Abstract
Allo-HSCT with CCR5Δ32/Δ32 donor cells is the only curative HIV-1 intervention. We investigated the impact of allo-HSCT on the viral reservoir in PBMCs and post-mortem tissue in two patients. IciS-05 and IciS-11 both received a CCR5Δ32/Δ32 allo-HSCT. Before allo-HSCT, ultrasensitive HIV-1 RNA quantification; HIV-1-DNA quantification; co-receptor tropism analysis; deep-sequencing and viral characterization in PBMCs and bone marrow; and post-allo-HSCT, ultrasensitive RNA and HIV-1-DNA quantification were performed. Proviral quantification, deep sequencing, and viral characterization were done in post-mortem tissue samples. Both patients harbored subtype B CCR5-tropic HIV-1 as determined genotypically and functionally by virus culture. Pre-allo-HSCT, HIV-1-DNA could be detected in both patients in bone marrow, PBMCs, and T-cell subsets. Chimerism correlated with detectable HIV-1-DNA LTR copies in cells and tissues. Post-mortem analysis of IciS-05 revealed proviral DNA in all tissue biopsies, but not in PBMCs. In patient IciS-11, who was transplanted twice, no HIV-1-DNA could be detected in PBMCs at the time of death, whereas HIV-1-DNA was detectable in the lymph node. In conclusion, shortly after CCR5Δ32/Δ32, allo-HSCT HIV-1-DNA became undetectable in PBMCs. However, HIV-1-DNA variants identical to those present before transplantation persisted in post-mortem-obtained tissues, indicating that these tissues play an important role as viral reservoirs.
Keywords: HIV-1, HIV persistence, reservoir, cure, tissue, CCR5Δ32, allo-HSCT
1. Introduction
Although antiretroviral therapy (ART) successfully averts HIV-1-related disease progression and saves lives of millions of HIV-infected individuals, the ongoing inflammation and costs urgently call for curative strategies [1]. Current antiretroviral compounds target several steps in the viral life cycle but do not target the integrated provirus nor suppress HIV-1 transcription and production from the cellular reservoir. This integrated provirus forms a stable viral reservoir and is the major barrier to HIV-1 eradication [1].
Several approaches are being explored to eliminate the viral reservoir and ultimately convert HIV-1 infection into a curable disease. Successful viral remission is known in four cases of treatment interruption after allogeneic hematopoietic stem cell transplantation (allo-HSCT) with homozygous CCR5Δ32 donor cells: “The Berlin patient”, two participants in the IciStem cohort; IciS-36, IciS-19—also named “The London patient” and “The Düsseldorf patient”; and recently in a woman of mixed race in New York. The cells used for transplantation lack expression of surface CCR5, rendering them resistant to infection by CCR5-tropic HIV-1 [2,3,4,5,6,7]. Though no viral rebound was observed in these patients after treatment interruption (ATI), traces of proviral HIV-1-DNA could still be found in some samples [3,5,7,8]. Also, in patients transplanted with CCR5-expressing donor cells (CCR5WT), a profound reduction in the viral reservoir was observed. However, in these patients, therapy interruption resulted in a delayed rebound of viruses comparable to the pre-allo-HSCT PBMC population in “the Boston patients”, but not in “the Minnesota patient” [9,10,11,12].
These data highlight the knowledge gap on how allo-HSCT impacts the viral reservoir and which cellular or anatomical compartments fuel HIV-1 rebound. IciStem is an international collaboration to guide and investigate the potential for HIV-1 cure after allo-HSCT and has compiled the largest cohort of HIV-1-infected individuals receiving allo-HSCT (www.icistem.org, accessed on 15 August 2022). Two IciStem CCR5Δ32 patients (IciS-05 and IciS-11) died after allo-HSCT. Post-mortem biopsies of these patients provided a unique opportunity to broadly investigate the viral reservoir after allo-HSCT in multiple tissues, which would never be feasible in living patients.
1.1. Cases
1.1.1. Patient IciS-05
A 38-year-old male presented in 1998 with a CD4+ T-cell count of 46 cells/μL whole blood and an HIV-1 viral load (VL) >500,000 RNA copies/mL plasma. Despite several changes in his therapy regimen, full suppression was not reached until 2008 (Table 1). Extensive drug resistance was selected and genotypic analyses of the viral envelope predicted infection with CCR5-tropic virus. In 2011, poor-risk myelodysplastic syndrome (MDS) was diagnosed, necessitating allo-HSCT in May 2012. Because no adult HLA-matched unrelated CCR5Δ32 donor was found, a cord-blood transplant of a homozygous CCR5Δ32 donor (HLA-match 4/6) combined with an infusion of CD34+-cells of an HLA-unmatched haploidentical family donor (CCR5wt) was performed, to aim for HIV-1 cure but also to ensure faster engraftment (Table 1). Using general diagnostic assays, viral load was 20 copies/mL on the day of transplant and remained suppressed (<50 copies/mL) after transplantation. Cord-blood donor chimerism in peripheral blood lymphocytes (PBLs) was 57% on day +16 and 100% on day +36. On day +54, chimerism reduced to 95%, and further down to 85% at day +65. Unfortunately, the patient died of severe sepsis on day +68.
Table 1.
Baseline clinical characteristics of patients.
