Abstract
We are currently investigating the role of ToxR-mediated gene regulation in Photobacterium profundum strain SS9. SS9 is a moderately piezophilic (“pressure loving”) psychrotolerant marine bacterium belonging to the family Vibrionaceae. In Vibrio cholerae, ToxR is a transmembrane DNA binding protein involved in mediating virulence gene expression in response to various environmental signals. A homolog to V. cholerae ToxR that is necessary for pressure-responsive gene expression of two outer membrane protein-encoding genes was previously found in SS9. To search for additional genes regulated by ToxR in SS9, we have used RNA arbitrarily primed PCR (RAP-PCR) with wild-type and toxR mutant strains of SS9. Seven ToxR-activated transcripts and one ToxR-repressed transcript were identified in this analysis. The cDNAs corresponding to these partial transcripts were cloned and sequenced, and ToxR regulation of their genes was verified. The products of these genes are all predicted to fall into one or both of two functional categories, those whose products alter membrane structure and/or those that are part of a starvation response. The transcript levels of all eight newly identified genes were also characterized as a function of hydrostatic pressure. Various patterns of pressure regulation were observed, indicating that ToxR activation or repression cannot be used to predict the influence of pressure on gene expression in SS9. These results provide further information on the nature of the ToxR regulon in SS9 and indicate that RAP-PCR is a useful approach for the discovery of new genes under the control of global regulatory transcription factors.
In Vibrio cholerae, the transmembrane DNA binding protein ToxR and the associated membrane protein ToxS are best known for their central role in the environmental regulation of virulence gene expression (15, 37, 38, 50). However, both ToxR and ToxS are widely distributed proteins among the Vibrionaceae, and their homologs have been found in eleven members of the family (32, 33, 41, 44, 55). Many of these additional species are human or fish pathogens. Vibrio parahaemolyticus, for example, is a major cause of gastroenteritis associated with seafood consumption (26). Other ToxR-containing bacteria are Vibrio fischeri, a bioluminescent bacterium associated with the light organs of certain fish and squid (16, 46), and Photobacterium profundum strain SS9, a deep-sea bacterium originally isolated from amphipod crustaceans (4, 14). Since many of the species containing ToxR are nonpathogenic to mammals, and since the ToxR-regulated virulence genes in V. cholerae (but not the toxRS operon) are largely acquired by horizontal gene transfer (27, 51), the ToxR regulatory system is not likely to have first evolved for mammalian colonization. However, little is known about its role outside of virulence in higher organisms.
Alkaline phosphatase gene fusion studies and toxR mutant analyses indicate that the V. cholerae ToxR regulon includes over 20 genes (20, 25, 29, 37, 42, 43). All but one of these genes require ToxR for expression, with the remaining gene being repressed by ToxR. These genes can be divided into those that do or do not depend on the transcription factor ToxT, whose gene expression is activated by ToxR and ToxS, along with TcpP and TcpH, another pair of membrane-localized proteins related to ToxR and ToxS, respectively (21). The ToxT-dependent branch of the regulon includes the expression of toxin and colonization genes associated with the CTXφ genome (51) and the vibrio pathogenicity island, which has also been proposed to represent a filamentous phage genome (27). The ToxT-independent branch of the ToxR regulon consists of two outer membrane protein (OMP)-encoding genes, ToxR-activated ompU and ToxR-repressed ompT (13, 33, 37, 47).
Outside of V. cholerae, much less is known about the ToxR regulon. In V. parahaemolyticus and Vibrio vulnificus, ToxR is needed for hemolysin gene expression (32, 34). SS9 ToxR and V. vulnificus ToxR, like V. cholerae ToxR, regulate the inverse expression of two OMPs (32, 55). In the case of SS9, the genes encoding these OMPs have been isolated. SS9 ompL is ToxR activated, and its product is 50% identical and 63% similar to V. cholerae OmpU (47, 54), whereas SS9 ompH is ToxR repressed, and its product is 30% identical and 53% similar to V. cholerae OmpT (5, 33). Western analysis indicates that proteins similar to OmpL are present in V. fischeri, Vibrio furnissii, Photobacterium angustum, and Photobacterium leiognathi, as well as in V. cholerae (55). So a minimum conserved unit for the ToxR regulon among diverse members of the Vibrionaceae could include ToxRS and one or two omp genes.
The ToxR regulation of omp gene expression contains similarities and differences with the well-characterized ompFC control by OmpR and EnvZ in Escherichia coli (19, 30). The cytoplasmic amino-terminal DNA binding domain of ToxR shares significant identity with the OmpR family of regulatory proteins (38), although, unlike OmpR, ToxR lacks a phosphoacceptor domain and is not believed to function as a two-component system response regulator. Also like ToxR, OmpR has been found to be a global regulator of gene expression in both E. coli and Salmonella enterica (see, for example, references 3, 31, 49, and 57).
