Abstract
Salmonella enterica degrades 1,2-propanediol by a pathway dependent on coenzyme B12 (adenosylcobalamin [AdoCb1]). Previous studies showed that 1,2-propanediol utilization (pdu) genes include those for the conversion of inactive cobalamins, such as vitamin B12, to AdoCbl. However, the specific genes involved were not identified. Here we show that the pduO gene encodes a protein with ATP:cob(I)alamin adenosyltransferase activity. The main role of this protein is apparently the conversion of inactive cobalamins to AdoCbl for 1,2-propanediol degradation. Genetic tests showed that the function of the pduO gene was partially replaced by the cobA gene (a known ATP:corrinoid adenosyltransferase) but that optimal growth of S. enterica on 1,2-propanediol required a functional pduO gene. Growth studies showed that cobA pduO double mutants were unable to grow on 1,2-propanediol minimal medium supplemented with vitamin B12 but were capable of growth on similar medium supplemented with AdoCbl. The pduO gene was cloned into a T7 expression vector. The PduO protein was overexpressed, partially purified, and, using an improved assay procedure, shown to have cob(I)alamin adenosyltransferase activity. Analysis of the genomic context of genes encoding PduO and related proteins indicated that particular adenosyltransferases tend to be specialized for particular AdoCbl-dependent enzymes or for the de novo synthesis of AdoCbl. Such analyses also indicated that PduO is a bifunctional enzyme. The possibility that genes of unknown function proximal to adenosyltransferase homologues represent previously unidentified AdoCbl-dependent enzymes is discussed.
Salmonella enterica catabolizes 1,2-propanediol via a pathway that is dependent upon adenosyl cobalamin (AdoCbl), a metabolically active form of vitamin B12 (18). Since 1,2-propanediol is formed by the fermentation of the common plant sugars rhamnose and fucose, its catabolism may provide a selective advantage in anaerobic environments (20, 25). Studies employing in vivo expression technology and competitive index assays have suggested that 1,2-propanediol degradation may also provide a growth advantage in host tissues (10, 15). A number of these aspects of Salmonella biology have been reviewed recently (27).
The genes for 1,2-propanediol utilization (pdu) are found at centisome 44 of the S. enterica genetic map (18). They are adjacent to and coregulated with 20 genes for de novo AdoCbl synthesis (1, 5, 26, 28). These genes are absent from Escherichia coli, and evidence indicates they were acquired by S. enterica via a single horizontal gene transfer (6, 28). Surprisingly, there are 23 pdu genes (6). Six of these probably encode enzymes of the degradative pathway, and four are thought to be involved in regulation, transport, and diol dehydratase reactivation (5, 6, 9). Perhaps five to seven pdu genes are needed for formation of a polyhedral body that is associated with the diol dehydratase, and the remaining six are of unknown function (6).
The proposed pathway of 1,2-propanediol degradation begins with its conversion to propionaldehyde by AdoCbl-dependent diol dehydratase (25, 33). Next, the aldehyde is disproportionated to either propanol or propionic acid. Alcohol dehydrogenase, aldehyde dehydrogenase, phosphotransacylase, and propionate kinase are thought to catalyze this process. The pathway yields ATP, an electron sink, and an intermediate (propionyl-coenzyme A [CoA]), which can feed into central metabolism via the methyl-citrate pathway (16).
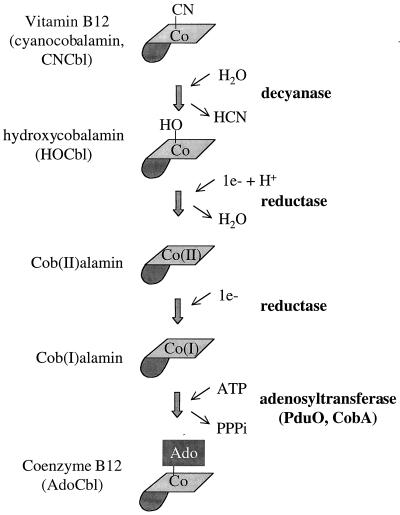
The AdoCbl needed for 1,2-propanediol degradation can be obtained either by de novo synthesis or by the assimilation of an exogenous cobalamin (Cbl). In S. enterica, de novo synthesis occurs only under strictly anaerobic conditions (19). However, Cbls, such as cyanocobalamin (vitamin B12 [CNCb]) and hydroxycobalamin (HOCbl), can be assimilated both aerobically and anaerobically (18). The conversion of CNCbl and HOCbl to AdoCbl is thought to proceed as shown in Fig. 1: CNCbl is decyanated to HOCbl, reduced to cob(II)alamin, further reduced to cob(I)alamin, and finally adenosylated to AdoCbl (14, 17). The cobA gene (which maps far from the pdu/cob locus at centisome 34) encodes an ATP:corrinoid adenosyltransferase (32). This enzyme participates in Cbl assimilation by adenosylation of cob(I)alamin and also in de novo AdoCbl synthesis by the adenosylation of an intermediate prior to Cbl (13). Genetic tests have indicated that Cbl assimilation genes are also found in the pdu operon, but the specific genes involved have not been identified (37; T. A. Bobik, unpublished results).
FIG. 1.
Proposed vitamin B12 adenosylation pathway. Coenzyme B12 (a required cofactor for a number of enzymes) is thought to be produced from vitamin B12 by the series of reactions shown. The corrin ring of vitamin B12 is shown lightly shaded, the nucleotide loop is darkly shaded, and the upper ligand of the cobalt is indicated as follows: CN, cyano group; HO, hydroxy group; Ado, 5′-deoxyadenosyl group.
Here we report that the pduO gene encodes an ATP:cob(I)alamin adenosyltransferase used in Cbl assimilation for 1,2-propanediol degradation by S. enterica. We also present the results of a functional genomic analysis of adenosyltransferases that indicates the following: (i) particular adenosyltransferases tend to be specialized for particular AdoCbl-dependent enzymes, (ii) the PduO adenosyltransferase is a bifunctional enzyme, and (iii) unknown genes proximal to adenosyltransferase homologues may represent previously unidentified AdoCbl-dependent enzymes.