| IciS-05 | IciS-11 | |
|---|---|---|
| Hematological Data | ||
| Hematological diagnosis | MDS | AML |
| Donor type/graft source | Cord blood + CD34+-cells third party donor | HLA-matched unrelated donor |
| Donor CCR5 | Homozygous CCR5Δ32 | First donor: homozygous CCR5Δ32, second donor: heterozygous CCR5Δ32/WT |
| Recipient HLA | HLA-A*03:01;24:02; HLA-B*07:02;35:01; HLA-Cw*04:01;07:02; HLA-DRB1*01:01;04:04; HLA-DQB1*03:02;05:01 |
HLA-A*01:01;02:01; HLA-B*07:02;-; HLA-Cw*07:02;-; HLA-DRB1*15:01;-; HLA-DQB1*06:02; - |
| Donor-recipient HLA match | 4/6 cord blood donor (A*03:01; -; B*07:02; 35:02; C*07:02; 04;01; DRB1*01:01; 11:04); 50% haploidentical family member (A*24:02;26:01;B*07:02; 14:01;C*07:02;08:02; DRB1*04:04; 14:54; DQB1*03:02; 05:03) |
10/10; 10/10 |
| Pre-transplant chemotherapy | None | 2 induction cycles with cytarabine and idarubicin |
| Conditioning | ATG, fludarabine with busulvex | ATG, fludarabine, and low-dose TBI before initial transplant; ATG fludarabine and treosulfan before second transplant |
| GvHD prophylaxis | Cyclosporine, prednisone | Cyclosporine, prednisone, and mycophenolic acid mofetil |
| GvHD severity | Acute skin GvHD grade 1 | No GvHD |
| Virological data | ||
| Time from HIV-1 diagnosis to allo-HSCT | 14 years | 22 years |
| Time from start cART to allo-HSCT | 14 years | 19 years |
| Host CCR5 | CCR5WT/WT | CCR5WT/WT |
| Predicted HIV-1 co-receptor tropism | CCR5-tropic virus (FPR 68.0–96.2%) | CCR5-tropic virus (FPR 9.7–77.3%) |
| Phenotypic HIV-1 co-receptor tropism | CCR5-tropic virus | CCR5-tropic virus |
| HIV-1 subtype | HIV-1 subtype B | HIV-1 subtype B |
| cART History | 1998: AZT/3TC, NFV. 2005: TNF, EFV, ATV/r 2006: TNF, LPV/r, NVP 2006: TNF, SQV/r, NVP 2008: AZT/3TC, TNF, SQV/r |
1996: AZT, DDI. 1996: D4T, 3TC, SQV 1997: D4T, 3TC, SQV/r 1999: D4T, 3TC, NVP 2003: TNF, 3TC, NVP |
| cART during allo-HSCT procedure | Day -152: TNF/FTC, DRV/r, RAL Day -34 until +68: TNF/FTC, RAL, MVC, ENF Day -5 until +37: ETR added |
Day -19: TNF, FTC, DTG Day +20 until +107: ABC/3TC, DTG |
| Plasma HIV RNA load at allo-HSCT | 20 copies/ml | <50/TND copies/ml |
| HCV | Anti-HCV negative | Anti-HCV negative |
| HBV | HbsAg negative, anti-HBc positive, anti-Hbs negative | HbsAg negative, anti-HBc positive, anti-Hbs positive |
| CMV status pre-SCT | Positive | Positive |
| Donor CMV status | Positive | Positive |
3TC, lamivudine. ABC, abacavir. Allo-HSCT, allogeneic hematopoietic stem cell transplantation. AML, acute myeloid leukemia. Anti-HBc, antibody against hepatitis B core antigen. Anti-HBs, antibody against hepatitis B surface antigen. ATG, anti-thymocyte globulin. ATV/r, ritonavir boosted atazanavir. AZT, zidovudine. cART, combined antiretroviral therapy. CCR5, C-C chemokine receptor type 5. CCR5Δ32, C-C chemokine receptor type 5 delta 32 mutation. CMV, cytomegalovirus. DDI, didanosine. DRV/r, ritonavir boosted darunavir. DTG, dolutegravir. D4T, stavudine. EBV, Epstein-Barr virus. EBV Vca, EBV viral capsid antigen antibody. EBV EBNA, EBV nuclear antigen antibody. EFV, efavirenz. ENF, enfuvirtide. ETR, etravirine. FTC, emtricitabine. FPR, False Positive Ratio. GvHD, Graft-versus-host disease. HBV, hepatitis B virus. HbsAG, hepatitis B surface antigen. HCV, hepatitis C virus. HLA, human leukocyte antigen. LPV/r, ritonavir boosted lopinavir. MDS, myelodysplastic syndrome. MVC, maraviroc. NFV, nelfivnavir. NVP, nevirapine. RAL, raltegravir. RS, respiratory syncytial virus. RTV or r, ritonavir. SQV/r, ritonavir boosted saquinavir. TBI, total body irradiation. TDF, tenofovir disoproxil fumarate. TND, target not detected. TPHA, Treponema pallidum haemagluttination test. WT, wild type.
1.1.2. Patient IciS-11
A 38-year-old male was diagnosed with HIV-1 in 1993 with a CD4+ T-cell count of 440 cells/. In March 1996, when CD4+ T-cell count fell below 200 cells/μL whole blood, the patient started dual therapy, quickly followed by triple therapy (Table 1). In December 2014, when VL was suppressed and CD4+ T-cells were around 700 cells/μL, he was diagnosed with poor-risk AML. Genotypic analyses of the viral envelope predicted infection with CCR5-tropic virus.
In April 2015, the patient received allo-HSCT with cells of a 10/10 HLA-matched unrelated donor homozygous for the CCR5Δ32 mutation (Table 1). Before allo-HSCT, CD4 count was 283 cells/μL and plasma VL remained suppressed. Donor chimerism decreased from 60% to 0% between day +27 and +55. On day +41, donor chimerism in bone marrow (BM) was only 18%, further indicating graft failure. On day +71, the patient received a second transplant of a heterozygous CCR5Δ32/WT unrelated donor (HLA-match 10/10); the first donor was not available. A month after the second allo-HSCT, on day +100 after the first allo-HSCT, full donor chimerism was reached. On day +108 after the first allo-HSCT and day +37 after the second allo-HSCT, the patient died due to respiratory failure.
2. Materials and Methods
2.1. Patients and Patient Material
This study included two IciStem patients. IciS-05 was included after signing a general waiver for the use of body material for future research, according to the regulations of the UMC Utrecht HSCT Biobank (HSCT study number 3440). IciS-11 was included after written informed consent was acquired. Ethical approval by the Institutional Medical Ethics Committee of the UMCU was obtained under protocol number NL53114.041.15. Before allo-HSCT, plasma, serum, PBMCs, and BM were obtained and a leukapheresis was performed. From the leukapheresis, 2–3 × 108 PBMCs were sorted into TN (CD3+CD4+CD45RO-CCR7+CD27+Fas-), TSCM (CD3+CD4+CD45RO-CCR7+CD27+Fas+), TCM (CD3+CD4+CD45RO+CCR7+CD27+), TTM (CD3+CD4+CD45RO+CCR7-CD27+Fas+) and TEM (CD3+CD4+CD45RO+CCR7-CD27-Fas+). CD4+ T-cells were obtained from baseline PBMCs and (CD4+-cell Isolation Kit, negative selection) (Miltenyi Biotec). At different time points after, allo-HSCT plasma and PBMCs were obtained. Post-mortem biopsies were obtained the day after death from terminal ileum, lung, liver, and spleen. From patient IciS-11, also brain biopsies were obtained and CD4+-cells were isolated from fresh ileum and a mediastinal lymph node.
