Abstract
The rugose colonial variant of Vibrio cholerae O1 El Tor produces an exopolysaccharide (EPSETr) that enables the organism to form a biofilm and to resist oxidative stress and the bactericidal action of chlorine. Transposon mutagenesis of the rugose variant led to the identification of vpsR, which codes for a homologue of the NtrC subclass of response regulators. Targeted disruption of vpsR in the rugose colony genetic background yielded a nonreverting smooth-colony morphotype that produced no detectable EPSETr and did not form an architecturally mature biofilm. Analysis of two genes, vpsA and vpsL, within the vps cluster of EPSETr biosynthesis genes revealed that their expression is induced above basal levels in the rugose variant, compared to the smooth colonial variant, and requires vpsR. These results show that VpsR functions as a positive regulator of vpsA and vpsL and thus acts to positively regulate EPSETr production and biofilm formation.
Vibrio cholerae O1, the causative agent of Asiatic cholera, has been isolated from coastal and estuarine environmental samples, both as free-living bacteria and in association with phytoplankton, zooplankton, crustacea, and mollusks (15–17). These distribution studies and its capacity to adaptively respond to changes in salinity, temperature, and the availability of nutrients (30, 31) have led to the idea that V. cholerae O1 can successfully occupy one or more ecological niches in a variety of aquatic habitats.
Laboratory microcosm studies conducted with V. cholerae O1 have shown that the duration of its survival in seawater is decreased if particulate matter is removed by filtration before inoculation of the filtrate with the organism. One interpretation of this result is that attachment to surfaces is important for long-term survival of the organism in marine environments. Growth on surfaces is believed to be advantageous because surfaces adsorb and thus concentrate scarce nutrients in the fluid phase. In addition, biotic surfaces, such as chitin, can be degraded by attached bacteria releasing assimilable sources of carbon and nitrogen (9). Thus, the surface mode-of-growth is likely to be preferred by V. cholerae in natural aquatic habitats.
Biofilms are a specialized and highly adapted form of surface growth characterized by assemblages of bacteria that form pillars or mushroom-like structures separated by fluid-filled channels. Pillars, in turn, consist of an extracellular polysaccharide (EPS) matrix and the bacteria that secrete it. Since EPS accounts for about 85% of biofilm depth, the production of EPS is critical for the development of a mature biofilm (4, 21). V. cholerae O1, biotype El Tor, in common with many aquatic bacterial species, forms a typical three-dimensional biofilm on a variety of abiotic surfaces (36, 39). Investigation of this phenotype showed that the rugose colonial variant forms a thicker and more differentiated biofilm than the smooth colonial variant (39). This capacity was found to be associated with production of a glucose- and galactose-rich EPS by the rugose form. Designated EPSETr, this compound was also shown to inactivate chlorine (39) and protect the organism from the bactericidal action of hydrogen peroxide (35). Open reading frames (ORFs) required for EPSETr synthesis are clustered in a 30.7-kb segment on the V. cholerae O1 chromosome. Because their putative protein products are homologous to capsular or EPS biosynthetic enzymes of other species, the corresponding Vibrio genes were designated vps, for Vibrio polysaccharide (39).
Phase transition occurs between the rugose and smooth colonial variants of V. cholerae O1 El Tor. Unlike the rugose form, the smooth-colony type of the strain A1552 does not produce EPS, forms only low-profile biofilms, and is rapidly killed by chlorine (39). Phenotype differences of this kind between the two colonial variants suggest that the rugose form may be better adapted for growth and survival in natural aquatic habitats and that transition frequencies between the two types may be governed by environmental signals.
