Abstract
Penicillin-binding protein 4a (PBP4a) from Bacillus subtilis was overproduced and purified to homogeneity. It clearly exhibits dd-carboxypeptidase and thiolesterase activities in vitro. Although highly isologous to the Actinomadura sp. strain R39 DD-peptidase (B. Granier, C. Duez, S. Lepage, S. Englebert, J. Dusart, O. Dideberg, J. van Beeumen, J. M. Frère, and J. M. Ghuysen, Biochem. J. 282:781–788, 1992), which is rapidly inactivated by many β-lactams, PBP4a is only moderately sensitive to these compounds. The second-order rate constant (k2/K) for the acylation of the essential serine by benzylpenicillin is 300,000 M−1 s−1 for the Actinomadura sp. strain R39 peptidase, 1,400 M−1 s−1 for B. subtilis PBP4a, and 7,000 M−1 s−1 for Escherichia coli PBP4, the third member of this class of PBPs. Cephaloridine, however, efficiently inactivates PBP4a (k2/K = 46,000 M−1 s−1). PBP4a is also much more thermostable than the R39 enzyme.
The Bacillus subtilis genome (16) contains a gene that encodes a putative 491-residue protein with sequence similarities to low-molecular-weight penicillin-binding proteins (PBPs). Successively referred to as pbp (25) and dacC (19), this gene was overexpressed in Escherichia coli by Pedersen et al. (19), who showed that dacC does indeed encode a membrane-bound PBP migrating between PBP4 and PBP5 of B. subtilis on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). This protein is now referred to as PBP4a. The phenotypic effect of dacC mutations and the role of PBP4a in the synthesis of the B. subtilis spore peptidoglycan and its influence on the properties of the spore were investigated, but no physiological role or enzymatic activity could be assigned to the protein (20).
As already reported for Actinomadura sp. strain R39 PBP4 (in short, the R39 dd-peptidase) (14), E. coli PBP4 (17), and a Haemophilus influenzae putative PBP (4), the B. subtilis PBP4a possesses a large insert (175 residues) between the first and second active-site-defining motifs, and it can therefore be classified as a class C low-molecular-weight PBP (14). Importantly, although preliminary X-ray data for E. coli PBP4 are available (24), no three-dimensional structure of a protein belonging to this class of proteins has been presently established.
In this work, we describe the overexpression of PBP4a in E. coli, its purification, and its kinetic characterization as an in vitro dd-carboxypeptidase. Values for the parameter of inactivation (k2/K, the second-order rate constant characterizing the acylation reaction) by some penicillins and cephalosporins were also determined and compared to those for the R39 dd-peptidase, to which PBP4a has the highest degree of sequence similarity (46% identity and 61% similarity) (8, 14).
MATERIALS AND METHODS
Recombinant DNA techniques, bacterial strains, plasmids, and growth conditions.
B. subtilis 168 1A1 (Genetic Stock Center) was grown at 37°C in Luria-Bertani (LB) medium, and its genomic DNA was extracted with the Qiagen genomic tip 20/G kit. (Westburg, Leusden, The Netherlands).
The cloning and expression experiments were performed with E. coli TG1 (Amersham Pharmacia Biotech, Roosendaal, The Netherlands), JM109, JM110 (Stratagene, Amsterdam Zuidoost, The Netherlands), Top 10, and LMG194 (Invitrogen, Groningen, The Netherlands) using pMK4 (22), a shuttle Bacillus-E. coli plasmid, or the pBAD/Myc-HisA (Invitrogen) vectors. The ampicillin resistance gene of the last plasmid was replaced by the kanamycin resistance gene using the following strategy. The vector (4.1 kb) was digested with BspHI, generating two fragments. The 3.1-kb fragment was purified from an agarose gel with the help of the Geneclean spin kit (Bio 101, Vista, Calif.), blunt-ended by filling with the Klenow DNA polymerase, and ligated to the 1.3-kb kanamycin resistance fragment isolated from the pYZ4 plasmid (3) by an NlaIV digestion, followed by the filling of the recessed 3′ ends. The resulting vector was named pBAD/Myc-HisAKr.
