substrate preparation and determination of extinction coefficients.
Extinction coefficents for HOPDA and monochlorinated HOPDAs were reported previously (29). Extinction coefficients for diCl HOPDAs were determined by addition of excess quantities of DHBD to solutions containing 100 mM phosphate buffer (pH 7.5) and weighed amounts of diCl DHBs, followed by the measurement of their absorbance spectrophotometrically on the instrument described below.
Kinetic measurements.
Enzymatic activity was measured by following the consumption of the yellow substrate on a Varian Cary 3 spectrophotometer equipped with a thermojacketed cuvette holder. The spectrophotometer was interfaced to a microcomputer and controlled by Cary OS/2 multitasking software. The amount of enzyme used in each assay was adjusted so that the progress curve was linear for at least 2 min. Initial velocities were determined from a least-squares analysis of the linear portion of the progress curves using the kinetics module of the Cary software.
The standard activity assay was performed in a total volume of 1.0 ml of 100 mM ionic-strength potassium phosphate (pH 7.5) containing 10 μM HOPDA at 25.0 ± 0.1°C. The reaction was initiated by adding between 5 to 10 μl of an appropriately diluted enzyme preparation to the reaction cuvette. A reaction mixture prepared without the hydrolytic enzyme served as a reference. The reaction was monitored at 434 nm. One unit of enzymatic activity is defined as the quantity of enzyme required to consume 1 μmol of HOPDA per min.
Specificity experiments were carried out in a total volume of 1.0 ml of 100 mM ionic-strength potassium phosphate (pH 7.5) at 25.0 ± 0.1°C. Reactions were monitored at the wavelength of maximum absorbance of each HOPDA (29) (see Results). For specificity experiments, initial velocities were determined at substrate concentrations that ranged from 0.2 to 10 times the Km for that substrate. For inhibition experiments, HOPDA was used as a substrate and the concentration of inhibitor was varied from at least 0.5 to 3 times the Ki for that compound. At each concentration of inhibitor, the concentration of HOPDA was varied from 0.2 to 10 times its apparent Km. A total volume of 1 ml was used in each assay. Appropriate equations were fitted to the initial velocities determined at different substrate and inhibitor concentrations using the least-squares and dynamic weighting options of LEONORA (11). Best-fit parameters were calculated using a minimum of 10 independent data points. The validity of the kinetic models was evaluated using residual plots.
RESULTS
Construction of the expression vector.
Amplification of the bphD gene from BamHI-digested genomic DNA from R. globerulus P6 yielded a DNA fragment of about 900 bp, as estimated from agarose gels. This fragment was digested with PstI and NdeI, extracted from agarose gels after electrophoresis, purified using a gel extraction kit, and then ligated into appropriately digested pT7-7 plasmid to produce plasmid pSS76. The entire sequence of the gene was determined from both strands of two separate clones. The sequences were identical to that of the bpdF gene of Rhodococcus sp. strain M5 except that the GGC codons corresponding to glycines 31 and 115 in the M5 sequence were replaced by the alanine codon GCC in the P6 sequence. The bphD gene, together with an upstream ribosomal binding site of T7 phage gene 10 protein of the T7-7 vector, was isolated from pSS76 as an XbaI/PstI fragment and inserted into two plasmids, pEMBL18 (13) and pVLT31 (12), a broad-host-range expression vector. These constructs were designated pSS186 and pSS316, respectively.
Expression, purification, and evaluation of molecular mass.
