Abstract
Neofusicoccum parvum is a fungal plant pathogen of a wide range of hosts but knowledge about the virulence factors of N. parvum and host–pathogen interactions is rather limited. The molecules involved in the interaction between N. parvum and Eucalyptus are mostly unknown, so we used a multi-omics approach to understand pathogen–host interactions. We present the first comprehensive characterization of the in vitro secretome of N. parvum and a prediction of protein–protein interactions using a dry-lab non-targeted interactomics strategy. We used LC-MS to identify N. parvum protein profiles, resulting in the identification of over 400 proteins, from which 117 had a different abundance in the presence of the Eucalyptus stem. Most of the more abundant proteins under host mimicry are involved in plant cell wall degradation (targeting pectin and hemicellulose) consistent with pathogen growth on a plant host. Other proteins identified are involved in adhesion to host tissues, penetration, pathogenesis, or reactive oxygen species generation, involving ribonuclease/ribotoxin domains, putative ricin B lectins, and necrosis elicitors. The overexpression of chitosan synthesis proteins during interaction with the Eucalyptus stem reinforces the hypothesis of an infection strategy involving pathogen masking to avoid host defenses. Neofusicoccum parvum has the molecular apparatus to colonize the host but also actively feed on its living cells and induce necrosis suggesting that this species has a hemibiotrophic lifestyle.
Keywords: Botryosphaeriaceae, Neofusicoccum parvum, plant fungal interaction, secretome, LC-MS, Eucalyptus globulus
1. Introduction
Eucalyptus species are native to Australia but due to their enormous economic significance are planted in many countries around the world. Eucalyptus species were introduced in Portugal more than 100 years ago and are nowadays the most representative forest tree species. Eucalyptus globulus is the most abundant species in Portugal, occupying ca. 8500 km2, the equivalent to ca. 9% of the country (26% of the forest area of Portugal), mostly in Central and Northwest Portugal [1,2]. This species is well adapted to the Mediterranean-like climate and is exploited mainly due to the commercial interests of the pulp and paper industries. Unfortunately, they are commonly susceptible to diseases/infections caused by various species of the family Botryosphaeriaceae.
Botryosphaeriaceae are well-known fungal opportunistic pathogens that elicit disease symptoms in plants under stress conditions, resulting in high economic losses [3,4,5]. In addition, these species are known to occur in asymptomatic plant tissues as commensals or latent pathogens in a variety of tree species including Eucalyptus [5,6,7,8]. Botryosphaeriaceae have been associated with Eucalyptus canker and dieback in Portugal [9,10] and are considered a significant threat to the productivity and sustainability of Eucalyptus spp. plantations. In a survey conducted in 2015, and again in 2018, the predominant isolates collected from Eucalyptus were identified as belonging to the genus Neofusicoccum [9,10]. Several studies have reported a diverse assemblage of Neofusicoccum species occurring on Eucalyptus spp. both as disease-causing agents and as commensals [6,11,12].
Neofusicoccum parvum is a vascular aggressive pathogen that causes severe decline and dieback symptoms in a wide range of hosts [7,13], being also common in many Eucalyptus species [4,6,9,14,15]. In general, the fungus penetrates through wounds and colonizes the host tissues, causing shoot dieback, stem canker, cane bleaching, bud necrosis, and graft failure. Neofusicoccum parvum is an endophyte (i.e., it colonizes the interior of plants) that switches from a ‘no- or not visible’ inducing host damage status to a clear pathogen. In fact, not much is known about the strategies that this fungus employs to infect its hosts, or about the molecules it expresses during infection. Several studies have suggested that N. parvum pathogenicity could be related to the ability of this fungus to colonize woody tissue combined with the production of several phytotoxins [16,17,18,19] and also the expression of extracellular proteins with phytotoxic properties [20]. A study of genes encoding necrosis and ethylene-inducing proteins (NLPs) in N. parvum showed that they are functional genes encoding proteins toxic both to plant and mammalian cells, being most probably involved in virulence or cell death during N. parvum infection [21]. Recent genomic and transcriptomic analyses have shown that this pathogen has evolved special adaptive mechanisms to infect woody plants [22,23]. These mechanisms include a significant expansion of gene families associated with virulence and nutrient uptake, including cellular transporters, cell-wall-degrading enzymes (CWDEs), cytochrome P450s, putative effectors, and biosynthesis of secondary metabolites. The interaction between grapevine and N. parvum was also studied at the transcriptomic level [22,24]. Host plant stems and leaves underwent extensive transcriptomic reprogramming, but woody stems reacted earlier than leaves to infection. Gene expression analysis showed that N. parvum co-expresses genes associated with secondary metabolism and plant cell wall degradation in a dependent manner on the growth substrate and the stage of plant infection. Overall, these studies have shed light on the interactions between plants and N. parvum. However, a full understanding of the pathogenicity mechanism is still far from being accomplished. To investigate the mechanisms of pathogenicity of this fungus, we centered our analysis on the secretome [25,26,27,28,29], due to its relevance to the infection mechanism and to fungus–plant interactions. Proteomics data from the species of the family Botryosphaeriaceae are limited. So far, the proteome of Diplodia seriata [30], Diplodia corticola [31], and, most recently, Lasiodiplodia theobromae [32,33,34] have been made available. Proteins identified in these studies suggest differences in the infection strategies of these fungi. Although the genome of N. parvum was sequenced and released in 2013 [13], no proteomics studies have been carried out until now.
The aim of this study was to characterize the secretome of N. parvum, evaluate its response to an in vitro host mimicry, and predict interactions of the secretome proteins with host proteins.
2. Materials and Methods
2.1. Fungal Strains, Plant Material, and Culture Conditions
The strain used in this study, N. parvum CAA704, was recovered from E. globulus displaying symptoms of dieback and decline in Portugal [9]. This strain also proved to be pathogenic to E. globulus in artificial inoculation trials [9]. The strain was grown on Potato Dextrose Agar (PDA, Merck, Germany) at 25 °C for 7 days prior to the inoculations. The 3-months-old E. globulus (MB43, obtained from Altri, SGPS, S.A.) seedlings were watered weekly and kept at room temperature under natural light.
Two conditions were tested: control and infection-like. For the control condition, two mycelium plugs (5 mm diameter) were inoculated into a 250 mL flask containing 50 mL of Potato Dextrose Broth (PDB, Merck, Germany) and incubated in triplicate at 25 °C for 12 days. For the infection-like condition, a sterilized piece of E. globulus stem (±2 g) was added to the PDB, as described elsewhere [31]. The culture supernatant of each condition was harvested through filter paper and immediately stored at −80 °C for extracellular protein extraction. Mycelia obtained from both conditions were collected by filtration, washed with sterile water, and frozen with N2(L) for DNA and RNA extraction.
2.2. RNA Extraction and cDNA Synthesis
Total RNA was extracted from 12-days-old mycelium ground in liquid nitrogen (three biological replicates from each condition) using the Spectrum Plant Total RNA kit (Sigma-Aldrich, St. Louis, MO, USA), according to the manufacturer’s instructions. Samples were treated with DNase I digestion set (RNase-Free DNase Set, QIAGEN, Hilden, Germany) for 15 min to remove genomic DNA. The quality and quantity of RNA were checked by gel electrophoresis and NanoDrop™ 1000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA). cDNA was generated using the Nzy First-Strand cDNA Synthesis Kit (Nzytech, Lisboa, Portugal), according to the manufacturer’s instructions.
2.3. Quantitative PCR
Target genes were selected according to their pattern of expression and functional annotation (Table 1). All reactions were performed in a CFX96 Real-Time thermocycler (BioRad, Hercules, CA, USA) using the NzySpeedy quantitative PCR (qPCR) Green Master Mix (2×) (NZYtech, Lisboa, Portugal). For each reaction, 5 µL of the Master Mix, 0.5 µL (10 µM) of each primer, 4.2 µL of nuclease-free water, and 0.5 µL of template cDNA were used. The PCR program used was: 95 °C—3 min, 40 cycles of 95 °C—15 s, and 60 °C—30 s. After this step, the fluorescence intensity was measured and, at the end of the program, the temperature was increased from 65 °C to 95 °C at a rate ramp of 0.1 °C/s, allowing the melting curves elaboration. Cq values were calculated with BIO-RAD CFX Manager software and used to compare the expression between reference and target genes.
Table 1.
Reference and target genes and respective primers.
| Protein Name | Gene | Expression Condition | Primer Sequence (5′-3′) |
Amplicon Length (bp) |
Reference |
|---|---|---|---|---|---|
| Elongation factor 1-α | EF1α | Reference gene | FW: CGGTCACTTGATCTACAAGTGC RV: CCTCGAACTCACCAGTACCG |
302 | [35] |
| Putative exo-beta protein (PL3) | UCRNP2_317 | Up-regulated | FW: ATTCAGCACTCCGGTACCAC RV: GCCGTCCACGGACTTGAT |
255 | Present study |
| Putative aspartic endopeptidase PEP1 protein | UCRNP2_6229 | Up-regulated | FW: AGCTCCAGCTATGGTGGCTA RV: GACGATAGAGAAGCCGATGC |
172 | Present study |
2.4. Extracellular Protein Extraction
Secreted proteins were extracted using TCA/acetone according to the method described by Fernandes, et al. [31]. To discard precipitated polysaccharides, 35 mL of the culture supernatant was centrifuged (48,400× g, 1 h at 4 °C). One volume of cold TCA/acetone (20%/80% (w/v)) supplemented with 0.14% (w/v) dithiothreitol (DTT) was added to the supernatant and incubated at −20 °C for 1 h. Precipitated proteins were collected by centrifugation (15,000× g, 20 min, 4 °C) and the supernatant was removed. Precipitated proteins were washed with 10 mL of ice-cold acetone (twice) (15,000× g, 15 min, 4 °C) and 10 mL of ice-cold 80% acetone (v/v) (15,000× g, 15 min, 4 °C) to discard the excess of TCA from the precipitate. Residual acetone was air-dried, and the protein pellet was resuspended in 0.1 M Tris HCl pH 8 and stored at −80 °C.
2.5. Protein Sample Cleaning
To remove salts, detergents, and phenolic compounds, the protein extract was cleaned with the water/chloroform/methanol protein precipitation method (adapted from [36]). Briefly, a mixture of methanol, chloroform, and water (4:1:3 (v/v/v)) was added to the sample and thoroughly vortexed. Then, the mixture was centrifuged at 14,000× g for 1 min and the top aqueous methanol layer was removed (the proteins being in the interphase). Four volumes of methanol were added, and the mixture was vortexed and centrifuged at 14,000× g for 5 min. The supernatant was removed without disturbing the pellet. The air-dried pellet was finally resuspended in 0.1 M Tris HCl pH 8 and stored at −80 °C.
2.6. Protein Quantification
Protein concentration assay was carried out with the Pierce® 660 nm Protein Assay kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s instructions, using Bovine Serum Albumin (BSA) as standard. All samples were quantified in triplicate.
2.7. Protein Quality Evaluation by Electrophoresis
The quality of protein samples was assessed by SDS–PAGE. Briefly, 3 µg of protein were denaturated and separated in a 12.5% SDS-PAGE gel electrophoresis, for 120 min at 120 V, in a Mini-PROTEAN 3 Cell (Bio-Rad, Hercules, CA, USA), according to Laemmli’s protocol [37]. The running buffer contained 100 mM Tris, 100 mM Bicine, and 0.1% (w/v) SDS. Gels were stained with Coomassie Brilliant Blue G-250. After staining, gels were scanned on a GS-800 Calibrated Densitometer (Bio-Rad, Hercules, CA, USA).
2.8. Tryptic Digestion, Mass Spectrometry Analysis, and Protein Identification
Ten μg of the protein sample were diluted in NH4HCO3 50 mM buffer (in 30 μL). Twenty μL of BSA 0.002 μg/mL was added and the solution was incubated at 80 °C for 10 min. Samples were reduced with 5 μL of DTT 50 mM/NH4HCO3 50 mM (incubation at 60 °C for 10 min) and alkylated with 5 μL of iodoacetamide (IAA) 150 mM/NH4HCO3 50 mM (incubation in the dark for 20 min). Proteins were digested with 2 μL of trypsin 0.1 μg/μL. Afterward, samples were acidified with 1% formic acid and incubated at 37 °C for 30 min. After centrifugation (16,000× g, 30 min), the supernatant was transferred to new vials and a peptide purification step was performed using C18 Omix tips. The peptides were dried in a vacuum concentrator (SpeedVac, ThermoFisher Scientific, Waltham, MA, USA) and stored at −20 °C until analysis.
