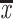Abstract
The common precursor to all tetrapyrroles is 5-aminolevulinic acid (ALA), and in Rhodobacter sphaeroides its formation occurs via the Shemin pathway. ALA synthase activity is encoded by two differentially regulated genes in R. sphaeroides 2.4.1: hemA and hemT. In our investigations of hemA regulation, we applied transposon mutagenesis under aerobic conditions, followed by a selection that identified transposon insertion mutants in which hemA expression is elevated. One of these mutants has been characterized previously (J. Zeilstra-Ryalls and S. Kaplan, J. Bacteriol. 178:985–993, 1996), and here we describe our analysis of a second mutant strain. The transposon inserted into the coding sequences of hbdA, coding for S-(+)-β-hydroxybutyryl–coenzyme A dehydrogenase and catalyzing an NAD-dependent reaction. We provide evidence that the hbdA gene product participates in polyhydroxybutyrate (PHB) metabolism and, based on our findings, we discuss possibilities as to how defective PHB metabolism might alter the level of hemA expression.
Rhodobacter sphaeroides has a diverse array of catabolic pathways at its command, including aerobic and anaerobic respiration and photosynthesis (for reviews, see references 24 and 51). These metabolisms are supported by the ability of R. sphaeroides to synthesize several metallotetrapyrroles, including hemes, bacteriochlorophyll (bchl), and corrinoids (reviewed in reference 23). Organisms commanding such metabolic versatility must have regulatory mechanisms that confer the ability to choose among these pathways in order to maximize energy production with the resources available and according to the prevailing environmental conditions. A readily observable indication of the presence of these mechanisms is the variation in absolute and relative levels of the different tetrapyrroles present in R. sphaeroides according to the catabolic state of the cell. The level of bchl undergoes a dramatic increase under conditions of lowering oxygen tension in preparation for anoxygenic photosynthetic energy production (29). On the other hand, heme levels must be maintained for aerobic and anaerobic respiration, as well as for photosynthesis (reviewed in references 23 and 29). The corrinoid vitamin B12 is an essential cofactor in methionine synthesis in this organism (10; for a general review, see reference 43) and thus is required under all conditions. One control point for regulated production of tetrapyrroles is at the level of formation of their common precursor, 5-aminolevulinic acid (ALA).
It is well established in R. sphaeroides that the levels of ALA formation parallel the amount of tetrapyrroles in the cell (29). However, the presence of more than one gene coding for ALA synthase activity in R. sphaeroides 2.4.1 (37, 48) requires knowledge of the expression of each gene in order to achieve a full understanding of how ALA levels are regulated. R. sphaeroides is the only known prokaryote possessing duplicate genes for ALA synthase, and the genes coding for the two isozymes, hemA and hemT, are differentially regulated (37, 53, 54). Under the laboratory conditions examined thus far, hemA appears to provide most, if not all of the ALA synthase activity present in cells, while hemT is transcriptionally off (37, 54). However, hemT encodes a fully functional enzyme and, when expressed, it is capable of fulfilling the cellular requirements for ALA under all of the conditions examined (36, 37).
Our understanding of the regulation of hemA expression is far from complete. We have learned that it is complex and involves at least transcriptional regulation (52, 53), as well as feedback inhibition of HemA activity by heme (reviewed in reference 29). Neidle and Kaplan (37) reported that the relative amount of total hemA message is approximately threefold higher in photosynthetically grown cells than in aerobically grown cells. However, based on both mRNA studies (37) and recent in vivo investigations (L. Fales, L. Kryszak, K. Nowosielski, and J. Zeilstra-Ryalls, unpublished data), hemA is transcribed from more than one promoter, and the levels of transcription from each vary according to the growth conditions. The presence of a consensus binding sequence for Escherichia coli Fnr in the upstream sequences of hemA had predicted the existence of an R. sphaeroides Fnr homolog (37), which we have since identified as FnrL, and determined that it is indeed involved in mediating an increase in hemA transcription in response to lowering oxygen tensions (52).
Among the genetic approaches we have used to identify loci that affect hemA expression is the application of transposon mutagenesis together with a selection demanding increased expression from hemA upstream sequences. We have previously described in detail one such trans-acting locus, har-1 (hemA regulatory locus 1 [53]). We describe here our characterization of another trans-acting locus, har-3, identified from this same selection. The two transposon mutant strains have similar characteristics that are not limited to increased transcription from hemA sequences but also include an increased presence of photosynthetic membranes under highly aerobic conditions. We consider these similarities to be highly significant, and we suggest that they are indicative of the participation of both trans-acting loci in the same regulatory network.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The growth of R. sphaeroides in Sistrom's succinic acid minimal medium A (14, 17, 45) and E. coli in Luria-Bertani media (44) have been described previously. For anaerobic-dark–dimethyl sulfoxide (DMSO) growth of R. sphaeroides, Sistrom's media was supplemented with DMSO to a final concentration of 0.06 M and yeast extract to a final concentration of 0.1% (wt/vol). Anaerobic-dark–DMSO growth took place in screw-capped tubes or bottles that were completely filled for 8 to 11 days at 30°C. Final cell densities were approximately 5 × 108 cells/ml. Aerobic growth of R. sphaeroides was carried out by sparging liquid cultures with a mixture of the following gases: nitrogen (68%), oxygen (30%), and carbon dioxide (2%) to a final cell density not exceeding 3 × 108 cells/ml, unless otherwise indicated. Photosynthetic growth conditions were achieved by sparging liquid cultures that were placed in front of the light (ca. 10 W/m2) with 98% nitrogen and 2% carbon dioxide. In all cases, final cell densities represented a minimum of five doublings from the initial inoculum. For plasmid selection and maintenance, media were supplemented with antibiotics. For E. coli, the final concentrations were 15 μg/ml for tetracycline and 50 μg/ml each for kanamycin, spectinomycin-streptomycin, and trimethoprim; for trimethoprim resistance selection, the use of minimal M63 medium (44) was required. The final concentrations for R. sphaeroides were 0.8 μg/ml for tetracycline and 50 μg/ml each for kanamycin, spectinomycin-streptomycin, and trimethoprim. Reagent-grade antibiotics were used and were purchased from Sigma Chemical Co. (St. Louis, Mo.).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| E. coli | ||
| DH5αphe | F− φ80dlacZ Δ(lacZYA−argF)U169 recA1 endA1 hsdR17 | 18, 22 |
| HB101 | F− Δ(gpt-proA)62 leuB6 supE44 ara-14 galK2 lacY1 | 16 |
| R. sphaeroides | ||
| 2.4.1 | Wild type | W. Sistrom |
| JZ724 | har-3 | 53 |
| Plasmids | ||
| pBSIISK+ | Apr | Stratagene |
| pBBRMCS2 | Knr | 26 |
| pCF1010 | Derivative of RSF1010; Tcr Spr/Str; used for creating lacZ transcriptional fusions | 30 |
| pJZ20 | 9.5-kb BamHI fragment from R. sphaeroides JZ724 in pUI1087 | This study |
| pLE20 | 4.5-kb ClaI fragment from P. denitrificans in pBSIISK+ | 5 |
| pLF14 | 11.5- and 3.3-kb SalI fragments from R. sphaeroides JZ724 in pUI1087 | This study |
| pLF15 | Amplified hbdA sequences ligated into MscI-treated pUI1087 | This study |
| pLF16 | XbaI-EcoRI fragment containing hbdA from pLF15 in pBBRMCS2 | This study |
| pRK2013 | ColE1 replicon; Tra+ of RK2; Knr | 16 |
| pUI1087 | pBSIISK+ with modified polylinker | 54 |
| pUI1088 | ΩSpr/Str-orfA2-hemA::lacZYA′ in RSF1010 | 52 |
Apr, ampicillin resistant; Knr, Kanamycin resistant; Spr/Str, spectinomycin resistant and streptomycin resistant.
