Abstract
Lateral organ boundaries domain (LBD) proteins are plant-specific transcription factors that play important roles in organ development and stress response. However, the function of LBD genes has not been reported in Euphorbiaceae. In this paper, we used Jatropha curcas as the main study object and added rubber tree (Hevea brasiliensis), cassava (Manihot esculenta Crantz) and castor (Ricinus communis L.) to take a phylogenetic analysis of LBD genes. Of LBD, 33, 58, 54 and 30 members were identified in J. curcas, rubber tree, cassava and castor, respectively. The phylogenetic analysis showed that LBD members of Euphorbiaceae could be classified into two major classes and seven subclasses (Ia-Ie,IIa-IIb), and LBD genes of Euphorbiaceae tended to cluster in the same branch. Further analysis showed that the LBD genes of Euphorbiaceae in the same clade usually had similar protein motifs and gene structures, and tissue expression patterns showed that they also have similar expression profiles. JcLBDs in class Ia and Ie are mainly expressed in male and female flowers, and there are multiple duplication genes with similar expression profiles in these clades. It was speculated that they are likely to play important regulatory roles in flower development. Our study provided a solid foundation for further investigation of the role of LBD genes in the sexual differentiaion of J. curcas.
Keywords: LBD, Jatropha curcas, gene family, phylogenetic analysis
1. Introduction
Transcription factors (TFs) can play significant roles in plant development by influencing gene expression. The LBD gene family, also known as the asymmetric leaves2-like (ASL) gene family, is a group of plant-specific transcription factors that encode proteins containing lateral organ boundaries (LOB) domains [1,2]. Previous studies have shown that LBD genes were predominantly expressed in cells located on the base of the adaxial axis in all lateral organs formed by the shoot apical meristem, as well as at the base of the lateral roots. Overexpression of AtLOB/AtASL4 results in a smaller plant type and changes in the shape and size of flower organs, implying that LBD genes play important roles in the development of lateral organs [3]. The LBD genes can be split into two major classes and seven subclasses (Ia-e, IIa and IIb) based on sequence similarity and phylogenetic trees. The N-terminal of each LBD gene is largely conserved, and the C-terminal is varied. The LOB domain is at the N-terminal end of the protein, which contains a conserved CX2CX6CX3C zinc finger-like motif. Class I, which most LBD genes belong to, contains a glycine-alanine-serine (GAS-block) region and a LX6LX3LX6L leucine zipper-like helical coiled-coil structure behind the CX2CX6CX3C zinc finger region [1,4,5].
Previous studies have identified that LBD genes participate in the development of many lateral organs [3,4]. For example, the AtLOB gene can regulate the development of young leaves [3]; homologs of AtLOB have been identified in other species: Romaso2 in maize regulates the development of ears [6], OsRA2 in rice is involved in modifying panicle architecture through regulating pedicel length [7], HvRA2 (Vrs4) in barley can regulate lateral spikelet fertility, the vrs4 mutant displays a loss of spikelet determinacy [8]; AtLBD10 and AtLBD27 can regulate pollen development and all pollen grains aborted in the lbd10 lbd27 double mutant [9]. Recent studies have revealed that in addition to their role in lateral organs, they are also involved in many other biological processes: MdLBD13 in apple can inhibit anthocyanin synthesis and nitrogen utilization via the flavonoid pathway [10]; EgLBD29 and EgLBD37 in Eucalyptus grandis can affect phloem fibre production and secondary xylem, respectively [11]; SlLBD40 is a negative regulator of drought tolerance, and sllbd40 knockout mutants with higher drought tolerance than widetype tomato [12]. Based on the functional reports of AtLBD proteins, it was found that LBD genes from the same phylogenetic branch tend to have similar functions: Class Ia proteins may regulate aboveground organs development [3,9,13]; Class Ib proteins mainly regulate lateral root formation [14,15]; and Class II LBD proteins play an important function in nitrogen response and anthocyanin synthesis [16,17].
The LBD gene family has been studied in several species, but so far there is no report about the function of LBD genes in Euphorbiaceae. Jatropha curcas is an important cash crop in Euphorbiaceae. All the parts of the J. curcas plant have important economic value, especially its seeds. The seeds of J. curcas are considered as important material for biodiesel production because they contain about 40% oil and do not produce harmful substances after burning [18]. However, the yield of J. curcas seeds is low, which is less than 1 ton per hectare a year under normal growth conditions. This limits the application of J. curcas as a biodiesel. J. curcas is a dioecious plant, and the ratio of female to male flowers is low, generally 1:29-1:13. The low ratio of female to male of J. curcas is an improtant factor that leads to a low yield of seeds. J. curcas have two sex determination patterns: male flowers are unisexual from early development; female flowers are bisexual first, and after the sixth stage of development, stamens appear abortive and eventually produce mature fertile female flowers [19,20,21]. Rubber tree, castor and cassava in Euphorbiaceae are also very important cash crops. Both rubber tree and castor have high oil content, while cassava is an important source of starch in the world. Unfortunately, each inflorescence in these species produces only a small number of female flowers, too [22,23,24]. As mentioned earlier, LBD genes have very important roles in pollen development, inflorescence development, and they affect a variety of organ development. Thus, it is important to identify and analyze the LBD gene family of Euphorbiaceae.
With the continuous development of sequencing technology, the draft genome of several species of Euphorbiaceae have been released [25,26,27,28]. It is now easy for us to further understand the physiological and biochemical processes of Euphorbiaceae plants through gene family analysis. In this study, we first identified 33, 58, 54 and 30 LBD members of J. curcas, rubber tree, cassava and castor, respectively. Next, we conducted a comprehensive analysis of basic physicochemical information, phylogenetic analysis, gene structure and protein motif analysis, cis-element prediction, miRNA target site prediction and gene duplication event analysis on them. Finally, we focused on J. curcas for further expression analysis, and downstream target gene prediction and annotation to provide theoretical reference for the study of the role of LBD genes in the sexual differentiation of J. curcas.
2. Results
2.1. Genome-Wide Identification of LBD Genes in J. curcas, Rubber Tree, Cassava and Castor
Based on hmmsearch and BLASTP, 33, 58, 54 and 30 LBD genes were identified in J. curcas, rubber tree, cassava and castor, respectively, and named as JcLBD1-JcLBD33, HbLBD1-HbLBD58, MeLBD1-MeLBD54 and RcLBD1-RcLBD30 according to their positions on chromosomes or scaffolds.
Basic information about Euphorbiaceae LBD proteins were calculated (listed in Table S1). The proteins encoded by the 33 LBD genes of J. curcas with amino acid lengths ranged from 119 (JcLBD2) to 332 (JcLBD31); molecular weights ranged from 13,481.67 (JcLBD2) to 37,487.12 (JcLBD31), except the JcLBD3, which had a length of 1077 amino acids and a molecular weight of 117,788.7 Da; the predicted isoelectric points of the JcLBD proteins ranged from 5.1 (JcLBD17) to 9.28 (JcLBD9); the fatty amino acid index ranged from 61.38 to 95.21, indicating a small difference in their thermal stability; and the minimum instability index was 43.75 (JcLBD10), indicating that they were all unstable in vitro; the hydrophilicity scores of all proteins were smaller than 0, indicating that they were all hydrophilic. The lengths of the proteins encoded by the LBD genes of rubber tree, cassava and castor ranged from 116–333, 116–302, 117–310 amino acids, respectively; the molecular weights ranged from 13,029.18–37,240.79, 13,138.24–33,889.14, 13,033.05–34,490.75; the isoelectric points ranged from 5.02–9.32, 4.67–9.02, 4.53–9.45; fatty amino acid index (A.I.) indices ranged from 51.94–90.86, 60.76–88.06, 57.78–91.89, respectively; all proteins had instability indices greater than 40, except for MeLBD35 and RcLBD13, which had instability indices of 36.04 and 38.72, and were unstable in vitro; the hydrophilicity scores of all proteins were less than 0, indicating that they were all hydrophilic. These results showed that LBD proteins are relatively conserved in these four species of Euphorbiaceae in terms of physical properties.
