Abstract
The Pseudomonas aeruginosa las (lasR-lasI) and rhl (rhlR-rhlI) quorum-sensing systems regulate the expression of several virulence factors, including elastase and rhamnolipid. P. aeruginosa strain PR1-E4 is a lasR deletion mutant that contains a second, undefined mutation which allows production of elastase and rhamnolipid despite a nonfunctional las system. We have previously shown that this strain accomplishes this by increasing the expression of the autoinducer synthase gene rhlI. In this report, we show that the elastolytic phenotype of mutant PR1-E4 can be complemented with a P. aeruginosa homologue of the Escherichia coli dnaK mutation suppressor gene dksA. When supplied in trans on a multicopy plasmid, this gene completely suppressed elastase production by mutant PR1-E4. Cloning and Northern blot analysis revealed that dksA was neither mutated nor less transcribed in mutant PR1-E4. When overexpressed, dksA also reduced rhamnolipid production by both mutant PR1-E4 and the wild type, PAO1. Using Northern blot analysis and lacZ reporter fusions, we show that dksA inhibits rhlI, rhlAB, and lasB transcription. Exogenous N-butyryl–l-homoserine lactone overcame the reduced expression of rhlI and restored rhlAB and lasB expression, as well as elastase production. Our results suggest that the overproduction of the P. aeruginosa DksA homologue inhibits quorum-sensing-dependent virulence factor production by downregulating the transcription of the autoinducer synthase gene rhlI.
Pseudomonas aeruginosa is a major opportunistic human pathogen. In P. aeruginosa the quorum-sensing circuitry, composed of the las and the rhl quorum-sensing systems, regulates the expression of numerous genes, including lasB (elastase) and rhlAB (rhamnosyltransferase, required for rhamnolipid production) (35). The transcriptional activator LasR and the autoinducer molecule 3-oxo-C12-HSL [N-(3-oxododecanoyl)-l-homoserine lactone] constitute the las quorum-sensing system (21, 23). Similarly, the transcriptional activator RhlR and the autoinducer molecule C4-HSL (N-butyryl-l-homoserine lactone) constitute the rhl system (18, 24). The lasI and the rhlI genes encode the autoinducer synthases that synthesize the autoinducer molecules 3-oxo-C12-HSL and C4-HSL, respectively. In cell-to-cell signaling, or quorum sensing, the concentration of the autoinducer molecule increases with bacterial cell density until a threshold concentration is reached. At this point the autoinducer binds to its corresponding transcriptional activator. The autoinducer-protein complex then activates the transcription of specific target genes (7, 10). The las and rhl quorum-sensing systems interact with each other, as the complex 3-oxo-C12-HSL–LasR activates rhlR transcription (14, 26), and the complex C4-HSL–RhlR is necessary for optimal expression of lasB (3). P. aeruginosa strain PAO-R1 is a lasR deletion mutant (8) which is unable to produce elastase and rhamnolipid (36) and is significantly less virulent than the parent wild-type strain PAO1 (27, 33). We have previously described a strain PAO-R1-derived mutant, PR1-E4, which produces elastase and rhamnolipid despite the absence of a functional las quorum-sensing system (36). The precise site of the mutation in strain PR1-E4 is unknown; however, because of the extensive deletion in the lasR gene, a simple reversion is impossible. An increased expression of the autoinducer synthase encoding gene rhlI seems to compensate for the loss of the las quorum-sensing system in this strain.
To further characterize mutant PR1-E4, we complemented this strain with a wild-type P. aeruginosa gene bank and screened transformants for loss of elastase production. A complementing gene was identified and found to be a homologue of the Escherichia coli multicopy mutation suppressor gene dksA (13). In trans, dksA suppressed the elastase production of mutant PR1-E4 and reduced the rhamnolipid production of both PR1-E4 and PAO1. We demonstrate that overexpression of dksA inhibits the expression of the autoinducer synthase gene rhlI, leading to a secondary reduction in the production of quorum-sensing-dependent virulence factors.
MATERIALS AND METHODS
Media and culture conditions.
Cultures were grown, with the appropriate antibiotics, at 37°C with shaking. PTSB medium (20) was used for P. aeruginosa cultures, and LB medium (28) was used for E. coli cultures. The defined, nitrogen-limited Guerra-Santos (GS) medium used for rhamnolipid determinations (11) was supplemented with 20% glycerol instead of 0.1 M glucose (17). Elastin-agar plates contained 0.5% elastin and 0.8% nutrient broth (20). For β-galactosidase (β-Gal) determinations, A medium (23) supplemented with 0.05% yeast extract, 0.4% glucose, and 1 mM MgSO4 was used. Antibiotics were used at the following concentrations when required: for P. aeruginosa, carbenicillin (200 μg/ml), tetracycline (100 or 50 μg/ml in solid or liquid medium, respectively), and gentamicin (100 μg/ml); for E. coli, ampicillin (100 μg/ml), tetracycline (20 μg/ml), and gentamicin (15 μg/ml). 3-oxo-C12-HSL and C4-HSL autoinducers were synthesized previously (22, 24).
Bacterial strains and plasmids.
