Abstract
A five-gene cluster encoding four nonheme iron proteins and a flavoprotein from the thermophilic anaerobic bacterium Clostridium thermoaceticum (Moorella thermoacetica) was cloned and sequenced. Based on analysis of deduced amino acid sequences, the genes were identified as rub (rubredoxin), rbo (rubredoxin oxidoreductase), rbr (rubrerythrin), fprA (type A flavoprotein), and a gene referred to as hrb (high-molecular-weight rubredoxin). Northern blot analysis demonstrated that the five-gene cluster is organized as two subclusters, consisting of two divergently transcribed operons, rbr-fprA-hrb and rbo-rub. The rbr, fprA, and rub genes were expressed in Escherichia coli, and their encoded recombinant proteins were purified. The molecular masses, UV-visible absorption spectra, and cofactor contents of the recombinant rubrerythrin, rubredoxin, and type A flavoprotein were similar to those of respective homologs from other microorganisms. Antibodies raised against Desulfovibrio vulgaris Rbr reacted with both native and recombinant Rbr from C. thermoaceticum, indicating that this protein was expressed in the native organism. Since Rbr and Rbo have been recently implicated in oxidative stress protection in several anaerobic bacteria and archaea, we suggest a similar function of these proteins in oxygen tolerance of C. thermoaceticum.
Several studies indicate that anaerobic and microaerophilic bacteria can tolerate varying degrees of O2 exposure. Superoxide dismutase (SOD) and catalase, which are known to relieve oxidative stress in aerobes, are often absent in anaerobic bacteria. Recently, nonheme iron proteins such as rubrerythrin (Rbr) and rubredoxin oxidoreductase (Rbo) (also known as desulfoferrodoxin) have been implicated in oxidative stress protection in anaerobes (1, 23, 27, 33, 42). So far, Rbr and Rbo or their genes have been found only in anaerobic or microaerophilic bacteria and archaea. The active sites of these proteins include a rubredoxin-type [Fe(SCys)4] center in both Rbo and Rbr (3, 5, 11, 12, 22, 23, 35), a mononuclear [Fe(NHis)4SCys] center in Rbo (6), and a nonsulfur, oxo-bridged di-iron center in Rbr (8, 17, 19, 23, 34). The latter two sites in their reduced forms react rapidly with superoxide in the case of Rbo (27) and with hydrogen peroxide in the case of Rbr (7, 8).
Clostridium thermoaceticum is a thermophilic gram-positive, obligately anaerobic bacterium that produces acetate from virtually any carbon source, including sugars, aromatic compounds, and C1 compounds (25, 36, 44). It is not known how this bacterium responds to oxygen toxicity during growth. Determinations of catalase and SOD activities in C. thermoaceticum have been inconclusive. In this study we show that C. thermoaceticum contains genes encoding Rbo and Rbr and that these two genes are present in a cluster with three additional genes encoding a rubredoxin (Rub), a high-molecular-weight rubredoxin (Hrb) and a type A flavoprotein (FprA). All these genes have been expressed in Escherichia coli, and recombinant Rbr, Rub, and FprA were purified and partially characterized. Except for rub, none of these genes have previously been reported to be present in any acetogenic bacterium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. C. thermoaceticum was grown at 58°C in the presence of glucose (1%) under 100% CO2, as previously described (26). All E. coli strains were routinely grown at 37°C in Luria-Bertani medium. All plasmids used in this study (Table 1) carry ampicillin resistance genes and were maintained in E. coli hosts in the presence of 100 μg of ampicillin per ml.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Genotype or description | Source |
|---|---|---|
| Bacterial strains | ||
| C. thermoaceticum (Moorella thermoacetica) ATCC 39073 | Wild type | |
| E. coli | ||
| XL1-Blue MRA | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 gyrA96 relA1 lac | Stratagene |
| XL1-Blue MRA(P2) | XL1-Blue MRA (P2 lysogen) | |
| BL21(DE3) | E. coli B; F−ompT rB− mB− (λDE3) | Novagen |
| INVαF′ | F′ endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 φ80lacZΔM15 Δ(lacZYA-argF)U169 λ− | |
| Plasmids | ||
| pCRII | Apr Kmr; vector for cloning PCR product | Invitrogen |
| pBluescript SK(+) | Apr, ColE1 origin, cloning vector | Stratagene |
| pET-21b(+) | Apr, T7 promoter expression vector | Novagen |
| pCRRub | pCRII derivative containing 96-bp PCR amplified Rub probe | This study |
| pRb5 | pBluescript derivative containing rbr, rbo, rrb, and a part of fprA in a 3.0-kb PstI fragment | This study |
| pRb48 | pBluescript derivative containing fprA, rbr, rbo, rub, and a part of hrb in a 4.8-kb KpnI fragment | This study |
| pRbo/Rub | pET-21(b+) derivative containing rbo and rub | This study |
| pFprA | pET-21(b+) derivative containing fprA | This study |
| pHrb | pET-21(b+) derivative containing hrb | This study |
DNA and RNA sources and purification.
Chromosomal DNA of C. thermoaceticum was isolated and purified as described previously (31). Plasmid DNA was isolated and purified using a QIAprep Spin Miniprep Kit from Qiagen Inc., Studio City, Calif. Lambda DNA was purified using the Wizard Lambda Preps DNA purification system from Promega, Madison, Wis. Total RNA was isolated from C. thermoaceticum harvested at exponential growth phase and purified using a RNeasy mini kit from Qiagen. Routine DNA manipulations were performed as described by Sambrook et al. (39).
DNA fragments to be cloned into plasmids were purified from 1% agarose gels after briefly staining with ethidium bromide using the QIAquick gel extraction kit from Qiagen. Plasmids used in cloning reactions were digested with the desired restriction enzymes and similarly gel purified. Purified, linearized plasmids were dephosphorylated by treatment with shrimp alkaline phosphatase (Roche Molecular Biochemicals, Indianapolis, Id.) prior to ligation with target DNA fragments. The DNA ligation reactions were carried out using T4 DNA ligase from New England Biolabs (Beverly, Mass.) using the conditions outlined by the manufacturer. Synthesis of oligonucleotides used in PCR and in DNA sequencing experiments was carried out at the Molecular Genetics and Instrumentation Facility of the University of Georgia.
Cloning and sequencing strategy.