2.2. DNA Isolation
Total DNA was isolated from frozen tissue biopsies using two different methods because of size diffence of the biopsy size and the maximum input in the the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany). Biopsy 1 was sliced into small pieces and cells were lysed using the MagNALyser (Roche, Basel, Switzerland) and purified using QIAquick (Qiagen, Hilden, Germany). Biopsy 2 was lysed and purified using the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany). Total DNA from PBMCs or CD4+-cells was isolated using the DNeasy Blood & Tissue kit.
2.3. Patient and Donor CCR5 Genotype Determination
To determine the CCR5 genotype, 30 ng total cellular DNA was amplified using CCR5forward2 5′-GATAGGTACCTGGCTGTCGTCCAT-3′, and CCR5d 5′-CCTGTGCCTCTTCTTCTCATTTCG-3′. As a control, plasmids containing the wild type CCR5 sequence or the CCR5∆32 sequence were also amplified and lengths of all amplified fragments were compared by gel-electrophoresis and the Bioanalyzer (Agilent, Santa Clara, United States). Deletions in CCR5 were confirmed by sequence analysis.
2.4. Ultra-Sensitive Viral Reservoir Quantification
Ultra-sensitive HIV-1 proviral DNA quantification was performed using primers in the conserved HIV-LTR region and pol region and total cell DNA was quantified using an RPP30 (Ribonuclease P/MRP Subunit P30) primer and probe set as described in Bosman et al. [13]. HIV-1 DNA was quantified using de droplet digital PCR (ddPCR, Bio-Rad). The threshold was set manually at a conservative fluorescence level. Water was used as a no-template control and DNA isolated from PBMCs of HIV-negative donors was used as a DNA template control. U1 cells were tested as a positive control. In 3/100 assays, a single positive droplet was observed. Thus, measurement of a single droplet is interpreted as a trace.
2.5. Micro-Chimerism Assessment
In addition to routine clinical chimerism testing, ultra-sensitive chimerism PCR was performed on peripheral blood (day +55, +106), post-mortem lymph node, and CD4+-cells from the lymph node of patient IciS-11 using the Mentype DIPscreen and Mentype DIPquant assays (Biotype), with a sensitivity of 0.01–0.001%, depending on quality and quantity of the purified DNA.
2.6. Ultra-Sensitive Plasma HIV-1 RNA Determination
Ultra-sensitive VL was measured using Cobas Ampliprep/Cobas TaqMan HIV-1 Test v2.0 (Roche Molecular Systems, Inc. Pleasanton, CA, USA) after ultracentrifugation of 4–9 mL of plasma [14].
2.7. HIV-1 Co-Receptor Prediction and Determination
Genotypic HIV-1 co-receptor tropism was predicted by deep sequence analysis of the V3 loop of the viral envelope (gp160-V3). Total cellular DNA was amplified in triplicate in a nested approach using primers envF1.1 5′-GGATATAATCAGYYTATGGGA-3′, envF1.2 5′-GAGGATATAATCAGTTTATGG′, envR1.1 5′-GGTGGGTGCTAYTCCYAITG-3′, envR1.2 5′-GGTGGGTGCTATTCCTAATGG-3′ for the first amplification and primers envF2.1 5′-GATCAAAGCCTAAARCCATGT-3′, envF2.2 5′-GATCAAAGCCTAAAGCCATG-3′, envR2.1 5′-CTCCAATTGTCCYTCATHTYTCC-3′, envR2.2 5′-ACTTCTCCAATTGTCCCTCATAT-3′ in the nested amplification. The DNA concentration of the purified amplicons was determined using Quant-iT PicoGreen dsDNA Assay (Thermo Fisher Scientific, Waltham, MA, USA) and a sequence library was formed using the Nextera DNA Library Preparation Kit (Illumina, San Diego, CA, United States). Finally, the DNA concentration of the library was measured (Quant-iT PicoGreen dsDNA Assay Kit, Thermo Fisher Scientific, MA, USA), diluted to 2 ng/mm3 denatured, and sequenced using the MiSeq Reagent Kit v2 (500-cycles) (Illumina, San Diego, USA). Viral tropism was predicted in silico using geno2pheno (454) with a 3.5% cut-off indicating the probability of classifying an R5-tropic virus falsely as CXCR4-tropic virus (FPR; false-positive-rate) [15].
Phenotypic HIV-1 co-receptor was determined using cells and virus culture. Cells: U373-MAGI-CCR5E, U373-MAGICXCR4CEM [16], MT2 cell lines [17] (NIH AIDS Reagent Program), and SupT1R5X4 (personal gift from J. Hoxie) were maintained as recommended. PBMCs from five healthy donors (homozygous for CCR5WT) were prepared by Ficoll–Paque density gradient centrifugation of heparinized blood. The mix was stimulated for 3 days with phytohaemagglutinin (2 mg/L) in culture medium [RPMI1640 with L-glutamine (BioWhittaker, Lonza, Basel, Switzerland), 10% fetal bovine serum (FBS; Biochrom AG, Berlin, Germany) and 10 mg/L gentamicin (Gibco)]. Virus culture: PBMCs from healthy donors were co-cultured with purified CD4+-cells from patients IciS-05 and IciS-11 for 2 h at 37 °C, after which cells were washed twice. Subsequently, cells were cultured with 25 U/mL IL-2 in 10 mL CM. Virus was cultured for 3 weeks, and twice-weekly half of the culture was replaced with fresh culture medium with 25 U/mL recombinant human IL-2 (Thermo Fisher Scientific, Massachusetts, United States) and once weekly freshly stimulated PBMCs from healthy donors were added. Twice weekly HIV-1 replication was monitored by CA-p24 ELISA and culture supernatant was stored for analyses of viral co-receptor tropism in MT2 cells (expressing CD4 and CXCR4) and as a control SupT1R5 × 4 cells (expressing CD4, CCR5 and CXCR4).