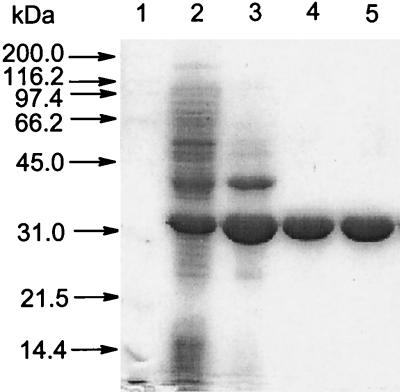
Among the expression systems tested, including various E. coli strains containing each of the three described plasmid constructions, BphDP6 was overexpressed only in P. putida KT2442 containing pSS316. The enzyme constituted about 15% of the total cellular protein as estimated from SDS-PAGE analysis and was therefore purified from this system. A total of 14 mg of BphDP6, representing a yield of approximately 8%, was purified from 27 g (wet weight) of cells by anion-exchange, hydrophobic interaction, and gel filtration chromatographies. The final preparation of BphDP6 was judged to be greater than 99% pure as judged from a Coomassie blue-stained denaturing gel (Fig. 2), and had a specific activity of 13.2 U/mg. HPLC-ESMS analysis revealed the subunit molecular mass of BphDP6 to be 32,583.0 Da. This agrees well with the predicted molecular mass of the protein with the N-terminal methionine removed (32,576.3 Da). The concentrated enzyme preparation (9 mg/ml) was stable for at least 12 months when stored frozen at −80°C. Diluted enzyme preparations (less than 0.1 mg/ml) were stabilized by adding bovine serum albumin to a final concentration of 1 mg/ml.
FIG. 2.
Coomassie-blue-stained SDS-polyacrylamide gel of purified BphDP6. The gel was loaded with samples of BphD from R. globerulus P6: crude extract (37 μg, lane 2); preparation after anion exchange (30 μg, lane 3); preparation after hydrophobic interaction (10 μg, lane 4); and preparation after gel filtration (11 μg, lane 5). The molecular masses of the proteins in the standard (lane 1) are indicated at the left.
Properties of HOPDAs.
The reported extinction coefficients of HOPDA and monochlorinated HOPDAs (29) were used in this study. The coefficients for 9,10-diCl and 9,11-diCl HOPDAs were 27.5 mM−1 cm−1 (λmax = 439 nm) and 27.0 mM−1 cm−1 (λmax = 438nm), respectively. The half lives of these compounds were 38 and 51 h, respectively, which are comparable to those of 9-Cl and 10-Cl HOPDAs (50 and 48 h, respectively [29]).
Steady-state kinetic analysis.
BphDP6 was found to obey classic Michaelis-Menten kinetics for all of the substrates tested here. Interestingly, the Km of BphDP6 for HOPDA was over twice that of BphDLB400, although the specificity constant (kcat/Km) of BphDP6 for this compound was within 70% of that of BphDLB400 (Table 1).
TABLE 1.
Steady-state kinetic parameters of BphD with different chlorinated substratesa
| HOPDA substituent | BphDP6
|
BphDLB400
|
||||
|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (10−5) (M−1 s−1) | km (μM) | kcat (s−1) | kcat/Km (10−5) (M−1 s−1) | |
| None | 0.47 (0.02) | 7.6 (0.1) | 162 (3) | 0.19 (0.01) | 4.18 (0.03) | 225 (6) |
| 3-Cl | 6.9 (0.3) | 0.0172 (0.0004) | 0.0252 (0.0007) | 0.54 (0.03) | 0.0089 (0.0001) | 0.16 (0.01) |
| 4-Cl | 0.40 (0.02)b | NDc | 3.6 (0.2)b | 0.00059 (6 × 10−5)d | 0.0016 (0.0002)d | |
| 4-OH | 4.4 (0.2)b | ND | 0.95 (0.04)b | 0.0055 (0.0008)d | 0.058 (0.008)d | |
| 5-Cl | 18 (1) | 1.03 (0.04) | 0.57 (0.03) | 4.9 (0.1) | 1.53 (0.01) | 3.11 (0.05) |
| 8-Cl | 1.29 (0.04) | 1.74 (0.02) | 13.5 (0.3) | 0.33 (0.02) | 2.10 (0.03) | 64 (3) |
| 9-Cl | 0.43 (0.01) | 15.3 (0.1) | 360 (4) | 0.46 (0.03) | 3.9 (0.1) | 85 (4) |
| 10-Cl | 0.65 (0.01) | 33.7 (0.2) | 517 (8) | 0.13 (0.01) | 3.13 (0.04) | 246 (13) |
| 9,10-diCl | 0.75 (0.03) | 29.0 (0.3) | 387 (13) | 0.154 (0.009) | 2.23 (0.03) | 143 (7) |
| 9,11-diCl | 1.04 (0.01) | 11.2 (0.2) | 108 (3) | 0.031 (0.001) | 0.144 (0.001) | 47 (2) |
Experiments were performed in 100 mM potassium phosphate buffer (pH 7.5) at 25°C. Kinetic parameters of BphDLB400 for HOPDA and monosubstituted HOPDAs have been previously published (29). Values in parentheses indicate standard errors.
