Abstract
Bacterial spores are protected from the environment by a proteinaceous coat and a layer of specialized peptidoglycan called the cortex. In Bacillus subtilis, the attachment of the coat to the spore surface and the synthesis of the cortex both depend on the spore protein SpoIVA. To identify functionally important amino acids of SpoIVA, we generated and characterized strains bearing random point mutations of spoIVA that result in defects in coat and cortex formation. One mutant resembles the null mutant, as sporulating cells of this strain lack the cortex and the coat forms a swirl in the surrounding cytoplasm instead of a shell around the spore. We identified a second class of six mutants with a partial defect in spore assembly. In sporulating cells of these strains, we frequently observed swirls of mislocalized coat in addition to a coat surrounding the spore, in the same cell. Using immunofluorescence microscopy, we found that in two of these mutants, SpoIVA fails to localize to the spore, whereas in the remaining strains, localization is largely normal. These mutations identify amino acids involved in targeting of SpoIVA to the spore and in attachment of the coat. We also isolated a large set of mutants producing spores that are unable to maintain the dehydrated state. Analysis of one mutant in this class suggests that spores of this strain accumulate reduced levels of peptidoglycan with an altered structure.
All cells have the capacity to direct proteins to discrete subcellular locations, an activity that is essential for the formation of complex cellular structures. In bacteria, correct placement of the cell division apparatus at the midcell and the polar location of the flagellum, in organisms such as Caulobacter crescentus (7, 16, 32), rely on the accurate targeting of proteins to specific cellular destinations. Subcellular localization also plays a readily observable role in the formation of the bacterial endospore, a dormant cell type produced in response to starvation by a variety of species of bacilli and clostridia (10, 33, 37). At an intermediate point in spore formation the developing spore, known as the forespore, is a double-membraned protoplast that resides within a larger cell called the mother cell. A specialized peptidoglycan, called the cortex, is then deposited between these membranes, and the entire spore is surrounded by a multilayered shell, called the coat, that protects the spore by blocking entry of large molecules (Fig. 1A). The coat is formed by the deposition of coat proteins, which are synthesized in the mother cell, onto the forespore surface (6). A critical outstanding question is how the assembly of the coat and other spore structures is directed to occur exclusively at the developing spore. The fully mature spore is then released into the environment by mother cell lysis. When environmental conditions once again become favorable, the spore germinates, during which time the interior of the spore rehydrates and metabolism initiates (11, 22). This is followed by outgrowth, during which the spore fully sheds the cortex and coat and begins vegetative growth.
FIG. 1.
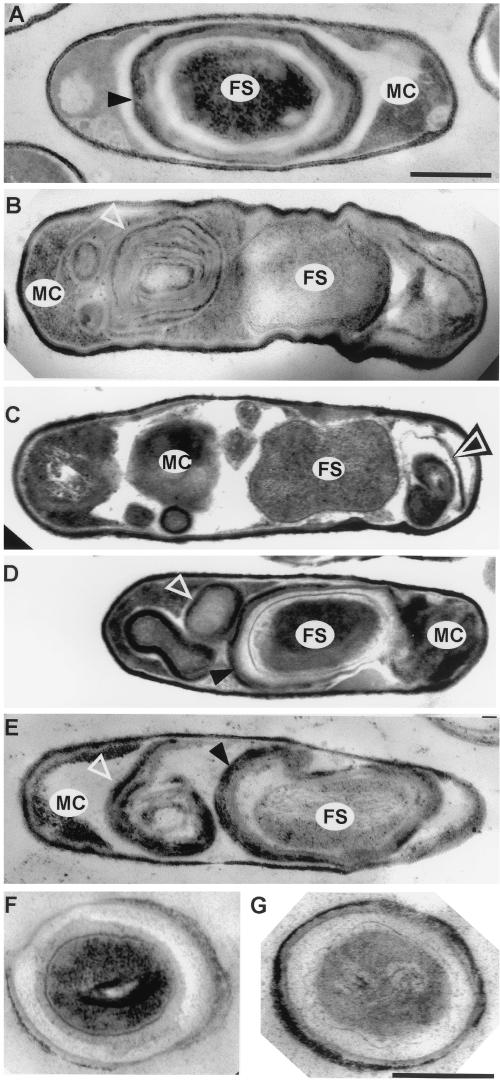
Electron microscopic examination of wild-type and spoIVA mutant sporulating cells. Thin-section electron micrographs of cells after 10 h of sporulation are shown. (A) PY79 (wild type); (B) AD18 (spoIVAΔ::neo); (C) FC336 (L393P); (D and E) FC329 (I400P, E418V, I457R); (F and G) FC297 (L59P) rehydrated spores. FS, forespore; MC, mother cell. Open triangles indicate swirls of coat; arrowheads indicate forespore-associated coat. Bars in panels A (for panels A to E) and G (for panels F and G), 500 nm.
Of the known sporulation genes, spoIVA has a particularly central and unique role in directing the assembly of spore protective structures. In a spoIVA null mutant, the forespore lacks a cortex and the coat does not surround the forespore; instead it appears as swirls in the cytoplasm of the mother cell (Fig. 1B) (23). SpoIVA is produced in the mother cell immediately after the septum appears (27, 34), at which time it localizes at or very near the forespore surface (8, 24, 26). Apparently, SpoIVA marks the outer surface of the forespore as the site of future coat protein deposition and marks the region between the membranes as the site of cortex formation. The mechanism that directs SpoIVA to the forespore surface is unknown, but it has been shown that SpoIVA localization requires the mother cell protein SpoVM as well as at least one other factor (26). Sequences within the C-terminal five amino acids of SpoIVA direct localization, and SpoIVA assembly further involves sequences elsewhere in the protein that mediate direct or indirect interactions between SpoIVA molecules.
To identify amino acids within SpoIVA with roles in forespore localization, cortex formation, and attachment of the coat, we generated and analyzed a set of random point mutants of spoIVA. In this work, we identify residues with roles in these functions.
MATERIALS AND METHODS
General methods, recombinant DNA procedures, western blot analysis, and microscopy.