Bacterial strains and plasmids are listed in Table 1. The P. aeruginosa gene bank, GB24, was kindly provided by U. Ochsner. This 95% complete gene bank contains a Sau3A partial digestion of the P. aeruginosa wild-type strain PAO1 genome, cloned into the BamHI site of the multicopy vector plasmid pUCP24 (30). It is composed of approximately 5,000 independent DNA fragments with a median size of 3.3 kb (2 to 7 kb) (U. Ochsner, personal communication). Plasmid pVD was constructed by cloning a dksA-containing 960-bp SmaI fragment from pVD99.3 into the Klenow-repaired XhoI site of pLPRI (36). Plasmid pDECP60 was constructed by cloning the 960-bp SmaI fragment of pVD99.3 into the SmaI site of pECP60 (26). The orientation of dksA in pDECP60 is opposite that of the adjacent rhlA′-lacZ fusion. Plasmid pPBL25 was constructed by ligating a Klenow enzyme-treated 3-kb Eco0109 lasB′-lacZ-containing fragment obtained from pKDT37 (22) into the SmaI site of pUCP18. Plasmid pPBLD26 was obtained by ligating a 960-bp SmaI dksA-containing fragment of pVD99.3 into the Klenow enzyme-repaired XbaI site of pPBL25.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| E. coli DH5α | F′/endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 deoR [φ80 dlacZΔM15 recA1] | 40 |
| P. aeruginosa | ||
| PAO1 | Wild-type, elastolytic, prototroph | 12 |
| PAO-R1 | ΔlasR::Tcr, nonelastolytic derivative of strain PAO1 | 8 |
| PR1-E4 | ΔlasR::Tcr, casein incubation-induced elastolytic mutant of strain PAO-R1 | 36 |
| PDO100 | ΔrhlI::Tn501-2 derivative of PAO1 | 3 |
| Plasmids | ||
| pBluescript II SK+ | General-purpose cloning vector, ori (ColE1), Apr | Stratagene |
| pUC18, pUC19 | General-purpose cloning vectors, ori (ColE1) | Gibco |
| pUCP18, pUCP19, pUCP24 | E. coli-P. aeruginosa shuttle vectors | 30 |
| pRL1500 | pUCP19 containing rhlA on a 1.5-kb BglI-NcoI fragment from strain PG201 | 18 |
| pPAO-2 | pUC19 containing a 280-bp Sau3A fragment of pilA from strain PAO1 | 29 |
| pJPP41 | pBluescript II SK+ containing rhlI on a 950-bp BglII-SphI fragment from strain PAO1 | 25 |
| pRB1801 | pUC18 containing a 1.75-kb EcoRI-SmaI fragment of lasB from strain PAO1 | 2 |
| GB24 | P. aeruginosa gene bank: partial Sau3A digest of strain PAO1 DNA inserted into the BamHI site of pUCP24 | U. Ochsner |
| pVD99.0 | pUCP24 containing a 2.5-kb Sau3A genome fragment from strain PAO1 | This study |
| pVD99.1 | pUCP24 containing a 1.98-kb EcoRI fragment from pVD99.0 | This study |
| pVD99.3 | pUCP24 containing dksA on a 960-bp SmaI fragment from pVD99.0 | This study |
| pVD99.4 | pUCP24 containing a 523-bp EcoRI fragment from pVD99.0 | This study |
| pVD99.5 | pUCP24 containing a 520-bp SalI fragment from pVD99.0 | This study |
| pECP60 | pSW205 containing a rhlA′-lacZ translational fusion | 26 |
| pDECP60 | pECP60 containing dksA on a 960-bp SmaI fragment from pVD99.3 | This study |
| pLPR1 | pLP170 containing a rhlI′-lacZ transcriptional fusion | 36 |
| pVD | pLPRI containing dksA on a 960-SmaI fragment from pVD99.3 | This study |
| pPBL25 | pUCP18 containing lasB′-lacZ on a 3-kb Eco0109 fragment from pKDT37 (22) | This study |
| pPBLD26 | pPBL25 containing dksA on a 960-bp SmaI fragment from pVD99.3 | This study |
DNA techniques.
We used standard techniques for DNA manipulations (28). Restriction endonucleases and DNA-modifying enzymes were purchased from Gibco/BRL or New England Biolabs. Plasmids were introduced into E. coli by transformation (28) and into P. aeruginosa by electroporation (31).
Cloning of the dksA gene from mutant PR1-E4.
A BamHI-NotI digest of mutant PR1-E4 DNA was separated by gel electrophoresis and screened for the presence of the dksA gene, using a 32P-radiolabeled 350-bp SmaI-SphI probe obtained from pVD99.3. A 2,500-bp BamHI-NotI chromosomal DNA fragment of mutant PR1-E4 containing the dksA gene was cloned into the BamHI-NotI site of pBluescript SKII+. Colony blot hybridization on nylon membranes was performed using a 350-bp SmaI-SphI probe for dksA obtained from pVD99.3, using E. coli strain DH5α as a host. The dksA gene recovered from strain PR1-E4 was sequenced using a Li-Cor 4000L electrophoresis apparatus and Ladderman dideoxy sequencing kit.
RNA preparation and Northern blot analysis.
Total cellular RNA was prepared as previously described (5). In brief, P. aeruginosa cells were grown at 37°C in PTSB medium to stationary phase (optical density at 660 nm [OD660] = 2.0 ± 0.2), when the las and rhl systems are active, collected, and lysed in 3.5% sodium dodecyl sulfate. RNA was recovered after centrifugation of cell lysates on a 5.7 M CsCl cushion and further purified by two phenol-chloroform extractions. Northern blot analyses were performed following standard protocols. After electrophoresis on a 2.2 M formaldehyde–1.2% agarose gel, RNA was transferred to Hybond nylon membranes and hybridized according to the Amersham protocol to 32P-labeled double-stranded DNA probes. DNA probes were obtained as follows. A 560-bp BamHI internal fragment of rhlA was obtained from pRL1500 (18), a 630-bp SalI internal fragment of lasB was obtained from pRB1801 (2), and a 250-bp KpnI-EcoRI internal fragment of rhlI was obtained from pJPP41 (25). A 350-bp SmaI-SphI fragment from pVD99.3 was used as a dksA probe. For Northern analysis of dksA expression, total cellular RNA was obtained from PTSB cultures of strains PAO1, PAO-R1, and PR1-E4 in the early exponential phase of growth (OD660 = 0.8) and late stationary phase of growth (OD660 = 2.0). 32P labeling was performed by nick translation, and probes were separated from unincorporated nucleotides using NucTrap push columns (Stratagene). RNA experiments were performed twice with independent RNA preparations. To ascertain that the RNA of each strain was intact and loaded equally, we used a probe for the P. aeruginosa pilA gene (a 280-bp EcoRI-BamHI internal fragment of pilA from pPAO-2 [29]), which encodes the structural subunit of pilin. Previous studies have shown that pilA mRNA is not affected by the lasR deletion in strain PAO-R1 (9).
