Abstract
By transforming N2O to N2, the multicopper enzyme nitrous oxide reductase provides a periplasmic electron sink for a respiratory chain that is part of denitrification. The signal sequence of the enzyme carries the heptameric twin-arginine consensus motif characteristic of the Tat pathway. We have identified tat genes of Pseudomonas stutzeri and functionally analyzed the unlinked tatC and tatE loci. A tatC mutant retained N2O reductase in the cytoplasm in the unprocessed form and lacking the metal cofactors. This is contrary to viewing the Tat system as specific only for fully assembled proteins. A C618V exchange in the electron transfer center CuA rendered the enzyme largely incompetent for transport. The location of the mutation in the C-terminal domain of N2O reductase implies that the Tat system acts on a completely synthesized protein and is sensitive to a late structural variation in folding. By generating a tatE mutant and a reductase-overproducing strain, we show a function for TatE in N2O reductase translocation. Further, we have found that the Tat and Sec pathways have to cooperate to produce a functional nitrite reductase system. The cytochrome cd1 nitrite reductase was found in the periplasm of the tatC mutant, suggesting export by the Sec pathway; however, the enzyme lacked the heme D1 macrocycle. The NirD protein as part of a complex required for heme D1 synthesis or processing carries a putative Tat signal peptide. Since NO reduction was also inhibited in the tatC mutant, the Tat protein translocation system is necessary in multiple ways for establishing anaerobic nitrite denitrification.
The Sec apparatus has long been viewed as the sole system for translocation of proteins across the inner bacterial membrane. However, recently it became clear that in addition to the Sec system many bacteria have the Tat system for the export of proteins which seem to be transported in a folded form (reviewed in references 4, 38, and 51). Evidence for a novel pathway of bacterial protein translocation came from studying protein import into chloroplast thylakoids. A ΔpH-driven pathway was shown to require a signal peptide with an arginine pair (11), and the maize protein Hcf106, involved in this pathway, was found to have homologs in bacteria (45). This fostered the discovery of the new secretory pathway in Escherichia coli, termed mtt, for membrane targeting and transport (48), or tat, for twin-arginine translocation (43).
Supportive evidence for the Tat pathway also came independently from observations of a new type of bacterial signal peptide. On determining several primary structures of nitrous oxide reductase (N2OR), a sequence motif with an arginine pair in an unusually long signal peptide of about 50 amino acids (aa) was recognized to be conserved not only in this enzyme but also in hydrogenase (29, 56). We reported the existence of the novel motif in a variety of exported and cofactor-carrying proteins (2, 17, 18). As initially found with hydrogenase (34), substitution of the first arginine residue in the conserved sequence prevented translocation of N2OR to the periplasm in an R20D mutant (17, 18). The significance of the twin-arginine motif has been studied now for other enzymes (16, 26, 27, 40), and allowed variations have been specified by site-directed mutagenesis (46). A data bank survey for the twin-arginine type of signal peptide revealed a large number of redox proteins with cofactors and led to the suggestion that these proteins may be folded in the cytoplasm and acquire their cofactors prior to transport. A heptameric consensus motif in Tat-specific signal peptides was defined as S(T)RRXFLK (3).
Current sequence information from genome projects has revealed a wide, though not ubiquitous, distribution of tat genes among prokaryotes, chloroplasts, and plant mitochondria (51). However, with only a few exceptions (5, 18, 26, 27), studies of the Tat system in bacteria have been confined to enzymes of E. coli. The tatC gene encodes an integral membrane protein composed of six transmembrane helices, which is thought to form the functional export complex in association with other tat-encoded components (4). A mutation in tatC blocks protein translocation (6). Recently, a 600-kDa complex has been described as consisting of TatA, TatB, and presumably the TatC component (7). TatC is rapidly degraded in the absence of TatB (44), suggesting that TatB stabilizes TatC, which provides another, albeit indirect, argument for the presence of TatC in the translocation complex. A further tat locus of E. coli, tatD, encodes a cytoplasmic protein with DNase activity but apparently is not an obligatory factor of the translocation pathway (50).
Utilization of N2O is a respiratory mode, found usually as part of bacterial denitrification. N2OR provides a terminal electron sink by reducing N2O to N2 and is essential for global N cycling by preventing the accumulation of the greenhouse gas N2O. In the denitrifying cell, N2O is generated by the consecutive actions of the periplasmic respiratory nitrite reductase (cytochrome cd1 in Pseudomonas stutzeri) (53) and the membrane-bound NO reductase, a structural and functional homolog of subunit II of cytochrome c oxidase (24, 31). N2OR carries six copper atoms per subunit, which are arranged in two types of centers: the binuclear electron transfer site, CuA (13, 33), and the tetranuclear catalytic site, CuZ (10). The latter was recently identified as the first example of a biologically active copper-sulfide cluster (37). Cytochrome cd1 and N2OR both have to be translocated across the cytoplasmic membrane to reach their functional sites in the periplasm (reviewed in reference 53).
In this study we have investigated N2OR translocation by the Tat system and the possibility of Cu cofactor insertion in the cytoplasm when the translocation pathway is interrupted. Towards these objectives, we have isolated tat loci from P. stutzeri and generated mutations in tatC and tatE. Since E. coli does not denitrify, it cannot provide the physiological environment to study the requirements for the translocation of denitrification enzymes. Our data support conformational constraints imposed by the Tat system on N2OR transport. However, metal cofactor insertion into the enzyme, contrary to a widely held view, did not take place in the cytoplasm of P. stutzeri. We have also investigated a broader effect of tatC on denitrification, which led to the finding that heme D1 synthesis or processing to establish a functional cytochrome cd1 nitrite reductase is Tat dependent.
(A preliminary report of this work appeared previously [28].)