Strains, plasmids, and primers are listed in Tables 1 and 2. Recombinant DNA procedures were carried out as described previously (28) unless otherwise indicated. Manipulations of Bacillus subtilis, β-galactosidase assays, and sporulation by exhaustion in Difco Sporulation Medium or by resuspension (for immunofluorescence microscopy experiments) were performed as described previously (5). Measurement of spore peptidoglycan (20) and determinations of glucose dehydrogenase and dipicolinic acid levels and heat resistance (5) were performed as described previously. We used anti-CotA (S. Little and A. Driks, unpublished data) and anti-CotE (4) antibodies at a dilution of 1:10,000 in Western blots. To generate anti-CotD antibodies, we first used primers OL85 and OL86, and DNA from strain PY79 as a template, in a PCR to generate a DNA fragment harboring cotD. We restriction digested the product and plasmid pET14b (Novagen) with XhoI and BamHI, joined them by ligation, and used the resulting plasmid to transform Escherichia coli strain BL21(DE3). We induced expression, purified the overproduced product, and injected the material into rabbits as described previously (36). We used the resulting antiserum at a dilution of 1:10,000.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or description | Reference and/or source |
|---|---|---|
| B. subtilis | ||
| PY79 | Wild type | 39 |
| AD18 | spoIVAΔ::neo | 8 |
| TB1 | cotEΔ::cat::spec | 4 |
| FC74 (I210A, I367V, I383R) | spoIVAΔ::neo amyE::spoIVA74 (cat) | This study |
| FC77 (L59P, H256P) | spoIVAΔ::neo amyE::spoIVA77 (cat) | This study |
| FC114 (C98S, V283N) | spoIVAΔ::neo amyE::spoIVA114 (cat) | This study |
| FC133 | spoIVAΔ::neo amyE::spoIVA133 (cat) | This study |
| FC297 (L59P) | spoIVAΔ::neo amyE::spoIVA297 (cat) | This study |
| FC329 (I400P, E418V, I457R) | spoIVAΔ::neo amyE::spoIVA329 (cat) | This study |
| FC333 (I210M, I367V, I383R) | spoIVAΔ::neo amyE::spoIVA333 (cat) | This study |
| FC334 (V241G) | spoIVAΔ::neo amyE::spoIVA334 (cat) | This study |
| FC335 (G416R) | spoIVAΔ::neo amyE::spoIVA335 (cat) | This study |
| FC336 (L393P) | spoIVAΔ::neo amyE::spoIVA336 (cat) | This study |
| FC341 (M69L, R230S) | spoIVAΔ::neo amyE::spoIVA341 (cat) | This study |
| FC351 (V453G) | spoIVAΔ::neo amyE::spoIVA351 (cat) | This study |
| FC533 | cotEΔ::cat::spec gerE36 | AD17 (36) × DNA TB1 |
| FC535 | cwlD::cat spoIVAΔ::neo | AD18 × DNA PS2307 (25) |
| FC549 | cotEΔ::cat::spec gerE36 spoIVAΔ::neo amyE::spoIVA297 (cat) | (AD17 × DNA TB1) × DNA FC297 (L59P) |
| FC618 | cwlD::cat::spec spoIVAΔ::neo amyE::spoIVA297 (cat) | (FC535 × pJL62) × DNA FC297 (L59P) |
| FC713 (M69L) | spoIVAΔ::neo amyE::spoIVA713 (cat) | This study |
| FC714 (R230S) | spoIVAΔ::neo amyE::spoIVA714 (cat) | This study |
| FC715 | α− β− pUB110 (cat) gpr+kan spoIVAΔ::neo | PS578 (29) × DNA FC297 (L59P) (selection for Neor) |
| FC729 | spoIVAΔ::neo amyE::spoIVA297 α− β− pUB110 (cat) gpr+kan | FC715 × DNA FC297 (L59P) (selection for Cmr) |
| E. coli | ||
| BL21(DE3) | Overexpression host | Laboratory stock |
| DH5α | Cloning host | Laboratory stock |
| Plasmids | ||
| pJL62 | Converts cat to spec | 15 |
| pDG364 | Permits recombination into the chromosome at amyE | 12 |
| pFC1 | pDG364::spoIVA | This study |
TABLE 2.
Primers
| Primer | Sequencea | Enzymeb | Positionc |
|---|---|---|---|
| OL12 | AAAATTCTCCAGTCTTCACATC | NAd | −117 |
| OL13 | CGACTAAGAAAATGCCGTC | NA | +5226 |
| OL81 | TTTTAAGCTTCCCGGGACTGAAATC | HindIII | +745 |
| OL82 | TTTTTGGATCCAAAGAGGTCTACCGG | BamHI | +2645 |
| OL85 | AAAAACTCGAGATGCATCACTGCAGA | XhoI | +1 |
| OL86 | AAAAAGGATCCTTAGTAGTCGCAGCAAGG | BamHI | +210 |
The restriction endonuclease site in each oligonucleotide is underlined.
Enzyme that cuts the sequence.
Position bound by the first nucleotide of each primer. The translational start site of cotD and the first nucleotide of the amyE open reading frame in pFC1 are designated +1.
NA, not applicable.
We performed electron microscopy (19) and immunofluorescence microscopy (4) as previously, with the following modifications for immunofluorescence microscopy. We used anti-SpoIVA antibodies at a dilution of 1:200, goat anti-rabbit fluorescein isothiocyanate-conjugated whole immunoglobulin G (Sigma) at a dilution of 1:200, and Hoechst 33342 (Sigma) (5 μg/ml) to counterstain the chromosomes. Data were collected on a Sensys 1400 cryocooled charge-coupled device camera and were not subjected to deconvolution analysis. We performed image processing using Adobe Photoshop.
Generation of spoIVA mutants.
To generate a DNA fragment to serve as the template for PCR mutagenesis of spoIVA, we first digested the amyE replacement vector pDG364 with HindIII and BamHI and purified the larger fragment by agarose gel electrophoresis. We then used PCR to amplify a fragment containing spoIVA and its known regulatory sequences (27), using oligonucleotides OL81 and OL82 and DNA of strain PY79 as the template. We next purified the product by agarose gel electrophoresis, digested it with HindIII and BamHI, and ligated it with the restriction-digested pDG364 to generate pFC1. Finally, we used oligonucleotide OL12, which anneals to the 5′ end of the amyE gene, and oligonucleotide OL13, which anneals to the 3′ end of the amyE gene (downstream of spoIVA in pFC1) in a PCR with pFC1 as the template to generate a 5-kb product containing (in order) the 5′ portion of the amyE gene, a potentially mutant allele of spoIVA, cat, and the 3′ portion of the amyE gene. We pooled these PCR products and used them to transform strain AD18 (spoIVAΔ::neo). We then selected for Cmr, amylase-negative colonies, patched these onto Difco Sporulation Medium agar plates, and incubated for 3 days at 37°C. We identified spoIVA mutants by using the tetrazolium overlay assay (5) to screen for germination defects, which are an expected consequence of most mutations affecting coat or cortex formation and therefore of most mutations in spoIVA (1, 4, 6).