Here we describe the identification, cloning, and characterization of vpsR, a gene that regulates the expression of the vps biosynthetic gene cluster. VpsR exhibits homology to the response element of two-component regulatory systems, which are involved in responding to environmental stimuli. Construction of a vpsR knockout mutation in a V. cholerae O1 El Tor rugose genetic background disclosed the role of this gene in colony morphology, EPS production, and biofilm formation. Studies of the smooth-colony type of a second strain of V. cholerae O1 El Tor (N16961) showed that the expression of vpsR and the genes it controls within the vps biosynthetic cluster exhibit interstrain differences that correspond to differences in biofilm forming capacity.
MATERIALS AND METHODS
Media and growth conditions.
All strains were maintained at −80°C in Luria-Bertani (LB) broth supplemented with glycerol (15%, vol/vol). For the experiments described below, the cells were grown aerobically at 30°C in LB broth, unless specified otherwise. The following antibiotics were added as appropriate: ampicillin (100 μg/ml) and chloramphenicol (5 μg/ml).
Bacterial strains.
Escherichia coli strains DH5α and S17-1 were used for standard DNA manipulations and mating, respectively. The V. cholerae strains used were smooth and rugose variants of 92A1552 (wild type, El Tor, Inaba, and Rifr) and vpsR and rpoN mutants of these strains, as well as strain N16961 (wild type, El Tor, and Inaba).
Scanning electron microscopy.
Pieces of agar containing a number of colonies were excised and processed sequentially as follows: 3% glutaraldehyde in 0.1 M sodium cacodylate for 60 min, 0.1 M sodium cacodylate for 5 min, 2% osmium tetroxide in 0.1 M sodium cacodylate for 45 min, and 0.1 M sodium cacodylate for 5 min. These samples were incubated with increasing concentrations of ethanol (15, 30, 50, 70, 90, and 100%) each for 15 min and then subjected to critical point drying, sputter coated, and analyzed by scanning electron microscopy (Philips XL40).
Transmission electron microscopy.
Bacterial cells were grown on a sterile dialysis membrane placed on the surface of LB plates, and the plates were incubated at 30°C for 24 h. Membrane-attached bacteria were processed sequentially as follows: 1% glutaraldehyde in 0.05 M sodium cacodylate containing 0.375% ruthenium red for 60 min, 0.05 M sodium cacodylate for 15 min, 2% osmium tetroxide in 0.2 M sodium cacodylate containing 1.5% ruthenium red for 60 min, and 0.2 M sodium cacodylate for 5 min. The cells were then incubated with ascending concentrations of ethanol (15, 30, 50, 70, 90, and 100%) each for 15 min and embedded according to standard procedures. The thin sections were stained with 1% uranyl acetate and then with lead citrate and then visualized using a Philips CM12 electron microscope.
CSLM.
Biofilms were formed by incubating 200 μl of a 1/100 dilution of an overnight LB broth culture grown on borosilicate cover chambers. The chambers were then incubated at 30°C for 24 h, rinsed with phosphate-buffered saline and analyzed by confocal scanning laser microscopy (CSLM) (MultiProbe 2010; Molecular Dynamics) using 488- and 510-nm excitation and emission wavelengths, respectively. The horizontal (xy) sections were obtained and the images were reconstructed using a maximum intensity algorithm.
Quantitative biofilm assay.
Biofilm formation was quantitatively monitored by incubating 200 μl of a 1/100 dilution of an overnight LB broth culture in microtiter plates (Falcon 3911). The microtiter plates were then incubated at 30°C for 24 h, and biofilm formation was quantified by crystal violet staining and ethanol solubilization as described elsewhere (27).
Transposon mutagenesis.
Tn5 mutagenesis of V. cholerae O1 El Tor, strain 92A1552, was performed by conjugation between the donor E. coli S-17λpir containing a pool of ∼40,000 signature-tagged Tn5Km2 DNAs on plasmids encoding resistance to ampicillin and the recipient, a rifampin-resistant V. cholerae O1 El Tor 92A1552 rugose variant. Conjugation was performed by mixing equal volumes of the donor and recipient, followed by filtration through a 0.45-μm-pore-size filter (Nalgene). The filters were then placed on LB agar plates, incubated for 5 h at 37°C, and suspended in LB, and the bacteria were recovered by vortexing. The exconjugants were selected by plating the suspension onto LB plates supplemented with 100 μg of rifampin and 150 μg of kanamycin per ml. A total of 15,000 exconjugants were screened visually to isolate mutants exhibiting smooth colonial morphology.