Purified peptides were re-dissolved in loading solvent (0.1% trifluoroacetic acid (TFA) in water/acetonitrile (ACN) (98:2, v/v)) and injected into an Ultimate 3000 RSLC nano system in-line connected to a Q Exactive HF mass spectrometer (Thermo, Waltham, MA, USA). Trapping was performed at 10 μL/min for 4 min in loading solvent A on a 20 mm trapping column (made in-house, 100 μm internal diameter (I.D.), 5 μm beads, C18 Reprosil-HD, Dr. Maisch, Ammerbuch, Germany) and the sample was loaded onto a 400 mm analytical column (made in-house, 75 µm I.D., 1.9 µm beads, C18 Reprosil-HD, Dr. Maisch, Ammerbuch, Germany). Peptides were eluted by a non-linear gradient from 2 to 56% solvent B [0.1% formic acid in water/acetonitrile (2:8, v/v)] over 145 min at a constant flow rate of 250 nL/min, followed by a 10 min wash reaching 97% MS solvent B and re-equilibration with solvent A (0.1% formic acid in water) for 20 min. The column temperature was kept constant at 50 °C by a column oven (Sonation COControl). The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition for the 16 most abundant ion peaks per MS spectrum. Full-scan MS spectra (375–1500 m/z) were acquired at a resolution of 60,000 in the Orbitrap analyzer after accumulation to a target value of 3 × 106. The 16 most intense ions above a threshold value of 1.3 × 104 were isolated for fragmentation at a normalized collision energy of 28% after filling the trap at a target value of 1 × 105 for a maximum of 80 ms. MS/MS spectra (200–2000 m/z) were acquired at a resolution of 15,000 in the Orbitrap analyzer.
The raw data generated from LC-MS was further inputted in Max-Quant (version 1.6.2.1, https://maxquant.net/maxquant/, accessed on 1 April 2017, Max Planck Institute, Martinsried, Germany), a quantitative proteomics software developed by Cox and Mann [38]. MS1 spectra were searched with the Andromeda peptide database engine [39] against a FASTA database of proteins from the N. parvum genome from UniProt (July, 2017) [40] and analyzed for label-free quantification of the peptides present in the samples. The peptide database was constructed from in-silico prediction of tryptic peptides with up to two missed cleavages, carbamidomethylation of cysteines as fixed modifications, and oxidation of methionines and N-terminal acetylation as variable modifications. Peptide spectral matches were validated using a percolator based on q-values at a 1% false discovery rate (FDR). Identified peptides were assembled into protein groups according to the law of parsimony and filtered to 1% FDR. Perseus software (version 1.6.1.3, https://maxquant.net/perseus/, accessed on 1 April 2017, Max Planck Institute, Martinsried, Germany) [41] enabled the affiliation of the protein groups into identified proteins. Identified proteins were filtered and only considered for analysis if present in 3 replicates and using at least 3 peptides for identification. Reverse proteins and proteins identified only by site were filtered out. A multi-scatter plot and hierarchical clustering were performed to assess the quality of the experiment. To identify interactor proteins, a two-sample t-test between control and infection-like samples was performed with minimal fold change (s0) of 1.8 and 1% FDR. A scatter plot, volcano plot, and profile plot were used to visualize the results (Figures S2–S4).
2.9. Bioinformatic Analysis
Identified proteins were classified according to the GO (biological process). Whenever necessary, the protein’s family and domain were determined by the identification of conserved domains in the InterPro database (http://www.ebi.ac.uk/interpro, accessed on 1 April 2017) [42]. Cell-wall-degrading enzymes were classified according to the carbohydrate-active enzymes database CAZy (http://www.cazy.org, accessed on 1 April 2017) [43].
All proteins were analyzed for subcellular localization using the BaCelLo fungi-specific predictor [44], SignalP v4.1 (https://services.healthtech.dtu.dk/service.php?SignalP, accessed on 1 April 2017) [45], and SecretomeP predictor [46].
2.10. Interactomics Analysis
The OralInt algorithm [47] was used to predict the interactions between all the proteins of Eucalyptus grandis reference proteome (Uniprot UP000030711, 44,150 proteins) with the differentially secreted proteins of N. parvum identified in this study (117 proteins). OralInt is a computational prediction method based on an ensemble methodology combining five distinct protein–protein interactions (PPI) prediction techniques, namely: literature mining, primary protein sequences, orthologous profiles, biological process similarity, and domain interactions [47]. Since the sequence is the feature with the best overall performance, this method of predicting interactions can be applied independently of the organisms under study.
Interactions with a score ≥ 0.900 are represented using yFiles Organic Layout with Cytoscape 3.7.2 or in an edge bundling structure built using R (v4.1.2) [48] with packages ggraph (v2.0.5) [49], igraph (v1.2.11) [50], and tidyverse (v1.3.1) [51].
3. Results
3.1. Secretome Analysis
Prior to LC-MS, protein extracts were analyzed for quality control by SDS-PAGE (Figure S1). The secretomes of N. parvum grown in the absence (control) and presence of a Eucalyptus stem (infection-like condition) were analyzed. In total, 471 proteins were identified in both control and infection-like secretomes, of which 131 proteins were significantly different in abundance between the two conditions (t-test, difference cutoff of 1.8). Most of these proteins are extracellular (Table 2 and Table S1), except for 14 proteins predicted as intracellular proteins (10.7%, Table S1).
Table 2.
Summary of the proteins differentially secreted by Neofusicoccum parvum (CAA704). Protein localization was predicted using SignalP [52], SecretomeP 2.0 [46], and BaCeILO [44] tools.
| Protein Name | Accession Number a |
Fold Change b | p-Value c | Unique Peptides d |
PEP e | Intensity f | Localization g,i |
|---|---|---|---|---|---|---|---|
| Cellulose degradation | |||||||
| GH5—Putative glycoside hydrolase family 5 protein | R1GZQ9 | 2.1 | 1.833 | 6 | 1.87 × 109 | 169.3 | Extracellular |
| GH5—Putative endoglucanase II protein | R1GLD6 | 1.9 | 1.554 | 6 | 8.42 × 108 | 97.74 | Extracellular |
| GH5—Putative cellulase family protein | R1G7G3 | −2.7 | 3.403 | 12 | 2.36 × 1010 | 323.3 | Extracellular |
| GH5—Putative endo-beta-protein | R1GDK9 | −3.9 | 2.961 | 18 | 9.7 × 109 | 323.3 | Extracellular |
| GH3—Putative beta-d-glucoside glucohydrolase protein | R1EK26 | −3.5 | 3.584 | 8 | 1.06 × 109 | 98.06 | Extracellular |
| GH3—Putative beta-glucosidase 1 protein | R1G324 | −2 | 2.565 | 11 | 3.43 × 108 | 84.3 | Extracellular |
| GH7—Glucanase | R1GZN3 | −2.5 | 2.374 | 10 | 1.49 × 1010 | 323.3 | Extracellular |
| AA9/GH61/CBM1—Putative fungal cellulose-binding domain protein | R1GHV2 | −2.1 | 3.779 | 7 | 2.05 × 109 | 204.1 | Extracellular |
| GH12—Putative glycoside hydrolase family 12 protein | R1GQP5 | −3.9 | 3.605 | 8 | 1.05 × 1010 | 120.04 | Extracellular |
| Hemicellulose degradation | |||||||
| GH35—Putative beta-galactosidase B protein | R1E7W9 | −2.5 | 3.430 | 21 | 4.23 × 109 | 255.29 | Extracellular |
| GH43—Putative glycosyl family protein | R1EP04 | −2.9 | 2.330 | 6 | 5.29 × 108 | 73.74 | Extracellular |
| GH10—Beta-xylanase | R1FWZ0 | −3.1 | 3.513 | 14 | 2.31 × 1010 | 323.31 | Extracellular |
| GH43—Putative xylosidase: arabinofuranosidase protein | R1G299 | −2.1 | 3.289 | 6 | 3.79 × 108 | 140.27 | Extracellular |
| GH43—Putative xylosidase glycosyl hydrolase protein | R1G5Y4 | −1.9 | 3.227 | 13 | 3.51 × 1010 | 323.31 | Extracellular |
| GH27—Alpha-galactosidase | R1G8C1 | −2.6 | 4.883 | 12 | 1.8 × 1010 | 286.19 | Extracellular NN h (0.861) |
| GH43—Putative galactan-beta-galactosidase protein | R1GG59 | −5.5 | 3.216 | 14 | 3.67 × 109 | 323.31 | Extracellular |
| GH43—Arabinan endo-1,5-alpha-L-arabinosidase | R1GAB3 | −6.4 | 5.780 | 10 | 4.97 × 109 | 117.45 | Extracellular |
| GH51—Putative alpha-l-arabinofuranosidase a protein | R1EVS4 | −3.1 | 3.133 | 10 | 7.22 × 108 | 190.73 | Extracellular |
| CE5—Putative acetylxylan esterase protein | R1EWW2 | −2.3 | 1.059 | 2 | 1.54 × 109 | 323.31 | Extracellular NN h (0.898) |
| GH11—Endo-1,4-beta-xylanase | R1GCT8 | −2.4 | 1.534 | 7 | 3.41 × 108 | 144.76 | Extracellular |
| GH43/CBM6—Putative glycosyl hydrolase family 43 protein | R1GE80 | −2.2 | 3.527 | 16 | 8.42 × 109 | 307.72 | Extracellular |
| Lignin degradation | |||||||
| AA5—Putative glyoxal oxidase protein | R1EDI4 | 2.1 | 2.596 | 10 | 1.32 × 109 | 86.505 | Extracellular |
| AA1—Putative laccase-1 protein | R1G4L9 | 1.9 | 3.262 | 11 | 1.03 × 1010 | 227.45 | Extracellular |
| AA7—Putative FAD-dependent oxidoreductase protein | R1FVT8 | 2.2 | 1.428 | 14 | 9.82 × 108 | 123.3 | Extracellular |
| AA3—Putative alcohol dehydrogenase protein | R1EH41 | −2.7 | 2.253 | 3 | 2.58 × 108 | 35.849 | Extracellular NN h (0.648) |
| Lignin/celulose degradation | |||||||
| AA3/CBM1—Putative cellobiose dehydrogenase protein | R1H3M7 | 2 | 1.856 | 16 | 1.72 × 109 | 157.89 | Extracellular |
| AA3—Putative GMC oxidoreductase protein | R1FVG2 | 1.8 | 3.233 | 23 | 5.79 × 1010 | 323.31 | Extracellular NN h (0.655) |
| Pectin degradation | |||||||
| GH53—Arabinogalactan endo-beta-1,4-galactanase | R1G7Y3 | −7 | 3.424 | 9 | 6.29 × 109 | 161.62 | Extracellular |
| CE12—Putative rhamnogalacturonan acetylesterase protein | R1GFP8 | −6.4 | 4.594 | 9 | 9.04 × 109 | 155.37 | Extracellular |
| GH53—Arabinogalactan endo-beta-1,4-galactanase | R1GVP5 | −2.3 | 2.391 | 5 | 5.89 × 108 | 56.599 | Extracellular |
| PL3—Putative pectate lyase protein | R1EWA7 | −6.6 | 3.224 | 9 | 6.54 × 109 | 297.03 | Extracellular |