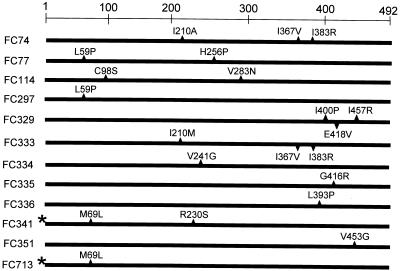
PCR mutagenesis resulted in several multiply mutant versions of spoIVA (Fig. 2). Therefore, we used recombinant DNA techniques to generate alleles bearing only one or two of each of the nucleotide changes present in the multiply mutant alleles spoIVA74 (I210A, I367V, and I383R), spoIVA77 (L59P and, H256P), spoIVA114 (C98S and V283N), and spoIVA341 (M69L and R230S). First, we generated two constructs bearing only one or the other of the mutations in spoIVA77. We used PCR with oligonucleotides OL81 and OL82 and chromosomal DNA from strain FC77 (with L59P and H256P mutations) to generate a DNA fragment containing spoIVA77. After gel purification, we digested this fragment with BamHI and HindIII and ligated it with similarly digested pUC19 to generate pFC57. Concurrently, we used PCR, with primers OL81 and OL82 and chromosomal DNA of strain PY79 as the template, to create a DNA fragment containing wild-type spoIVA. We digested this DNA and pUC19, as described above, and joined the products by ligation to make plasmid pFC59. We then linearized pFC59 and pFC57 with ScaI, which digests each within the bla gene. The products were gel purified and digested with SphI, which cuts once between the two mutations in spoIVA77. The larger fragment of pFC57 was ligated to the smaller fragment of pFC59 to generate pFC63 (bearing spoIVA297). We also joined the smaller fragment of pFC57 to the larger fragment of pFC59 to generate pFC62. Finally, we digested pFC63 and pFC62 with BamHI and HindIII, purified the smaller DNA fragments, and ligated them to similarly digested pDG364 to generate pFC68 and pFC75, respectively. We linearized pFC68 and pFC75 with XhoI and used them to transform strain AD18, creating strains FC297 (L59P) and FC316 (H256P), respectively. We used essentially the same procedure to generate separate strains containing spoIVAC98S or spoIVAV283N (from spoIVA114 [C98S, V283N]) and strains bearing spoIVAI210A and both spoIVAI367V and spoIVAI383R (from spoIVA74 [I210A, I367V, I383R]). We also generated strains (FC713 [M69L] and FC714 [R230S]) bearing spoIVAM69L (including the mutation in the untranslated region) and spoIVAR230S, respectively (both from spoIVA341). To generate merodiploid strains bearing both a wild-type and a mutant allele of spoIVA, we transformed strain PY79 with DNAs from strains bearing the appropriate spoIVA mutant alleles.
FIG. 2.
Amino acids altered in spoIVA mutants. The positions and changes in amino acids are indicated. The asterisks by FC341 (M69L, R230S) and FC713 (M69L) indicate the presence of a nucleotide change in the untranslated region of the mRNAs.
Western blot analysis of SpoIVA steady-state levels.
We collected 1-ml samples from a liquid culture 5 h after the initiation of sporulation, centrifuged the aliquots and stored the pellets at −20°C. For sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis, we resuspended the pellets in 30 μl of 50 mM EDTA–0.1 M NaCl–1 mg of lysozyme per ml and incubated the samples at 37°C for 15 minutes. We determined protein concentrations using the bicinchoninic acid kit (Pierce) and then added a volume of lysate containing 1 mg of protein to 8 μl of 4× SDS loading buffer (28) and 4 μl of 1 M dithiothieitol, vortexed, and boiled for 10 min. Finally, we centrifuged the samples and loaded them onto an SDS–10% polyacrylamide gel. Western blot analysis and comparison of relative levels of protein were performed as described previously (35). Relative levels of SpoIVA were calculated from the means of three quantitative Western blot experiments. Standard errors of these means varied from 0.09 to 0.006. We used anti-SpoIVA antibodies (4) at a dilution of 1:10,000.
Analysis of spore resistance and outgrowth.
First, we centrifuged 5-ml aliquots of culture, which were harvested 48 h after the initiation of sporulation. To measure heat resistance, we resuspended each pellet in 5 ml of 1 M Tris-HCl (pH 7.5), incubated at 80°C for 30 min, and determined the numbers of viable bacteria by plating on solid Luria-Bertani (LB) medium. To measure outgrowth, we resuspended each pellet in 5 ml of LB medium, incubated the samples with shaking at 37°C, and counted the cells under light microscopic examination at 15-min intervals.
RESULTS
Identification of novel alleles of spoIVA.
To identify functionally important amino acids within SpoIVA, we used PCR to generate DNA fragments bearing mutant versions of spoIVA and placed these genes at the amyE locus of a strain bearing a null allele of spoIVA (see Materials and Methods). To identify spoIVA mutants, we sporulated about 4,000 candidates and tested the resulting spores for their capacity to germinate using the tetrazolium overlay assay (5). Because germination is impaired by most defects in coat or cortex formation, this assay should readily detect even minor deficiencies in SpoIVA function (1, 4, 6). We identified 48 candidates, of which all but 18 produced a version of SpoIVA of the expected mass, as determined by Western blot analysis using anti-SpoIVA antibodies. Of the remaining 30 mutants, 2 assemble a largely normal spore but possess a subtle defect in the coat and will be described in a separate report. One appears to be completely defective in localization of SpoIVA to the forespore, 6 are significantly defective in cortex and coat formation, and 21 appear to spontaneously rehydrate (see below). Here, we analyze 11 mutants that are representative of the last three groups (Fig. 2).
To learn if steady-state levels of SpoIVA are altered in these 11 strains, we determined the relative amounts of SpoIVA at hourly intervals between the second and eighth hours of sporulation by quantitative Western blot analysis using an anti-SpoIVA antibody. The levels of SpoIVA in these mutants was between 0.7- and 1.1-fold that of the wild type (with the exception of strain FC133 [see below]) and therefore are unlikely to be the major cause of the mutant phenotypes (data not shown).
A spoIVA null mutation significantly reduces the level of processing of inactive pro-ςK to active ςK (the major postengulfment, mother cell ς factor [14]) (18) and lowers the expression of at least one ςK-dependent gene (cotD) by about 78% (40). It was therefore possible that mother cell gene expression is significantly altered and that this, in turn, contributes to the mutant phenotypes. Therefore, we introduced cotD-lacZ (40) into each spoIVA mutant strain and measured β-galactosidase activity during sporulation. We found that the activity was between 30 and 50% of the wild-type level (data not shown), a reduction in expression that is unlikely to be a major cause of the mutant phenotypes (see below).
Mutations that alter cortex and coat formation.
We identified seven strains (FC329 [I400P, E418V, I457R], FC333 [I210M, I367V, I383R], FC334 [V241G], FC335 [G416R], FC336 [L393P], FC341 [M69L, R230S], and FC351 [V453G]) (Fig. 2) which entered sporulation but did not produce refractile endospores and lysed after 12 h of sporulation, as judged by phase-contrast light microscopy (data not shown). At h 6 of sporulation in a wild-type culture, almost all cells possessed refractile forespores. In contrast, the forespores in the mutant cells appeared gray, indicating that dehydration was incomplete or had entirely failed to take place (17). Consistent with the sporulation defects revealed by light microscopic analysis, all of these strains resembled the null mutant in sensitivity to heat and lysozyme (data not shown), indicating major defects in the cortex and coat, respectively (9, 21). Six mutants have changes solely in the open reading frames. In addition to changes in the coding sequence, in FC341 (M69L, R230S), a guanine is changed to adenine at nucleotide +47 relative to the start site of transcription specified by promoter P2 (27).
A mutation that prevents localization of SpoIVA.
By electron microscopy, we observed that after 10 h of sporulation, cells from strain FC336 (L393P) possessed swirls of coat in the mother cell cytoplasm and did not possess a cortex (Fig. 1C), similar to spoIVA null mutant cells (Fig. 1B). To determine the location of SpoIVA in these cells, we performed immunofluorescence microscopy, using anti-SpoIVA antibodies, on cells harvested at h 3 and 5 of sporulation. In wild-type cells, SpoIVA appears as a ring or caps around the forespore (24, 26) (Fig. 3A and data not shown). We detected little or no fluorescence in cells bearing a null mutation in spoIVA (spoIVAΔ::neo, in strain AD18) (data not shown). In FC336 (L393P) cells, the signal corresponding to SpoIVA was dispersed throughout the mother cell cytoplasm (Fig. 3B and data not shown), indicating that SpoIVA was largely unassociated with the forespore.