DNA manipulation and analysis.
Plasmid DNA and chromosomal DNA preparation, DNA ligation, bacterial transformation, agarose gel electrophoresis, PCR, and Southern blotting were carried out by the standard techniques described by Sambrook et al. (29). Restriction enzymes and DNA modification enzymes were purchased from New England Biolabs, Inc.
Cloning of the transposon insertion site.
Chromosomal DNA was isolated from the mutants using standard procedures; digested singly with EcoRI, SalI, KpnI, and PstI, the fragments resolved on an agarose gel; transferred to Hybond N membranes; and hybridized to the kanamycin-resistance gene of pUT mini-Tn5-Km2. Restriction enzymes that yielded hybridizing fragments of between 3 and 8 kb were used to digest chromosomal DNA, the resulting fragments were ligated into pBluescript KS that had been digested with the corresponding enzyme, and the plasmids were transformed into DH5α, followed by selection for kanamycin- and ampicillin-resistant transformants.
Construction of cosmid library.
A V. cholerae O1 El Tor 92A1552 gene bank was constructed by partially digesting wild-type genomic DNA with 0.016 U of Sau3A per μg for 5 min, which yielded mainly 25- to 40-kb fragments. These partially digested, dephosphorylated fragments were ligated into the vector SuperCos/Mob-II that had been digested with XbaI and BamHI. This vector had been constructed by introducing a 1,700-bp BamHI mobilization fragment (Mob) from GP704 into the BglII site of pSuperCos (Strategene). The ligation mixture was packaged into bacteriophage lambda particles using the Gigapack III gold packaging extract (Stratagene). The titer of the library was determined, and the library was amplified using E. coli XL1-Blue MR as a host cell and stored at −80°C in 25% glycerol.
PCR amplification of vpsR.
A 2,890-bp fragment containing vpsR was amplified using the following primers: VpsR2-F (GTTCTATGATGCCGACTACA) and VpsR2-R (ACGCTTCTCACGCTACTTT). The amplified fragment includes 431 nucleotides of upstream sequence, the entire vpsR coding sequence, and 1,130 nucleotides of sequence downstream of vpsR. BamHI and SalI restriction sites were added to forward and reverse primers, respectively, to facilitate cloning into pACYC184. The PCR products were generated using a high-fidelity PCR kit (Clontech), digested with BamHI and SalI, and the resulting fragment was ligated into pACYC184 that had been digested with the corresponding enzymes to generate p1. The ligation mix was then transformed into DH5α, and the transformants were selected on LB plates supplemented with chloramphenicol. The plasmids were introduced into V. cholerae mutant strains by electroporation. The ability of the plasmid to complement the mutations was initially tested by a change in the colony morphology from smooth to rugose and then tested by expression analysis.
Generation of null mutants.
Mutants with insertions into the vpsR and rpoN genes were regenerated in a new rugose strain background. For rpoN, primers rpoN1-F (CCCTTTAGCGATGTGGAT) and rpoN1-R (CAGCCATTTTGCCTCTTG) were used to amplify an internal 558-nucleotide region of rpoN corresponding to amino acids 144 to 339. For vpsR, primers vpsR1-F (GGGGAATCTATGCCTATGAAG) and vpsR1-R (CGTCTCCACAGTCCCTTCTTG) were utilized to generate a 351-nucleotide internal fragment corresponding to amino acids 148 to 264. XbaI and EcoRI restriction sites were added to forward and reverse primers, respectively, to facilitate the cloning into pGP704. The plasmids were introduced to V. cholerae O1 El Tor wild-type rugose and smooth colonial variants by biparental mating, and exconjugants were selected on LB plates supplemented with 100 μg of ampicillin and rifampin per ml. Insertion into the correct site was confirmed by Southern and PCR analysis.