The expression experiments with B. subtilis were performed with partly protease-deficient strain 1A751 (from the Bacillus Genetic Stock Center) grown at 37°C in LB medium supplemented with 7 μg of chloramphenicol ml−1. Cultures of E. coli TG1, JM109, or JM110 transformed by the recombinant plasmid derived from pMK4 were grown at 30°C in LB medium supplemented with 10 μg of chloramphenicol ml−1. E. coli LMG194 cells transformed with the plasmid derived from pBAD/Myc-HisAKr were grown in minimal RM medium (Invitrogen) supplemented with 0.2% glucose and 50 μg/of kanamycin ml−1.
The oligonucleotides were purchased from Eurogentec (Seraing, Belgium) or from Amersham Pharmacia Biotech.
PCR amplifications were carried out with a DNA thermal cycler (Biometra-Trio Thermoblock; Westburg, Leusden, The Netherlands) using Vent DNA polymerase (New England Biolabs, Beverly, Mass.) or Platinum DNA polymerase (Gibco BRL, Merelbeke, Belgium). The PCR products were cloned into pUCBM20 (Boehringer, Mannheim, Germany), and their sequences were completely verified before insertion in the expression vectors.
Purification of PBP4a.
E. coli JM110 cells transformed with the expression vector derived from pMK4 were grown overnight at 30°C in 1 liter of LB medium. The cells were collected by centrifugation and resuspended in 30 mM Tris-HCl, pH 8.0, containing 27% (wt/vol) sucrose. The periplasmic content was isolated as described previously (9), dialyzed against 100 mM Tris-HCl, pH 7.8, and loaded onto a DEAE-cellulose column equilibrated with the same buffer. The PBP4a was eluted with a curvilinear gradient of NaCl (the mixing flask held a constant volume of 300 ml; the added solution was 175 mM NaCl in the same buffer). The fractions exhibiting the highest activity against Nα-acetyl-l-Lys-d-Ala-d-Ala (AcKAA) were concentrated by ultrafiltration on an Amicon PM-30 membrane and loaded onto a molecular-sieve Sephacryl S100 column equilibrated in 50 mM Tris-HCl, pH 7.8–300 mM KCl–10% ethyleneglycol. The latter step was repeated to obtain homogeneous PBP4a. The active fractions were collected and concentrated by centrifugation in a Millipore Ultrafree-15 filter.
Since the enzyme was sensitive to freeze-and-thaw sequences, it was stored at 4°C in the presence of 0.02% sodium azide or at −20°C in 100 mM Tris, pH 7.8, containing 20% ethyleneglycol or 30% glycerol.
Substrates.
AcKAA was a gift from UCB Bioproducts (Braine-l'Alleud, Belgium). Nα-Acetyl-l-Lys-d-Ala-d-thiolactate (AcKAThl) was a gift from Hoechst Marion Roussel (Romainville, France). The thiolesters benzoyl-Gly-thioglycolate (Bz-Gly-Thg) and benzoyl-d-Ala-thioglycolate (Bz-d-Ala-Thg) were obtained as previously described (1, 2). Benzylpenicillin, ampicillin, and cephaloridin were from Sigma (Bornem, Belgium). Cefuroxime was a gift from Glaxo Wellcome (Verona, Italy). β-Iodopenicillanate was obtained as described previously (6, 11). Ceftiofur (also named Exenel) was from Upjohn (Crawley, United Kingdom).
Kinetic measurements.