FIG. 3.
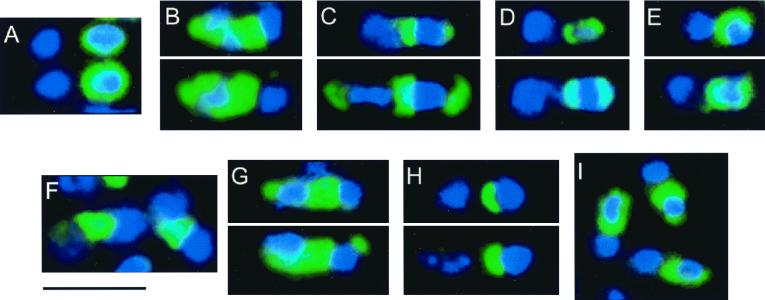
Immunofluorescence microscopic localization of SpoIVA. Cells were prepared for immunofluorescence microscopy at h 5 of sporulation and treated with anti-SpoIVA antibodies (green) and a DNA counterstain (blue). In all cells in which localization was evident, the pattern of fluorescence was similar, with the exception of strain FC713 (panel I; see text). In each panel, the forespore is oriented towards the right. (A) PY79 (wild type); (B) FC336 (L393P); (C) FC333 (I210M, I367V, I383R); (D) FC334 (V241G); (E) FC335 (G416R); (F) FC329 (I400P, E418V, I457R); (G) FC351 (V453G); (H) FC341 (M69L, R230S); (I) FC713 (M69L). Bar, 3 μm.
Mutations with a novel effect on coat assembly.
By electron microscopy, the majority of cells in sporulating cultures of strains FC329 (I400P, E418V, I457R), FC333 (I210M, I367V, I383R), FC334 (V241G), FC335 (G416R), FC341 (M69L, R230S), and FC351 (V453G) appeared wild type after 10 h of growth, while a small percentage (about 5%) were indistinguishable from a spoIVA null mutant (data not shown). Strikingly, in each strain, we also identified a significant number of cells (ranging from about 5 to 30%, depending on the sample) that possessed both a coat around the forespore and swirls of coat, unattached to the forespore, in the mother cell cytoplasm (Fig. 1D and E and data not shown). We detected at least some cortex-like material in the forespores of most of these cells. We never saw cells with an apparently normal cortex but no coat. FC333 (I210M, I367V, I383R) has two amino acid changes in common with FC74 (I210A, I367V, I383R) (Fig. 2). As discussed below, a strain bearing only the alterations in amino acids 367 and 383 is phenotypically wild type. Therefore, the change in amino acid 210 in FC333 (I210M, I367V, I383R) is required for the mutant phenotype and possibly is solely responsible.
The changes in SpoIVA in strains FC329 (I400P, E418V, I457R), FC333 (I210M, 1367V, I383R), FC334 (V241G), FC335 (G416R), and FC351 (V453G) are within the C-terminal 282 residues. In contrast, strain FC341 (M69L, R230S) possesses an N-terminal change, in addition to a more C-terminal alteration and a nucleotide change in the untranslated region of the message. To learn if all of these changes are required to produce the mutant phenotype, we built a strain bearing only the change at amino acid 230 (FC714 [R230S]) and a strain bearing only the two upstream mutations (FC713 [M69L]). Strain FC714 (R230S) appeared wild type by light microscopy, but strain FC713 (M69L) was indistinguishable from the other mutants in this group (data not shown). To learn if the mutation in the untranslated portion of the message altered the steady-state level of SpoIVA, we used quantitative Western blot analysis, with anti-SpoIVA antibodies, to determine the levels of SpoIVA at h 5, 6, and 7 of sporulation. This experiment indicated that the steady-state levels of SpoIVA in strains FC341 (M69L, R230S) and FC713 (M69L) were similar to those in the wild type (data not shown). Most likely, therefore, the phenotype of strain FC713 (M69L) is largely due to the alteration of amino acid 69.
The presence of a coat around the forespore as well as in a swirl in strains FC329 (I400P, E418V, I457R), FC333 (I210M, I367V, I383R), FC334 (V241G), FC335 (G416R), FC341 (M69L, R230S), and FC351 (V453G) implies that the total area of coat assembled by these cells is greater than that in wild-type cells. Indeed, inspection of electron micrographs of these mutants confirmed this assumption and further indicated that a coat of greater surface area is assembled in a spoIVA null mutant as well (Fig. 1A, B, D, and E). It was possible, therefore, that a mutation in spoIVA increases coat protein steady-state levels and, as a consequence, drives assembly of extra coat. To test this, we determined the relative levels of three representative coat proteins, CotA, CotD, and CotE, in wild-type and spoIVA null mutant strains (PY79 and AD18, respectively) at h 7 or, in the case of CotE, h 5 of sporulation, using quantitative Western blot analysis. At each time point, the ratios of coat protein steady-state levels in the mutant to that in the wild type were 0.9, 0.7, and 0.9 for CotA, CotD, and CotE, respectively. The presence of additional assembled coat in spoIVA mutant strains, therefore, is unlikely to be the result of a change in coat protein steady-state levels.
To determine whether SpoIVA is localized to the forespore in these mutant strains, we carried out immunofluorescence microscopy using anti-SpoIVA antibodies. The pattern of fluorescence in cells of strains FC333 (I210M, I367V, I383R), FC334 (V241G), and FC335 (G416R) at h 3 or 5 of sporulation resembled that of wild-type cells (Fig. 3C, D, and E and data not shown). In contrast, in cells of strains FC329 (I400P, E418V, I457R) and FC351 (V453G), fluorescence was not confined to the forespore surface but rather was present in the space between the forespore and mother cell chromosomes, indicating that it was largely mislocalized (Fig. 3F and G and data not shown). We note that the inability to detect SpoIVA at the forespore by immunofluorescence does not exclude the possibility that some SpoIVA is present at this location. For example, a spoVM mutant prevents most but, almost certainly, not all SpoIVA deposition (26). The minimal amount of SpoIVA that apparently remains at the forespore is undetectable by even the most sensitive current methods (26).
Unlike the case for the rest of the mutants, in cells of strain FC341 (M69L, R230S) SpoIVA was present as an arc on the side of the forespore facing the mother cell chromosome (Fig. 3H). Strikingly, for cells of strain FC713 (M69L), we found that in about 30% of cells SpoIVA was now localized in a wild-type pattern (Fig. 3I). Therefore, although the mutation at residue 230 is insufficient to cause the mutant phenotype, it contributes to a defect in SpoIVA localization in combination with the change at amino acid 69.
Mutations that affect germination.