ToxR regulation in SS9 is responsive to environmental cues different from those for V. cholerae ToxR. Whereas the V. cholerae ToxR circuit is controlled by medium salinity and amino acid content and also by temperature and pH in the case of the ToxT branch (37), SS9 ToxR modulation of ompL and ompH expression is controlled by hydrostatic pressure. ompL transcription is optimal at atmospheric pressure, while at 28 MPa ompL expression is low and ompH transcription is derepressed (4, 11, 12, 54, 55). Thus, the ToxR system in the piezophilic deep-sea bacterium SS9 appears to have evolved mechanisms for responding to the changes in pressure that the organism encounters as it is moved up or down within the water column.
In order to further assess the nature of the ToxR regulon in SS9, we have chosen to use an RNA arbitrarily primed PCR (RAP-PCR) approach. This technique uses random oligonucleotide primers to create a unique cDNA fingerprint for a given microorganism in a particular physiological state, thus providing a powerful tool for assessing differential gene expression in prokaryotes. It has recently been used to examine microbial responses to stress induction, soil growth conditions, and host infection (7, 10, 18, 56). Here we report on using RAP-PCR to reveal differences in gene expression between wild-type and toxR mutant strains of SS9 both at atmospheric pressure and at 28 MPa, the SS9 pressure optimum. These two different conditions were selected because we were interested in uncovering additional genes influenced by ToxR in SS9 as well as examining the nature of high-pressure gene regulation by ToxR. Eight additional SS9 ToxR-regulated genes are described along with their significance to understanding the nature of the SS9 ToxR regulon.
MATERIALS AND METHODS
Bacterial strains and plasmids.
P. profundum SS9 strains DB110 and TW30 were used in these studies. DB110 is a rifampin-resistant derivative of wild-type SS9 (11), and TW30 contains a 478-bp deletion within the toxR gene of DB110 (55). Strains were aerobically cultured in 2216 medium (28 g/liter; Difco Laboratories, Detroit, Mich.) at 15°C. Anaerobic growth of SS9 cultures was performed at 0.1 or 28 MPa (9°C) in 2216 medium supplemented with glucose (22 mM) and 0.1 M HEPES, pH 7.5. High-pressure growth of SS9 cultures was performed as previously described (8). Plasmid pDB1231 contains the SS9 toxRS operon from pTW15 (50) cloned into the BamHI-XhoI site of the broad-host-range vector pKT231 (2). The antibiotic kanamycin was used at a concentration of 200 μg/ml during the cultivation of TW30 with pDB1231.
RNA preparations.
Mid-exponential-phase cultures were harvested and RNA was extracted using RNAzol B (Tel-Test; Friendswood, Tex.), a guanidinium thiocyanate-phenol-based reagent, according to the manufacturer's instructions. RNA was quantitated using a UV spectrophotometer, and its quality was assessed on an agarose gel prior to experiments. When necessary, RNA was treated with DNase I (Ambion; Austin, Tex.) according to the manufacturer's recommendations.
RAP-PCR.
RAP-PCR was performed essentially as described by Fleming et al. (18). Arbitrary 10-mer primer kits with G+C contents of 50% (GEN1-50) and 60% (GEN1-60) were obtained from Genosys Biotechnologies (The Woodlands, Tex.). First-strand cDNA synthesis was performed using 200 ng of heat-denatured (10 min, 65°C) RNA in 20-μl reaction mixtures containing the following: 200 μM concentrations of each deoxynucleoside triphosphate, 5 mM dithiothreitol, 50 U of Moloney murine leukemia virus-reverse transcriptase (RT) (Ambion), 1× RT reaction buffer, and 0.4 μM arbitrary primer. The first-strand reaction was performed in an MJ Research minicycler (PTC-150) as follows: touchdown from 50 to 30°C in 45-s increments followed by 1 h at 37°C. Second-strand synthesis was performed in 30-μl reactions containing the following; 3 μl of first-strand reaction, 0.3 U of Taq polymerase (Gibco BRL, Rockville, Md.), 20 μM concentrations of each deoxynucleoside triphosphate, 6% dimethyl sulfoxide, 2 μM concentrations of each primer (primer 1 is from first-strand synthesis; primer 2 is a different randomly chosen primer), 2.5 μCi of [α-32P]dCTP, 1× PCR buffer, and 1.5 mM MgCl2. Cycles for RAP-PCR were 94°C for 30 s, 40°C for 2 min, and 72°C for 1 min for 40 cycles, with a 10-min extension at 72°C on the final cycle. All products were stored at −20°C for up to 1 day prior to electrophoresis. RAP-PCR samples were heated at 92°C for 2 min after the addition of loading dye (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol FF). Samples were then loaded onto a 5% denaturing acrylamide gel (20 by 42 cm) containing 7 M urea and were electrophoresed at 1,700 V for ∼2 h until the xylene cyanol band reached the bottom of the gel. The gels were subsequently dried, marked asymmetrically with a phosphorescent pen for orientation, and exposed to film (Eastman Kodak, Rochester, N.Y.) for 1 to 3 days at −80°C.
ompL RAP-PCR.