Hydrolysis of AcKAA was followed by monitoring the amount of d-alanine released using the d-amino acid oxidase method (12). Hydrolysis of Bz-Gly-Thg and Bz-d-Ala-Thg was monitored at 250 nm on a Uvikon 860 apparatus using a Δɛ value of −2,000 M−1 cm−1. kcat and Km values were derived from the complete time courses of hydrolysis of the substrate as described by De Meester et al. (7). Hydrolysis of AcKAThl was monitored at 412 nm in the presence of 1.2 mM 5,5′-dithiobis(2-nitrobenzoic acid) using a Δɛ value of 13,600 M−1 cm−1. kcat/Km was obtained from initial rates of hydrolysis at substrate concentrations [S] <Km. For the R39 enzyme, kinetic parameters for AcKAThl were derived from the complete hydrolysis time course. Unless otherwise stated, all experiments were performed at 30°C in 0.1 M HEPES pH 7.5.
The interaction between the enzyme (E) and the various β-lactam compounds (C) was analyzed on the basis of the following model:
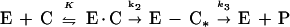 |
where E · C is the Henri-Michaelis complex, E-C* is the inactive acyl-enzyme, P is the degraded β-lactam, and k3 is a first-order rate constant characterizing the deacylation reaction. Hydrolysis of E-C* to restore the active enzyme (k3) was slow and was thus neglected. The apparent first-order acylation rate constant (ka) was measured at different β-lactam concentrations by recording the progressive decrease in the rate of hydrolysis of 100 μM AcKAThl. In all cases, the β-lactam concentration was at least 10 times greater than that of the enzyme. In the presence of the substrate, ka is given by
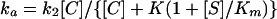 |
1 |
where [C] is the β-lactam concentration. In equation 1, Km refers to the substrate, and under our experimental conditions [S] was well below Km. In addition, the direct proportionality between ka and [C] showed that [C] was negligible compared to K(1 + [S]/Km). Therefore equation 1 could be simplified to give ka = k2[C]/K. Values for k2/K were obtained from the slope of the linear plots of ka versus [C].
The intrinsic fluorescence of the R39 dd-peptidase decreases as a result of its interaction with various cephalosporins. This property was used to determine k2/K for cephaloridine. Fluorescence quenching caused by cephaloridine was monitored at 20°C in 0.1 M Tris-HCl buffer, pH 7.8, containing 5 mM MgCl2 with the help of a Bio-Logic SFM-3 stopped-flow apparatus (excitation wavelength = 280 nm; emission wavelength > 320 nm), and k2/K was calculated from the slope of the linear plot of the apparent rate constant for acylation versus the β-lactam concentration, as previously described (13).
pH dependence of the activity.
Hydrolysis of AcKAA was measured at 37°C at different pHs ranging from 3 to 12. The universal buffer system described by Teorell and Stenhagen (23) was used at all pHs.
Thermal unfolding curves.
Thermal-denaturation melting curves were obtained as described by Vanhove et al. (26). The sample was heated at 1°C/min using a thermostatically regulated circulating water bath. Excitation wavelength was 284 nm and emission wavelengths were 335 and 330 nm for the R39 enzyme and PBP4a, respectively. Experiments were performed in 0.1 M Tris buffer, pH 7.8, using an SLM-AMINCO 8100 spectrofluorometer (Spectronic Instruments).
N-terminal sequencing.
The N-terminal sequence was determined with 160 pmol of protein with the help of an Applied Biosystems Procise sequencer (PE Biosystems, Nieuwerkerk a/d Ijssel, The Netherlands).
RESULTS
Expression of dacC from the B. subtilis-E. coli shuttle plasmid pMK4 and purification of PBP4a.