In cultures of 21 mutants, about 90% of the spores were phase gray and swollen after 48 h of growth, as determined by light microscopy, indicating that these spores had rehydrated (17, 22) even though no germinant was added to the medium. In contrast, in cultures of wild-type cells grown under the same conditions, fewer than 0.1% of the spores had this appearance. We further analyzed four of these strains (FC74 [I210A, I367V, I383R], FC77 [L59P, H256P], FC114 [C98S, V283N], and FC133). Cells of strains FC74 (I210A, I367V, I383R), FC77 (L59P, H256P), and FC114 (C98S, V283N) had advanced in sporulation beyond the production of phase-gray forespores by the fourth hour of sporulation, as determined by phase-contrast light microscopy (data not shown), indicating that at least some cortex had formed and that the forespores had at least partially dehydrated. In spite of becoming phase gray and swollen, only 5 to 15% of the cells (depending on the strain) resumed vegetative growth, indicating a defect in germination.
Like FC74 (I210A, I367V, I383R), FC77 (L59P, H256P), and FC114 (C98S, V283N) spores, FC133 spores swell and become phase gray without the addition of germinants to the medium. In contrast to the case for these mutants, however, nucleotide sequencing revealed that the sole mutation in strain FC133 is the change of the adenine at nucleotide −29, relative to promoter P1 (27), to a guanine. Even though this change introduces a nonconsensus nucleotide at the −35 site of the promoter (13), transcription is apparently upregulated, as steady-state levels of SpoIVA in strain FC133 were 5.7-fold greater than those in the wild type as determined by quantitative Western blot analysis (data not shown). SpoIVA localization in FC133, as judged by immunofluorescence microscopy, is indistinguishable from that in the wild type (data not shown). It is unlikely, therefore, that overproduction of SpoIVA severely disrupts SpoIVA localization.
Strains FC77 (L59P, H256P) and FC114 (C98S, V283N) each have two, and FC74 (I210A, I367V, I383R) has three, changes in the spoIVA open reading frame. We used recombinant DNA methods to generate four spoIVA alleles, each containing only one of the nucleotide changes found in FC77 (L59P, H256P) or FC114 (C98S, V283N). We built two derivatives of FC74 (I210A, I367V, I383R), one containing the change at residue 210 and the other bearing the changes at residues 367 and 383. All of the strains generated in this way were phenotypically wild type, with the exception of FC297 (L59P), which was indistinguishable from FC74 (I210A, I367V, I383R), FC77 (L59P, H256P), FC114 (C98S, V283N), or FC133 by light microscopy (data not shown). It is notable that the change of amino acid 210 from isoleucine to alanine (derived from FC74 [I210A, I367V, I383R]) has an impact on germination (at least when in combination with changes at residues 367 and 282), as this same residue is altered in FC333 (I210M, I367V, I383R), resulting in mislocalized coat. Possibly this amino acid is directly involved with both germination and coat assembly. Because FC297 (L59P) harbored a single nucleotide change, we characterized it in more detail.
Using thin-section electron microscopy of FC297 (L59P) cells prepared at h 10 of sporulation, we identified many released spores that were swollen and had thin coats and punctate structures in the cores (Fig. 1F and G), indicating that these spores had rehydrated (30). Spores still within mother cells appeared largely normal: we readily identified the cortex and coat, and in most cells, the spore core appearance was consistent with a significant degree of dehydration (17), although as judged by light microscopy, the forespores never became completely refractile (data not shown). Although most of the sporulating cells were indistinguishable from the wild type, we occasionally detected forespores with partially detached sections of coat and, in a few percent of cells, with entirely disconnected swirls of coat (data not shown). This appearance differs significantly from that of a spoIVA null mutant, in which every forespore is devoid of coat and missing the cortex.
The most straightforward explanation for the apparent spontaneous rehydration in FC297 (L59P) spores is a cortex defect. However, we had not ruled out the alternative possibility that cortex lytic enzymes (which lyse the cortex during germination, thereby rehydrating the spore core) are activating inappropriately in the absence of germinant molecules. To test this, we took advantage of a mutation in cwlD which prevents the formation of δ-lactam residues in the spore peptidoglycan. In a cwlD strain, the δ-lactam-recognizing cortex lytic enzymes fail to degrade the cortex, and therefore the spore does not swell during germination (3, 25, 31). If the defect in FC297 (L59P) spores is an inability to prevent triggering of these lytic enzymes, the combination of the spoIVA mutation in this strain (spoIVA297) and cwlD should result in stable dehydrated spores. By light microscopy, we found that a cwlD::cat::spec spoIVA297 strain (FC618) had the same germination phenotype as FC297 (L59P) (data not shown), indicating that the spoIVA297 phenotype does not require the action of at least this class of cortex lytic enzymes. It is unlikely, therefore, that the germination deficiency in FC297 (L59P) is due to a defect in the activation of cortex lytic enzymes.
From our data, we could not exclude the possibility that the effect of spoIVA297 on germination was largely an indirect consequence of the partial defect in spore coat attachment. Therefore, we generated a strain bearing spoIVA297 and lacking both the inner and outer coat layers (due to the mutations gerE36 cotE::cat::spec [8] in strain FC533). A strain harboring mutations in gerE and cotE produces wild-type levels of stable spores (8). Not surprisingly, spores bearing mutations in gerE and cotE, as well as spoIVA297 were highly unstable, resulting in an approximately 97% reduction in spore titer. Although there were too few spores to perform a quantitative analysis, light microscopic examination indicated that the few spores present 48 h after the beginning of sporulation had swollen and become phase gray, consistent with a lack of involvement of a coat defect in this phenotype.
Characterization of outgrowth of strain FC297 (L59P).
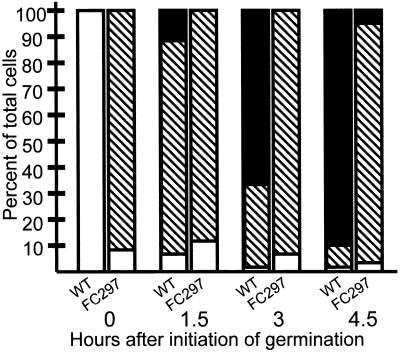
To characterize the fate of spores of strain FC297 (L59P) after release from the mother cell, we collected wild-type and FC297 (L59P) spores 48 h after the initiation of sporulation and resuspended them in LB medium. We then used light microscopy to determine the percentages of refractile (dormant) and swollen, phase-gray (rehydrated) spores, and vegetative cells, over time (Fig. 4). We observed <1% of swollen, phase-gray spores in the wild-type strain after 15 min, and fewer than 10% of the spores were still refractile after 90 min. By 4.5 h, over 90% of the wild-type cells were vegetative and thus had completed germination and outgrowth. In contrast, spores of FC297 (L59P) were swollen and phase gray at the onset of the experiment, and even after 4.5 h, only about 5% of the FC297 (L59P) spores had become vegetative cells. These data imply that although FC297 (L59P) spore cores rehydrate, they are then unable to return to vegetative growth at a rate comparable to that of the wild type, indicating a defect in germination and/or outgrowth.
FIG. 4.
Outgrowth of FC297 (L59P) spores. Spores were resuspended in LB medium, monitored over time, and classified by light microscopic examination. The numbers of phase-bright cells (dormant spores) (open bars), swollen phase-gray cells (rehydrated spores) (hatched bars), and rod-shaped cells (vegetative cells) (solid bars) were counted. At least 400 cells were counted in each experiment, and each measurement was carried out in triplicate. For each time point, the left bar indicates wild type (WT) and the right bar indicates FC297 (L59P).
