Abstract
The UspA1 protein of Moraxella catarrhalis has been shown to function as an adhesin that mediates adherence to human epithelial cell lines in vitro (E. R. Lafontaine, L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen, J. Bacteriol. 182:1364–1373, 2000). In the present study, cell lysates prepared from individual colonies of several M. catarrhalis wild-type strains were analyzed by Western blot analysis using monoclonal antibodies (MAbs) specific for the UspA1 protein. Expression of UspA1 was shown to exhibit phase variation that was correlated with both adherence ability in vitro and the number of guanine (G) residues contained within a homopolymeric [poly(G)]tract located upstream of the uspA1 open reading frame (ORF). Nucleotide sequence analysis revealed that isolates expressing relatively high levels of UspA1 had 10 G residues in their uspA1 poly(G)tracts, whereas isolates that expressed much lower levels of UspA1 had 9 G residues. This poly(G) tract was located 30 nucleotides (nt) upstream of the uspA1 ORF and 168 nt downstream of the uspA1 transcriptional start site. Primer extension experiments, RNA slot blot analysis, and cat reporter constructs were used to demonstrate that M. catarrhalis isolates with 10 G residues in their uspA1 poly(G) tracts expressed two-to threefold more uspA1 mRNA than did isolates which had 9 G residues in their poly(G)tracts. Northern hybridization analysis revealed that an intact uspA1 mRNA was readily detectable in RNA from M. catarrhalis isolates that had 10 G residues in their uspA1 poly(G) tracts, whereas no full-length uspA1 mRNA was observed in isolates whose poly(G)tracts contained 9 G residues. M. catarrhalis strain O35E uspA1 genes that contained wild-type and mutated poly(G) tracts were expressed in Haemophilus influenzae to demonstrate that the length and composition of the poly(G)tract affected expression of UspA1.
Moraxella (Branhamella) catarrhalis is an unencapsulated gram-negative bacterium that can cause both upper and lower respiratory tract infections (14, 33). It has been estimated that M. catarrhalis causes approximately 20% of cases of acute bacterial otitis media in infants and young children (5) and is associated with nearly 30% of infectious exacerbations of chronic obstructive pulmonary disease in adults (17). The significant morbidity associated with M. catarrhalis infections as well as the substantial health care costs of these infections have prompted recent interest in the development of an M. catarrhalis vaccine (37).
Proteins present in or closely associated with the outer membrane of M. catarrhalis strains obtained from diverse geographic and clinical sources display highly similar patterns when analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (4) and have received the most attention as potential vaccine candidates. Several of these cell surface-exposed proteins have been characterized in some detail, including UspA1, UspA2 (HMWP), and UspA2H (24, 26, 32); OMP CD (21, 34); the iron-regulated CopB protein (3, 8); the LbpA and LbpB proteins (6); and the TbpA and TbpB proteins (7, 28, 35).
Little is known about the regulation of expression of M. catarrhalis outer membrane proteins. Campagnari et al. (8) were the first to show that the availability of iron in the growth medium affected expression of several M. catarrhalis outer membrane proteins. A spontaneous mutant of M. catarrhalis that lacked the ability to express several different outer membrane antigens was described by Murphy and coworkers (25). In addition, it was reported that one strain of M. catarrhalis could give rise to variants that expressed a truncated UspA2 protein (K. R. VanDerMeid, S. M. Baker, and J. C. McMichael, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. D/B-289, p. 256, 1999). Most recently, it was reported that the 200-kDa surface protein of this organism, which may be involved in hemagglutination (15), underwent phase-variable expression that involved apparent slipped-strand mispairing in a homopolymeric nucleotide repeat located within the open reading frame (ORF) encoding this protein. (K. Sasaki, L. Myers, S. M. Loosmore, and M. H. Klein, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., 1999, abstr. B/D-306, p. 89, 1999).
The UspA1 surface protein of M. catarrhalis is synthesized as an 80- to 90-kDa monomer that forms very large aggregates or complexes that are relatively resistant to heating in the presence of SDS (12, 32). This protein also has been shown to mediate attachment of this bacterium to Chang conjunctival epithelial cells in vitro (26). In the present study, expression of the M. catarrhalis UspA1 protein was found to exhibit phase variation. Nucleotide sequence analysis indicated that this phenotypic switch could be correlated with changes in the length of a homopolymeric nucleotide [poly(G)] tract located upstream of the uspA1 ORF. Primer extension, RNA slot blot, and Northern hybridization experiments revealed that UspA1 expression was regulated at the level of transcription. Cloning and expression of uspA1 genes in Haemophilus influenzae revealed that the changes in the length of the poly(G) tract were sufficient to account for the phase-variable expression of UspA1.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids are listed in Table 1. M. catarrhalis (2) and H. influenzae (16) strains were cultured as described earlier. Recombinant H. influenzae and E. coli strains were selected with chloramphenicol (2 and 10 μg/ml, respectively). The M. catarrhalis mutants containing the chloramphenicol acetyltransferase (CAT) reporter gene (cat) were selected with chloramphenicol (0.6 μg/ml). For adherence assays, RNA isolation experiments, and measurement of cat reporter activity, strains were grown in broth without antibiotic supplementation for several (three to four) generations.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Phenotype or description | Source or reference |
|---|---|---|
| M. catarrhalis | ||
| O35E | Wild-type strain | 19 |
| O35E.1 | Isogenic uspA1 mutant of O35E; expresses no UspA1 | 2 |
| O35E.98 | O35E isolate with poly(G) tract containing 10 G residues; expresses wild-type levels of UspA1 | This study |
| O35E.118 | O35E isolate with poly(G) tract containing 10 G residues; expresses wild-type levels of UspA1 | This study |
| O35E.117 | O35E isolate with poly(G) tract containing 9 G residues; expresses very low levels of UspA1 | This study |
| O35E.121 | O35E isolate with poly(G) tract containing 9 G residues; expresses very low levels of UspA1 | This study |
| O35E.135 | O35E isolate with poly(G) tract containing 9 G residues; expresses very low levels of UspA1 | This study |
| O35E.137 | O35E isolate with poly(G) tract containing 9 G residues; expresses very low levels of UspA1 | This study |
| O35E.118ATT1 | Derived from O35E.118 by selection for attachment to Chang cell monolayers; poly(G) tract has 10 G residues | This study |
| O35E.118ATT2 | Derived from O35E.118 by selection for attachment to Chang cell monolayers; poly(G) tract has 10 G residues | This study |
| O35E.118ATT3 | Derived from O35E.118 by selection for attachment to Chang cell monolayers; poly(G) tract has 10 G residues | This study |
| O35E.135ATT1 | Derived from O35E.135 by selection for attachment to Chang cell monolayers; poly(G) tract has 9 G residues | This study |
| O35E.135ATT2 | Derived from O35E.135 by selection for attachment to Chang cell monolayers; poly(G) tract has 9 G residues | This study |
| O35E.135ATT3 | Derived from O35E.135 by selection for attachment to Chang cell monolayers; poly(G) tract has 9 G residues | This study |
| O35E.118CAT | uspA1::cat reporter strain; poly(G) tract contains 10 G residues | This study |
| O35E.135CAT | uspA1::cat reporter strain; poly(G) tract contains 9 G residues | This study |
| O12E | Wild-type strain | 12 |
| O12E.44 | O12E isolate with poly(G) tract containing 10 G residues; expresses wild-type levels of UspA1 | This study |
| O12E.77 | O12E isolate with poly(G) tract containing 9 G residues; expresses very low levels of UspA1 | This study |
| O12E.1 | Isogenic uspA1 mutant of O12E; expresses no UspA1 | 26 |
| TTA37 | Wild-type isolate | 26 |
| O46E | Wild-type isolate | 26 |
| E. coli DH5α | Host strain for cloning experiments | New England Biolabs |
| H. influenzae DB117 | Host strain for cloning experiments | 41 |
| Plasmids | ||
| pACYC184 | Cloning vector | New England Biolabs |
| pELU1-10 G | pACYC184 containing the M. catarrhalis O35E.118 uspA1 gene with 10 G residues in the poly(G) tract | This study |
| pELU1-8 G | pACYC184 containing the M. catarrhalis O35E.118 uspA1 gene with 8 G residues in the poly(G) tract; obtained by site-directed mutagenesis of pELU1-10 G | This study |
| pELU1-11 G | pACYC184 containing the M. catarrhalis O35E.118 uspA1 gene with 11 G residues in the poly(G) tract; obtained by site-directed mutagenesis of pELU1-10 G | This study |
| pELU1-3G4R3G | pACYC184 containing the M. catarrhalis O35E.118 uspA1 gene with the poly(G) tract replaced by GGGACTAGGG; obtained by site-directed mutagenesis of pELU1-10 G | This study |
| pELU1-3G4R2G | pACYC184 containing the M. catarrhalis O35E.118 uspA1 gene with the poly(G) tract replaced by GGGACTAGG; obtained by site-directed mutagenesis of pELU1-10 G | This study |
| pELU1-9 G | pACYC184 containing the M. catarrhalis O35E.135 uspA1 gene with 9 G residues in the poly(G) tract | This study |
| pELU1-NOG | pACYC184 containing the M. catarrhalis O35E.135 uspA1 gene lacking the poly(G) tract and upstream DNA | This study |
| pUSPA1 | pBluescript containing an incomplete uspA1 gene from M. catarrhalis O35E | 2 |
| pELU1CAT | pUSPA1 with a promoterless cat cartridge inserted into the incomplete uspA1 gene | This study |
| pSL1 | Source of the promoterless cat cartridge | 29 |
Recombinant DNA techniques.