Conjugation and transformation.
Mobilizations of plasmids into R. sphaeroides were performed by previously described protocols (14). Transformation of E. coli was performed using CaCl2-treated cells prepared according to standard methodologies (44).
DNA manipulations and DNA sequence analysis.
DNA isolation, restriction endonuclease treatment, and other enzymatic treatment of DNA fragments and plasmids were done according to standard protocols (44) or manufacturers' instructions, with enzymes purchased from New England BioLabs, Inc. (Beverly, Mass.), Gibco-BRL/Life Technologies, Inc. (Gaithersburg, Md.), and Promega (Madison, Wis.). DNA was analyzed by standard electrophoretic techniques (44), and isolation of DNA from agarose was performed using the Prep-A-Gene DNA Purification System from Bio-Rad (Hercules, Calif.). Chromosomal DNA used for cloning was isolated from R. sphaeroides according to the procedure of Ausubel et al. (3).
DNA sequencing was performed in part by the DNA Sequencing Facility of the Department of Molecular Biology, Iowa State University, Ames, Iowa. Other DNA sequencing was performed on site, using an ABI Prism 310 Genetic Analyzer and the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Inc., Foster City, Calif.). Sequencing reactions were prepared according to manufacturers' instructions. To improve primer extensions due to the high G+C content of the R. sphaeroides genome, DMSO was added to the sequencing reactions prior to thermal cycling at a final concentration of 5%. Most of the oligonucleotides used for priming standard sequencing reactions were synthesized at Gibco-BRL/Life Technologies. Our laboratory was also a beta test site for CleanCut primers (Integrated DNA Technologies, Inc., Coralville, Iowa), which were used according to the manufacturer's instructions.
DNA sequences were analyzed by using the BLAST server at the National Center of Biotechnology Information (Bethesda, Md.) and the BLASTX program (1). Other analyses were performed by using Wisconsin Package version 9.1 software (Genetics Computer Group, Madison, Wis.).
Cloning wild-type hbdA from R. sphaeroides 2.4.1.
The hbdA sequences were amplified from chromosomal DNA using the primers HBD-UP (5′-GCG GAA CAT CAG ACG AGA CCC GCC ATA CCG-3′) and HBD-DOWN (5′-GTA TCT CTG GGG CTG GCC GGT GCA GAT GCT-3′) with Pfu Turbo (Stratagene, La Jolla, Calif.) in the following reaction: (i) denaturation at 95°C for 3 min and (ii) annealing and extension at 65°C for 5 min, repeated 35 times, followed by a final incubation of 5 min at 72°C. The DNA was purified, following electrophoresis through an agarose gel, using the Prep-A-Gene purification kit (Bio-Rad) according to the manufacturer's instructions and ligated to pUI1087 (54) restricted with MscI.
Spectral analysis of membrane fractions and quantitation of pigments.
Crude cell-free lysates from R. sphaeroides 2.4.1, JZ722, and JZ724 were prepared by according to previously described protocols (53). Spectra were recorded over a range of 400 to 900 nm. The B875 and B800-850 pigment-protein complex levels were determined by the method of Meinhardt et al. (33) from the spectral data.
Enzyme assays.
Cells used for assaying β-galactosidase, ALA synthase, and acetoacetyl-coenzyme A (CoA) reductase activity were grown under aerobic, anaerobic-dark–DMSO, and photosynthetic conditions. In all cases, the protein synthesis was halted by the addition of chloramphenicol (0.3 mg/ml, final concentration) to the cultures, which were then chilled on ice. Cleared cell lysates were prepared by passaging cells through an SLM-Aminco French pressure cell (Spectronic Instruments, Inc., Rochester, N.Y.) at 700 lb/in2, followed by centrifugation at 18,000 × g for 10 min at 4°C. All assays were performed immediately on freshly prepared lysates. All spectrophotometric measurements were made using a U-2010 UV/Vis Spectrophotometer (Hitachi Instruments, Inc.), with the exception of the cell density determinations, which were made using a Klett colorimeter (1 Klett unit = 107 cells/ml).
Assays for β-galactosidase activity were performed using the cell lysates prepared in 100 mM NaPO4 buffer, as described elsewhere (47). Reagent-grade o-nitrophenyl-β-d-galactopyranoside, purchased from Sigma Chemical Co., was used as the substrate.
ALA synthase activity levels present in cleared cell lysates in 100 mM NaPO4 buffer was determined using the method of Burnham (9), which relies on endogenous succinyl-CoA synthetase activity to convert succinate to succinyl-CoA substrate for ALA synthesis. Assays were also performed in which succinyl-CoA was directly added to the reaction mix; CoA, ATP, and succinate were then omitted.
Assays for acetoacetyl-CoA reductase activity were performed essentially as described by Chohan and Copeland (12). Cleared cell lysates were prepared in 0.05 M Tris (pH 8.5). Enzyme activity was assayed at 25°C in a total volume of 1 ml containing 50 mM Tris-HCl (pH 8.5), 15 mM MgCl2, 250 μM NAD(P)H, and 100 μM acetoacetyl-CoA. The sample was monitored spectrophotometrically at 340 nm over a total of 4 min, during which NADH or NADPH oxidation was linear. In all cases, nonspecific activity was corrected for by monitoring the decrease in absorbance at 340 nm for 2 min prior to the addition of acetoacetyl-CoA.
Immunoblot analysis.
Samples of cleared cell lysates prepared as described above were subjected to immunoblot analysis according to standard procedures (20), using as primary antiserum a 1:10,000 dilution of HemA rabbit antisera (6) and alkaline phosphatase-conjugated goat anti-rabbit antisera (Sigma Chemical Co.) as the secondary antibody. The substrate used for detection of immunocomplexes was an equimolar mixture of nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate p-toluidine (Gibco-BRL/Life Technologies, Inc.).
Protein determinations.
The concentrations of protein in extracts subjected to spectral analysis, assayed for enzyme activity, or analyzed by immunoblotting were determined with the Pierce BCA Protein Assay Reagents (Rockford, Ill.). In all cases, bovine serum albumin was used as a reference.
Nile red colony stain.