Initially, we targeted to sequence the Rub gene. Two highly conserved regions of the amino acid sequences of Rub proteins from different origins were used to design primers for PCR. The forward primer 5′-GTITG(TC)GG(TC)TA(TC)AT(TC)TA(TC)(AG)A(TC)C-3′ was designed from a conserved amino acid sequence, VCGYIYN/D, at the N-terminal ends of Rub proteins and the reverse primer 5′-G(AG)CAIACCCA(AG)TC(AG)TC(GC)G-3′ was designed from a conserved amino acid sequence, PCVWDDP, at the C-terminal end (29). The PCR was carried out with these primers using C. thermoaceticum genomic DNA as a template for 25 cycles under the following conditions for each cycle: denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and elongation at 72°C for 1 min. A 96-bp PCR product was amplified. It was sequenced after cloning into the pCR 2.1 vector (Invitrogen, Carlsbad, Calif.). The deduced amino acid sequence of the PCR product was found to be highly homologous with those of Rub proteins from different sources (not shown). The PCR product was labeled with digoxigenin (DIG)–11-dUTP (Roche Molecular Biochemicals) and used as a probe to screen a genomic library of C. thermoaceticum constructed in λFIX II by Stratagene (La Jolla, Calif.) according to a method described previously (9). A Rub-positive clone designated λRd 2 was purified from the library, and its DNA was analyzed by Southern hybridization using the same 96-bp DIG-labeled PCR product as a probe. A 3.0-kb PstI fragment and a 4.8-kb KpnI fragment from λRd 2 were found to hybridize to the Rub probe. These two fragments were purified and cloned into pBluescript (Stratagene), and constructs designated pRb 5 (3.0-kb PstI insert) and pRb 48 (4.8-kb KpnI insert) were obtained (Table 1). The two constructs (pRb 5 and pRb 48) have the 3.0-kb PstI fragment (see above) in common. The nucleotide sequences reported in this study were derived from these two clones, except for some sequences (described below) which were obtained directly from λRd 2.
Hybridization techniques.
The genomic library of C. thermoaceticum in λFIX II was screened with the Rub probe (see above) by plaque hybridization experiments using the Genius system from Roche Molecular Biochemicals; Southern and Northern hybridization experiments were also carried out using the same Genius system as previously described (9). Individual genes were amplified by PCR with C. thermoaceticum genomic DNA as a template in the presence of DIG–11-dUTP and used as probes in Northern hybridization experiments.
Heterologous expression of rbr, fprA, rbo, rub, and hrb in E. coli.
The plasmid constructs used for the expression of the genes are listed in Table 1. Except for rbr, the genes were cloned into pET-21b (Novagen, Inc., Madison, Wis.) and expressed in E. coli. The rbr gene was expressed directly from its clone pRb5, which is a derivative of pBluescript. For cloning into pET-21b, the genes were amplified by PCR from C. thermoaceticum genomic DNA using specific primers. The primers were designed to have unique restriction sites at their 5′ ends, NdeI for forward primers and EcoRI for reverse primers. The PCR products were purified using a QIAquick PCR purification kit from Qiagen, digested with NdeI and EcoRI, and ligated into the corresponding restriction sites of pET-21b. E. coli strain BL21(DE3) was used for expression of rbo, rub, fprA, and hrb, and E. coli DH5α was used for expression of rbr. E. coli strains carrying recombinant plasmids were grown in 1- or 2-liter volumes of either Luria-Bertani medium or MZ9 salt medium (pET manual, Novagen). The MZ9 salt medium was supplemented with 20 mM ferrous sulfate for the expression of recombinant Rbr, Rbo, and Rub. After expression, recombinant Rbo (CthRbo) and Hrb (CthHrb) formed inclusion bodies, while recombinant Rbr (CthRbr), Rub (CthRub), and FprA (CthFprA) remained soluble upon cell lysis. The purification steps for the recombinant proteins are described below.
Purification of recombinant CthRbr, CthRub, and CthFprA.
CthRbr was expressed without any inducer, while CthFprA and CthRub were expressed following induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The IPTG was added after the optical density at 600 nm of the cultures reached 0.6. Optimum expression of recombinant proteins occurred after 4 h of IPTG induction. After being harvested by centrifugation, cells were washed, resuspended in buffer A (20 mM Tris-HCl, pH 7.5) (1 g of cell paste in 3 ml of buffer), and sonicated. Crude cell lysates were centrifuged (100,000 × g for 1 h), and the supernatants were collected for purification of recombinant proteins.
For purification of CthRbr, supernatants were subjected to heat treatment at 65°C for 30 min. Most E. coli proteins precipitated in this step were removed by centrifugation at 25,000 × g. Crystalline ammonium sulfate was added to the clear light-red supernatant to 60% of saturation at room temperature. Proteins precipitated at this step were discarded, and ammonium sulfate was added to the supernatant to obtain 80% of saturation. Proteins, including CthRbr, precipitated at this step were collected by centrifugation and dissolved in 2 ml of buffer A. The CthRbr was purified from this suspension by repeated gel filtration on a TSK gel G 3000 SW column (Tosahaas) using the fast protein liquid chromatography system of Amersham-Pharmacia, Piscataway, N.J. The elution buffer was buffer A plus 0.1 M NaCl. Colored fractions from each gel filtration step were analyzed for the presence of Rbr by UV-visible spectroscopy. The characteristic spectra of oxidized Rbr proteins include peaks at 280, 370, and 492 nms. Fractions of Rbr collected after the second gel filtration were found to be pure based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Recombinant CthRub and CthFprA were purified from the supernatants using anion-exchange chromatography on DEAE–Sepharose CL 6B (Pharmacia) followed by size exclusion chromatography on a TSK gel G 3000 SW column (Tosahaas). In the first step, supernatants containing soluble recombinant proteins were passed through a DEAE–Sepharose CL 6B column (300 by 25 cm) preequilibrated with buffer A. Proteins bound to the column were eluted by a salt gradient of 0 to 1 M NaCl in buffer A. Light-colored fractions (red for CthRub and yellow for CthFprA) eluted from the columns were analyzed for the presence of recombinant proteins by recording their UV-visible absorption spectra. Characteristic spectra for oxidized CthRub include peaks at 280, 492, and 380 nm, and those for CthFprA include peaks at 280, 350, and 450 nm. Fractions having these spectral properties were pooled, concentrated by Amicon ultrafiltrations, and applied to a TSK 3000 gel filtration column preequilibrated with buffer A plus 0.1 M NaCl. Fractions containing CthRub and CthFprA collected from the latter column were found to be more than 90% pure based on SDS-PAGE.
Spectral analysis and analytical methods.