Kic, determined from inhibition experiments as described in Results.
ND, not determined.
Calculated from specific activities and/or assuming Kic ∼ Km as described previously(29).
The specificity of BphDP6 for chlorinated HOPDAs displayed a marked dependence on the position of the chloro substituents (Table 1). In general, HOPDAs possessing a chloro substituent on the phenyl moiety (positions 8, 9, and 10) were very good substrates for BphDP6. In contrast, HOPDAs possessing a chloro substituent on the dienoate moiety (positions 3, 4, and 5) were very poor substrates for BphDP6. As summarized in Table 1, this is generally similar to the reported specificity of BphDLB400 (29).
Careful examination of the specificity data reveals at least two significant differences between BphDP6 and BphDLB400. The first of these concerns substrates with chloro substituents in the 9 or 10 position. Thus, BphDP6 catalyzed the hydrolysis of 9-Cl, 10-Cl, 9,10-diCl, and 9,11-diCl HOPDAs two- to fourfold more specifically than BphDLB400 did (Table 1). Moreover, with the exception of 9,11-diCl HOPDA, these compounds were all substantially better substrates for BphDP6 than nonchlorinated HOPDA. Comparisons of kcat values are even more striking. For example, BphDP6 turned over 9,11-diCl HOPDA almost 2 orders of magnitude faster than BphDLB400 did. The corollary of this is that BphDLB400 had very low Km values for some of these HOPDAs. Thus, the Km of BphDLB400 was sixfold lower for 9,11-diCl HOPDA than for nonchlorinated HOPDA. The second difference between BphDP6 and BphDLB400 concerns their respective specificities for 5- and 8-Cl HOPDAs: BphDLB400 is approximately fivefold more specific for these substrates than BphDP6.
BphDP6 had an exceptionally low specificity for 3-Cl HOPDA and no detectable activity toward either 4-Cl HOPDA or its spontaneous transformation product, 4-OH HOPDA. The low reactivity of BphDLB400 toward these same compounds was largely due to their slow catalytic turnover. Accordingly, each of these three compounds competitively inhibits the BphDLB400-catalyzed hydrolysis of HOPDA (29). Therefore, the inhibition of the BphDP6-catalyzed hydrolysis of HOPDA by 4-Cl and 4-OH HOPDAs was investigated. When the data obtained using freshly prepared 4-Cl HOPDA were fit to an equation describing competitive inhibition, random trends in the residuals were observed and an analysis of variance indicated an insignificant lack of fit. An analysis of variance of the fit of the data to an equation describing mixed inhibition also indicated an insignificant lack of fit. However, a negative value for the uncompetitive inhibition constant, Kiu, was obtained. Finally, fits of the data to an equation describing uncompetitive inhibition yielded nonrandom trends in the residuals and a highly significant lack of fit (results not shown). This analysis indicated that the BphDP6-catalyzed cleavage of HOPDA was inhibited by 4-Cl HOPDA in a competitive fashion with a Kic of 0.40 ± 0.02 μM.