Elastase and rhamnolipid production assays.
Elastase production was measured by elastin Congo red assays as previously described (25). Rhamnolipid concentration in P. aeruginosa culture fluids was determined as previously described by orcinol assays (25).
β-Gal activity assays.
β-Gal activity was measured as previously described (16), with the following modifications. P. aeruginosa cultures were grown for 18 h at 37°C with vigorous shaking in PTSB medium supplemented with carbenicillin (200 μg/ml) and subcultured into the same medium to a starting OD660 of 0.15. Cultures were assayed for β-Gal activity at regular intervals during growth. Cells were washed twice and resuspended in A medium prior to β-Gal activity determinations. All experiments were done in triplicates and performed at least two times.
Determination of C4-HSL concentrations.
Culture supernatants were extracted with ethyl acetate, and C4-HSL concentrations were determined in a previously described (25) bioassay using PAO-JP2(pECP61.5).
Nucleotide sequence accession number.
The sequence of dksA is accessible in GenBank (accession number AF062653).
RESULTS
Isolation of a P. aeruginosa dksA homologue that suppresses elastase production of mutant PR1-E4.
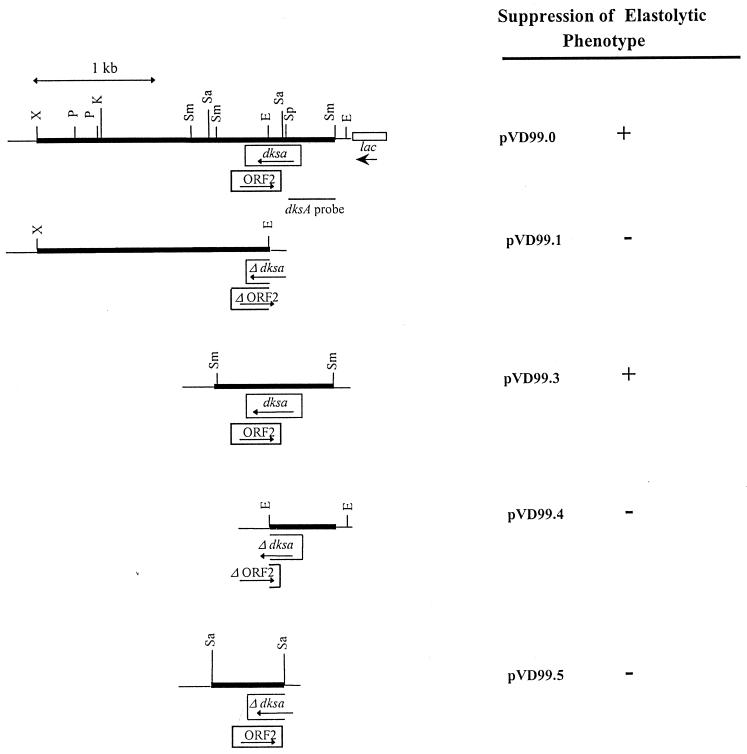
We wanted to determine whether the mutation restoring elastase production of the lasR-deficient strain PR1-E4 could be complemented by a wild-type gene or suppressed by overexpression of a different gene. We therefore electroporated the wild-type P. aeruginosa gene bank GB24 into the elastase-producing mutant PR1-E4; 6,700 isolated clones were screened on elastin-agar plates for the absence of elastase production. We found one non-elastase-producing clone, PR1-E4(pVD99.0). Isolation of pVD99.0 revealed a 2.5-kb fragment from the wild-type PAO1 genome (Fig. 1). Neither plasmid pVD99.1, which contained a 1.98-kb fragment of pVD99.0, nor plasmid pVD99.4, which contained the other 523-bp fragment of pVD99.0, suppressed elastase production by mutant PR1-E4. Plasmid pVD99.3 was obtained during subcloning of the 2.5-kb fragment by ligating a 960-bp SmaI fragment of pVD99.0 into the SmaI site of pUCP24. This plasmid, when electroporated into PR1-E4, still completely suppressed its elastase production. Sequencing of the 960-bp insert of pVD99.3 revealed two complete putative open reading frames (ORFs), one from bases 292 to 735 (ORF1) and the second, in the opposite orientation, from bases 805 to 464 (ORF2). Subcloning a 520-bp SalI fragment of pVD99.0 created pVD99.5, which contains an intact ORF2 but lacks the first 153 bp of ORF1. Plasmid pVD99.5 did not suppress elastase production by mutant PR1-E4. This result, together with the finding that ORF2 exhibits codon usage unusual (38) for P. aeruginosa, makes it unlikely that ORF2 is responsible for the suppression of elastase production by mutant PR1-E4. In contrast, the 443-bp ORF1 had a codon usage typical for P. aeruginosa, and accordingly the Genetics Computer Group program CODON PREFERENCE (38) indicated a high coding probability throughout its entire sequence. A search of the GenBank-European Molecular Biology Laboratory sequence database revealed amino acid homology between the gene product of this ORF and the E. coli dnaK mutation suppressor protein DksA (76% identity and 87% similarity) (13). We had therefore identified a new P. aeruginosa gene, which was homologous to the E. coli gene dksA and suppressed elastase production by mutant PR1-E4.