Primers ompLf (5′ CATCACTGAAGATTTCTAC) and ompLr (5′ CTACAGTGTTACGTGA) were synthesized (Integrated DNA Technologies, Coralville, Iowa) and were designed to amplify a 513-bp internal fragment of the ompL gene. ompLr was used in first-strand cDNA synthesis, and both primers were used in second-strand synthesis as described above.
Isolation and cloning of RAP-PCR fragments.
Following identification of regulated fragments, bands of interest were isolated in the following manner. The developed film was placed on the dried gel, with phosphorescent marks used for alignment. Bands were excised from the dried gel using a scalpel and were placed into a microcentrifuge tube containing 50 μl of Tris-EDTA for overnight elution at room temperature. A portion of the eluted fragment was used in a subsequent PCR containing the original primers used in RAP-PCR amplification. Following secondary PCR, fragments were cloned using the TOPO-TA cloning kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Clones verified for the correctly sized insert were then sequenced to determine identity.
DNA sequencing.
DNA sequences were determined by thermal cycle dideoxy sequencing with an ABI373A automated sequencer using fluorescently labeled terminators (Perkin-Elmer, Branchburg, N.J.) and T7 primers. Global similarity searches were performed with the BLAST network service (1).
Confirmation of regulation of RAP-PCR fragments by ToxR.
To verify differential expression of the fragments identified in RAP-PCR, the approach described by Benson et al. was used (7). This is a quantitative RAP-PCR method that relies first on the creation of random cDNA molecules from an RNA population, followed by gene-specific PCR. Briefly, first-strand cDNA was created using RNA from SS9 strains following the same RT conditions stated above, with the exception that 2 μM concentrations of random hexamers were used in place of a single arbitrary primer. RAP-PCR was then performed, under the same conditions as those already described, on the first-strand cDNA population by using custom-made 18-mer primer pairs specific for each sequence being tested. Synthesis of a PCR product of the correct size and expression pattern was considered verified for differential expression of the original RAP-PCR result. All quantitative RAP-PCR experiments were performed in duplicate to ensure reproducibility.
Nucleotide sequence accession numbers.
The sequences of the partial gene fragments identified in this work are deposited in GenBank under accession no. AF307974 to AF307981.
RESULTS AND DISCUSSION
RAP-PCR analysis of ToxRS regulated sequences.
Differential gene expression in ToxR+ and ToxR− SS9 strains was examined in cells grown aerobically in a nutrient-rich medium at 15°C and atmospheric pressure. These conditions were employed because the previously identified ToxR-regulated ompH and ompL genes are well expressed in either ToxR+ (ompL) or ToxR− (ompH) strains under these growth conditions (54, 55).
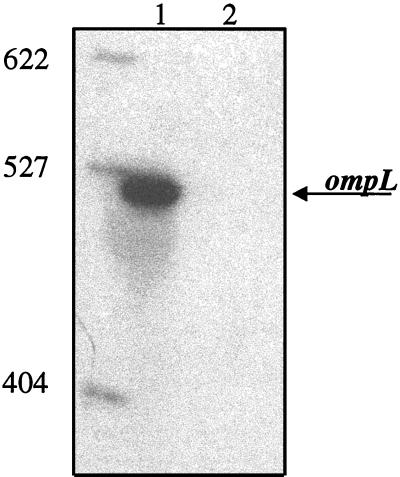
Before the start of the RAP-PCR survey of ToxR-regulated genes in SS9, a positive control for the technique was first performed using the ToxR-regulated ompL gene. Reactions were carried out to attempt to amplify an internal fragment of ompL from DB110 (ToxR+) and TW30 (ToxR−) SS9 strains grown at 0.1 MPa by using RAP-PCR with primers specific to an internal portion of the ompL sequence. As expected, a 513-bp fragment of ompL was expressed in DB110 but not in TW30 (Fig. 1). A second control testing for reproducibility of the technique was also performed. The first RAP-PCR experiment was performed in duplicate using the same primer sets with RNA that was isolated, from strains of DB110 and TW30 grown aerobically at atmospheric pressure (0.1 MPa), on separate occasions. This experiment yielded nearly identical results (data not shown); therefore, to increase the numbers of primer sets examined, all subsequent RAP-PCR experiments were performed only once.
FIG. 1.
RAP-PCR performed using ompL-specific primers. Lane 1, DB110; lane 2, TW30. Molecular size standards are given at the left in base pairs. The arrow indicates the 518-bp ompL fragment.