R. sphaeroides was grown on Sistrom's minimal medium or Sistrom's minimal medium without aspartate and glutamate but with either low (2.5 mM) or high (7.5 mM) concentrations of (NH4)2SO4. The added carbon source was either succinate or malate (30 mM). Plates were affixed to supports and placed upright in anaerobic jars in front of the light (ca. 10 W/m2) at 28°C. Incubation continued until well-developed colonies had formed (4 to 5 days). The plates were then removed from the jars, and Nile red dye (Sigma Chemical Co.) was applied immediately, according to the protocol of Kranz et al. (28), using a 0.1% (wt/vol) solution in acetone. After the evaporation of acetone solvent had occurred, the colony fluorescence was visualized by placing the plates on a UV (300 nm) transilluminator (Fotodyne, Hartland, Wis.).
Nucleotide sequence accession numbers.
The DNA sequences presented here are accessible from the GenBank database under accession numbers AF212163 for the R. sphaeroides 2.4.1 sequences and AF212164 for the Paracoccus denitrificans DNA sequences.
RESULTS
Defining the har-3 locus.
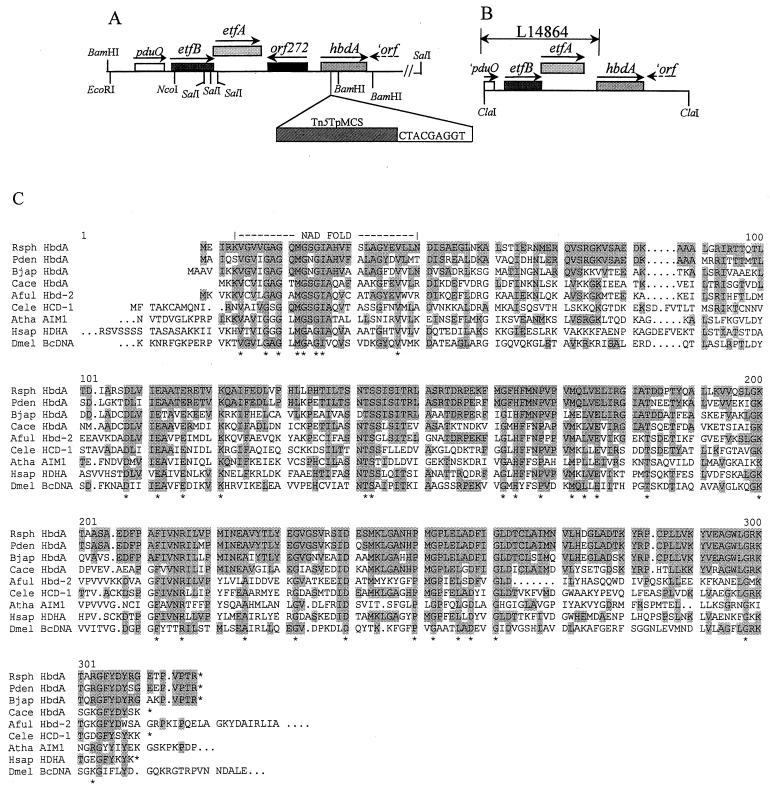
R. sphaeroides mutant strain JZ724 is one of several mutant strains isolated as previously described, and the position of the transposon insertion site was mapped to 1,408 kbp clockwise from the zero position on chromosome I (53). We cloned two chromosomal DNA fragments from mutant strain JZ724 using the trimethoprim resistance gene within the transposon as a marker. Plasmid pJZ20 contains an approximately 9.5-kbp BamHI fragment, and plasmid pLF14 contains an approximately 11.5-kbp SalI fragment. Based on our DNA sequence analysis of these clones, a gene map of the transposon insertion locus has been constructed and is shown in Fig. 1. The precise location of the transposon insertion, also indicated in Fig. 1, is located immediately following nucleotide 86, in the coding region of sequences that predict a protein with approximately 67% identity with the product of the Bradyrhizobium japonicum hbdA gene (49). The har-3 locus of R. sphaeroides resembles a region already partially described previously (5) in the closely related, but metabolically quite different organism P. denitrificans. Our analysis of the DNA sequences downstream of the P. denitrificans etf genes in plasmid pLE20 (kindly provided by F. Frerman, Denver, Colorado) revealed that the arrangement of several genes within the region is similar in both organisms (Fig. 1). In fact, the proximity of hbdA homologues to etf genes is conserved among both gram-negative and gram-positive bacteria, including B. japonicum (49) and Clostridium acetobutylicum ATCC 824 (7).
FIG. 1.
Genetic and physical map of the har-3 locus of R. sphaeroides 2.4.1 and the corresponding region in P. denitrificans and alignment of the predicted amino acid sequence of the hbdA gene products of these organisms with similar proteins from other organisms. (A) The position of the transposon insertion in mutant strain JZ724 and the duplicated sequence generated through transposition are indicated. (B) The region spanning the DNA sequences previously determined for P. denitrificans are identified by an arrow and the GenBank accession number. (C) Alignments were generated using the PILEUP algorithm of the Genetics Computer Group program (version 9.1). The predicted protein product of the R. sphaeroides 2.4.1 hbdA gene aligned with similar proteins from several organisms that were identified using BLASTX. Asterisks indicate amino acid residues that are absolutely conserved between all of the proteins. Displayed are the following: Rsph HbdA, R. sphaeroides 2.4.1 HbdA; Pden HbdA, P. denitrificans HbdA, Bjap HbdA, B. japonicum HbdA (3287857); Cace HbdA, C. acetobutylicum ATCC 824 HbdA (1708125); Aful Hbd-2, Archaeoglobus fulgidus 3-hydroxyacyl-CoA dehydrogenase (hbd-2) (2650351); Cele HCD-1, Caenorhabditis elegans probable 3-hydroxyacyl-CoA dehydrogenase (462252); Atha AIM1, Arabidopsis thaliana AIM1 protein (4337025); Hsap HDHA (residues 10 to 314), human short-chain l-3-hydroxyacyl-CoA dehydrogenase (4240477); Dmel BcDNA (residues 362 to 667), Drosophila melanogaster BcDNA protein (5901852). All sequence numbers are GI numbers (GenInfo Identifier) from NCBI.
The predicted product of hbdA belongs to the hydroxyacyl-CoA dehydrogenase family of proteins which are not only highly conserved not only among species of Bacteria but also among virtually all cells in which these sequences have been identified, including Bacteria, Archaea, plants, insects, and animals. An alignment of the amino acid sequences of several members is shown in Fig. 1. The function commonly ascribed to this family of proteins is the stereospecific oxidation of S-(+)-β-hydroxybutyryl-CoA and related compounds in the iterative process of β-oxidation of fatty acids. However, homologs participate in other metabolisms as well (7, 8).