UV-visible absorption spectra of the recombinant proteins were obtained on a Shimadzu model UV 2100PC spectrophotometer. The electron paramagnetic resonance (EPR) spectra of recombinant proteins were obtained on a Bruker ESP-300E spectrometer as described previously (17). The molecular masses of the recombinant proteins were determined by gel filtration on Superose 12 column using the fast protein liquid chromatography system of Amersham-Pharmacia. The molecular mass standards used were chymotrypsin (25 kDa), egg albumin (45 kDa), and bovine serum albumin (68 kDa). Protein concentrations were determined by the Lowry method, as described previously (9). Desulfovibrio vulgaris Rbr (17) and Rub (4) were used as protein standards for quantitation of CthRbr and CthRub, respectively. The concentrations of these protein standards were determined using their well-established extinction coefficients at 492 nm, i.e., 5,400 M−1 cm−1 for D. vulgaris Rbr monomer (17) and 8,700 M−1 cm−1 for D. vulgaris rubredoxin (4). SDS-PAGE was carried out by the method of Laemmli (20) using 12% acrylamide in resolving gels and 4% acrylamide in stacking gels. Western blotting experiments were carried out according to Bio-Rad (Hercules, Calif.). Antibodies against D. vulgaris Rbr (17) were raised in rabbits at the Animal Care and Use Facility of the University of Georgia.
Identification of the flavin cofactor of recombinant CthFprA was carried out as follow. Recombinant CthFprA (25 mg per ml) was heated at 100°C for 20 min and then centrifuged at 14,000 × g to remove precipitated proteins. Four microliters of the supernatant was subjected to thin-layer chromatography on silica gel-coated glass plates along with flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) (Sigma, St. Louis, Mo.) as standards as described by Fetzner et al. (14). The molar content of FMN was determined spectrophotometrically using ɛ450 = 12,200 M−1 cm−1 (2).
For reconstitution, recombinant CthFprA (25 mg/ml) in a 100-μl reaction volume was treated with a 10-molar excess of FMN (Sigma) for 30 min at room temperature. The sample was diluted to 2 ml with 50 mM MOPS (morpholinepropanesulfonic acid) (pH 7.0) and concentrated to ∼100 μl using an Ultra-free-15 protein concentrator from Fisher (Pittsburgh, Pa.). This dilution and reconcentration were repeated until the flowthrough contained no detectable FMN. The FMN content of reconstituted CthFprA was determined as described above.
Metal analysis of the recombinant proteins was carried out by inductively coupled plasma atomic emission at the Chemical Analysis Laboratory of the University of Georgia.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study has been has been deposited at the GenBank, EMBL, and DDJB libraries with accession number AF202316.
RESULTS
Cloning, sequencing, and identification of the genes.
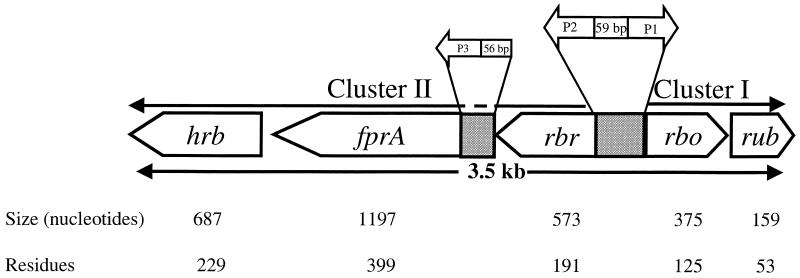
Details of the cloning and sequencing strategy are described in Materials and Methods. The rub gene was sequenced from pRb5 carrying a 3.0-kb PstI fragment isolated from a rub-positive lambda clone, λRd2. The latter clone was isolated from a C. thermoaceticum genomic library in λFIX II (9) after screening with a rub probe. Extended sequencing from both ends of rub, including the sequences of the 3.0-kb PstI insert in pRb5 and the 4.8-kb KpnI insert in pRb48, revealed the presence of four additional open reading frames (ORFs) at the 5′ end of the rub gene. Analysis of the deduced amino acid sequences of the five ORFs by FASTA identified them as homologs of rubredoxin (Rub), rubredoxin oxidoreductase (Rbo), rubrerythrin (Rbr), type A flavoprotein (FprA), and a protein with a C-terminal rubredoxin sequence motif which we referred to as high-molecular-weight rubredoxin (Hrb). The organization of the genes and the relationships between the genes and their products are summarized in Fig. 1. Except for Hrb, the predicted molar masses of the deduced proteins are in close agreement with those of the homologous proteins from other sources. The C. thermoaceticum FprA was identified by its sequence homology (between 28 and 48%) to FprA proteins from other microorganisms. The C. thermoaceticum Rub, Rbo, and Rbr were readily identified by their sequence identities (between 29 and 70%) to their homologs in other microorganisms and by their characteristic iron binding sequence motifs. Those motifs are CX2CX29CX2C for the single iron site in Rub, EX30–34EX2HXnEX30–34EX2H and CX2CX12CX2C for the diiron and rubredoxin sites, respectively, in Rbr, and CX2CX15CC and HX15HX9HX40CX2H for the [Fe(SCys)4] and [Fe(NHis)4SCys] centers, respectively, in Rbo (not shown). The deduced amino acid sequences of CthRub (70% identity), CthRbr (66% identity), and CthRbo (63% identity) are more homologous with the amino acid sequences of the corresponding proteins from D. vulgaris than with those from other sources.
FIG. 1.
Organization of the five ORFs identified as rbr, fprA, hrb, rbo, and rub and of the putative promoters P1, P2, and P3, and relationship between the genes and their products.
C. thermoaceticum contains two rubredoxins, designated Rd I and Rd II (46). Rd I had a molar mass of 7.4 kDa, and Rd II had a molar mass of 6 kDa. Analysis of the amino acid compositions of the two rubredoxins shows the presence of six cysteine residues in Rd I and four cysteine residues in Rd II. The presence of four cysteine residues and the predicted molar mass of the protein encoded by C. thermoaceticum rub (5,745 Da [see Table 2]) are both in close agreement with the corresponding values for Rd II. Therefore, the protein encoded by rub is presumed to be Rd II. However, the deduced amino acid sequence of the 96-bp PCR product used as a Rub probe in this study does not match the corresponding amino acid sequence deduced from either rub or hrb (Fig. 2), indicating that the rub probe belongs to another rub gene, which may encode Rd I.
TABLE 2.