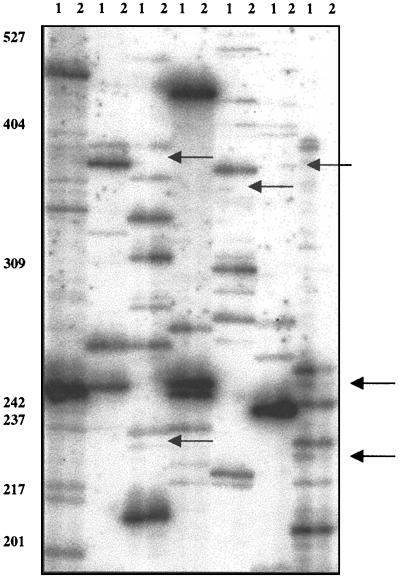
By use of 22 different primer combinations in this RAP-PCR analysis, a total of eight unique differentially regulated gene fragments was obtained from an approximate total of 800 PCR bands examined. Fragment sizes generally ranged from 650 to 100 bp, although bands smaller than 180 bp were not considered. The initial results of our RAP-PCR survey yielded 20 differentially expressed bands between strains DB110 and TW30. However, either due to the inability to reamplify the excised band (eight clones) or confirm differential expression of the fragment (four clones), our analysis considered only eight of the original bands. One example of a RAP-PCR autoradiograph displaying a differential gene expression fingerprint from the ToxR+ and ToxR− strains can be seen in Fig. 2. Differentially expressed RAP-PCR fragments were purified from each gel and were reamplified using the appropriate primers to both verify that the correct fragment size was isolated and to facilitate cloning.
FIG. 2.
Example of a RAP-PCR autoradiograph from these studies. Lane 1, DB110; lane 2, TW30. Arrows indicate differentially expressed gene fragments. Size standards are shown at the left in base pairs.
Verification of differential gene expression by ToxR in SS9.
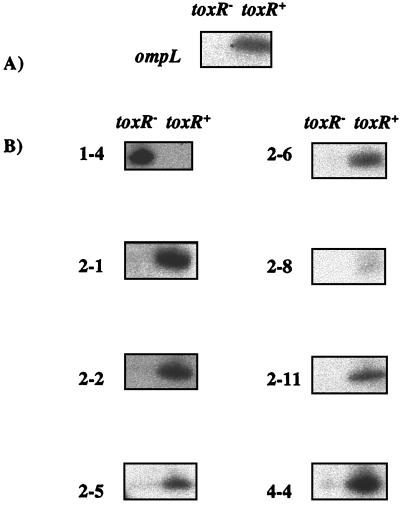
RAP-PCR fragments were cloned and sequence analysis was performed for further transcription analysis, as well as gene characterization. The ToxR regulation of each of the eight identified genes was then confirmed using gene-specific PCR primers and a new set of ToxR+ and ToxR− strains. cDNA was prepared from the strain TW30 (with a deletion of toxR) containing either the vector pKT231 (2) or pKT231 harboring the entire toxRS operon (pDB1231). As a control the regulation of ompL was examined in these two strains, and as expected, ompL amplification was detected in TW30(pDB1231) but not in TW30(pKT231) (Fig. 3A). When all of the RAP-PCR clones were examined using this method, the pattern of toxR-regulated gene expression matched that seen in the original RAP-PCR analyses (Fig. 3B). Therefore, these experiments confirmed that all eight genes are indeed regulated, either directly or indirectly, by ToxR.
FIG. 3.
Confirmation of differential gene expression of RAP-PCR fragments in TW30(pKT231) (toxR−) and TW30(pDB1231) (toxR+) using ompL-specific primers (A) and gene-specific primers (B) from each of the eight RAP-PCR clones. Clone designations are given at the left.
Characterization of the ToxR-regulated genes.
Table 1 presents a summary of related genes found within the National Center for Biotechnology Information nucleic acid sequence databases available through the BLAST network service (1) which showed greatest similarity to the ToxR-regulated partial gene sequences. The P values (28) obtained from BLASTX ranged from 6 × 10−45 to 0.41.
TABLE 1.