The portion of the gene encoding the mature PBP4a was cloned in phase with the signal sequence of the Bacillus licheniformis 749i β-lactamase under the control of the B. licheniformis blaP promoter (15, 18). The resulting vector was first cloned in E. coli TG1, isolated, and used to transform B. subtilis 1A751, which was grown in LB medium for 16 h at 37°C. Under these conditions, the expected secretion of PBP4a into the culture medium was not observed. An SDS-PAGE analysis of the culture supernatant also did not reveal the presence of the target protein after staining with Coomassie brilliant blue, and no hydrolysis of AcKAA was detected. The reasons of the latter failure to express PBP4a remain unclear, although protease degradation of the PBP4a and inefficient processing by the signal peptidase are possible explanations. This was not further investigated since the same recombinant plasmid led to periplasmic production of PBP4a in E. coli TG1. Among the other transformed E. coli strains, DH5α, JM109, and JM110, the last yielded the best production of PBP4a. Although not highly overproduced, PBP4a was highly toxic when secreted into the periplasm of E. coli, and the significant cell lysis observed resulted in the detection of 50% of the total PBP4a in the culture supernatant. Pedersen et al. (19) also mentioned the toxicity of this protein for E. coli, at least when produced in a soluble processed form.
With this expression system, a low periplasmic production of the TEM β-lactamase was also obtained, even in the absence of ampicillin in the culture medium. The proteins exhibit similar isoelectric pH values (pIs of 5.14 for PBP4a and 5.9 for the TEM enzyme), giving rise to overlapping elution profiles during the DEAE-cellulose purification step (see Materials and Methods). Other ion exchanger (DEAE- and Q-Sepharose) and hydrophobic-interaction (RESSOURCE ISO and RESSOURCE PHE; Pharmacia Biotech) columns did not allow the recovery of active PBP4a. The Ni2+-nitrilotriacetic acid agarose, on which the TEM enzyme is naturally retained, was also tested, but PBP4a could not be eluted in an active form.
Homogeneous, β-lactamase-free PBP4a was eventually obtained by loading the sample obtained after the DEAE-cellulose purification step onto a molecular-sieve Sephacryl S100 column. This step had to be repeated in order to obtain the desired purity. Active enzyme could only be recovered when 10% ethyleneglycol was added to the buffer, suggesting that the protein interacts with the matrix in the absence of ethyleneglycol.
PBP4a exhibits an apparent molecular mass of 59 kDa on SDS-PAGE; the theoretical mass deduced from the protein sequence was 49.7 kDa.
Cloning of the dacC gene into pBAD/Myc-HisAKr.
The dacC gene without its signal sequence was amplified from genomic DNA by PCR using the following primers: 5′ CATGCCATGGCTGAAAAACAAGATGCACTTTCCGG3′ (the underlined sequence corresponds to an NcoI restriction site and contains a translation initiation codon in phase with the triplet coding for the first residue of the mature PBP4a) and 5′CGGAATTCGGTTCGACAAAGCGTTATTACAG3′ (the underlined sequence corresponds to an EcoRI site, and the last 23 bases are complementary to nucleotides 1683 to 1705 in the sequence with accession no. Z34883 in the EMBL database). Therefore, the amplified fragment contains the sequence encoding the mature PBP4a with an additional N-terminal methionine as well as the endogenous stop codons and putative terminator sequence (25).
After verification of the sequence, the amplified fragment was inserted into expression plasmid pBAD/Myc-HisAKr between the NcoI and EcoRI restriction sites without fusion to the polyhistidine coding region since preliminary assays of purification by affinity chromatography on Ni2+-nitrilotriacetic acid agarose were unsuccessful.
Overproduction of PBP4a is detrimental to the viability of E. coli when the enzyme is secreted into the periplasm (our first expression system and reference 19). Therefore, a maximal repression of arabinose promoter pBAD was obtained by growing the transformed LMG194 cells in minimal RM medium containing 0.2% glucose. Different inducer concentrations were tested, and the best production was obtained with 0.2% arabinose and 4 h of culture at 37°C.