FIG. 1.
Restriction endonuclease map and subcloning strategy for plasmid pVD99.0. Closed and open boxes represent complete and truncated genes, respectively. Important restriction endonuclease sites, the dksA probe, and the position and orientation of the lac promoter are indicated. The elastolytic phenotype suppressor activity of the different subclones was determined on elastin-agar plates. Abbreviations: E, EcoRI; K, KpnI; P, PstI; Sa, SalI; Sm, SmaI; Sp, SphI; X, XbaI.
Cloning and transcription levels of the dksA gene from mutant PR1-E4.
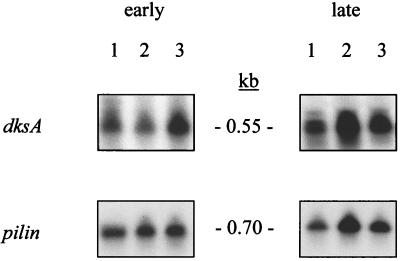
To examine whether strain PR1-E4 bears a mutated dksA gene that had been complemented by the wild-type dksA gene on plasmid pVD99.3, we cloned the dksA gene from mutant PR1-E4. The dksA gene from mutant PR1-E4 was recovered and sequenced, and its nucleotide sequence was compared with that of the wild-type P. aeruginosa dksA gene as described in Materials and Methods. We found no mutation in either the dksA ORF or the 350 bp of DNA upstream from dksA (data not shown). To determine whether a reduction of dksA transcription restored the elastase production of mutant PR1-E4, we also examined the expression of dksA in both exponential (early) and stationary (late) growth phases. Using Northern blot analysis, we could not detect a difference in dksA mRNA levels between the wild-type strain PAO1 and mutants PAO-R1 and PR1-E4 (Fig. 2). (Note that the difference seen for dksA between lanes 1 and 2 for late RNA is identical to that seen with the pilin probe and is therefore presumed to be caused by loading differences.) Consequently, the dskA gene in strain PR1-E4 does not contain a mutation, nor is its expression downregulated. The suppression of elastase production of mutant PR1-E4 by dksA supplied in trans is therefore a phenotypic complementation, similar to the mutation suppressor effect of the E. coli dksA gene on chaperone mutations (13).
FIG. 2.
dksA transcription in strains PAO1, PAOR-1, and PR1-E4. Total cellular RNA (12.5 μg) from early (exponential)- and late (stationary)-phase cultures were hybridized to a 32P-labeled dksA probe. A pilA probe was used to ascertain equal loading and transfer of RNA. Lanes: 1, strain PAO1; 2, strain PAO-R1; 3, strain PR1-E4. Molecular sizes are indicated in the center.
dksA affects the production of virulence factors in both PR1-E4 and PAO1.
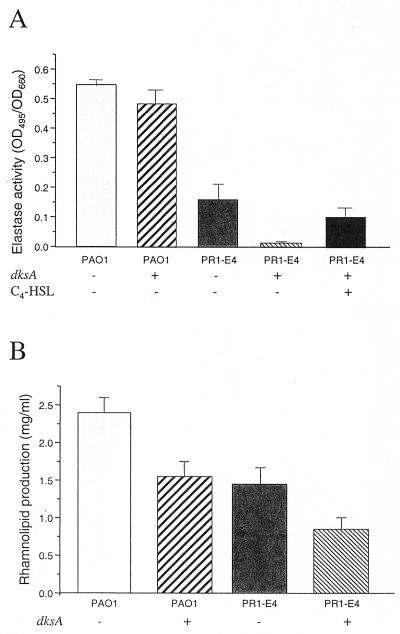
To quantify the inhibition of elastase and rhamnolipid production by dksA, we performed elastin Congo red and orcinol assays in the presence of the dksA-containing plasmid pVD99.3 or the vector control pUCP24 (see Materials and Methods). dksA completely abolished the elastase production of mutant PR1-E4 (Fig. 3A) and reduced its rhamnolipid production by 40% (Fig. 3B). We also wondered whether multiple copies of dksA could inhibit the elastase and/or rhamnolipid production of the wild-type strain PAO1. dksA only slightly reduced elastase production by strain PAO1 (Fig. 3A). However, the production of rhamnolipid was reduced by 35% when dksA was overexpressed in PAO1 (Fig. 3B). Therefore, dksA suppressed both elastase and rhamnolipid production in the absence of a functional las quorum-sensing system but mainly affected the production of rhamnolipid in a wild-type background.
FIG. 3.
Effect of dksA on elastase and rhamnolipid production in PAO1 and PR1-E4. Elastase (A) and rhamnolipid (B) production in wild-type strain PAO1 and mutant PR1-E4 were measured by the elastin Congo red and orcinol assays, respectively, in the presence of either the dksA-containing plasmid pVD99.3 (dksA +), or the vector control pUCP24 (dksA −). Supernatants were obtained from stationary-phase cultures from cells growing for 20 h in PTSB medium (OD660 = 2.0 ± 0.2) for determination of elastase production in the presence or absence of 10 μM C4-HSL and from cells growing for 72 h in modified GS medium (OD660 = 3.2 ± 0.4) for determination of rhamnolipid production. All experiments were performed in triplicate and repeated at least two times.
dksA inhibits the expression of genes belonging to the quorum-sensing circuitry.