Products of the P. profundum ToxR-regulated genes identified using RAP-PCR along with their expression characterizationa
| Clone | Fragment length (bp) | Closest relative | P | ToxR conditionb | High-pressure gene expression (28 MPa/0.1 MPa)
|
|
|---|---|---|---|---|---|---|
| ToxR+ | ToxR− | |||||
| 1-4 | 344 | E. coli YaiC integral membrane protein | 6 × 10−45 | R | −/− | +/+ |
| 4-4 | 249 | V. cholerae SMP integral membrane protein | 0.52 | A | −/− | +/− |
| 2-5 | 279 | E. coli ATP-binding cassette transporter | 2 × 10−24 | A | ±/− | ++/± |
| 2-1 | 395 | E. coli cyclopropane fatty acid synthase | 0.41 | A | +/++ | −/− |
| 2-11 | 223 | P. falciparum acyl carrier protein | 0.028 | A | ++/± | +/± |
| 2-2 | 365 | E. coli ChpA | 6 × 10−04 | A | −/++ | ++/+ |
| 2-8 | 378 | V. cholerae HF-1 | 4 × 10−28 | A | +/++ | ++/+ |
| 2-6 | 238 | V. cholerae PepD histidine dipeptidase | 2 × 10−23 | A | ++/+ | +/+ |
The ToxR-regulated genes all belonged to one or both of two categories: genes whose products influence membrane structure and those whose products are part of a starvation response. The product of the one ToxR-repressed gene (clone 1-4) is similar to the integral membrane protein AdrA from S. enterica (YaiC from E. coli) (45). The adrA gene is transcriptionally regulated by agfD, which is required for multicellular aggregation, a behavior commonly seen in both environmental and pathogenic bacteria. Mutations within S. enterica adrA result in loss of an unknown extracellular substance and weakening of some cell-cell interactions. The expression of adrA is indirectly dependent on OmpR, a global response regulator that directs the transcription of agfD (45). Like ToxR, OmpR is required for inverse expression of omp genes in response to environmental cues (19, 30). Due to the similarity between ToxR and OmpR, it is not surprising that the AdrA homolog in SS9 is not expressed in the ToxR mutant.
The remaining genes encode putative proteins that are likely to alter membrane structure. These genes include a protein with weak similarity to the integral membrane protein SMP from V. cholerae and its homolog in E. coli (40), a protein similar to a family of ATP-binding cassette transporters, and two genes whose products have limited similarity to the cyclopropane fatty acid synthase from E. coli (53) and an acyl carrier protein from Plasmodium falciparum (52). These last two genes could be involved in fatty acid biosynthesis.
The last three clones appear to participate in starvation responses. Their products exhibit highest similarity to E. coli ChpA (35) and to V. cholerae HF-1 and PepD (22). The chpA gene is a chromosomal homolog of a gene required for plasmid R100 maintenance and is within the relA operon that is responsible for the stringent response during amino acid starvation (36). No data are available for the two indicated V. cholerae genes, but there is information on their counterparts in E. coli. E. coli HF-1 is an abundant RNA-binding protein required for the efficient translation of mRNA encoding the stationary-phase sigma factor RpoS (39). In this regard it is noteworthy that the previously mentioned adrA and cyclopropane fatty acid synthase genes are positively regulated by RpoS (17, 45). And finally, in E. coli expression of the peptidase gene pepD is induced by phosphate limitation and is dependent on the pho regulon (23, 24). Another ToxR-regulated gene in SS9, ompH, is also inducible by starvation, in this case carbon and energy starvation (6).
The ompL and ompH transcripts, which are known to be ToxR regulated, were not identified in this study. However, this is not surprising, since it is estimated that only a portion of the SS9 genome was surveyed by RAP-PCR, which is limited by the number of arbitrary 10-mer primers available for use. Peterson and Mekalanos previously screened TnphoA insertion mutants of V. cholerae for translational fusions in which alkaline phosphatase activity was dependent on ToxR (43). In their study no ompU or ompT phoA fusions were obtained, and more recent data suggest that mutations in these genes are deleterious (32, 47), a situation which is not the case for the ompL and ompH genes (11, 55).
Examination of ToxR regulation of the identified genes at high pressure.
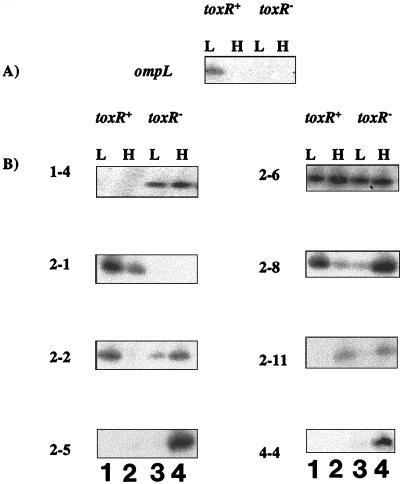
Because ToxR modulates high-pressure expression of two OMP genes, ompH and ompL (55), it was of interest to determine whether any of the genes identified here are also pressure regulated. RNA was prepared from ToxR+ and ToxR− strains (DB110 and TW30, respectively) cultured anaerobically at 0.1 and 28 MPa and subjected to gene-specific PCR. Once again, the expression of ompL was tested as a control, and as expected, based on previous work (55), it was found that only DB110 (ToxR+) cells grown at 0.1 MPa expressed ompL (Fig. 4A). However, when gene expression of the RAP-PCR clones was examined as a function of pressure and strain type, a variety of gene expression patterns emerged (Fig. 4B and Table 1). For example, the expression of the ToxR-repressed gene (clone 1-4) was absent in the ToxR+ strain but was present in the ToxR− strain regardless of pressure. And curiously, except for clone 2-1, all of the ToxR-activated genes were expressed in the ToxR− strain, and in most cases this expression was highest at elevated pressure. So the pattern of both ToxR regulation and pressure regulation was distinct for most of the genes identified in this study from the previously characterized ToxR-regulated ompH and ompL genes (54, 55). These results suggest that the regulation of many of the identified genes is complex. It is worth noting that our original RAP-PCR analyses were performed using cells cultured aerobically, while the pressure study was performed using cells cultured under anaerobic conditions, owing to oxygen toxicity at high pressure, regardless of pressure regimen. Under these culture conditions, the strains were incubated in sealed plastic bulbs which contain little oxygen but contain glucose to allow for enhanced fermentative growth. It may be that under these different physiological conditions the reliance of many of the identified genes on ToxR is lost.