2.2. Phylogenetic Analysis
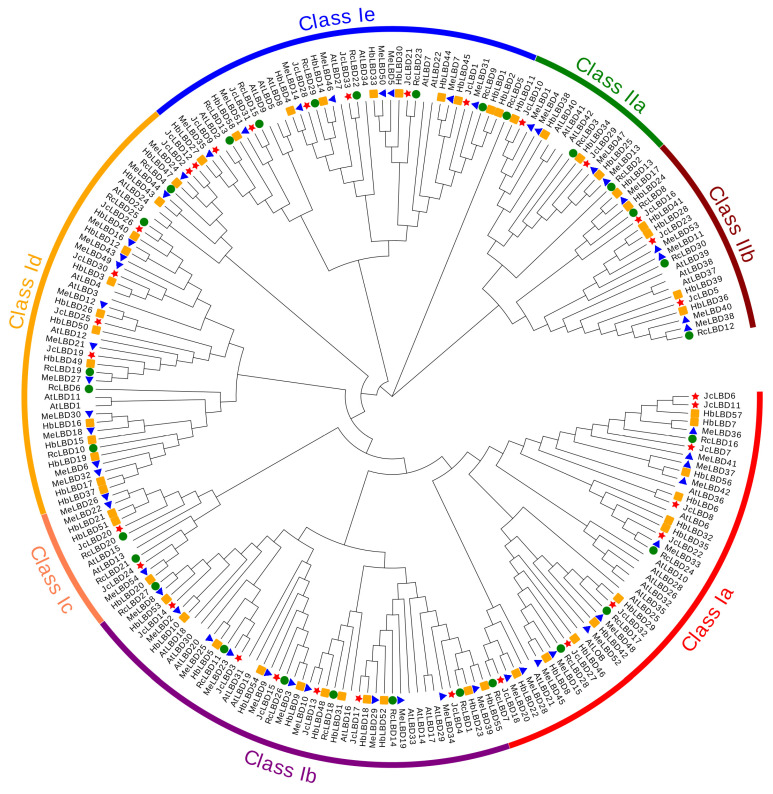
To better understand the evolutionary trajectory and function of the Euphorbiaceae LBD gene family, we constructed a phylogenetic tree using 175 identified LBD proteins in Euphorbiaceae and 43 known LBD proteins in A. thaliana (Figure 1). The Euphorbiaceae LBD gene family can be divided into two subfamilies according to the presence or lack of the motif LX6LX3LX6L. Most of the LBD genes belong to Class I, which contained 183 (83.94%) LBD genes, while Class II contained only 35 (16.06%) LBD genes. Further analysis found that LBD genes of Euphorbiaceae could also be divided into seven subclasses, Class Ia-Ie, Class IIa and Class IIb. Among these subclasses, Class Ia contained the largest number of LBD gene family with 45 members, and Class Ic had the least with only 12 genes. Each subclass contained LBD genes of all species, suggesting that they may share a common ancestor. Most of the LBD members of Euphorbiaceae were clustered together. LBD members of A. thaliana were clustered together as well, such as AtLBD10, AtLBD26, AtLBD28, AtLBD32 and AtLBD35 of Class Ia. This result suggests that LBD genes in Euphorbiaceae and A. thaliana diverged during evolution.
Figure 1.
The phylogenetic tree of LBD genes of J. curcas, rubber tree, cassava, castor and A. thaliana was constructed by MEGA 10 using the maximum likelihood (ML) model. Red stars, orange rectangles, blue triangles, and green circles indicate J. curcas, rubber tree, cassava, and castor sequences, respectively.
2.3. Gene Structure and Protein Motif Analysis
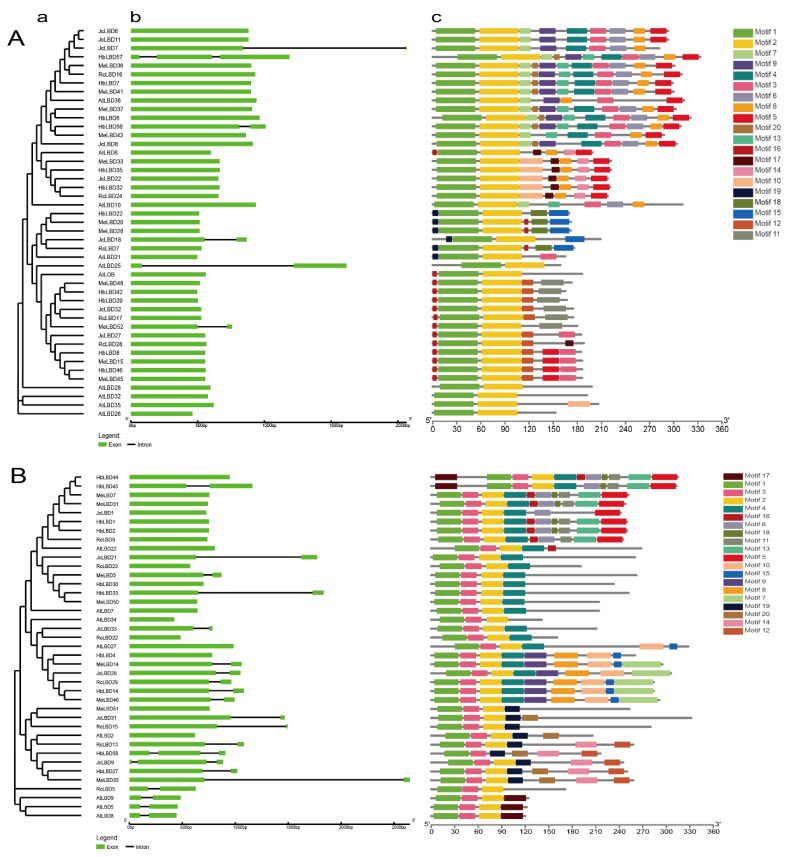
To further explore the possible evolutionary relationship and function of LBDs in Euphorbiaceae, we performed gene structures and protein motifs analyses of them (Figure 2 and Figure S1). The number of introns in the LBD genes of Euphorbiaceae ranged from 0 to 14. Of those, 43 (24.57%) LBD genes of Euphorbiaceae did not contain a intron, 113 genes (64.57%) contained 1 intron, 18 genes (10.29%) contained 2 introns, and only JcLBD3 contained 14 introns. LBD genes close in the phylogenetic tree tended to exhibit similar exon-intron structures: most Class Ia LBD genes did not contain an intron; most Class Ib, Id, IIa and IIb LBD genes contained only one intron; all Class Ic genes contained two introns; about half of Class Ie genes contained no intron, and another half contained one intron.
Figure 2.
The phylogenetic relationship, gene structure, and conserved protein motifs of Euphorbiaceae LBD genes. (A,B) are phylogenetic tree, gene structure and conserved motif of Class Ia and Ie of Euphorbiaceae, respectively. (a) A maximum likelihood tree was constructed by MEGE 10 with 1000 bootstrap replicates; (b) Exon-introns structure of LBDs performed by GSDS2; (c) The conserved motifs of Euphorbiaceae species LBD proteins analysed by MEME suit.
An analysis of protein motif results showed that all Euphorbiaceae LBD proteins contained CX2CX6CX3C zinc finger motif (motif 1 in Ia-Ic, Ie and IIa; motif 2 for IIb; motif 1 and motif 3 for Id). The Euphorbiaceae LBD proteins in the same clade contained similar motifs, indicating that the classification of the Euphorbiaceae LBD gene family in this study is reliable. The LBD proteins of Euphorbiaceae and A. thaliana in the same clade usually contain different motifs, indicating that the LBD genes of Euphorbiaceae and A. thaliana may be functionally differentiated.