RNA preparation.
Overnight cultures of V. cholerae in LB medium at 37°C were diluted 1:100 in fresh LB medium. At the mid-log-phase growth point, 10-ml aliquots of culture were collected. Cell pellets were frozen with liquid nitrogen and stored at −80°C. RNA was isolated from frozen pellets using Trizol reagent (Gibco-BRL). For biofilm RNA isolation, overnight LB culture was diluted 1:100, and 25 ml of this culture was incubated in petri plates at 30°C for 24 h. The plates were rinsed with LB three times, the attached bacteria were resuspended in 2 ml of Trizol reagent, and the total RNA was isolated according to the manufacturer's instructions. To remove contaminating DNA, 50 μg of total RNA was incubated with 4 U of RNase-free DNase I (Ambion) for 30 min at 37°C. The RNeasy Mini Kit (Qiagen) was used to clean up RNA after DNase digestion, and the RNA was stored at −80°C.
cDNA synthesis.
Reverse transcription (RT) of RNA was performed with TaqMan RT reagents (Perkin-Elmer) and nuclease-free water (Ambion). Five hundred nanograms of total RNA was used for RT in a final volume of 50 μl containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 5.5 mM MgCl2, 500 μM concentrations of each deoxynucleotide triphosphate (dNTP), 2.5 μM concentrations of random hexamers, 20 U of RNase inhibitor, and 62.5 U of Multiscribe Reverse Transcriptase. The RT-negative control reactions (without RT) were performed to determine how much contaminating genomic DNA was present in each total RNA sample. The reactions were incubated in GeneAmp PCR System 9700 (Perkin-Elmer) by using parameters of 25°C for 10 min, 48°C for 30 min, 95°C for 5 min, and cooling at 4°C. The cDNA samples were stored at −20°C.
Primers and probes for real-time RT-PCR.
Primers and probes for the three genes of interest, vpsR, vpsA, and vpsL, and for rRNA were chosen using Primer Express software (Perkin-Elmer). Fluorogenic probes were used to monitor PCR product formation continuously during PCR. The probe is an oligonucleotide, dually labeled with a reporter dye (FAM; 6-carboxyfluorescein) covalently attached at the 5′ end and a quencher dye (TAMRA; 6-carboxytetramethylrhodamine) covalently attached at the 3′ end. Primers and probes were synthesized and purified by Biosearch Technologies. The primers and probes used for real-time PCR were as follows: vpsR-F (21-mer), GTCTCAGCTCGATCTTCCCAA; vpsR-R (20-mer), CGTTCCCGAATGCTTTTCAG; vpsA-F (19-mer), TTCCCCTTGGCCTGAAGAG; vpsA-R (21-mer) AGGTGCAAAGTGGTACTGCGT; vpsL-F (21-mer), ATCGCACCATAGTGAATCGCT; vpsL-R (21-mer), TCTGTGCCCATCCAGTAATGC; rDNA-F (ribosomal DNA [rDNA]; 18-mer), GAGCGGCAGCACAGAGGA; rDNA-R (21-mer), TTTCCCAGGCATTACTCACCC; vpsR TaqMan probe (20-mer), 5′FAM-CTGCGACGGCCATCACTGCG-TAMRAp3′; vpsA TaqMan probe (20-mer), 5′FAM-CCGCAAACTCACGGCCGCAC-TAMRAp3′; vpsL TaqMan probe (25-mer), 5′FAM-CATGCTGCGTCACAAAGTGAAGCCC-TAMRAp3′; and rDNA TaqMan probe (22-mer), 5′FAM-CGCTCGCCACCCAAGGAACAAG-TAMRAp3′. The amplicon lengths generated using these primers were 67, 69, 63, and 67 bp for vpsA, vpsL, vpsR, and rDNA, respectively.