Standard molecular biology methods were performed as described previously (38). The H. influenzae Rd strain DB117 was used as the host strain for most recombinant DNA manipulations; Escherichia coli DH5α was used where noted. Electroporation was carried out as described earlier (26). DNA fragments used in cloning experiments, as templates for nucleotide sequence analysis, or as probes in hybridization experiments were purified with the Wizard PCR Preps system (Promega Corp., Madison, Wis.) after electrophoresis in agarose gels. Plasmid DNA was purified with the Wizard Plus Minipreps or Midipreps systems (Promega).
PCR.
DNA probes for Northern and RNA slot blot hybridization experiments were generated with Taq DNA polymerase (Promega). The 579-bp uspA1 probe was amplified from M. catarrhalis O35E with the oligonucleotide primers P1 (5′-AGGGATCCCCGTCCCCCTAATAAGTGAG-3′; BamHI site underlined) and P2 (5′-AACTCGAGTTGAACCTGTACCTGTGGCTTGG-3′; XhoI site underlined), whereas primers P3 (5′-AGGCATGCTTATGCTGGCTTTTGTC-3′; SphI site underlined) and P4 (5′-ACCTCGAGTTTAGCACTCTCTTTTGG-3′; XhoI site underlined) were used to generate the 503-bp uspA2 probe. The oligonucleotide primers P5 (5′-CGGCATGCCGGGTGACTAACTAGAGG-3′) and P6 (5′-CGGCATGCTCCTTCCAGAAATTACGC-3′) were used to amplify a 695-bp probe for the cat gene from pSL1. The Tm for the uspA1 probe was 78.4°C, while that of the uspA2 probe was 78.1°C, and that of the cat probe was 77.4°C. DNA fragments used for nucleotide sequence analysis were amplified with the Gene Amp XL PCR kit (Perkin-Elmer Biosystems, Branchburg, N.J.) according to the manufacturer's specifications. Amplicons used for cloning of uspA1 genes in H. influenzae were generated with Pfu DNA polymerase (Stratagene, La Jolla, Calif.) using the primers P7 (5′-AGGGATCCAACGACGGTCCAAGATGG-3′; BamHI site underlined) and P8 (5′-AGGGATCCCCTGCCACCTAAAGCCTTG-3′; BamHI site underlined) or P9 (5′-AGGGATCCGGAGACCCCAGTCATTTATTAG-3′; BamHI site underlined) and P8.
Construction of uspA1::cat reporter fusions.
Plasmid pUSPA1, which contains an incomplete uspA1 ORF from M. catarrhalis O35E (2) inserted into the multiple cloning site downstream from and in the same orientation as the plasmid-based lac promoter, was digested with BglII to remove a 0.6-kb internal fragment, and the BglII ends were then filled in with Pfu DNA polymerase using the manufacturer's recommended conditions. The resulting blunt-ended pUSPA1 plasmid was then ligated with a 706-bp SmaI fragment containing the promoterless cat gene from plasmid pSL1 (29). The ligation mixture was used to electroporate E. coli DH5α, and recombinant clones were selected for resistance to chloramphenicol; the plasmids in these recombinants had the cat insert oriented in the same direction as the plasmid-based lac promoter. Nucleotide sequence analysis of one of these, designated pELU1CAT, confirmed that the promoterless cat gene had been introduced in the direction of transcription of the uspA1 gene. Plasmid pELU1CAT was used to electroporate M. catarrhalis isolates O35E.118 and O35E.135. Chloramphenicol-resistant transformants were screened by PCR to identify those containing a uspA1::cat transcriptional fusion in the chromosome. Allelic exchange was confirmed by Southern blot analysis of two of these transformants, O35E.118CAT and O35E.135CAT. Nucleotide sequence analysis of the entire uspA1::cat gene of M. catarrhalis O35E.118CAT (including 300 nucleotides [nt] upstream of the translational start codon) revealed that it was identical to that in O35E.135CAT except that the poly(G) tract of the former contained 10 G residues, whereas the poly(G) tract of the latter had only 9 G residues.
Northern and slot blot hybridization analysis.
Total RNA was isolated from bacterial cells by use of the RNAWIZ reagent (Ambion, Austin, Tex.). For Northern hybridization analysis, RNA samples (20 μg) were electrophoresed into a denaturing (i.e., formaldehyde) agarose gel and transferred to a HybondN+ membrane (Amersham Pharmacia) as described elsewhere (38). For slot blot hybridization experiments, RNA samples were prepared as described previously (38) and transferred to a Nytran SuperCharge membrane (Schleicher & Schuell, Keene, N.H.) using a MINIFOLD 1 microsample filtration manifold (Schleicher & Schuell) as specified by the manufacturer. Hybridization was performed at 42°C for 18 h in ULTRAhyb hybridization buffer (Ambion). mRNAs were detected by autoradiography. The uspA1, uspA2, and cat probes were radioactively labeled with [α-32P]dCTP (NEN, Boston, Mass.) by use of a random-primed DNA labeling kit (Boehringer-Mannheim GmbH) according to the manufacturer's protocol. For one experiment, uspA1 and cat riboprobes were generated with the MaxiScript T7 kit (Ambion).
Primer extension analysis.
Primer extension experiments were performed with the avian myeloblastosis virus (AMV) reverse transcriptase (RT) Primer Extension System (Promega) according to the manufacturer's recommended procedure with the exception that the AMV RT supplied with this system was replaced by the RNA-dependent Moloney murine leukemia virus (MMLV) RT enzyme (Promega) for the synthesis of cDNA. The oligonucleotide primers P10 and P11 (Fig. 2B) were radioactively labeled with [γ-32P]dATP (NEN). Nucleotide sequence analysis was carried out with the AmpliCycle sequencing kit (Perkin-Elmer) and the radioactively labeled primers P10 and P11.
FIG. 2.
Primer extension analysis of the uspA1 gene of M. catarrhalis isolates O35E.118 and O35E.135. (A) The radioactively-labeled primer P10 was extended with MMLV-RT by using 20 μg of total RNA isolated from O35E.135 (lane 1) or O35E.118 (lane 2) and was also used to analyze the nucleotide sequence of the uspA1 gene (lanes A, C, G, and T); the arrowhead indicates the nucleotide at which transcription of the uspA1 gene originates. (B) Sequence of 290 nt of the sense strand located upstream of the M. catarrhalis O35E.118 uspA1 ORF (italic and bold), which contains 10 residues in its poly(G) tract (box). The arrows represent the binding sites for the oligonucleotide primers P7, P9, P10, and P11, and the arrowhead indicates the nucleotide at which transcription of the uspA1 gene was determined to originate in panel A. Sequences exhibiting homology to the −10 and −35 consensus sequences for the bacterial promoters are underlined.
Nucleotide sequence analysis.
The nucleotide sequence of the PCR products and recombinant plasmids described in this study was determined with an Applied Biosystems model 373A automated DNA sequencer (Applied Biosystems, Foster City, Calif.). Nucleotide sequence information was analyzed with the SeqEd v1.0.3 (Applied Biosystems) and AssemblyLIGN and Mac Vector (version 6.5; Oxford Molecular, Ltd., Campbell, Calif.) software packages.
CAT ELISA assay.
Expression of CAT by the M. catarrhalis reporter strains O35E.118CAT and O35E.135CAT was quantitatively measured using an enzyme immunoassay (CAT enzyme-linked immunosorbent assay [ELISA]) according to the manufacturer's recommended protocol (Roche Diagnostics GmbH, Mannheim, Germany). Cell extracts were prepared from 1 ml of broth culture grown to a density of 125 Klett units. Serial dilutions of the cell extracts were used in the CAT ELISA to measure the amount of CAT expressed by the reporter strains. Results were normalized with respect to the total protein concentration of the cell extracts as determined by the Markwell modification of the Lowry assay (30).
Cloning and mutagenesis of M. catarrhalis uspA1 genes in H. influenzae.
PCR products of approximately 3 kb containing the uspA1 genes from M. catarrhalis isolates O35E.118 and O35E.135 were amplified with the oligonucleotide primers P7 and P8 and Pfu polymerase. A PCR product containing the uspA1 gene lacking the poly(G) tract and upstream DNA was similarly amplified from O35E.135 using the oligonucleotide primers P9 and P8. These amplicons were digested with BamHI and ligated into the BamHI site of the vector pACYC184 (New England Biolabs, Inc., Beverly, Mass.). The ligation reactions were subsequently used to electroporate H. influenzae DB117. Chloramphenicol-resistant colonies were screened for reactivity with the UspA1-reactive monoclonal antibody (MAb) 17C7 in the colony blot radioimmunoassay (RIA). Site-directed mutagenesis of the poly(G) tract in plasmid pELU1-10G was accomplished using the QuikChange Site-Directed Mutagenesis system (Stratagene).