FIG. 4.
Examination of RAP-PCR fragments for differential gene expression at low (L) and high (H) pressure using ompL-specific primers (A) and gene-specific primers (B) from each of the eight RAP-PCR clones. toxR+: L, DB110 at 0.1 MPa; H, DB110 at 28 MPa. toxR−: L, TW30 at 0.1 MPa; H, TW30 at 28 MPa.
Concluding remarks.
This study used RAP-PCR to identify genes under the control of the global transcriptional regulator ToxR. We have identified eight additional ToxR-regulated genes in SS9, bringing the total of known ToxR-regulated genes in this microorganism to 10. To our knowledge, this is the first application of this technique for use in identifying genes under the control of a particular transcriptional regulator. Under circumstances when genome sequence information is not available and it is not possible to explore global regulation of gene expression using DNA microarrays (9) or macroarrays (48), RAP-PCR should be considered.
Many of the identified genes have similarity to genes present in the recently completed V. cholerae genome (22), although none of these genes are known to be ToxR regulated in V. cholerae. Because there is no well-conserved consensus ToxR binding sequence (13, 33) and because many of the genes identified could be indirectly controlled by ToxR, it is not known at this time whether the related genes in V. cholerae are also subject to ToxR control. Further comparisons of ToxR-regulated genes among SS9 and V. cholerae are needed to better understand the nature and evolution of this important regulon. However, it is already clear that the SS9 ToxRS system should be viewed in the context of membrane structure and nutrient acquisition and that alteration of gene expression by ToxR and hydrostatic pressure need not always be coordinated in the manner previously reported for ompH and ompL (55).
ACKNOWLEDGMENT
We are grateful to the National Science Foundation Metabolic Biochemistry program (NSF 9974528) for support of this research.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bagdasarian M, Lurz R, Ruckert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 3.Bang I, Kim B, Foster J, Park Y. OmpR regulates the stationary-phase acid tolerance response of Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:2245–2252. doi: 10.1128/jb.182.8.2245-2252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartlett D, Wright M, Yayanos A, Silverman M. Isolation of a gene regulated by hydrostatic pressure. Nature. 1989;342:572–574. doi: 10.1038/342572a0. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett D H, Chi E, Wright M E. Sequence of the ompH gene from the deep-sea bacterium Photobacterium SS9. Gene. 1993;131:125–128. doi: 10.1016/0378-1119(93)90680-2. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett D H, Welch T J. ompH gene expression is regulated by multiple environmental cues in addition to high pressure in the deep-sea bacterium Photobacterium species strain SS9. J Bacteriol. 1995;177:1008–1016. doi: 10.1128/jb.177.4.1008-1016.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson N R, Wong R M-Y, McClelland M. Analysis of the SOS response in Salmonella enterica serovar Typhimurium using RNA fingerprinting by arbitrarily primed PCR. J Bacteriol. 2000;182:3490–3497. doi: 10.1128/jb.182.12.3490-3497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bidle K A, Bartlett D H. RecD function is required for high pressure growth in a deep-sea bacterium. J Bacteriol. 1999;181:2330–2337. doi: 10.1128/jb.181.8.2330-2337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown P O, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 10.Chakrabortty A, Das S, Majumdar S, Mukhopadhyay K, Roychoudhury S, Chaudhuri K. Use of RNA arbitrarily primed-PCR fingerprinting to identity Vibrio cholerae genes differentially expressed in the host following infection. Infect Immun. 2000;68:3878–3887. doi: 10.1128/iai.68.7.3878-3887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi E, Bartlett D H. Use of a reporter gene to follow high-pressure signal transduction in the deep-sea bacterium Photobacterium sp. strain 559. J Bacteriol. 1993;175:7533–7540. doi: 10.1128/jb.175.23.7533-7540.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi E, Barlett D H. An rpoE-like locus controls outer membrane protein synthesis and growth at cold temperatures and high pressures in the deep-sea bacterium Photobacterium SS9. Mol Microbiol. 1995;17:713–726. doi: 10.1111/j.1365-2958.1995.mmi_17040713.x. [DOI] [PubMed] [Google Scholar]