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli DH10B and JM110 were grown in Luria-Bertani (LB) medium at 37°C and 240 rpm on a gyratory shaker. For recombinant DNA work, Pseudomonas strains were grown in LB medium at 30°C in a gyratory shaker at 240 (aerobic growth) or 120 (O2-limited growth) rpm. For strain maintenance, when necessary, kanamycin (Km), ampicillin (Ap), or streptomycin (Str) was added at a final concentration of 50, 100, or 200 μg ml−1, respectively. Growth studies were done in an asparagine- and citrate-containing synthetic medium (AC) (13); when necessary, AC medium was supplemented with the appropriate N oxide. Cells for subcellular fractionation were grown as described previously (18).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| P. stutzeri | ||
| MK21 | Spontaneous Strr mutant of P. stutzeri Zobell (ATCC 14405) | 55 |
| MK401 | MK21; nosD::Kmr Strr | 55 |
| MK4211 | MK21; ΔnosZ::Kmr Strr | 19 |
| MK4T4 | MK21; ΔtatC::Kmr Strr | This study |
| MK4E1 | MK21; tatE::Kmr Strr | This study |
| P. aeruginosa PAO | Strain used to generate tatC probes | DSM 1707 |
| E. coli | ||
| DH10B | Host for cloning | Gibco BRL |
| JM110 | Host for cloning | 52 |
| Plasmids | ||
| cDEN1 | Cosmid clone carrying nos genes of a Sau3A genomic library in pJA1 | 8 |
| pBluescript II SK(+) | Cloning vector; AprlacZ | Stratagene |
| pBTAT3 | 3-kb PstI fragment carrying tatABC of MK21 cloned into pBluescript II SK(+) | This study |
| pBTAT3K | pBTAT3; ΔtatC(BglII)::Kmr | This study |
| pBTE1 | 1,017-bp PCR fragment carrying tatE of MK21 cloned into pBluescript II SK(+) | This study |
| pBTE1K | pBTE1; tatE(BclI)::Kmr | This study |
| pBSL15 | Source of Kmr cassette | 1 |
| pSZ | 3.57-kb XhoI-SmaI fragment carrying nosZ cloned into pSUP104 | 19 |
| pSZ(C618V) | pSZ derivative; cysteine 618 of CuA in NosZ replaced by valine | This study |
| pSZ(C618D) | pSZ derivative; cysteine 618 of CuA in NosZ replaced by aspartic acid | 13 |
| pSZ(C622V) | pSZ derivative; cysteine 622 of CuA in NosZ replaced by valine | This study |
| pSZ(H583G) | pSZ derivative; histidine 583 of CuA in NosZ replaced by glycine | 13 |
| pSZ(R20D) | pSZ derivative; arginine 20 of signal peptide of NosZ replaced by aspartic acid | 18 |
| pUC-HFH | 1.08-kb HindIII-HinfI fragment of nosZ cloned into pUC18 | 13 |
| pUCP22 | Cloning vector; Apr Gmr | 49 |
| pUCP22RE | 8.8-kb Eco47III-XbaI fragment carrying the nos gene cluster and tatE cloned into pUCP22 | This study |
| pUCP22RL | 8.6-kb Eco47III-XbaI fragment carrying the nos gene cluster of MK21 cloned into pUCP22 | This study |
DNA techniques.
Genomic DNA was extracted from cells by the one-step chloroform method (14); plasmid DNA was prepared by alkaline cell lysis (22). For cloning purposes, plasmid DNA was purified in a preparative agarose gel with crystal violet (36) combined with the QIAquick gel extraction method (Qiagen). Standard procedures were used for agarose gel electrophoresis, dephosphorylation, ligation, PCR amplification of DNA fragments, and transformation of E. coli by electroporation (41). Restriction enzymes were used as recommended by the manufacturers.
Gene probes.
Probes for tatC were deduced from sequence information of Pseudomonas aeruginosa. Data were obtained from the web site of the Pseudomonas Genome Project (http://www.pseudomonas.com). The probe TC1 was amplified from genomic DNA of P. aeruginosa PAO with the primers 5′-GGTCTGGGGCTTCATCGC-3′ and 5′-CCATGCCGACCACGAAAC-3′, which have the start positions 5707528 and 5707874, respectively, in the P. aeruginosa chromosome. The probe TC2 was generated with the primers 5′-GGCGGCGATCTTCCTGATCT-3′ and 5′-AGGAACAGCGCCACCACCAT-3′ at positions 5707330 and 5707503, respectively. The annealing temperature was 57°C. Labeling was done with digoxigenin (DIG) by PCR (Boehringer Mannheim).
Probe TE1 was used to screen for tatE. A 170-bp fragment, almost covering the complete tatE gene (174 bp) of P. stutzeri, was amplified by PCR with plasmid pBTE1 as the template and DIG-labeled simultaneously using the primers 5′-GTATCAGCGTCTGGCAACTCC-3′ (forward primer; start position, 9602) and 5′-TCAGCTCCTGCTTGACGC-3′ (reverse primer; start position, 9433) (25). DIG Easy Hyb granules (Roche) were used for Southern hybridization with this probe at 45°C.
Subcloning and sequencing of the tatC fragment.
A P. stutzeri cosmid library (8) was screened for tatC by Southern hybridization with the probe TC1. DNA was blotted to nitrocellulose membranes by downward alkaline capillary transfer (15); hybridization was done at 55°C (20). A 3-kb PstI fragment was subcloned into pBluescript II SK(+) with E. coli DH10B as the host to give plasmid pBTAT3. A 1,831-bp region of this plasmid was sequenced on both strands by primer walking using a dye terminator kit or Cy5-labeled primers with an ALFexpress sequencer according to the instructions of the manufacturer (Amersham Pharmacia Biotech).
Mutagenesis procedures.
A 150-bp BglII fragment, covering the sequence positions 1078 to 1227, was excised from the tatC gene in plasmid pBTAT3 and replaced by a Kmr cassette derived from the BamHI-digested plasmid pBSL15. The orientation of the cassette in the construct pBTAT3K was verified to be opposite to that of tatC by sequencing. pBTAT3K was transferred to P. stutzeri MK21 by electroporation (21). The transformed cells were plated on LB plates containing 200 μg of kanamycin ml−1 and 200 μg of streptomycin ml−1. After 2 days of incubation at 30°C, colonies were picked and tested for ampicillin sensitivity (200 μg ml−1). Genomic DNA from Aps strains was prepared, digested with PstI, and analyzed by Southern hybridization with the TC1 probe. The tatC strain MK4T4 was obtained by this procedure.
The tatE gene (formerly orf57) was mutagenized by insertion of a Kmr cassette. A 1,017-bp fragment with tatE was amplified by PCR from genomic DNA of P. stutzeri with the primers 5′-CTGCTCGATGCCAAGCTC-3′ and 5′-AGCCGCCGGTTGGTTGAA-3′, targeting the positions 308 (19) and 8950 (25), respectively. To generate blunt ends for cloning, Pwo DNA polymerase (Boehringer Mannheim) was used in the PCR according to the instructions of the manufacturer. The annealing temperature was 50°C.
The 1,017-bp fragment was cloned blunt ended into the SmaI site of pBluescript II SK(+) to give plasmid pBTE1, which was propagated in JM110. For mutagenesis of tatE, plasmid pBTE1 was cleaved with BclI and ligated with a BamHI-digested Kmr cassette. The orientation of the cassette in the resulting plasmid, pBTE1K, was verified by sequencing to be opposite to the direction of the tatE gene. The construct was transformed into MK21 by electroporation, and the recombination event was verified by Southern hybridization. The tatE strain MK4E1 was obtained by this procedure.
The CuA mutants C618V and C622V of N2OR were generated as described previously (19). The mutagenic primers were 5′-ACTGGTACTACgtCAGCTGG-3′ and 5′-GCAGCTGGTTCgtgCAcGCGCTGCA-3′ (the nucleotides altered from the wild-type sequence are in lowercase). The alterations led to the loss of a PstI site and the gain of an ApaL1 site, respectively, which was used for screening the PCR product. The mutation was introduced by PCR using the mutagenic primer and plasmid pUC-HFH as the template together with a universal pUC18-derived primer. pUC-HFH is a pUC18 derivative carrying a 1.08-kb HindIII-HinfI fragment of nosZ. The annealing temperatures were 53 (C618V primer) and 61°C (C622V primer). Reconstruction of the nosZ gene in the expression vector pSZ was done as described previously (13). The mutations were verified by sequencing. Recombinant N2OR was expressed from vector pSZ-C618V or -C622V in trans in the nosZ deletion mutant MK4211.
Construction of nos gene expression vectors.