While the overall arrangement of the pduO, etf, and hbdA genes are similar between R. sphaeroides 2.4.1 and P. denitrificans, a considerably longer intervening sequence is present in R. sphaeroides 2.4.1, 1,040 bp, compared to the 198 bp of the DNA sequence between the nonsense codon of EtfA and the translation initiation codon of HbdA in P. denitrificans. Codon usage analysis of these sequences based on a R. sphaeroides 2.4.1 codon preference profile suggests the presence of an open reading frame divergently arranged relative to hbdA. Searches of the sequence databases did not identify any predicted homologues to this putative open reading frame. Our analysis of DNA sequences downstream of hbdA suggest that the presence of an open reading frame with similarity to NodX of Rhizobium leguminosarum (see reference 41), oriented in an end-on (→←) direction with hbdA (Fig. 1). An analysis of DNA sequences downstream of hbdA in P. denitrificans suggests the presence of a similarly arranged open reading frame, but the predicted product is most similar to a conserved hypothetical protein in Vibrio cholerae (GenBank accession number AAF93691). Based on the convergent arrangement of these putative open reading frames with respect to hbdA, as well as our complementation analysis results (see below), we conclude that the hbdA gene is monocistronic in both R. sphaeroides and P. denitrificans, as it is in B. japonicum (49).
Analysis of hemA expression in JZ724 using hemA::lacZ.
Using plasmids bearing transcriptional fusions between the hemA upstream sequences and the lacZ gene of E. coli (52) we quantified the effect of the transposon insertion mutation. Table 2 lists the levels of β-galactosidase activity present in extracts of mutant strain JZ724 versus the wild-type 2.4.1 strain bearing the hemA::lacZ transcription fusion plasmid pUI1088. The assays were performed on extracts of cells grown aerobically (30% oxygen) or anaerobically in the dark with DMSO. The greatest difference in activity was measured in extracts of mutant JZ724 versus wild-type 2.4.1 cells grown under aerobic conditions, in which the β-galactosidase activity was approximately fourfold higher in the extracts of mutant cells relative to that present in the extracts of wild-type cells with plasmid pUI1088. Also, at least some level of oxygen control remains intact in mutant strain JZ724, since the activity is approximately twofold higher in extracts of anaerobically grown mutant cells versus aerobically grown mutant cells.
TABLE 2.
Levels of β-galactosidase activity present in extracts of R. sphaeroides
| Strain | Features | Growth conditionsa | β-Galactosidase
activityb
|
|
|---|---|---|---|---|
|
ςn | |||
| 2.4.1(pUI1088) | Wild type (hemA::lacZ) | +O2 | 912 | 142 |
| JZ724(pUI1088) | Tn5Tp::hbdA (hemA::lacZ) | +O2 | 3,709 | 224 |
| 2.4.1(pUI1088) | Wild type (hemA::lacZ) | −O2 | 2,293 | 130 |
| JZ724(pUI1088) | Tn5Tp::hbdA (hemA::lacZ) | −O2 | 6,394 | 241 |
Aerobic growth (30% oxygen, 68% nitrogen, 2% carbon dioxide), +O2; anaerobic-dark growth with DMSO, −O2.
Values indicated are from at least three independent assays.
Assays of ALA synthase activity in cell extracts also indicate that hemA expression is approximately fourfold higher in mutant strain JZ724 (102 ± 5 U/mg of protein) than in wild-type 2.4.1 (24 ± 5 U/mg of protein) under aerobic conditions. The activity levels are higher in extracts of photosynthetically grown mutant JZ724 (204 U/mg of protein) and wild-type 2.4.1 (158 U/mg of protein) cells than in aerobically grown cells. We also determined that the ALA synthase activities in extracts of both aerobically (113 ± 16 U/mg of protein) and photosynthetically (204 U/mg of protein) grown cells of mutant strain JZ722 are similar to those in cell extracts of mutant strain JZ724, suggesting that hemA expression is similar in the two mutant strains.
Phenotype of mutant strain JZ724.
Visual inspection of liquid cultures and colonies of JZ724 revealed that the cells are consistently more pigmented than wild-type cells. We suspected that, as has previously been demonstrated for mutant strain JZ722 (53), this pigmentation indicated that photosynthetic membranes were present at elevated levels relative to wild-type cells under repressing aerobic conditions. Figure 2 shows the spectra of membrane extracts of wild-type 2.4.1 and mutant strains JZ722 and JZ724 cells grown in parallel at 30% oxygen; the levels of pigment-protein complexes are also indicated. As shown in Fig. 2, even when the wild-type cell density is approaching twofold higher than that of the mutant strains JZ722 and JZ724, the pigment-protein complex levels are approximately threefold less. These results suggest that, as in mutant strain JZ722, the cellular consequences of the hbdA disruption in mutant strain JZ724 are not limited to increasing hemA expression.
FIG. 2.
Spectral analysis of membrane extracts of aerobically grown (30% oxygen, 68% nitrogen, and 2% carbon dioxide) wild-type 2.4.1 and mutant strains JZ722 and JZ724 and quantitation of pigment-protein complexes. Spectra were generated using equivalent amounts of total protein.
Complementation analysis of mutant strain JZ724.
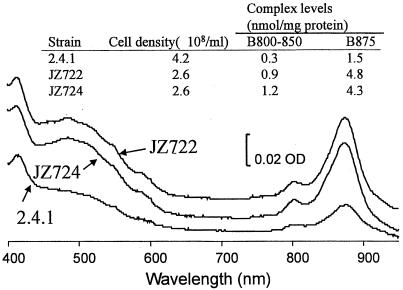
In order to determine whether or not the transposon disruption alone is responsible for the phenotypes described above, hbdA was cloned from wild-type strain 2.4.1. Using a pair of oligonucleotides corresponding to DNA sequences flanking the gene (see Materials and Methods), the wild-type hbdA sequences were amplified by PCR from genomic DNA and then cloned into plasmid vector pUI1087 (see Table 1 and reference 54). DNA sequence analysis confirmed the integrity of the hbdA sequences present. The intact gene was then moved into pBBR1MCS2 (26), which is capable of replicating in R. sphaeroides, resulting in plasmid pLF16 (see Table 1). Finally, plasmid pLF16 was introduced into mutant strain JZ724, and the characteristics of these exconjugants were compared to those of mutant strain JZ724 bearing the plasmid vector pBBR1MCS2.
As shown in Fig. 3, both direct assays of ALA synthase activity and immunoblot analysis, using antisera raised against HemA (6), indicate that the levels of HemA correlate with the presence or absence of an intact hbdA gene. We also compared the level of spectral complexes in mutant strain JZ724 with plasmid pLF16 to that present in cells with the vector and found, as shown in Fig. 3, that the levels present in JZ724 with pLF16 were similar to those in wild-type cells. These results indicate that the disruption in hbdA is responsible for the increase in detectable complexes under aerobic conditions in mutant strain JZ724.
FIG. 3.
Complementation analysis of mutant strain JZ724 with hbdA. (A) ALA synthase activities in extracts of R. sphaeroides wild-type 2.4.1 or mutant JZ724 cells with either pBBR1MCS2 or the hbdA clone pLF16. Growth occurred aerobically (30% O2, 2% CO2, 68% N2) or anaerobically in the dark with 60 mM DMSO. The results are the mean values from assays of at least three independent cultures. Vertical bars represent the standard deviations from the mean, and the numerical values for each result are provided. (B) Immunoblot and spectral analysis of extracts of R. sphaeroides cells grown under aerobic conditions. The blot was probed with anti-HemA antisera (6). In lanes 1 to 4, The samples contained equal amounts of total protein. For further details regarding sample preparation and analysis, see Materials and Methods.