- 13.Crawford J A, Kaper J B, DiRita V J. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol Microbiol. 1998;29:235–246. doi: 10.1046/j.1365-2958.1998.00925.x. [DOI] [PubMed] [Google Scholar]
- 14.DeLong E F. Adaptations of deep-sea bacteria to the abyssal environment. Ph.D. thesis. University of California, San Diego; 1986. [Google Scholar]
- 15.DiRita V J, Mekalanos J J. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991;64:29–37. doi: 10.1016/0092-8674(91)90206-e. [DOI] [PubMed] [Google Scholar]
- 16.Dunlap P V. Quorum regulation of luminescence in Vibrio fischeri. J Mol Microbiol Biotechnol. 1999;1:5–12. [PubMed] [Google Scholar]
- 17.Eichel J, Chang Y-Y, Riesenberg D, Cronan J E., Jr Effect of ppGpp on Escherichia coli cyclopropane fatty acid synthesis is mediated through the RpoS sigma factor (ςs) J Bacteriol. 1999;181:572–576. doi: 10.1128/jb.181.2.572-576.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming J T, Yao W-H, Sayler G S. Optimization of differential display of prokaryotic mRNA: application to pure culture and soil microcosms. Appl Environ Microbiol. 1998;64:3698–3706. doi: 10.1128/aem.64.10.3698-3706.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall M N, Silhavy T J. Genetic analysis of the ompB locus in Escherichia coli K-12. J Mol Biol. 1981;151:1–15. doi: 10.1016/0022-2836(81)90218-7. [DOI] [PubMed] [Google Scholar]
- 20.Harkey C W, Everiss K D, Peterson K M. Isolation and characterization of a Vibrio cholerae gene (tagA) that encodes a ToxR-regulated lipoprotein. Gene. 1995;153:81–84. doi: 10.1016/0378-1119(94)00811-6. [DOI] [PubMed] [Google Scholar]
- 21.Hase C H, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heidelberg J F, Eisen J A, Clayton R A, Gwinn M L N.W.C.; and others. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–484. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henrich B, Backes H, Klein J, Plapp R. The promoter region of the Escherichia coli pepD gene: deletion analysis and control by phosphate limitation. Mol Gen Genet. 1992;232:117–125. doi: 10.1007/BF00299144. [DOI] [PubMed] [Google Scholar]
- 24.Henrich B, Monnerjahn U, Plapp R. Peptidase D gene (pepD) of Escherichia coli K-12: nucleotide sequence, transcript mapping, and comparison with other peptidase genes. J Bacteriol. 1990;172:4641–4651. doi: 10.1128/jb.172.8.4641-4651.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes K J, Everiss K D, Harkey C W, Peterson K M. Identification of a Vibrio cholerae ToxR-activated gene (tagD) that is physically linked to the toxin-coregulated pilus (tcp) gene cluster. Gene. 1994;148:97–100. doi: 10.1016/0378-1119(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 26.Joseph S W, Colwell R R, Kaper J B. Vibrio parahaemolyticus and related halophilic Vibrios. Crit Rev Microbiol. 1982;10:77–124. doi: 10.3109/10408418209113506. [DOI] [PubMed] [Google Scholar]
- 27.Karaolis D K R, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 28.Karlin S, Altschul S F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci USA. 1990;87:2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovach M E, Hughes K J, Everiss K D, Peterson K M. Identification of a ToxR-activated gene, tagE, that lies within the accessory colonization factor gene cluster of Vibrio cholerae 0395. Gene. 1994;148:91–95. doi: 10.1016/0378-1119(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 30.Lan C Y, Igo M M. Differential expression of the OmpF and OmpC porin proteins in Escherichia coli K-12 depends upon the level of active OmpR. J Bacteriol. 1998;180:171–174. doi: 10.1128/jb.180.1.171-174.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee A, Detweiler C, Falkow S. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J Bacteriol. 2000;182:771–781. doi: 10.1128/jb.182.3.771-781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S E, Shin S H, Kim S Y, Kim Y R, Shin D H, Chung S S, Lee Z H, Lee J Y, Jeong K C, Choi S H, Rhee J H. Vibrio vulnificus has the transmembrane transcriptional activator ToxRS stimulating the expression of the hemolysin gene vvhA. J Bacteriol. 2000;182:3405–3415. doi: 10.1128/jb.182.12.3405-3415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C C, Crawford J A, DiRita V J, Kaper J B. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol Microbiol. 2000;35:189–203. doi: 10.1046/j.1365-2958.2000.01699.x. [DOI] [PubMed] [Google Scholar]