| PL3—Putative pectate lyase protein | R1GN84 | −6.2 | 4.605 | 6 | 4.57 × 109 | 103.84 | Extracellular |
| PL1—Putative pectate lyase a protein | R1GII6 | −4.4 | 4.962 | 13 | 1.75 × 1010 | 323.31 | Extracellular |
| PL4—Putative rhamnogalacturonan lyase protein | R1GJ02 | −5.5 | 4.999 | 18 | 3.48 × 109 | 227.89 | Extracellular |
| PL1—Putative pectate protein | R1GSQ1 | −4.8 | 3.712 | 5 | 1.4 × 109 | 91.554 | Extracellular NN h (0.592) |
| PL3—Putative exo-beta-protein | R1H382 | −2.2 | 2.235 | 24 | 1.8 × 1011 | 323.31 | Extracellular NN h (0.798) |
| GH28—Putative extracellular exo-protein | R1GW72 | −3.2 | 3.359 | 5 | 8.03 × 108 | 57.212 | Extracellular |
| PL4—Rhamnogalacturonate lyase | R1EPI5 | −2 | 2.077 | 6 | 4.03 × 108 | 107.86 | Extracellular |
| PL4—Rhamnogalacturonate lyase | R1GGA5 | −7.6 | 4.898 | 23 | 1.38 × 1010 | 323.31 | Extracellular |
| Chitin degradation | |||||||
| CE4—Putative chitin deacetylase protein | R1E7G7 | −5.6 | 2.993 | 6 | 3.73 × 109 | 53.089 | Extracellular |
| GH75—Endo-chitosanase | R1GTL6 | −4.1 | 1.099 | 4 | 4.37 × 109 | 59.996 | Extracellular |
| Other CAZY | |||||||
| GH16—Putative glycoside hydrolase family 16 protein | R1EVI7 | −2.7 | 2.462 | 3 | 2.12 × 109 | 34.095 | Extracellular |
| Esterase | |||||||
| Carboxylic ester hydrolase | R1GKX8 | 2.7 | 4.271 | 7 | 5.44 × 108 | 70.895 | Extracellular |
| Putative GDSL-like lipase acylhydrolase protein | R1E852 | −4.1 | 2.117 | 6 | 3.13 × 109 | 323.31 | Extracellular |
| Carboxylic ester hydrolase | R1E8C5 | −2.8 | 2.209 | 9 | 7.04 × 108 | 79.717 | Extracellular |
| Putative GDSL-like lipase acylhydrolase protein | R1GK66 | −2.6 | 3.165 | 6 | 4.2 × 108 | 40.614 | Extracellular |
| Carboxylic ester hydrolase | R1GSL8 | −2.1 | 2.550 | 6 | 1.91 × 108 | 83.343 | Extracellular |
| Putative carboxylesterase protein | R1EIK3 | −3.7 | 1.530 | 4 | 7.19 × 108 | 37.138 | Extracellular NN h (0.768) |
| Carboxylic ester hydrolase | R1G8E3 | −5.8 | 3.503 | 9 | 2.21 × 109 | 171.03 | Extracellular |
| Putative carboxylesterase family protein | R1G9C5 | −2.1 | 3.136 | 5 | 1.71 × 108 | 39.971 | Extracellular |
| Putative GDSL lipase acylhydrolase family protein | R1EIF4 | −1.8 | 2.818 | 6 | 3.13 × 109 | 134.94 | Extracellular NN h (0.756) |
| Carboxylic ester hydrolase/tannase family | R1GJW0 | −1.9 | 3.095 | 20 | 4.01 × 109 | 323.31 | Extracellular |
| Protease | |||||||
| Peptidase S1 family—putative carboxypeptidase S1 protein | R1FV38 | 1.9 | 2.351 | 7 | 4.06 × 109 | 175.54 | Extracellular |
| Peptidase S8 family—putative peptidase S8 S53 subtilisin kexin sedolisin protein | R1EAW3 | 2.2 | 1.289 | 5 | 1.54 × 109 | 157.52 | Extracellular |
| Peptidase A1 family—Putative aspartic endopeptidase PEP1 protein | R1GM42 | −3.2 | 4.661 | 4 | 4.3 × 109 | 98.628 | Extracellular |
| Peptidase M43—Putative metalloprotease protein | R1FXE7 | −5.1 | 3.311 | 5 | 3.6 × 109 | 134.74 | Extracellular |
| Peptidase M28 family—peptide hydrolase | R1GBR8 | −2.7 | 2.039 | 6 | 1.35 × 109 | 209.59 | Extracellular |
| Peptidase M35 family—neutral protease 2 | R1EL46 | −2.3 | 0.943 | 5 | 1.62 × 109 | 102.51 | Extracellular |
| Oxidoreductase | |||||||
| Putative FMN-dependent dehydrogenase protein | R1E6X7 | 2.9 | 1.290 | 16 | 6.56 × 108 | 127.23 | Extracellular |
| Putative FAD-binding domain-containing protein | R1E8E1 | 3.6 | 3.973 | 11 | 4.38 × 109 | 264.43 | Extracellular |
| Putative cyclohexanone monooxygenase protein | R1EF40 | −3.7 | 1.628 | 2 | 9.32 × 109 | 20.921 | Extracellular |
| Putative tyrosinase central domain protein | R1ERX8 | −2.4 | 2.164 | 9 | 8.84 × 108 | 90.821 | Extracellular NN h (0.817) |
| Putative FAD FMN-containing dehydrogenase protein | R1GB06 | −3.4 | 3.369 | 16 | 9.58 × 108 | 192.5 | Extracellular |
| Putative berberine-like protein | R1GD68 | −5 | 2.241 | 13 | 2.88 × 109 | 323.31 | Extracellular |
| Putative GMC protein | R1ELQ0 | −2.1 | 0.517 | 6 | 5.13 × 109 | 40.919 | Extracellular |
| Lyase | |||||||
| Putative pectate lyase protein | R1H2U7 | −2.3 | 3.013 | 4 | 2.56 × 108 | 28.73 | Extracellular |
| Putative-secreted protein | R1GFS9 | −3 | 3.218 | 20 | 2.59 × 1011 | 323.31 | Extracellular |
| Putative pectate lyase protein | R1G436 | −5.7 | 4.498 | 17 | 3.09 × 109 | 217.8 | Extracellular |
| Uncharacterized protein | R1GU06 | −1.9 | 2.289 | 7 | 2.48 × 1010 | 323.31 | Extracellular |
| Protein–protein interaction | |||||||
| Putative six-bladed beta-propeller-like protein | R1ENG6 | 2.3 | 2.942 | 3 | 4.52 × 108 | 36.528 | Extracellular |
| Putative six-bladed beta-propeller-like protein | R1E9S0 | −2.3 | 1.704 | 2 | 4.15 × 108 | 21.736 | Extracellular |
| Putative SMP-30 gluconolaconase LRE-like region protein | R1GCJ5 | −2.1 | 0.903 | 5 | 1.13 × 109 | 92.83 | Extracellular NN h (0.754) |
| Carbohydrate binding | |||||||
| Putative alpha-mannosidase family protein | R1EYI5 | 1.8 | 1.196 | 2 | 6.81 × 108 | 23.805 | Extracellular |
| Putative ricin B lectin protein | R1GAK8 | −4.3 | 2.336 | 6 | 1.66 × 109 | 97.147 | Extracellular |
| RNA binding | |||||||
| Putative ribonuclease T2 protein | R1ERG2 | 2.8 | 2.404 | 2 | 5.88 × 108 | 62.528 | Extracellular |
| Uncharacterized protein | R1FZX2 | −4.1 | 1.575 | 6 | 1.9 × 109 | 43.942 | Extracellular |
| Putative extracellular guanyl-specific ribonuclease protein | R1H1L9 | −2.1 | 0.586 | 3 | 4.24 × 109 | 48.559 | Extracellular |
| Other function | |||||||
| Putative allergen V5 Tpx-1-related protein | R1EAF3 | 2.2 | 3.639 | 5 | 5.58 × 109 | 181.15 | Extracellular |
| Putative ethanolamine utilization protein | R1G1U2 | 2 | 2.760 | 5 | 4.1 × 108 | 54.96 | Extracellular NN h (0.223) |
| Putative ABC-type Fe3+ transport system protein | R1FV21 | 2.9 | 1.484 | 6 | 4.15 × 108 | 58.409 | Extracellular |
| Putative major royal jelly protein | R1FVG4 | 2.7 | 1.896 | 15 | 5.75 × 1010 | 323.31 | Extracellular |
| Putative ABC-type Fe3+ transport system protein | R1GBA7 | 2.8 | 1.611 | 15 | 7.33 × 1010 | 323.31 | Extracellular |
| Putative alpha beta hydrolase protein | R1EGT1 | 2.7 | 3.692 | 11 | 3.31 × 109 | 118.1 | Extracellular |
| Putative glutaminase protein | R1GV87 | 2.7 | 3.603 | 10 | 1.15 × 109 | 215.65 | Extracellular |
| Putative fasciclin domain family protein | R1EWZ5 | −2 | 2.878 | 12 | 2.37 × 109 | 129.86 | Extracellular |
| Uncharacterized protein | R1GDV3 | −4.1 | 2.108 | 5 | 3.67 × 1010 | 323.31 | Extracellular |
| Putative BNR Asp-box repeat domain protein | R1GKT0 | −2.2 | 2.950 | 11 | 2.45 × 1010 | 323.31 | Extracellular |
| Putative extracellular aldonolactonase protein | R1E681 | −1.8 | 0.892 | 5 | 2.15 × 109 | 183.99 | Extracellular |
| Unknown | |||||||
| Putative extracellular serine-threonine rich protein | R1E9T1 | 2.9 | 3.242 | 3 | 8.48 × 108 | 78.551 | Extracellular |
| Putative membrane-spanning 4-domains subfamily a member 14 protein | R1EE60 | 2.9 | 3.242 | 3 | 8.48 × 108 | 78.551 | Extracellular |
| Uncharacterized protein | R1EBL8 | 2.1 | 3.282 | 11 | 1.26 × 1010 | 323.31 | Extracellular |
| Putative GPI anchored cell wall protein | R1G7D5 | 2.2 | 1.914 | 4 | 1.35 × 109 | 24.812 | Extracellular |
| Uncharacterized protein | R1GMX5 | 2.3 | 3.894 | 6 | 2.11 × 1010 | 185.31 | Extracellular |
| Uncharacterized protein | R1GRM4 | 2.4 | 1.314 | 4 | 7.62 × 108 | 37.391 | Extracellular |
| Uncharacterized protein | R1G5W7 | 2.1 | 0.681 | 2 | 8.78 × 108 | 20.89 | Extracellular |
| Uncharacterized protein | R1ESR7 | −4 | 1.737 | 3 | 1.4 × 1010 | 48.44 | Extracellular |
| Putative-secreted protein | R1G8U3 | −3.4 | 3.421 | 6 | 4.85 × 108 | 49.633 | Extracellular |
| Uncharacterized protein | R1GYB0 | −5.3 | 1.742 | 7 | 6.21 × 109 | 132.23 | Extracellular |
| Putative GPI anchored cell wall protein | R1ENT4 | −2.4 | 0.948 | 4 | 1.24 × 109 | 40.251 | Extracellular |
| Putative 34-dihydroxy-2-butanone 4-phosphate synthase protein | R1EY60 | −2.1 | 1.093 | 2 | 3.23 × 108 | 44.267 | Extracellular |
| Uncharacterized protein | R1GLY2 | −2.1 | 1.562 | 6 | 3.69 × 108 | 67.175 | Extracellular |
| Putative exo-beta-glucanase protein | R1G5R2 | −2.6 | 1.282 | 7 | 1.19 × 1011 | 323.31 | Extracellular |
a Protein accession provided by the UniProtKB database [40]; b Fold change: the difference between the average intensities of two groups (log ratio control vs infection-like); Negative fold change values indicate proteins are more abundant in the infection-like secretome and positive fold change values indicate proteins are more abundant in the control secretome; c p-value: displaying significance which is expressed as -log values; d Unique peptides: The total number of unique peptides associated with the protein group (i.e., these peptides are not shared with another protein group); e PEP: Posterior Error Probability of the identification. This value essentially operates as a p-value, where smaller is more significant; f Intensity: Summed up extracted ion current (XIC) of all isotopic clusters associated with the peptide sequence, and protein intensities summed the intensities of all peptides assigned to the protein group; g Signal prediction calculated by using the SignalP [52]; h NN: Non-classically secreted proteins analyzed with SecretomeP 2.0 [46]; Proteins with NN score ≥ 0.5 were considered unconventionally secreted; i Protein localization was predicted by the BaCeILO predictor [44].
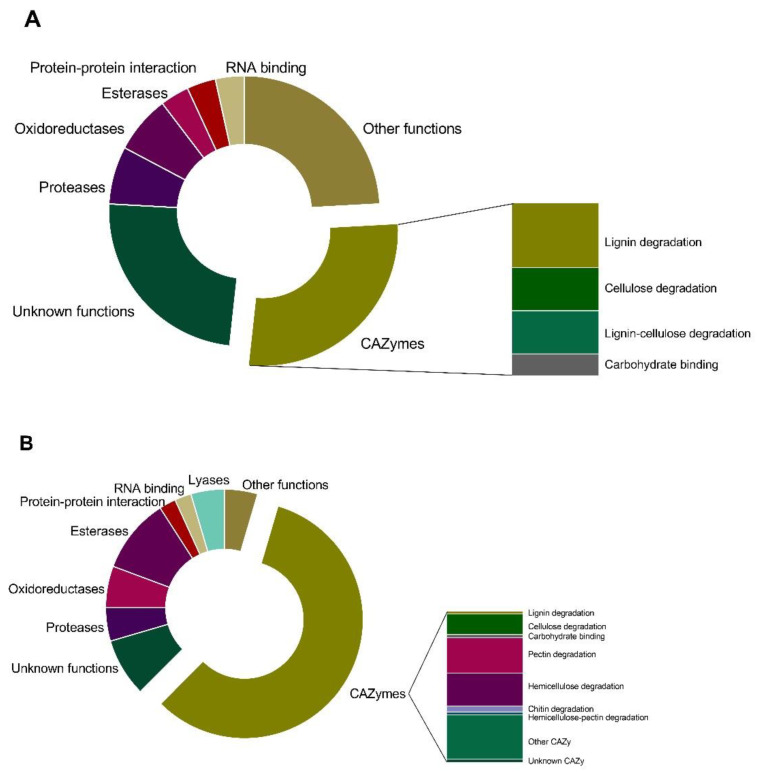
Proteins were classified according to their gene ontology (GO) (Molecular Function), and into 10 protein families: CAZymes, hydrolases, proteases, oxidoreductases, lyases, protein–protein interaction, carbohydrate-binding proteins, RNA-binding proteins, and proteins with other functions and unknown functions (Figure 1).
Figure 1.