Adherence assays.
Adherence assays were performed with Chang conjunctival epithelial cells as previously described (1).
Colony blot RIA and characterization of protein antigens.
The colony blot-RIA was performed as described elsewhere (36). Preparation of whole-cell lysates, SDS-PAGE, and Western blot analysis were accomplished as described previously (36). It should be noted that the heating of whole-cell lysates at 100°C for 3 to 5 min will convert the very-high-molecular-weight UspA1 aggregates to a form that has an apparent molecular weight of 120 to 130 kDa in SDS-PAGE (12). The indirect antibody-accessibility assay was performed as described earlier (1).
Densitometric measurements.
Densitometric measurements of band intensities in autoradiographs was accomplished by the use of an Alpha Imager 2000 documentation and analysis system (Alpha Innotech Corp., San Leandro, Calif.), together with Alpha Imager software (version 4.0).
MAbs.
The UspA1- and UspA2-reactive MAb 17C7 (2), the UspA1-specific MAb 24B5 (12), the UspA2-specific MAb 17H4 (1), and the CopB-specific MAb 10F3 (19) have been described. To obtain a second UspA1-specific MAb, the synthetic peptide ETNNRQDQKIDQLGYALKEQGQHFNNR (PEP1) was synthesized by the Biopolymers Facility at the University of Texas Southwestern Medical Center and covalently bound to keyhole limpet hemocyanin (Sigma Chemical Co., St. Louis, Mo.) using glutaraldehyde. The sequence of PEP1 corresponds to amino acid residues 723 to 749 of the M. catarrhalis O35E UspA1 protein and is highly conserved in all UspA1 proteins characterized to date (data not shown). The PEP1-KLH conjugate was used to immunize mice for hybridoma production as previously described (1); MAb 33B5 was shown by ELISA to bind PEP1 which had been covalently linked to ovalbumin (Sigma) using glutaraldehyde and to specifically bind the UspA1 protein of M. catarrhalis O35E in Western blot analysis. Rat polyclonal antiserum raised against the 39-kDa P2 major outer membrane protein of H. influenzae type b strain DL42 has been described elsewhere (18).
RESULTS
Variable expression of the M. catarrhalis UspA1 protein.
During construction of isogenic uspA2 mutants in previous studies (1, 26), it was noted that a few of these uspA2 mutants underwent an apparent spontaneous change which resulted in reduced expression of the UspA1 protein (data not shown). To investigate this phenomenon, whole-cell lysates were generated from individual colonies (isolates) of the wild-type M. catarrhalis strain O35E and probed in Western blot analysis with the UspA1-reactive MAbs 33B5 and 24B5. These two different UspA1-specific MAbs were used independently to detect possible epitope variation. Some of these individual isolates (O35E.118 and O35E.98; Fig. 1A and B, lanes 2 and 3, respectively) expressed UspA1 at levels indistinguishable from that observed with the wild-type parent strain (Fig. 1A and B, lanes 1). In contrast, expression of the UspA1 protein was dramatically decreased in other individual isolates (Fig. 1A and B, lanes 4 to 7 containing isolates O35E.117, O35E.121, O35E.135, and O35E.137, respectively). MAb 10F3, specific for the CopB outer membrane protein (19), was used to demonstrate that equivalent amounts of whole-cell lysate had been analyzed (Fig. 1C). In addition, the use of the indirect antibody accessibility assay indicated that there was significantly more UspA1 expressed on the surface of the strain O35E.118 than on the surface of strain O35E.135 (Table 2).
FIG. 1.
Characterization of selected proteins expressed by M. catarrhalis isolates derived from strain O35E and nucleotide sequence analysis of their uspA1 poly-G tract. (A to C) Western blot analysis of proteins present in whole-cell lysates prepared from M. catarrhalis O35E (lane 1), O35E.118 (lane 2), O35E.98 (lane 3), O35E.117 (lane 4), O35E.121 (lane 5), O35E.135 (lane 6), O35E.137 (lane 7), and the isogenic uspA1 mutant O35E.1 (lane 8) was performed with the UspA1-specific MAbs 24B5 (A) and 33B5 (B), as well as with the CopB-specific MAb 10F3 (C). Molecular weight markers are shown to the left in kilodaltons. (D) Whole cells of the aforementioned isolates were spotted in duplicate on filter paper and tested in the colony blot- RIA with the UspA1-specific MAb 24B5. Panel E shows the nucleotide sequence of the poly(G) tract (bold) that is located 30 nt upstream of the uspA1 predicted translational start codon (underlined and italicized) of the isolates described above.
TABLE 2.
Adherence of M. catarrhalis strains and recombinant H. influenzae cells to Chang monolayers and detection of UspA1 on the bacterial cell surface
| Strain | Length of poly(G) tract (nt) | UspA1 expressionc | Mean % adherence ± SD to Chang monolayersd | Mean detection (cpm) ± SD of UspA1 on cell surfacee |
|---|---|---|---|---|
| O35E.118 | 10 | ++++ | 72.4 ± 17.4 | 113,000 ± 14,300 |
| O35E.135 | 9 | + | 15.6 ± 10.3 | 13,780 ± 4,065 |
| O35E.1 | NDa | − | 1.6 ± 0.2 | 1,731 ± 315 |
| O35E.135CAT | 9 | − | 1.5 ± 0.5 | ND |
| O12E.44 | 10 | ++++ | 126.4 ± 9.6 | 103,032 ± 7,844 |
| O12E.77 | 9 | + | 31.7 ± 14.6 | 17,078 ± 1,732 |
| O12E.1 | ND | − | 4.3 ± 0.7 | 1,604 ±183 |
| DB117(pELU1-10G) | 10 | +++ | 26.6 ± 8.6 | 7,558 ± 304 |
| DB117(pELU1-9G) | 9 | + | 1.4 ± 0.3 | 2,149 ± 885 |
| DB117(pACYC184) | NAb | − | 0.1 ± 0.1 | 898 ± 198 |
ND, not determined.
NA, not applicable.
The level of UspA1 expression was evaluated by colony blot RIA and Western blot analysis.
Adherence to Chang conjunctival epithelial cells is expressed as the mean percentage of the inoculum which adhered to the monolayers. A total of six independent experiments were performed to obtain these data.
Expressed as the mean counts per minute of radioiodinated goat anti-mouse immunoglobulin bound to MAb 24B5 (for M. catarrhalis isolates) or MAb 17C7 (for recombinant H. influenzae clones) on the bacterial cell surface.
The use of the colony blot RIA reinforced the fact that M. catarrhalis isolates O35E.117, O35E.121, O35E.135, and O35E.137 still bound MAb 24B5 (Fig. 1D, lanes 4, 5, 6, and 7), although the level of reactivity was much reduced relative to that observed with the wild-type strain (Fig. 1D, lane 1) and the variants O35E.118 and O35E.98 (Fig. 1D, lanes 2 and 3, respectively). The uspA1 mutant O35E.1 was included in these experiments as a negative control strain unable to express UspA1 (Fig. 1A to D, lanes 8). Individual colonies of the wild-type M. catarrhalis strains O46E, O12E, and TTA37 were also analyzed as described above, and variants that expressed very low levels of UspA1 were readily identified (data not shown). These results indicated that individual cells of these wild-type strains of M. catarrhalis did not consistently express uniform levels of the UspA1 protein as detected by reactivity with UspA1-directed MAbs.
Variation in expression of UspA1 can be correlated with the length of a poly-G tract.
The nucleotide sequences of the several different uspA1 genes characterized to date have a stretch of consecutive guanine (G) residues located 30 nt upstream of the predicted translational start codon (2, 12, 26). Such homopolymeric nucleotide tracts have been previously shown to be directly involved in phase variation of expression of surface-exposed antigens of gram-negative pathogens (20). Thus, the nucleotide sequence of the DNA located directly upstream of the uspA1 ORF was determined for individual M. catarrhalis isolates that expressed different levels of UspA1. The two individual isolates (O35E.118 and O35E.98) that expressed levels of UspA1 equivalent to that of the wild-type strain had 10 G residues in their respective poly(G) tracts (Fig. 1E, lines 2 and 3). In contrast, the four individual isolates that expressed greatly reduced levels of UspA1 (O35E.117, O35E.121, O35E.135, and O35E.137) all had nine G residues in this same region (Fig. 1E, lines 4 to 7). Analysis of individual colonies of M. catarrhalis strains O46E, O12E, and TTA37 revealed the same correlation between relative levels of expression of the UspA1 protein and the number of G residues in the poly(G) tract upstream of the uspA1 ORF (data not shown).