The complete nos gene cluster was assembled in vitro from an Eco47III-SmaI fragment of cosmid cDEN1 (8), carrying nosRZD′, and a fragment amplified by PCR. The latter was designed to carry the missing 3′ part of nosD together with either nosYL or the nosD′LY part and, additionally, the tatE gene. The genes were assembled in plasmid pUCP22, whose replicon is functional in Pseudomonas species. The first step consisted of cloning the cDEN1 fragment into Ecl136II- and SmaI-digested pUCP22 to yield plasmid pUCP22RZ. Next, PCR amplification was done with cDEN1 as the template. The forward primer, 5′-ACGTGCGCAGATCAGCAATAACC-3′, was located 34 bp upstream of the SmaI site in nosD used for the construction of pUCP22RZ. The reverse primers, 5′-gtactatctagaGCCGAACAGCATGACGAC-3′ and 5′-gtactatctagaCGCGCAGTCTTGTAGAGG-3′, were designed to add XbaI sites (lowercase) 174 and 45 bp downstream of nosL and tatE, respectively. The PCR products were digested with SmaI and XbaI and ligated into the likewise digested pUCP22RZ, yielding pUCP22RL (nosRZDFYL) and pUCP22RE (nosRZDFYL tatE).
Purification of cytochrome cd1.
After high-speed centrifugation, cell extract was loaded onto a DE-52 cellulose column (2.5 by 12 cm; Whatman) and eluted with a linear gradient (300 ml; 0 to 0.35 M NaCl). The buffer was 25 mM Tris-HCl, pH 7.5, throughout the procedure. Cytochrome cd1-containing fractions were detected immunochemically, concentrated, and applied to a fast-protein liquid chromatography Superdex 200 column (1.6 by 60 cm; Amersham Pharmacia Biotech), which was developed at 0.5 ml/min with buffer (0.2 M in NaCl). The concentrated cytochrome cd1 was applied to a 1-ml High-Trap anion-exchange column (Amersham Pharmacia Biotech), eluted with a linear gradient (30 ml; 0 to 0.25 M NaCl), and concentrated by ultrafiltration (Centricon 10,000).
Cell fractionation, gel electrophoresis, and enzyme assays.
The periplasm, cytoplasm, and membrane fraction were prepared by an osmotic-shock procedure (32). The protein concentration was determined by the Lowry method. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis with a 12.5% acrylamide gel was used for protein separation. Immunochemical detection of N2OR and nitrite reductase was done with polyclonal antisera and protein A-horseradish peroxidase conjugate (18). The activities of nitrate, nitrite, NO, and N2O reductases of whole cells and cell extract were measured by gas chromatography or by colorimetric determination of nitrite (23, 35).
Nucleotide sequence accession number.
The nucleotide sequence data reported here have been deposited in the EMBL nucleotide sequence data bank under the accession number AJ299712.
RESULTS
Isolation of tat genes from P. stutzeri.
As a starting point, we used the sequence information provided by the Pseudomonas Genome Project to search the P. aeruginosa genome for tat homologs with E. coli sequences. A putative tatABC gene cluster was found in the P. aeruginosa PAO1 chromosome at the sequence position 5706551 to 5708038 (Fig. 1). Tat-mediated protein translocation in this bacterium has not been addressed; however, the high degree of positional identity of the predicted P. aeruginosa Tat proteins with the corresponding E. coli components allowed a clear assignment to tat genes (Fig. 2). The annotated sequence of the P. aeruginosa chromosome was published recently; a TatE component is not apparent in this sequence (47).
FIG. 1.
Physical maps of the DNA regions carrying tat genes in P. stutzeri and P. aeruginosa. Transcriptional directions are indicated by the open arrows. The effected deletion in tatC, strain MK4T4, and the insertion site of the Kmr cassette in tatE, strain MK4E1, are illustrated. Downstream of tatE, the nnrS gene, formerly orf396, shows similarity to nnrS of Rhodobacter sphaeroides (accession number AF016258). The bars indicate the positions of the gene probes used in this work.
FIG. 2.
Multiple sequence alignments showing the similarity of Tat proteins from P. stutzeri (Pst) and P. aeruginosa (Pae) to their homologs from E. coli (Eco). The alignments were done by the BESTFIT algorithm of the Genetics Computer Group programs. Identical residues are shaded.
Next, a cosmid library of P. stutzeri was screened by Southern hybridization with the tatC probes TC1 and TC2 based on the P. aeruginosa sequence (see Materials and Methods). Cosmid c31 was found to carry tat genes. An ≈3-kb PstI fragment which hybridized with both probes, indicating the likelihood that it harbored the entire tatC gene, was subcloned into pBluescript II SK(+) to give plasmid pBTAT3. Sequence analysis of a 1.8-kb region of this plasmid revealed a tatABC gene cluster (Fig. 1). The stop and start codons of tatB and tatC overlap by 4 nucleotides, indicative of translational coupling. An alignment of the deduced Tat proteins of P. stutzeri with their homologs in P. aeruginosa and E. coli is shown in Fig. 2. The TatB proteins of P. stutzeri and P. aeruginosa have the same two deletions compared to the E. coli sequence. The protein has a strongly conserved N-terminal moiety but is C-terminally rather variable. This may imply specificity for interaction with a further component residing C terminally and a common function in the N-terminal part. The positional identities of TatA, TatB, and TatC of P. stutzeri with their homologs in P. aeruginosa are 58, 61, and 77%, and with those of E. coli, they are 55, 34, and 62%, respectively.
The tatE locus of P. stutzeri (accession number Z73914) was identified previously as orf57 by sequencing the nos region downstream of nosL (Fig. 1) (25). orf57 had been found to be a homolog of E. coli ybeC, now tatE. A 1,017-bp fragment with orf57 was amplified and subcloned into pBluescript II SK(+) to give plasmid pBTE1. We consider the orf57 product to be TatE, even though its positional identity (67%) with TatA of E. coli is higher than with TatE (51%). The deduced 57-aa protein of orf57 corresponds to the size of TatE, which is significantly smaller than that of TatA (76 aa), and the tatE genes of P. stutzeri and E. coli are both loci separate from the tatABC clusters.
TatC is required for N2O respiration.