Function of the hbdA gene product.
The product of the phaB gene converts acetoacetyl-CoA to the R-(−) stereoisomer of β-hydroxybutyryl-CoA, which can then be polymerized by the phaC gene product (or a homologue) to form polyhydroxybutyrate (or more generally referred to as polyhydroxyalkanoate [PHA]; see reference 46). In contrast, the β-hydroxyacyl-CoA dehydrogenase family of proteins, to which HbdA most likely belongs, catalyzes the formation of the S-(+) isomers. In addition to substrate stereospecificity, the enzyme activities have been distinguished from one another in Rhizobium (Cicer) sp. strain CC1192 by the absolute requirement for NADH in the case of the S-(+) stereospecific enzyme and NADPH in the case of the R-(−) specific enzyme.
We assayed mutant strain JZ724 and wild-type strain 2.4.1 for acetoacetyl-CoA reductase activity in aerobically grown cells by following the oxidation of NADH. While cell extracts of wild-type 2.4.1 (4.5 × 108 cells/ml) had activities of 20 ± 4 U/mg of protein, the levels of activity present in cell extracts of mutant strain JZ724 (3.2 × 108 cells/ml) were below detection.
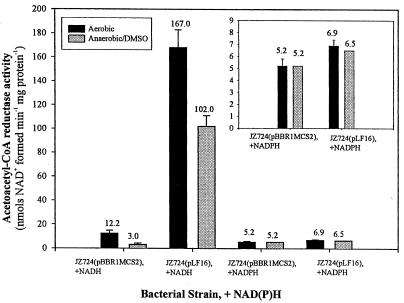
To confirm the identity of the product of the hbdA gene, we compared the levels of activity in extracts of the mutant strain JZ724 with pLF16 to that in cell extracts of mutant strain with plasmid vector pBBR1MCS2. The pyridine nucleotide requirement of HbdA was also evaluated by comparing the activity in the presence of NADH to that in the presence of NADPH. As shown in Fig. 4, the results of this analysis reveal that the levels of NADH-dependent enzyme activity are approximately 14-fold lower in extracts of aerobically grown mutant strain JZ724 with plasmid pBBR2MCS1 than in extracts of cells with plasmid pLF16 and approximately 34-fold lower in extracts of anaerobically (with DMSO) grown cells. In contrast to the NADH results, NADPH-associated enzymatic activity is the same in extracts of mutant strain JZ724 with either plasmid under aerobic or anaerobic (with DMSO) conditions. The protein alignment presented in Fig. 1, suggesting that HbdA codes for a β-hydroxybutyryl-CoA dehydrogenase, is fully supported by these results.
FIG. 4.
Acetoacetyl-CoA reductase activities in extracts of R. sphaeroides mutant strain JZ724 with either pBBR1MCS2 or the hbdA clone pLF16. Each sample was assayed in the presence of NADH and NADPH, as indicated. Growth occurred aerobically (30% O2, 2% CO2, 68% N2) or anaerobically in the dark with 60 mM DMSO. The results are the mean values from assays of at least two independent cultures. Vertical bars represent the standard deviations from the mean, and the numerical values for each result are provided. For a description of the assay protocol, see Materials and Methods.
Analysis of PHA accumulation.
In R. capsulatus, inactivation of phaB has no effect on PHA accumulation (27). Other evidence suggests that R. sphaeroides also possesses more than one pathway for the synthesis of monomer substrate for PHA polymerase (21). It may be that in these bacteria, the S-(+) stereoisomer of β-hydroxybutyryl-CoA produced by hbdA can be converted to the R-(−) stereoisomer in a manner that has been described previously for Rhodospirillum rubrum catalyzed by two enoyl-CoA hydratases (34). We therefore reasoned that disabling hbdA might affect PHA metabolism.
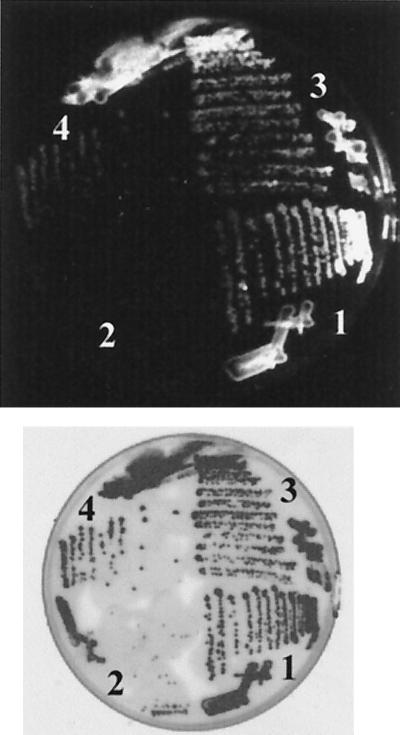
Using a colony-staining method developed by Kranz et al. (28), we compared PHA accumulation in wild-type 2.4.1 to that in mutant strain JZ724 grown under photosynthetic conditions. In contrast to colonies of wild-type 2.4.1 with either the vector or pLF16, we observed less fluorescence for mutant strain JZ724 with the vector pBBR1MCS2, indicating that PHA accumulation is lower in the mutant strain compared to the wild type. Fluorescence was comparable to the wild type for colonies of mutant strain JZ724 with plasmid pLF16, indicating that hbdA on the plasmid had restored PHA accumulation in mutant strain JZ724 to levels similar to that in wild-type 2.4.1. This difference in fluorescence was observed for colonies formed on a variety of different media, but the greatest difference was observed on medium containing malate and high levels of nitrogen, which is shown in Fig. 5. On the basis of these observations, we conclude that the transposon insertion in the hbdA gene in mutant strain JZ724 does affect PHA metabolism.
FIG. 5.
Colonies of R. sphaeroides wild-type 2.4.1 and mutant JZ724 exconjugants of either pBBR1MCS2 or pLF16. (Top) Photograph of fluorescent colonies illuminated by UV (300-nm) light after Nile red dye was applied. (Bottom) Scan of colonies by visible light. Cells shown were grown on Sistrom's modified (no aspartate or glutamate) minimal medium with 30 mM malate as the carbon source, with 7.5 mM (NH4)2SO4, and with kanamycin for plasmid maintenance. Numbered sections refer to colonies of the following strains. Section 1, 2.4.1(pBBR1MCS2); section 2, JZ724(pBBR1MCS2); section 3, 2.4.1(pLF16); section 4, JZ724(pLF16).
DISCUSSION
Using transposon mutagenesis and applying a selection for increased expression of hemA in the presence of oxygen, we isolated a set of mutants. Three of the trans-acting mutations represented among the mutant strains are known to be at widely distributed locations on Chromosome I (53) of R. sphaeroides 2.4.1. The present investigation reveals that a transposon insertion in hbdA results in an increase in hemA expression and, based on the increased presence of photosynthetic membranes under highly aerobic conditions, may alter the expression of other photosynthesis genes as well. We have also demonstrated that hbdA participates in PHA metabolism. Are these observations directly related? Our hypothesis is that they are. We first present our model as to how hbdA participates in PHA synthesis, and then we suggest how altering PHA metabolism might affect hemA expression at the level of transcription.