The nucleotide sequences of the uspA1 genes of four isolates, derived from two different M. catarrhalis wild-type strains, were determined in their entirety. Isolates O35E.118 (Fig. 1A and B, lanes 2) and O12E.44 (data not shown) both expressed wild-type levels of UspA1, whereas isolates O35E.135 (Fig. 1A and B, lanes 6) and O12E.77 (data not shown) produced greatly reduced amounts of this protein. Other than differing in the number of residues in their respective poly(G) tracts, the nucleotide sequence of the uspA1 gene of strain O35E.118 was identical to that of O35E.135. Similarly, the sequences of the uspA1 genes of isolates O12E.44 and O12E.77 were identical to each other except in the poly-G tract, where O12E.44 contained 10 G residues, whereas that of O12E.77 contained 9 G residues.
The poly-G tract is part of the uspA1 mRNA.
Primer extension experiments were performed to determine the transcriptional start site of the uspA1 gene. The use of the oligonucleotide primer P10 (Fig. 2B) in primer extension analysis of total RNA isolated from M. catarrhalis O35E.118 yielded a cDNA fragment that was 86 nt in length (Fig. 2A, lane 2). Nucleotide sequence analysis using P10 mapped the transcriptional start site of uspA1 to a G residue (Fig. 2A) located 208 nt upstream from the translational start codon of the uspA1 ORF (Fig. 2B). Primer extension analysis with the oligonucleotide primer P11 (Fig. 2B) identified the same G residue as the transcriptional start site (data not shown). These results indicate that, in an isolate expressing wild-type levels of UspA1 (i.e., O35E.118), the poly(G) tract is part of the uspA1 mRNA and is positioned 168 nt downstream of the transcriptional start site (Fig. 2B). Nucleotide sequences exhibiting homology to the −10 and −35 consensus sequences for bacterial promoters were observed upstream of the uspA1 transcriptional start site (Fig. 2B). It should be noted that a putative uspA1 translational initiation codon (ATG) was located 34 nt 5′ from the poly(G) tract. However, there is no obvious Shine-Dalgarno sequence preceding this ATG and there is an in-frame TGA stop codon located immediately before the poly(G) repeat, so it is highly unlikely that translation was initiated from this ATG codon. Furthermore, when site-directed mutagenesis was used to change the predicted ATG start codon (located downstream from the poly(G) tract) to ACG, this change abolished expression of UspA1 (data not shown).
Primer extension analysis of the uspA1 gene of isolate O35E.135, which expressed greatly reduced levels of UspA1 (Fig. 1A and B, lanes 6) and possessed only nine G residues in its poly(G) tract (Fig. 1E, row 6), identified the same G as the start of transcription for uspA1 using either P10 (Fig. 2A, lane 1) or P11 (data not shown). Primer extension experiments performed with total RNA isolated from isolates O12E.44 (10 G) and O12E.77 (9 G) showed that transcription of uspA1 also originated at a G residue located 208 and 207 nt, respectively, upstream of the ATG translational start codon in these isolates (data not shown). Thus, the poly(G) tract located upstream of the M. catarrhalis uspA1 ORF is included in the uspA1 mRNA. Furthermore, the uspA1 mRNA originated at the same residue regardless of the length of the homopolymeric G tract or the amount of UspA1 being produced by the isolates. It should also be noted that the amount of cDNA that was generated upon primer extension of the uspA1 gene of O35E.135 (Fig. 2A, lane 1) was 1.9-fold less than that obtained with isolate O35E.118 (Fig. 2A, lane 2). This relative difference in cDNA products was also noted in the primer extension analyses of isolates O12E.44 (10 G) and O12E.77 (9 G), respectively (data not shown).
Slot blot and Northern hybridization analysis of uspA1 transcription in M. catarrhalis.
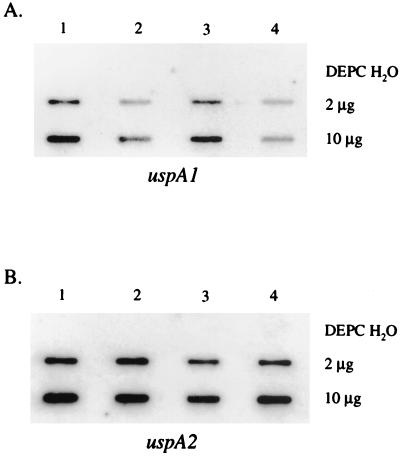
Slot blot and Northern hybridization analyses were performed using total RNA obtained from the M. catarrhalis isolates described above. The DNA probes for uspA1 and uspA2 mRNA were specific for the 5′ end of each gene. Slot blot hybridization experiments showed that 2.2-fold more uspA1 mRNA was detected in O35E.118 (10 G) (Fig. 3A, lanes 1 and 3) than in O35E.135 (9 G) (Fig. 3A, lanes 2 and 4). This effect was observed both in the early (Fig. 3A, lanes 1 and 2) and mid-logarithmic (Fig. 3A, lanes 3 and 4) phases of growth. A uspA2-specific probe was used to demonstrate that equivalent amounts of RNA were analyzed at both time points (Fig. 3B). Similar results were obtained from slot blot hybridization analysis of total RNA isolated from M. catarrhalis O12E.44 (10 G) and O12E.77 (9 G) (data not shown).
FIG. 3.
Slot blot hybridization of M. catarrhalis RNA. Portions (2 and 10 μg) of total RNA isolated from broth cultures of M. catarrhalis O35E.118 (lanes 1 and 3) and O35E.135 (lanes 2 and 4) at cell densities of 50 (lanes 1 and 2) and 125 (lanes 3 and 4) Klett units were analyzed in slot blot hybridization experiments with DNA probes that were specific for either the 5′ end of the uspA1 gene (panel A) or the 5′ end of the uspA2 gene (panel B). Diethyl pyrocarbonate-H2O was used as a negative control.
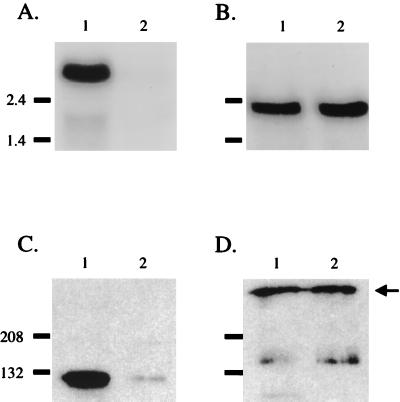
Northern hybridization analysis of total RNA isolated from O35E.118 (10 G) with the uspA1-specific probe identified a transcript of approximately 2.7 kb (Fig. 4A, lane 1), which is consistent with transcription of the 2.5-kb uspA1 ORF originating 208 nt upstream of the gene's predicted translational start codon. The full-length uspA1 transcript, however, was not detected in isolate O35E.135 (9 G) (Fig. 4A, lane 2). Detection of equivalent levels of a 2.3-kb uspA2 mRNA in both O35E.118 and O35E.135 indicated that comparable amounts of RNA had been analyzed (Fig. 4B). Western blot analysis of these same cells with a UspA1-specific MAb (Fig. 4C) and a UspA2-specific MAb (Fig. 4D) confirmed that UspA2 expression was similar in these two isolates whereas UspA1 expression was much less in O35E.135 than in O35E.118. The length of the uspA1 poly(G) tract similarly affected the amount of intact uspA1 mRNA detected in O12E.44 (10 G) and O12E.77 (9 G) but not the level of the uspA2 transcripts (data not shown).
FIG. 4.
Northern hybridization of M. catarrhalis RNA and corresponding protein expression. Portions (20 μg) of total RNA isolated from strains O35E.118 (lane 1) and O35E.135 (lane 2) were analyzed in Northern blot experiments with probes specific for the 5′ end of the uspA1 gene (A) and the 5′ end of the uspA2 gene (B). Molecular size markers (in kilobases) are shown to the left of panels A and B. Whole-cell lysates of these same two strains were probed in Western blot analysis with the UspA1-specific MAb 24B5 (C) and the UspA2-specific MAb 17H4 (D). The arrow on the right side of panel D indicates the position of the high-molecular-weight form of UspA2. Molecular mass markers (in kilodaltons) are present on the left side of panels C and D.
Construction of uspA1::cat reporter strains.
In order to quantitatively determine the effect of the poly(G) tract on transcription of uspA1, a promoterless cat gene was introduced into the uspA1 gene of the M. catarrhalis isolates O35E.118 (10 G) and O35E.135 (9 G). The resulting reporter strains were designated O35E.118CAT and O35E.135CAT, respectively. Nucleotide sequence analysis confirmed that the promoterless cat gene was inserted in the proper orientation in these two strains and that there were no mutations in the 5′ untranslated region of the uspA1 gene.