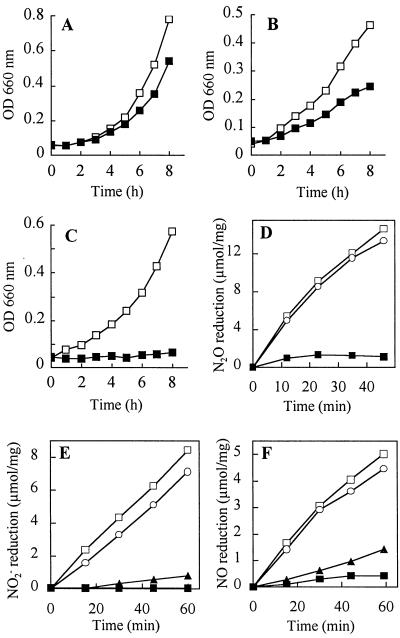
If N2OR were translocated by the Tat system, the mutational inactivation of tatC should prevent export of the reductase, since inactivation of tatC blocks translocation of proteins with the twin-arginine leader peptide in E. coli (6). We have generated a tatC deletion strain and studied its growth behavior under conditions where different respiratory substrates are utilized. The activities of the denitrification enzymes of whole cells and, when appropriate, of cell extract were also followed (Fig. 3). The following traits of the tatC deletion strain MK4T4 of P. stutzeri were observed. Since the aerobic growth rate of the mutant was only slightly decreased, we conclude that no vital component of aerobic respiration had been affected (Fig. 3A). When cells were grown under denitrifying conditions, i.e., with nitrate under O2 limitation, growth was clearly retarded, indicating that one or more functions of denitrification had been targeted by mutation of tatC (Fig. 3B). Nevertheless, nitrate reductase, the first enzyme of the anaerobic denitrification pathway, was active, and nitrite accumulated in a nitrate-containing medium. Hence, the cause of growth inhibition had to be elsewhere in the further reduction of nitrite. The most striking effect was found when cells were grown anaerobically with N2O as the electron acceptor. While the wild type, represented by MK21 (Strr), grew readily on N2O, MK4T4 had lost this capability (Fig. 3C). We also assayed cells for the reduction of the denitrification substrates, nitrite, NO, and N2O. The tatC mutation caused the loss of N2O reductase activity (Fig. 3D). Cell extract was not assayed in this case because N2OR is usually inactive in crude extract due to an unspecified interference (55). There was also a striking loss of nitrite reductase activity both in vivo and in vitro (Fig. 3E). The underlying causes of the observed Nos− and Nir− phenotypes of MK4T4 were studied in more detail.
FIG. 3.
Growth characteristics of the tatC mutant and denitrification activities in vivo and in vitro of tatC and tatE mutants. (A to C) Growth curves. ■, MK4T4; □, wild-type behavior represented by MK21. (A) Aerobic conditions; 150 ml of AC medium in 500-ml flasks shaken on a gyratory incubator at 240 rpm and 30°C. (B) O2-limited, denitrifying conditions; AC medium containing nitrate (1 g/liter); shaker speed, 120 rpm. (C) Incubation with N2O; 100 ml of AC medium in 150-ml flasks inoculated with overnight cultures to give a beginning optical density at 660 nm (OD660) of ≈0.05. The flasks were sparged with filter-sterilized N2O and incubated at 30°C. (D to F) Activity measurements of MK4T4 (■; ▴), MK4E1 (○), and MK21 (□). (D) N2OR activity of whole cells. (E) Nitrite reductase activity of whole cells and additionally MK4T4 in vitro (▴). (F) NO reductase activity of whole cells and additionally MK4T4 in vitro (▴). For the assay conditions, see Materials and Methods. The reaction was started by the addition of the substrate. Gases were analyzed by gas chromatography.
NO reductase, which was detected by immunoblotting in MK4T4, did not have in vivo activity but was active in cell extract (Fig. 3F). The enzyme is an integral membrane protein. Processing of NO reductase is limited to removal of the terminal methionine residues from its two subunits, NorC and NorB (54). The conditional nature of the defect in vivo and in vitro could be caused by various means, such as an assembly factor, an electron donor, or an indirect effect related to the defect in nitrite reduction, and will require a separate study. NO is the inducer for the nir and nor genes, which is reflected in the down-regulation of nor expression in nir mutants lacking NO production (35). However, in the previously studied nir mutants, the activity of NO reductase was up-regulated, whereas this was not the case for the tatC mutant.
tatC inactivation results in a mislocated, cytoplasmic N2OR lacking the Cu chromophores.
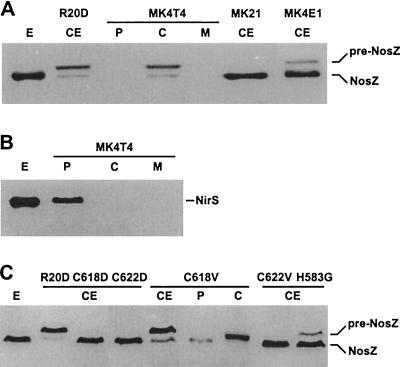
The location of N2OR in MK4T4 was investigated by fractionating cells into cytoplasm, periplasm, and membranes and analyzing them for the enzyme immunochemically. N2OR of MK4T4 was found to be mislocated in the cytoplasmic cell compartment, mainly in its unprocessed precursor form (Fig. 4A). No N2OR was detected in the periplasm or bound to membranes of MK4T4. Under all growth conditions, only one band, corresponding to processed enzyme, was detected in the cell extract of MK21 representing wild-type character. Thus, translocation and processing of N2OR was not rate limiting in relation to synthesis.
FIG. 4.
Localization and processing of N2OR in tat and CuA mutants. Immunoblot analysis with polyclonal antisera was done for cell extract (CE) and cells fractionated into periplasm (P), cytoplasm (C), and membranes (M). Enzyme (E) purified from MK21 was used as a size marker for the mature forms of N2OR (NosZ) and cytochrome cd1 (NirS). Cell extract from the signal peptide mutant R20D provided unprocessed N2OR (pre-NosZ); cell extract from MK21 shows the wild-type situation. (A) Cytoplasmic location of a predominantly unprocessed N2OR in MK4T4; cell extract from MK21 shows only processed N2OR, whereas extract from MK4E1 also shows unprocessed enzyme. (B) Processing and periplasmic location of cytochrome cd1 nitrite reductase in MK4T4. The cell fractions of MK4T4 were the same as those used in panel A. The cytoplasmic fraction was free of contamination by periplasm, as is evident from the absence of a cytochrome cd1 signal. In panels A and B, no N2OR or cytochrome cd1 was found bound to membranes. (C) Localization and processing of N2OR in CuA mutants. N2OR in the cell extracts of C618D and C622D was completely processed. C618V synthesized a predominantly unprocessed, cytoplasmic N2OR; the small contribution of processed enzyme was periplasmic. No unprocessed material was found in the cell extract of C622V; H583 harbored both species, with the mature form being predominant.
As the reference material for unprocessed enzyme, we used mutant R20D, in which the twin-arginine motif of the signal peptide had been targeted by site-directed mutagenesis. We had shown previously by electrospray mass spectrometry that the mass of N2OR from the R20D mutant is increased by 5 kDa compared with that of the wild-type enzyme due to the uncleaved signal peptide. The lack of processing was concomitant with the mislocation of N2OR in the cytoplasm (18). In both MK4T4 and R20D, a small portion of processed material was seen which, however, was not located in the periplasm. Cytochrome cd1 nitrite reductase, which has a Sec-type signal peptide, was transported to the periplasm in the tatC mutant and thus was used as a control to make certain that the fraction representing the cytoplasm was free of contamination by periplasm (Fig. 4B).