2.4. Cis-Element Prediction
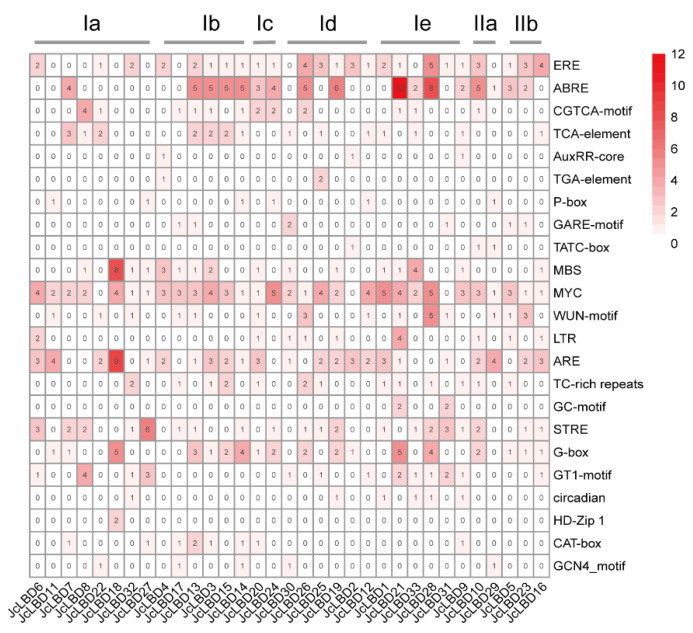
The analysis of cis-elements is the key to understanding gene regulation patterns, and provides important information for further understanding the function of genes. We have submitted the 1500 bp upstream sequences of Euphorbiaceae LBD genes to the plantCARE database for cis-element search. Of these cis-elements, 23 representative elements were extracted for display (Figure 3 and Figure S2). These 23 cis-elements were related to hormone response, stress response and plant development. The type and number of cis-acting elements in Euphorbiaceae and Arabidopsis LBD genes did not differ significantly. Among these 23 components, the ARE element associated with hypoxic stress was the most abundant. Almost all LBD promoters contain ARE element: 34, 23, 45, 34 and 22 in A. thaliana, J. curcas, rubber tree, cassava and castor, respectively. Ethylene (ERE), abscisic acid (ABRE), drought (MBS), stress (STRE), and light- (G-box) related response elements also appeared in the promoter regions of several genes. In addition, the LBD promoters contained the hormone-related response elements of jasmonic acid (CGTCA-motif), salicylic acid (TCA-element), auxin (AuxRR-core and TGA-element), gibberellin (P-box, GARE-motif and TATC-box); stress response elements of wound (WUN-motif), low temperature (LTR), defense and stress (TC-rich repeats), anoxic inducibility (GC-motif), and light (GT1-motif); and developmental regulatory elements: circadian, HD Zip 1, CAT-box and GCN4_motif. The types and numbers of cis-elements of different LBD genes in the same species are quite different, even for genes located in the same clade. These results suggested that LBD genes may respond to different signaling pathways due to different cis-acting elements in its promoter region.
Figure 3.
Cis-acting element matrix identified in the 1500-bp upstream promoters of each JcLBDs.
2.5. Prediction of miRNA Target Sites
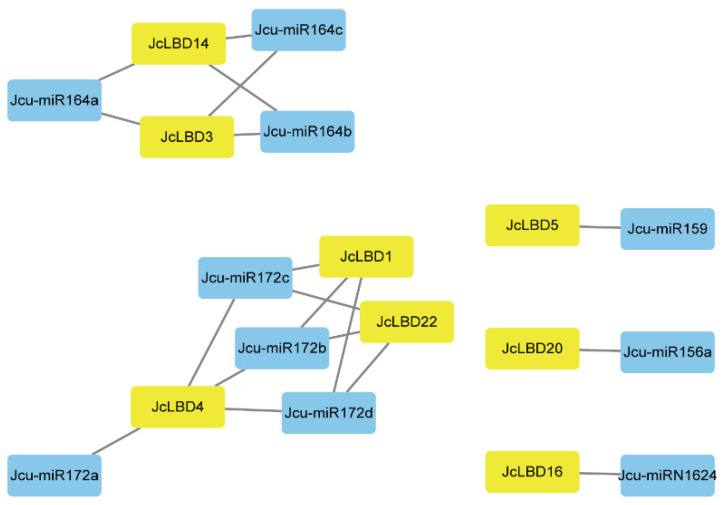
The prediction of miRNA target sites of genes would provide further understanding of the regulation model of the genes. We submitted the CDS sequences of LBD genes of each species together with mature miRNAs to psRNATarget to predict miRNA target sites. It was predicted that 30, 8, 24, 19 and 6 LBD genes are regulated by miRNA in A. thaliana, J. curcas, rubber tree, cassava, and castor, respectively. And they are regulated by several different miRNA families (Figure 4 and Figure S3 and Table S2). All species have members of the LBD gene family regulated by miRNA172, in which miRNA172 mainly regulated members of the Class Ia, Id, and Ie LBD genes in five species. In addition, miRNA156, miRNA159, and miRNA164 can also act on multiple LBD genes in Euphorbiaceae. Genes in the same subfamily may be regulated by different miRNAs. These results suggested that the interaction between LBD genes and miRNAs in Euphorbiaceae is not conservative, and the homologous genes may have evolved in different regulatory patterns.
Figure 4.
Netwrok between the Jcu-miRNAs and their targeted JcLBD genes. The blue and yellow rectangles indicate miRNAs and target genes, respectively.
2.6. Gene Duplication and Selective Pressure Analysis
To further investigate the evolution of LBD genes in the Euphorbiaceae, we performed gene duplication analysis and selection pressure analysis. We first used MCScanX software to analyze duplicated LBD genes of each species in Euphorbiaceae. As listed in Table S3, all LBD genes in J. curcas, rubber tree, cassava and castor were duplication genes. Among them, 0, 2, 0, 2 of the LBD genes in J. curcas, rubber tree, cassava, and castor were proximal duplication genes; 5, 6, 4, 1 were tandem duplication genes; 15, 12, 42, 7 were segmental duplication genes respectively. The number of segmental duplication genes was significantly higher than that of proximal duplication genes. This suggests that segmental duplication may be important for the LBD gene family.
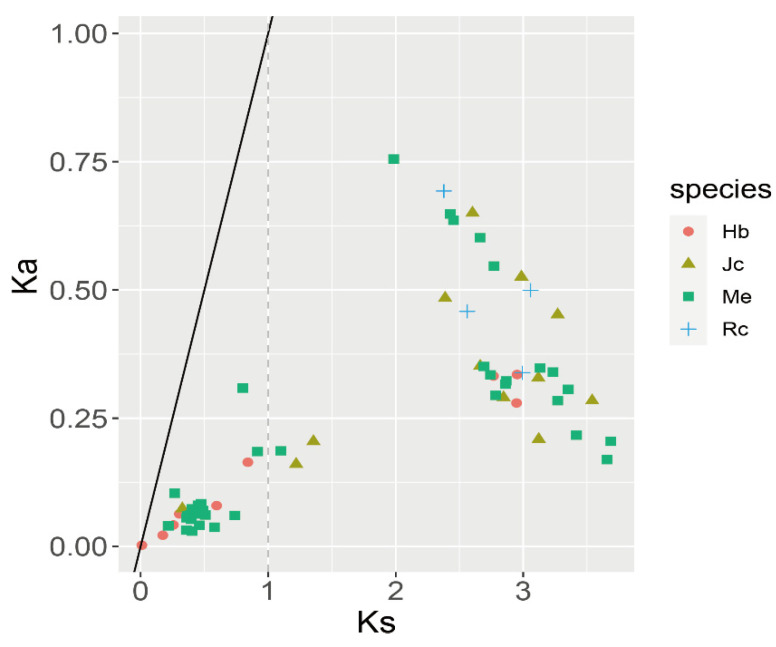
Next, we analyzed the selection pressure by calculating the Ka and Ks of gene duplication pairs (Figure 5 and Table S4). We identified 64 duplication gene pairs in Euphorbiaceae: 3, 3, 4 and 0 tandem duplication genes and 9, 6, 35 and 4 segmental duplication gene pairs in J. curcas, rubber tree, cassava, and castor, respectively. The Ka/Ks of all duplicated gene pairs ranged from 0.046269 to 0.389515, which was much less than 1, suggesting that they were subjected to purifying selection during the evolutionary process. From Ks value, we can see that the LBD genes of Euphorbiaceae experience two large-scale duplication events. The most recent duplication event mainly involved the duplication of HbLBDs and MeLBDs. Previous studies discovered that rubber tree and cassava experienced another whole genome duplication event compared to other Euphorbiaceae species. And our results are consistent with this conclusion [29].
Figure 5.
Selective pressure of Euphorbiaceae species LBD duplication genes. Red circles, yellow triangles, green rectangles, and blue crosses represent LBD duplication gene pairs of rubber tree, J. curcas, cassava, and castor, respectively.