The cells from 25 ml of culture were treated as described previously (9), and the different cellular contents were analyzed by SDS-PAGE; as expected from the genetic construction, the cytoplasm contained a significantly overproduced soluble protein (data not shown). Moreover, the cytoplasmic extract exhibited a clear hydrolytic activity on AcKAA, which indicated a correct folding of PBP4a. The enzyme was purified as described above from 1 liter of culture after breaking the cells in cell disruption equipment (Constant Systems Ltd., Warwick, United Kingdom). The final yield was 8 mg of pure enzyme per liter of culture. The N-terminal sequence, AEKQDALS, confirmed the identity of the purified enzyme as PBP4a, with the loss of the first theoretical methionine residue. The enzyme produced with the help of the second expression system (pBAD/Myc-HisAKr) was used for the biochemical characterization.
Kinetic characterization of PBP4a. (i) dd-peptidase activity and interaction with thiolester substrates.
PBP4a clearly exhibits a dd-carboxypeptidase activity on AcKAA. Surprisingly, however, the enzyme is inhibited by an excess of substrate (data not shown) and the Km value could not be determined. By contrast, kcat/Km could be determined by measuring the initial rate of hydrolysis at low concentrations of AcKAA (Table 1). Thiolesters are also efficiently hydrolyzed by PBP4a (Table 1). Km values for AcKAThl, Bz-Gly-Thg, and Bz-d-Ala-Thg are, however, much higher that those reported for the R39 dd-peptidase.
TABLE 1.
Hydrolysis parameters (kcat/Km and Km) of the thiolesters for the R39 dd-peptidase and for B. subtilis PBP4aa
| Thiolester | R39 dd-peptidase
|
B. subtilis PBP4a
|
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | |
| AcKAA | 18c | 280c | 64,000c | N.D.e | N.D. | 6,700 |
| AcKAThl | 5.4 | 42 | 130,000 | N.D. | >400b | 35,000 |
| Bz-Gly-Thg | 0.3d | 30d | 10,000d | 2 | 470 | 4,300 |
| Bz-d-Ala-Thg | 5.6d | 15d | 336,000d | 23 | 380 | 61,000 |
| Bz-d-Ala-Thg + 10 mM d-Ala | N.D. | N.D. | N.D. | 28 | 430 | 65,000 |
| Bz-d-Ala-Thg + 50 mM d-Ala | N.D. | N.D. | N.D. | 42 | 540 | 78,000 |
(ii) Transpeptidase activity.
The kinetic parameters for the hydrolysis of Bz-d-Ala-Thg in the presence of 10 and 50 mM d-alanine are included in Table 1. If d-alanine can be used as an acceptor in a transpeptidation reaction, and assuming that the deacylation step is rate limiting, increases in both kcat and Km are expected. This was indeed observed, although the increases in kcat and Km are relatively modest. In addition, similar to what was reported for the R39 enzyme (27), a slight but significant increase in kcat/Km was also measured.
(iii) Inactivation by β-lactam antibiotics.
Table 2 shows values of k2/K determined with various β-lactam compounds. Despite a very high similarity to the R39 dd-peptidase, which is extremely sensitive to β-lactams, B. subtilis PBP4a is significantly more resistant to these compounds with the exception of cephaloridine, by which it is efficiently inactivated.
TABLE 2.
Comparison of the k2/K values for the B. subtilis PBP4a and R39 dd-peptidase with various penicillins and cephalosporins
| β-lactam |
k2/K (M−1 s−1)a for:
|
|
|---|---|---|
| B. subtilis PBP4a | R39 dd-peptidase | |
| Benzylpenicillin | 1,400 | 300,000b |
| Ampicillin | 4,800 | 70,000b |
| Cephaloridine | 46,000 | 270,000 |
| Cefuroxime | 740 | 3,900b |
| Ceftiofur | 860 | 45,000c |
| β-Iodopenicillanate | 160 | 7,600b |
Standard deviations did not exceed 10%.
From reference 10.
Sophie Lepage, personal communication.
pH dependence of the activity of PBP4a.
The rate of hydrolysis of AcKAA was measured at 37°C between pH 3 and 12. The activity of PBP4a was undetectable between pH 3 and 5.5. From pH 6 to 12, however, the activity increased linearly (not shown). Despite a maximum of activity at pH 12, all the kinetic measurements were performed at physiological pH values.