Analysis of HIV-1 co-receptor usage in U373-MAGI cells: Infection of these cells was done using 2 ng of CA-p24 of the patient-derived viral isolate or the control viruses [6].
Phylogenetic sequence analysis; viral evolution and compartmentalization: HIV-1 subtype was determined using REGA HIV Subtyping Tool [18]. The evolutionary history was inferred using Maximum Likelihood (MEGA 7) (500 bootstraps) [19]. The tree with the highest log likelihood is shown. Initial tree (s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated.
3. Results
3.1. Patient IciS-05
Twenty days before allo-HSCT, only an unquantifiably low amount of HIV-1 RNA could be detected with routine diagnostics. Ultra-sensitive plasma viral load was 15 copies/mL (Figure 1a). The viral reservoir was quantified in BM, PBMCs, naïve T-cells, and all memory T-cell subsets with HIV-1-DNA being most abundant in the more differentiated T-cell subsets (1135-6924 HIV-1-LTR copies/106 cells) (Table 2).
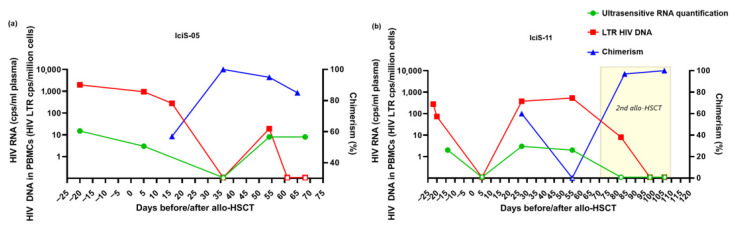
Figure 1.
Clinical and virological data of IciS-05 (a) and IciS-11 (b) pre- and post-allo-HSCT. HIV-1 proviral DNA expressed as LTR DNA copies/million PBMC, and HIV-1 RNA as copies/mL for plasma and chimerism. Open symbols represent values under the level of quantification. Abbreviations: allo-HSCT, allogeneic hematopoietic stem cell transplantation. PBMCs, peripheral blood mononuclear cells.
Table 2.
Results of HIV-1-DNA quantification and characterization in patient IciS-05.
| Time Point (Days) | Chimerism (%) | Ultrasensitive RNA Quantification (Copies/mL) | Patient Material | ddPCR (Copies/106 Cells) |
Gp120V3-Sequence (Dominant FPRs; Range, %) | |
|---|---|---|---|---|---|---|
| HIV-1 LTR | HIV-1 Pol | |||||
| Pre allo-HSCT | ||||||
| −20 | 15 | PBMCs | 1967 | 432 | 87.2, 89.7 (68.8–96.2) | |
| Tn | 1284 | 167 | 87.2, 89.7 (87.2–89.7) | |||
| Tcm | 3074 | 609 | 87.2, 89.7 (87.2–91.0) | |||
| Ttm | 5600 | 1622 | 87.2, 89.7 (68.0–89.7) | |||
| Tem | 6924 | 1886 | 87.2, 89.7 (68.8–89.7) | |||
| −19 | BM | 1135 | 167 | 87.2, 89.7(73.7–89.7) | ||
| Post allo-HSCT | ||||||
| +5 | 3 | PBMCs | 949 | ND | ||
| +16 | 57 | PBMCs | 278 | <21 | ||
| +36 | 100 | 0 | PBMCs | <5 | ND | |
| +54 | 95 | 8 | PBMCs | 19 | ND | |
| +61 | PBMCs | <20 | <20 | |||
| +65 | 85 | |||||
| +68 (died) | 8 | PBMCs | <7 | <22 | ||
| Post-mortem biopsies (one biopsy from same site is separated in two parts; 1,2) | ||||||
| +69 | Liver, biopsy 1; 2 | 54; trace | ND; <8 | No amplification | ||
| Lung left, biopsy 1; 2 | 36; 49 | ND; trace | 89.7, 87.2 (87.2–89.7) | |||
| Lung right, biopsy 1; 2 | 90; 32 | ND; trace | 89.7, 87.2 (71.4–92.2) | |||
| Spleen, biopsy 1; 2 | 43; 67 | ND; 34 | No amplification | |||
| Terminal Ileum, biopsy 1; 2 | 549; 81 | ND; 89 | 89.7, 87.2 (64.4–89.7) | |||
Allo-HSCT, allogeneic hematopoietic stem cell transplantation. BM, bone marrow. ddPCR, droplet digital PCR. FPR, false-positive-rate. LTR, long terminal repeat DNA sequence. ND, not done. NTC, no template control. PBMCs, peripheral blood mononuclear cells. pol, DNA polymerase DNA sequence. Tcm, central memory T-cell. Tem, effector memory T-cell. Tn, naive T-cell. Ttm, transitional memory T-cell.
In-depth genotypic analyses of the viral reservoir showed that the population was rather homogeneous with two viral variants dominating all cell types (Supplementary data Figure S1a). Genotypic analyses predicted CCR5 co-receptor dependency for both variants (FPR 87.2% and 89.7%) (Table 2). Viruses cultured from PBMCs also showed CCR5-tropism and were unable to infect MAGI-X4 cells (Supplementary data Figure S1b) and MT2 cells.
After allo-HSCT with CCR5Δ32 donor cells, a decrease in proviral DNA was observed in PBMCs (Figure 1a, Table 2). On day +36, when full chimerism was observed in PBLs, proviral DNA in PBMCs and plasma VL were both below the limit of quantification. Subsequently, a decrease in chimerism was observed in peripheral T-cells, and HIV-1-DNA in PBMCs and plasma VL both became detectable on day +54. On day +68, when the patient deceased, plasma VL was detectable (8 copies/mL) but proviral DNA in PBMCs was still undetectable. Interestingly, at the same time, proviral DNA could be detected in all post-mortem biopsies (Table 2). Viral envelope could be amplified and sequenced from lung tissue and terminal ileum and revealed the dominance of the same CCR5-tropic viral variants as observed in peripheral blood and BM prior to allo-HSCT (Supplementary data Figure S1a).
3.2. Patient IciS-11
Shortly before allo-HSCT, low-level plasma HIV-1 RNA was detectable by ultrasensitive viral load assay (2 copies/mL) (Figure 1b). HIV-1-DNA could be detected in BM, PBMCs, naïve T-cells, and all memory T-cell subsets with most HIV-1-LTR DNA in the differentiated subsets (73-4629 HIV-1-LTR copies/106 cells) (Table 3).