Functional classification (GO, Molecular Function) of the extracellular proteins secreted by N. parvum whose abundance was significantly different (p < 0.05) between the two conditions. (A) Proteins less abundant in the presence of the Eucalyptus stem, and (B) Proteins more abundant in the infection-like condition. For each category, the number of proteins is reflected by the size of the pie slice. The classification was obtained from the GO annotation at the UniProt database [40]. When lacking exact functional annotations in UniProt, the family and domain databases InterPro and Pfam [42,53] were used to reveal annotations of the identified proteins of conserved domains.
Among differentially secreted proteins, 74.6% were more abundant in infection-like conditions, while 24.5% were more abundant in control conditions (Table 2, Figure 1). Among induced proteins in the presence of Eucalyptus, we identified mainly CAZy proteins (50 proteins), esterases (9 proteins), proteases (4 proteins), oxidoreductases (5 proteins), and proteins with lyase activity (4 proteins) (Figure 1, Table 2).
Among the CAZy proteins, whose abundance is affected by the interaction with the Eucalyptus stem, glycosyl hydrolases (GH) are the most abundant group (68% of CAZymes), followed by proteins with auxiliary activities (AAs, 4 proteins), polysaccharide lyases (PLs, 8 proteins), carbohydrate esterases (CEs, 3 proteins) and unknown CAZy proteins (1 protein) (Table 2 and Table S1). Esterases (EC 3.1.1.x) were more abundant in the infection-like conditions (Figure 1B).
A variety of proteases (endo and exoproteases) were also identified. The aspartic endopeptidase PEP1 (R1GM42) was more abundant in the infection-like condition (although its mRNA was as abundant as in control conditions, Figure S6) along with a variety of other metallopeptidases (M28, M35, and M43, Figure 1B and Table 2). In contrast, serine peptidases (S8 (R1EAW3) and S10 (R1FV38)) were less abundant in the infection-like conditions (Figure 1 and Table 2).
A putative berberine-like protein (R1GD68)—an oxidoreductase with a FAD-binding domain—was the most abundant protein in the infection-like condition (Table 2).
Other functional categories—proteins involved in carbohydrate binding (R1EYI5 and R1GAK8), RNA binding (R1ERG2, R1FZX2, and R1H1L9), protein–protein interactions (R1ENG6, R1E9S0, and R1GCJ5), and proteins with other functions (R1EGT1, R1GV87, R1EAF3, R1G1U2, R1FV21, R1FVG4, R1GBA7, R1EWZ5, R1GDV3, R1GKT0, R1E681)—were also identified (Figure 1 and Table 2).
3.2. Protein—Protein Interaction
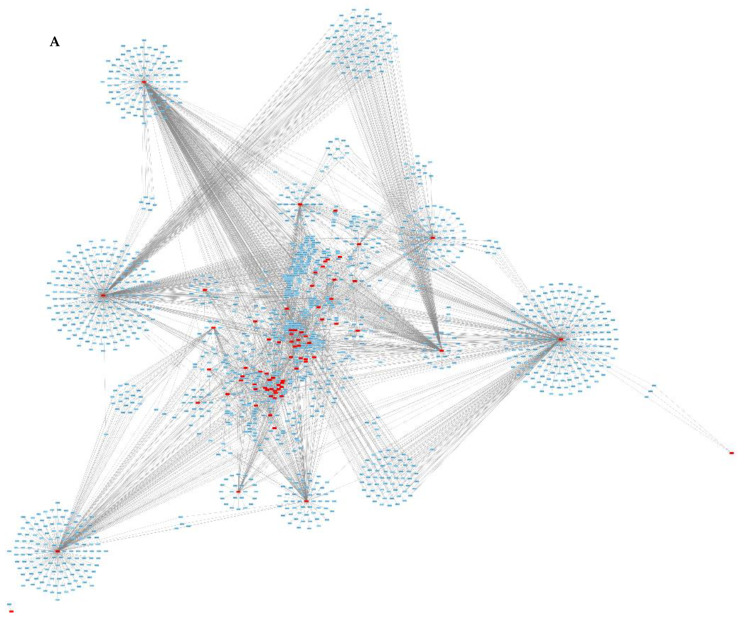
Due to the lack of data available on the proteome sequence of E. globulus, the reference proteome for E. grandis (a closely related species) was used. Protein–protein interaction (PPI) networks between all the proteins of E. grandis (44,150 proteins, blue) and the extracellular proteins of N. parvum (117 proteins, red) were predicted using the OralInt algorithm (Figure 2). OralInt is based on high-quality experimental PPIs that feeds an artificial intelligence algorithm that is later validated. In previous studies, OralInt was applied to predict interactions between the Zika virus and the host [54] and between the oral microbiome and the host [55].
Figure 2.
PPIs network prediction between secreted proteins from N. parvum (red) and the reference proteome of Eucalyptus globulus (blue). (A)—PPI interactions, the figure produced using Cytoscape v3.7.2. (B)—Visualization of the interactions between N. parvum proteins and the Biological Processes of the Eucalyptus-interacting proteins. The opacity of dots for each protein/protein category reflects the observed number of interactions.
A total of 3201 interactions were predicted involving 76 proteins of N. parvum and 1591 proteins of E. grandis. Some proteins, both in Eucalyptus and in the fungus, showed a high number of interactions (Figure 2B, Table 3 and Table S3). The functional analysis of Eucalyptus proteins on which the fungus acts is provided in the Supplementary file (Figure S5). Neofusicoccum parvum hub proteins—those that center a high number of interactions—include mainly enzymes (Table 3). Neofusicoccum parvum proteins interact mainly with proteins involved in biosynthetic processes, nucleobase-containing compound metabolic processes, signal transduction, cell communication, response to endogenous stimulus, and response to stress, which fits well with a response to a foreign attack.
Table 3.
Summary of the proteins with the highest number of PPI between proteins differentially secreted by Neofusicoccum parvum (CAA704) and Eucalyptus grandis proteins. The “Degree” stands for the number of interactions.
| Protein Name | Accession Number |
Degree | Organism |
|---|---|---|---|
| Putative gmc protein | R1ELQ0 | 419 | Neofusicoccum parvum (strain UCR-NP2) |
| Uncharacterized protein | R1G5W7 | 406 | Neofusicoccum parvum (strain UCR-NP2) |
| Uncharacterized protein | R1FZX2 | 367 | Neofusicoccum parvum (strain UCR-NP2) |
| Putative metalloprotease protein | R1FXE7 | 258 | Neofusicoccum parvum (strain UCR-NP2) |
| Putative alpha-mannosidase family | R1EYI5 | 225 | Neofusicoccum parvum (strain UCR-NP2) |
| Putative cyclohexanone monooxygenase | R1EF40 | 166 | Neofusicoccum parvum (strain UCR-NP2) |
| Putative GDSL-like lipase acylhydrolase | R1GK66 | 154 | Neofusicoccum parvum (strain UCR-NP2) |
| Putative alcohol dehydrogenase protein | R1EH41 | 117 | Neofusicoccum parvum (strain UCR-NP2) |
| Endo-chitosanase | R1GTL6 | 69 | Neofusicoccum parvum (strain UCR-NP2) |
| Uncharacterized protein | R1ESR7 | 69 | Neofusicoccum parvum (strain UCR-NP2) |
| Auxin response factor | A0A059ACB3 | 33 | Eucalyptus grandis |
| Histone H3 | A0A059AF37 | 28 | Eucalyptus grandis |
| Histone H3 | A0A059BQE5 | 20 | Eucalyptus grandis |
| Protein kinase domain-containing protein | A0A059CUY0 | 19 | Eucalyptus grandis |
| HATPase_c domain-containing protein | A0A059DD44 | 17 | Eucalyptus grandis |
| Glyco_transf_20 domain-containing protein | A0A059CZ70 | 17 | Eucalyptus grandis |
| Uncharacterized protein | A0A059CUY2 | 17 | Eucalyptus grandis |
| Protein kinase domain-containing protein | A0A059CBV7 | 16 | Eucalyptus grandis |
| ERCC4 domain-containing protein | A0A059C0I5 | 16 | Eucalyptus grandis |
| Na_H_Exchanger domain-containing protein | A0A059DJ06 | 15 | Eucalyptus grandis |
4. Discussion
Plant infection by phytopathogens, such as N. parvum, is a complex process that starts with the attachment of the infective propagule to the plant surface followed by penetration and infection. The infection mechanism of Neofusicoccum species relies on a myriad of molecules, mainly secondary metabolites, and proteins. It is known that species like N. parvum are able to express metabolites with phytotoxic activities such as Cyclohexenones, 5,6-Dihydro-2-pyrones, Melleins, Isosclerone, Hydroxypropyl- and methyl-salicylic acid, Tyrosol, Ethyl linoleate, Stearic acid, and Naphthalenones (Botryosphaerones A and D and 3,4,5-Trihydroxy-1-tetralone (for a review on the metabolites produced by Neofusicoccum spp. see [19]). These compounds were identified in isolates pathogenic of Vitis vinifera, and although it is expected that some of them will be present in other pathogen–host systems, it is not known. Understanding how other pathogen–host function is vital to fully understanding the mechanism of infection of N. parvum. The choice of the Eucalyptus-N. parvum system was based on the following reasons: (1) there are no reports on any molecules involved in the infection of Eucalyptus by N. parvum and (2) Eucalyptus is an economically vital crop in many countries, such as Portugal, and (3) there are no reports on the proteins involved in the infection mechanism of N. parvum.
Neofusicoccum parvum is a common pathogen of grapevine that infects many other hosts. In fact, the strain used in this work, N. parvum CAA704, was recovered from E. globulus displaying symptoms of dieback and decline and later it was shown to be pathogenic to E. globulus [9]. We compared the protein profiles of N. parvum in the control and infection-like conditions and identified and quantified proteins whose abundance changes in response to the Eucalyptus stem, to highlight proteins involved in the interaction with Eucalyptus during fungal infection. Although simple, and lacking the influence that molecules expressed by the host in response to the fungus attack have on the infection mechanism, the in vitro infection-like system used has been successfully used for other systems to mimic the infection mechanism of fungal pathogens [56,57].
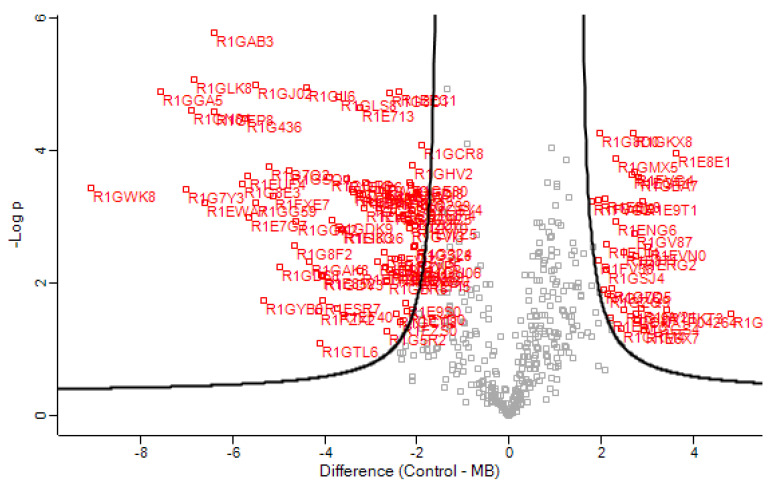
Secreted proteins were visualized in a volcano plot (Figure 3) to have a quick visual identification of proteins that display large magnitude fold change and high statistical significance. The most significant proteins are discussed below.
Figure 3.
First volcano scatterplot of the samples under analysis. Proteins with a fold change <−2 or >2 are presented in red, and the remaining ones are in grey.
Most of the proteins (86.3%) were predicted to contain a Signal P motif and are supposed to traverse the classical Golgi and endoplasmic reticulum secretion pathway. The possible implementation of the non-classical pathway for the proteins lacking signal peptide (13.7%) was confirmed using the SecretomeP predictor [46] (Table 2). Such proteins, known as leaderless-secreted proteins (LCPs), have been identified in several other studies involving the secretome [30,58,59]. Of these LCPs proteins, a putative ethanolamine utilization protein (R1G1U2) and a chitin-binding protein (R1EW80) showed low SecretomeP scores (NN score = 0.223 and 0.417, respectively) (Table 2). However, the NN score of a chitin-binding protein is relatively close to the 0.5 threshold, suggesting that the protein may in fact be secreted. The presence of intracellular proteins in the secretome is common and can result from cell death during culture, cell lysis during protein extraction, or secretion through non-common mechanisms. The number of cellular proteins identified in this study is similar to that identified in the secretome of D. seriata (16 proteins, [30]), D. corticola (12 proteins, [56]), and L. theobromae (16 proteins, [32]) the closest Botryosphaeriaceae species whose secretomes were studied.