Thermostability of PBP4a.
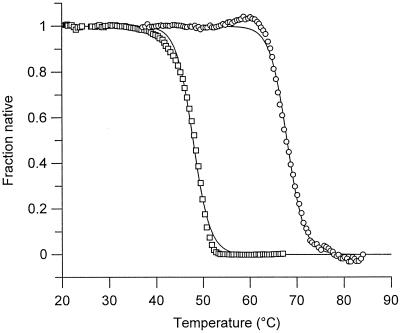
Thermal unfolding curves for PBP4a and the R39 dd-peptidase are shown in Fig. 1. Activity measurements at the end of the transition indicated that unfolding was not reversible, and therefore only apparent melting temperatures (Tm) could be computed. Apparent Tms were 47.9 and 67.9°C for the R39 and PBP4a dd-peptidases, respectively, highlighting a very significantly higher stability for the latter enzyme.
FIG. 1.
Thermal unfolding curves for the R39 dd-peptidase (squares) and PBP4a from B. subtilis (circles). Apparent Tms deduced from the curves are 47.9 and 67.9°C for the R39 and the PBP4a enzymes, respectively.
DISCUSSION
PBP4a from B. subtilis, a new member of the class C low-molecular-weight PBP family, was overexpressed in a soluble and active form in E. coli. The first expression system using the B. subtilis-E. coli shuttle vector directed the PBP4a to the periplasm of E. coli. This resulted in significant lysis of the different strains tested, indicating a high toxicity of the protein. By contrast, the expression of PBP4a from the pBAD/Myc-HisAKr plasmid resulted in cytoplasmic production without any observable cell lysis in E. coli LMG194.
The purification of PBP4a was particularly difficult since the protein was rapidly inactivated on several ion exchangers and was not eluted from molecular-sieve columns in the absence of ethyleneglycol. However, PBP4a was successfully purified to homogeneity by chromatography on a weak ion exchanger (DEAE-cellulose) followed by filtration on Sephacryl S100 in a buffer supplemented with 10% ethyleneglycol. The final yield was 8 mg per liter of culture.
In vitro, the protein clearly exhibited dd-carboxypeptidase and thiolesterase activities on short compounds exhibiting d-Ala-d-Ala and d-Ala-thiolactate or d-Ala-thioglycolate C termini. Interestingly, the enzyme is inhibited by an excess of AcKAA, but inhibition occurs at relatively high substrate concentrations (>10 mM), and it is not clear whether this observation has any physiological relevance. Kinetic parameters for Bz-d-Ala-Thg measured in the presence of d-alanine indicate that the PBP4a can use the latter as an acceptor in a transpeptidation reaction. By contrast to the R39 dd-peptidase, which is very sensitive to inactivation by β-lactams, the B. subtilis PBP4a is only weakly inhibited by many of these compounds, although the two proteins exhibit a high degree of homology (61% similarity and 46% strict identity). This low affinity of PBP4a for penicillin explains the observation of Pedersen et al. (19), who failed to visualize the protein in membranes of B. subtilis incubated with fluorescein-hexanoic-6-aminopenicillanic acid.
We also show here that the B. subtilis PBP4a is much more thermostable than the related R39 enzyme (ΔTm = 20°C). Many analyses to relate the protein conformational characteristics to thermostability have been carried out (for a review, see reference 21); however, no unambiguous rules can be established to predict how substituted amino acids improve the thermostability of a protein, particularly in the absence of three-dimensional structures. The prolyl contents and the percentages of hydrophobic residues are quite similar for the two enzymes. The only significant difference is in the percentage of positively charged residues (mainly due to the number of lysine residues): 8.4% in PBP4a versus 1% in the R39 dd-peptidase. The percentage of negatively charged residues being almost similar in both proteins, there is an unfavorable balance of charged amino acids in the R39 enzyme (ratio of positive to negative charges: 0.31 versus 0.79 for PBP4a), resulting in a particularly low isoelectric pH value for the R39 enzyme (<3.6). As pointed out by Vanhove et al. (26), this unfavorable balance might contribute to the particularly low stability of the R39 dd-peptidase.