Slot blot hybridization analysis of total RNA with a uspA1-specific probe (described above) indicated the presence of a two- threefold-greater amount of specific mRNA in O35E.118 (10 G) compared to the amount detected in O35E.135 (9 G) (Fig. 5A, lanes 1 and 2, respectively). A two- to threefold-greater level of uspA1::cat mRNA was also observed in O35E.118CAT (10 G) compared to the level detected in O35E.135CAT (9 G), and this effect was seen using both a uspA1-specific probe (Fig. 5A, lanes 3 and 4, respectively) and a probe corresponding to the promoterless cat cartridge (Fig. 5C, lanes 3 and 4, respectively). A uspA2-specific probe was used to demonstrate that an equivalent amount of RNA from each of the respective strains was analyzed (Fig. 5B, lanes 1 to 4).
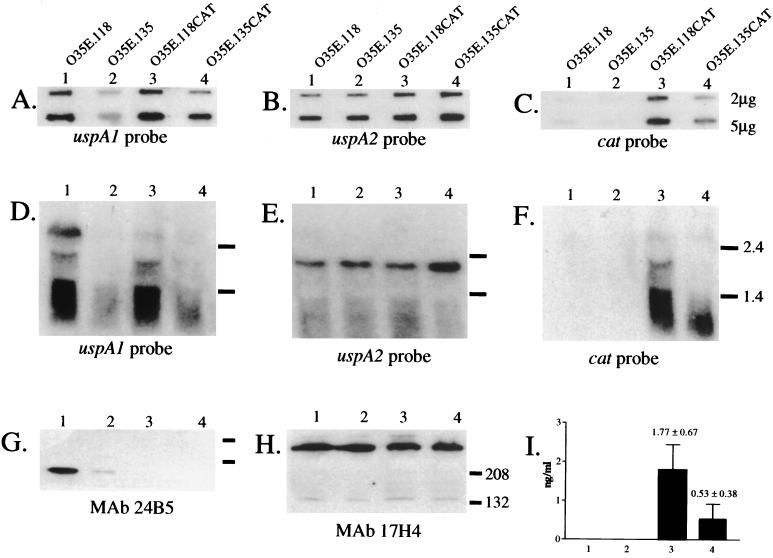
FIG. 5.
Use of a uspA1::cat reporter system to measure transcription of uspA1. Total RNA from M. catarrhalis strain O35E.118 (10 G) (lane 1), M. catarrhalis strain O35E.135 (9 G) (lane 2), the uspA1::cat reporter strain O35E.118CAT (lane 3), and the uspA1::cat reporter strain O35E.135 CAT (lane 4) was subjected to slot blot analysis (A to C) and Northern blot analysis (D to F) using a uspA1-specific probe (A and D), a uspA2-specific probe (B and E), and a cat-specific probe (C and F). Two different amounts of RNA (2 and 5 μg) were used in the slot blot analysis. Size markers (in kilobases) for panels D, E, and F are listed on the right side of panel F. Whole-cell lysates of these four strains were subjected to Western blot analysis using the UspA1-specific MAb 24B5 (G) and the UspA2-specific MAb 17H4 (H). Size markers (in kilodaltons) for panels G and H are listed on the right side of panel H. Panel I contains the CAT enzyme activity expressed by all four strains.
Northern hybridization analysis of total RNA with a uspA1-specific probe identified the full-length uspA1 mRNA of 2.7 kb in O35E.118 (10 G; Fig. 5D, lane 1); this message was not detected in O35E.135 (9 G) (Fig. 5D, lane 2). Nucleotide sequence analysis of the uspA1::cat reporter constructs of strains O35E.118CAT and O35E.135CAT predicted the uspA1::cat mRNA to be approximately 2.0 kb in length (data not shown), and a transcript of that size was detected in O35E.118CAT (10 G) with both the uspA1- and cat-specific probes (lane 3 in Fig. 5D and F, respectively). A uspA1::cat mRNA was not detected in O35E.135CAT (9 G) (lane 4 in Fig. 5D and F). Northern hybridization was also performed with a uspA2-specific probe to confirm that an equivalent amount of RNA from each of the four strains was analyzed (Fig. 5E, lanes 1 to 4).
Western blot analysis of whole-cell lysates of O35E.118 and O35E.135 with the UspA1-specific MAb 24B5 indicated that the difference in the levels of UspA1 expression between these two isolates (Fig. 5G, lanes 1 and 2, respectively) was sixfold. The use of the indirect antibody accessibility assay determined that the amount of UspA1 on the surface of O35E.118 was significantly greater than that on the surface of O35E.135 (Table 2). Due to the insertion of the cat cartridge within the uspA1 gene of the reporter strains O35E.118CAT and O35E.135CAT, the latter two strains did not express a UspA1 protein that bound the UspA1-specific MAb 24B5 (Fig. 5G, lanes 3 and 4). Western blot analysis with the UspA2-specific MAb 17H4 demonstrated that equivalent amounts of protein were analyzed (Fig. 5E, lanes 1 and 2). Determination of the levels of CAT enzyme produced by the reporter strains indicated an approximately threefold reduction in the amount of CAT expressed by O35E.135CAT (9 G) (Fig. 5I, lane 4) compared to that of O35E.118CAT (10 G) (Fig. 5I, lane 3).
Effect of UspA1 phase variation on the adherence of M. catarrhalis to human epithelial cells in vitro.
Isolate O35E.135 (9 G) exhibited a fourfold decrease in attachment to Chang cells relative to the level obtained with O35E.118 (10 G) (Table 2). By comparison, the lack of expression of UspA1 in the isogenic mutant strain O35E.1 caused a 45-fold decrease in adherence to Chang monolayers (Table 2). Proof that the observed attachment ability of isolate O35E.135 (9 G) was the result of the low-level expression of UspA1 was obtained by using the reporter construct O35E.135CAT which has nine G residues in its poly(G) tract but cannot express any functional UspA1 protein; this latter strain had an attachment level similar to that obtained with the uspA1 mutant O35E.1 (Table 2). Similar results were obtained when the strain O12E.44 (10 G) and its UspA1 phase variant O12E.77 (9 G) were analyzed in this same manner (Table 2).
To eliminate the possibility that phase variants expressing higher levels of UspA1 were the organisms that actually attached to the Chang monolayers incubated with isolate O35E.135 (9 G), bacteria attached to the Chang cells were recovered as individual colonies which were then passaged once on a brain heart infusion agar plate. Whole-cell lysates, as well as uspA1 amplicons, were generated from these colonies. All three of the adherent O35E.118 (10 G) isolates that were tested in this manner produced wild-type levels of UspA1 (Fig. 6A and B, lanes 1 to 3), and the uspA1 poly(G) tract in each of these three isolates contained 10 G residues (Fig. 6D, lines 1 to 3). All of the adherent O35E.135 (9 G) isolates that were analyzed, however, still expressed greatly reduced amounts of UspA1 (Fig. 6A and B, lanes 4 to 6) and their uspA1 poly(G) tracts contained 9 G residues (Fig. 6D, lines 3 to 6). The reactivity of MAb 10F3 with the CopB outer membrane protein was again used to confirm that equivalent amounts of cell lysate were analyzed (Fig. 6C).
FIG. 6.
Characterization of selected proteins expressed by M. catarrhalis isolates recovered from Chang monolayers and nucleotide sequence analysis of their uspA1 poly(G) tract. Western blot analysis of proteins present in whole-cell lysates (A to C) prepared from M. catarrhalis O35E.118ATT1 (lane 1), O35E.118ATT2 (lane 2), O35E.118ATT3 (lane 3), O35E.135ATT1 (lane 4) O35E.135ATT2 (lane 5), and O35E.135ATT3 (lane 6) which had been recovered from Chang monolayers and from the uspA1 isogenic mutant O35E.1 (lane 7) was performed with the UspA1-specific MAbs 24B5 (A) and 33B5 (B), as well as with the CopB-specific MAb 10F3 (C). Molecular mass markers are shown to the left in kilodaltons. (D) Nucleotide sequence of the poly(G) tract (bold) that is located 30 nt upstream of the uspA1 predicted translational start codon (underlined and italicized) of the isolates described above.
Expression of wild-type and mutated uspA1 genes in H. influenzae.
To determine whether the length of the poly(G) tract would affect expression of the uspA1 gene in a heterologous genetic background, the uspA1 genes from isolates O35E.118 (10 G) and O35E.135 (9 G) were cloned into H. influenzae DB117. These two cloned PCR products, derived from the use of the oligonucleotide primers P7 and P8 (Fig. 2B), did not contain the native uspA1 promoter region and were inserted in the same orientation into the tetracycline resistance gene in pACYC184.