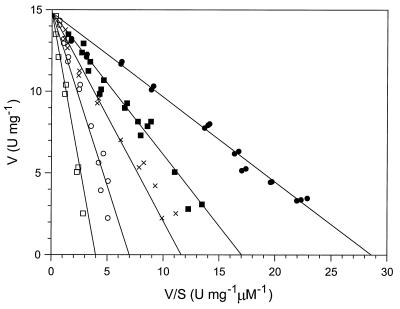
Inhibition experiments were also performed using a solution that contained a 19:1 mixture of 4-OH and 4-Cl HOPDAs, based on electronic absorption spectra. An analysis of the data revealed that this mixture competitively inhibited the BphD-catalyzed hydrolysis of HOPDA less strongly (Kic = 4.2 ± 0.1 μM [Fig. 3]) than did freshly prepared 4-Cl HOPDA. Considering this value, the Kic of 4-Cl HOPDA, and the relative concentrations of 4-Cl and 4-OH HOPDAs, the Kic of the 4-OH HOPDA was calculated to be 4.4 ± 0.2 μM.
FIG. 3.
Woolf-Augustinsson-Hofstee plot of the inhibition of the BphDP6-catalyzed hydrolysis of HOPDA by 4-OH HOPDA. Experiments were performed using 0 μM (●), 2.9 μM (■), 6.1 μM (×), 13.0 μM (○), and 26.0 μM (□) 4-substituted HOPDA (100 mM phosphate [pH 7.5], 25°C). The solution of 4-substituted HOPDA contained a mixture of approximately 5% 4-Cl HOPDA and 95% 4-OH HOPDA. The lines represent a best fit of the data to an equation describing competitive inhibition using the least-squares and dynamic weighting options of LEONORA. The fitted parameters are Kic = 4.2 ± 0.2 μM, Km = 0.52 ± 0.01 μM, and Vmax = 14.81 ± 0.01 U/mg.
These inhibition data reveal a further difference in the reactivities of the hydrolases: 4-Cl HOPDA is a more potent inhibitor of BphDP6 than 3-Cl HOPDA, whereas 3-Cl HOPDA is a more potent inhibitor of BphDLB400 than 4-Cl HOPDA. In particular, the Kic of BphDP6 for 4-Cl HOPDA was very similar to Km of the enzyme for HOPDA, whereas the Kic of BphDP6 for 3-Cl HOPDA was approximately 15-fold higher (Table 1; note that Kic ∼ Km for these poorly transformed compounds [29]). In contrast, in BphDLB400, it is the Kic for 4-Cl HOPDA that is most similar to the Km of the enzyme for HOPDA. It is further noted that the relative magnitude of Kic for 4-Cl and 4-OH HOPDA is reversed in the two enzymes (Table 1).
DISCUSSION
This work describes the efficient heterologous expression and purification of BphD of R. globerulus P6, a serine hydrolase involved in the catabolism of biphenyl and some PCB congeners. Mass spectrometry analysis of the purified recombinant protein indicates that the N-terminal methionine is removed. Interestingly, heterologous expression of BphDP6 was observed only in a pseudomonad host and not in those E. coli-based systems that worked well for BphDLB400 (30). The heterologous expression of biphenyl dioxygenase of R. globerulus P6 was also observed only in P. putida KT2442 (25). Inefficient expression of the dioxygenase in E. coli was attributed to posttranscriptional host factors. Inspection of the nucleotide sequence of bphDP6 revealed the presence of eight arginine codons (AGG and CGA), and six proline codons (CCC) that occur at a frequency of less than 1% in E. coli (19). However, mutation of a cluster of four of these codons, corresponding to residues 28, 33, 35, and 36, to frequently used codons resulted in only a slight improvement of the expression of BphDP6 in E. coli (results not shown). Interestingly, a histidine-tagged variant of the rhodococcal dioxygenase is well expressed in E. coli (10). However, the recombinant dioxygenase did not contain its complement of Rieske-type iron sulfur cluster.