2.7. Collinearity Analysis
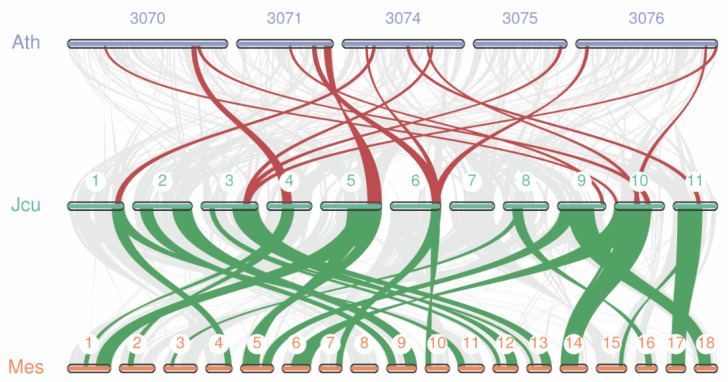
To infer the evolutionary relationship of LBD genes among different species, we performed a collinearity analysis on the Euphorbiaceae genomes. Since the rubber tree and castor genomes were not assembled to the chromosome level, only J. curcas, cassava and A. thaliana genomes were selected for analysis. As shown in Figure 6, there were many collinear blocks between them. J. curcas and A. thaliana have 15 collinear blocks containing LBD genes, and they contained a total of 17 LBD gene pairs; J. curcas and cassava have 25 collinear blocks containing a total of 40 LBD gene pairs. In addition, most of J. curcas had more than two orthologs in cassava, suggesting that cassava has undergone an additional whole genome duplication event during its evolution.
Figure 6.
LBD genes synteny analysis among the three species J. curcas, cassava, and A. thalina. The grey lines on the background indicate the collinear blocks within J. curcas and other species genomes, and the hilighted lines are the collinear blocks containg the LBD genes. Ath, Jcu, and Mes represents A. thalina, J. curcas, and cassava, respectively. 3070, 3071, 3074, 3075, and 3076 respresnt chromsomes NC_003070.9, NC_003071.7, NC_003074.8, NC_003075.7, and NC_003076.8, respectively. 1–11 in green represent chromosomes chr1–chr11, respectively. 1–18 in orange represent chromosomes Chromosome01–Chromosome18, respectively.
2.8. Gene Expression Analysis of JcLBD Genes
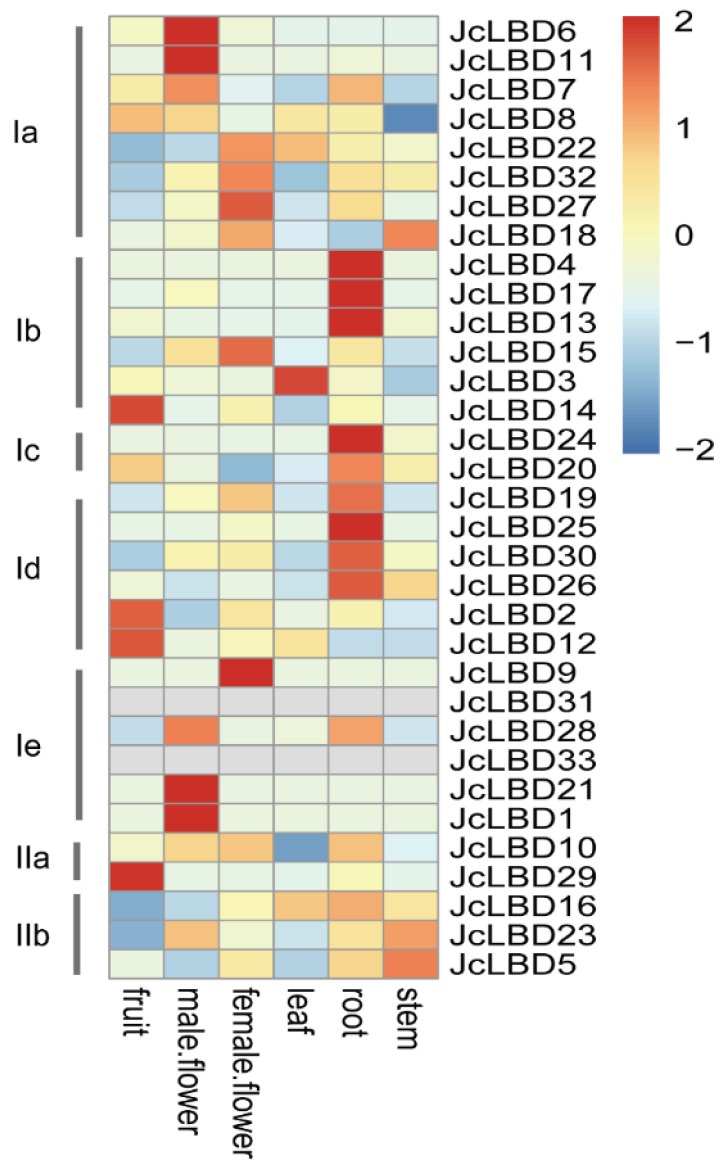
To further investigate the potential functions of each LBD gene in J. curcas, we analyzed the expression of JcLBDs in different tissues and stress treatments. Figure 7 and Table S5 show the tissue expression profile of JcLBD genes. We found that the genes of the same subclass usually have similar expression profiles. Class Ia LBD genes were mainly expressed in flowers; class Ib and Ic genes were mainly expressed in roots; class Id genes are mainly expressed in roots and fruits; class Ie genes were mainly expressed in flowers, especially male flowers; and in class II, there were no significant differences in gene expression among the organs, except JcLBD29, which was highly expressed in fruits. JcLBD6, JcLBD7, and JcLBD8 are tandem duplication genes, and they were extremely close on the phylogenetic tree. JcLBD6 and JcLBD7 have samilar expression profiles and both expressed highly in male flowers; JcLBD11 clustered on the same phylogentic branch with JcLBD6 and JcLBD7, and it was also expressed in male flowers; however, JcLBD8 was expressed mainly in fruit, and it also expressed in male flower, suggesting that genes on this evolutionary branch may be extremely important for male flower development. JcLBD1 and JcLBD21 are two proteins produced by segmental duplications, both of which have high expression in male flowers, suggesting that they are likely to play a very important role for male flower development. JcLBD18, JcLBD27, and JcLBD32 are close on the phylogenetic tree, and JcLBD27 and JcLBD32 are segmental duplication genes that are significantly expressed in female flowers, indicating that they may be associated with female flower development. Moreover, we found these genes have similar expression profiles and protein motifs, but their cis-acting elements and miRNA interactions are relatively different. These results indicate that these genes play extremely important roles in the growth and development of J. curcas, and they have evolved different regulatory patterns despite their conserved functions.
Figure 7.
Tissue expression patterns of JcLBDs. Heatmap of expression levels of 33 JcLBD genes in different tissues. fruit: immature fruit; male flower: fully open; female flower: fully open; leaf: fully mature; root: fully expanded; stem: fully mature. Heatmap was drawn by pheatmap using log10(TPM) values.
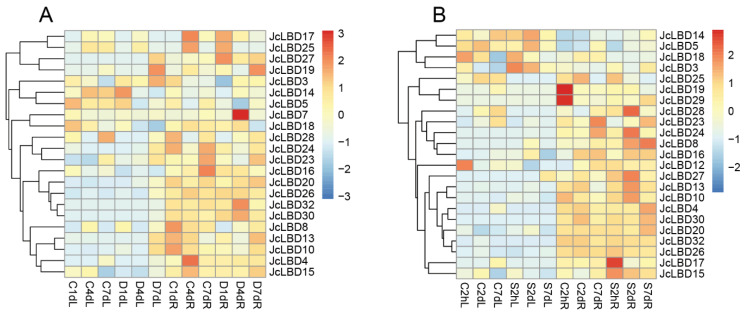
The Figure 8A,B are the expression profiles of the JcLBD genes after drought and salt stress treatments, respectively. The heatmap reveals that neither salt stress nor drought treatments obviously changed the expression of the majority of JcLBDs in the leaves. But in root, the situation is very different; the response of LBDs to stress gets more complex. Since there are only two replicates per sample, our expression analysis below was based on fold change.