To find out whether the reductase synthesized by MK4T4 was holo-N2OR with its Cu cofactors, a qualitative test was applied that has been used in the past to show the cochromatography of Cu with N2OR by gel filtration (13, 18, 55). Cell extract was passed over a column of Sephacryl S-300 HR, and the effluent was analyzed for Cu and N2OR. Figure 5 shows a comparison of the elution profiles obtained with MK4T4 and MK21. In clear contrast to the wild-type situation, no Cu peak was found to migrate with N2OR in the tatC mutant extract. An estimate of Cu in the peak N2OR fractions showed that the small amount present was not enough even to occupy the binuclear CuA site. Cu was associated with a low-molecular-mass protein, eluting around fraction no. 105, which is also found in the wild type but has not been characterized functionally. N2OR from the mutant eluted at the position of the enzyme from MK21, which indicated that the dimeric quaternary structure was also attained by the reductase in the mutant case. If Cu were bound only loosely to cytoplasmic N2OR, it might be lost during gel filtration. In a previously studied case of an M629C site-directed exchange in the CuA center, Cu bonding apparently was weakened, which resulted in a low Cu content of the purified protein. Nevertheless, the initial association of Cu with the M629C derivative could be demonstrated clearly by the gel filtration technique (13). In conclusion, N2OR of the tatC mutant accumulated in the incorrect cell compartment in its unprocessed form, with empty CuA and CuZ centers. Considering the two periplasmic reductases of denitrification, N2OR and cytochrome cd1, translocation of N2OR was clearly dependent on the Tat pathway.
FIG. 5.
Lack of incorporation of Cu into N2OR of the tatC mutant. (A) Cell extract of MK4T4 was separated on a Sephacryl S-300 HR gel column (2.5 by 90 cm) equilibrated with 25 mM Tris-HCl (pH 7.5)–0.1 M KCl (flow rate, 25 ml/h; 2.5-ml fractions). (B) The same experiment using extract from MK21. The eluate was analyzed immunochemically for N2OR (■) and for Cu (○) by atomic absorption spectroscopy (18). The data were normalized with respect to the highest concentrations of N2OR and Cu in each data set.
Role for TatE in N2OR translocation.
tatE (formerly orf57) of P. stutzeri is part of the nos region encoding assembly factors. We have generated a tatE mutant (see Materials and Methods) and studied in the knockout strain MK4E1 whether TatE has a role specific for the translocation or maturation of its N2OR substrate. Mutant MK4E1 grew aerobically with the growth rate of strain MK21. It also grew on N2O like the wild type and, consequently, intact cells had N2OR activity (Fig. 3D). We also found nitrite reductase and NO reductase activities, which were only slightly reduced, if at all, compared to those of MK21 (Fig. 3E and F). However, cell extract of MK4E1 consistently showed a small fraction of the unprocessed form of N2OR (Fig. 4A), whereas in MK21, no unprocessed enzyme was detectable.
We attempted to demonstrate a role for TatE in N2OR translocation and/or processing more clearly by a complementation study of a mutant with a defect in Cu insertion (57). For this purpose, the nosD strain, MK401, was used as the host to express N2OR from a plasmid carrying the nos gene cluster nosRZDFYL with or without tatE (Fig. 6). The genes nosR, nosZ, and nosD were kept under the control of their own promoters. Both vectors resulted in the overexpression of a functional N2OR in the P. stutzeri wild type or the nosD background. When tatE was not present in trans in the nosD strain, a large portion of unprocessed N2OR was found, indicating that the chromosomally encoded Tat translocation system had become saturated. The unprocessed form, however, disappeared when the mutant was also complemented with tatE (Fig. 6B). This clearly demonstrated the participation of TatE in the translocation of N2OR.
FIG. 6.
TatE supports transport and processing of N2OR. (A) Expression vector, pUCP22RE, for the entire nos region, including tatE; vector pUCP22RL was identical but without tatE. The rep function is required for stable maintenance of the vector in Pseudomonas spp. (49). (B) Immunoblot analysis for N2OR of cell extracts of MK21 (lane 1), nosD mutant MK401 (lane 2), MK401 complemented with pUCP22RL (nosRZDFL) (lane 3), and MK401 complemented with pUCP22RE (nosRZDFYtatE) (lane 4). The protein standard was Seeblue (Novex). (C) Southern blot evidence for a tatE homolog in P. stutzeri. Genomic DNA was digested by PstI and hybridized at 45°C stringency by the downward capillary method; detection was done with DIG Easy Hyb granules (Roche). The observed fragments correspond to the tatABC cluster (3 kb), tatE (1.6 kb), and a putative homolog of tatE (0.35 kb).
The phenotype of the tatE mutant was intermediate compared with MK4T4. The mutation affected N2OR transport but, other than inactivation of tatC, did not impede it completely. This suggested the presence of a functional homolog. The genome of P. stutzeri was therefore searched by Southern hybridization with probe TE1 (Fig. 6C). Of the three signals observed, that with the strongest intensity was due to the 1.6-kb PstI fragments carrying tatE, whereas the weakest signal, at 3 kb, represented tatA. The intensity of the third signal, at 0.35 kb, was greater than that due to tatA (TatA and TatE have 71% positional identity) and hence is tentatively considered to represent a further tatE homolog.
The Tat system is sensitive to mutation of the CuA domain of N2O reductase.
The Tat system is thought to transport folded proteins, in contrast to the Sec system (3). We asked, therefore, whether the Tat system would be sensitive to a conformational alteration of the exported protein. We addressed this with site-directed mutants of the CuA center. The previously generated CuA mutants, H583G, C618D, C622D, H626G, and M629C, displayed a loss of catalytic activity, but all of these recombinant N2OR derivatives were exported to the periplasm (13). Figure 4C shows cell extracts of C618D and C622D, which contained only mature N2OR consistent with the periplasmic location. Both cysteine mutations affected the bridging residues of the CuA center. Replacement of the same residues by valine resulted in a differential positional effect for cysteine 618. Cell extract from the C618V mutant showed a predominance of the unprocessed N2OR species and a small amount of processed material (Fig. 4C). Unprocessed and processed forms of enzyme C618V were located in the cytoplasm and periplasm, respectively. In contrast, the C622V recombinant form exhibited wild-type behavior. This indicated for the C618V exchange a strong qualitative and positional effect. The H583G mutation of the CuA center also consistently showed a small percentage of unprocessed N2OR (Fig. 4C), and occasionally we have observed this with the M629C derivative. Thus, there are distinct structural alterations in the CuA domain of N2OR to which the Tat translocation pathway is sensitive.
The synthesis of a functional cytochrome cd1 is TatC dependent.
A conspicuous trait of MK4T4 was related to cytochrome cd1 nitrite reductase. This enzyme was detected immunochemically in MK4T4, where it was exported to the periplasm (Fig. 4B). However, the cells did not reduce nitrite, and the weak reductase activity detected in vitro could not be due to cytochrome cd1 for the following reason: we have isolated cytochrome cd1 from MK4T4 and found it to carry only heme C but no heme D1, as is evident from the electronic spectrum (Fig. 7). We attribute this to the dependence of the NirD protein, involved in heme D1 biosynthesis, on the Tat translocation pathway (see Discussion). In contrast, cytochrome cd1 from MK4E1 exhibited wild-type properties and was synthesized as an active holoenzyme with both types of heme groups.
FIG. 7.
Electronic absorption spectrum of purified cytochrome cd1 from tatC and tatE mutants. a, dithionite-reduced semiapo-cytochrome cd1 from MK4T4 (tatC); absorption bands of only the heme C moiety are present. b, dithionite-reduced holoenzyme from MK4E1 (tatE); the arrows indicate absorption bands due to heme D1.