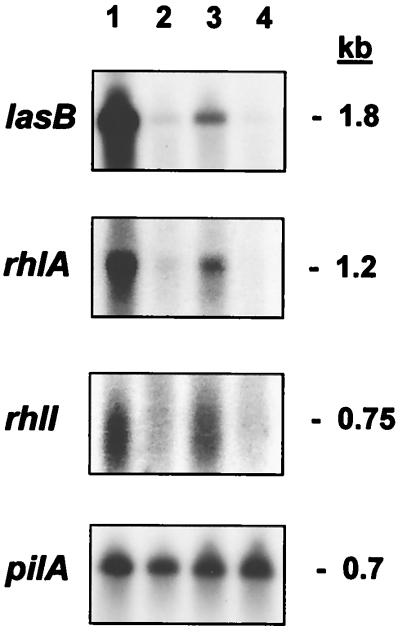
Expression of the elastase gene (lasB), the rhamnosyltransferase genes (rhlAB), and the rhl autoinducer synthase gene (rhlI) is partially restored in mutant PR1-E4 compared to its parent strain, PAO-R1 (36). To determine whether overexpression of dksA affects the transcription of these genes, we performed Northern blot analysis of strains PAO1, PAO-R1, PR1-E4, and PR1-E4(pVD99.3), using lasB, rhlA, and rhlI probes, as described in Materials and Methods. lasB, rhlA, and rhlI mRNAs gave intense signals in the wild-type strain PAO1, contrasting with mutant PAO-R1, in which the mRNAs of these three genes were barely visible (Fig. 4, lanes 1 and 2, respectively). All three mRNAs were detected in mutant PR1-E4 (Fig. 4, lane 3). These results confirm our previous report that lasB, rhlA, and rhlI are barely transcribed in the absence of LasR in mutant PAO-R1 and that their expression is partially restored in mutant PR1-E4 (36). However, when dksA was supplied in trans on pVD99.3 to mutant PR1-E4, neither lasB, rhlA, nor rhlI message was detected (Fig. 4, lane 4). In contrast, the pilA message, used as a control, was not affected by the presence of dksA.
FIG. 4.
Northern blot analysis of lasB, rhlA, and rhlI mRNA. Total cellular RNA (10 to 15 μg) was hybridized to 32P-labeled lasB, rhlA, and rhlI probes. Equal loading and transfer of RNA were verified using a pilA probe. Lanes: 1, strain PAO1(pUCP24); 2, strain PAO-R1(pUCP24); 3, strain PR1-E4(pUCP24); 4, strain PR1-E4(pVD99.3). pVD99.3 contains the P. aeruginosa dksA gene cloned in the pUCP24 vector plasmid. Molecular sizes are indicated to the right.
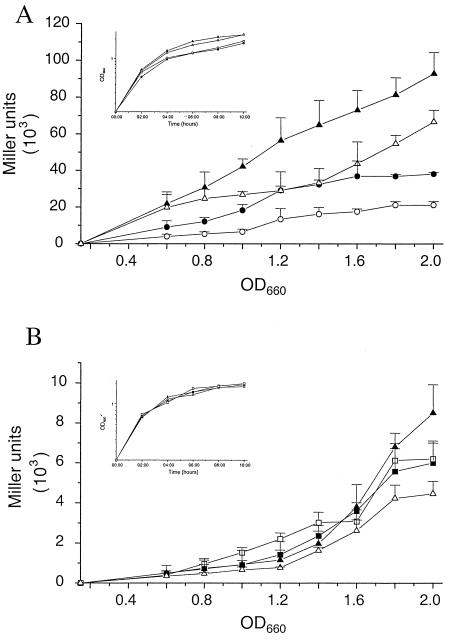
To confirm the suppressive effect of dksA on rhlI mRNA levels, we measured the expression of rhlI′-lacZ in the presence and absence of multiple copies of dksA, using plasmids pLPRI (rhlI′-lacZ) and pVD (rhlI′-lacZ dksA). The expression of rhlI′-lacZ in mutant PR1-E4 was reduced to 17% in the presence of dksA [PR1-E4(pLPRI), 18,201 ± 909 Miller units; PR1-E4(pVD), 3,135 ± 586 Miller units; measured at an OD660 of 1.8]. As PR1-E4 is an undefined mutant, we wanted to confirm the inhibition of rhlI expression in a defined genetic background. We therefore compared the expression of rhlI′-lacZ from plasmid pLPRI and pVD in the defined ΔlasR mutant PAO-R1 and in the wild type, PAO1. rhlI is barely transcribed in mutant PAO-R1 because of a nonfunctional las quorum-sensing system (36). To compensate for this defect, we measured the expression of rhlI in mutant PAO-R1 in the presence of 10 μM C4-HSL. In both strains, the presence of dksA reduced the expression of rhlI′-lacZ (Fig. 5A). Therefore, dksA interferes with the transcription of rhlI not only in the undefined mutant PR1-E4 but also in the ΔlasR mutant PAO-R1, despite constant C4-HSL levels, and in a wild-type background.
FIG. 5.
Effect of dksA on the expression of rhlI and rhlAB. (A) Expression of rhlI was monitored by β-Gal determinations during growth of the ΔlasR mutant PAO-R1 in the presence of 10 μM C4-HSL (circles) and the wild-type strain PAO1 (triangles), using either plasmid pLPRI (rhlI′-lacZ; solid symbols) or plasmid pVD (rhlI′-lacZ dksA; open symbols). (B) Expression of rhlAB was determined during the growth of PAO1 (triangles) and the ΔrhlI mutant PDO100 in the presence of 10 μM C4-HSL (squares) by β-Gal assays, using either plasmid pECP60 (rhlA′-lacZ; solid symbols) or plasmid pDECP60 (rhlA′-lacZ dksA; open symbols). Growth of the strains was not influenced by the presence of either pVD or pDECP60, as shown in the insets.