4.1. Proteins Involved in Carbohydrate Metabolic Processes
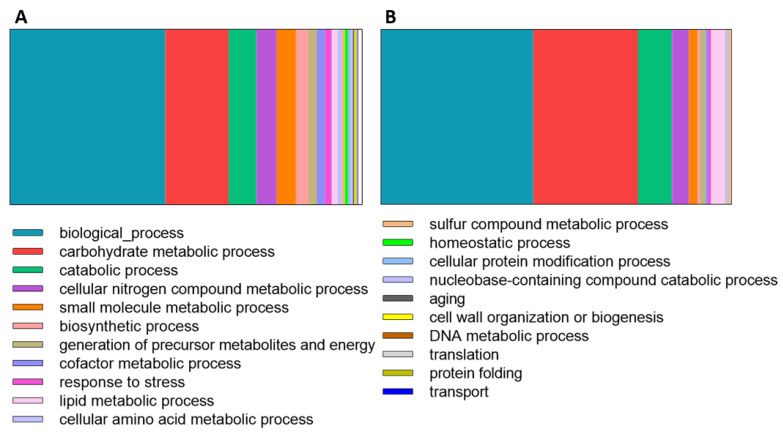
Being a plant pathogen, it is not surprising that the N. parvum strain CAA704 secretome is mainly constituted by proteins involved in carbohydrate metabolic processes (18% and 30% of the proteins identified in control and in the presence of Eucalyptus, respectively) and in catabolic processes (8% and 9.7%; Figure 4). As a plant pathogen, N. parvum uses its host as a nutrient source and for that the carbohydrate-degrading enzymes are essential (Figure 4). Although there are very few secretomes of Botryosphaeriaceae fungi available, data shows that the trend is similar with a major component of the secretomes being carbohydrate-related enzymes [30,31,60,61]. Botryosphaeriaceae species can shift between a pathogenic and non-pathogenic lifestyle when triggered (by conditions not yet fully understood) and therefore the characterization of the secretomes under controlled in vitro conditions is of the utmost relevance to understanding the nature of these organisms.
Figure 4.
Gene Ontology (GO-Biological processes) of Neofusicoccum parvum proteins identified in (A)—control conditions and (B)—differentially expressed in the presence of the Eucalyptus stem.
Plant cell-wall-degrading enzymes (PCWDEs) play significant roles in plant colonization and are typical of necrotrophic life-style fungi [62], allowing them to perceive weak regions of plant epidermal cells and penetrate the plant’s primary cell wall. Our results indicate that N. parvum is equipped with an army of extracellular PCWDEs expressed even in the absence of plant tissue, but induced by the presence of the Eucalyptus stem (Table 2). Pectic enzymes (in multiple forms) are the first cell-wall-degrading enzymes induced by pathogens when cultured on isolated plant cell walls and the first produced in infected tissues [63,64]. Pectic enzymes induce the modification of the cell wall structure, exposing cell wall components for degradation by other enzymes [65]. Pectin is also present in Eucalyptus cell walls (15.2–25.8 mg g−1 pectin, [66]) and the secretion of pectin-degrading enzymes by N. parvum upon interaction with the Eucalyptus stem surely promotes the close interaction between the fungus and plant. All identified pectin-degrading enzymes [pectinases (GH53 and CE12) and pectate lyases (PL1, P3, PL4), Table 2 and Table S1] are more abundant in the presence of host material, suggesting that this fungus is more adapted to degrade intact or living plants than decaying biomass (where pectin is not present and is already decayed), which is in consonance with the fungus being a biotroph. Kang and Buchenauer [67] and Tomassini, et al. [68] demonstrated that wheat infection by Fusarium culmorum and F. graminearum depends on the production of CWDE at the early stages of infection which results in the facilitation of a rapid colonization of wheat spikes. Moreover, the up-regulation of these enzymes in lethal isolates of Verticilium albo-atrum compared to mild isolates was also described by Mandelc and Javornik [28], having implied its hypothetical contribution for plant vascular system colonization. We also identified cellulose-degrading enzymes mainly in the presence of the Eucalyptus stem. Putative GH12 protein (R1GQP5) raises special attention due to a high increase in response to Eucalyptus (3.9-fold up, Table 2). Recently, the xyloglucan-specific endo-β-1,4-glucanase (GH12 family) isolated from P. sojae culture filtrates induced cell death in dicot plants [69]. Gui, et al. [70] demonstrated that two of the six GH12 proteins in the fungus Verticillium dahliae Vd991 (VdEG1 and VdEG3) acted as virulence factors and as Pathogen-Associated Molecular Patterns (PAMPs), inducing cell death and triggering PAMP-triggered immunity in Nicotiana benthamiana. A glucanase (R1GZN3, GH7) was more abundant in the presence of the Eucalyptus stem than in control conditions. Cellulases belonging to GH6 and GH7 families are related to fungal virulence in the phytopathogenic fungus Magnaporthe oryzae, where they seem to be involved in the penetration of the host epidermis and further invasion [71].
Hemicellulases are generally involved in the degradation of hemicellulose from plant cell walls, helping in the colonization and in the acquisition of nutrients during infection. The up-regulation of two endoxylanases [beta-xylanase GH10 (R1FWZ0) and endo-1,4-beta-xylanase GH11 (R1GCT8)] was observed in N. parvum secretome in response to the Eucalyptus stem. GH10 and GH11 endoxylanases play significant roles in both vertical penetration of cell walls and horizontal expansion of the rice pathogen M. oryzae in infected leaves [72]. A recent study showed that two genes encoding GH10 xylanases are crucial for the virulence of the oomycete plant pathogen Phytophthora parasitica [73]. In B. cinerea, Xyn11A encodes an endo-β-1,4-xylanase Xyn11A, and the disruption of this gene resulted in reduced virulence of the pathogen [74]. However, several reports failed to show an essential role of endoxylanases in fungal pathogenicity [75,76,77]. Therefore, the role of xylanases in fungal pathogenesis may vary depending on the characteristics of the pathosystem and awaits further investigation.
The most abundant CAZy proteins in the control secretome of N. parvum are involved in lignin degradation (AA1, AA5, AA7) (Table 2 and Table S1). These enzymes belong to the oxidoreductase family which can produce the H2O2 required for the action of extracellular peroxidases. Usually, N. parvum is not considered a major lignin-depolymerizing fungus, like white-rot fungi [78], but, in our study, several extracellular lignin-degrading enzymes were identified. However, most of those enzymes (five out of six) were less abundant in the presence of the Eucalyptus stem, indicating that they may have another role in N. parvum rather than a direct role in lignocellulose deconstruction during infection. A similar observation was described for the white-rot fungus Lentinula edodes when exposed to microcrystalline cellulose, cellulose with lignosulfonate, and glucose [79]. CAZymes involved in lignin degradation were repressed by cellulose. Cai, Gong, Liu, Hu, Chen, Yan, Zhou, and Bian [79] suggested that laccases may have a role in increasing fungal resistance to oxidative stress rather than being involved in lignocellulosic degradation.
4.2. Defense from Host
Oxidoreductases, important virulence factors induced during plant infection [80,81], are over-represented in the secretome of the N. parvum supplemented with the Eucalyptus stem (5-fold), where they may contribute to the alkaloid biosynthesis and production of hydrogen peroxide through the oxidation of metabolites [82].
Besides host degradation, fungal cell wall degradation plays a fundamental role in fungal development during infection, facilitating fungal branching and elongation [83]. The putative chitin-binding protein (R1EW80) contains a chitin deacetylase domain which catalyzes the conversion of chitin into chitosan required for appressorium formation [84]. Interestingly, the up-regulation of this protein in response to the host mimicry reinforces the hypothesis that this protein might play a role in host colonization. Some pathogens produce chitin-binding proteins that mask chitin, avoiding host recognition by shielding, or by modifying it [85,86]. Similarly, we identified a putative chitin deacetylase (CE4, R1E7G7) which is significantly up-regulated (5.6-fold) in the presence of the Eucalyptus stem. Chitin deacetylases are also involved in the protection of fungi from host plant chitinases by converting the fungal cell wall chitin into chitosan [87,88]. An endo-chitosanase (GH75, R1GTL6) is also up-regulated (4.1-fold) during the interaction with the Eucalyptus stem, suggesting that chitosan generation by chitin deacetylase enhanced the chitosanolytic activity of the fungus. We predicted that this endo-chitosanase (GH75, R1GTL6) has numerous interactions with other Eucalyptus proteins (69 proteins, Table 3 and Table S4), some of which share important functions in Eucalyptus [auxin response factor (A0A059ACB3), delta-1-pyrroline-5-carboxylate synthase (A0A059BIT2), alpha-1,4 glucan phosphorylase (A0A059D8I8)], suggesting a central role for this protein in N. parvum infection mechanism of Eucalyptus (Table 3 and Table S4).
4.3. Virulence
Most extracellular proteases identified—several of which were described as virulence factors in fungal necrotrophs [89,90,91,92,93]—were more abundant upon induction by Eucalyptus. Specifically, metalloproteases such as deuterolysin are also induced in a virulent strain of D. corticola upon challenge by the host (Quercus suber) [56]. It has been suggested that deuterolysin targets proteins in the plant cell wall [94], being directly involved in the infection mechanism.
The putative ricin B lectin protein (R1GAK8), involved in carbohydrate binding, contains a pectin_lyase_fold/virulence domain (InterPro IPR011050) considered a virulence factor in several species [95,96,97]. Ricin B lectins inhibit protein synthesis [98] and are highly expressed during infection [99,100]. In N. parvum, the putative ricin B lectin protein was induced in response to the host mimicry (4.3-fold).
Proteins containing ribonuclease/ribotoxin domains are also more abundant in the secretome of N. parvum supplemented with the Eucalyptus stem when compared to the axenic culture. Ribonucleases perform a variety of functions, serving as extra- or intracellular cytotoxins, and modulating host immune responses [101,102]. Ribotoxins are fungal extracellular ribonucleases that are highly toxic due to their ability to enter host cells and their effective ribonucleolytic activity against the ribosome [103]. Extracellular ribonucleases have been related to biotrophic fungi defenses, inhibiting the action of plant ribosome-inactivating proteins that would otherwise lead to host cell death, and pathogen death [102]. Secretion of low-molecular-weight guanyl-preferring ribonucleases (RNases) has also been reported in the secretome of the D. corticola [56]. Nonetheless, we predicted that the uncharacterized protein with ribonuclease activity (R1FZX2) establishes more than 360 interactions with Eucalyptus proteins.
Neofusicoccum parvum secretome contains necrotic elicitors like the necrosis-inducing protein (R1FZC0) and the putative epl1 protein (R1G1Q3) containing cerato-platanin domain, both in the control and infection-like conditions (Table S2), suggesting that these phytotoxins are constitutively expressed by N. parvum. Interestingly, earlier we showed that NLP genes coding necrosis-inducing proteins in N. parvum are functional genes. NLP genes of N. parvum encode proteins toxic both to plant and mammalian cells, most probably involved in virulence or cell death during infection by N. parvum [21].
Proteins from the fasciclin family have been identified as cell adhesion molecules in various organisms [104,105,106,107]. In this study, the accumulation of the putative fasciclin domain family protein (R1EWZ5) in the infection-like conditions could be responsible for the attachment of fungal hyphae to the host material. In the rice blast fungus, Magnaporthe oryzae, MoFLP1 null mutants generated by targeted fasciclin gene disruption showed a significant reduction of conidiation, conidial adhesion, and appressorium turgor, resulting in overall decreased fungal pathogenicity [108]. But the knowledge on the role of fasciclin domain-containing proteins on fungi pathogenesis is still scarce and, according to Seifert [109], “cell adhesion” might be a result of turgor pressure and a buildup of adhesive materials such as pectin and not a direct function of fasciclin domain family proteins [109].
4.4. Protein–Protein Interactions
We predicted PPIs between N. parvum secreted proteins and proteins of E. grandis (used as a reference, since E. globulus genome sequence is not available). Some proteins identified display a high number of PPIs with host proteins, suggesting that those proteins may function as cross-talkers in biological functions between the fungus and the host. Among these, an auxin response factor (A0A059ACB3) in Eucalyptus raises special attention: not only it has a high number of interactions (33, Table S5) but most proteins (27 out of 33 proteins), with whom the auxin response factor interacts, are more abundant in the infection-like condition. Furthermore, virulence factors like ricin B lectin protein (R1GAK8), putative exo-beta protein (PL3, R1H382), glucanase (R1GZN3, GH7), and putative pectate lyase (R1H2U7) were identified as the proteins interacting with auxin response factor (Table S5). Auxin response factors (ARFs) family proteins are key players in auxin signaling [110]. Indole-3-acetic acid (IAA), the major form of auxin in plants, is the most important phytohormone with the main effects on plant growth, development, and on the regulation of plant senescence [111]. Pathogens may promote auxin accumulation or auxin signaling in the host through the action of virulence factors that have evolved to modulate host auxin biology. So far, studies on Arabidopsis imply that auxin reduces (hemi) biotroph resistance but enhances plant defenses toward necrotrophic pathogens [112]. Consistent with IAA promoting hemibiotroph susceptibility, auxin, and, more specifically, IAA, also act as virulence factors of the hemibiotrophic rice pathogens Magnaporthe oryzae, Xanthomonas oryzae pv. oryzae, and X. oryzae pv. oryzicola [113,114,115]. Like many other microorganisms, these pathogens produce and secrete IAA themselves and increase IAA biosynthesis and signaling upon infection [115].