The steady-state kinetic data indicate that the respective specificities of BphDP6 and BphDLB400 are in some respects quite similar (Table 1), consistent with the notion that both enzymes evolved to degrade biphenyl. Accordingly, the specificity of BphDP6 for HOPDA is similar to that of BphDLB400. Moreover, to a first approximation, the two hydrolases displayed the same specificity toward monochlorinated HOPDAs: those bearing chloro substituents on the phenyl moiety are good substrates, while those bearing chloro substituents on the dienoate moiety are poor substrates if not competitive inhibitors. Considering that BphDP6 and BphDLB400 share 42% sequence identity, it seems likely that their specificities are representative of the majority of BphDs identified to date. However, it would be of interest to verify the specificity of even more divergent BphDs, such as that of Rhodococcus sp. strain RHA1 (23), which shares less than 30% sequence identity with either BphDLB400 or BphDP6.
The reactivity of BphDP6 did, however, differ from that of BphDLB400 in at least three respects. First, BphDP6 catalyzed the hydrolysis of 9-Cl and 10-Cl HOPDAs at least twofold more specifically than BphDLB400 did. Second, BphDLB400 was approximately fivefold more specific for 5- and 8-Cl HOPDAs than BphDP6. Finally, 4-Cl HOPDA was a more potent inhibitor of BphDP6 than 3-Cl HOPDA, whereas 3-Cl HOPDA was a more potent inhibitor of BphDLB400 than 4-Cl HOPDA. Thus, the respective specificities of BphDP6 and BphDLB400 for chlorinated metabolites are complementary to a certain extent.
In a recently published study, the specificity of BphDLB400 for chlorinated HOPDAs was found to be largely consistent with the results of in vivo studies in which Burkholderia sp. strain LB400 was incubated with specific congeners. Accordingly, the accumulation of yellow-colored HOPDAs was observed when Burkholderia sp. strain LB400 was incubated with congeners such as 4,4′-diClB, 2,4,4′-triClB, and 2,5,4′-triClB, which are predicted to yield 3-Cl HOPDAs (5). Similarly, chloroacetophenones were detected when this strain was incubated with congeners such as 2,3′-diClB and 2,3,3′-triClB, which are predicted to yield unstable 4-Cl HOPDAs.
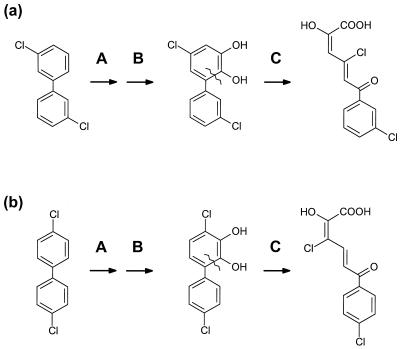
The specificity of BphDP6 for chlorinated HOPDAs extends the interpretation of in vivo studies and raises some important questions. Consistent with the low specificity of this enzyme for 3-Cl HOPDAs, yellow-colored HOPDAs accumulate when R. globerulus P6 is incubated with 2,4,4′-triClB and 2,5,4′-triClB (15, 16). However, chlorobenzoates were reported to be the principal metabolites when the strain was incubated with 3,3′-diClB and 4,4′-diClB (15), which would be predicted to give rise to 4,9-diCl and 3,10-diCl HOPDA, respectively (Fig. 4). As benzoates are not formed spontaneously from HOPDAs (29), the in vivo data suggest that R. globerulus P6 harbors an enzyme which hydrolyzes 3,10-diCl, and 4,9-diCl HOPDA. It is possible that this enzyme is not the BphDP6 characterized in this study. Significantly, R. globerulus P6 contains three DHBD isoenzymes, the genes of which map to different locations in the genome (4). Moreover, the dibenzofuran-degrading strain Sphingomonas sp. strain RW1 contains at least three BphD-type hydrolases (1, 8).