Table 3.
Results of HIV-1-DNA quantification and characterization in patient IciS-11.
| Time Point (Days) | Chimerism (%) | Ultrasensitive RNA Quantification (Copies/mL) | Patient Material | ddPCR (Copies/106 Cells) | Gp120V3-Sequence (Dominant FPR; Range, %) | |
|---|---|---|---|---|---|---|
| HIV-1 LTR | HIV-1 Pol | |||||
| Pre allo-HSCT | ||||||
| −22 | PBMCs | 279 | 21 | 18.0 (18.0–70.5) | ||
| −20 | BM | 73 | <14 | 70.5 (39.4–70.5) | ||
| −14 | 2 | Tn | 571 | 69 | 31.8 (18.0–70.5) | |
| Tscm | 490 | ND | 9.7 (9.7–50.3) | |||
| Tcm | 2222 | 544 | 42.4 (22.1–77.3) | |||
| Ttm | 2780 | 838 | 42.4 (18.0–70.5) | |||
| Tem | 4629 | 882 | 21.8 (18.0–39.4) | |||
| Post allo-HSCT | ||||||
| +5 | 0 | PBMCs | trace | <5 | ||
| +27 | 60 | 3 | PBMCs | 378 | ND | |
| +55 | 0 | 2 | PBMCs | 534 | 72 | 31.8 (31.8–70.5) |
| Post 2nd allo-HSCT (days post 1st allo-HSCT) | ||||||
| +11 (+82) | 0 | PBMCs | 8 | ND | ||
| +27 (+98) | 0 | PBMCs | <2 | <4 | ||
| +35 (+106) | 100 | 0 | PBMCs | <2 | ND | |
| Post-mortem biopsies (one biopsy from same site is separated in two parts; 1,2) | ||||||
| +37 (+108) | Liver, biopsy 1; 2 | 60; <7 | ND; trace | No amplification | ||
| Lung left, biopsy 1; 2 | 28; <4 | ND; <4 | No amplification | |||
| Lung right, biopsy 1; 2 | trace; <3 | ND; <3 | No amplification | |||
| Spleen, biopsy 1; 2 | 60; <6 | ND; <6 | No amplification | |||
| Brain, biopsy 1; 2 | <4; <14 | <4; <4 | No amplification | |||
| 38 | LN CD4+ cells | 10 | <4 | No amplification | ||
Allo-HSCT, allogeneic hematopoietic stem cell transplantation. BM, bone marrow. ddPCR, droplet digital PCR. FPR, false-positive-rate. LN, lymph node. LTR, long terminal repeat DNA sequence ND, not done. NTC, no template control. PBMCs, peripheral blood mononuclear cells. pol, DNA polymerase DNA sequence. Tcm, central memory T-cell. Tem, effector memory T-cell. Tn, naive T-cell. Tscm, stem memory T-cell. Ttm, transitional memory T-cell.
While proviral DNA levels were lower in comparison to IciS-05, in-depth genotypic analyses of the viral envelope sequence demonstrated more inter- and intra-sample variation (Table 3, Supplementary data Figure S1a). All variants were predicted for the use of the CCR5 co-receptor for viral entry (FPR 9.7–77.3%) (Supplementary data Figure S1a). HIV-1 variants cultured from BM and PBMCs were also CCR5-tropic and unable to use the alternative CXCR4 co-receptor as present in MAGI-X4 cells (Supplementary data Figure S1b) and MT2 cells.
Shortly after allo-HSCT, plasma HIV-1 RNA could no longer be detected and also PBMC proviral DNA rapidly declined (Table 3, Figure 1b). Over time, with decreasing chimerism, proviral DNA increased to pre-transplantation levels and plasma ultrasensitive VL became detectable (3 copies/mL). On day +55 when the graft was rejected, proviral DNA in PBMCs was twice the pre-transplant level. Considering the relatively low cell counts after allo-HSCT, a relatively big increase in proviral DNA was observed. In-depth sequence analyses revealed the same viral sequences as seen before transplantation, and thus no signs of viral evolution or escape of CCR5 co-receptor usage were observed.
On day +71, IciS-11 received a second allo-HSCT with heterozygous CCR5Δ32/WT donor cells. In the first week after transplantation, a rapid decrease of proviral DNA was observed. On day +35, when chimerism was 100%, PBMC proviral DNA and plasma VL were undetectable. Two days later the patient died of respiratory failure. Multiple post-mortem biopsies were obtained from liver, lung, spleen, brain, and lymph node. In contrast to patient IciS-05, detection of HIV-1-DNA in these tissues varied between the different tissues and biopsies, with lung and brain being negative and other tissues being positive in only one of the biopsies. The viral envelope could not be successfully amplified from any of the biopsies (Table 3).
4. Discussion
To date, allo-HSCT is the only strategy to profoundly reduce the HIV-1 reservoir irrespective of whether CCR5 wild-type stem cells or stem cells lacking expression of the CCR5 co-receptor for HIV-1 entry are used [2,3,4,5,7,8,9,11,20,21,22].
Autopsy of patients IciS-05 and IciS-11 provided a unique opportunity to investigate the impact of allo-HSCT on the dynamics and composition of the viral reservoir in PBMCs as well as in different tissue compartments. In patient IciS-05 who was transplanted in a non-myeloablative procedure with CCR5Δ32/Δ32 donor cells, no HIV-1-DNA could be detected by ddPCR in PBMCs but HIV-1-DNA could readily be detected in all biopsies taken from lung, liver, spleen, and ileum. A study describing post-mortem HIV-1 reservoir analysis in a perinatally infected child who also underwent a myeloablative procedure using cord blood CCR5Δ32 donor cells could not detect HIV-1-DNA in blood but could detect viral DNA in some of the tissues analyzed using sensitive in-situ hybridization techniques [21]. The sensitivity of the ddPCR as employed in the study of Rothenberger et al. was likely affected by the smaller number of cells that could be assessed from the tissue biopsies as compared to our study.