MATERIALS AND METHODS
Chemical and reagents.
Titanium(III) citrate was prepared as previously described (7). Vitamin B12, coenzyme B12, and HOCbl were from Sigma Chemical Company, St. Louis, Mo. Isopropyl-β-d-thiogalactopyranoside (IPTG), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were from Diagnostic Chemicals Limited, Charlottetown, Canada. Restriction enzymes and T4 DNA ligase were from New England Biolabs, Beverly, Mass. Other chemicals were from Fisher Scientific, Norcross, Ga.
Bacterial strains, media, and growth conditions.
The bacterial strains used in this study are listed in Table 1. The minimal medium used was NCE (4, 36) supplemented with 0.4% 1,2-propanediol, 1 mM MgSO4, and 3 mM (each) valine, isoleucine, leucine, and threonine. LB (Luria-Bertani) medium was the rich medium used (22). Ampicillin was used at 100 μg/ml unless otherwise indicated. Kanamycin was used at 25 μg/ml, IPTG at was used at 1 mM or as indicated, and X-Gal was used at 20 μg/ml. MacConkey–1,2-propanediol–CNCbl indicator plates were composed of MacConkey agar base (Difco, Detroit, Mich.) supplemented with 1% 1,2-propanediol and 200 ng of vitamin B12/ml. Aldehyde indicator plates were prepared according to the method of Conway et al. (11) with the following modifications: pararosaniline was added to sterile medium as a fine powder, 200 ng of CNCbl/μl was included, and ethanol was replaced by 1% 1,2-propanediol.
TABLE 1.
Bacterial strains
| Species and strain | Genotype |
|---|---|
| E. coli | |
| DH5α | F−λ−endA1 hsdR17 relA1 supE44 thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 (φ80dlacZΔM15) |
| BL21DE3 RIL | BF−ompT hsdS (rB− mB−) dcm+ Tetrgal λ(DE3) endA Hte[argU ileY leuW Camr] |
| S17.1 λ pir | recA (RP4-2-Tc::Mu)λ pir |
| BE11 | (E. coli ER2267) F− e14− (McrA−) endA1 supE44 thi-1 relA? rfbD1? spoT? Δ(mcrC-mrr)114::IS10 Δ(argF-lac)U169 recA1/F′ ProA+B+lacIq Δ(lacZ)M15 zzf::mini-Tn10(Kanr)/pMGS2 |
| BE115 | DH5α/pTA925 |
| BE116-BE118 | BL21DE3 RIL/pTA926 (T7 expression vector with pduO insert) |
| BE119 | BL21DE3 RIL/pTA925 (T7 expression vector without insert) |
| S. enterica serovar Typhimurium LT2 | |
| TR6579 | metA22 metE551 trpD2 ilv-452 hsdLT6 hsdSA29 HsdB−strA120 GalE− Leu− Pro− |
| BE47 | thr-480::Tn10dCam |
| BE83 | cobA366::Tn10dCam |
| BE111 | ΔpduO651 |
| BE112 | metE205 ara-9 Δ299/pDP1 (entire cob and pdu operons are deleted, and plasmid contains pduO under plac control) |
| BE113 | metE205 ara-9 Δ299/placIqPOBglII |
| BE114 | ΔpduO651 cobA366::Tn10dCam/pDP1 (pduO under lacp control) |
| BE121 | ΔpduO651 cobA366::Tn10dCam |
| BE122 | LT2/pDP1 (pduO under plac control) |
| BE129 | ΔpduO651 cobA366::Tn10dCam/placIqPOBglII |
General molecular methods.
Agarose gel electrophoresis was performed as described previously (21). Plasmid DNA was purified by the alkaline lysis procedure (21) or by using Qiagen (Chatsworth, Calif.) products according to the manufacturer's directions. Following restriction enzyme digestion or PCR amplification, DNA was purified with Qiagen PCR purification and gel extraction kits or by using phenol-chloroform extraction followed by ethanol precipitation (21). Restriction digestions were carried out using standard protocols (21). For ligation of DNA fragments, T4 DNA ligase (New England Biolabs) was used according to the manufacturer's directions. Electroporation was used for bacterial transformation. A Gene Pulser (Bio-Rad, Richmond, Calif.) was used according to the manufacturer's directions and at the following settings: capacitance, 25 μF; capacitance extender, 250 μF; pulse controller, 200 Ω; and voltage, 2.5 kV. LB medium containing the appropriate antibiotic(s) was used to select for transformed cells, and prior to the analysis of transformants, pure cultures were prepared.
General protein methods.
Polyacrylamide gel electrophoresis (PAGE) was performed using Bio-Rad Redigels and Mini-protean II electrophoresis cells. PAGE was run at 200 V for 45 min using a Bio-Rad Power Pac 300. Following gel electrophoresis, Coomassie brilliant blue R-250 was used to stain proteins. The protein concentrations of solutions were determined using Bio-Rad protein assay reagent according to the manufacturer's directions.
P22 transduction.
Transductional crosses were performed as described previously (12) using P22 HT105/1 int-210, a mutant phage that has high transducing ability (29). For the preparation of P22 transducing lysates from strains having galE mutations, overnight cultures were grown on LB medium supplemented with 0.2% glucose and 0.2% galactose.
Cloning of the pduO gene for complementation studies.
Vector placIqPO-BglII was used for cloning the pduO gene so that its expression could be induced by IPTG (8). The DNA used for cloning the pduO gene was obtained via PCR amplification of plasmid pMGS2 using the primers GGAATTCAGATCTTATGGCGATTTATACCCGAAC and GGAATTCAAGCTTGGTTTCGAGTTCAGAAGTATTC. Pfu DNA polymerase was employed for the amplification because of its high fidelity of replication. The amplified DNA was purified and ligated to placIqPO-BglII that had been digested with the same restriction enzymes and purified. The ligation mixture was used to transform S. enterica TR6579 by electroporation. Five of six transformants carried plasmids containing inserts of the expected size. Of these, four of five had identical DNA sequences. The majority consensus sequence was compared to the pduO DNA sequence we previously reported (6). There were three differences; however, reexamination of our prior data revealed errors in our previously published DNA sequence. Clones with a pduO gene having the majority consensus sequence were used for further study.