Figure 8.
Stress response analysis of LBDs in Jatropha curcas. (A) Expression of JcLBDs in drought treatment. C and D represent control and drought treatments, respectively, and 1d, 4d and 7d correspond to one, four, and seven days post-treatment sampling, respectively, and L and R represent leaf and root tissues, respectively. (B) Expression of JcLBDs under salt stress. C and S represent control and salt stress treatments, respectively, 2h, 2d and 7d correspond to 2 h, two days and seven days post-treatment sampling, respectively, and L and R represent leaf and root tissues, respectively. Heatmap was drawn by pheatmap using log10(TPM) values.
As shown in Figure 8A and Table S6, the expression of JcLBD14 increased in leaves at day 1 after drought treatment (2 fold change); JcLBD7 and JcLBD32 were highly expressed in roots at day 4 after drought treatment (70 and 7 fold change in JcLBD7 and JcLBD32, respectively); JcLBD19 showed increased expression in both leaves and roots at day seven after drought treatment (34 fold change in leaves and 50 fold change in roots); and drought treatment results in altered timing of JcLBD17 and JcLBD25 expression in roots (both highly expressed in C4dR and D1dR).
As shown in Figure 8B and Table S7, JcLBD25 had low expression in control leaves (2.168, 15.5785, and 11.4697 in C2hL, C2dL, and C7dL, respectively), but after salt stress treatment, its expression in leaves was 0. JcLBD19 and JcLBD29 also showed decreased expression in roots after salt stress treatment (20 and nine-fold change after 2 h in JcLBD19 and JcLBD29, respectively); most salt response genes in the roots showed increased expression after salt treatment, such as JcLBD8, JcLBD15, and JcLBD17 (five- and three-fold change after two days and seven days in JcLBD8, three- and four-fold change after 2 h and two days in JcLBD15, and a five-fold change after 2 h in JcLBD17).
These results indicated that the JcLBDs are mainly expressed in roots, flowers and fruits, and that JcLBDs barely respond to drought and salt stress in leaves, while some JcLBDs expressions altered in roots.
2.9. Prediction and Annotation of Target Genes
Finally, we screened the downstream target genes that may be regulated by JcLBD transcription factors, and annotated the function of these target genes with GO enrichment and expression analysis in order to further investigate the potential roles of LBDs in J. curcas. Table 1 showed the best possible binding sites for three known JcLBDs and their corresponding motif information. Using the binding motif scanning method, we identified 148, 18, and 69 target genes that may be regulated by JcLBD27, JcLBD24 and JcLBD22, respectively (Table S8).
Table 1.
Best possible binding sites for JcLBDs.
| Transcription Factor | Motif | Best Possible Match |
|---|---|---|
| JcLBD27 | Jcr4S00009.60 | TCCGCCGCCGCCTCCGCCGCC |
| JcLBD24 | Jcr4S00803.60 | CGGCGGAAATTGCGGCG |
| JcLBD22 | Jcr4S13769.10 | TCTCCGCCGCCTTCTCCGCCG |
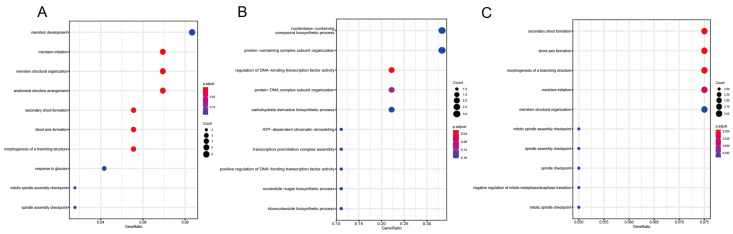
As shown in Figure 9, the GO annotation results showed that JcLBD27 target genes’ biological processes were mainly meristem initiation (GO:0010014), meristem structural organization (GO:0009933), secondary shoot formation(GO:0010223), shoot axis formation (GO:0010346) and morphogenesis of a branching structure (GO:0001763), anatomical structure arrangement (GO:0048532); and the biological processes of JcLBD24 genes were mainly the regulation of DNA-binding transcription factor activity (GO:0051090), protein-DNA complex subunit organization (GO:0071824); and the JcLBD22 were mainly secondary shoot formation (GO:0010223), shoot axis formation (GO:0010346), morphogenesis of a branching structure (GO:0001763), and meristem initiation (GO:0010014). The tissue expression profiles of the JcLBD target genes were displayed in Figure S4, and we can see that these genes were highly expressed primarily in flowers, leaves and roots. These results indicate that JcLBDs may play a very important regulatory role in plant growth and development.
Figure 9.
GO enrichment of JcLBD target genes. (A–C) are the downstream target genes of JcLBD27, JcLBD24 and JcLBD22, respectively.
3. Discussion
The LBD gene family is a plant-specific transcription factor family which plays an important role in the growth and development of various lateral organs and stress responses [4,16]. LBD genes are widely distributed in the plant kingdom, from green algae to angiosperms, and have been identified and studied in a variety of plants including A. thaliana, rice, moso bamboo, wheat, ginkgo, potato, fassion fruit, and so on [3,30,31,32,33,34,35]. However, the LBD genes have not been studied in Euphorbiaceae yet. The high-quality genomes of J. curcas, cassava, castor and rubber tree make it possible for us to explore the function of LBD genes in Euphorbiaceae. In this study, we identified J. curcas, rubber tree, cassava and castor containing 33, 58, 54 and 30 LBD family members, respectively. Previous studies identified 28 LBD genes in Physcomitrium patens, 71 in Picea abies, 44 in maize, 43 in A. thaliana, 42 in grapes, and 55 in Eucalyptus grandis [11,36]. This suggested that the LBD gene family retained largely function in the genetic evolution of the angiosperms species. The LBD gene family of Euphorbiaceae can be divided into seven subclasses (Ia-Ie and IIa-IIb). Although the phylogenetic tree classification of this family of Euphorbiaceae is similar to that of A. thaliana, members of the family of Euphorbiaceae appear clustered to one branch. The results of protein motif analysis showed that proteins close on the phylogenetic tree have similar structures, but LBD proteins of Euphorbiaceae have evolved new motifs compared to A. thaliana. Gene structure analysis showed that the intron numbers and length of LBD genes of Euphorbiaceae is more variable than those of A. thaliana. For example, JcLBD3 of the Ib subfamily contains 14 introns, and the intron length of HbLBD3-HbLBD26 of Ie subfamily is significant longer than others. In addition, there are 64 pairs of LBD duplication genes in Euphorbiaceae, of which 10 pairs are tandem duplication genes and 54 pairs are fragment duplication genes. The collinearity blocks between J. curcas and cassava LBD genes were significantly more than those between J. curcas and A. thaliana. These results suggested that the LBD genes function may be conserved in Euphorbiaceae.
Promoter cis-acting element prediction, expression pattern analysis, and target gene prediction and annotation can better explain the possible functions of JcLBDs. The results of cis-element analysis showed that the elements contained in Euphorbiaceae LBD genes are mostly related to ethylene, drought and hypoxia responses. LBD genes located in the same clade of J. curcas usually contained different cis-acting element types and quantities. The miRNA prediction results showed that miRNA156, miRNA159, miRNA164, and miRNA172 can target multiple LBD genes in Euphorbiaceae, and they all can regulate plant development [37,38,39]. Except these four miRNAs, the miRNA families interacting with LBD genes in different species of Euphorbiaceae were quite different, and LBD genes in the same subclass may be regulated by different miRNA family members. Target gene prediction and annotation results showed that the downstream target genes of JcLBDs play roles in various biological processes. These results suggest that LBD family members of J. curcas may play regulatory roles in various signaling pathways through complex synergistic effects, thereby participating in various physiological processes.