Properties of purified recombinant Rbr, Rub, and FprA from C. thermoaceticum
| Parameter | Value for:
|
||
|---|---|---|---|
| Rbr | FprA | Rub | |
| Molecular mass (Da) | |||
| Deduced | 21,329 | 44,296 | 5,745 |
| Native | 44,000, 66,000a | 90,000 | 6,000 |
| Denatured | 22,000 | 44,000 | 6,000 |
| Content/mol | |||
| Fe | 3–4/dimer | ∼0.2/monomer | 0.7/monomer |
| Zn | 1–2/dimer | ∼0.1/monomer | 0.2/monomer |
| FMN | 0.5/monomer (reconstituted) | ||
| UV-visible absorption maxima (nm) | 280, 372, 492 | 280, 350, 456 | 280, 378, 492 |
Two oligomers of Rbr were detected (see text).
FIG. 2.
Alignment of the deduced amino acid sequence of the 96-bp PCR-amplified rub probe with the corresponding sequences deduced from rub (CthRub) and hrb (CthHrb).
No protein that is homologous to full-length CthHrb was found in the database. The carboxyl-terminal sequence of Hrb is homologous to those of rubredoxins (Fig. 3). The amino-terminal sequence of Hrb is homologous (28 to 44% sequence identities) to several proteins, including nitrilotriacetate monooxygenase component B from Chelatobacter heinzii (45), actinorhodin polyketide dimerase-related proteins from Streptomyces coelicolor (13) and Thermotoga maritima (accession no. C72410), a probable monooxygenase designated as b1007 from E. coli (accession no. E64842), a probable FMN:NADH oxidoreductase from Streptomyces violaceoruber (accession no. T46545), and a 63.5-kDa FprA (accession no. S75748) COOH-terminal sequence of Synechocystis sp. strain PCC 6803. The proteins listed above are also homologous among themselves (30 to 38% identical residues). The component B of nitrilotriacetate monooxygenase from C. heinzii was shown to have NADH:FMN oxidoreductase activity (45).
FIG. 3.
Deduced amino acid sequence of high-molecular-weight rubredoxin (Hrb). The rubredoxin domain of the protein (in boldface) and the cysteine ligands for the rubredoxin iron center (designated by asterisks) are marked as shown.
All of the newly identified genes of C. thermoaceticum were found to have an AUG start codon except the gene encoding Hrb, which has a UUG start codon. The five genes were organized in two divergently oriented subclusters separated by an AT-rich region, presumably containing regulatory sequences (Fig. 1). Subcluster I consists of rbo-rub (5′→3′), and subcluster II consists of rbr-fprA-hrb (5′→3′).
Northern blot analysis and regulatory sequences.
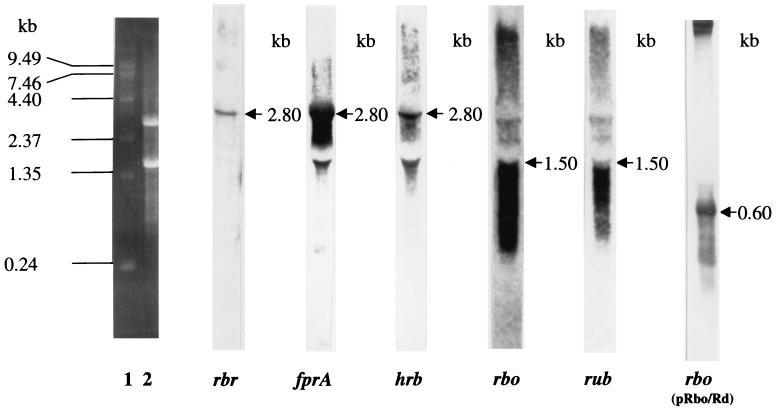
Northern blot hybridization experiments on total RNA isolated from C. thermoaceticum confirmed the two predicted polycistronic operons within the five-gene cluster. The DIG-labeled DNA probes of rbr, fprA, and hrb were each found to hybridize to a 2.8-kb transcript (Fig. 4). Therefore, these three genes must be co-transcribed from a promoter located upstream of rbr. Similarly, the DIG-labeled DNA probes for rbo and rub each hybridized to a 1.5-kb transcript (Fig. 4), indicating an rbo-rub operon with a promoter apparently located upstream of rbo. We ruled out the possibility of any secondary promoter upstream of rub due to a very short intergenic region (20 bp) between the two genes. The 1.5-kb transcript hybridizing to rbo and rub is much larger than the total size of rbo and rub plus their intergenic region, 550 bp. No ORF or transcription terminator was apparent within the 300-bp sequence downstream of rub. In order to verify the cotranscription of rbo and rub, Northern hybridization experiments were carried out on total RNA isolated from E. coli BL21(DE3) harboring pRbo/Rub. The DIG-labeled rbo probe hybridized to a 0.6-kb transcript (Fig. 4), a size which is in close agreement with the expected size of the rbo-rub operon (550 bp). A similar size (∼0.6 kb) of transcript was also found to hybridize DIG-labeled rub probe when a replicate RNA blot from this clone was used (not shown). The cotranscription of rbo and rub was further supported by their coexpression from pRbo/Rub in E. coli (Fig. 5).
FIG. 4.
Northern blots of total RNAs isolated from C. thermoaceticum and E. coli BL21(DE3)(pRbo/Rub) (far right lane) after hybridization with DIG-labeled rub, rbo, rbr, fprA, and hrb, as indicated below each lane. Lanes 1 and 2, positions of RNA markers and ribosomal RNA, respectively, in the ethidium bromide-stained gel.
FIG. 5.
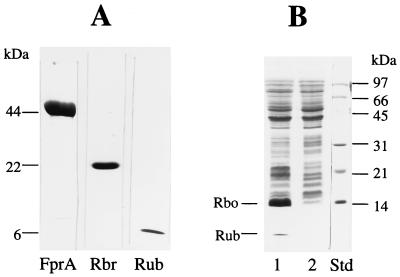
(A) SDS-PAGE of the purified recombinant FprA (10 μg), Rbr (10 μg), and Rub (10 μg) expressed in E. coli. (B) Lane 1, SDS-PAGE of the extracts (40 μg) of E. coli harboring pRbo/Rub grown in MZ9 salt medium after induction with IPTG; lane 2, same as in lane 1 but without IPTG induction. The protein standards (std) and relative positions of the recombinant proteins are shown.
Putative promoter sequences.
Consistent with the Northern blot results, putative promoter sequences, P1 and P2, occur upstream of rbo and rbr, respectively, as diagramed in Fig. 1. The nucleotide sequences of P1 (5′-ATGACG-N15—TAATAAT-N12-AGGAG-3′) and P2 (5′-TTGACT-N17—TACAAT–N21-AGGAG-3′) are homologous to that of the E. coli consensus ς70 promoter (cTTGACa-N15–21-TATAaT-Nx-AGGAG) (18), as we have shown previously for promoters of several C. thermoaceticum genes (30). A third putative promoter, P3 (5′-TTGATA-N21—TATAAT–N32-GGAGG-3′), was also found within the 79-bp rbr-fprA intergenic region, but no promoter-like sequence was found in the 133-bp fprA-hrb intergenic region. The presence of P3 upstream of fprA suggests that fprA might be subjected to secondary regulation of its own.