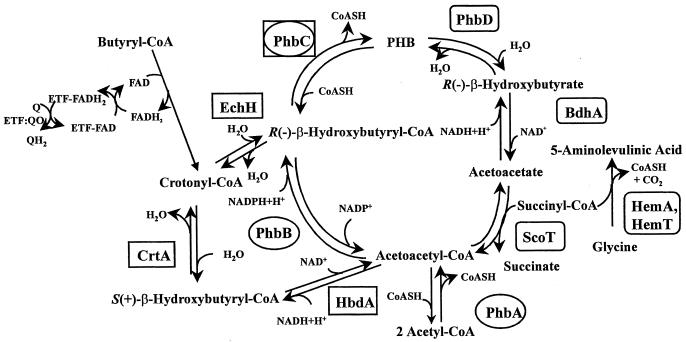
Figure 6 represents a synthesis of what is known or inferred from the literature about the metabolism of R. sphaeroides pertaining to both hbdA and hemA. A key feature is that two pathways are proposed for PHA synthesis from acetoacetyl-CoA in R. sphaeroides. We believe that a similar scheme for PHA metabolism, presented in 1978 by Merrick (see Fig. 7 [p. 211] in reference 13), represented a composite diagram of two separate pathways that were known to occur in separate organisms. One pathway involves reactions catalyzed by pha/phb gene products (surrounded by ovals in Fig. 6). The phaA and B genes have been cloned from Rhodobacter capsulatus (27) and P. denitrificans (50), and we have identified sequences that code for proteins that are approximately 75 and 86% identical to PhaA and -B of P. denitrificans, respectively, among the preliminary DNA sequences of the R. sphaeroides 2.4.1 genome (from the Department of Energy Joint Genome Institute, http://spider.jgi-psf.org/JGI_microbial/html). The gene for the last enzyme in the pathway, phaC, has been cloned and sequenced from several wild-type strains of R. sphaeroides (25, 46; see also GenBank accession no. X97200). The second pathway for PHA synthesis involves the enzymes surrounded by boxes in Fig. 6. Anderson and Dawes have pointed out that the S-(+) stereoisomer of β-hydroxybutyryl-CoA, produced by the β-oxidation of fatty acids, could be converted to the R-(−) isomer via acetoacetyl-CoA by the concerted action of HbdA and PhaB (2). However, this would not explain the observation by Kranz et al. (27) that inactivation of phaB does not affect PHA synthesis in R. capsulatus, since that alternate route would still require PhaB activity (see Fig. 6). We propose that, in both R. sphaeroides and R. capsulatus, PHA can be synthesized from either stereoisomer of β-hydroxybutyryl-CoA according to the diagram shown in Fig. 6. Recently, Reiser et al. (42) reported that R. capsulatus could encode an R-specific β-hydroxybutyryl-CoA hydratase. An earlier report (4) described a different orf (ORF257) in R. capsulatus predicted to encode a polypeptide with similarity to enoyl-CoA hydratases involved in fatty acid β-oxidation. Inspection of the DNA sequence of the R. sphaeroides 2.4.1 genome (http://spider.jgi-psf.org/JGI_microbial/html) predicts genes that encode polypeptides with approximately 77 and 66% identity, respectively, to the R. capsulatus polypeptides. Thus, together with hbdA, DNA sequences that predict all of the enzymatic requirements for both pathways have now been identified in species of Rhodobacter. The lack of any discernible effect on PHA synthesis in a PhaB− mutant of R. capsulatus (27), compared to the decrease in PHA accumulation we observe when hbdA is disabled in R. sphaeroides, suggests that a significant amount of PHA precursor is provided by the activity of HbdA in both organisms.
FIG. 6.
Schematic diagram of proposed dual pathway for polyhydroxybutyrate biosynthesis in R. sphaeroides 2.4.1. Enzymes catalyzing the reactions have been abbreviated as follows: PhbA, 3-ketothiolase; PhbB, acetoacetyl-CoA reductase; PhbC, polyhydroxybutyrate synthase; CrtA, S-(+)-β-hydroxybutyryl-CoA dehydratase (crotonase); EchH, enoyl CoA hydratase; HbdA, S-(+)-β-hydroxybutyryl-CoA dehydrogenase; PhbD, polyhydroxybutyrate depolymerase; BdhA, R-(−)-β-hydroxybutyrate dehydrogenase; ScoT, 3-oxoacid CoA-transferase (succinyl-CoA–acetoacetate transferase); HemA and HemT, ALA synthases.
Clues as to the relationship between PHA metabolism and hemA transcription come from studies in which all PHA synthesis has been abolished by genetically disabling the PHA synthase gene, phaC, in several α-proteobacteria. These studies reveal that a number of cellular processes are affected (11, 21, 32). A common feature appears to be that abolishing PHA accumulation leads to a decrease in the NAD+/NADH ratio (11, 32). Although PHA accumulation is not abolished in the absence of HbdA activity in R. sphaeroides, our results indicate that it is clearly reduced. We propose that the loss of HbdA activity resembles the PhaC− mutants by lowering PHA accumulation, thereby altering the NAD+/NADH ratio.
While recent studies have shown that the ratio between oxidized and reduced pyridine nucleotides can affect transcription (15, 31), the molecular details as to how this occurs are not yet understood in any organism. However, in light of the similarity of phenotypes of mutant strains JZ724 and JZ722, with respect to both hemA expression and the increased presence of photosynthetic membranes under aerobic conditions, we suggest that downstream events are related. A description of possible events has been presented by Oh and Kaplan (40), who have recently documented the existence of signal transduction systems involving the electron transport chain and several associated regulatory proteins. One signal emanates from the rate of electron flow through the cbb3 cytochrome c oxidase, coded for by the ccoNOQP operon. Mutant strain JZ722 has a transposon insertion between the coding sequences of ccoN and ccoO (53), which therefore lacks a functional cbb3 oxidase. This is considered to abolish an inhibitory signal that normally exists when the electron flow through the oxidase is high and which is then transduced to the two-component regulatory proteins PrrB, the sensor partner, and PrrA, the response regulator (40). A second signal is believed to emanate from the redox state of the quinone pool, which is transduced to regulatory proteins other than Prr, possibly the antirepressor protein AppA, and its repressor partner PpsR (for details, see reference 40). If PHA metabolism affects the NAD+/NADH ratio, as NAD+ becomes limiting, the donation of electrons from NADH to the quinone pool would be favored. To ascertain whether increased expression of hemA proceeds from an altered redox state of the quinone pool or a variation in the rate of electron flow through the cbb3 oxidase will require additional experimentation. Reasons to think the signal is at the level of the cbb3 oxidase include: (i) mutant strains JZ722 and JZ724 have very similar phenotypes; (2) AppA (19) and PpsR do not appear to influence hemA expression (J. H. Zeilstra-Ryalls, unpublished results), whereas recent evidence suggests that the Prr proteins do (39); and (iii) other than the transposon insertion in mutant strain JZ724, its genome is considered to be intact and, thus, FnrL regulation of gene expression is unaltered. Under aerobic conditions (30% oxygen; see reference 35), the level of expression of the ccoNOQP operon is low, resembling the situation that exists in mutant strain JZ722 which lacks a functional cbb3 oxidase. We suggest that in both cases, insufficient inhibitory signal is transduced from cbb3 oxidase to the Prr two-component system, and the expression of Prr-regulated genes is higher in the presence of oxygen, compared to wild-type 2.4.1.