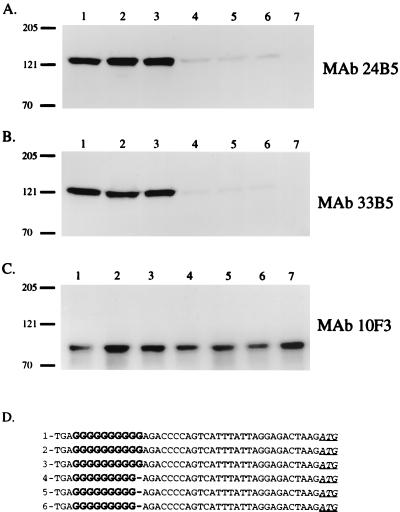
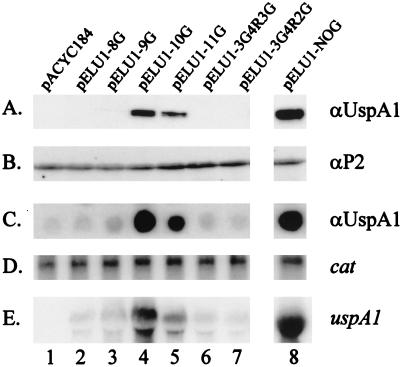
Recombinant H. influenzae clones containing pELU1-10 G (Fig. 7C, lane 4; encodes O35E.118 uspA1) and pELU1-9 G (Fig. 7C, lane 3; encodes O35E.135 uspA1) were isolated on the basis of their reactivity with the UspA1-reactive MAb 17C7 in the colony blot RIA. In this assay, a significant difference was observed in the relative levels of UspA1 expressed by the two recombinant strains (Fig. 7C, lanes 3 and 4). Western blot analysis of whole-cell lysates prepared from H. influenzae DB117 (pELU1-10 G) demonstrated that MAb 17C7 bound an approximately 125-kDa UspA1 antigen (Fig. 7A, lane 4). In contrast, expression of UspA1 by H. influenzae DB117(pELU1-9 G) was not detected by Western blot analysis (Fig. 7A, lane 3). The reactivity of polyclonal antibodies with the H. influenzae P2 major outer membrane protein was used to demonstrate that equivalent amounts of cell lysate were analyzed (Fig. 7B). Northern blot analysis of these two strains revealed less uspA1 mRNA in H. influenzae DB117(pELU1-9 G) (Fig. 7E, lane 3) than in H. influenzae DB117(pELU1-10 G) (Fig. 7E, lane 4). Detection of mRNA from the vector-based cat gene was used to confirm that equivalent amounts of total RNA were analyzed in these experiments (Fig. 7D). Finally, H. influenzae DB117(pACYC184) was included in these experiments as a negative control (Fig. 7, lane 1).
FIG. 7.
Analysis of H. influenzae DB117 recombinant clones containing wild-type and mutated uspA1 genes. The recombinant plasmids contained by each strain are listed at the top of this figure and are described in detail in Table 1. (A) Western blot analysis using the UspA1-reactive MAb 17C7. (B) Western blot analysis using polyclonal rat antibody to the H. influenzae P2 major outer membrane protein. (C) Colony blot RIA using the UspA1-reactive MAb 17C7. (D) Northern blot analysis using a cat riboprobe. (E) Northern blot analysis using a uspA1 riboprobe.
These recombinant H. influenzae strains were also tested for their ability to adhere to Chang monolayers (Table 2). DB117(pELU1-10 G) displayed a 250-fold increase in its attachment capability compared to DB117 containing only the vector pACYC184. DB117(pELU1-9 G), however, only exhibited about a 10-fold increase in adherence relative to DB117(pACYC184), a finding which was consistent with the very low level of expression of the UspA1 adhesin by the former strain (Table 2 and Fig. 7C, lane 3).
To further investigate the effect of the poly(G) tract on expression of uspA1, site-directed mutagenesis was used both to alter the number of G residues and the composition of the nucleotides in the poly(G) tract of plasmid pELU1-10 G. Reduction in the number of G residues from 10 to 8 in pELU1-8 G (Fig. 7, lane 2) resulted in very little or no expression of uspA1 mRNA and UspA1 protein, a finding similar to that seen with pELU1-9 G. Increasing the number of G residues to 11 in pELU1-11 G (Fig. 7, lane 5) caused a slight reduction in both uspA1 mRNA and UspA1 protein levels relative to those obtained with pELU1-10 G (Fig. 7, lane 4). When the sequence of the poly(G) tract was changed from GGGGGGGGGG to either GGGACTAGGG in pELU1-3G4R3G (Fig. 7, lane 6) or GGGACTAGG in pELU1-3G4R2G (Fig. 7, lane 7), uspA1 expression was again very low or undetectable, similar to that observed with the 9 G construct (Fig. 7, lane 3).
A third recombinant H. influenzae strain was constructed with a uspA1 gene that lacked both the poly(G) tract and the M. catarrhalis DNA immediately upstream from this tract. This PCR product was obtained from the use of the oligonucleotide primers P9 and P8 and was cloned into pACYC184 as described above. The level of UspA1 expressed by the recombinant clone DB117(pELU1-NOG) (Fig. 7, lane 8) was 2.9-fold greater than that expressed by DB117(pELU1-10 G). Similarly, the level of uspA1 mRNA expressed by the recombinant clone DB117(pELU1-NOG) (Fig. 7E, lane 8) was greater than that expressed by any of the other clones.
DISCUSSION
The data presented in this study indicate that expression of the M. catarrhalis UspA1 protein is subject to phase variation, resulting in greatly reduced levels of this antigen on the surface of the organism. This phenomenon could be correlated with changes in the number of G residues contained in a region [i.e., the poly(G) tract] located 30 nt upstream from the uspA1 translational start codon. The uspA1 poly(G) tracts of most M. catarrhalis wild-type strains characterized to date (i.e., O35E, O12E, TTA37, and O46E) were found to contain 10 G residues. All isolates of these four strains which expressed reduced levels of UspA1 had 9 G residues in their uspA1 poly(G) tracts. Colony blot RIA combined with Western blot and nucleotide sequence analyses of isolates derived from O35E.118 (10 G) and O35E.135 (9 G) indicated the rate of switching from 10 G→9 G residues to be 3.7 × 10−2 and the frequency of the 9 G→10 G conversion to be 9.7 × 10−3 (data not shown). It should also be noted, with all of the wild-type M. catarrhalis strains used in this study, colonies which completely lacked expression of the UspA1 protein were never detected regardless of the length of the uspA1 poly(G) tract. Therefore, variable expression of the UspA1 protein appeared to oscillate only between HIGH and LOW phases.
Extended homogeneous stretches of purines or pyrimidines have been shown to be involved in the molecular mechanisms by which expression of surface-exposed structures of several gram-negative pathogens undergoes phase variation (20). These homopolymeric tracts appear to be more prone to transitory base-pair misalignment, also referred to as slipped-strand mispairing, during DNA replication, which results in the addition or the removal of one or more of the repeated nucleotide residues. Depending on its location within a gene, variation in the length of a homopolymeric nucleotide tract can alter translation of ORFs or influence the transcription of genes (20).
With regard to homopolymeric nucleotide tracts involved in controlling expression of surface antigens, N. meningitidis possesses genes whose expression is controlled by these elements at either the transcriptional or translational levels. For example, biosynthesis of the lipopolysaccharide terminal structure lacto-N-neotetraose is subject to high-frequency ON-OFF phase variation, controlled at the level of translation, that involves a homopolymeric G tract located inside the first ORF of the meningococcal lgtABE locus (22, 23). Poly(G) tracts containing either 5 or 14 G residues maintain an intact lgtA ORF and can be correlated with the presence of lacto-N-neotetraose on the surface of N. meningitidis isolates. In contrast, lgtA alleles containing 9, 10, 12, or 13 G residues were predicted to be out of frame and isolates containing these poly(G) tracts did not express lipopolysaccharide molecules containing lacto-N-neotetraose. Interestingly, meningococcal isolates containing shorter lgtA poly(G) tracts consisting of five residues were found to constitutively express lacto-N-neotetraose. It was inferred from this observation that shorter homopolymeric tracts are less sensitive to slipped-strand mispairing and that expression of genes containing shorter tracts is more stable (23). Other examples of phase-variable expression of antigens controlled by a translational frameshift mechanism and involving a poly(G) tract include the N. meningitidis (27) and N. gonorrhoeae (9) outer membrane hemoglobin-binding protein HpuA, the N. meningitidis outer membrane hemoglobin-binding protein HmbR (27), and a 200-kDa outer membrane protein of M. catarrhalis (Sasaki et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999).
Variation in the length of homopolymeric nucleotide tracts can also control gene expression at the level of transcription. Expression of the class 1 outer membrane porin (PorA) of N. meningitidis was shown to display three distinct levels which could be correlated with the number of G residues present in a poly(G) tract located directly between the −35 and −10 regions of the porA promoter (42). The presence of 11, 10, or 9 G residues in the porA promoter resulted in high-level, medium-level, or no expression of porA mRNA, respectively, which in turn resulted in corresponding levels of PorA in the outer membrane of N. meningitidis. Other examples of phase-variable expression of antigens controlled by a transcriptional mechanism through slipped-strand mispairing within a homopolymeric tract include the N. meningitidis Opc outer membrane protein (40); the Bordetella pertussis Fim2, Fim3, and FimX fimbrial subunits (44); and the Mycoplasma hyorhinis Vlp lipoproteins (45). It should be noted that in all of the aforementioned examples, the homopolymeric tracts are positioned upstream from the transcriptional start site of each gene and between the −35 and −10 regions of their respective promoters.