It was possible that the failure to resume vegetative growth is the consequence of an inability to degrade the α- and β-type small, acid-soluble proteins (SASP), abundant forespore proteins that must be proteolyzed by the forespore protease GPR before germinated spores can begin vegetative growth (29). Therefore, we tested whether deletion of the α and β SASP genes would suppress the spoIVA297 defect (in strain FC729), similarly to the suppression of a gpr phenotype by deletion of the α and β SASP genes (29). We found that outgrowth of strain FC729 was indistinguishable from that of FC297 (L59P), arguing that a defect in α and β SASP proteolysis is unlikely to be the cause of the FC297 (L59P) phenotype.
Spore peptidoglycan synthesis in strain FC297 (L59P).
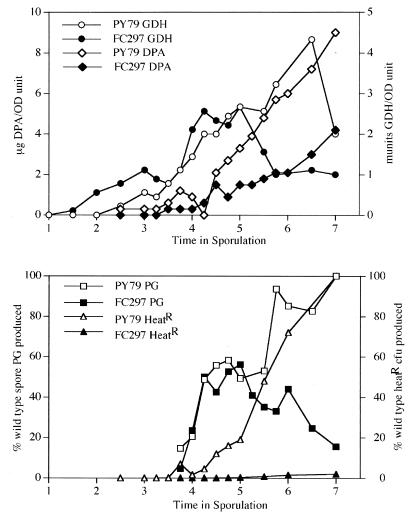
To test whether levels of spore peptidoglycan are altered in FC297 (L59P) sporulating cells, we first monitored the progress of sporulation by measuring glucose dehydrogenase (GDH) and dipicolinic acid (DPA) levels and the acquisition of heat resistance (5). These assays indicated that sporulation proceeds essentially normally in FC297 (L59P) until about the fifth hour of sporulation, when GDH and DPA levels become lower than those of the wild type (Fig. 5). Although the timings of appearance of heat resistance in both FC297 (L59P) and wild-type spores follow similar kinetics, the degree of heat resistance in FC297 (L59P) is about 50-fold lower than that in the wild type and remains so at least until 24 h after sporulation. Spore peptidoglycan synthesis is initiated and proceeds with similar kinetics in the wild type and FC297 (L59P) until about 50% of the wild-type spore peptidoglycan has been produced (Fig. 5). However, analysis of the structure of the developing forespore peptidoglycan indicates that synthesis is abnormal in FC297 (L59P). Structural parameters of the forespore peptidoglycan produced in the wild-type strain, PY79, are similar to those observed in another wild-type B. subtilis 168 strain (reference 20 and data not shown). FC297 (L59P) forespore peptidoglycan contains significant amounts of tripeptide side chains in which the diaminopimelic acid residues are amidated (data not shown). This modification is normally found only in B. subtilis vegetative cell peptidoglycan (2, 38). We verified the identities of two major amidated muropeptides by cochromatography with the same muropeptides derived from vegetative peptidoglycan (muropeptides 3 and 21 described in reference 2, disaccharide-tripeptide with 1 amidation and disaccharide-tripeptide-disaccharide-tetrapeptide with 2 amidations, respectively) in two different gradient systems and using amino acid analysis (data not shown). This was not simply contamination of our FC297 (L59P) forespore peptidoglycan preparation by mother cell wall peptidoglycan. Mother cell wall peptidoglycan is completely degraded during preparation of the forespore peptidoglycan (20). We detected these novel FC297 (L59P) forespore muropeptides in three separate experiments and never in parallel samples from the wild-type strain. In the first spore peptidoglycan produced in FC297 (L59P), 60% of the peptide side chains were amidated, and this level dropped to 35% as more peptidoglycan was synthesized up until h 5 of sporulation. In addition, these initial layers of FC297 (L59P) spore peptidoglycan contained fewer muramic-δ-lactam residues, fewer single l-Ala side chains, and slightly less cross-linking than the peptidoglycan made in the wild type during the same time period. Simultaneously with the decline in levels of GDH, DPA, and heat resistance, the levels of spore peptidoglycan begin to decrease in FC297 (L59P) cells, suggesting that the cortex was actively degraded. This degradation did not result in the production of the major muropeptides produced during spore germination (reference 3a and data not shown). Therefore, spoIVA297 results in abnormal initiation of spore peptidoglycan synthesis and prevents the accumulation of normal levels of spore peptidoglycan.
FIG. 5.
Progression of sporulation events and spore peptidoglycan levels. Cells of strains PY79 and FC297 (L59P) were induced to sporulate and samples were collected for analysis of GDH activity, DPA, heat-resistant (HeatR) CFU, and amount of spore peptidoglycan (PG) produced. Spore PG produced is expressed as a percentage of the amount of spore PG detected in the wild type (PY79) at h 7. Heat-resistant CFU are expressed as a percentage of the value found for the wild type at h 7. OD, optical density.
DISCUSSION
To understand how SpoIVA localizes at the forespore surface early in sporulation, directs cortex synthesis, and attaches the coat to the spore, we used random mutagenesis to isolate a set of spoIVA alleles that impair these functions. These alleles fell into four phenotypic groups. The single mutant in the first group (strain FC336 [L393P]) is indistinguishable from a true null mutant (such as a spoIVAΔ::neo mutant [8]) by light or electron microscopy. Each strain from the second group of mutants (FC329 [I400P, E418V, I457R], FC333 [I210M, I367V, I383R], FC334 [V241G], FC335 [G416R], FC341 [M69L, R230S], and FC351 [V453G]) produces a variety of cell types. Among these are cells that resemble a spoIVA null mutant as well as the wild type. Strikingly, in each strain we also found cells with both a swirl of coat and a coat around the forespore. The third group of mutants appeared to rehydrate spontaneously and to be defective in germination. Finally, we also found two mutants with only a subtle defect, most likely in the coat, which will be discussed elsewhere. We did not identify mutants that assembled a normal coat but were unable to build any cortex or mutants that assembled a normal cortex but no coat.
All of the open reading frame mutations are recessive, as judged by the ability of the appropriate merodiploid strains to produce a wild-type color reaction in the tetrazolium overlay assay and to produce wild-type-appearing spores as judged by light microscopy (data not shown). This suggests that these are partial-loss-of-function alleles and that they identify amino acids critical for specific functions of SpoIVA. For example, residue 416 (altered in FC335 [G416R]) may be critical for proper coat attachment, as swirls of coat are present in the mother cell cytoplasm of some cells while SpoIVA is present at the forespore. Amino acid 59 (altered in FC297 [L59P]) plays a role in cortex formation, and amino acids 400, 418, and/or 457 (altered in FC329 [I400P, E418V, I457R]) and amino acid 453 (altered in FC351 [V453G]) affect localization. Residue 210, which is altered in both SpoIVA74 (I210A, I367V, I383R) and SpoIVA333 (I210M, I367V, I383R), appears to affect both coat assembly and, most likely, cortex formation. Residue 393 (altered in FC336 [L393P]) also is involved in some manner in localization of SpoIVA to the forespore. However, as we do not detect any SpoIVA function in this allele, it is possible that this mutation does not identify a localization region of SpoIVA but rather interferes with protein folding.