The effect of dksA on rhlAB and lasB transcription is indirect.
dksA could primarily affect the expression of rhlI, leading to an indirect reduction of rhlAB and lasB expression. However, it could also directly inhibit the expression of these three genes. To address this question, we cloned dksA under its own promoter on plasmid pECP60, which contains a rhlA′-lacZ reporter fusion, to obtain plasmid pDECP60 (see Materials and Methods). In wild-type strain PAO1, the presence of dksA in trans reduced the expression of rhlA′-lacZ (Fig. 5B). These results confirm those of the orcinol assays that showed reduced rhamnolipid production in the presence of dksA (Fig. 3B). To determine whether the inhibition of rhlI transcription by dksA is responsible for this effect, we determined the effect of dksA on rhlAB expression in the defined rhlI mutant PDO100 (3). As rhlAB is normally not transcribed in this mutant, these experiments had to be performed in the presence of 10 μM exogenous C4-HSL (26). These experimental conditions allow distinction of a direct effect of dksA on rhlAB expression from an indirect effect, due primarily to an inhibition of rhlI expression. Indeed, if rhlAB transcription is not directly affected by dksA, then its expression should not be diminished in this strain, as the source of the C4-HSL autoinducer is exogenous. As shown in Fig. 5B, rhlAB was expressed similarly from plasmids pECP60 (rhlA′-lacZ) and pDECP60 (rhlA′-lacZ dksA), suggesting that dksA does not directly affect rhlAB expression. To demonstrate that the inhibitory effect of dksA can be overcome by exogenous C4-HSL, we also determined the expression of rhlAB in the presence of exogenous C4-HSL in PAO1 and PR1-E4 (Table 2). The addition of 10 μM exogenous C4-HSL restored the expression of rhlAB in both strains PAO1(pDECP60) and PR1-E4(pDECP60). These results show that exogenous C4-HSL can compensate for the inhibition of rhlI expression by dksA and confirm the hypothesis that dksA does not affect rhlAB expression directly. The expression of rhlR is reduced to 18% of the wild-type level in both strains PAO1 and PR1-E4 (36) and is not further reduced in PR1-E4 by the addition of dksA in trans (assayed by Northern blotting [data not shown]). Apparently the concentration of RhlR is high enough to support, in the presence of an adequate C4-HSL concentration, the level of expression of rhlAB observed in mutant PR1-E4.
TABLE 2.
Restoration of rhlA and lasB expression by exogenous C4-HSLa
| Fusion | Enzyme activity (Miller units)
|
|||||
|---|---|---|---|---|---|---|
| PAO1
|
PR1-E4
|
|||||
| −dksA | +dksA | +dksA + 10 μM C4-HSL | −dksA | +dksA | +dksA + 10 μM C4-HSL | |
| rhlA′-lacZ | 6,830 ± 625 | 4,200 ± 600 | 7,350 ± 650 | 390 ± 45 | 54 ± 8 | 372 ± 32 |
| lasB′-lacZ | ND | ND | ND | 379 ± 40 | 110 ± 10 | 594 ± 42 |
Cultures of PAO1 and mutant PR1-E4 carrying the indicated reporter fusions were grown for 8 h (OD660 = 1.8 ± 0.1), and β-Gal activity was assayed. Data are the mean ± 1 standard deviation of at least two separate experiments performed in triplicate. −dksA, pECP60 (rhlA′-lacZ) or pPBL25 (lasB′-lacZ); +dksA, pDECP60 (rhlA′-lacZ dksA) or pPBLD26 (lasB′-lacZ dksA). ND, not determined.
To determine whether dksA inhibits elastase production directly by inhibition of lasB expression, we used a similar strategy and cloned dksA under its own promoter on plasmid pPBL25, which contains a lasB′-lacZ reporter fusion to obtain plasmid pPBLD26 (see Materials and Methods). To avoid the interference of a reduced expression of rhlI, these experiments were also first performed in the rhlI mutant PDO100. Similar to the case for rhlAB, lasB expression is reduced in mutant PDO100 due to the lack of C4-HSL production (3). The transcription of lasB in mutant PDO100 was therefore measured in the presence of 10 μM exogenous C4-HSL. In these experimental conditions, the expression of lasB was not suppressed by dksA [PDO100(pPBL25), 16,640 ± 885 Miller units; PDO100(pPBLD26), 15,347 ± 697 Miller units; mean of five independent experiments ± standard errors, measured at an OD660 of 2.0). To confirm these results, we determined the expression of lasB in mutant PR1-E4, using the reporter fusions pPBL25 and pPBLD26, in the absence and the presence of exogenous C4-HSL (Table 2). As expected, the expression of lasB was reduced in the presence of dksA. This inhibition was overcome by the addition of 10 μM exogenous C4-HSL. These results suggest that dksA does not inhibit lasB expression directly but reduces the production of the C4-HSL autoinducer, which leads indirectly to a reduction of las B and rhlAB expression. To support this hypothesis, we measured the C4-HSL concentrations in the culture supernatants obtained from the same experiments (Table 3). Not surprisingly, mutant PR1-E4 produced less C4-HSL autoinducer than PAO1, confirming the partial restoration of rhlI expression observed previously in this strain (36). The concentration of C4-HSL in supernatants of mutant PAO-R1 was at the limit of detection of our bioassay (the sensitivity of our bioassay was 0.04 μM). In the presence of dksA, no C4-HSL was detected by our bioassay in supernatants of strain PR1-E4. Therefore, dksA in trans severely reduced the production of the C4-HSL autoinducer, as expected from the inhibition of rhlI expression by dksA. To finally confirm that exogenous C4-HSL can overcome the inhibitory effect of dksA, we also measured the production of elastase by strain PR1-E4 in the presence of dksA and exogenous C4-HSL. As shown in Fig. 3A, the addition of 10 μM C4-HSL in elastin Congo red assays restored the production of elastase by strain PR1-E4(pVD99.3). It therefore appears that dksA does not inhibit the expression of lasB and rhlAB directly but affects both elastase and rhamnolipid production indirectly by the inhibition of rhlI transcription, leading to reduced C4-HSL levels.