5. Conclusions
Multi-omics approaches to pathogen–host interactions are becoming more common, but still largely rely on the existence of genome data. Nonetheless, even in the absence of such information, using similar genome data (e.g., from the same genus) has been widely used with success. We used proteomics (focused on the secretome) and computational interactomics to shed light on the infection mechanism of N. parvum on Eucalyptus. Most of the secretome proteins induced under host mimicry are, as in the case of other Botryosphaeriaceae fungi, cell-wall-degrading enzymes (CWDEs), especially those targeting pectin and hemicellulose, allowing the fungus to invade host tissues and extract nutrients for its own growth. Additionally, the degradation of xylan (hemicellulose) and pectin is required for fungal pathogens to invasively penetrate and proliferate inside host cells. We also found the up-regulation of chitosan synthesis and chitin degradation proteins during interaction with the Eucalyptus stem, suggesting that the pathogen masks itself to avoid plant defenses.
Surely not of negligible relevance, N. parvum proteins predicted to be involved in the largest number of interactions with Eucalyptus proteins are degradative enzymes. But besides attacking the integrity of the host, N. parvum appears to mask or modify its own cell surface avoiding plant defenses, which would allow the fungus to colonize the host while actively releasing enzymes and toxins (such as proteins containing ribonuclease/ribotoxin domains, putative ricin B lectins, putative epl1 proteins containing cerato-platanin domain and necrosis inducing proteins). The isolate used in this study is able to infect and cause disease in E. globulus [9]. Like many other species of Botryosphaeriaceae fungi, N. parvum can change between a commensal and a pathogenic lifestyle.
The distinction between an organism that colonizes living plants without causing symptoms of disease and that, given a certain stimulus, becomes a pathogen and an hemibiotroph (organisms that switch from an initial biotroph to necrotroph behavior) is not easy. Neofusicoccum parvum can colonize (infect?) its hosts without causing (visible) damages (compatible with being a non-pathogenic endophyte or—we believe this is a more accurate adjective—a commensal organism), but it can also become a pathogen, causing necrosis and ultimately the death of its host. The physiology of this shift is not known, although the literature refers that host stress can induce the shift.
It is our belief that the main issue is: is N. parvum a plant commensal that shifts to a pathogen? Or is it a pathogen that, for some time, does not express its pathogenic traits? Or is it a biotroph, feeding on its host, but without causing major harm?
What do we know for sure? We know that N. parvum can cause necrosis, so, at a given point of its life cycle, it is a necrotroph: it can express active necrosis elicitors able to induce phytotoxicity and necrosis [21]. The secretome of N. parvum in the presence of eucalyptus is also compatible with a necrotrophic lifestyle: it expresses CAZymes that are as important in establishing infection as in accessing nutrients during necrotrophic growth [116].
But is N. parvum a commensal or a (non-obligate) biotroph? The molecular evidence that we have indicates (“suggests”) that N. parvum is in fact a biotroph: it has the molecular machinery that allows it to colonize, spread, and feed on living plants (e.g., the overexpression of chitosan’s synthesis proteins). Furthermore, N. parvum expresses pathogenesis-related proteases and proteins containing ribonuclease/ribotoxin domains (known as toxic due to their ability to enter host cells and their effective ribonucleolytic activity against the ribosome) even in the absence of the plant host.
Last, the N. parvum genome contains some of the proteins involved in the shift between the biotroph and the necrotrophic phases of a typical hemibiotroph. One example is the 4-phosphopantetheinyl transferase protein (CgPPT1). CgPPT1 is functionally involved in and required for the biotrophy–necrotrophy transition of Colletotrichum graminicola [117]. In accordance with the hypothesis that N. parvum is an hemibiotroph, its genome contains a CgPPT1 gene. Also, in Colletotrichum, the shift from biotrophy to necrotrophy is defined by induction of the degradome, mirroring necrotrophic pathosystems [118]. Neofusicoccum parvum has the enzymes (CAZYmes, proteases, …) typical of these ‘degradomes’.
There are still many questions to be answered, but N. parvum secretome is more closely related to that of an hemibiotroph than to a plant commensal. It lives inside plants, feeds on them, and causes damage, eventually killing them. Why it remains “dormant” or why initial damages are not seen are questions that still need to be addressed.
Acknowledgments
LC-MS were performed via the Ghent University and the VIB Proteomics Expertise Centers (Proteogent, VIB-PEC).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8090971/s1, Figure S1: SDS-PAGE of N. parvum-secreted proteins; Figure S2: Histograms of the LFQ intensity values of the nine samples under analysis; Figure S3. Multi-scatter plot with Pearson correlation values of the nine samples against each other; Figure S4: Hierarchical clustering of the nine samples under analysis; Figure S5: Enriched functions of Eucalyptus grandis proteins on which the fungus acts; Figure S6: Relative quantification by RT-qPCR of mRNA; Table S1: Differentially expressed proteins in the control and infection-like secretome of N. parvum; Table S2: Common proteins identified in the control and infection-like secretome of N. parvum; Table S3: PPI prediction involving proteins of N. parvum and proteins of E. grandis; Table S4: List of proteins of Eucalyptus interacting with the endo-chitosanase protein of N. parvum; Table S5: List of proteins of N. parvum interacting with auxin response factor (ARF) protein of Eucalyptus.
Author Contributions
Conceptualization, A.C.E. and A.A.; Investigation, F.N.P., B.P., M.O., G.V.D., C.F. (Carina Félix) and N.R.; Validation, A.S.D., A.A., B.D. and A.C.E.; Formal Analysis, F.N.P., B.P., C.F. (Carina Félix), G.V.D., B.D., C.F. (Cátia Fidalgo) and A.C.E.; Methodology, B.P., A.S.D., N.R., B.D., A.A. and A.C.E.; Resources, B.D. and A.A.; Data Curation, N.R. and A.C.E.; Writing—Original, F.N.P. and C.F.; Writing— Review and Editing, all authors; Visualization, F.N.P., A.S.D., B.D., A.A. and A.C.E.; Project Administration, A.C.E. and A.A.; Supervision, A.C.E. and A.A.; Funding Acquisition, B.D. and A.A. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
The data presented in this study are available in Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by Portuguese Foundation for Science and Technology (FCT) through national funds to CESAM (UIDP/50017/2020+UIDB/50017/2020+LA/P/0094/2020) and to Center for Interdisciplinary Research in Health (UIDB/04279/2020+UIDP/04279/2020). Thanks are also due to FCT for CEEC financing of C Fidalgo (CEECIND/01373/2018) and UCP for the CEEC institutional financing of AS Duarte (CEECINST/00137/2018/CP1520/CT0013).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Deus E., Silva J.S., Castro-Díez P., Lomba A., Ortiz M.L., Vicente J. Current and future conflicts between eucalypt plantations and high biodiversity areas in the Iberian Peninsula. J. Nat. Conserv. 2018;45:107–117. doi: 10.1016/j.jnc.2018.06.003. [DOI] [Google Scholar]
- 2.ICNF . 6 Inventário Florestal Nacional. Instituto da Conservação da Natureza e das Florestas; Lisboa, Portugal: 2019. p. 284. [Google Scholar]
- 3.Li G., Arnold R.J., Liu F., Li J., Chen S. Identification and pathogenicity of Lasiodiplodia species from Eucalyptus urophylla × grandis, Polyscias balfouriana and Bougainvillea spectabilis in Southern China. J. Phytopathol. 2015;163:956–967. doi: 10.1111/jph.12398. [DOI] [Google Scholar]
- 4.Slippers B., Roux J., Wingfield M.J., van der Walt F.J.J., Jami F., Mehl J.W.M., Marais G.J. Confronting the constraints of morphological taxonomy in the Botryosphaeriales. Persoonia. 2014;33:155–168. doi: 10.3767/003158514X684780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slippers B., Wingfield M.J. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: Diversity, ecology and impact. Fungal Biol. Rev. 2007;21:90–106. doi: 10.1016/j.fbr.2007.06.002. [DOI] [Google Scholar]
- 6.Pérez C.A., Wingfield M.J., Slippers B., Altier N.A., Blanchette R.A. Endophytic and canker-associated Botryosphaeriaceae occurring on non-native Eucalyptus and native Myrtaceae trees in Uruguay. Fungal Divers. 2010;41:53–69. doi: 10.1007/s13225-009-0014-8. [DOI] [Google Scholar]
- 7.Phillips A.J., Alves A., Abdollahzadeh J., Slippers B., Wingfield M.J., Groenewald J.Z., Crous P.W. The Botryosphaeriaceae: Genera and species known from culture. Stud. Mycol. 2013;76:51–167. doi: 10.3114/sim0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith H., Wingfield M.J., Petrini O. Botryosphaeria dothidea endophytic in Eucalyptus grandis and Eucalyptus nitens in South Africa. Forest Ecol. Manag. 1996;89:189–195. doi: 10.1016/S0378-1127(96)03847-9. [DOI] [Google Scholar]
- 9.Barradas C., Phillips A.J.L., Correia A., Diogo E., Bragança H., Alves A. Diversity and potential impact of Botryosphaeriaceae species associated with Eucalyptus globulus plantations in Portugal. Eur. J. Plant Pathol. 2016;146:245–257. doi: 10.1007/s10658-016-0910-1. [DOI] [Google Scholar]
- 10.Batista E., Lopes A., Alves A. Botryosphaeriaceae species on forest trees in Portugal: Diversity, distribution and pathogenicity. Eur. J. Plant Pathol. 2020;158:693–720. doi: 10.1007/s10658-020-02112-8. [DOI] [Google Scholar]
- 11.Slippers B., Burgess T., Pavlic D., Ahumada R., Maleme H., Mohali S., Rodas C., Wingfield M.J. A diverse assemblage of Botryosphaeriaceae infect Eucalyptus in native and non-native environments. South For.-J. For. Sci. 2009;71:101–110. doi: 10.2989/SF.2009.71.2.3.818. [DOI] [Google Scholar]
- 12.Mohali S., Slippers B., Wingfield M.J. Identification of Botryosphaeriaceae from Eucalyptus, Acacia and Pinus in Venezuela. Fungal Divers. 2007;25:103–125. [Google Scholar]
- 13.Blanco-Ulate B., Rolshausen P., Cantu D. Draft genome sequence of Neofusicoccum parvum isolate UCR-NP2, a fungal vascular pathogen associated with grapevine cankers. Genome Announc. 2013;1:e00339-13. doi: 10.1128/genomeA.00339-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S.F., Pavlic D., Roux J., Slippers B., Xie Y.J., Wingfield M.J., Zhou X.D. Characterization of Botryosphaeriaceae from plantation-grown Eucalyptus species in South China. Plant Pathol. 2011;60:739–751. doi: 10.1111/j.1365-3059.2011.02431.x. [DOI] [Google Scholar]
- 15.Pavlic D., Slippers B., Coutinho T.A., Wingfield M.J. Botryosphaeriaceae occurring on native Syzygium cordatum in South Africa and their potential threat to Eucalyptus. Plant Pathol. 2007;56:624–636. doi: 10.1111/j.1365-3059.2007.01608.x. [DOI] [Google Scholar]