- 34.Lin Z, Kumagai K, Baba K, Mekalanos J J, Nishibuchi M. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J Bacteriol. 1993;175:3844–3855. doi: 10.1128/jb.175.12.3844-3855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metzger S, Dror I B, Aizenman E, Schreiber G, Toone M, Friesen J D, Cashel M, Glaser G. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J Biol Chem. 1988;263:15699–15704. [PubMed] [Google Scholar]
- 37.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 39.Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-1, known as a host factor for Qb RNA replication, is essential for RpoS translation in Escherichia coli. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 40.Neuwald A F, Stauffer G V. An Escherichia coli membrane protein with a unique signal sequence. Gene. 1989;82:219–228. doi: 10.1016/0378-1119(89)90047-4. [DOI] [PubMed] [Google Scholar]
- 41.Osorio C R, Klose K E. A region of the transmembrane regulatory protein ToxR that tethers the transcriptional activation domain to the cytoplasmic membrane displays wide divergence among Vibrio species. J Bacteriol. 2000;182:526–528. doi: 10.1128/jb.182.2.526-528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parsot C, Taxman E, Mekalanos J J. ToxR regulates the production of lipoproteins and the expression of serum resistance in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:1641–1645. doi: 10.1073/pnas.88.5.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson K M, Mekalanos J J. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun. 1988;56:2822–2829. doi: 10.1128/iai.56.11.2822-2829.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reich K A, Schoolnik G K. The light organ symbiont Vibrio fischeri possesses a homolog of the Vibrio cholerae transmembrane transcriptional activator ToxR. J Bacteriol. 1994;176:3085–3088. doi: 10.1128/jb.176.10.3085-3088.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Römling U, Rohde M, Olsen A, Normark S, Reinkoster J. AgfD, the checkpoint of multicellular and aggregative behavior in Salmonella typhimurium, regulates at least two independent pathways. Mol Microbiol. 2000;36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- 46.Ruby E G. The Euprymna scolopes-Vibrio fischeri symbiosis: a biomedical model for the study of bacterial colonization of animal tissue. J Mol Microbiol Biotechnol. 1999;1:13–21. [PubMed] [Google Scholar]
- 47.Sperandio V, Bailey C, Giron J A, DiRita V J, Silveira W D, Vettore A L, Kaper J B. Cloning and characterization of the gene encoding the OmpU outer membrane protein of Vibrio cholerae. Infect Immun. 1996;64:5406–5409. doi: 10.1128/iai.64.12.5406-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waldor M K, Mekalanos J J. ToxR regulates virulence gene expression in non-01 strains of Vibrio cholerae that cause epidemic cholera. Infect Immun. 1994;62:72–78. doi: 10.1128/iai.62.1.72-78.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 52.Waller R F, Keeling P J, Donald R G, Striepen B, Handman E, Lang-Unnasch N, Cowman A F, Besra G S, Roos D S, McFadden G I. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc Natl Acad Sci USA. 1998;95:12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang A Y, Grogan D W, Cronan J E., Jr Cyclopropane fatty acid synthase of Escherichia coli: deduced amino acid sequence, purification, and studies of the enzyme active site. Biochemistry. 1992;31:11020–11028. doi: 10.1021/bi00160a011. [DOI] [PubMed] [Google Scholar]
- 54.Welch T J, Bartlett D H. Isolation and characterization of the structural gene for OmpL, a pressure-regulated porin-like protein from the deep-sea bacterium Photobacterium species strain SS9. J Bacteriol. 1996;178:5027–5031. doi: 10.1128/jb.178.16.5027-5031.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welch T J, Bartlett D H. Identification of a regulatory protein required for pressure-responsive gene expression in the deep-sea bacterium Photobacterium species strain SS9. Mol Microbiol. 1998;27:977–985. doi: 10.1046/j.1365-2958.1998.00742.x. [DOI] [PubMed] [Google Scholar]
- 56.Wong K K, McClelland M. Stress-inducible gene of Salmonella typhimurium identified by arbitrarily primed PCR of RNA. Proc Natl Acad Sci USA. 1994;91:639–643. doi: 10.1073/pnas.91.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamamoto K, Nagura R, Tanabe H, Fujita N, Ishihama A, Utsumiand R. Negative regulation of the bolA 1p of Escherichia coli K-12 by the transcription factor OmpR for osmolarity response genes. FEMS Microbiol Lett. 2000;186:257–262. doi: 10.1111/j.1574-6968.2000.tb09114.x. [DOI] [PubMed] [Google Scholar]