FIG. 4.
Proposed transformations of 3,3′-diClB to 4,9-diCl HOPDA (a) and 4,4′-diClB to 3,10-diCl HOPDA (b) in R. globerulus P6. Enzyme-catalyzed reactions are identified as A (biphenyl dioxygenase), B (2,3-dihydro-DHB dehydrogenase), and C (DHBD). The absolute configuration of the HOPDAs is not intended.
The transformation of 3,3′-diClB and 4,4′-diClB to chlorobenzoates by R. globerulus P6 might alternatively be explained by the influence of chloro substituents on the phenyl ring. Thus, BphDP6 may be able to hydrolyze 3,10-diCl and 4,9-diCl HOPDA more efficiently than 3-Cl and 4-Cl HOPDA. Significantly, 9-Cl and 10-Cl HOPDAs are better substrates for the enzyme than unchlorinated HOPDA (Table 1). The reactivity of HOPDAs that are chlorinated on both the dienoate and phenyl moieties is particularly intriguing in light of the low Km of BphDLB400 for 9,11-diCl HOPDA. Should meta- and para-chloro HOPDA substituents increase the affinity of the enzyme for these compounds, then compounds such as 3,9,11-triCl HOPDA may be a exceptionally potent inhibitors of BphDLB400. Further studies on the specificity of polychlorinated HOPDAs are required to resolve these issues.
The specificity and inhibition data point to significant electronic or structural differences in the respective active sites of BphDP6 and BphDLB400. For example, the higher Kic of BphDLB400 for 4-Cl HOPDA than for 4-OH HOPDA was suggested to arise from the configurations of these compounds (29). Thus, the proposed configuration of 4-OH HOPDA is more similar to that of the trans transoid enol tautomer of the unchlorinated substrate. However, the Kic of BphDP6 for 4-Cl HOPDA is an order of magnitude lower than that for 4-OH HOPDA, suggesting that there is a fundamental difference in how BphDP6 and BphDLB400 recognize the 4-substituted HOPDAs. Differences in these enzymes' active sites are also suggested by their different specificities for 5-Cl and 8-Cl HOPDA. Chloro substituents at these positions are relatively close to the site of nucleophilic attack at C-6 of HOPDA and thus could sterically hinder the binding of the substrate or its intermediates to the enzyme. Regardless of the origin of these deleterious effects, they are more significant in BphDP6 than in BphDLB400. The relevant differences in the active sites of these proteins may be revealed by the structure of BphDLB400 (29).
Commercial mixtures of PCBs typically contain up to 60 different congeners. The effective microbial degradation of these environmental pollutants thus requires enzymes of broad or complementary specificities. The present study of BphDP6, and its comparison to BphDLB400, identifies this hydrolase as a key determinant of PCB degradation in evolutionarily divergent biphenyl-degrading microorganisms. In particular, the predicted competitive inhibition of BphD by 3-Cl and 4-Cl HOPDAs will slow the degradation of all PCB congeners at this step. Judicious screening of naturally occurring microorganisms may identify enzymes that are able to efficiently hydrolyze these compounds. Alternatively, in vitro evolution strategies may yield enzymes of the requisite specificity.
ACKNOWLEDGMENTS
This research was funded by Strategic Grant STP0193182 from the Natural Sciences and Engineering Research Council of Canada (to L.D.E.) and by contract EV5V-CT92-0192 from the European Union (to W.R.).
We thank Shouming He for performing the HPLC-ESMS analyses.