DISCUSSION
In gram-negative bacteria, complete nitrite respiration leading to N2 is a periplasmic process, with nitrite reductase and N2OR both located in the outer cell compartment together with electron carriers and ancillary proteins necessary for denitrification (53). For the extant topology of the denitrification apparatus, it is necessary to understand how the principal components are transported to their functional site. Our strategy to isolate tat genes was based on sequence information from the chromosome of P. aeruginosa and a comparison with their E. coli homologs. This enabled us to design specific probes for identification of the P. stutzeri counterparts based on the considerable sequence similarity between the two species. We anticipate that the findings with P. stutzeri have relevance for its close relative.
A brief survey among pseudomonads indicated that the Tat system occurs frequently in this bacterial group. We found tatC homologs in EcoRI-digested genomic DNA by Southern hybridization with probe TC1 at 50°C stringency in Pseudomonas aureofaciens (ATCC 13985); Pseudomonas chlororaphis (ATCC 9446); Pseudomonas fluorescens biotypes A (DSM 50091), C (DSM 50117), and F (ATCC 12983); Pseudomonas putida (DSM 50906); and Ralstonia (formerly Pseudomonas) solanacearum (DSM 50905) irrespective of whether these strains are denitrifiers or utilize N2O (data not shown). In agreement with our evidence from Southern hybridization, we found a tatABC cluster in contig 10775 at positions 40584 to 42019 in the ongoing genome project of P. putida KT2440 (The Institute for Genomic Research website [http://www.tigr.org]).
TatC and TatB of E. coli play crucial roles in the translocation mechanism, because deletion of either tat gene is sufficient to block protein export (6, 44). Likewise, a deletion in tatC of P. stutzeri blocked N2OR export and caused the unprocessed enzyme to accumulate in the cytoplasm. In the denitrifier Ralstonia eutropha, a putative tatA locus has also been implicated in N2OR translocation based on the observation of a mutational loss of N2O-reducing activity of whole cells (5). The P. stutzeri tatC mutant exhibited the phenotype of the R20D mutant studied previously (18). Although the unprocessed N2OR of mutant R20D lacked Cu, the electron transfer center, CuA, could be occupied by incubating the isolated protein with Cu. We assume this to be indicative of a correctly folded form of the enzyme in spite of its cytoplasmic location. Although a rather specific conformation seems to be required to explain the observation regarding the C618V and H583G recombinant forms of the enzyme, Cu insertion into N2OR is not a prerequisite for the Tat pathway to function. This is different from nickel-iron hydrogenase 2, where Ni has to be incorporated into the protein prior to export (39). Our results with N2OR are in contrast to the view that cofactor insertion prior to export is a general trait of Tat-dependent proteins. Preexport cofactor insertion may simply reflect protein folding (42) and serve metabolic efficiency by dispensing with a separate transport system for the cofactor.
On substituting the polar side chain of cysteine 618 for the hydrophobic one of valine, the Tat translocation pathway strongly rejected N2OR. Valine fits into the site of cysteine in the N2OR structure (10), but it is unknown precisely which structural change is imposed by the mutation on the electron transfer domain and the overall enzyme conformation. Certain changes in the CuA center affect the juxtaposed CuZ domain, as deduced from altered spectroscopic properties (13). Our observations indicate that the Tat system is constrained by distinct conformations in the folded form of N2OR. Since cysteine 618 is positioned only 20 aa away from the C terminus of N2OR (overall, the enzyme comprises 638 aa), it means that the Tat system recognizes the mutationally induced structural change in the completely or nearly completely synthesized N2OR.
When a Tat system is present, the genome usually encodes one TatC protein and at least two homologs of the TatA or -E and TatB groups. It has been suggested that TatA, TatB, and TatE may function as membrane receptors for different subsets of redox proteins (12), although no difference in substrate specificity between TatA and TatE has been established (43, 44). Our data do not support a specific receptor role of TatE for N2OR, in spite of the location of tatE in the nos gene cluster. The view that TatC provides the membrane receptor for the signal peptide and that TatE or TatA forms the translocase pore (4) is more plausible from our observations. A TatE transport channel may be the usual route for N2OR, but a homolog can fulfill this role when TatE is missing in mutant MK4E1.
The periplasmic cytochrome cd1 nitrite reductase of P. stutzeri has a Sec-type signal peptide (30). The translocation of cytochrome cd1 was not affected by the tatC mutation, which strongly suggests export of this enzyme via the Sec pathway. Since nitrite reductase was not active in the tatC strain, at least one Tat-dependent factor is required to establish a functional cytochrome cd1. We suggest that this factor is NirD. Heme D1 is found only in the cytochrome cd1-type nitrite reductase of denitrifying bacteria. The nirD locus is involved in heme D1 biosynthesis or processing (35). The presequence of the NirD protein, MHIDALSRRLIDRYQHGMPLCAEPYRAMA (critical residues are shown in boldface), exhibits the characteristics of a Tat-specific signal peptide. It has an H region significantly less hydrophobic than that of a Sec signal peptide. The heptameric motif with an arginine pair overlaps the boundary of the N and H regions, and even though this motif varies from the suggested consensus for Tat-dependent transport, most of the variant positions are conservative substitutions and consistent with evidence from site-directed mutagenesis (9, 46). A critical proline residue which is commonly found in Tat signal sequences (16) occupies position −6 of the putative signal peptidase cleavage site, further supporting the notion of a Tat-type signal peptide. Cleavage may be at 27-AMA↓E, and a basic residue, arginine 26, is next to the consensus sequence, features which are frequent in Tat signal peptides. The likely dependence of NirD export on the Tat pathway raises interesting questions as to whether there are steps in the synthesis of heme D1 and its delivery to cytochrome cd1 that proceed in the periplasm or at the periplasmic side of the inner membrane and precisely which function is performed by the NirD protein. Our data on cytochrome cd1 present an intriguing case where the Tat and Sec systems have to cooperate for the assembly of a periplasmic enzyme.
ACKNOWLEDGMENTS
We thank H. Körner for performing computational tasks and S. Berker for assistance with nitrite reductase purification. M.P.H. is indebted to K.-U. Vollack for helpful advice. We acknowledge sequence information from the Pseudomonas Genome Project prior to publication.
The work was supported by the Deutsche Forschungsgemeinschaft, Fonds der Chemischen Industrie, and Deutscher Akademischer Austauschdienst.