In patient IciS-11, who was also transplanted with CCR5Δ32 donor cells, an initial decrease of proviral DNA in PBMCs was followed by an increase in proviral DNA perPBMC. Eberhard et al. showed an HIV-1-specific T-cell activation in IciStem patients after allo-HSCT and the risk of viral escape from the HIV-1 reservoir shortly after transplantation [23]. In-depth sequence analyses of the viral envelope gene revealed identical viral sequences as seen before transplantation of patient IciS-11 and as such showed no signs of viral evolution or escape of CCR5 co-receptor usage. As patient IciS-11 experienced cytomegalovirus (CMV) reactivation after allo-HSCT during incomplete chimerism, we hypothesize that the increase of proviral DNA could be due to the expansion of particular T-cell populations after allo-HSCT. This observation is in line with the described expansion of HIV-1-infected CD4+ T-cells in response to CMV or Epstein–Barr viral antigens, contributing to sustain the HIV-1-DNA reservoir following chemotherapy [12].
Patient IciS-11 did not suffer from GvHD and, like the Berlin patient, was transplanted twice. HIV-1-DNA copy number in the tissues was at the detection level of our ddPCR assays with discrepancies between different tissue biopsies and target regions. These discrepancies show the difficulties analyzing proviral DNA in high-volume post-mortem tissues and emphasize the challenges that we face in the accurate detection and quantification of proviral DNA in low-volume biopsies of patients undergoing curative strategies. Though with a successful transplantation full chimerism in the peripheral blood is reached in the weeks after allo-HSCT, little is known about the turnover and donor chimerism in tissues. In the Berlin patient, CCR5-positive macrophages were still detected in rectal tissue more than 5 months after allo-HSCT [4]. In patient IciS-11, who died 1 month after his second transplant, chimerism in PBLs was 100% and no HIV-1-DNA was detected in blood. Simultaneously, in CD4+-cells obtained from the lymph node, where donor chimerism only reached 38%, HIV-1-DNA was detected, most likely in the recipient cells.
In patient IciS-05, proviral DNA from the different tissue samples was genotypically characterized and phylogenetic analysis indicated that all tissue-derived sequences intermingled with pre-transplantation viral populations as present in the different T-cell subsets and BM. These data show no evident compartmentalization between viruses obtained from peripheral blood and BM before the transplant compared to viruses obtained from lung and ileum afterwards. Unlike in the Berlin patient, sequences obtained from the different post-mortem tissues after allo-HSCT showed no evidence of persistence of macrophage-tropic viral variants. In general, our data are in agreement with post-mortem studies demonstrating the lack of compartmentalization of HIV-1 envelope sequences observed in lung, lymph nodes, and colon [24,25]. Previous ATI studies performed after allo-HSCT suggested that the observed viral rebound was caused by pre-existing viral variants present in the pre-transplant PBLs [12,26,27]. However, these patients’ tissues were not characterized. Since no HIV-1-DNA-positive cells could be found in blood, the rebounding viruses post allo-SCT most likely originated from tissue compartments.
A limitation of this study that should be addressed is the short follow-up of the patients after allo-HSCT. Both patients died shortly after allo-HSCT, hampering the follow-up of the dynamics of the donor cells in different tissues. Also because sampling occurred shortly after allo-HSCT, only limited cells were available due to the allo-HSCT procedure. Analyses of the proviral DNA could only be performed on PBMCs and not on a more concentrated CD4+ T-cell subset, which could have underestimated the true size of the peripheral reservoir after allo-HSCT.
In conclusion: This is the first report that performed in-depth post-mortem tissue quantification and genotypic characterization after allo-HSCT with CCR5Δ32/Δ32 donor cells of HIV-1-infected individuals. We demonstrate that the detection of proviral DNA in tissues was prolonged compared to PBMCs. Tissues clearly play an essential role as a long-standing viral reservoir and routine sampling in living HIV-1-individuals will be insufficient to represent the extent of this reservoir. Before conducting ATI, the role of the tissue reservoir has to be taken into consideration.
Acknowledgments
The authors thank the following members of the IciStem Consortium who were not named as authors: Vanderson Rocha (Churchill Hospital, Oxford, United Kingdom); Julià Blanco and Jorge Carillo (IrsiCaixa AIDS Research Institute and Institute for Health Science Research Germans Trias i Pujol (IGTP), Badalona, Spain); Johanna Eberhard and Maximilian Christopeit (University Medical Center Hamburg-Eppendorf, Hamburg, Germany); Rebeca Bailén and Pascual Balsalobre (Hospital Gregorio Marañón, Madrid, Spain); Linos Vandekerckhove Marie-Angélique de Scheerder, and Eva Steel (University of Ghent, Ghent, Belgium); Lisa Barrett, Sharon Oldford, Jill Moore, and Clarissa Brisseau (NSHA/Dalhousie University, Halifax, Canada); Dieter Häussinger, Guido Kobbe, and Björn Jensen (University Hospital Düsseldorf, Düsseldorf, Germany); Rolf Kaise and, Elena Knops (University of Cologne, Cologne, Germany); Marek Widera (University Hospital Essen, Essen, Germany); Alessandra Bandera, Antonio Muscatello, and Alessandro Soria (San Gerardo Hospital, Monza, Italy); Gabriella Scarlatti and Simona Piemontese (Istituto Scientifico San Raffaele, Milan, Italy); Jon Badiola and Manuel Jurado Chacón (Complejo Hospitalario Universitario de Granada, Granda, Spain); Raquel Saldana (Hospital General de Jerez de la Frontera, Jerez de la Frontera, Spain); Luz Martín Carbonero (La Paz Hospital, Madrid, Spain); Ildefonso Espigado (University Hospital Virgen del Rocío, Seville, Spain); Piotr Nowak and Anders Sonnerborg (Karolinska Institutet, Solna, Sweden); Mitja Nabergoj and Alexandra Calmy (Hôpitaux Universitaires de Genève, Genève, Switzerland); Kavita Raj, Fabio Cruciani, Varun Mehra, and Carmel Rice (Kings College Hospital, London, United Kingdom); Angela Bailey (Imperial College Healthcare NHS Trust, London, United Kingdom); Waseem Qasim (Institute of Child Health & Great Ormond Street Hospital, London, United Kingdom); Ravindra Gupta (University College London Hospital, London, United Kingdom); and Koen van Besien (New York Presbyterian Hospital, New York, United States). The authors also thank Agueda Hernández Rodríguez, from the Microbiology Department of the Hospital Universitari Germans Trias i Pujol.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14092069/s1, Figure S1: Phylogenetic analysis and relative coreceptor preference.