Expression of the C. thermoaceticum genes in E. coli and properties of the recombinant proteins.
C. thermoaceticum Rbo and Rub were expressed in E. coli from the same plasmid, pRbo/Rub, while C. thermoaceticum Rbr, FprA, and Hrb were expressed from pRb5, pFprA, and pHrb, respectively (Table 1). The recombinant Rbo and Hrb formed inclusion bodies and were not further purified. Figure 5 shows SDS-PAGE of crude preparations of recombinant Rbo and Rub and of the purified recombinant Rbr, FprA, and Rub, all expressed in E. coli. Table 2 summarizes the properties of the recombinant proteins. The molar masses of the recombinant Rub and FprA were estimated by gel filtration as 6 and 90 kDa, respectively, indicating that, under nondenaturing conditions, Rub is present as a monomer and FprA is present as a dimer. Purification of Rbr by gel filtration yielded two major fractions with molar masses of about 66 and 44 kDa, respectively. On SDS-polyacrylamide gels, the two fractions ran as a single band both with an approximate molar mass of 22 kDa, which is in close agreement with the predicted molar mass of 21,348 Da calculated from the deduced amino acid sequence. The two fractions gave identical UV-visible absorption spectra (Fig. 6), indicating that they are the same protein. These results suggest that in solutions recombinant CthRbr is present as a mixture of dimers and trimers in a molar ratio of ∼2:1.
FIG. 6.
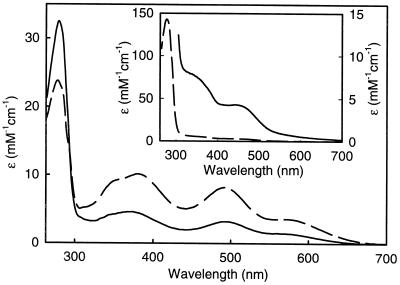
UV-visible absorption spectra of as-isolated recombinant CthRub (57 μM monomer) (dashed line) and CthRbr (108 μM monomer) (solid line). Inset, UV-visible absorption spectra of as-isolated CthFprA (22 μM monomer [dashed line] and 206 μM monomer [solid line]). All spectra were recorded at room temperature in 50 mM MOPS, pH 7.0.
Metal composition of recombinant C. thermoaceticum Rbr and Rub.
The metal analyses of recombinant Rbr and Rub indicate the presence of iron and zinc in significant amounts in both proteins (Table 2). A mixture of iron and zinc forms is typically obtained when rubredoxins are overexpressed in E. coli (4). The iron-plus-zinc contents indicate that the single metal site in CthRub is occupied predominantly (∼70%) by iron, with the remainder occupied by Zn2+. The UV-visible absorption spectra of the recombinant CthRub and CthRbr are shown in Fig. 6, and the absorption maxima are listed in Table 2. The absorption features are typical of the respective homologous proteins from other species (17, 22, 27, 29, 42) and reflect their Fe3+ contents, i.e., any Zn2+-occupied sites would have no optical absorption (4). The 492-nm absorption feature in the spectrum of Rbr is due almost entirely to the oxidized rubredoxin-type iron site (17). Comparing the ɛ492 of 10,800 M−1 cm−1 for the D. vulgaris Rbr homodimer to the ɛ492 of 3,200 M−1 cm−1 determined for CthRbr (using D. vulgaris Rbr as the protein standard), it was estimated that approximately 30% (i.e., 3,200/10,800) of the CthRbr rubredoxin sites contain iron. Since the metal analyses indicated that zinc was the only other heavy metal present in significant amounts (Table 2), it is assumed that the remaining 70% of CthRbr rubredoxin sites are occupied by Zn2+. This interpretation is consistent with the higher-than-expected A280/A492 absorbance ratio for CthRbr (∼10) compared to that for D. vulgaris Rbr (∼5.5), in which all rubredoxin-type sites are occupied by iron (17). The Rbr di-iron(III) site absorbs most intensely between 300 and 400 nm, but this feature is obscured by overlapping and more intense absorption from the rubredoxin-type site (17). Therefore, optical absorption cannot be used to quantitate iron occupancy of the Rbr di-iron sites. However, the estimated 70% Zn2+ occupancy of the rubredoxin-type sites, together with the metal analysis of purified CthRbr, namely, 0.5 to 1 Zn atom per monomer and 1.5 to 2 iron atoms per monomer (Table 2), implies that most of the di-iron sites must be occupied by iron and not zinc. The recombinant CthRbr shows an oxidized rubredoxin-type EPR signal and a relatively weak, mixed-valent di-iron EPR signal (not shown), both of which closely resemble the corresponding signals in D. vulgaris Rbr and provide additional evidence for the metal site occupancies (17).
Cofactor content of recombinant C. thermoaceticum FprA.
Thin-layer chromatography identified FMN and not FAD as a cofactor in the recombinant CthFprA. The FMN/FprA monomer molar ratio was quantitated as 0.2 in as-purified recombinant CthFprA and as 0.54 in the FMN-treated CthFprA (Table 2). No detectable FAD was bound to FAD-treated CthFprA. The A280/A450 absorbance ratio of the as-purified CthFprA (Fig. 6, inset) is consistent with the substoichiometric FMN. Substoichiometric but significant levels of iron and zinc were also detected in the recombinant FprA (Table 2).
Antigenic relationship between Rbr proteins from C. thermoaceticum and D. vulgaris.
Figure 7 shows that antibodies against D. vulgaris Rbr (17) reacted with both the recombinant CthRbr (dimer) and a protein present in crude cell extracts of C. thermoaceticum of the size expected for CthRbr. Both the dimer and trimer of recombinant CthRbr reacted similarly with the antibodies against D. vulgaris Rbr (not shown).
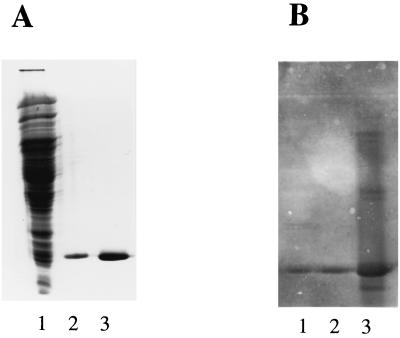
FIG. 7.