Construction of in-frame pduO deletions.
PCR primers were designed to delete bases 19 to 981 of the pduO coding sequence. The first 18 and last 27 bases remained intact, as did all predicted translational start and stop sites of pdu genes. To improve fidelity, the PCRs employed Pfu polymerase and a high concentration of template (1 ng of pMGS2/μl) and were limited to 30 cycles. The following four primers were used to generate the deletions: primer 1, GCGCGCTCTAGATATTCACCGATGAGCACGGACTGC; primer 2, GATGAGTTCCCACGTTAATAGCCGCTCGGGTATAAATCGCCATAACCG; primer 3, GCGGCTATTAACGTGGGAACTCATC; and primer 4, GGAATTCAGGCTAATCAGCTTCAGAGAGACC. Primers 1 and 2 were used to amplify 480 bases of DNA upstream of the pduO gene. Primers 3 and 4 were used to amplify 430 bp of DNA downstream of the pduO gene. The upstream and downstream amplification products were purified and fused by a PCR that included 1 ng of each product/μl and primers 1 and 4. Fusion was possible because the 5′ end of primer 2 was the reverse complement of primer 3. After the fusion product was obtained, it was restricted with XbaI and SphI (these sites were designed into primers 1 and 4) and ligated to pCVD442. The ligation reaction was used to transform E. coli S17/1 via electroporation. One clone (plasmid pAP6) containing an insert of the expected size (910 bp) was used to introduce the deletion into the S. enterica chromosome using the procedure of Miller and Mekalanos (23). For the conjugation step, BE47 was used as the recipient and Ampr and Camr were selected. Following the sucrose selection step, replica printing was used to identify Amps colonies. Deletion strains were identified by PCR using whole cells as a source of template.
Cloning the pduO gene for high-level expression.
For use in high-level expression of the PduO protein, a DNA linker was used to modify the T7 expression vector pET41a (Novagen, Milwaukee, Wis.). The DNA linker was prepared by annealing two oligonucleotides, CTAGAATGCATAAATTTTGTTAACTTAAGAAGGAAGATCTCA and TATGAGATCTTCCTTCTTAAGTTAACAAAATTTATGCATT. A 1-ml solution of 50 μM (each) oligonucleotide, 100 mM Tris · HCl (pH 8), and 10 mM MgCl2 was placed in a 1.5-ml microcentrifuge tube, heated in a boiling water bath for 5 min, removed to the bench top, and allowed to cool to room temperature. Plasmid pET41a was restricted with XbaI and NdeI, purified, and ligated to the DNA linker described above. A portion of the ligation reaction mixture was used to transform E. coli DH5α. Restriction and DNA sequence analyses of selected clones verified that the ligation reaction produced the expected product. The pET41a derivative containing the DNA linker described above was named pTA925.
The pduO coding sequence was subcloned from plasmid pDP1 to the T7 expression plasmid pTA925 using BglII and HindIII restriction sites. E. coli DH5α was used as the host. Following transformation, five of six isolates contained plasmids that released the expected 1,046-bp fragment when restricted with BglII and HindIII. DNA from one such plasmid was used to transform the expression strain E. coli BL21 DE3 RIL (Stratagene, La Jolla, Calif.). Three isolates obtained from this transformation (BE116 to BE118) were used for high-level expression of the PduO protein.
Growth of PduO expression strains and preparation of cell extracts.
PduO expression strains were grown in 50-ml cultures prepared in 250-ml baffled Erlenmeyer flasks. The medium used was LB medium supplemented with 25 μg of kanamycin/ml, and the cultures were incubated at 37°C with shaking at 275 rpm. The cells were grown to an optical density of 0.6 to 0.8 at 600 nm. At that time, expression of the PduO protein was induced by the addition of IPTG to a final concentration of 1 mM. The cells were incubated at 37°C with shaking at 275 rpm for an additional 3 h. The cultures were removed from the shaker, placed on ice for 5 min, and collected by centrifugation for 5 min at 7,740 × g (maximum) using a Beckman (Fullerton, Calif.) J2-HS centrifuge and a Beckman JA20 rotor. The cells were then resuspended in 40 ml of ice-cold 20 mM Tris · HCl and again collected by centrifugation as described above. The cell pellet was frozen at −80°C. Bacterial Protein Extraction Reagent II (B-PERII; Pierce, Rockford, Ill.) was used to prepare extracts of soluble proteins and inclusion bodies from cell pellets that had been stored one to several days at −80°C. B-PERII was used according to the “midi-prep” sample protocol provided by the supplier with the following modifications. The B-PERII solution used for the first extraction was supplemented with the protease inhibitor phenylmethylsulfonylfluoride at a concentration of 100 μg/ml. The B-PERII solution used for the second extraction was supplemented with DNase at a concentration of 20 μg/ml. Inclusion bodies were washed twice with 10 ml of a 1-to-20 dilution of B-PERII and then resuspended in 20 mM Tris · HCl (pH 8.0).
Growth curves.
Cells were grown in 125-ml baffled Erlenmeyer flasks containing 10 ml of the appropriate medium. The cultures were incubated at 37°C in a New Brunswick model C-24 water bath with the shaking speed set to 7. Cell growth was determined by measuring the optical density at 600 nm using a Beckman model DU640 spectrophotometer. The inoculum for growth curves was obtained as follows: bacterial strains were grown overnight at 37°C with shaking in LB medium or LB medium supplemented with 100 μg of ampicillin/ml for strains that carried plasmids; cells from 1.5 ml of overnight culture were pelleted by centrifugation and resuspended in 1 ml of growth curve medium, and 0.25 ml was used to inoculate 10-ml cultures.
ATP:cob(I)alamin adenosyltransferase assays.