Author Contributions
Conceptualization, A.M.J.W., M.N., J.M.-P.; A.M.J.W., and A.S. were involved in the ethical approval. A.B., P.E., J.H.E.K. and J.T.M.v.d.M. were involved in the clinical process and provided clinical knowledge. L.A.A.B. performed the autopsy. D.d.J., N.A.N., S.B.H.G., K.T., K.B., M.S. and T.M.d.K. generated and analyzed the data. L.E.P.H., A.M.J.W., M.N. and N.A.N. were involved in draft preparation. L.E.P.H. editing the manuscript. M.S., G.H., M.K., J.D.M., J.T.M.v.d.M., A.S.-C., J.J.B., J.S.z.W., J.M.-P., J.H.E.K., A.M.J.W. and M.N. provided scientific guidance and domain knowledge. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Medical Ethics Committee of the UMCU (NL35448.041.11 on 27 November 2012 for IciS-05 and NL53114.041.15. on 14 April 2015 for IciS-11).
Informed Consent Statement
IciS-05 was included after signing a general waiver for the use of body material for future research, according to the regulations of the UMC Utrecht HSCT Biobank (HSCT study number 3440). IciS-11 was included after written informed consent was acquired.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed. AJS resports a grant form Dutch Government (ZonMw) and is a committee member for revision Dutch guideline on Sexually Transmitted Diseases in primary care. JJB reports consultancy fees for the following companies: Avrobio, Omeros, Advanced Clinical, Bluebird Bio, Equillium, Sobi, and SmartImmune. MS reports grants or contracts from Spanisch Ministry of research and private TV fundraising funding (Marató TV3) and payments or honoraria for lectures, presentations, speakers bureaus, manuscript writing, or educational events from Autonomous University of Barcelona and GESIDA Spain. ASC report grants or contracts from ANRS, NIH, and MSDAVENIR; payments from MSD, ViiV healthcare, and Gilead, and a patent for a method for reprogramming CD8+ T-cells to enhance their therapeutic potential and applications thereof; U.S. Patent application # 63/301, 532 and chair of the Sidaction Scientific and Medical Committee. JHEK reports grants or contracts from Novartis, Miltenyi, and Gadeta. MN reports grants from Gilead: Rosetta (Global INI resistance); Dutch Government (ZonMw/NWO): VIMP2, Bridge project; COVIDex-vivo models, Health Holland: Clear COVID organoids2cureHIV, amfAR: Epigenetic engineering of HIV, NSF: SARS-CoV-2 mutation frequency and evolution and honoraria for lectures from Virology Education. AW reports grants from Gilead, Aidfsonds, Dutch Government (ZonMw/NOW), and Health Holland/NLF4 cure, as well as consulting fees from Gilead, Janssen, Viiv/GSK, and honoraria for lectures from Virology Education and Southern African HIV Clinicians Society.
Funding Statement
This work was supported by amfAR, the Foundation for AIDS Research through the ARCHE program [108930-56-RGRL, 109293-59-RGRL, 109552-61-RGRL], and by the Dutch Aidsfonds [P-2013034, P-60802, P-13204].
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Deeks S.G., Archin N., Cannon P., Collins S., Jones R.B., de Jong M.A.W.P., Lambotte O., Lamplough R., Ndung’u T., Sugarman J., et al. Research priorities for an HIV cure: International AIDS Society Global Scientific Strategy 2021. Nat. Med. 2021;12:2085–2098. doi: 10.1038/s41591-021-01590-5. [DOI] [PubMed] [Google Scholar]
- 2.Gupta R.K., Abdul-Jawad S., McCoy L.E., Mok H.P., Peppa D., Salgado M., Martinez-Picado J., Nijhuis M., Wensing A.M.J., Lee H., et al. HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature. 2019;7751:244–248. doi: 10.1038/s41586-019-1027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta R.K., Peppa D., Hill A.L., Gálvez C., Salgado M., Pace M., McCoy L.E., Griffith S.A., Thornhill J., Alrubayyi A., et al. Evidence for HIV-1 cure after CCR5Δ32/Δ32 allogeneic haemopoietic stem-cell transplantation 30 months post analytical treatment interruption: A case report. Lancet HIV. 2020;7:e340–e347. doi: 10.1016/S2352-3018(20)30069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hütter G., Nowak D., Mossner M., Ganepola S., Müssig A., Allers K., Schneider T., Hofmann J., Kücherer C., Blau O., et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 5.Jensen B., Haussinger D., Knops E. CCR5Δ32 SCT-induced HIV remission: Traces of HIV-DNA but fading immune reactivity [abstract 348]; Presented at The Annual Conference on Retroviruses and Opportunistic Infections 2020; Boston, MA, USA. 8–11 March 2020. [Google Scholar]
- 6.Symons J., Vandekerckhove L., Hütter G., Wensing A.M., van Ham P.M., Deeks S.G., Nijhuis M. Dependence on the CCR5 coreceptor for viral replication explains the lack of rebound of CXCR4-predicted HIV variants in the Berlin patient. Clin. Infect. Dis. 2014;59:596–600. doi: 10.1093/cid/ciu284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu J., Van Besien K., Glesby M.J. HIV-1 remission with CCR5Δ32Δ32 haplo-cord transplant in a US woman: IMPAACT P1107 [abstract 65]; Proceedings of the Conference on Retroviruses and Opportunistic Infections (CROI); Virtual. 12–24 February 2022. [Google Scholar]
- 8.Yukl S.A., Boritz E., Busch M., Bentsen C., Chun T.W., Douek D., Eisele E., Haase A., Ho Y.C., Hütter G., et al. Challenges in Detecting HIV Persistence during Potentially Curative Interventions: A Study of the Berlin Patient. PLoS Pathog. 2013;9:e1003347. doi: 10.1371/journal.ppat.1003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avettand-Fenoel V., Mahlaoui N., Chaix M.L., Milliancourt C., Burgard M., Cavazzana-Calvo M., Rouzioux C. Failure of bone marrow transplantation to eradicate HIV reservoir despite efficient HAART. AIDS. 2007;6:776–777. doi: 10.1097/QAD.0b013e3280b01836. [DOI] [PubMed] [Google Scholar]