Western blots showing interactions between recombinant CthRbr and antibodies against D. vulgaris Rbr. (A) SDS-PAGE of extracts of C. thermoacetcicum (lane 1) (50 μg) and of recombinant CthRbr (lane 2) (0 μg) and recombinant D. vulgaris Rbr (lane 3) (8 μg). (B) Western blot of a replica of the SDS gel shown in panel A after reaction with antibodies against D. vulgaris Rbr.
DISCUSSION
We have characterized a unique five-gene cluster consisting of two divergently transcribed operons, namely, rbo-rub and rbr-fprA-hrb. Anti-D. vulgaris Rbr antibodies reacted strongly with CthRbr, and this cross-reaction confirmed that the C. thermoaceticum rbr gene is expressed in the native organism. In other air-sensitive bacteria and archaea, Rub, Rbr, Rbo, and FprA have all been implicated in oxidative stress protection, and their genes often occur in tandem pairs (15, 28). However, these four genes have not previously been found to occur within the same cluster. The results described in this paper provide the first evidence for the presence of Rbr, Rbo, and FprA in any acetogenic bacterium. The C. thermoaceticum Hrb appears to be a unique protein with at least two domains. The sequence homologies of the C-terminal end to rubredoxins suggests a redox-linked function for Hrb. Since we were unable to express a recombinant Hrb in soluble form, we could not further characterize its properties.
The spectroscopic and physical properties of recombinant C. thermoaceticum Rub and Rbr expressed in E. coli were found to be very similar to those of the corresponding proteins from other sources (17, 22, 23, 32, 43). The metal analysis and spectroscopic properties indicate that the purified recombinant CthRbr contains significant amounts of both zinc and iron, with the majority of the zinc being in the rubredoxin-type site. Zinc can be incorporated into both the rubredoxin-type and di-iron sites of D. vulgaris Rbr (8, 41). However, Rbr and a closely related protein, nigerythrin, as isolated from D. vulgaris, each contain predominantly iron in both types of sites (22, 34).
The recombinant C. thermoaceticum FprA, when overexpressed in E. coli, contained substoichiometric amounts of FMN, iron, and zinc. In Methanobacterium thermoautotrophicum the function of FprA (designated FpaA) has been proposed to be an intermediate electron carrier between H2 and CO2 during methanogenesis (32). The M. thermoautotrophicum fpaA gene occurs in a cluster with two additional genes, organized in the order fpaA-orfX-rdxA (32). The deduced protein encoded by orfX has a 60-residue region that contains the di-iron site sequence motif found in Rbr, and the deduced protein encoded by rdxA contains a rubredoxin-type CX2CXnCX2C sequence motif. Thus, the M. thermoautotrophicum fpaA-orfX-rdxA gene cluster bears some resemblance to the C. thermoaceticum rbr-fprA-hrb operon. The FprA proteins have been previously reported for only two other bacterial species, E. coli and Desulfovibrio gigas (43). The 479-residue FprA from E. coli is sequentially homologous to archaeal FprA proteins but contains in addition a rubredoxin domain at the C-terminal end. In addition to flavin, the D. gigas FprA was reported to contain a di-iron site and to function as a rubredoxin:oxygen oxidoreductase (15, 16). The di-iron site ligands identified in the D. gigas FprA homolog are conserved in C. thermoaceticum FprA. The presence of a putative promoter structure, P3 upstream of C. thermoaceticum fprA (Fig. 2) suggests that its expression could be regulated independently from that of of rbr. Independent regulation of flavoproteins and a reverse relationship between the expression of flavoproteins and iron proteins (namely ferredoxins) were previously reported for methanogens (32) and acetogens (37). In any case, our results suggest that the expressions of Rbr, Rbo, and FprA are coregulated, and their homologies to known proteins suggest a cooperative role in oxidative stress protection.
The widespread occurrence of Rub in anaerobes is already well established, and while it is presumed to function as an intermediary electron carrier in various enzymatic reactions (21, 40, 47), its exact role(s) in anaerobes has never been established. In acetogens Rub has been proposed as an electron acceptor in the carbon monoxide dehydrogenase reaction (38) and as a terminal electron acceptor to the membrane electron transport chain (10, 24). Rub has been proposed to be a redox partner to D. gigas FprA, mostly on the basis that its genes are cotranscribed (15). However, the demonstration that rbo and rub genes are cotranscribed in D. vulgaris (4) and now in C. thermoaceticum (this study) suggests a functional relationship between Rbo and Rub.
Both Rbo and Rbr have been reported to restore aerobic growth to sod mutant strains of E. coli (1, 23, 27, 33). Furthermore, deletion of rbo led to increased dioxygen and superoxide sensitivities of D. vulgaris (28, 42). Evidence for superoxide reductase and NADH peroxidase activities for Rbo and Rbr, respectively, in vitro has been presented (7, 8, 27). Recently it was shown that C. thermoaceticum, which lacks any detectable catalase or SOD activities, could grow in the presence of trace amounts of O2 in liquid media without any reducing agent (A. Karnholz, K. Küsel, and M. Drake, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. I-91, p. 401, 2000). This oxygen tolerance of C. thermoaceticum could result from expression of a novel five-gene cluster consisting of two divergently transcribed but coregulated operons, one containing Rbo and the other containing Rbr. With the addition of acetogenic bacteria to the list, it is becoming increasingly evident that, whatever their functions may be, Rbr, Rbo, and FprA are widespread in air-sensitive microorganisms.
ACKNOWLEDGMENTS
This work was funded by grant DE-FG02-93ER20127 from the Department of Energy (to L.G.L.) and grant GM40388 from the National Institutes of Health (to D.M.K.).
We thank Bijoy Mohanty for helpful suggestions on RNA work and Mike Clay for assistance in obtaining EPR spectra.