Adenosyltransferase assays were carried out using a modification of a previously published protocol (35). The assay mixtures were incubated at 37°C under strict anaerobic conditions in 1-cm-wide glass cuvettes modified for sealing with 13-mm-diameter gray butyl rubber stoppers and aluminum crimp seals. The total assay volume was 2 ml, and the assay mixtures contained the following components: 200 mM Tris · HCl (pH 8), 0.4 mM ATP, 1.6 mM KH2PO4, 2.8 mM MgCl2, 0.05 mM HOCbl, and 1 mM titanium(III) citrate. The assay components [except for titanium(III) citrate and the component used to initiate the reaction] were combined within an anaerobic chamber (Coy Laboratory Products, Grass Lake, Mich.), dispensed into cuvettes, sealed, removed from the chamber, and flushed with N2 for 30 s. The cuvettes were placed in a 37°C water bath for 5 min, and then 20 μl of Ti(III) citrate was added from a 100-mM stock solution. To allow reduction of the HOCb1 to cob(I)alamin, the reaction mixtures were kept at 37°C for an additional 3 to 5 min. The reactions were initiated by adding a source of enzyme or a particular assay component using the following procedures to minimize the introduction of oxygen. The assay component to be added was placed within a sealed serum vial and flushed with N2 for 2 min. Additions were made using a Hamilton syringe that had been flushed with anoxic H2O just prior to use. Two methods were used to quantitate the AdoCbl formed: (i) the decrease in absorbance at 388 nm was followed, and the equation Δɛ388 = 24.9 cm−1 mM−1 was used for calculations; (ii) the AdoCbl formed was photolyzed by exposure of the assay mixtures to a 100-W incandescent light at a distance of 15 cm for 20 min, and then the decrease in absorbance at 525 nm was measured; the equation Δɛ525 = 4.9 cm−1 mM−1 was used for calculations. Prior methods used the equation Δɛ525 = 4.8 mM−1 Cm−1 for calculations following photolysis (35). However, those assays employed borohydride as a reductant, and cob(II)alamin was the product of photolysis. When titanium(III) citrate is used as described here, the cob(II)alamin formed by photolysis is reduced to cob(I)alamin. Accordingly, the Δɛ value used for calculation reflects the difference in A525 between AdoCbl and cob(I)alamin. One unit of ATP:cob(I)alamin adenosyltransferase activity was defined as 1 nmol of AdoCbl formed per min per mg of protein.
DNA sequencing and analysis.
DNA sequencing was carried out by the University of Florida Interdisciplinary Center for Biotechnology Research DNA Sequencing Core Facility using Applied Biosystems Inc. automated sequencing equipment (Perkin-Elmer, Norwalk, Conn.). The template for DNA sequencing was plasmid DNA purified using Qiagen tip 100 columns. BlastP and Ψ-Blast software were used to search the nonredundant database of the National Center for Biotechnology Information for homologous protein sequences (2, 3).
Electron microscopy.
For electron microscopy, cells were grown on minimal succinate (1%) medium supplemented with 0.4% 1,2-propanediol. Cultures (100 ml) were incubated at 37°C with shaking at 275 rpm. The inoculum was 1 ml of an LB overnight culture. Fixation and staining were performed as previously described (6).
Nucluotide sequence accession number.
The pduO sequence determined here has been submitted to GenBank for the update of accession number AF026270.
RESULTS
The cobA and pduO genes have similar functions.
To examine the effects of cobA and pduO null mutations on 1,2-propanediol degradation, two qualitative tests were employed. MacConkey–1,2-propanediol medium was used to detect acids produced from the degradation of 1,2-propanediol, and aldehyde indicator medium was used to detect the production of propionaldehyde by AdoCbl-dependent diol dehydratase. For these experiments, both indicator media were supplemented with CNCbl. Thus, acid and aldehyde production relied not only on enzymes of the 1,2-propanediol degradative pathway but also on enzymes that convert CNCbl to AdoCbl. Strains BE111 (pduO) and BE83 (cobA) produced acid and aldehyde at levels similar to those of the wild-type strain; colonies were bright red on MacConkey indicator medium and dark red- brown on aldehyde indicator medium. However, under similar conditions, acid and aldehyde production by BE121 (pduO cobA), were undetectable. Except for the mutations under study, the strains used were isogenic, and two different pduO deletion mutations (in conjunction with a well-characterized cobA mutation) gave similar results in analogous tests.
Given that the pduO and cobA single mutants were essentially wild type in these tests but the pduO cobA double mutant was unable to degrade 1,2-propanediol, we infer that the cobA and pduO genes have similar functions. The cobA gene was previously shown to encode an ATP:corrinoid adenosyltransferase that functions in the assimilation of CNCbl to AdoCbl (13). Hence, the pduO gene is likely to have a similar function.
Complementation of the ΔpduO651 mutation by plasmid pDP1.
P22 transduction was used to move an expression plasmid containing a pduO minimal clone (pDP1) from strain BE112 to strain BE121 (cobA pduO). Transduction mixtures were plated on aldehyde indicator medium supplemented with 1,2-propanediol, CNCbl, and ampicillin. Fifty-six transductant colonies were observed, and all were red-brown. This indicated complementation of the ΔpduO651 mutation by the pduO minimal clone carried on plasmid pDP1. As a negative control, a similar experiment was carried out using the vector lacking the pduO insert. In this case, complementation was not observed; all 110 transductant colonies were white. This confirmed that the loss of aldehyde production in the double mutant was the consequence of the pduO mutation and not a consequence of another mutation inadvertently introduced during strain construction.
Effects of cobA and pduO null mutations on the growth of S. enterica on minimal 1,2-propanediol medium.
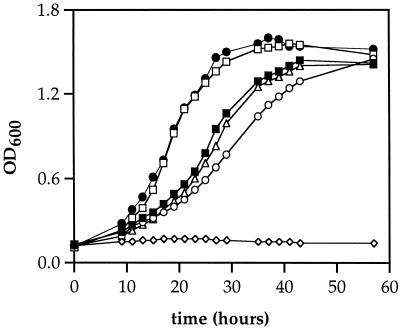
The effects of cobA and pduO null mutations on the growth of S. enterica on minimal 1,2-propanediol–CNCbl medium were examined (Fig. 2). The wild-type strain and a cobA mutant strain (BE83) both grew with generation times of about 8.4 h, and both reached a maximum optical density at 600 nm of about 1.4. The pduO mutant (BE111) showed a small growth impairment. Its generation time was 12.2 h, and it reached a maximum density of 1.45. However, BE121 (pduO cobA) was unable to grow on 1,2-propanediol minimal medium supplemented with CNCbl. These findings provided further evidence that the pduO and cobA genes have similar functions. The growth curves were repeated three times with similar results except that the lag times varied by several hours.