Most of the Euphorbiaceae species are dioecious, and the ratio of female to male flowers is very low. Therefore, it is very important to study the regulation of flower development in Euphorbiaceae. According to the tissue expression pattern in Figure 7, we found that multiple members of the Ia and Ie subfamily are specifically expressed in male or female flowers. Previous studies have suggested that the Ia subfamily has an important function in the formation of aerial organs, while the Ie subfamily function has not yet been summarized [16]. JcLBD6, JcLBD7, and JcLBD8 in Class Ia are close in the phylogenetic tree, and they are tandem duplication genes. JcLBD6 and JcLBD7 are clustered in one clade, and JcLBD11 is on this clade, as well. JcLBD6 and JcLBD11 are maily expressed in male flower, and JcLBD7 is expressed in male flower and root. According to the phylogenetic analysis, JcLBD6, JcLBD7, JcLBD8 and JcLBD11 are homologous genes of AtLBD36/AS1. AS1 can regulate flower development [40,41]. This indicates that the function of AtLBD36 may be separated in J. curcas, and JcLBD6, JcLBD7 and JcLBD11 may be extremely important for the male flower development of J. curcas. JcLBD1 and JcLBD21 are segmental duplication genes in Class Ie, and they are both highly expressed in male flowers. Their homologous protein AtLBD27 has been confirmed to play an extremely important role in pollen development. The lbd27 mutant causes pollen abortion (the abortion rate was high as 70%), and all pollen aborted in lbd10 lbd27 double mutant [9]. This clade contains another A. thaliana gene, AtLBD22, which has also been shown to play a role in pollen development [42]. In addition, the types and numbers of cis-acting elements of JcLBD1 and JcLBD21 are quite different, and JcLBD1 is a target site of miRNA172, so it is speculated that JcLBD1 and JcLBD21 are also involved in the development of pollen. It is speculated that the Euphorbiaceae LBD proteins of this clade may play a regulatory function on pollen development through different pathways. JcLBD18, JcLBD27 and JcLBD32 are close in the phylogenetic tree, and JcLBD27 and JcLBD32 are segmental duplication genes, which are significantly expressed in female flowers. The phylogenetic tree shows that JcLBD27 and JcLBD32 are homologous of AtLOB, which has been confirmed to play an important role in the development of A. thaliana lateral organs, and several important homologous proteins identified in other species. OsRA2, rasoma2, and HvRA2 are homologous of AtLOB, which is involved in the regulation of floral development, so it is speculated that JcLBD27 and JcLBD32 may also regulate flower development [3,6,7,8]. In addition, JcLBD9 and JcLBD28 of the Class Ie are divided into two major branches on the phylogenetic tree, but they are all significantly expressed in flower. These results suggest that Class Ia and Class Ie proteins in J. curcas are likely to play an important role in the regulation of flower development in plants, and appear to be more delicately regulated than AtLBDs. The functional differentiation of JcLBDs may occur due to transcriptional regulation and post-transcriptional modifications. For example, JcLBD6, JcLBD7, JcLBD8 and JcLBD11 promoters contain different cis-acting elements; JcLBD5; JcLBD16 and JcLBD23 are regulated by miRNAs in different ways: JcLBD5 and JcLBD16 are regulated by miR159 and miRN1624, respectively, while JcLBD23 is not regulated by miRNAs.
4. Materials and Methods
4.1. Collection of Sequencing Data
Genome data and protein data of Jatropha curcas come from the Giga database (http://gigadb.org/dataset/view/id/100689, accessed on 8 December 2021); cassava data was downloaded from the Phytozome V13 database (Manihot esculenta v6.1; https://www.ncbi.nlm.nih.gov/, accessed on 29 August 2021); the gene family information of A. thaliana was downloaded from the TAIR database (https://www.arabidopsis.org/, accessed on 2 June 2021); and the genome and protein data of A. thaliana (TAIR10.1), rubber tree (ASM165405v1), and castor (JCVI_RCG_1.1) were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 29 August 2021).
4.2. Identification of LBD Genes
In order to identify the LBD gene family members of J. curcas, rubber tree, cassava and castor, a hidden Markov model of the LOB domain (PF03195) was obtained from the Pfam database (http://pfam.xfam.org/, accessed on 28 March 2022) and used as the seed model for an HMMER3 search of the local Euphorbiaceae protein database [43]. In addition, 43 published A. thaliana protein sequences containing the LOB domain were used as the original alignment sequence of BLASTP [44]. The sequences identified by these two methods were submitted to NCBI CD-Search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 29 March 2022) to confirm the conserved domain [45]. Genes with incomplete LOB domain and redundant genes were removed to produce the confirmed LBD genes.
LBD genes of Euphorbiaceae were renamed according to their positions on the chromosomes, and then the ExPASY tool (https://web.expasy.org/protparam/, accessed on 18 April 2022) was used to predict protein physicochemical parameters such as protein size (aa), molecular weight (MW), isoelectric point (PI), stability, fatty amino acid index (A.I.), and the grand average of hydropathicity (GRAVY) [46].
4.3. Phylogenetic Analyses
Multiple sequence alignment of all LBD protein sequences of Euphorbiaceae and A. thaliana were by Clustal W. And based on the alignment results, maximum likelihood (ML) trees were constructed using MEGA X software with the JTT model, and the Bootstrap was set to 1000 times [47]. The phylogenetic tree was visualized and modified using the online website Evolview (https://www.evolgenius.info/evolview/#/treeview, accessed on 14 June 2022) [48].
4.4. Motif and Gene Structure Analysis
The MEME suite was used for conservative motif prediction with the following parameters: maximum width 50, minimum width 6, and the number of motifs set to 20 [49]. The results were then further modified with the TBtools software [50]. To analyze gene structure, we first extract exon and intron positions from the gene annotation file and then submit them to the online Gene Structure Display Server (GSDS: http://gsds.gao-lab.org/, accessed on 13 April 2022) for visualization [51].
4.5. Cis-Acting Element Analysis
The 1500 bp sequence upstream the LBD genes were extracted as promoter sequences based on the gene annotation and chromosome sequence, and the plantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 4 May 2022) was used to predict the cis-element [52]. The elements related to hormone response, stress response and plant developmrnt were then extracted. R package pheatmap was used for further visualization [53].
4.6. miRNA Target Gene Analysis
Mature miRNA sequences of the Euphorbiaceae species were downloaded from the plant microRNA Encyclopedia Database (PmiREN; https://www.pmiren.com/, accessed on 18 April 2022), and CDs sequences of the LBD gene were inputted into the psRNATarget online tool (https://www.zhaolab.org/psRNATarget/analysis, accessed on 28 April 2022) to predict the target sites with the default parameters [54,55]. The interacting miRNAs and target genes with expectation values greater than 4.5 were extracted, and Cytoscape software was used to construct the interaction network between these miRNAs and the target genes [56].
4.7. Collinearity and Selective Pressure Analysis
Collinearity of LBD genes in Euphorbiaceae was analyzed by JCVI [57]. Since the genome of the rubber tree and castor were only assembled to the scaffold level, we only analyzed the collinearity between species for J. curcas, cassava, and A. thaliana.
Gene duplication includes the tandem duplication and segmental duplication. First, BLASTP was used to all-against-all BLAST, and the results were used to identify gene duplication events using MCScanX v1.1. Finally, KaKs_ Calculator2.0 was used to calculate the synonymous substitution rate (Ks), the nonsynonymous substitution rate (Ka), and the Ka/Ks ratio between homologous gene pairs [58].
4.8. Tissue Expression Analysis and Stress Response Analysis of JcLBDs
To further explore the potential function of JcLBDs in J. curcas, we examined the expression of JcLBDs through public transcriptome data. Raw data of six different tissues including fruit, male flower, female flower, leaf, root, and stem were obtained for tissue expression analysis (listed in Table S9; Accession number: PRJNA399175). Raw expression data of J. curcas treated by drought and salt were used to analysis, too (listed in Table S10; Accession number: PRJNA257901 and PRJNA244896). Hisat2 and StringTie were used for comparison and quantitative analysis, respectively [59,60]. GCEN softwere were then used for normalization by quantile normalization algorithm [61]. For samples with duplicates, we calculated the avarage of TPM values. A heatmap of different tissues and stress treatment of the JcLBDs was drawn by R package pheatmap using log10(TPM) values [53].
4.9. Identification and Annotation of Downstream Genes
To obtain potential downstream regulatory genes for the LBD protein, we used bedtools to extract 1500 bp of the J. curcas promoter sequence as potential target sites for binding.
The DNA binding sites of three members of the JcLBD genes are already known, and the base sequences of these three binding sites were downloaded from the database (http://planttfdb.gao-lab.org/index.php, accessed on 2 June 2022) [62]. Then the Motif FIMO program (5.3.0) were used to detect the binding site with the P < 1 × 10−7 [49]. Genes containing binding sites were then considered to be downstream target genes of LBD.