REFERENCES
- 1.Alban P S, Popham D L, Rippere K E, Krieg N R. Identification of a gene for a rubrerythrin/nigerythrin-like protein in Spirillum volutans by using amino acid sequence data from mass spectrometry and NH2-terminal sequencing. J Appl Microbiology. 1988;85:875–882. doi: 10.1046/j.1365-2672.1998.00602.x. [DOI] [PubMed] [Google Scholar]
- 2.Batie C J, LaHaie E, Ballou D P. Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J Biol Chem. 1987;262:1510–1518. [PubMed] [Google Scholar]
- 3.Beeumen J J V, Driesschet G V, Liu M-Y, LeGall J. The primary structure of rubrerythrin, a protein with inorganic pyrophosphatase activity from Desulfovibrio vulgaris. Comparison with hemerythrin and rubredoxin. J Biol Chem. 1991;266:20645–20653. [PubMed] [Google Scholar]
- 4.Bonomi F, Iametti S, Ragg E, Richie K A, Kurtz D M., Jr Direct metal ion substitution at the [M(Cys)4]2− site of rubredoxin. J Biol Inorg Chem. 1998;3:595–605. [Google Scholar]
- 5.Brumlik M J, Voordouw G. Analysis of the transcriptional unit encoding the genes for rubredoxin (rub) and a putative rubredoxin oxidoreductase (rbo) in Desulfovibrio vulgaris Hildenborough. J Bacteriol. 1989;171:4996–5004. doi: 10.1128/jb.171.9.4996-5004.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coelho A V, Matias P, Fulop V, Thompson A, Gonzalez A, Carrondo M A. Desulfoferrodoxin structure determined by MAD phasing and refinement to 1.9-angstrom resolution reveals a unique combination of a tetrahedral FeS4 center with square pyramidal FeSN4 center. J Biol Inorg Chem. 1997;2:680–689. [Google Scholar]
- 7.Coulter E D, Shenvi N V, Kurtz D M. NADH peroxidase activity of rubrerythrin. Biochem Biophys Res Commun. 1999;255:317–323. doi: 10.1006/bbrc.1999.0197. [DOI] [PubMed] [Google Scholar]
- 8.Coulter E D, Shenvi N V, Beharry Z M, Smith J J, Prickril B C, Kurtz D M. Rubrerythrin-catalyzed substrate oxidation by dioxygen and hydrogen peroxide. Inorg Chim Acta. 2000;297:231–241. [Google Scholar]
- 9.Das A, Ljungdahl L G. Composition and primary structure of the F1F0 ATP synthase from the obligatory anaerobic bacterium Clostridium thermoaceticum. J Bacteriol. 1997;179:3746–3755. doi: 10.1128/jb.179.11.3746-3755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das A, Hugenholtz J, van Halbeek H, Ljungdahl L G. Structure and function of a menaquinone involved in electron transport in membranes of Clostridium thermoautotrophicum and Clostridium thermoaceticum. J Bacteriol. 1989;171:5823–5829. doi: 10.1128/jb.171.11.5823-5829.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMare F, Kurtz D M, Jr, Nordlund P. The structure of Desulfovibrio vulgaris rubrerythrin reveals a unique combination of rubredoxin-like FeS4 and ferritin-like diiron domains. Nat Struct Biol. 1996;6:539–546. doi: 10.1038/nsb0696-539. [DOI] [PubMed] [Google Scholar]
- 12.Devreese B, Tavares P, Lampreia J, Van Damme N, LeGall J, Moura J J G, Van Beeumen J, Moura I. Primary structure of desulfoferrodoxin from Desulfovibrio desulfuricans ATCC 2774, a new class of non-heme iron proteins. FEBS Lett. 1996;385:138–142. doi: 10.1016/0014-5793(96)00364-x. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Moreno M A, Martinez E, Boto L, Hopwood D A, Malpartida F. Nucleotide sequence and deduced functions of a set of co-transcribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J Biol Chem. 1992;267:19278–19290. [PubMed] [Google Scholar]
- 14.Fetzner S, Müller R, Lingens F. Purification and some properties of 2-halobenzoate 1,2-dioxygenase, a two-component enzyme system from Pseudomonas cepacia 2CBS. J Bacteriol. 1992;174:270–290. doi: 10.1128/jb.174.1.279-290.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frazao C, Silva G, Gomes C M, Matias P, Coelho R, Sieker L, Macedo S, Liu M Y, Oliveira S, Teixeira M, Xavier A V, Rodrigues-Pousada C, Carrondo M A, Le Gall L. Structure of a dioxygen reduction enzyme from Desulfovibrio gigas. Nat Struct Biol. 2000;7:1041–1045. doi: 10.1038/80961. [DOI] [PubMed] [Google Scholar]
- 16.Gomes C M, Silva G, Oliviera S, LeGall J, Liu M-Y, Xavier A V, Rodrigues-Pousada C, Teieira M. Studies on the redox centers of the terminal oxidase from Desulfovibrio gigas and evidence for its interaction with rubredoxin. J Biol Chem. 1997;272:22502–22508. doi: 10.1074/jbc.272.36.22502. [DOI] [PubMed] [Google Scholar]
- 17.Gupta N, Bonomi F, Kurtz D M, Jr, Ravi N, Wang D L, Huynh B H. Recombinant Desulfovibrio vulgaris rubrerythrin. Isolation and characterization of the diiron domain. Biochemistry. 1995;14:3310–3318. doi: 10.1021/bi00010a021. [DOI] [PubMed] [Google Scholar]
- 18.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurtz D M, Jr, Prickril B C. Intrapeptide sequence homology in rubrerythrin from Desulfovibrio vulgaris: identification of potential ligands to the diiron site. Biochem Biophys Res Commun. 1991;181:337–341. doi: 10.1016/s0006-291x(05)81423-8. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lee H J, Basran J, Scrutton N S. Electron transfer from flavin to iron in the Pseudomonas oleovorans rubredoxin reductase-rubredoxin electron transfer complex. Biochemistry. 1998;37:15513–15522. doi: 10.1021/bi981853v. [DOI] [PubMed] [Google Scholar]
- 22.LeGall J, Prickril B C, Moura I, Xavier A V, Moura J J G, Huynh B-H. Isolation and characterization of rubrerythrin, a non-heme iron protein from Desulfovibrio vulgaris that contains rubredoxin centers and a hemerythrin-like binuclear iron cluster. Biochemistry. 1988;27:1636–1642. doi: 10.1021/bi00405a037. [DOI] [PubMed] [Google Scholar]
- 23.Lehmann Y, Meile L, Teuber M. Rubrerythrin from Clostridium perfringens: cloning of the gene, purification of the protein, and characterization of its superoxide dismutase function. J Bacteriol. 1996;178:7152–7158. doi: 10.1128/jb.178.24.7152-7158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ljungdahl L G. The acetyl-CoA pathway and the chemiosmotic generation of ATP during acetogenesis. In: Drake H L, editor. Acetogenesis. New York, N.Y: Champman and Hall; 1994. pp. 63–87. [Google Scholar]