FIG. 2.
Effects of pduO and cobA mutations on growth of S. enterica on 1,2-propanediol–CNCbl minimal medium. ■, Wild type (S. enterica serovar Typhimurium LT2); ▵, cobA; ○, pduO; ◊, pduO cobA; □, pduO cobA/pDP1; ●, LT2/pDP1. The strains used were BE83, BE111, BE121, and BE114. The cells were cultured as described in Materials and Methods. OD600, optical density at 600 nm.
The growth of strain BE114 (pduO cobA/pDP1) on 1,2-propanediol–CNCbl minimal medium was also measured. This strain grew faster than the wild-type strain (Fig. 2). Its generation time was 5.8 h compared to 8.4 h for the wild-type strain. However, strain BE114 grew similarly to the wild-type when glucose, succinate, propionate, or acetate was used in place of 1,2-propanediol (data not shown). These results confirm the results of the complementation tests described above and, somewhat surprisingly, also indicate that pduO expression is limiting for growth on 1,2-propanediol minimal medium. Strain BE122 (wild type/pDP1) also grew faster than the wild-type strain on 1,2-propanediol minimal medium but grew similarly to the wild type on glucose, succinate, propionate, or acetate minimal medium. This is further evidence that pduO expression is limiting for growth on 1,2-propanediol minimal medium. The growth tests were repeated several times with essentially similar results. For the experiments shown, the media were supplemented with 0.02 mM IPTG. As an additional control, similar experiments were performed using the expression vector placIqPO-BglII without the pduO insert. This control plasmid had no discernible effect on the growth of a pduO cobA double mutant or the wild-type strain on 1,2-propanediol–CNCbl minimal medium.
AdoCbl supplementation partially corrects the phenotype of cobA pduO double mutants.
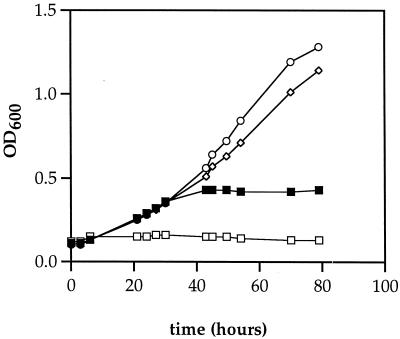
Growth of the wild-type strain was similar with either AdoCbl or CNCbl at concentrations between 0.2 and 20 μM. Generation times were about 8.5 h, and final cell densities reached about 1.5 at 600 nm. In contrast, strain BE121 (pduO cobA) was unable to grow with CNCbl at concentrations between 0.2 and 20 μM but was capable of growth with AdoCbl (Fig. 3). At a concentration of 20 μM AdoCbl, BE121 had a generation time of 23 h, about three times longer than that of the wild-type strain. Thus, AdoCbl supported significant growth of the pduO cobA double mutant. This indicates that the PduO protein is involved in the conversion of CNCbl to AdoCbl, providing further evidence that it has ATP:corrinoid adenosyltransferase activity. The reason AdoCbl did not restore growth of the pduO cobA mutant to the full wild-type level is likely the instability of AdoCbl (30). During catalysis, AdoCbl breaks down to Cbls with one of several upper ligands (XCbl). XCbls are both substrates for adenosylation and potent inhibitors of diol dehydratase. Since, the pduO cobA double mutant is incapable of the adenosylation of XCbl, these inhibitors are likely to accumulate and slow growth.
FIG. 3.
Growth of S. enterica strain BE121 (pduO cobA) on 1,2-propanediol minimal medium supplemented with either AdoCbl or CNCbl. □, 2 μg of CNCbl/ml; ■, 0.2 g of AdoCbl/ml; ◊, 2 μg of AdoCbl/ml; ○, 20 μg of AdoCbl/ml. Strain BE121 did not grow with CNCbl at concentrations from 0.2 to 20 μg/ml (not shown). The cells were cultured as described in Materials and Methods. OD600, optical density at 600 nm.
High-level expression of the PduO protein.
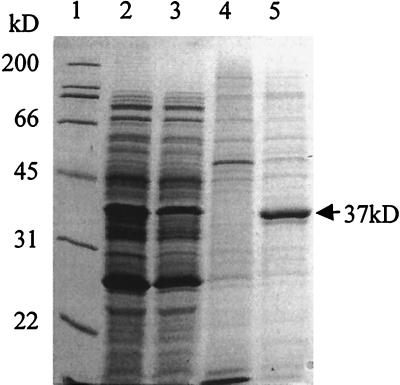
Protein expression from three E. coli strains (BE116 to BE118) constructed to produce high levels of the PduO protein via a T7 expression system was analyzed by sodium dodecyl sulfate (SDS)-PAGE. With or without induction by IPTG, all three strains produced large amounts of a protein with a molecular mass of approximately 37 kDa (data not shown); this is close to the predicted mass of the protein encoded by the entire pduO coding sequence (36.6 kDa). To confirm that the observed 37-kDa protein was expressed from the pduO coding sequence, protein expression by E. coli strains BE118 (pduO insert) and BE119 (no insert) was analyzed. Extracts of both soluble proteins and inclusion bodies were prepared and analyzed by SDS-PAGE (Fig. 4). Large amounts of a 37-kDa protein were found only in the inclusion bodies from cells containing the pduO T7 expression plasmid (Fig. 4, lane 5). Cells containing the T7 expression plasmid without an insert did not express detectable amounts of a 37-kDa protein (lanes 1 and 4). This indicated that the observed 37-kDa protein expressed by E. coli BE118 was the PduO protein.
FIG. 4.
SDS-PAGE analysis of pduO expression strains. Lane 1, molecular mass markers; lanes 2 and 3, soluble extracts of strain BE119 (T7 expression vector without insert) and strain BE118 (T7 expression vector with pduO insert), respectively; lanes 4 and 5, inclusion body extracts from strains BE119 and BE118, respectively. The 37-kDa protein band is indicated.