Previous studies demonstrate that the N-terminal 64 amino acids are not essential, and that the C-terminal 5 residues are required, for SpoIVA localization to the forespore (26). Consistent with this, mutants with defective localization had amino acid changes at residues 400, 418, and 457 (in strain FC329 [I400P, E418V, I457R]) and 453 (in strain FC351 [V453G]). Analysis of strains FC341 (M69L, R230S) and FC713 (M69L) indicates that changes in amino acids spread widely apart in the primary sequence (at positions 69 and 230) can work together to produce a localization phenotype. Likewise, changes in both residues 98 and 283 (in strain FC114 [C98S, V283N]) can generate a germination defect. It is possible, therefore, that regions of SpoIVA involved in localization and germination are spread over relatively large stretches of primary sequence. Some amino acid changes that affect germination fall relatively close to changes affecting localization and the formation of swirls of coat, and they can even be in the same residue, as in the case of amino acid 210, which is altered in strains FC74 (I210A, I367V, I383R) and FC333 (I210M, I367V, I383R). Therefore, amino acids with differing roles in spore assembly are not necessarily confined to separate regions of the SpoIVA sequence.
It is noteworthy that our mutagenesis did not generate any mutants whose sole defect was the lack of a cortex. One possible explanation for this is that cortex formation is an indirect consequence of one of the other functions of SpoIVA and, therefore, that a mutation that prevents cortex formation will also impair SpoIVA localization or coat formation. Nonetheless, as our mutagenesis was not carried out to saturation, we may have missed mutants with this phenotype. It is also notable that we identified more mutants whose primary defect is in germination and not in coat assembly than any other type. Most likely, all of these mutants are unable to build a normal cortex. One explanation for the size of this class is that slight alterations in the structure of SpoIVA are sufficient to partially disrupt cortex formation without producing a readily detected coat assembly defect. It is surprising that a mutation resulting in an increase in SpoIVA steady-state levels (in strain FC133) is a member of this class. Possibly an excess of SpoIVA accumulates at the forespore in this strain, interfering with SpoIVA's ability to interact with other proteins or to adopt the normal conformation.
The identification of mutants with coats both around the spore and in swirls was unexpected, and its implications for SpoIVA function are not yet entirely clear. Nonetheless, we propose a speculative model in which SpoIVA guides an as-yet-unidentified coat protein nucleating factor to the forespore. Normally, SpoIVA sequesters all of the nucleator at the forespore surface. Because the nucleator is confined to a closed surface with no edges (the forespore), coat assembly is similarly restricted. However, when SpoIVA is absent or defective, at least some nucleator fails to bind to the forespore, causing sheets of coat to appear in the cytoplasm. The presence of free edges on the sheet permits coat formation to continue beyond the usual closed surface area of the forespore. This view is consistent with the observation that spoIVA null mutants assemble more coat than the wild type. We speculate that in cultures of strains FC329 (1400P, E418V, 1457R), FC333 (I210M, I367V, 1383R), FC334 (V241G), FC335 (G416R), FC341 (M69L, R230S), and FC351 (V453G), the defects in SpoIVA prevent it from entirely sequestering the nucleator in some cells and cause it to fail to do so entirely in others (resulting in a spoIVA null mutant-like phenotype). In cells with both a swirl of coat and a coat around the forespore, some nucleator would have deposited around the forespore but enough would have remained in the mother cell cytoplasm to drive the assembly of swirls. In this view, the versions of SpoIVA in FC333 (I210M, I367V, I383R), FC334 (V241G), and FC335 (G416R) are defective primarily in attaching to the nucleator. The versions of SpoIVA in the remaining strains have significant defects in localization of SpoIVA to the forespore.
This model (and, indeed, the spoIVA null phenotype) argues that, in principle, coat can assemble in the mother cell cytoplasm. Therefore, in wild-type cells, some mechanism must ensure that coat assembly does not initiate in the mother cell cytoplasm before the coat proteins have reached the shell of SpoIVA at the forespore surface. One possible mechanism would be a more rapid assembly of the sheet of nucleator when in contact with SpoIVA. In support of this notion, formation of swirls occurs several hours later in sporulation than does coat assembly in wild-type cells (A. Driks, unpublished observations).
We identified a group of mutants, bearing nucleotide changes throughout spoIVA, whose spores form largely normal coats and possess at least some cortex but cannot remain dehydrated after mother cell lysis. From an analysis of one of these strains, FC297 (L59P), we propose that this phenotype is due, in large part, to abnormal synthesis and an insufficient level of spore peptidoglycan. FC297 (L59P) spore peptidoglycan contained amidated diaminopimelic acid, which is normally found only in vegetative cell peptidoglycan (2, 38), as well as other structural alterations indicative of a failure to properly shift from vegetative and germ cell wall synthesis to cortex synthesis. It is not clear if these peptidoglycan structure alterations are the direct cause of the failure of FC297 (L59P) spores to achieve a dormant state or if they are one aspect of a larger problem. The role of SpoIVA in cortex formation raises the possibility that in much the same way that SpoIVA directs coat protein localization, it also guides enzymes controlling peptidoglycan synthesis to their correct positions within the forespore. The FC297 (L59P) phenotype could be the result of the failure of one or more of these proteins to localize correctly. The finding of vegetative muropeptides in FC297 (L59P) spore peptidoglycan suggests that one function of these proteins may be the inhibition of vegetative peptidoglycan synthesis.
In summary, we have identified a set of spoIVA mutants that identify amino acids with roles in localization, coat assembly, and cortex formation. These results will permit further analysis of the functions of these residues and identification of proteins in the coat and at the forespore surface that interact with SpoIVA. We found that amino acids throughout SpoIVA participate in SpoIVA localization and/or coat assembly and cortex formation, suggesting that these functions are not necessarily each encoded in separate stretches of primary sequence. Although SpoIVA has at least three functions (localization, cortex formation, and coat attachment), we did not find mutants with defects in each of these activities. Instead, we found only one mutant with a possible defect in localization and several novel and unexpected classes of mutants, including cells with swirls of coat as well as coat around the forespore and cells that cannot remain dormant after mother cell lysis. These results suggest that localization, cortex formation, and/or coat attachment do not necessarily represent distinct, genetically separable functions of SpoIVA.
ACKNOWLEDGMENTS
We thank Margarita Correa for generating the anti-SpoIVA antibodies and Shawn Little for generating the anti-CotD antibodies and for very helpful comments on the manuscript. We also thank Lionel Coignet (Cardinal Bernardine Cancer Center, Loyola University) for use of his fluorescent in situ hybridization facility and Peter Setlow (University of Connecticut) for kindly providing strains.
This work was supported by Public Health Service grants GM53989 (to A.D.) and GM56695 (to D.L.P.) from the National Institutes of Health.