- 16.Abou-Mansour E., Débieux J.-L., Ramírez-Suero M., Bénard-Gellon M., Magnin-Robert M., Spagnolo A., Chong J., Farine S., Bertsch C., L’Haridon F., et al. Phytotoxic metabolites from Neofusicoccum parvum, a pathogen of Botryosphaeria dieback of grapevine. Phytochemistry. 2015;115:207–215. doi: 10.1016/j.phytochem.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Andolfi A., Mugnai L., Luque J., Surico G., Cimmino A., Evidente A. Phytotoxins produced by fungi associated with grapevine trunk diseases. Toxins. 2011;3:1569–1605. doi: 10.3390/toxins3121569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masi M., Cimmino A., Reveglia P., Mugnai L., Surico G., Evidente A. Advances on Fungal Phytotoxins and Their Role in Grapevine Trunk Diseases. J. Agric. Food Chem. 2018;66:5948–5958. doi: 10.1021/acs.jafc.8b00773. [DOI] [PubMed] [Google Scholar]
- 19.Salvatore M.M., Alves A., Andolfi A. Secondary metabolites produced by Neofusicoccum species associated with plants: A review. Agriculture. 2021;11:149. doi: 10.3390/agriculture11020149. [DOI] [Google Scholar]
- 20.Bénard-Gellon M., Farine S., Goddard M.L., Schmitt M., Stempien E., Pensec F., Laloue H., Mazet-Kieffer F., Fontaine F., Larignon P., et al. Toxicity of extracellular proteins from Diplodia seriata and Neofusicoccum parvum involved in grapevine Botryosphaeria dieback. Protoplasma. 2015;252:679–687. doi: 10.1007/s00709-014-0716-y. [DOI] [PubMed] [Google Scholar]
- 21.Nazar Pour F., Cobos R., Rubio Coque J.J., Serôdio J., Alves A., Félix C., Ferreira V., Esteves A.C., Duarte A.S. Toxicity of recombinant Necrosis and Ethylene-Inducing Proteins (NLPs) from Neofusicoccum parvum. Toxins. 2020;12:235. doi: 10.3390/toxins12040235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massonnet M., Figueroa-Balderas R., Galarneau E.R.A., Miki S., Lawrence D.P., Sun Q., Wallis C.M., Baumgartner K., Cantu D. Neofusicoccum parvum colonization of the grapevine woody stem triggers asynchronous host responses at the site of infection and in the leaves. Front. Plant Sci. 2017;8:1117. doi: 10.3389/fpls.2017.01117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan J.Y., Zhao W.S., Chen Z., Xing Q.K., Zhang W., Chethana K.W.T., Xue M.F., Xu J.P., Phillips A.J.L., Wang Y., et al. Comparative genome and transcriptome analyses reveal adaptations to opportunistic infections in woody plant degrading pathogens of Botryosphaeriaceae. DNA Res. 2018;25:87–102. doi: 10.1093/dnares/dsx040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massonnet M., Morales-Cruz A., Figueroa-Balderas R., Lawrence D.P., Baumgartner K., Cantu D. Condition-dependent co-regulation of genomic clusters of virulence factors in the grapevine trunk pathogen Neofusicoccum parvum. Mol. Plant Pathol. 2018;19:21–34. doi: 10.1111/mpp.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.González-Fernández R., Valero-Galván J., Gómez-Gálvez F.J., Jorrín-Novo J.V. Unraveling the in vitro secretome of the phytopathogen Botrytis cinerea to understand the interaction with its hosts. Front. Plant Sci. 2015;6:839. doi: 10.3389/fpls.2015.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jami M.S., Barreiro C., García-Estrada C., Martín J.F. Proteome analysis of the penicillin producer Penicillium chrysogenum: Characterization of protein changes during the industrial strain improvement. Mol. Cell Proteom. 2010;9:1182–1198. doi: 10.1074/mcp.M900327-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyu X., Shen C., Fu Y., Xie J., Jiang D., Li G., Cheng J. Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development. Sci. Rep. 2015;5:15565. doi: 10.1038/srep15565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandelc S., Javornik B. The secretome of vascular wilt pathogen Verticillium albo-atrum in simulated xylem fluid. Proteomics. 2015;15:787–797. doi: 10.1002/pmic.201400181. [DOI] [PubMed] [Google Scholar]
- 29.Shah P., Atwood J.A., Orlando R., El Mubarek H., Podila G.K., Davis M.R. Comparative proteomic analysis of Botrytis cinerea secretome. J. Proteome Res. 2009;8:1123–1130. doi: 10.1021/pr8003002. [DOI] [PubMed] [Google Scholar]
- 30.Cobos R., Barreiro C., Mateos R.M., Coque J.J. Cytoplasmic- and extracellular-proteome analysis of Diplodia seriata: A phytopathogenic fungus involved in grapevine decline. Proteome Sci. 2010;8:46. doi: 10.1186/1477-5956-8-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandes I., Alves A., Correia A., Devreese B., Esteves A.C. Secretome analysis identifies potential virulence factors of Diplodia corticola, a fungal pathogen involved in cork oak (Quercus suber) decline. Fungal Biol. 2014;118:516–523. doi: 10.1016/j.funbio.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Félix C., Duarte A.S., Vitorino R., Guerreiro A.C., Domingues P., Correia A.C., Alves A., Esteves A.C. Temperature modulates the secretome of the phytopathogenic fungus Lasiodiplodia theobromae. Front. Plant Sci. 2016;7:1096. doi: 10.3389/fpls.2016.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Félix C., Meneses R., Gonçalves M.F.M., Tilleman L., Duarte A.S., Jorrín-Novo J.V., Van de Peer Y., Deforce D., Van Nieuwerburgh F., Esteves A.C., et al. A multi-omics analysis of the grapevine pathogen Lasiodiplodia theobromae reveals that temperature affects the expression of virulence- and pathogenicity-related genes. Sci. Rep. 2019;9:13144. doi: 10.1038/s41598-019-49551-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uranga C.C., Ghassemian M., Hernández-Martínez R. Novel proteins from proteomic analysis of the trunk disease fungus Lasiodiplodia theobromae (Botryosphaeriaceae) Biochim. Open. 2017;4:88–98. doi: 10.1016/j.biopen.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alves A., Crous P.W., Correia A., Phillips A. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Divers. 2008;28:1–13. [Google Scholar]
- 36.Wessel D., Flügge U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.Cox J., Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- 39.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 40.Consortium T.U. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 42.Blum M., Chang H.-Y., Chuguransky S., Grego T., Kandasaamy S., Mitchell A., Nuka G., Paysan-Lafosse T., Qureshi M., Raj S., et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2020;49:D344–D354. doi: 10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drula E., Garron M.-L., Dogan S., Lombard V., Henrissat B., Terrapon N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2021;50:D571–D577. doi: 10.1093/nar/gkab1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pierleoni A., Martelli P.L., Fariselli P., Casadio R. BaCelLo: A balanced subcellular localization predictor. Bioinformatics. 2006;22:e408–e416. doi: 10.1093/bioinformatics/btl222. [DOI] [PubMed] [Google Scholar]
- 45.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 46.Bendtsen J.D., Jensen L.J., Blom N., Von Heijne G., Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004;17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 47.Coelho E.D., Arrais J.P., Matos S., Pereira C., Rosa N., Correia M.J., Barros M., Oliveira J.L. Computational prediction of the human-microbial oral interactome. BMC Syst. Biol. 2014;8:24. doi: 10.1186/1752-0509-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Team R.C. R: A Language and Environment for Statistical Computing. [(accessed on 1 April 2022)]. Available online: https://www.R-project.org/
- 49.Pedersen T.L. ggraph: An Implementation of Grammar of Graphics for Graphs and Networks. R package Version 2.0.5. [(accessed on 1 April 2022)]. Available online: https://CRAN.R-project.org/package=ggraph.
- 50.Csardi G., Nepusz T.T. The igraph software package for complex network research. InterJournal Complex Syst. 2006;1695:1–9. [Google Scholar]
- 51.Wickham H., Averick M., Bryan J., Chang W., McGowan L., François R., Grolemund G., Hayes A., Henry L., Hester J., et al. Welcome to the tidyverse. J. Open. Source Softw. 2019;4:1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- 52.Nielsen H., Tsirigos K.D., Brunak S., von Heijne G. A brief history of protein sorting prediction. Protein J. 2019;38:200–216. doi: 10.1007/s10930-019-09838-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Esteves E., Rosa N., Correia M.J., Arrais J.P., Barros M. New targets for Zika Virus determined by human-viral interactomic: A bioinformatics approach. Biomed Res. Int. 2017;2017:1734151. doi: 10.1155/2017/1734151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosa N., Campos B., Esteves A.C., Duarte A.S., Correia M.J., Silva R.M., Barros M. Tracking the functional meaning of the human oral-microbiome protein-protein interactions. Adv. Protein Chem. Struct. Biol. 2020;121:199–235. doi: 10.1016/bs.apcsb.2019.11.014. [DOI] [PubMed] [Google Scholar]
- 56.Fernandes I.O. Ph.D. Thesis. Universidade de Aveiro; Aveiro, Portugal: 2015. [(accessed on 21 July 2022)]. Infection Mechanism of Diplodia corticola. Available online: http://hdl.handle.net/10773/15487. [Google Scholar]
- 57.Salvatore M.M., Félix C., Lima F., Ferreira V., Naviglio D., Salvatore F., Duarte A.S., Alves A., Andolfi A., Esteves A.C. Secondary Metabolites Produced by Macrophomina phaseolina Isolated from Eucalyptus globulus. Agriculture. 2020;10:72. doi: 10.3390/agriculture10030072. [DOI] [Google Scholar]
- 58.Jung Y.H., Jeong S.H., Kim S.H., Singh R., Lee J.E., Cho Y.S., Agrawal G.K., Rakwal R., Jwa N.S. Secretome analysis of Magnaporthe oryzae using in vitro systems. Proteomics. 2012;12:878–900. doi: 10.1002/pmic.201100142. [DOI] [PubMed] [Google Scholar]
- 59.Sibbald M.J., Ziebandt A.K., Engelmann S., Hecker M., de Jong A., Harmsen H.J., Raangs G.C., Stokroos I., Arends J.P., Dubois J.Y., et al. Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol. Mol. Biol. Rev. 2006;70:755–788. doi: 10.1128/MMBR.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagel J.H., Wingfield M.J., Slippers B. Increased abundance of secreted hydrolytic enzymes and secondary metabolite gene clusters define the genomes of latent plant pathogens in the Botryosphaeriaceae. BMC Genom. 2021;22:589. doi: 10.1186/s12864-021-07902-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sinha N., Patra S.K., Ghosh S. Secretome analysis of Macrophomina phaseolina identifies an array of putative virulence factors responsible for charcoal rot disease in plants. Front. Microbiol. 2022;13:847832. doi: 10.3389/fmicb.2022.847832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Silva N., Lumyong S., Hyde K., Bulgakov T., Phillips A., Yan J. Mycosphere Essays 9: Defining biotrophs and hemibiotrophs. Mycosphere. 2016;7:545–559. doi: 10.5943/mycosphere/7/5/2. [DOI] [Google Scholar]
- 63.Martínez M.J., Alconada M.T., Guillén F., Vázquez C., Reyes F. Pectic activities from Fusarium oxysporum f. sp. melonis: Purification and characterization of an exopolygalacturonase. FEMS Microbiol. Lett. 1991;81:145–149. [Google Scholar]
- 64.Niture S.K., Kumar A.R., Pant A. Role of glucose in production and repression of polygalacturonase and pectate lyase from phytopathogenic fungus Fusarium moniliforme NCIM 1276. World J. Microbiol. Biotechnol. 2006;22:893–899. doi: 10.1007/s11274-006-9119-3. [DOI] [Google Scholar]
- 65.Panda T., Nair S.R., Kumar M.P. Regulation of synthesis of the pectolytic enzymes of Aspergillus niger. Enzym. Microb. Technol. 2004;34:466–473. doi: 10.1016/j.enzmictec.2003.12.004. [DOI] [Google Scholar]
- 66.Coetzee B., Schols H.A., Wolfaardt F. Determination of Pectin Content of Eucalyptus Wood. pp. 327–331. [(accessed on 21 July 2022)]. Available online: https://www.degruyter.com/document/doi/10.1515/hf.2011.054/html.