In B. subtilis, PBP4a expression is initiated at the end of the stationary phase. The data from Pedersen et al. (19) and Popham et al. (20) indicate that the PBP4a product has no significant role in spore peptidoglycan production. Data for insertional dacC mutants indicate also that this gene is dispensable under normal growth conditions.
Although a dd-carboxypeptidase activity was clearly assigned in vitro to the PBP4a product, the physiological role of the whole protein and the function of the additional domain remain to be determined.
ACKNOWLEDGMENTS
This work was supported by the Belgian program on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Services fédéraux des affaires scientifiques, techniques et culturelles (PAI no. P4/03). C.D. and M.V. are respectively Chercheur qualifié and Chargé de Recherche of the Fonds National de la Recherche Scientifique (Brussels, Belgium).
REFERENCES
- 1.Adam M, Damblon C, Plaitin B, Christiaens L, Frère J M. Chromogenic depsipeptide substrates for β-lactamases and penicillin-sensitive DD-peptidases. Biochem J. 1990;270:525–529. doi: 10.1042/bj2700525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam M, Damblon C, Jamin M, Zorzi W, Dusart J, Galleni M, El Kharroubi A, Piras G, Spratt B, Keck W, Coyette J, Ghuysen J M, Nguyen-Distèche M, Frère J M. Acyltransferase activities of the high-molecular-mass essential penicillin-binding proteins. Biochem J. 1991;279:601–604. doi: 10.1042/bj2790601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broome-Smith J K, Tadayyon M, Zhang Y. β-Lactamase as a probe of membrane protein assembly and protein export. Mol Microbiol. 1990;4:1637–1644. doi: 10.1111/j.1365-2958.1990.tb00540.x. [DOI] [PubMed] [Google Scholar]
- 4.Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 5.Damblon C, Zhao G H, Jamin M, Ledent P, Dubus A, Vanhove M, Raquet X, Christiaens L, Frère J M. Breakdown of the stereospecificity of DD-peptidases and β-lactamases with thiolester substrates. Biochem J. 1995;309:431–436. doi: 10.1042/bj3090431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Meester F, Frère J M, Piette J L, Vanderhaeghe H. Synthesis of (β-methyl 3H)-6-β-iodopenicillanic acid. J Label Compd Radiopharm. 1985;22:415–425. [Google Scholar]
- 7.De Meester F, Joris B, Reckinger G, Bellefroid-Bourguignon C, Frère J M, Waley S G. Automated analysis of enzyme inactivation phenomena. Application to β-lactamases and DD-peptidases. Biochem Pharmacol. 1987;36:2393–2403. doi: 10.1016/0006-2952(87)90609-5. [DOI] [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraipont C, Adam M, Nguyen-Distèche M, Keck W, van Beeumen J, Ayala J A, Granier B, Hara H, Ghuysen J M. Engineering and overexpression of periplasmic forms of the penicillin-binding protein 3 of Escherichia coli. Biochem J. 1994;298:189–195. doi: 10.1042/bj2980189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frère J M, Joris B. Penicillin-sensitive enzymes in peptidoglycan biosynthesis. Crit Rev Microbiol. 1985;11:299–396. doi: 10.3109/10408418409105906. [DOI] [PubMed] [Google Scholar]
- 11.Frère J M, Dormans C, Duyckaerts C, De Graeve J. Interaction of β-iodopenicillanate with the β-lactamases of Streptomyces albus G and Actinomadura R39. Biochem J. 1982;207:437–444. doi: 10.1042/bj2070437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frère J M, Leyh-Bouille M, Ghuysen J M, Nieto M, Perkins H R. Exocellular DD-peptidases-transpeptidases from Streptomyces. Methods Enzymol. 1976;45:610–636. doi: 10.1016/s0076-6879(76)45054-1. [DOI] [PubMed] [Google Scholar]