We have established a relationship between hbdA, PHA metabolism, and hemA expression. We have also proposed a tentative explanation as to how altering PHA metabolism might affect the transcription of hemA, which is testable and provides a clear direction for future investigation. The value of the hemA gene for examining gene expression in response to environmental conditions is confirmed by our previous (52, 53) and present studies. Since we first identified a relationship between expression of the ccoNOQP genes and photosynthesis gene expression (53) the role of this operon in R. sphaeroides has been extensively investigated (see references 38 to 40, among others) and has lead to the formulation of an integrated model for redox signaling in this organism (40). It also led to the identification of the global regulator, fnrL (52). We believe additional features of hemA expression exist, as we have yet other transposon mutant strains to be characterized (53).
ACKNOWLEDGMENTS
This work was supported by grant 9805556 from the National Science Foundation and funds from the Research Excellence Program in Biotechnology at Oakland University. Preliminary R. sphaeroides 2.4.1 genome sequence data was obtained from The DOE Joint Genome Institute (JGI) at http://spider.jgi-psf.org/JGI_microbial/html/.
We thank F. Frerman (University of Colorado) for generously providing pLE20, A. Sinskey and G. York (MIT) for helpful suggestions regarding PHA analysis, and S. Kaplan (University of Texas Health Sciences Center–Houston) for useful discussions and for support during the early stages of this work.
REFERENCES
- 1.Altschul S, Madden T, Schäffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson A, Dawes E. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 4.Beckman D, Kranz R. A bacterial homolog to the mitochondrial enoyl-CoA hydratase. Gene. 1991;107:171–172. doi: 10.1016/0378-1119(91)90313-z. [DOI] [PubMed] [Google Scholar]
- 5.Bedzyk L A, Escudero K W, Gill R E, Griffin K J, Frerman F E. Cloning, sequencing, and expression of the genes encoding subunits of Paracoccus denitrificanselectron transfer flavoprotein. J Biol Chem. 1993;268:20211–20217. [PubMed] [Google Scholar]
- 6.Bolt E, Kryszak L, Zeilstra-Ryalls J, Shoolingin-Jordan P, Warren M. Characterisation of the Rhodobacter sphaeroides 5-aminolevulinic acid synthase isoenzymes, ALAS-A and ALAS-T, isolated from recombinant Escherichia coli. Eur J Biochem. 1999;265:290–299. doi: 10.1046/j.1432-1327.1999.00730.x. [DOI] [PubMed] [Google Scholar]
- 7.Boynton Z, Bennett G, Rudolph F. Cloning, sequencing, and expression of clustered genes encoding β-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryrl-CoA dehydrogenase from Clostridium acetobutylicumATCC 824. J Bacteriol. 1996;178:3015–3024. doi: 10.1128/jb.178.11.3015-3024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryan E M, Beall B W, Moran C P., Jr A ςE-dependent operon subject to catabolite repression during sporulation in Bacillus subtilis. J Bacteriol. 1996;178:4778–4786. doi: 10.1128/jb.178.16.4778-4786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnham B F. [14]δ-Aminolevulinic acid synthase (Rhodopseudomonas spheroides) Methods Enzymol. 1970;17:195–204. [Google Scholar]
- 10.Cauthen S, Pattison J, Lascelles J. Vitamin B12 in photosynthetic bacteria and methionine synthesis in Rhodopseudomonas spheroides. Biochem J. 1967;102:774–781. doi: 10.1042/bj1020774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cevallos M, Encarnacion S, Leija A, Mora Y, Mora J. Genetic and physiological characterization of a Rhizobium etlimutant strain unable to synthesize poly-β-hydroxybutyryate. J Bacteriol. 1996;178:1646–1654. doi: 10.1128/jb.178.6.1646-1654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chohan S N, Copeland L. Acetoacetyl Coenzyme A reductase and polyhydroxybutyrate synthesis in Rhizobium (Cicer) sp. strain CC1192. Appl Environ Microbiol. 1998;64:2859–2863. doi: 10.1128/aem.64.8.2859-2863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clayton R, Sistrom W, editors. The photosynthetic bacteria. New York, N.Y: Plenum Press; 1978. [Google Scholar]
- 14.Davis J, Donohue T J, Kaplan S. Construction, characterization, and complementation of a puf mutant of Rhodobacter sphaeroides. J Bacteriol. 1988;170:320–329. doi: 10.1128/jb.170.1.320-329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Graef M, Alexeeva S, Snoep J, Teixeira de Mattos M. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J Bacteriol. 1999;181:2351–2357. doi: 10.1128/jb.181.8.2351-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ditta G, Stanfield S, Corbin D, Helinski D. Broad host range DNA cloning system for gram negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donohue T J, McEwan A G, Kaplan S. Cloning, DNA sequence, and expression of the Rhodobacter sphaeroides cytochrome c2gene. J Bacteriol. 1986;168:962–972. doi: 10.1128/jb.168.2.962-972.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eraso J M, Kaplan S. prrA, a putative response regulator involved in oxygen regulation in photsynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomelsky M, Kaplan S. appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides2.4.1. J Bacteriol. 1995;177:4609–4618. doi: 10.1128/jb.177.16.4609-4618.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Using antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 21.Hustede E, Steinbuchel A, Schlegel H. Relationship between the photoproduction of hydrogen and the accumulation of PHB in non-sulphur purple bacteria. Appl Microbiol Biotechnol. 1993;39:87–93. [Google Scholar]
- 22.Jessee J. New subcloning efficiency competent cells: >1 × 106transformants/μg. Focus. 1986;8:9. [Google Scholar]
- 23.Jordan P, editor. Biosynthesis of tetrapyrroles. Amsterdam, The Netherlands: Elsevier; 1991. [Google Scholar]
- 24.Kiley P J, Kaplan S. Molecular genetics of photosynthetic membrane biosynthesis in Rhodobacter sphaeroides. Microbiol Rev. 1988;52:50–69. doi: 10.1128/mr.52.1.50-69.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J-H, Lee J. Cloning, nucleotide sequence and expression of gene coding for poly-3-hydroxybutyric acid (PHB) synthase of Rhodobacter sphaeroides2.4.1. J Microbiol Biotechnol. 1997;7:229–236. [Google Scholar]