The PduO protein has ATP:cob(I)alamin adenosyltransferase activity.
The four extracts used for SDS-PAGE (Fig. 4) were also assayed for ATP:cob(I)alamin adenosyltransferase activity. Inclusion bodies from BE118 had very high specific activity (312 nmol/min/mg of protein), but the other cell extracts tested had no detectable activity. This indicated that the PduO protein has ATP:cob(I)alamin adenosyltransferase activity. ATP, HOCbl, titanium(III) citrate, and enzyme were all required for adenosyltransfer, indicating that ATP and cob(I)alamin were substrates for the reaction. The inclusion body preparations used for these assays were active without solubilization.
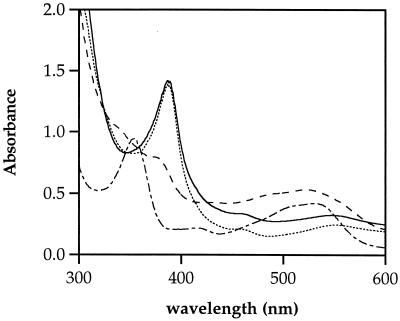
UV-visible (Vis) spectroscopy was used to follow the major corrinoids present during the course of adenosyltransferase assays (Fig. 5). The spectrum of the assay mixture prior to the addition of titanium(III) citrate and enzyme indicated that HOCbl was the major corrinoid present. The addition of titanium(III) citrate resulted in the quantitative reduction of HOCbl to cob(I)alamin. After a source of PduO protein (inclusion body extracts) was added to the reaction mixture, the UV-Vis spectrum changed over time to that of AdoCbl. The reaction mixtures were then exposed to a 100-W incandescent light at a distance of 15 cm for 20 min to photolyze the carbon-cobalt bond of AdoCbl. The UV-Vis spectrum taken after exposure to light was that of cob(I)alamin, indicating photolysis had occurred: the expected product of photolysis is cob(II)alamin, but in the presence of titanium(III) citrate, the reduction of cob(II)alamin to cob(I)alamin is expected. These results indicated that cob(I)alamin was a reaction substrate and that AdoCbl was a reaction product
FIG. 5.
Absorption spectra of adenosyltransferase assay mixtures. The spectra indicate the predominant corrinoid present in the assay mixture: prior to the addition of titanium(III) citrate, the spectrum is that of HOCbl (–·–·); after the addition of titanium(III) citrate, the spectrum is that of cob(I)alamin (…………); 10 min after the addition of an inclusion body preparation of the PduO enzyme, the spectrum is that of AdoCbl (–––); after photolysis, the spectrum is that of cob(I)alamin (——).
An improved ATP:cob(I)alamin adenosyltransferase assay.
In this study, adenosyltransferase activity was quantitated by determining the disappearance of cob(I)alamin by continuous measurement of the A388. The use of titanium(III) citrate as a reductant made this method possible. Previously reported adenosyltransferase assays employed borohydride for the reduction of HOCbl to cob(I)alamin (35). During such assays, oxidation of cob(I)alamin to cob(II)alamin made quantitation based on the disappearance of cob(I)alamin inaccurate (35). Therefore, it was necessary to photolyze AdoCbl to cob(II)alamin and to base quantitation on the difference in A525 between AdoCbl and cob(II)alamin. Moreover, we found that the use of borohydride in adenosyltransferase assays resulted in the formation of gas bubbles that interfered with continuous spectrophotometric measurements. When titanium(III) citrate was used as a reductant, no gas bubbles were formed and no oxidation of cob(I)alamin to cob(II)alamin was observed even after several hours of incubation at 37°C. To test the accuracy of continuous measurement at 388 nm, this method was compared to the previously published method; the two assays gave similar results (data not shown). Additional tests showed that the continuous assay was reproducible and that photolysis of AdoCbl by light from the spectrophotometer beam was insignificant under the conditions used (data not shown). The continuous method is faster and about five times more sensitive than the previously reported method (Δɛ388 = 24.9 cm−1 mM−1).
Functional genomic analysis of the pduO gene and its homologues.
Database searches were used to analyze genes that cluster with the pduO gene and its homologues (Table 2). Among proteins related to the N-terminal portion of the PduO protein, the genes encoding 3 of 12 cluster with genes encoding enzymes known to use AdoCbl as a cofactor. ORFW of Citrobacter freundii and ORF2c of Klebsiella pneumoniae are found associated with genes for AdoCbl-dependent glycerol dehydratases. ORF AF1290 of Archaeoglobus fulgidus is adjacent to sequences encoding an AdoCbl-dependent methylmalonyl CoA mutase homologue. Among the seven amino acid sequences related to the C-terminal portion of the PduO protein, the genes encoding two are found clustered with genes for AdoCbl-dependent glycerol dehydratases. These are ORFY of C. freundii and ORF2a of K. pneumoniae. The organization of pduO and related genes with genes encoding Ado-Cbl-dependent enzymes is consistent with a role in Cbl assimilation and has some further implications that are addressed in Discussion below.
TABLE 2.
Instances in which genes encoding proteins with homology to PduO are proximal to genes for AdoCbl-dependent enzymes
| Organism | Gene | Accession no. and GI | Region of homology to PduO | Degree of amino acid homology (identity/length) (% identity) | Protein encoded by nearby genes |
|---|---|---|---|---|---|
| S. enterica | pduO | AF026270 5069458 | Entire length | 336/336 (100) | AdoCbl-dependent diol dehydratase |
| K. pneumoniae | ORF2C | U30903 940442 | N terminal | 73/167 (43) | AdoCbl-dependent glycerol dehydratase |
| C. freundii | ORFW | U09771 1175767 | N terminal | 69/167 (41) | AdoCbl-dependent glycerol dehydratase |
| A. fulgidus | Af1290 | AE001015 2649297 | N terminal | 35/159 (22) | AdoCbl-dependent methylmalonyl CoA mutase homologue |
| C. freundii | ORFY | U09771 1175769 | C terminal | 45/120 (37) | AdoCbl-dependent glycerol dehydratase |
| K. pneumoniae | ORF2a | U30903 940439 | C terminal | 42/120 (35) | AdoCbl-dependent glycerol dehydratase |
DISCUSSION
The experiments presented here show that the pduO gene encodes an enzyme with ATP:cob(I)alamin adenosyltransferase activity. Partially purified preparations of the PduO protein had ATP:cob(I)alamin adenosyltransferase activity higher than that reported for purified CobA enzyme: 312 nmol/min/mg of protein compared to 53 nmol/min/mg of protein (31). The primary role of the PduO enzyme is apparently the assimilation of exogenous corrinoids for 1,2-propanediol degradation. The function of the pduO gene was partially replaced by that of the cobA gene, which encodes a corrinoid adenosyltransferase that functions both in the assimilation of exogenous corrinoids and in the de novo synthesis of AdoCbl (13). However, the optimal growth of S. enterica on 1,2-propanediol required expression of the pduO gene.