- 67.Kang Z., Buchenauer H. Ultrastructural and cytochemical studies on cellulose, xylan and pectin degradation in wheat spikes infected by Fusarium culmorum. J. Phytopathol. 2000;148:263–275. doi: 10.1046/j.1439-0434.2000.00489.x. [DOI] [Google Scholar]
- 68.Tomassini A., Sella L., Raiola A., D’Ovidio R., Favaron F. Characterization and expression of Fusarium graminearum endo-polygalacturonases in vitro and during wheat infection. Plant Pathol. 2009;58:556–564. doi: 10.1111/j.1365-3059.2008.02019.x. [DOI] [Google Scholar]
- 69.Ma Z., Song T., Zhu L., Ye W., Wang Y., Shao Y., Dong S., Zhang Z., Dou D., Zheng X., et al. A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during Soybean infection and is recognized as a PAMP. Plant Cell. 2015;27:2057–2072. doi: 10.1105/tpc.15.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gui Y.J., Chen J.Y., Zhang D.D., Li N.Y., Li T.G., Zhang W.Q., Wang X.Y., Short D.P.G., Li L., Guo W., et al. Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ. Microbiol. 2017;19:1914–1932. doi: 10.1111/1462-2920.13695. [DOI] [PubMed] [Google Scholar]
- 71.Van Vu B., Itoh K., Nguyen Q.B., Tosa Y., Nakayashiki H. Cellulases belonging to glycoside hydrolase families 6 and 7 contribute to the virulence of Magnaporthe oryzae. Mol. Plant Microbe Interact. 2012;25:1135–1141. doi: 10.1094/MPMI-02-12-0043-R. [DOI] [PubMed] [Google Scholar]
- 72.Nguyen Q.B., Itoh K., Van Vu B., Tosa Y., Nakayashiki H. Simultaneous silencing of endo-β-1,4 xylanase genes reveals their roles in the virulence of Magnaporthe oryzae. Mol. Microbiol. 2011;81:1008–1019. doi: 10.1111/j.1365-2958.2011.07746.x. [DOI] [PubMed] [Google Scholar]
- 73.Lai M.W., Liou R.F. Two genes encoding GH10 xylanases are essential for the virulence of the oomycete plant pathogen Phytophthora parasitica. Curr. Genet. 2018;64:931–943. doi: 10.1007/s00294-018-0814-z. [DOI] [PubMed] [Google Scholar]
- 74.Brito N., Espino J.J., González C. The endo-beta-1,4-xylanase xyn11A is required for virulence in Botrytis cinerea. Mol. Plant Microbe Interact. 2006;19:25–32. doi: 10.1094/MPMI-19-0025. [DOI] [PubMed] [Google Scholar]
- 75.Apel-Birkhold P.C., Walton J.D. Cloning, disruption, and expression of two endo-beta 1, 4-xylanase genes, XYL2 and XYL3, from Cochliobolus carbonum. Appl. Environ. Microbiol. 1996;62:4129–4135. doi: 10.1128/aem.62.11.4129-4135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Calero-Nieto F., Di Pietro A., Roncero M.I., Hera C. Role of the transcriptional activator xlnR of Fusarium oxysporum in regulation of xylanase genes and virulence. Mol. Plant Microbe Interact. 2007;20:977–985. doi: 10.1094/MPMI-20-8-0977. [DOI] [PubMed] [Google Scholar]
- 77.Gómez-Gómez E., Ruíz-Roldán M.C., Di Pietro A., Roncero M.I., Hera C. Role in pathogenesis of two endo-beta-1,4-xylanase genes from the vascular wilt fungus Fusarium oxysporum. Fungal Genet. Biol. 2002;35:213–222. doi: 10.1006/fgbi.2001.1318. [DOI] [PubMed] [Google Scholar]
- 78.Stempien E., Goddard M.L., Wilhelm K., Tarnus C., Bertsch C., Chong J. Grapevine Botryosphaeria dieback fungi have specific aggressiveness factor repertory involved in wood decay and stilbene metabolization. PLoS ONE. 2017;12:e0188766. doi: 10.1371/journal.pone.0188766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cai Y., Gong Y., Liu W., Hu Y., Chen L., Yan L., Zhou Y., Bian Y. Comparative secretomic analysis of lignocellulose degradation by Lentinula edodes grown on microcrystalline cellulose, lignosulfonate and glucose. J. Proteom. 2017;163:92–101. doi: 10.1016/j.jprot.2017.04.023. [DOI] [PubMed] [Google Scholar]
- 80.Raffaele S., Win J., Cano L.M., Kamoun S. Analyses of genome architecture and gene expression reveal novel candidate virulence factors in the secretome of Phytophthora infestans. BMC Genom. 2010;11:637. doi: 10.1186/1471-2164-11-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seidl M.F., Van den Ackerveken G., Govers F., Snel B. A domain-centric analysis of oomycete plant pathogen genomes reveals unique protein organization. Plant Physiol. 2011;155:628–644. doi: 10.1104/pp.110.167841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leferink N.G., Heuts D.P., Fraaije M.W., van Berkel W.J. The growing VAO flavoprotein family. Arch. Biochem. Biophys. 2008;474:292–301. doi: 10.1016/j.abb.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 83.Do Vale L.H., Gómez-Mendoza D.P., Kim M.S., Pandey A., Ricart C.A., Ximenes F.F.E., Sousa M.V. Secretome analysis of the fungus Trichoderma harzianum grown on cellulose. Proteomics. 2012;12:2716–2728. doi: 10.1002/pmic.201200063. [DOI] [PubMed] [Google Scholar]
- 84.Kuroki M., Okauchi K., Yoshida S., Ohno Y., Murata S., Nakajima Y., Nozaka A., Tanaka N., Nakajima M., Taguchi H., et al. Chitin-deacetylase activity induces appressorium differentiation in the rice blast fungus Magnaporthe oryzae. Sci. Rep. 2017;7:9697. doi: 10.1038/s41598-017-10322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rovenich H., Zuccaro A., Thomma B.P. Convergent evolution of filamentous microbes towards evasion of glycan-triggered immunity. New Phytol. 2016;212:896–901. doi: 10.1111/nph.14064. [DOI] [PubMed] [Google Scholar]
- 86.Sharp R.G. A review of the applications of chitin and its derivatives in agriculture to modify plant-microbial interactions and improve crop yields. Agronomy. 2013;3:757–793. doi: 10.3390/agronomy3040757. [DOI] [Google Scholar]
- 87.Kouzai Y., Mochizuki S., Saito A., Ando A., Minami E., Nishizawa Y. Expression of a bacterial chitosanase in rice plants improves disease resistance to the rice blast fungus Magnaporthe oryzae. Plant Cell Rep. 2012;31:629–636. doi: 10.1007/s00299-011-1179-7. [DOI] [PubMed] [Google Scholar]
- 88.Ride J.P., Barber M.S. Purification and characterization of multiple forms of endochitinase from wheat leaves. Plant Sci. 1990;71:185–197. doi: 10.1016/0168-9452(90)90008-C. [DOI] [Google Scholar]
- 89.Hislop E.C., Paver J.L., Keon J.P.R. An acid protease produced by Monilinia fructigena in vitro and in infected apple fruits, and its possible role in pathogenesis. Microbiology. 1982;128:799–807. doi: 10.1099/00221287-128-4-799. [DOI] [Google Scholar]
- 90.Movahedi S., Heale J.B. Purification and characterization of an aspartic proteinase secreted by Botrytis cinerea Pers ex. Pers in culture and in infected carrots. Physiol. Mol. Plant Pathol. 1990;36:289–302. doi: 10.1016/0885-5765(90)90060-B. [DOI] [Google Scholar]
- 91.Poussereau N., Gente S., Rascle C., Billon-Grand G., Fèvre M. aspS encoding an unusual aspartyl protease from Sclerotinia sclerotiorum is expressed during phytopathogenesis. FEMS Microbiol. Lett. 2001;194:27–32. doi: 10.1111/j.1574-6968.2001.tb09441.x. [DOI] [PubMed] [Google Scholar]
- 92.Urbanek H., Yirdaw G. Hydrolytic ability of acid protease of Fusarium culmorum and its possible role in phytopathogenesis. Acta Microbiol. Pol. 1984;33:131–136. [PubMed] [Google Scholar]
- 93.O’Connell R.J., Thon M.R., Hacquard S., Amyotte S.G., Kleemann J., Torres M.F., Damm U., Buiate E.A., Epstein L., Alkan N., et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 2012;44:1060–1065. doi: 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lakshman D.K., Roberts D.P., Garrett W.M., Natarajan S.S., Darwish O., Alkharouf N., Pain A., Khan F., Jambhulkar P.P., Mitra A. Proteomic investigation of Rhizoctonia solani AG 4 identifies secretome and mycelial proteins with roles in plant cell wall degradation and virulence. J. Agric. Food Chem. 2016;64:3101–3110. doi: 10.1021/acs.jafc.5b05735. [DOI] [PubMed] [Google Scholar]
- 95.González-Fernández R., Aloria K., Valero-Galván J., Redondo I., Arizmendi J.M., Jorrín-Novo J.V. Proteomic analysis of mycelium and secretome of different Botrytis cinerea wild-type strains. J. Proteom. 2014;97:195–221. doi: 10.1016/j.jprot.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 96.Ismail I.A., Able A.J. Secretome analysis of virulent Pyrenophora teres f. teres isolates. Proteomics. 2016;16:2625–2636. doi: 10.1002/pmic.201500498. [DOI] [PubMed] [Google Scholar]
- 97.Kubicek C.P., Starr T.L., Glass N.L. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 2014;52:427–451. doi: 10.1146/annurev-phyto-102313-045831. [DOI] [PubMed] [Google Scholar]
- 98.Endo Y., Tsurugi K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J. Biol. Chem. 1987;262:8128–8130. doi: 10.1016/S0021-9258(18)47538-2. [DOI] [PubMed] [Google Scholar]
- 99.Andersson K.-M., Kumar D., Bentzer J., Friman E., Ahrén D., Tunlid A. Interspecific and host-related gene expression patterns in nematode-trapping fungi. BMC Genom. 2014;15:968. doi: 10.1186/1471-2164-15-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Meerupati T., Andersson K.M., Friman E., Kumar D., Tunlid A., Ahrén D. Genomic mechanisms accounting for the adaptation to parasitism in nematode-trapping fungi. PLoS Genet. 2013;9:e1003909. doi: 10.1371/journal.pgen.1003909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Luhtala N., Parker R. T2 family ribonucleases: Ancient enzymes with diverse roles. Trends Biochem. Sci. 2010;35:253–259. doi: 10.1016/j.tibs.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pennington H.G., Jones R., Kwon S., Bonciani G., Thieron H., Chandler T., Luong P., Morgan S.N., Przydacz M., Bozkurt T., et al. The fungal ribonuclease-like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathog. 2019;15:e1007620. doi: 10.1371/journal.ppat.1007620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Olombrada M., Martínez-del-Pozo A., Medina P., Budia F., Gavilanes J.G., García-Ortega L. Fungal ribotoxins: Natural protein-based weapons against insects. Toxicon. 2014;83:69–74. doi: 10.1016/j.toxicon.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 104.Carr M.D., Bloemink M.J., Dentten E., Whelan A.O., Gordon S.V., Kelly G., Frenkiel T.A., Hewinson R.G., Williamson R.A. Solution structure of the Mycobacterium tuberculosis complex protein MPB70: From tuberculosis pathogenesis to inherited human corneal desease. J. Biol. Chem. 2003;278:43736–43743. doi: 10.1074/jbc.M307235200. [DOI] [PubMed] [Google Scholar]
- 105.Elkins T., Zinn K., McAllister L., Hoffmann F.M., Goodman C.S. Genetic analysis of a Drosophila neural cell adhesion molecule: Interaction of fasciclin I and Abelson tyrosine kinase mutations. Cell. 1990;60:565–575. doi: 10.1016/0092-8674(90)90660-7. [DOI] [PubMed] [Google Scholar]
- 106.Gaspar Y., Johnson K.L., McKenna J.A., Bacic A., Schultz C.J. The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol. Biol. 2001;47:161–176. doi: 10.1023/A:1010683432529. [DOI] [PubMed] [Google Scholar]
- 107.Kawamoto T., Noshiro M., Shen M., Nakamasu K., Hashimoto K., Kawashima-Ohya Y., Gotoh O., Kato Y. Structural and phylogenetic analyses of RGD-CAP/beta ig-h3, a fasciclin-like adhesion protein expressed in chick chondrocytes. Biochim. Biophys. Acta. 1998;1395:288–292. doi: 10.1016/S0167-4781(97)00172-3. [DOI] [PubMed] [Google Scholar]
- 108.Liu T.B., Chen G.Q., Min H., Lin F.C. MoFLP1, encoding a novel fungal fasciclin-like protein, is involved in conidiation and pathogenicity in Magnaporthe oryzae. J. Zhejiang Univ. Sci. B. 2009;10:434–444. doi: 10.1631/jzus.B0920017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Seifert G.J. Fascinating fasciclins: A surprisingly widespread family of proteins that mediate interactions between the cell exterior and the cell surface. Int. J. Mol. Sci. 2018;19:1628. doi: 10.3390/ijms19061628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li S.-B., Xie Z.-Z., Hu C.-G., Zhang J.-Z. A review of Auxin response factors (ARFs) in plants. Front. Plant Sci. 2016;7:47. doi: 10.3389/fpls.2016.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.De Vleesschauwer D., Xu J., Höfte M. Making sense of hormone-mediated defense networking: From rice to Arabidopsis. Front. Plant Sci. 2014;5:611. doi: 10.3389/fpls.2014.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fu J., Wang S. Insights into auxin signaling in plant-pathogen interactions. Front. Plant Sci. 2011;2:74. doi: 10.3389/fpls.2011.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ding X., Cao Y., Huang L., Zhao J., Xu C., Li X., Wang S. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell. 2008;20:228–240. doi: 10.1105/tpc.107.055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Domingo C., Andrés F., Tharreau D., Iglesias D.J., Talón M. Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant Microbe Interact. 2009;22:201–210. doi: 10.1094/MPMI-22-2-0201. [DOI] [PubMed] [Google Scholar]
- 115.Fu J., Liu H., Li Y., Yu H., Li X., Xiao J., Wang S. Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol. 2011;155:589–602. doi: 10.1104/pp.110.163774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gan P., Ikeda K., Irieda H., Narusaka M., O’Connell R.J., Narusaka Y., Takano Y., Kubo Y., Shirasu K. Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol. 2013;197:1236–1249. doi: 10.1111/nph.12085. [DOI] [PubMed] [Google Scholar]
- 117.Horbach R., Graf A., Weihmann F., Antelo L., Mathea S., Liermann J.C., Opatz T., Thines E., Aguirre J.S., Deising H.B. Sfp-Type 4′-Phosphopantetheinyl Transferase Is indispensable for fungal pathogenicity. Plant Cell. 2009;21:3379–3396. doi: 10.1105/tpc.108.064188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McDowell J.M. Genomic and transcriptomic insights into lifestyle transitions of a hemi-biotrophic fungal pathogen. New Phytol. 2013;197:1032–1034. doi: 10.1111/nph.12141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in Supplementary Material.