The eggNOG-mapper was used for GO annotation of all target genes of JcLBDs [63]. Then GO enrichment of downstream target genes for each of the three known binding site genes was performed using the R package clusterProfiler [64]. Tissue-specific expression analysis of these target genes was performed separately using the R package pheatmap (listed in Table S9; Accession number: PRJNA399175) [53].
5. Conclusions
In this study, the members of the Euphorbiaceae LBD gene family were identified and analyzed for the first time by means of bioinformatics. Jatropha curcas, rubber tree, cassava and castor contained 33, 58, 54 and 30 LBD members, respectively. Phylogenetic analysis found that Euphorbiaceae LBD genes could be divided into seven subgroups. The properties of LBD proteins in Euphorbiaceae species are similar, and similar proteins on the Euphorbiaceae phylogenetic tree have similar gene structures and protein motifs, indicating that the LBD genes of Euphorbiaceae are conserved in the evolutionary process. The number of LBD members in rubber tree and cassava is twice that of J. curcas and castor, and most of the replicative gene pairs are generated by fragment duplication. Further analysis of cis-acting elements, miRNA target sites, expression profiles, protein interactions, and target gene prediction and annotation analysis of JcLBDs showed that Class Ia and Ie genes have important regulatory effects on plant flower development. This study provides a reference for further exploration of the mechanism of sexual differentiation in Jatropha curcas.
Acknowledgments
We are very grateful to the members in the laboratory for their helpful discussions and technical assistance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11182397/s1, Figure S1: The phylogenetic relationship, gene structure, and conserved motifs of Euphorbiaceae LBD proteins. A, B, C, D, and E are phylogenetic tree, gene structure and conserved motif of Euphorbiaceae species LBDs grouped into Ib, Ic, Id, IIa and IIb, respectively, Figure S2: Cis-acting element matrinx identified in the 1500-bp upstream promoters of each LBD genes.A, B, C, and D represent rubber tree, cassava, castor, and A. thaliana, respectively, Figure S3: Netwrok between the miRNAs and their targeted LBD genes. The blue and yellow rectangles indicate miRNAs and target genes, respectively. A, B, and C represent rubber tree, cassava, and castor, respectively, Figure S4: Tissue expression patterns of JcLBD target genes. A-C are the downstream target genes of JcLBD27, JcLBD24 and JcLBD22, respectively, Table S1: Basic information of LBD genes of Euphorbiaceae, Table S2: Prediction of microRNA target sites for LBD genes in Euphorbiaceae, Table S3: Duplication analysis of LBD genes in Euphorbiaceae, Table S4: Analysis of selection pressure on LBD duplication gene pairs in Euphorbiaceae, Table S5: Tissue expression of JcLBD genes, Table S6: Expression of JcLBD genes in drought treatment, Table S7: Expression of JcLBD genes in salt treatment, Table S8: Target genes for JcLBD transcription factors, Table S9: RNA-seq samples in 6 tissues of J. curcas, Table S10: RNA-seq samples under stress of J. curcas.
Author Contributions
C.L. and Q.J. designed the research; Q.J. and Z.Y. performed the research, W.Y. and X.G. modified figures; Q.J. and C.L. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Funding Statement
This research was funded by National Natural Science Foundation of China (No. 31970609); Crop Varietal Improvement and Insect Pests Control by Nuclear Radiation; Startup Fund from Xishuangbanna Tropical Botanical Garden; ‘Top Talents Program in Science and Technology’ from Yunnan Province. Publication costs are funded by National Natural Science Foundation of China (No. 31970609).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Iwakawa H., Ueno Y., Semiarti E., Onouchi H., Kojima S., Tsukaya H., Hasebe M., Soma T., Ikezaki M., Machida C., et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002;43:467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- 2.Xu C., Luo F., Hochholdinger F. LOB domain proteins: Beyond lateral organ boundaries. Trends Plant Sci. 2016;21:159–167. doi: 10.1016/j.tplants.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Shuai B., Reynaga-Pena C.G., Springer P.S. The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 2002;129:747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majer C., Hochholdinger F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011;16:47–52. doi: 10.1016/j.tplants.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Matsumura Y., Iwakawa H., Machida Y., Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 2009;58:525–537. doi: 10.1111/j.1365-313X.2009.03797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bortiri E., Chuck G., Vollbrecht E., Rocheford T., Martienssen R., Hake S. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell. 2006;18:574–585. doi: 10.1105/tpc.105.039032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu H., Dai Z., Li L., Wang J., Miao X., Shi Z. OsRAMOSA2 shapes panicle architecture through regulating pedicel length. Front. Plant Sci. 2017;8:1538. doi: 10.3389/fpls.2017.01538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koppolu R., Anwar N., Sakuma S., Tagiri A., Lundqvist U., Pourkheirandish M., Rutten T., Seiler C., Himmelbach A., Ariyadasa R., et al. Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc. Natl. Acad. Sci. USA. 2013;110:13198–13203. doi: 10.1073/pnas.1221950110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim M.J., Kim M., Lee M.R., Park S.K., Kim J. LATERAL ORGAN BOUNDARIES DOMAIN (LBD) 10 interacts with SIDECAR POLLEN/LBD27 to control pollen development in Arabidopsis. Plant J. 2015;81:794–809. doi: 10.1111/tpj.12767. [DOI] [PubMed] [Google Scholar]
- 10.Li H.H., Liu X., An J.P., Hao Y.J., Wang X.F., You C.X. Cloning and elucidation of the functional role of apple MdLBD13 in anthocyanin biosynthesis and nitrate assimilation. Plant Cell Tissue Organ Cult. 2017;130:47–59. doi: 10.1007/s11240-017-1203-x. [DOI] [Google Scholar]
- 11.Lu Q., Shao F., Macmillan C., Wilson I.W., van der Merwe K., Hussey S.G., Myburg A.A., Dong X., Qiu D. Genomewide analysis of the lateral organ boundaries domain gene family in Eucalyptus grandis reveals members that differentially impact secondary growth. Plant Biotechnol. J. 2018;16:124–136. doi: 10.1111/pbi.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L., Zhang J., Xu J., Li Y., Guo L., Wang Z., Zhang X., Zhao B., Guo Y.-D., Zhang N. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 2020;301:110683. doi: 10.1016/j.plantsci.2020.110683. [DOI] [PubMed] [Google Scholar]
- 13.Semiarti E., Ueno Y., Tsukaya H., Iwakawa H., Machida C., Machida Y. The asymmetric leaves2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001;128:1771–1783. doi: 10.1242/dev.128.10.1771. [DOI] [PubMed] [Google Scholar]
- 14.Lee H.W., Cho C., Pandey S.K., Park Y., Kim M.J., Kim J. LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biol. 2019;19:46. doi: 10.1186/s12870-019-1659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Li Z., Ma B., Hou Q., Wan X. Phylogeny and functions of LOB domain proteins in plants. Int. J. Mol. Sci. 2020;21:2278. doi: 10.3390/ijms21072278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubin G., Tohge T., Matsuda F., Saito K., Scheible W.-R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009;21:3567–3584. doi: 10.1105/tpc.109.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan B.-Z., Xu Z.-F. Benzyladenine treatmentsignificantly increases the seed yield of the biofuel plant Jatropha curcas. J. Plant Growth Regul. 2011;30:166–174. doi: 10.1007/s00344-010-9179-3. [DOI] [Google Scholar]
- 19.Wu J., Liu Y., Tang L., Zhang F., Chen F. A study on structural features in early flower development of Jatropha curcas L. and the classification of its inflorescences. Afr. J. Agric. Res. 2011;6:275–284. [Google Scholar]