REFERENCES
- 1.Armengaud J, Happe B, Timmis K N. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J Bacteriol. 1998;180:3954–3966. doi: 10.1128/jb.180.15.3954-3966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asturias J A, Diaz E, Timmis K N. The evolutionary relationship of biphenyl dioxygenase from the gram-positive Rhodococcus globerulus P6 to multicomponent dioxygenase from gram-negative bacteria. Gene. 1995;156:11–18. doi: 10.1016/0378-1119(94)00530-6. [DOI] [PubMed] [Google Scholar]
- 3.Asturias J A, Moore E R B, Yakimov M M, Klatte S, Timmis K N. Reclassification of the polychlorinated biphenyl-degraders Acinetobacter sp. strain P6 and Corynebacterium sp. strain MB1 as Rhodococcus globerulus. Syst Appl Microbiol. 1994;17:226–231. [Google Scholar]
- 4.Asturias J A, Timmis K N. Three different 2,3-dihydroxybiphenyl 1,2-dioxygenase genes in the gram-positive polychlorobiphenyl-degrading bacterium Rhodococcus globerulus P6. J Bacteriol. 1993;175:4631–4640. doi: 10.1128/jb.175.15.4631-4640.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedard D L, Haberl M L. Influence of chlorine substitution pattern on the degradation of polychlorinated biphenyls by eight bacterial strains. Microb Ecol. 1990;20:87–102. doi: 10.1007/BF02543870. [DOI] [PubMed] [Google Scholar]
- 6.Bopp L H. Degradation of highly chlorinated PCBs by Pseudomonas strain LB400. J Ind Microbiol. 1986;1:23–29. [Google Scholar]
- 7.Borck K, Beggs J D, Brammar W J, Hopkins A S, Murray M E. The construction in vitro of transducing derivatives of phage lambda. Mol Gen Genet. 1976;146:199–207. doi: 10.1007/BF00268089. [DOI] [PubMed] [Google Scholar]
- 8.Bunz P V, Falchetto R, Cook A M. Purification of two isofunctional hydrolases (EC 3.7.1.8) in the degradative pathway for dibenzofuran in Sphingomonas sp. strain RW1. Biodegradation. 1993;4:171–178. doi: 10.1007/BF00695119. [DOI] [PubMed] [Google Scholar]
- 9.Catelani D, Colombi A, Sorlini C, Treccani V. 2-Hydroxy-6-oxo-6-phenylhexa-2,4-dienoate: the meta-cleavage product from 2,3-dihydroxybiphenyl by Pseudomonas putida. Biochem J. 1973;134:1063–1066. doi: 10.1042/bj1341063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chebrou H, Hurtubise Y, Barriault D, Sylvestre M. Heterologous expression and characterization of the purified oxygenase component of Rhodococcus globerulus P6 biphenyl dioxygenase and of chimeras derived from it. J Bacteriol. 1999;181:4805–4811. doi: 10.1128/jb.181.16.4805-4811.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornish-Bowden A. Analysis of enzyme kinetic data. New York, N.Y: Oxford University Press; 1995. [Google Scholar]
- 12.de Lorenzo V, Eltis L, Kessler B, Timmis K N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 13.Dente L, Cortese R. pEMBL: a new family of single-stranded plasmids for sequencing DNA. Methods Enzymol. 1987;155:111–119. doi: 10.1016/0076-6879(87)55011-x. [DOI] [PubMed] [Google Scholar]
- 14.Focht D D. Strategies for the improvement of aerobic metabolism of polychlorinated biphenyls. Curr Opin Biotechnol. 1995;6:341–346. [Google Scholar]
- 15.Furukawa K, Tomizuka N, Kamibayashi A. Effects of chlorine substitution on the bacterial metabolism of various polychlorinated biphenyls. Appl Environ Microbiol. 1979;38:301–310. doi: 10.1128/aem.38.2.301-310.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furukawa K, Tonomura K, Kamibayashi A. Metabolism of 2,4,4′-trichlorobiphenyl by Acinetobacter sp. P6. Agric Biol Chem. 1979;43:1577–1583. [Google Scholar]