Growth studies indicated that AdoCbl is limiting for growth on 1,2-propanediol. Overexpression of the PduO adenosyltransferase from a plasmid decreased the generation time of S. enterica on 1,2-propanediol from 8.4 to 5.8 h but did not affect the growth rate on glucose succinate, acetate, or propionate. In vitro studies have shown that AdoCbl breaks down during catalysis into inactive Cbls that are inhibitors of diol dehydratase (24, 34). Thus, it appears that the readenosylation of inactive Cbls generated during catalysis limits growth on 1,2-propanediol. This finding may be of importance in biotechnology, since overexpression of adenosyltransferase enzymes might be used to enhance AdoCbl-dependent processes of commercial importance.
Amino acid sequence similarity searching indicated that the PduO adenosyltransferase has two discrete domains. The N- and C-terminal regions of the PduO protein align with complete proteins encoded by different genes. The GenBank database currently contains 12 proteins homologous to the N-terminal region of PduO (Gene Identifiers [GIs] 940442, 1175767, 6459405, 2635812, 8568803, 3257079, 1722966, 699336, 5458793, 5103556, 6015885, and 2649297) and 7 proteins homologous to its C-terminal region (GIs 1175769, 940439, 4160465, 7672527, 4808412, 6624269, and 4235479). Among the C-terminal homologues, four of seven (GIs 4160465, 7672527, 6624269, and 4235479) are encoded by genes arranged with those for the degradation of aromatic compounds, suggesting an independent function. Hence, PduO could be a bifunctional enzyme. If so, it would likely catalyze sequential steps in the adenosylation pathway, namely, the reduction of cob(II)alamin to cob(I)alamin and the adenosylation of cob(I)alamin (Fig. 1).
Based on amino acid sequence similarity, adenosyltransferases and their homologues can be divided into three families: PduO type, CobA type, and EutT type. Analysis of the genomic context of the genes that encode each family suggests that particular types of adenosyltransferases are specialized for particular AdoCbl-dependent enzymes or for the de novo synthesis of AdoCbl. At present, in the GenBank nonredundant database there are two EutT proteins (GIs 3885914 and 6685444). The corresponding eutT genes are both organized with genes encoding AdoCbl-dependent ethanolamine ammonia lyases. In the CobA group, 12 proteins were identified in GenBank (GIs 399274, 115148, 78899, 7469288, 7469130, 231830, 3724050, 7471252, 7477928, 6117894, 7518352, and 7520977). Of these, seven are encoded by genes organized with genes predicted to function in the de novo synthesis of AdoCbl. Thus, the main role for the CobA-type of proteins is apparently as ATP:corrinoid adenosyltransferases that function in the de novo synthesis of AdoCbl. The PduO-type adenosyltransferases include 20 members (PduO, 12 proteins with homology to the N-terminal region of PduO, and 7 proteins with homology to the C-terminal region of PduO). In this group, the genes for six members are arranged with genes for AdoCbl-dependent diol or glycerol dehydratases, and the gene for one member is organized together with sequences similar to those encoding AdoCbl-dependent methylmalonyl CoA mutases (Table 2). The genes for the remaining 13 PduO group members are arranged with genes encoding proteins of unknown function or proteins not known to employ AdoCbl as a cofactor. It seems likely that PduO-type adenosyltransferases primarily support AdoCbl-dependent diol and glycerol dehydratases (two very closely related enzymes) and that some PduO group members have divergent functions. The preferential use of CobA-type adenosyltransferases for de novo AdoCbl synthesis might reflect the substrate specificity. De novo synthesis requires adenosylation of an early biosynthetic intermediate, whereas assimilation involves adenosylation of intact cobamides, such as CNCbl (13). The apparent preferential use of PduO-type adenosyltransferases with diol and glycerol dehydratases and EutT-type adenosyltransferases with ethanolamine ammonia lyases suggests protein-protein interactions between adenosyltransferases and the AdoCbl-dependent enzymes they support. Such interactions would be helpful if AdoCbl is limiting, and this appears to be the case during growth on 1,2-propanediol (see above).
Although particular groups of adenosyltransferases tend to be specific for certain AdoCbl-dependent enzymes, it is interesting that pduO group members are organized with both diol and glycerol dehydratase genes and, in one case, with sequences similar to those encoding methylmalonyl CoA mutases (Table 2). This suggests that adenosyltransferases within a given group can sometimes support AdoCbl-dependent enzymes that are unrelated in amino acid sequence and that have different substrate specificities. This raises the possibility of identifying previously unknown AdoCbl-dependent enzymes based on analysis of genes clustering with adenosyltransferase homologues. In this regard, we note that a number of PduO and CobA homologues cluster with genes of unknown function.
ACKNOWLEDGMENTS
This work was supported by grant GM59486 from the National Institutes of Health and by the Florida Agricultural Experiment Station.
We thank M. Rasche, K. T. Shanmugam, and J. Maupin-Furlow for their invaluable assistance.
Footnotes
Florida Agricultural Experiment Station Journal Series no. RO7931.