- 20.Adriano-Anaya M.D., Perez-Castillo E., Salvador-Figueroa M., Ruiz-Gonzalez S., Vazquez-Ovando A., Grajales-Conesa J., Ovando-Medina I. Sex expression and floral diversity in Jatropha curcas: A population study in its center of origin. Peerj. 2016;4:e2071. doi: 10.7717/peerj.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gangwar M., Shankar J. Molecular mechanisms of the floral biology of Jatropha curcas: Opportunities and challenges as an energy crop. Front. Plant Sci. 2020;11:609. doi: 10.3389/fpls.2020.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y., Xu J., Li Q., Mortimer P.E. Investigation of rubber seed yield in Xishuangbanna and estimation of rubber seed oil based biodiesel potential in Southeast Asia. Energy. 2014;69:837–842. doi: 10.1016/j.energy.2014.03.079. [DOI] [Google Scholar]
- 23.Hyde P.T., Guan X., Abreu V., Setter T.L. The anti-ethylene growth regulator silver thiosulfate (STS) increases flower production and longevity in cassava (Manihot esculenta Crantz) Plant Growth Regul. 2020;90:441–453. doi: 10.1007/s10725-019-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeboah A., Ying S., Lu J., Xie Y., Amoanimaa-Dede H., Boateng K.G.A., Chen M., Yin X. Castor oil (Ricinus communis): A review on the chemical composition and physicochemical properties. Food Sci. Technol. 2021;41:399–413. doi: 10.1590/fst.19620. [DOI] [Google Scholar]
- 25.Rivarola M., Foster J.T., Chan A.P., Williams A.L., Rice D.W., Liu X., Melake-Berhan A., Creasy H.H., Puiu D., Rosovitz M.J., et al. Castor bean organelle genome sequencing and worldwide genetic diversity analysis. PLoS ONE. 2011;6:e21743. doi: 10.1371/journal.pone.0021743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang C., Yang M., Fang Y., Luo Y., Gao S., Xiao X., An Z., Zhou B., Zhang B., Tan X., et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants. 2016;2:16073. doi: 10.1038/nplants.2016.73. [DOI] [PubMed] [Google Scholar]
- 27.Bredeson J.V., Lyons J.B., Prochnik S.E., Wu G.A., Ha C.M., Edsinger-Gonzales E., Grimwood J., Schmutz J., Rabbi I.Y., Egesi C., et al. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat. Biotechnol. 2016;34:562–570. doi: 10.1038/nbt.3535. [DOI] [PubMed] [Google Scholar]
- 28.Chen M.-S., Niu L., Zhao M.-L., Xu C., Pan B.-Z., Fu Q., Tao Y.-B., He H., Hou C., Xu Z.-F. De novo genome assembly and Hi-C analysis reveal an association between chromatin architecture alterations and sex differentiation in the woody plant Jatropha curcas. Gigascience. 2020;9:giaa009. doi: 10.1093/gigascience/giaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou Z., Yang J. Genome-wide comparison reveals divergence of cassava and rubber aquaporin family genes after the recent whole-genome duplication. BMC Genom. 2019;20:380. doi: 10.1186/s12864-019-5780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y., Yu X.B., Wu P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet. Evol. 2006;39:248–262. doi: 10.1016/j.ympev.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 31.Liu H., Cao M., Chen X., Ye M., Zhao P., Nan Y., Li W., Zhang C., Kong L., Kong N., et al. Genome-wide analysis of the lateral organ boundaries domain (LBD) gene family in Solanum tuberosum. Int. J. Mol. Sci. 2019;20:5360. doi: 10.3390/ijms20215360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J., Hu P., Tao Y., Song P., Gao H., Guan Y. Genome-wide identification and characterization of the Lateral Organ Boundaries Domain (LBD) gene family in polyploid wheat and related species. Peerj. 2021;9:e11811. doi: 10.7717/peerj.11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang B., Huang Z., Ma R., Ramakrishnan M., Chen J., Zhang Z., Yrjala K. Genome-wide identification and expression analysis of LBD transcription factor genes in Moso bamboo (Phyllostachys edulis) BMC Plant Biol. 2021;21:296. doi: 10.1186/s12870-021-03078-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang J.X., Hou Z.M., Liao J.Y., Qin Y., Wang L.L., Wang X.M., Su W.Q., Cai Z.Y., Fang Y.Y., Aslam M., et al. Genome-wide identification and expression analysis of LBD transcription factor genes in Passion Fruit (Passiflora edulis) Int. J. Mol. Sci. 2022;23:4700. doi: 10.3390/ijms23094700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian Y., Han X., Qu Y., Zhang Y., Rong H., Wu K., Xu L.a. Genome-wide identification of the Ginkgo (Ginkgo biloba L.) LBD transcription factor gene and characterization of its expression. Int. J. Mol. Sci. 2022;23:5474. doi: 10.3390/ijms23105474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong Y., Xu P., Jing X., Chen L., Li L., Li X. Decipher the ancestry of the plant-specific LBD gene family. Bmc Genom. 2017;18:951. doi: 10.1186/s12864-016-3264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reyes J.L., Chua N.-H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007;49:592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- 38.Sieber P., Wellmer F., Gheyselinck J., Riechmann J.L., Meyerowitz E.M. Redundancy and specialization among plant microRNAs: Role of the MIR164 family in developmental robustness. Development. 2007;134:1051–1060. doi: 10.1242/dev.02817. [DOI] [PubMed] [Google Scholar]
- 39.Wu G., Park M.Y., Conway S.R., Wang J.-W., Weigel D., Poethig R.S. The Sequential Action of miR156 and miR172 Regulates Developmental Timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu L., Xu Y., Dong A.W., Sun Y., Pi L.M., Xu Y.Q., Huang H. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development. 2003;130:4097–4107. doi: 10.1242/dev.00622. [DOI] [PubMed] [Google Scholar]
- 41.Gubert C.M., Christy M.E., Ward D.L., Groner W.D., Liljegren S.J. ASYMMETRIC LEAVES1 regulates abscission zone placement in Arabidopsis flowers. BMC Plant Biol. 2014;14:195. doi: 10.1186/s12870-014-0195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim M., Kim M.-J., Pandey S., Kim J. Expression and protein interaction analyses reveal combinatorial interactions of LBD transcription factors during Arabidopsis pollen development. Plant Cell Physiol. 2016;57:2291–2299. doi: 10.1093/pcp/pcw145. [DOI] [PubMed] [Google Scholar]
- 43.Schuster-Bockler B., Bateman A. An introduction to hidden Markov models. Curr. Protoc. Bioinform. 2007;Appendix 3:Appendix 3A. doi: 10.1002/0471250953.bia03as18. [DOI] [PubMed] [Google Scholar]
- 44.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST plus: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I., et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian B., Gao S., Lercher M.J., Hu S., Chen W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019;47:W270–W275. doi: 10.1093/nar/gkz357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey T.L., Johnson J., Grant C.E., Noble W.S. The MEME suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C.J., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y.H., Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 51.Hu B., Jin J.P., Guo A.Y., Zhang H., Luo J.C., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lescot M., Dehais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouze P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolde R. pheatmap: Pretty Heatmaps. [(accessed on 18 December 2021)]. R Package Version 1.0.12. Available online: https://CRAN.R-project.org/package=pheatmap.
- 54.Dai X.B., Zhuang Z.H., Zhao P.X.C. psRNATarget: A plant small RNA target analysis server (2017 release) Nucleic Acids Res. 2018;46:W49–W54. doi: 10.1093/nar/gky316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo Z., Kuang Z., Wang Y., Zhao Y., Tao Y., Cheng C., Yang J., Lu X., Hao C., Wang T., et al. PmiREN: A comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 2020;48:D1114–D1121. doi: 10.1093/nar/gkz894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smoot M.E., Ono K., Ruscheinski J., Wang P.-L., Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang H.B., Bowers J.E., Wang X.Y., Ming R., Alam M., Paterson A.H. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 58.Wang D., Zhang Y., Zhang Z., Zhu J., Yu J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen W., Li J., Huang S., Li X., Zhang X., Hu X., Xiang S., Liu C. GCEN: An Easy-to-Use Toolkit for Gene Co-Expression Network Analysis and lncRNAs Annotation. Curr. Issues Mol. Biol. 2022;44:1479–1487. doi: 10.3390/cimb44040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian F., Yang D.C., Meng Y.Q., Jin J.P., Gao G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020;48:D1104–D1113. doi: 10.1093/nar/gkz1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cantalapiedra C.P., Hernandez-Plaza A., Letunic I., Bork P., Huerta-Cepas J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021;38:5825–5829. doi: 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.