- 25.Ljungdahl L G. The autotrophic pathway of acetate synthesis in acetogenic bacteria. Annu Rev Microbiol. 1986;40:415–450. doi: 10.1146/annurev.mi.40.100186.002215. [DOI] [PubMed] [Google Scholar]
- 26.Ljungdahl L G, Wiegel J. Working with anaerobic bacteria. In: Demain A L, Soloman N E, editors. Manual of industrial microbiology and biotechnology. Washington, D.C.: American Society for Microbiology; 1988. pp. 84–96. [Google Scholar]
- 27.Lombard M, Fontecave M, Touati D, Niviere V. Reaction of the desulfoferrodoxin from Desulfoarculus baarsii with superoxide anion. Evidence for a superoxide reductase activity. J Biol Chem. 2000;275:115–121. doi: 10.1074/jbc.275.1.115. [DOI] [PubMed] [Google Scholar]
- 28.Lumppio H L, Shenvi N V, Summers A O, Voordouw G, Kurtz D M., Jr Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J Bacteriol. 2001;183:101–108. doi: 10.1128/JB.183.1.101-108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathieu I, Meyer J, Moulis M. Cloning, sequencing, and expression in Escherichia coli of the rubredoxin gene from Clostridium pasteurianum. Biochem J. 1992;285:255–262. doi: 10.1042/bj2850255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morton T A, Chou C-F, Ljungdahl L G. Cloning, sequencing, and expression of genes encoding enzymes of the autotrophic acetyl-CoA pathway in the acetogen Clostridium thermoaceticum. In: Sebald M, editor. Genetics and molecular biology of anaerobic bacteria. New York, NY: Springer Verlag; 1992. pp. 389–406. [Google Scholar]
- 31.Morton T A, Runquist J A, Ragsdale S W, Shanmugasundaram T, Wood H G, Ljungdahl L G. The primary structure of the subunits of carbon monoxide dehydrogenase/acetyl-CoA synthase from Clostridium thermoaceticum. J Biol Chem. 1991;266:23824–23828. [PubMed] [Google Scholar]
- 32.Nolling J, Ishii M, Koch J, Pihl T D, Reeve J N, Thauer R K, Hedderich R. Characterization of a 45 kDa flavoprotein and evidence for a rubredoxin, two proteins that could participate in electron transport from H2 to CO2 in methanogenesis in Methanobacterium thermoautotrophicum. Eur J Biochem. 1995;231:628–638. doi: 10.1111/j.1432-1033.1995.0628d.x. [DOI] [PubMed] [Google Scholar]
- 33.Pianzzola M J, Soubes M, Touati D. Overproduction of the rbo gene product from Desulfovibrio species suppresses all deleterious effects of lack of superoxide dismutase in Escherichia coli. J Bacteriol. 1996;178:6736–6742. doi: 10.1128/jb.178.23.6736-6742.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierik A J, Wolbert R B G, Portier G L, Verhagen M F G, Hagen W R. Nigerythrin and rubrerythrin from Desulfovibro vulgaris each contain two mononuclear iron centers and two dinuclear iron clusters. Eur J Biochem. 1993;212:237–245. doi: 10.1111/j.1432-1033.1993.tb17655.x. [DOI] [PubMed] [Google Scholar]
- 35.Prickril B C, Kurtz D M, Jr, LeGall J, Voordouw G. Cloning and sequencing of the gene for rubrerythrin from Desulfovibrio vulgaris (Hildenborough) Biochemistry. 1991;19:1118–1123. doi: 10.1021/bi00110a014. [DOI] [PubMed] [Google Scholar]
- 36.Ragsdale S W. Enzymology of acetyl-CoA pathway of CO2 fixation. Crit Rev Biochem Mol Biol. 1991;26:261–300. doi: 10.3109/10409239109114070. [DOI] [PubMed] [Google Scholar]
- 37.Ragsdale S W, Ljungdahl Lars G. Characterization of ferredoxin, flavodoxin, and rubredoxin from Clostridium formicoaceticum grown in media with high and low iron contents. J Bacteriol. 1984;157:1–6. doi: 10.1128/jb.157.1.1-6.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ragsdale S W, Ljungdahl L G, DerVartanian D. Isolation of carbon monoxide dehydrogenase from Acetobacterium woodii and comparison of its properties with those of the Clostridium thermoaceticum. J Bacteriol. 1983;155:1224–1237. doi: 10.1128/jb.155.3.1224-1237.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Seki Y, Seki S, Satoh M, Ikeda A, Ishimoto M. Rubredoxin from Clostridium perfringens: complete amino acid sequence and participation in nitrate reduction. J Biochem (Tokyo) 1989;106:336–341. doi: 10.1093/oxfordjournals.jbchem.a122854. [DOI] [PubMed] [Google Scholar]
- 41.Sieker L C, Holmes M, Le Trong I, Turley S, Liu M Y, LeGall J, Stenkamp R E. The 1.9 Å crystal structure of the “as isolated” rubrerythrin from Desulfavibrio vulgaris: some surprising results. J Biol Inorg Chem. 2000;5:505–513. doi: 10.1007/pl00021450. [DOI] [PubMed] [Google Scholar]
- 42.Voordouw J K, Voordouw G. Deletion of the rbo gene increases the oxygen sensitivity of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Appl Environ Microbiol. 1998;64:2882–2887. doi: 10.1128/aem.64.8.2882-2887.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wasserfallen A, Ragettli S, Jouanneau Y, Leisinger T. A family of flavoproteins in the domains archaea and bacteria. Eur J Biochem. 1998;254:325–332. doi: 10.1046/j.1432-1327.1998.2540325.x. [DOI] [PubMed] [Google Scholar]
- 44.Wood H G, Ljungdahl L G. Autotrophic character of the acetogenic bacteria. In: Shively J M, Barton L L, editors. Variations of autotrophic life. New York, N.Y: Academic Press; 1991. pp. 201–250. [Google Scholar]
- 45.Xu Y, Mortimer M W, Fisher T S, Kahn M L, Brockman F J, Xun L. Cloning, sequencing, and analysis of a gene cluster from Chelatobacter heinzii ATCC 29600 encoding nitrilotriacetate monooxygenase and NADH:flavin mononucleotide oxidoreductase. J Bacteriol. 1997;179:1112–1116. doi: 10.1128/jb.179.4.1112-1116.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang S-S, Ljungdahl L G, DerVartanian D V, Watt G D. Isolation and characterization of two rubredoxins from Clostridium thermoaceticum. Biochim Biophys Acta. 1980;590:24–33. doi: 10.1016/0005-2728(80)90143-7. [DOI] [PubMed] [Google Scholar]
- 47.Yoon K S, Hille R, Hemann C, Tabita F R. Rubredoxin from the green sulfur bacterium Chlorobium tepidum functions as an electron acceptor for pyruvate ferredoxin oxidoreductase. J Biol Chem. 1999;274:29772–29778. doi: 10.1074/jbc.274.42.29772. [DOI] [PubMed] [Google Scholar]