The present study contains the first report of phase-variable expression of an M. catarrhalis surface-exposed antigen that is controlled at the level of transcription by variation in a homopolymeric nucleotide tract. Specifically, a decrease in the number of G residues in the poly(G) tract (i.e., from 10 to 9) exerted a significant effect on the level of uspA1 mRNA that could be detected in Northern blot analysis (Fig. 4). Additional quantitative analyses using slot blot hybridization and a cat reporter construct (Fig. 5), as well as primer extension experiments (Fig. 2), confirmed the existence of a difference in the levels of transcription of the uspA1 genes containing 10 and 9 G residues in their respective poly(G) tracts. The apparent discrepancies between the results obtained with the latter three methods (i.e., two- to threefold differences between 10 G and 9 G isolates) and that obtained with Northern blot analysis (i.e., no detectable uspA1 mRNA in the 9 G isolate) are likely the result of differences in the relative sensitivities of these methods. It is worth nothing that the differences between the 10 G and 9 G isolates detected with Northern blot analysis (Fig. 4) were more similar to the protein expression data, where Western blot analysis (Fig. 4) indicated a sixfold difference and the indirect antibody accessibility assay (Table 2) suggested an eightfold difference in UspA1 expression.
The changes in the number of G residues within the poly(G) tract in these isolates is likely the result of slipped-strand mispairing, although no experiments were performed in this study to address this specific issue. The molecular mechanism by which the addition or removal of a single residue within the uspA1 poly(G) tract affects transcription of this gene, however, remains unclear. In related phase-variation systems, homopolymeric tracts were located between the −35 and −10 promoter regions of their respective genes. Variation in the length of these tracts has been proposed to affect binding of RNA polymerase to the promoter (11, 40, 42–45). In contrast, the M. catarrhalis uspA1 poly(G) tract was determined to be located 168 nt downstream of the transcriptional start site of the gene and 30 nt upstream of the ORF. Therefore, it is unlikely that variation in the length of the uspA1 poly(G) tract would directly affect the binding of RNA polymerase to the −35 and −10 sequences of the uspA1 gene. It is possible that the addition or removal of residues within the uspA1 poly(G) tract may affect the binding of a transcriptional activator [i.e., when the poly(G)tract contains 10 G residues] or repressor [i.e., when the poly(G)tract contains 9 G residues]. Alternatively, changes in the number of G residues could affect the stability of the uspA1 mRNA. However, attempts to address this last issue by using S1 nuclease protection assays after rifampin treatment of the M. catarrhalis cells were inconclusive (data not shown).
Site-directed mutagenesis of the poly(G) tract in a recombinant uspA1 gene in H. influenzae DB117 determined that not only changes in length but also changes in the composition of the poly(G) tract adversely affected expression of both uspA1 mRNA and UspA1 protein (Fig. 7). Our results also indicated that the absence of the poly(G) tract and upstream DNA in DB117(pELU1-NOG) resulted in expression of uspA1 mRNA and UspA1 protein at levels greater than those expressed by H. influenzae DB117(pELU1-10 G) (Fig. 7, lanes 8 and 4, respectively). At this time, we do not know whether this increase in expression is a consequence of eliminating the poly(G) tract or simply the result of placing the uspA1 ORF in closer proximity to a plasmid-based promoter.
While it is clear that a reduction in the number of G residues from 10 to 9 in the poly(G) tract of the uspA1 gene in strains O35E, O12E, TTA37, and O46E reduced the expression of UspA1, it must be noted that two other wild-type M. catarrhalis strains (ATCC 25238 and P44) were identified that had only 6 or 7 G residues, respectively, in their uspA1 poly(G) tracts and yet still readily expressed UspA1 (data not shown). This finding indicates that either relatively short poly(G) tracts (i.e., six or seven residues) have no deleterious effect on expression of UspA1 or that there is something in the genetic background of these two strains that allows high-level expression of UspA1 despite the presence of the very short poly(G) tract. Isolates expressing significantly lower levels of UspA1, however, were not identified among the approximately 10,000 colonies of both M. catarrhalis ATCC 25238 (6 G) and P44 (7 G) that were tested in the colony blot RIA (data not shown). The failure to detect isolates of ATCC 25238 and P44 that expressed reduced levels of UspA1 raises the possibility that phase variation of UspA1 expression may not occur in these two strains. Alternatively, the frequency of phase variation in these two strains may simply be too low to be detected by the method (i.e., colony blot RIA) used in this study.
The M. catarrhalis UspA1 protein has been shown to function as an adhesin in vitro (26). Since bacterial adherence is likely an important first step in colonization of the upper respiratory tract by M. catarrhalis, the UspA1 protein may play a pivotal role in the development of infection by this gram-negative pathogen. The UspA1 protein, however, has also been shown to be immunogenic and to stimulate the production of biologically relevant antibodies (10, 31, 32, 39). The data presented in this study clearly indicate that, even though phase variation of UspA1 (i.e., 10 G→9 G) resulted in greatly reduced levels of the protein being expressed on the surface of the bacterium, these phase variants were still capable of adhering to human epithelial cells in vitro, albeit at a reduced level (Table 2). Thus, phase-variable expression of UspA1 may enable a population of M. catarrhalis to establish a balance between the requirement for adherence to human epithelial cells in order to colonize its human host and the necessity to evade the host immune response in order to persist and subsequently cause infection. Phase variation of a bacterial surface antigen that is regulated at the level of transcription has been reported to occur in vivo; this involves the HMW1 and HMW2 adhesins of nontypeable H. influenzae (13). Whether phase variation of the M. catarrhalis UspA1 protein occurs in vivo remains to be determined.
ACKNOWLEDGMENTS
This study was supported by U.S. Public Health Service grant AI36344 to E.J.H.
We thank Jo Latimer and Sheryl Lumbley for technical assistance and Irene Rombel and Ross Chambers for helpful discussions.
REFERENCES
- 1.Aebi C, Lafontaine E R, Cope L D, Latimer J L, Lumbley S R, McCracken G H, Jr, Hansen E J. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis O35E. Infect Immun. 1998;66:3113–3119. doi: 10.1128/iai.66.7.3113-3119.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi C, Maciver I, Latimer J L, Cope L D, Stevens M K, Thomas S E, McCracken G H, Jr, Hansen E J. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect Immun. 1997;65:4367–4377. doi: 10.1128/iai.65.11.4367-4377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aebi C, Stone B, Beucher M, Cope L D, Maciver I, Thomas S E, McCracken G H, Jr, Sparling P F, Hansen E J. Expression of the CopB outer membrane protein by Moraxella catarrhalis is regulated by iron and affects iron acquisition from transferrin and lactoferrin. Infect Immun. 1996;64:2024–2030. doi: 10.1128/iai.64.6.2024-2030.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartos L C, Murphy T F. Comparison of the outer membrane proteins of 50 strains of Branhamella catarrhalis. J Infect Dis. 1988;158:761–765. doi: 10.1093/infdis/158.4.761. [DOI] [PubMed] [Google Scholar]
- 5.Bluestone C D, Stephenson J S, Martin L M. Ten-year review of otitis media pathogens. Pediatr Infect Dis J. 1992;11:S7–S11. doi: 10.1097/00006454-199208001-00002. [DOI] [PubMed] [Google Scholar]
- 6.Bonnah R A, Yu R H, Wong H, Schryvers A B. Biochemical and immunological properties of lactoferrin binding proteins from Moraxella (Branhamella) catarrhalis. Microb Pathog. 1998;24:89–100. doi: 10.1006/mpat.1997.0173. [DOI] [PubMed] [Google Scholar]