TABLE 3.
C4-HSL concentrations in cultures of wild-type PAO1 and mutant PR1-E4 carrying or not carrying dksA
| Culture density (OD660) | C4-HSL concn (μM)a
|
||
|---|---|---|---|
| PAO1/pPBL25 | PRI-E4 carrying:
|
||
| pPBL25 | pPBLD26 | ||
| 1.4 | 5.1 ± 0.2 | 0.4 ± 0.1 | <0.04 |
| 1.8 | 6.3 ± 0.4 | 1.2 ± 0.2 | <0.04 |
Mean ± 1 standard deviation of at least two separate experiments performed in triplicate.
DISCUSSION
An upregulation of rhlI expression has been suggested to be responsible for the partial restoration of elastase and rhamnolipid production by the P. aeruginosa starvation mutant PR1-E4 (36). In the present study, we have complemented mutant PR1-E4 with a wild-type P. aeruginosa gene bank carried on a multicopy plasmid. By screening for the loss of elastase production, we isolated a P. aeruginosa homologue to the E. coli dnaK multicopy suppressor gene dksA. We have shown that dksA is neither mutated nor expressed at a reduced level in mutant PR1-E4. dksA in trans not only abolishes elastase production in mutant PR1-E4 but also downregulates rhamnolipid production in mutant PR1-E4 and in the wild type, PAO1. Using Northern blot analysis and plasmids carrying both dksA and rhlI′-lacZ, rhlA′-lacZ, and lasB′-lacZ reporter fusions, we have shown that dksA reduces the expression of these three genes, as well as the production of the C4-HSL autoinducer. The addition of exogenous C4-HSL autoinducer compensates for the inhibitory effect of dksA and restores elastase production, as well as rhlAB and lasB transcription. These results suggest that overexpression of dksA inhibits rhlI expression, leading to a secondary reduction of rhamnolipid and elastase production.
dksA homologues with strikingly conserved sequences have been isolated from E. coli (13), Haemophilus influenzae (4), Salmonella enterica serovar Typhimurium (34), and now P. aeruginosa. The cellular localization and function of DksA are still unknown. DksA shares amino acid similarity with the E. coli TraR protein, a putative transcriptional regulator (6). Both proteins contain a zinc finger domain (Cx2Cx17Cx2C) that facilitates binding to DNA. Interestingly, the putative zinc finger motif is the region most conserved between the known DksA homologues, suggesting that these proteins might also function as gene expression regulators. In E. coli, dksA expressed from multicopy plasmids suppresses the temperature-sensitive phenotype associated with deletion mutations in the heat shock genes dnaK, dnaJ, and grpE280n (13) and in the mukB gene (41). It is also a multicopy suppressor of the conditional lethal phenotype associated with a prc null mutation in E. coli (1) and has been suggested to be involved in plasmid replication (19). The S. enterica serovar Typhimurium DksA homologue has been linked to virulence of this strain, as a ΔdksA S. enterica serovar Typhimurium mutant was impaired in the ability to colonize chickens (34). Both E. coli and S. enterica serovar Typhimurium dksA mutants show poor growth in minimal media and defects in glutamine-glutamate biosynthesis (13, 34). The S. enterica serovar Typhimurium dksA mutant also yielded a higher RNA amount in stationary-phase cultures than the wild type (34). For these reasons, DksA has been suggested to be involved in stress responses such as the stringent response (34). Recently DksA has also been shown to be required for optimal translation of the stationary-phase sigma factor rpoS, as well as for the expression of several other genes in S. enterica serovar Typhimurium (37). DksA might therefore regulate the expression of genes which products are required for the stabilization of proteins during the stringent response and entrance into stationary phase (34)
The relationship between rpoS and the rhl quorum-sensing system in P. aeruginosa is a matter of debate. Whereas previous data suggested that the transcription of rpoS might be regulated by the rhl system (14), recent experiments have suggested that RpoS might repress rhlI, rather than the rhl system influencing the expression of rpoS (39). Could an upregulation of RpoS, secondary to the overexpression of dksA, explain the effects observed in this study? The repression of rhlI by RpoS manifests essentially during early exponential growth, whereas the inhibition of rhlI by dksA occurs mainly during stationary growth. Moreover, an rpoS mutation results in increased production of pyocanin, a secondary metabolite dependent on the rhl quorum-sensing system (whether this mutation also affects rhamnolipid production is unknown), but more importantly also in a 20% reduction in the production of elastase (32). This effect of RpoS on elastase production is opposite what would be needed to explain the inhibition of elastase production by dksA. It is therefore unlikely that the results presented in this study are due to an upregulation of RpoS alone. The overexpression of dksA could also affect another regulator of rhlI expression that has been previously suspected (14). This regulator could be the recently described Pseudomonas quinolone signal, which increases the expression of rhlI (15). This third P. aeruginosa cell-to-cell signal seems to acts as a link between the las and rhl quorum-sensing systems.
Our data show for the first time that DksA can interfere with quorum sensing. Further work is required to determine the physiologic function of DksA and whether it plays a significant role in the regulation of virulence factor production by P. aeruginosa.