REFERENCES
- 1.Ailion M, Bobik T A, Roth J R. Two global regulatory systems (Crp and Arc) control the cobalamin/propanediol regulon of Salmonella typhimurium. J Bacteriol. 1993;175:7200–7208. doi: 10.1128/jb.175.22.7200-7208.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz D, Hushon J, Whitfield H, Jr, Roth J, Ames B. Procedure for identifying nonsense mutations. J Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobik T A, Ailion M E, Roth J R. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol. 1992;174:2253–2266. doi: 10.1128/jb.174.7.2253-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bobik T A, Havemann G D, Busch R J, Williams D S, Aldrich J C. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for the formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobik T A, Wolfe R S. Activation of formylmethanofuran synthesis in cell extracts of Methanobacterium thermoautotrophicum. J Bacteriol. 1989;171:1423–1427. doi: 10.1128/jb.171.3.1423-1427.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bobik T A, Xu Y, Jeter R M, Otto K E, Roth J R. Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J Bacteriol. 1997;179:6633–6639. doi: 10.1128/jb.179.21.6633-6639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen P, Andersson D, Roth J R. The control region of the pdu/cob regulon in Salmonella typhimurium. J Bacteriol. 1994;176:5474–5482. doi: 10.1128/jb.176.17.5474-5482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conner C P, Heithoff D M, Julio S M, Sinsheimer R L, Mahan M J. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci USA. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conway T, Sewell G W, Osman Y A, Ingram L O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol. 1987;169:2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 13.Escalante-Semerena J C, Suh S J, Roth J R. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J Bacteriol. 1989;72:273–280. doi: 10.1128/jb.172.1.273-280.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedmann H C. Biosynthesis of corrinoids. In: Babior B M, editor. Cobalamin. New York, N.Y: John Wiley and Sons; 1975. pp. 75–103. [Google Scholar]
- 15.Heithoff D M, Conner C P, Hentschel U, Govantes F, Hanna P C, Mahan M J. Coordinate intracellular expression of Salmonella genes induced during infection. J Bacteriol. 1999;181:799–807. doi: 10.1128/jb.181.3.799-807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horswill A, Escalante-Semerena J. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J Bacteriol. 1997;179:928–940. doi: 10.1128/jb.179.3.928-940.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huennekens F M, Viktols K S, Fujii K, Jacobsen D W. Biosynthesis of the cobalamin coenzymes. In: Dolophin D, editor. B12. New York, N.Y: John Wiley and Sons; 1982. pp. 145–167. [Google Scholar]
- 18.Jeter R M. Cobalamin-dependent 1,2-propanediol utilization by Salmonella typhimurium. J Gen Microbiol. 1990;136:887–896. doi: 10.1099/00221287-136-5-887. [DOI] [PubMed] [Google Scholar]
- 19.Jeter R M, Olivera B M, Roth J R. Salmonella typhimurium synthesizes cobalamin (vitamin B12) de novo under anaerobic growth conditions. J Bacteriol. 1984;159:206–213. doi: 10.1128/jb.159.1.206-213.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin E C C. Dissimilatory pathways for sugars, polyols, and carboxylates. In: Niedhardt F D, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 244–284. [Google Scholar]
- 21.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning; a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 23.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori K, Tobimatsu T, Hara T, Toraya T. Characterization, sequencing, and expression of the genes encoding a reactivating factor for glycerol-inactivated adenosylcobalamin-dependent diol dehydratase. J Biol Chem. 1997;272:32034–32041. doi: 10.1074/jbc.272.51.32034. [DOI] [PubMed] [Google Scholar]
- 25.Obradors N, Badía J, Baldomà L, Aguilar J. Anaerobic metabolism of the l-rhamnose fermentation product 1,2-propanediol in Salmonella typhimurium. J Bacteriol. 1988;170:2159–2162. doi: 10.1128/jb.170.5.2159-2162.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rondon M R, Escalante-Semerena J. The poc locus is required for 1,2-propanediol-dependent transcription of the cobalamin biosynthetic (cob) and propanediol utilization (pdu) genes of Salmonella typhimurium. J Bacteriol. 1992;174:2267–2272. doi: 10.1128/jb.174.7.2267-2272.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth J R, Lawrence J G, Bobik T A. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 28.Roth J R, Lawrence J G, Rubenfield M, Kieffer-Higgins S, Church G M. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J Bacteriol. 1993;175:3303–3316. doi: 10.1128/jb.175.11.3303-3316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmieger H. A method for detection of phage mutants with altered transducing ability. Mol Gen Genet. 1971;110:378–381. doi: 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- 30.Schneider Z, Stroinski A, editors. Comprehensive B12. Berlin, Germany: Walter de Gruyter; 1987. [Google Scholar]
- 31.Suh S-J, Escalante-Semerena J C. Purification and initial characterization of the ATP:corrinoid adenosyltransferase encoded by the cobA gene of Salmonella typhimurium. J Bacteriol. 1995;177:921–925. doi: 10.1128/jb.177.4.921-925.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suh S J, Escalante-Semerena J C. Cloning, sequencing and overexpression of cobA which encodes ATP:corrinoid adenosyltransferase in Salmonella typhimurium. Gene. 1993;129:93–97. doi: 10.1016/0378-1119(93)90701-4. [DOI] [PubMed] [Google Scholar]
- 33.Toraya T, Honda S, Fukui S. Fermentation of 1,2-propanediol and 1,2-ethanediol by some genera of Enterobacteriaceae, involving coenzyme B12-dependent diol dehydratase. J Bacteriol. 1979;139:39–47. doi: 10.1128/jb.139.1.39-47.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toraya T, Mori K. A reactivating factor for coenzyme B12-dependent diol dehydratase. J Biol Chem. 1999;274:3372–3377. doi: 10.1074/jbc.274.6.3372. [DOI] [PubMed] [Google Scholar]
- 35.Vitols E, Walker G A, Huennekens R M. Enzymatic conversion of vitamin B12s to a cobamide coenzyme, α-(5,6-dimethylbenzimidazolyl)deoxyadenosylcobamide (adenosyl-B12) J Biol Chem. 1965;241:1455–1461. [PubMed] [Google Scholar]
- 36.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 37.Walter D, Ailion M, Roth J. Genetic characterization of the pdu operon: use of 1,2-propanediol in Salmonella typhimurium. J Bacteriol. 1997;179:1013–1022. doi: 10.1128/jb.179.4.1013-1022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]