- 13.Fuad N, Frère J M, Ghuysen J M, Duez C. Mode of interaction between β-lactam antibiotics and the exocellular DD-carboxypeptidase from Streptomyces R39. Biochem J. 1976;155:623–629. doi: 10.1042/bj1550623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granier B, Duez C, Lepage S, Englebert S, Dusart J, Dideberg O, van Beeumen J, Frère J M, Ghuysen J M. Primary and predicted secondary structures of the Actinomadura R39 extracellular DD-peptidase, a penicillin-binding protein (PBP) related to the Escherichia coli PBP4. Biochem J. 1992;282:781–788. doi: 10.1042/bj2820781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi T, Zhu Y F, Nicholls N J, Lampen J O. A second regulatory gene, blaRI, encoding a potential penicillin-binding protein required for induction of β-lactamase in Bacillus licheniformis. J Bacteriol. 1987;169:3873–3878. doi: 10.1128/jb.169.9.3873-3878.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunst F, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 17.Mottl H, Terpstra P, Keck W. Penicillin-binding protein 4 of Escherichia coli shows a novel type of primary structure among penicillin-interacting proteins. FEMS Microbiol Lett. 1991;78:213–220. doi: 10.1016/0378-1097(91)90160-c. [DOI] [PubMed] [Google Scholar]
- 18.Neugebauer K, Sprengel R, Schaller H. Penicillinase from Bacillus licheniformis: nucleotide sequence of the gene and implications for the biosynthesis of a secretory protein in a gram-positive bacterium. Nucleic Acids Res. 1981;9:2577–2588. doi: 10.1093/nar/9.11.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedersen L B, Murray T, Popham D L, Setlow P. Characterization of dacC, which encodes a low-molecular-weight penicillin-binding protein in Bacillus subtilis. J Bacteriol. 1998;180:4967–4973. doi: 10.1128/jb.180.18.4967-4973.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popham D L, Gilmore M E, Setlow P. Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties. J Bacteriol. 1999;181:126–132. doi: 10.1128/jb.181.1.126-132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Querol E, Perez-Pons J A, Mozo-Villarias A. Analysis of protein conformational characteristics related to thermostability. Protein Eng. 1996;9:265–271. doi: 10.1093/protein/9.3.265. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan M A, Yasbin R E, Young F E. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
- 23.Teorell T, Stenhagen E. Ein universalpuffer für den pH-Bereich 2,0 bis 12,0. Biochem Z. 1938;299:416–419. [Google Scholar]
- 24.Thunnissen M M G M, Fusetti F, de Boer B, Dijkstra B W. Purification, crystallization and preliminary X-ray analysis of penicillin-binding protein 4 from Escherichia coli, a protein related to class A β-lactamases. J Mol Biol. 1995;247:149–153. doi: 10.1006/jmbi.1994.0128. [DOI] [PubMed] [Google Scholar]
- 25.Tognoni A, Franchi E, Magistrelli C, Colombo E, Cosmina P, Grandi G. A putative new peptide synthase operon in Bacillus subtilis: partial characterization. Microbiology. 1995;141:645–648. doi: 10.1099/13500872-141-3-645. [DOI] [PubMed] [Google Scholar]
- 26.Vanhove M, Houba S, Lamotte-Brasseur J, Frère J M. Probing the determinants of protein stability: comparison of class A β-lactamases. Biochem J. 1995;308:859–864. doi: 10.1042/bj3080859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao G, Duez C, Lepage S, Forceille C, Rhazi N, Klein D, Ghuysen J M, Frère J M. Site-directed mutagenesis of the Actinomadura R39 DD-peptidase. Biochem J. 1998;327:377–381. doi: 10.1042/bj3270377. [DOI] [PMC free article] [PubMed] [Google Scholar]