ACKNOWLEDGMENTS
We thank L. Passador and T. DeKievit for discussions, U. Ochsner for the gift of the GB24 gene bank, R. Comte for outstanding technical assistance, and L. Tabak, T. Barras, W. Kuhnert, and G. Campo for help with DNA sequencing.
This work was supported by NIH grant R01A133713-04 (to B.H.I.), NIH predoctoral training grant 5-T32 AI07362-09 (to J.P.P.), Cystic Fibrosis Foundation grant PESCI96FO (to E.C.P.), and a Wilmot Foundation grant and Swiss National Research Foundation grants 3231-051940.97 and 3200-052189.97 (to C.V.D.).
REFERENCES
- 1.Bass S, Gu Q, Christen A. Multicopy suppressors of prC mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoAB), DksA, and a truncated R1pA. J Bacteriol. 1996;178:1154–1161. doi: 10.1128/jb.178.4.1154-1161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bever R A, Iglewski B H. Molecular characterization and nucleotide sequence of the Pseudomonas aeruginosa elastase structural gene. J Bacteriol. 1988;170:4309–4314. doi: 10.1128/jb.170.9.4309-4314.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clifton S W, McCarthy D, Roe B A. Sequence of the rec-2 locus of Haemophilus influenzae: homologies to comE-ORF3 of Bacillus subtilis and msbA of Escherichia coli. Gene. 1994;146:95–100. doi: 10.1016/0378-1119(94)90840-0. [DOI] [PubMed] [Google Scholar]
- 5.Deretic V, Gill J F, Chakrabarty A M. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987;169:351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doran T J, Loh S M, Firth N, Skurray R A. Molecular analysis of the F plasmid traVR region: traV encodes a lipoprotein. J Bacteriol. 1994;176:4182–4186. doi: 10.1128/jb.176.13.4182-4186.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 8.Gambello M J, Iglewski B H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gambello M J, Kaye S, Iglewski B H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenberg E P. Quorum sensing in gram-negative bacteria. ASM News. 1997;63:371–377. [Google Scholar]
- 11.Guerra-Santos L H, Käppeli O, Fiechter A. Dependence of Pseudomonas aeruginosa continuous culture biosurfactant production on nutritional and environmental factors. Appl Microbiol Biotechnol. 1986;24:443–448. [Google Scholar]
- 12.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang P J, Craig E A. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 15.McKnight S L, Iglewski B H, Pesci E C. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 2000;182:2702–2708. doi: 10.1128/jb.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 17.Ochsner U A, Fiechter A, Reiser J. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem. 1994;269:19787–19795. [PubMed] [Google Scholar]
- 18.Ochsner U A, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohkubo S, Yamaguchi K. A suppressor of mutations in the region adjacent to iterons of pSC101 ori. J Bacteriol. 1997;179:2089–2091. doi: 10.1128/jb.179.6.2089-2091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohman D E, Cryz S J, Iglewski B H. Isolation and characterization of Pseudomonas aeruginosa PAO mutant that produces altered elastase. J Bacteriol. 1980;142:836–842. doi: 10.1128/jb.142.3.836-842.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 22.Passador L, Tucker K D, Guertin K R, Journet M P, Kende A S, Iglewski B H. Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J Bacteriol. 1996;178:5995–6000. doi: 10.1128/jb.178.20.5995-6000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson J P, Passador L, Iglewski B H, Greenberg E P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson J P, Pesci E C, Iglewski B H. Role of Pseudomonas aeruginosa las and rhl quorum-sensing systems in the control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rumbaugh K P, Griswold J A, Iglewski B H, Hamood A N. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect Immun. 1999;67:5854–5862. doi: 10.1128/iai.67.11.5854-5862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sastry P A, Finlay B P, Pasloske B L, Paranchych W, Pearlstone J R, Smillie L B. Comparative studies of the amino acid sequences of pilin derived from Pseudomonas aeruginosa PAK and PAO1. J Bacteriol. 1985;164:571–577. doi: 10.1128/jb.164.2.571-577.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 31.Smith A W, Iglewski B H. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 1989;17:10509–10509. doi: 10.1093/nar/17.24.10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suh S J, Silo-Suh L, Woods D E, Hassett D J, West S E, Ohman D E. Effect of rpoS Mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang H B, DiMango E, Bryan R, Gambello M, Iglewski B H, Goldberg J B, Prince A. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner A K, Lovell M A, Hulme S D, Zhang-Barber L, Barrow P A. Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect Immun. 1998;66:2099–2106. doi: 10.1128/iai.66.5.2099-2106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Delden C, Iglewski B H. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Delden C, Pesci E C, Pearson J P, Iglewski B H. Starvation selection restores elastase and rhamnolipid production in a Pseudomonas aeruginosa quorum-sensing mutant. Infect Immun. 1998;66:4499–4502. doi: 10.1128/iai.66.9.4499-4502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webb C, Moreno M, Wilmes-Riesenberg M, Curtiss R, Foster J W. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol Microbiol. 1999;34:112–123. doi: 10.1046/j.1365-2958.1999.01581.x. [DOI] [PubMed] [Google Scholar]
- 38.West S E, Iglewski B H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988;16:9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whiteley M, Parsek M R, Greenberg E P. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J Bacteriol. 2000;182:4356–4360. doi: 10.1128/jb.182.15.4356-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodcock D M, Crowther P J, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith S S, Michael M Z, Graham M W. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamanaka K, Mitani T, Ogura T, Niki H, Hiraga S. Cloning, sequencing, and characterization of multicopy suppressors of a mukB mutation in Escherichia coli. Mol Microbiol. 1994;13:301–312. doi: 10.1111/j.1365-2958.1994.tb00424.x. [DOI] [PubMed] [Google Scholar]