REFERENCES
- 1.Aronson A I, Fitz-James P. Structure and morphogenesis of the bacterial spore coat. Bacteriol Rev. 1976;40:360–402. doi: 10.1128/br.40.2.360-402.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atrih A, Bacher G, Allmaier G, Williamson M P, Foster S J. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J Bacteriol. 1999;181:3956–3966. doi: 10.1128/jb.181.13.3956-3966.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atrih A, Zollner P, Allmaier G, Foster S J. Structural analysis of Bacillus subtilis 168 endospore peptidoglycan and its role during differentiation. J Bacteriol. 1996;178:6173–6183. doi: 10.1128/jb.178.21.6173-6183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Atrih A, Zollner P, Allmaier G, Williamson M P, Foster S J. Peptidoglycan structural dynamics during germination of Bacillus subtilis 168 endospores. J Bacteriol. 1998;180:4603–4612. doi: 10.1128/jb.180.17.4603-4612.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer T, Little S, Stöver A G, Driks A. Functional regions of the B. subtilis spore coat morphogenetic protein CotE. J Bacteriol. 1999;181:7043–7051. doi: 10.1128/jb.181.22.7043-7051.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutting S M, Vander Horn P B. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1990. [Google Scholar]
- 6.Driks A. The Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 1999;63:1–20. doi: 10.1128/mmbr.63.1.1-20.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Driks A. Spatial and temporal control of gene expression in prokaryotes. In: Russo V E A, Cove D J, Edgar L G, Jaenisch R, Salamini F, editors. Development: genetics, epigenetics and environmental regulation. Berlin, Germany: Springer; 1999. [Google Scholar]
- 8.Driks A, Roels S, Beall B, Moran C P, Jr, Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994;8:234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- 9.Driks A, Setlow P. Morphogenesis and properties of the bacterial spore. In: Brun Y V, Shimkets L J, editors. Prokaryotic development. Washington, D.C.: American Society for Microbiology; 2000. pp. 191–218. [Google Scholar]
- 10.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster S J, Johnstone K. Pulling the trigger: the mechanism of bacterial spore germination. Mol Microbiol. 1990;4:137–141. doi: 10.1111/j.1365-2958.1990.tb02023.x. [DOI] [PubMed] [Google Scholar]
- 12.Karmazyn-Campelli C, Fluss L, Leighton T, Stragier P. The spoIIN279(ts) mutation affects the FtsA protein of Bacillus subtilis. Biochimie. 1992;74:689–694. doi: 10.1016/0300-9084(92)90141-z. [DOI] [PubMed] [Google Scholar]
- 13.Kodama T, Takamatsu H, Asai K, Kobayashi K, Ogasawara N, Watabe K. The Bacillus subtilis yaaH gene is transcribed by SigE RNA polymerase during sporulation, and its product is involved in germination of spores. J Bacteriol. 1999;181:4584–4591. doi: 10.1128/jb.181.15.4584-4591.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kroos L, Zhang B, Ichikawa H, Yu Y T. Control of sigma factor activity during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1285–1294. doi: 10.1046/j.1365-2958.1999.01214.x. [DOI] [PubMed] [Google Scholar]
- 15.LeDeaux J R, Grossman A D. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol. 1995;177:166–175. doi: 10.1128/jb.177.1.166-175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Losick R, Shapiro L. Changing views on the nature of the bacterial cell: from biochemistry to cytology. J Bacteriol. 1999;181:4143–4145. doi: 10.1128/jb.181.14.4143-4145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Losick R, Youngman P, Piggot P J. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- 18.Lu S, Halberg R, Kroos L. Processing of the mother-cell ς factor, ςK, may depend on events occurring in the forespore during Bacillus subtilis development. Proc Natl Acad Sci USA. 1990;87:9722–9726. doi: 10.1073/pnas.87.24.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margolis P S, Driks A, Losick R. Sporulation gene spoIIB from Bacillus subtilis. J Bacteriol. 1993;175:528–540. doi: 10.1128/jb.175.2.528-540.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meador-Parton J, Popham D L. Structural analysis of Bacillus subtilis spore peptidoglycan during sporulation. J Bacteriol. 2000;182:4491–4499. doi: 10.1128/jb.182.16.4491-4499.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milhaud P, Balassa G. Biochemical genetics of bacterial sporulation IV. Sequential development of resistance to chemical and physical agents during sporulation of Bacillus subtilis. Mol Gen Genet. 1973;125:241–250. doi: 10.1007/BF00270746. [DOI] [PubMed] [Google Scholar]
- 22.Moir A, Kemp E H, Robinson C, Corfe B M. The genetic analysis of bacterial spore germination. J Appl Bacteriol. 1994;77:9S–16S. [PubMed] [Google Scholar]
- 23.Piggot P J, Coote J G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pogliano K, Harry E, Losick R. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol Microbiol. 1995;18:459–470. doi: 10.1111/j.1365-2958.1995.mmi_18030459.x. [DOI] [PubMed] [Google Scholar]
- 25.Popham D L, Helin J, Costello C E, Setlow P. Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc Natl Acad Sci USA. 1996;93:15405–15410. doi: 10.1073/pnas.93.26.15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price K D, Losick R. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J Bacteriol. 1999;181:781–790. doi: 10.1128/jb.181.3.781-790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roels S, Driks A, Losick R. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Sanchez-Salas J L, Santiago-Lara M L, Setlow B, Sussman M D, Setlow P. Properties of Bacillus megaterium and Bacillus subtilis mutants which lack the protease that degrades small, acid-soluble proteins during spore germination. J Bacteriol. 1992;174:807–814. doi: 10.1128/jb.174.3.807-814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santo L Y, Doi R H. Ultrastructural analysis during germination and outgrowth of Bacillus subtilis spores. J Bacteriol. 1974;120:475–481. doi: 10.1128/jb.120.1.475-481.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekiguchi J, Akeo K, Yamamoto H, Khasanov F K, Alonso J C, Kuroda A. Nucleotide sequence and regulation of a new putative cell wall hydrolase gene, cwlD, which affects germination in Bacillus subtilis. J Bacteriol. 1995;177:5582–5589. doi: 10.1128/jb.177.19.5582-5589.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro L, Losick R. Dynamic spatial regulation in the bacterial cell. Cell. 2000;100:89–98. doi: 10.1016/s0092-8674(00)81686-4. [DOI] [PubMed] [Google Scholar]
- 33.Sonenshein A L. Endospore-forming bacteria: an overview. In: Brun Y V, Shimkets L J, editors. Prokaryotic development. Washington, D.C.: American Society for Microbiology; 2000. pp. 133–150. [Google Scholar]
- 34.Stevens C M, Daniel R, Illing N, Errington J. Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:586–594. doi: 10.1128/jb.174.2.586-594.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stöver A G, Driks A. Control of the synthesis and secretion of the B. subtilis protein YqxM. J Bacteriol. 1999;181:7065–7069. doi: 10.1128/jb.181.22.7065-7069.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stöver A G, Driks A. Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J Bacteriol. 1999;181:1664–1672. doi: 10.1128/jb.181.5.1664-1672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 38.Warth A D, Strominger J L. Structure of the peptidoglycan from cell walls of Bacillus subtilis. Biochemistry. 1971;10:4349–4358. doi: 10.1021/bi00800a001. [DOI] [PubMed] [Google Scholar]
- 39.Youngman P, Perkins J B, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 40.Zheng L, Losick R. Cascade regulation of spore coat gene expression in Bacillus subtilis. J Mol Biol. 1990;212:645–660. doi: 10.1016/0022-2836(90)90227-d. [DOI] [PubMed] [Google Scholar]