- 17.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 18.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:5557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kane J F. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–5000. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 20.Kaschabek S R. Ph.D. thesis. Chemische Mikrobiologie. Wuppertal, Germany: Bergische Universität—GH Wuppertal; 1995. [Google Scholar]
- 21.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Lau P C K, Garnon J, Labbé D, Wang Y. Location and sequence analysis of a 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase-encoding gene (bpdF) of the biphenyl/ polychlorinated biphenyl degradation pathway in Rhodococcus sp. M5. Gene. 1996;171:53–57. doi: 10.1016/0378-1119(96)00025-x. [DOI] [PubMed] [Google Scholar]
- 23.Masai E, Sugiyama K, Iwashita N, Shimizu S, Hauschild J E, Hatta T, Kimbara K, Yano K, Fukuda M. The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1. Gene. 1997;187:141–149. doi: 10.1016/s0378-1119(96)00748-2. [DOI] [PubMed] [Google Scholar]
- 24.McFarland V A, Clarke J U. Environmental occurrence, abundance, and potential toxicity of polychlorinated biphenyl congeners: considerations for a congener-specific analysis. Environ Health Perspect. 1989;81:225–239. doi: 10.1289/ehp.8981225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKay D B, Seeger M, Zielinski M, Hofer B, Timmis K N. Heterologous expression of biphenyl dioxygenase-encoding genes from a gram-positive broad-spectrum polychlorinated biphenyl degrader and characterization of chlorobiphenyl oxidation by the gene products. J Bacteriol. 1997;179:1924–1930. doi: 10.1128/jb.179.6.1924-1930.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nerdinger S, Kendall C, Marchhart R, Riebel P, Johnson M R, Yin C-F, Eltis L D, Snieckus V. Directed ortho metalation and Suzuki-Miyaura cross-coupling connections: regiospecific synthesis of all isomeric chlorodihydroxybiphenyls for microbial degradation studies of PCBs. Chem Commun. 1999;22:2259–2260. [Google Scholar]
- 27.Omori T, Sugimura K, Ishigooka H, Minoda Y. Purification and some properties of a 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid (HOPDA) reducing enzyme from Pseudomonas cruciviae S93 B1 involved in the degradation of biphenyl. Agric Biol Chem. 1986;50:931–937. [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Seah S Y K, Labbé G, Nerdinger S, Johnson M, Snieckus V, Eltis L D. Identification of a serine hydrolase as a key determinant in the microbial degradation of PCBs. J Biol Chem. 2000;275:15701–15708. doi: 10.1074/jbc.275.21.15701. [DOI] [PubMed] [Google Scholar]
- 30.Seah S Y K, Terracina G, Bolin J T, Riebel P, Snieckus V, Eltis L D. Purification and preliminary characterization of a serine hydrolase involved in the microbial degradation of polychlorinated biphenyls. J Biol Chem. 1998;273:22943–22949. doi: 10.1074/jbc.273.36.22943. [DOI] [PubMed] [Google Scholar]
- 31.Seeger M, Timmis K N, Hofer B. Conversion of chlorobiphenyls into phenylhexadienoates and benzoates by the enzymes of the upper pathway for polychlorinated biphenyl degradation encoded by the bph locus of Pseudomonas sp. strain LB400. Appl Environ Microbiol. 1995;61:2654–2655. doi: 10.1128/aem.61.7.2654-2658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 33.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timmis K N, Pieper D H. Bacteria designed for bioremediation. Trends Biotechnol. 1999;17:201–204. doi: 10.1016/s0167-7799(98)01295-5. [DOI] [PubMed] [Google Scholar]
- 35.Vaillancourt F H, Han S, Fortin P D, Bolin J T, Eltis L D. Molecular basis for the stabilization and inhibition of 2,3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol. J Biol Chem. 1998;273:34887–34895. doi: 10.1074/jbc.273.52.34887. [DOI] [PubMed] [Google Scholar]