- 10.Capoferri A.A., Redd A.D., Gocke C.D., Clark L.R., Quinn T.C., Ambinder R.F., Durand C.M. Rebound HIV viremia with meningoencephalitis following antiretroviral therapy interruption after allogeneic bone marrow transplant. J. Acquir. Immune Defic. Syndr. 2021;89:297–302. doi: 10.1097/QAI.0000000000002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cummins N.W., Rizza S., Litzow M.R., Hua S., Lee G.Q., Einkauf K., Chun T.W., Rhame F., Baker J.V., Busch M.P., et al. Extensive virologic and immunologic characterization in an HIV-infected individual following allogeneic stem cell transplant and analytic cessation of antiretroviral therapy: A case study. PLoS Med. 2017;14:e1002461. doi: 10.1371/journal.pmed.1002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henrich T.J., Hanhauser E., Marty F.M., Sirignano M.N., Keating S., Lee T.H., Robles Y.P., Davis B.T., Li J.Z., Heisey A., et al. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: Report of 2 cases. Ann. Intern Med. 2014;161:319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosman K.J., Wensing A.M., Pijning A.E., van Snippenberg W.J., van Ham P.M., de Jong D.M., Hoepelman A.I., Nijhuis M. Development of sensitive ddPCR assays to reliably quantify the proviral DNA reservoir in all common circulating HIV subtypes and recombinant forms. J. Int. AIDS Soc. 2018;21:e25185. doi: 10.1002/jia2.25185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salgado M., Kwon M., Gálvez C., Badiola J., Nijhuis M., Bandera A., Balsalobre P., Miralles P., Buño I., Martinez-Laperche C., et al. Mechanisms That Contribute to a Profound Reduction of the HIV-1 Reservoir After Allogeneic Stem Cell Transplant. Ann. Intern Med. 2018;169:674–683. doi: 10.7326/M18-0759. [DOI] [PubMed] [Google Scholar]
- 15.Lengauer T., Sander O., Sierra S., Thielen A., Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nat. Biotechnol. 2007;12:1407–1410. doi: 10.1038/nbt1371. [DOI] [PubMed] [Google Scholar]
- 16.Pineda-Peña A.C.P., Faria N.R., Imbrechts S., Libin P., Abecasis A.B., Deforche K., Gómez-López A., Camacho R.J., de Oliveira T., Vandamme A.M. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: Performance evaluation of the new REGA version 3 and seven other tools. Infect Genet. Evol. 2013;19:337–348. doi: 10.1016/j.meegid.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 17.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;7:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vodicka M.A., Goh W.C., Wu L.I., Rogel M.E., Bartz S.R., Schweickart V.L., Raport C.J., Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 19.Harada S., Koyanag Y., Yamamoto N. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985;713:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 20.Duarte R.F., Salgado M., Sánchez-Ortega I., Arnan M., Canals C., Domingo-Domenech E., Fernández-de-Sevilla A., González-Barca E., Morón-López S., Nogues N., et al. CCR5 Δ32 homozygous cord blood allogeneic transplantation in a patient with hiv: A case report. Lancet HIV. 2015;2:e236–e242. doi: 10.1016/S2352-3018(15)00083-1. [DOI] [PubMed] [Google Scholar]
- 21.Rothenberger M.K., Wagner J.E., Haase A., Richman D., Grzywacz B., Strain M., Lada S., Estes J., Fletcher C.V., Podany A.T., et al. Transplantation of CCR5∆32 Homozygous Umbilical Cord Blood in a Child With Acute Lymphoblastic Leukemia and Perinatally Acquired HIV Infection. Open Forum Infect. Dis. 2018;5:ofy090. doi: 10.1093/ofid/ofy090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koelsch K.K., Rasmussen T.A., Hey-Nguyen W.J., Pearson C., Xu Y., Bailey M., Marks K.H., Sasson S.C., Taylor M.S., Tantau R., et al. Impact of Allogeneic Hematopoietic Stem Cell Transplantation on the HIV Reservoir and Immune Response in 3 HIV-Infected Individuals. J. Acquir. Immune Defic. Syndr. 2017;75:328–337. doi: 10.1097/QAI.0000000000001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eberhard J.M., Angin M., Passaes C., Salgado M., Monceaux V., Knops E., Kobbe G., Jensen B., Christopeit M., Kröger N., et al. Vulnerability to reservoir reseeding due to high immune activation after allogeneic hematopoietic stem cell transplantation in individuals with HIV-1. Sci. Transl. Med. 2020;12:eaay9355. doi: 10.1126/scitranslmed.aay9355. [DOI] [PubMed] [Google Scholar]
- 24.Brese R.L., Gonzalez-Perez M.P., Koch M., O’Connell O., Luzuriaga K., Somasundaran M., Clapham P.R., Dollar J.J., Nolan D.J., Rose R., et al. Ultradeep single-molecule real-time sequencing of HIV envelope reveals complete compartmentalization of highly macrophage-tropic R5 proviral variants in brain and CXCR4-using variants in immune and peripheral tissues. J. Neurovirol. 2018;24:439–453. doi: 10.1007/s13365-018-0633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaillon A., Gianella S., Dellicour S., Rawlings S.A., Schlub T.E., De Oliveira M.F., Ignacio C., Porrachia M., Vrancken B., Smith D.M. HIV persists throughout deep tissues with repopulation from multiple anatomical sources. J. Clin. Investig. 2020;130:1699–1712. doi: 10.1172/JCI134815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kordelas L., Verheyen J., Beelen D.W., Horn P.A., Heinold A., Kaiser R., Trenschel R., Schadendorf D., Dittmer U., Esser S., et al. Shift of HIV tropism in stem-cell transplantation with CCR5 Delta32 mutation. N. Engl. J. Med. 2014;371:880–882. doi: 10.1056/NEJMc1405805. [DOI] [PubMed] [Google Scholar]
- 27.Verheyen J., Thielen A., Lübke N., Dirks M., Widera M., Dittmer U., Kordelas L., Däumer M., de Jong D.C.M., Wensing A.M.J., et al. Rapid Rebound of a Preexisting CXCR4-tropic Human Immunodeficiency Virus Variant After Allogeneic Transplantation With CCR5 Δ32 Homozygous Stem Cells. Clin. Infect. Dis. 2018;68:684–687. doi: 10.1093/cid/ciy565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article. Further inquiries can be directed to the corresponding author.