- 26.Kovach M, Phillips R, Elzer P, Roop II R, Peterson K. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 27.Kranz R, Gabbert K, Locke T, Madigan M. Polyhydroxyalkanoate production in Rhodobacter capsulatus: genes, mutants, expression, and physiology. Appl Environ Microbiol. 1997;63:3003–3009. doi: 10.1128/aem.63.8.3003-3009.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kranz R, Gabbert K, Madigan M. Positive selection systems for discovery of novel polyester biosynthesis genes based on fatty acid detoxification. Appl Environ Microbiol. 1997;63:3010–3013. doi: 10.1128/aem.63.8.3010-3013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lascelles J. Tetrapyrrole biosynthesis and its regulation. W. A. New York, N.Y: Benjamin, Inc.; 1964. [Google Scholar]
- 30.Lee J, Kaplan S. Transcriptional regulation of puc operon expression in Rhodobacter sphaeroides: analysis of the cis-acting downstream regulatory sequence. J Biol Chem. 1995;270:20453–20458. [PubMed] [Google Scholar]
- 31.Leonardo M, Dailly Y, Clark D. Role of NAD in regulating the adhE gene of Escherichia coli. J Bacteriol. 1996;178:6013–6018. doi: 10.1128/jb.178.20.6013-6018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandon K, Michel-Reydellet N, Encarnacíon S, Kaminski P, Leija A, Cevallos M, Elmerich C, Mora J. Poly-β-hydroxybutyrate turnover in Azorizobium caulinodans is required for growth and affects nifAexpression. J Bacteriol. 1998;180:5070–5076. doi: 10.1128/jb.180.19.5070-5076.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meinhardt S W, Kiley P J, Kaplan S, Crofts A R, Harayama S. Characterization of light-harvesting mutants of Rhodopseudomonas sphaeroides. I. Measurement of the efficiency of light energy transfer from light-harvesting complexes to the reaction center. Arch Biochem Biophys. 1985;236:130–139. doi: 10.1016/0003-9861(85)90612-5. [DOI] [PubMed] [Google Scholar]
- 34.Moskowitz G, Merrick J. Metabolism of poly-β-hydroxybutyrate. II. Enzymatic synthesis of D-(−)-β-hydroxybutyryl coenzyme A by an enoyl hydrase from Rhodospirillum rubrum. Biochemistry. 1969;8:2748–2755. doi: 10.1021/bi00835a009. [DOI] [PubMed] [Google Scholar]
- 35.Mouncey N J, Kaplan S. Oxygen regulation of the ccoN gene encoding a component of the cbb3 oxidase in Rhodobacter sphaeroides 2.4.1T: involvement of the FnrL protein. J Bacteriol. 1998;180:2228–2231. doi: 10.1128/jb.180.8.2228-2231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neidle E L, Kaplan S. 5-aminolevulinic acid availability and control of spectral complex formation in HemA and HemT mutants of Rhodobacter sphaeroides. J Bacteriol. 1993;175:2304–2313. doi: 10.1128/jb.175.8.2304-2313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neidle E L, Kaplan S. Expression of the Rhodobacter sphaeroides hemA and hemTgenes, encoding two 5-aminolevulinic acid synthase isozymes. J Bacteriol. 1993;175:2292–2303. doi: 10.1128/jb.175.8.2292-2303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Gara J, Eraso J, Kaplan S. A redox-responsive pathway for aerobic regulation of photosynthesis gene expression in Rhodobacter sphaeroides2.4.1. J Bacteriol. 1998;180:4044–4050. doi: 10.1128/jb.180.16.4044-4050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh J-I, Eraso J, Kaplan S. Interacting regulatory circuits involved in orderly control of photosynthesis gene expression in Rhodobacter sphaeroides2.4.1. J Bacteriol. 2000;182:3081–3087. doi: 10.1128/jb.182.11.3081-3087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oh J-I, Kaplan S. Redox signaling: globalization of gene expression. EMBO J. 2000;19:4237–4247. doi: 10.1093/emboj/19.16.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perret X, Staehelin C, Broughton W. Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev. 2000;64:180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reiser S, Mitsky T, Gruys K. Characterization and cloning of an (R)-specific trans-2,3-enoylacyl-CoA hydratase from Rhodospirillum rubrum and use of this enzyme for PHA production in Escherichia coli. Appl Microbiol Biotechnol. 2000;53:209–218. doi: 10.1007/s002530050010. [DOI] [PubMed] [Google Scholar]
- 43.Roth J, Lawrence J, Bobik T. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45.Sistrom W R. A requirement for sodium in the growth of Rhodopseudomonas sphaeroides. J Gen Microbiol. 1960;22:778–785. doi: 10.1099/00221287-22-3-778. [DOI] [PubMed] [Google Scholar]
- 46.Steinbuchel A, Hustede E, Liebergesell M, Pieper U, Timm A, Valentin H. Molecular basis for biosynthesis and accumulation of polyhydroxyalkanoic acids in bacteria. FEMS Microbiol Rev. 1992;9:217–230. doi: 10.1111/j.1574-6968.1992.tb05841.x. [DOI] [PubMed] [Google Scholar]
- 47.Tai T-N, Havelka W A, Kaplan S. A broad-host-range vector system for cloning and translational lacZfusion analysis. Plasmid. 1988;19:175–188. doi: 10.1016/0147-619x(88)90037-6. [DOI] [PubMed] [Google Scholar]
- 48.Tai T-N, Moore M D, Kaplan S. Cloning and characterization of the 5-aminolevulinate synthase gene(s) from Rhodobacter sphaeroides. Gene. 1988;70:139–151. doi: 10.1016/0378-1119(88)90112-6. [DOI] [PubMed] [Google Scholar]
- 49.Weidenhaupt M, Rossi P, Beck C, Fischer H-M, Hennecke H. Bradyrhizobium japonicum possesses two discrete sets of electron transfer flavoprotein genes: fixA, fixB and etfS, etfL. Arch Microbiol. 1996;165:169–178. doi: 10.1007/BF01692858. [DOI] [PubMed] [Google Scholar]
- 50.Yabutani T, Maehara A, Ueda S, Yamane T. Analysis of beta-ketothiolase and acetoacetyl-CoA reductase genes of a methylotrophic bacterium, Paracoccus denitrificans, and their expression in Escherichia coli. FEMS Microbiol Lett. 1995;133:85–90. doi: 10.1111/j.1574-6968.1995.tb07865.x. [DOI] [PubMed] [Google Scholar]
- 51.Zeilstra-Ryalls J, Gomelsky M, Eraso J, Yeliseev A, O'Gara J, Kaplan S. Control of photosystem formation in Rhodobacter sphaeroides. J Bacteriol. 1998;180:2801–2809. doi: 10.1128/jb.180.11.2801-2809.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeilstra-Ryalls J H, Kaplan S. Aerobic and anaerobic regulation in Rhodobacter sphaeroides 2.4.1: the role of the fnrLgene. J Bacteriol. 1995;177:6422–6431. doi: 10.1128/jb.177.22.6422-6431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeilstra-Ryalls J H, Kaplan S. Control of hemA expression in Rhodobacter sphaeroides2.4.1: regulation through alterations in cellular redox state. J Bacteriol. 1996;178:985–993. doi: 10.1128/jb.178.4.985-993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeilstra-Ryalls J H, Kaplan S. Regulation of 5-aminolevulinic acid synthesis in Rhodobacter sphaeroides2.4.1: the genetic basis of mutant H-5 auxotrophy. J Bacteriol. 1995;177:2760–2768. doi: 10.1128/jb.177.10.2760-2768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]