- 7.Campagnari A A, Ducey T F, Rebmann C A. Outer membrane protein B1, an iron-repressible protein conserved in the outer membrance of Moraxella (Branhamella) catarrhalis, binds human transferrin. Infect Immun. 1996;64:3920–3924. doi: 10.1128/iai.64.9.3920-3924.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campagnari A A, Shanks K L, Dyer D W. Growth of Moraxella catarrhalis with human transferrin and lactoferrin: expression of iron-repressible proteins without siderophore production. Infect Immun. 1994;62:4909–4914. doi: 10.1128/iai.62.11.4909-4914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C-J, Elkins C, Sparling P F. Phase variation of hemoglobin utilization in Neisseria gonorrhoeae. Infect Immun. 1998;66:987–993. doi: 10.1128/iai.66.3.987-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen D, Barniak V, VanDerMeid K R, McMichael J C. The levels and bactericidal capacity of antibodies directed against the UspA1 and UspA2 outer membrane proteins of Moraxella (Branhamella) catarrhalis in adults and children. Infect Immun. 1999;67:1310–1316. doi: 10.1128/iai.67.3.1310-1316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Citti C, Watson-McKown R, Droesse M, Wise K S. Gene families encoding phase- and size-variable surface lipoproteins of Mycoplasma hyorhinis. J Bacteriol. 2000;182:1356–1363. doi: 10.1128/jb.182.5.1356-1363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cope L D, Lafontaine E R, Slaughter C A, Hasemann C A, Jr, Aebi C, Henderson F W, McCracken G H., Jr Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J Bacteriol. 1999;181:4026–4034. doi: 10.1128/jb.181.13.4026-4034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawid S, Barenkamp S J, St. Geme J W., III Variation in expression of the Haemophilus influenzae HMW adhesins: a prokaryotic system reminiscent of eukaryotes. Proc Natl Acad Sci USA. 1999;96:1077–1082. doi: 10.1073/pnas.96.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright M C, McKenzie H. Moraxella (Branhamella) catarrhalis—clinical and molecular aspects of a rediscovered pathogen. J Med Microbiol. 1997;46:360–371. doi: 10.1099/00222615-46-5-360. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald M, Mulcahy R, Murphy S, Keane C, Coakley D, Scott T. A 200 kDa protein is associated with haemagglutinating isolates of Moraxella (Branhamella) catarrhalis. FEMS Immunol Med Microbiol. 1997;18:209–216. doi: 10.1111/j.1574-695X.1997.tb01047.x. [DOI] [PubMed] [Google Scholar]
- 16.Gulig P A, McCracken G H, Jr, Frisch C F, Johnston K H, Hansen E J. Antibody response of infants to cell surface-exposed outer membrane proteins of Haemophilus influenzae type b after systemic Haemophilus disease. Infect Immun. 1982;37:82–88. doi: 10.1128/iai.37.1.82-88.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hager H, Verghese A, Alvarez S, Berk S L. Branhamella catarrhalis respiratory infections. Rev Infect Dis. 1987;9:1140–1149. doi: 10.1093/clinids/9.6.1140. [DOI] [PubMed] [Google Scholar]
- 18.Hansen E J, Pelzel S E, Orth K, Moomaw C R, Radolf J D, Slaughter C A. Structural and antigenic conservation of the P2 porin protein among strains of Haemophilus influenzae type b. Infect Immun. 1989;57:3270–3275. doi: 10.1128/iai.57.11.3270-3275.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helminen M E, Maciver I, Latimer J L, Cope L D, McCracken G H, Jr, Hansen E J. A major outer membrane protein of Moraxella catarrhalis is a target for antibodies that enhance pulmonary clearance of the pathogen in an animal model. Infect Immun. 1993;61:2003–2010. doi: 10.1128/iai.61.5.2003-2010.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson I R, Owen P, Nataro J P. Molecular switches—the ON and OFF of bacterial phase variation. Mol Microbiol. 1999;33:919–932. doi: 10.1046/j.1365-2958.1999.01555.x. [DOI] [PubMed] [Google Scholar]
- 21.Hsiao C B, Sethi S, Murphy T F. Outer membrane protein CD of Branhamella catarrhalis—sequence conservation in strains recovered from the human respiratory tract. Microb Pathog. 1995;19:215–225. doi: 10.1016/s0882-4010(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 22.Jennings M P, Hood D W, Peak I R, Virji M, Moxon E R. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol Microbiol. 1995;18:729–740. doi: 10.1111/j.1365-2958.1995.mmi_18040729.x. [DOI] [PubMed] [Google Scholar]
- 23.Jennings M P, Srikhanta Y N, Moxon E R, Kramer M, Poolman J T, Kuipers B, van der L P. The genetic basis of the phase variation repertoire of lipopolysaccharide immunotypes in Neisseria meningitidis. Microbiology. 1999;145(Pt. 11):3013–3021. doi: 10.1099/00221287-145-11-3013. [DOI] [PubMed] [Google Scholar]
- 24.Klingman K L, Murphy T F. Purification and characterization of a high-molecular-weight outer membrane protein of Moraxella (Branhamella) catarrhalis. Infect Immun. 1994;62:1150–1155. doi: 10.1128/iai.62.4.1150-1155.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyd J M, Cripps A W, Murphy T F. Outer-membrane antigen expression by Moraxella (Branhamella) catarrhalis influences pulmonary clearance. J Med Microbiol. 1998;47:159–168. doi: 10.1099/00222615-47-2-159. [DOI] [PubMed] [Google Scholar]
- 26.Lafontaine E R, Cope L D, Aebi C, Latimer J L, McCracken G H, Jr, Hansen E J. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J Bacteriol. 2000;182:1364–1373. doi: 10.1128/jb.182.5.1364-1373.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis L A, Gipson M, Hartman K, Ownbey T, Vaughn J, Dyer D W. Phase variation of HpuAB and HmbR, two distinct haemoglobin receptors of Neisseria meningitidis DNM2. Mol Microbiol. 1999;32:977–989. doi: 10.1046/j.1365-2958.1999.01409.x. [DOI] [PubMed] [Google Scholar]
- 28.Luke N R, Campagnari A A. Construction and characterization of Moraxella catarrhalis mutants defective in expression of transferrin receptors. Infect Immun. 1999;67:5815–5819. doi: 10.1128/iai.67.11.5815-5819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukomski S, Hull R A, Hull S I. Identification of the O antigen polymerase (rfc) gene in Escherichia coli O4 by insertional mutagenesis using a nonpolar chloramphenicol resistance cassette. J Bacteriol. 1996;178:240–247. doi: 10.1128/jb.178.1.240-247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markwell M A K, Haas S M, Bieber L L, Tolbert N E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 31.Mathers K, Leinonen M, Goldblatt D. Antibody response to outer membrane proteins of Moraxella catarrhalis in children with otitis media. Pediatr Infect Dis J. 1999;18:982–988. doi: 10.1097/00006454-199911000-00010. [DOI] [PubMed] [Google Scholar]
- 32.McMichael J C, Fiske M J, Fredenburg R A, Chakravarti D N, VanDerMeid K R, Barniak V, Caplan J, Bortell E, Baker S, Arumugham R, Chen D. Isolation and characterization of two proteins from Moraxella catarrhalis that bear a common epitope. Infect Immun. 1998;66:4374–4381. doi: 10.1128/iai.66.9.4374-4381.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy T F. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol Rev. 1996;60:267. doi: 10.1128/mr.60.2.267-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy T F, Kirkham C, DeNardin A, Sethi S. Analysis of antigenic structure and human immune response to outer membrane protein CD of Moraxella catarrhalis. Infect Immun. 1999;67:4578–4585. doi: 10.1128/iai.67.9.4578-4585.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers L E, Yang Y-P, Du R P, Wang Q J, Harkness R E, Schryvers A B, Klein M H, Loosmore S M. The transferrin binding protein B of Moraxella catarrhalis elicits bactericidal antibodies and is a potential vaccine antigen. Infect Immun. 1998;66:4183–4192. doi: 10.1128/iai.66.9.4183-4192.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patrick C C, Kimura A, Jackson M A, Hermanstorfer L, Hood A, McCracken G H, Jr, Hansen E J. Antigenic characterization of the oligosaccharide portion of the lipooligosaccharide of nontypable Haemophilus influenzae. Infect Immun. 1987;55:2902–2911. doi: 10.1128/iai.55.12.2902-2911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pelton S I, Klein J O. The promise of immunoprophylaxis for prevention of acute otitis media. Pediatr Infect Dis J. 1999;18:926–935. doi: 10.1097/00006454-199910000-00018. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning—a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Samukawa T, Yamanaka N, Hollingshead S, Klingman K, Faden H. Immune responses to specific antigens of Streptococcus pneumoniae and Moraxella catarrhalis in the respiratory tract. Infect Immun. 2000;68:1569–1573. doi: 10.1128/iai.68.3.1569-1573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarkari J, Pandit N, Moxon E R, Achtman M. Variable expression of the Opc outer membrane protein in Neisseria meningitidis is caused by size variation of a promoter containing poly-cytidine. Mol Microbiol. 1994;13:207–217. doi: 10.1111/j.1365-2958.1994.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 41.Setlow J K, Brown D C, Boling M E, Mattingly A, Gordon M P. Repair of deoxyribonucleic acid in Haemophilus influenzae. J Bacteriol. 1968;95:546–558. doi: 10.1128/jb.95.2.546-558.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Ende A, Hopman C T, Zaat S, Essink B B, Berkhout B, Dankert J. Variable expression of class 1 outer membrane protein in Neisseria meningitidis is caused by variation in the spacing between the −10 and −35 regions of the promoter. J Bacteriol. 1995;177:2475–2480. doi: 10.1128/jb.177.9.2475-2480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Washburn L R, Weaver K E, Weaver E J, Donelan W, Al Sheboul S. Molecular characterization of Mycoplasma arthritidis variable surface protein MAA2. Infect Immun. 1998;66:2576–2586. doi: 10.1128/iai.66.6.2576-2586.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willems R, Paul A, van der Heide H, ter Avest A R, Mooi F R. Fimbrial phase variation in Bordetella pertussis: a novel mechanism for transcriptional regulation. EMBO J. 1990;9:2803–2809. doi: 10.1002/j.1460-2075.1990.tb07468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yogev D, Rosengarten R, Watson-McKown R, Wise K S. Molecular basis of Mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5′ regulatory sequences. EMBO J. 1991;10:4069–4079. doi: 10.1002/j.1460-2075.1991.tb04983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]