REFERENCES
- 1.Alexeyev M F. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques. 1995;18:52–55. [PubMed] [Google Scholar]
- 2.Beinert H. Copper in biological systems. A report from the 7th Manziana Conference, held at Santa Severa, September 11–15, 1995. J Inorg Biochem. 1996;64:79–135. doi: 10.1016/0162-0134(96)00083-9. [DOI] [PubMed] [Google Scholar]
- 3.Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 4.Berks B C, Sargent F, Palmer T. The Tat protein export pathway. Mol Microbiol. 2000;35:260–274. doi: 10.1046/j.1365-2958.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 5.Bernhard M, Friedrich B, Siddiqui R A. Ralstonia eutropha TF93 is blocked in Tat-mediated protein export. J Bacteriol. 2000;182:581–588. doi: 10.1128/jb.182.3.581-588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogsch E G, Sargent F, Stanley N R, Berks B C, Robinson C, Palmer T. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- 7.Bolhuis A, Bogsch E G, Robinson C. Subunit interaction in the twin-arginine translocase complex of Escherichia coli. FEBS Lett. 2000;472:88–92. doi: 10.1016/s0014-5793(00)01428-9. [DOI] [PubMed] [Google Scholar]
- 8.Braun C, Zumft W G. The structural genes of the nitric oxide reductase complex from Pseudomonas stutzeri are part of a 30-kilobase gene cluster for denitrification. J Bacteriol. 1992;174:2394–2397. doi: 10.1128/jb.174.7.2394-2397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brink S, Bogsch E G, Edwards W R, Hynds P J, Robinson C. Targeting of thylakoid proteins by the ΔpH-driven twin-arginine translocation pathway requires a specific signal in the hydrophobic domain in conjunction with the twin-arginine motif. FEBS Lett. 1998;434:425–430. doi: 10.1016/s0014-5793(98)01028-x. [DOI] [PubMed] [Google Scholar]
- 10.Brown K, Tegoni M, Prudêncio M, Pereira A S, Besson S, Moura J J, Moura I, Cambillau C. A novel type of catalytic copper cluster in nitrous oxide reductase. Nat Struct Biol. 2000;7:191–195. doi: 10.1038/73288. [DOI] [PubMed] [Google Scholar]
- 11.Chaddock A M, Mant A, Karnauchov I, Brink S, Herrmann R G, Klösgen R B, Robinson C. A new type of signal peptide: central role of a twin-arginine motif in transfer signals for the ΔpH-dependent thylakoidal protein translocase. EMBO J. 1995;14:2715–2722. doi: 10.1002/j.1460-2075.1995.tb07272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chanal A, Santini C-L, Wu L-F. Potential receptor function of three homologous components, TatA, TatB, and TatE, of the twin-arginine signal-sequence-dependent metalloenzyme pathway in Escherichia coli. Mol Microbiol. 1998;30:673–678. doi: 10.1046/j.1365-2958.1998.01095.x. [DOI] [PubMed] [Google Scholar]
- 13.Charnock J M, Dreusch A, Körner H, Neese F, Nelson J, Kannt A, Michel H, Garner C D, Kroneck P M H, Zumft W G. Structural investigations of the CuA centre of nitrous oxide reductase from Pseudomonas stutzeri by site-directed mutagenesis and X-ray absorption spectroscopy. Eur J Biochem. 2000;267:1368–1381. doi: 10.1046/j.1432-1327.2000.01131.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen W, Kuo T. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res. 1993;21:2260. doi: 10.1093/nar/21.9.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomczynski P. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal Biochem. 1992;201:134–139. doi: 10.1016/0003-2697(92)90185-a. [DOI] [PubMed] [Google Scholar]
- 16.Cristóbal S, de Gier J-W, Nielsen H, von Heijne G. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J. 1999;18:2982–2990. doi: 10.1093/emboj/18.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreusch A. Liganden der Kupferzentren der N2O-Reduktase von Pseudomonas stutzeri sowie Untersuchungen zur Biosynthese und Prozessierung des Enzyms. Ph.D. dissertation. Karlsruhe, Germany: Karlsruhe University; 1995. [Google Scholar]
- 18.Dreusch A, Bürgisser D M, Heizmann C W, Zumft W G. Lack of copper insertion into unprocessed cytoplasmic nitrous oxide reductase generated by an R20D substitution in the arginine consensus motif of the signal peptide. Biochim Biophys Acta. 1997;1319:311–318. doi: 10.1016/s0005-2728(96)00174-0. [DOI] [PubMed] [Google Scholar]
- 19.Dreusch A, Riester J, Kroneck P M H, Zumft W G. Mutation of the conserved Cys165 outside the CuA domain destabilizes nitrous oxide reductase but maintains its catalytic activity: evidence for disulfide bridges and a putative disulfide isomerase gene. Eur J Biochem. 1996;237:447–453. doi: 10.1111/j.1432-1033.1996.0447k.x. [DOI] [PubMed] [Google Scholar]
- 20.Engler-Blum G, Meier M, Frank J, Müller G A. Reduction of background problems in nonradioactive Northern and Southern blot analyses enables higher sensitivity than 32P-based hybridizations. Anal Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- 21.Farinha M A, Kropinski A M. High efficiency electroporation of Pseudomonas aeruginosa using frozen cell suspensions. FEMS Microbiol Lett. 1990;70:221–225. doi: 10.1111/j.1574-6968.1990.tb13982.x. [DOI] [PubMed] [Google Scholar]
- 22.Feliciello I, Chinali G. A modified alkaline lysis method for the preparation of highly purified plasmid DNA from Escherichia coli. Anal Biochem. 1993;212:394–401. doi: 10.1006/abio.1993.1346. [DOI] [PubMed] [Google Scholar]
- 23.Frunzke K, Zumft W G. Inhibition of nitrous-oxide respiration by nitric oxide in the denitrifying bacterium Pseudomonas perfectomarina. Biochim Biophys Acta. 1986;852:119–125. [Google Scholar]
- 24.Giuffrè A, Stubauer G, Sarti P, Brunori M, Zumft W G, Buse G, Soulimane T. The heme-copper oxidases of Thermus thermophilus catalyze the reduction of nitric oxide: evolutionary implications. Proc Natl Acad Sci USA. 1999;96:14718–14723. doi: 10.1073/pnas.96.26.14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glockner A B, Zumft W G. Sequence analysis of an internal 9.72-kb segment from the 30-kb denitrification gene cluster of Pseudomonas stutzeri. Biochim Biophys Acta. 1996;1277:6–12. doi: 10.1016/s0005-2728(96)00108-9. [DOI] [PubMed] [Google Scholar]
- 26.Gross R, Simon J, Kröger A. The role of the twin-arginine motif in the signal peptide encoded by the hydA gene of the hydrogenase from Wolinella succinogenes. Arch Microbiol. 1999;172:227–232. doi: 10.1007/s002030050764. [DOI] [PubMed] [Google Scholar]
- 27.Halbig D, Wiegert T, Blaudeck N, Freudl R, Sprenger G A. The efficient export of NADP-containing glucose-fructose oxidoreductase to the periplasm of Zymomonas mobilis depends both on an intact twin-arginine motif in the signal peptide and on the generation of a structural export signal induced by cofactor binding. Eur J Biochem. 1999;263:543–551. doi: 10.1046/j.1432-1327.1999.00536.x. [DOI] [PubMed] [Google Scholar]
- 28.Heikkilä M P, Honisch U, Zumft W G. Translocation of N2O reductase by the Tat system. Biospectrum. 2000;6:54. . (Abstract.) [Google Scholar]
- 29.Hoeren F U, Berks B C, Ferguson S J, McCarthy J E G. Sequence and expression of the gene encoding the respiratory nitrous-oxide reductase from Paracoccus denitrificans: new and conserved structural and regulatory motifs. Eur J Biochem. 1993;218:49–57. doi: 10.1111/j.1432-1033.1993.tb18350.x. [DOI] [PubMed] [Google Scholar]
- 30.Jüngst A, Wakabayashi S, Matsubara H, Zumft W G. The nirSTBM region coding for cytochrome cd1-dependent nitrite respiration of Pseudomonas stutzeri consists of a cluster of mono-, di-, and tetraheme proteins. FEBS Lett. 1991;279:205–209. doi: 10.1016/0014-5793(91)80150-2. [DOI] [PubMed] [Google Scholar]
- 31.Kannt A, Michel H, Cheesman M R, Thomson A J, Dreusch A B, Körner H, Zumft W G. The electron transfer centers of nitric oxide reductase: homology with the heme-copper oxidase family. In: Canters G W, Vijgenboom E, editors. Biological electron-transfer chains: genetics, composition and mode of operation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 279–291. [Google Scholar]
- 32.Körner H, Mayer F. Periplasmic location of nitrous oxide reductase and its apoform in denitrifying Pseudomonas stutzeri. Arch Microbiol. 1992;157:218–222. doi: 10.1007/BF00245153. [DOI] [PubMed] [Google Scholar]
- 33.Kroneck P M H, Antholine W A, Riester J, Zumft W G. The cupric site in nitrous oxide reductase contains a mixed-valence [Cu(II), Cu(I)] binuclear center: a multifrequency electron paramagnetic resonance investigation. FEBS Lett. 1988;242:70–74. doi: 10.1016/0014-5793(88)80987-6. [DOI] [PubMed] [Google Scholar]
- 34.Nivière V, Wong S-L, Voordouw G. Site-directed mutagenesis of the hydrogenase signal peptide consensus box prevents export of a β-lactamase fusion protein. J Gen Microbiol. 1992;138:2173–2183. doi: 10.1099/00221287-138-10-2173. [DOI] [PubMed] [Google Scholar]
- 35.Palmedo G, Seither P, Körner H, Matthews J C, Burkhalter R S, Timkovich R, Zumft W G. Resolution of the nirD locus for heme d1 synthesis of cytochrome cd1 (respiratory nitrite reductase) from Pseudomonas stutzeri. Eur J Biochem. 1995;232:737–746. [PubMed] [Google Scholar]
- 36.Rand, K. N. 1996. Crystal violet can be used to visualize DNA bands during gel electrophoresis and to improve cloning efficiency. Tech. Tips Online 1. [Online.] http://research.bmn.com/tto.
- 37.Rasmussen T, Berks B C, Sanders-Loehr J, Dooley D M, Zumft W G, Thomson A J. The catalytic center in nitrous oxide reductase, Cuz, is a copper sulfide cluster. Biochemistry. 2000;39:12753–12756. doi: 10.1021/bi001811i. [DOI] [PubMed] [Google Scholar]
- 38.Robinson C. The twin-arginine translocation system: a novel means of transporting folded proteins in chloroplasts and bacteria. Biol Chem. 2000;381:89–93. doi: 10.1515/BC.2000.013. [DOI] [PubMed] [Google Scholar]
- 39.Rodrigue A, Boxer D H, Mandrand-Berthelot M A, Wu L F. Requirement for nickel of the transmembrane translocation of NiFe-hydrogenase 2 in Escherichia coli. FEBS Lett. 1996;392:81–86. doi: 10.1016/0014-5793(96)00788-0. [DOI] [PubMed] [Google Scholar]
- 40.Sambasivarao D, Turner D J, Simala-Grant J L, Shaw G, Hu J, Weiner J H. Multiple roles for the twin arginine leader sequence of dimethyl sulfoxide reductase of Escherichia coli. J Biol Chem. 2000;275:22526–22531. doi: 10.1074/jbc.M909289199. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Santini C-L, Ize B, Chanal A, Müller M, Giordano G, Wu L-F. A novel Sec-independent periplasmic translocation pathway in Escherichia coli. EMBO J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sargent F, Bogsch E G, Stanley N R, Wexler M, Robinson C, Berks B C, Palmer T. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sargent F, Stanley N R, Berks B C, Palmer T. Sec-independent protein translocation in Escherichia coli, a distinct and pivotal role for the TatB protein. J Biol Chem. 1999;274:36073–36082. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- 45.Settles A M, Yonetani A, Baron A, Bush D R, Cline K, Martienssen R. Sec-independent protein translocation by the maize Hcf106 protein. Science. 1997;278:1467–1470. doi: 10.1126/science.278.5342.1467. [DOI] [PubMed] [Google Scholar]
- 46.Stanley N R, Palmer T, Berks B C. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J Biol Chem. 2000;275:11591–11596. doi: 10.1074/jbc.275.16.11591. [DOI] [PubMed] [Google Scholar]
- 47.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S L, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K-S, Wu Z, Paulsen I T, Reizer J, Saier M H, Hancock R E W, Lory S, Olson M V. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 48.Weiner J H, Bilous P T, Shaw G M, Lubitz S P, Frost L, Thomas G H, Cole J A, Turner R J. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 49.West S E H, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;128:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 50.Wexler M, Sargent F, Jack R L, Stanley N R, Bogsch E G, Robinson C, Berks B C, Palmer T. TatD is a cytoplasmic protein with DNase activity: no requirement for TatD family proteins in Sec-independent protein export. J Biol Chem. 2000;275:16717–16722. doi: 10.1074/jbc.M000800200. [DOI] [PubMed] [Google Scholar]
- 51.Wu L-F, Ize B, Chanal A, Quentin Y, Fichant G. Bacterial twin-arginine signal peptide-dependent protein translocation pathway: evolution and mechanism. J Mol Microbiol Biotechnol. 2000;2:179–189. [PubMed] [Google Scholar]
- 52.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 53.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zumft W G, Braun C, Cuypers H. Nitric oxide reductase from Pseudomonas stutzeri: primary structure and gene organization of a novel bacterial cytochrome bc complex. Eur J Biochem. 1994;219:481–490. doi: 10.1111/j.1432-1033.1994.tb19962.x. [DOI] [PubMed] [Google Scholar]
- 55.Zumft W G, Döhler K, Körner H. Isolation and characterization of transposon Tn5-induced mutants of Pseudomonas perfectomarina defective in nitrous oxide respiration. J Bacteriol. 1985;163:918–924. doi: 10.1128/jb.163.3.918-924.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zumft W G, Dreusch A, Löchelt S, Cuypers H, Friedrich B, Schneider B. Derived amino acid sequences of the nosZ gene (respiratory N2O reductase) from Alcaligenes eutrophus, Pseudomonas aeruginosa and Pseudomonas stutzeri reveal potential copper-binding residues: implications for the CuA site of N2O reductase and cytochrome-c oxidase. Eur J Biochem. 1992;208:31–40. doi: 10.1111/j.1432-1033.1992.tb17156.x. [DOI] [PubMed] [Google Scholar]
- 57.Zumft W G, Viebrock-Sambale A, Braun C. Nitrous oxide reductase from denitrifying Pseudomonas stutzeri: genes for copper-processing and properties of the deduced products, including a new member of the family of ATP/GTP-binding proteins. Eur J Biochem. 1990;192:591–599. doi: 10.1111/j.1432-1033.1990.tb19265.x. [DOI] [PubMed] [Google Scholar]