PCR amplification conditions for RT-PCR analysis.
PCR conditions were identical for all reactions. For each PCR run, a master mix was prepared on ice with the following components: 1× TaqMan Universal Master Mix containing AmpliTaq Gold DNA polymerase, Amp Erase UNG, dNTPs with UTP, passive reference dye, and optimized buffer components (Perkin-Elmer). To this were added 200 nM of the fluorogenic probe and 900 nM of the forward and reverse primers. Then, 5 μl of each appropriately diluted (250 pg, unless specified) RT sample was added to 20 μl of the PCR master mix. Experiments were performed with three replicas for each datum point, and negative-RT and no-template controls were run for each reaction. Reactions were performed in sealed MicroAmp Optical 96-well reaction plates (Perkin-Elmer) using an ABI Prism 7700 Sequence Detection System (Perkin-Elmer) for PCR amplification and the following cycle parameters: 2 min for 50°C and 10 min for 95°C, followed by 40 cycles of 15 s for 95°C and 1 min for 60°C.
Quantitative analysis of RT-PCR data.
Real-time PCR measures the degradation of a fluorescent oligonucleotide probe in real time concurrent with PCR amplification. The method uses a dually labeled probe with a reporter dye FAM at the 5′ end and the quencher dye TAMRA at the 3′ end. During PCR the 5′ nucleolytic activity of Taq polymerase separates the 5′ reporter dye from the 3′ quencher dye, resulting in an increase in the fluorescence of the reporter dye, which can be detected by the laser detector. In real-time PCR analysis, quantitation is based on cycle threshold (Ct) calculations (10, 12). Ct is defined as the number of cycles required for reporter dye fluorescence, resulting from the synthesis of PCR products, to become significantly higher than background fluorescence. There is an inverse exponential relationship between the logarithm of initial target copy number in the reaction and the corresponding Ct determinations by the model 7700 instrument. Validation experiments were performed for each PCR primer-probe set, and these yielded a linear relationship when Ct was plotted against the logarithm of a varying amount of cDNA reverse-transcribed from total RNA in the range of 10 pg to 1 ng. Twofold serial dilutions of cesium chloride-purified chromosomal DNA of V. cholerae, corresponding to 101 to 106 theoretical copies (the molecular weight of V. cholerae genomic DNA was estimated to be 4.1 Mbp), were used in real-time PCR. For each set of TaqMan amplicons, the standard curve was constructed by using the equation y = a + b log x, where x is the starting DNA copy number, y is the Ct obtained by amplifying x copies, a is the y intercept of the standard curve line, and b is the slope of the standard curve line. The results of this calculation were used to estimate the number of cDNA copies present in the different samples.
RESULTS
Isolation of the vpsR regulatory mutant and cloning of the corresponding gene.
To identify genes required for the rugose colony type, EPS production, and biofilm formation, the rugose colonial variant of V. cholerae O1 El Tor was subjected to transposon mutagenesis; transconjugants that exhibited smooth colonial morphology were selected for subsequent analysis. Chromosomal genes interrupted by transposon insertion were isolated by marker rescue, and DNA sequences flanking these insertion sites were determined. The majority of genes identified by this method were located within the aforementioned 30.7-kb segment of the chromosome that contains the vps biosynthetic cluster. However, sequence analysis of other tagged genes also disclosed two independent insertions in another locus situated elsewhere on the chromosome; analysis of these sequences revealed an ORF predicted to encode a protein homologous to the NtrC subclass of response regulators. This ORF was denoted VpsR. A sequence tag corresponding to this locus was used as a probe to isolate a complete copy of vpsR from a cosmid library of the smooth colonial variant. Two such clones were obtained, returned to the mutant, and found to cause conversion from the smooth colonial morphology of the mutant to the rugose colonial morphology of the original wild-type parent.
DNA sequence analysis of the vpsR region.
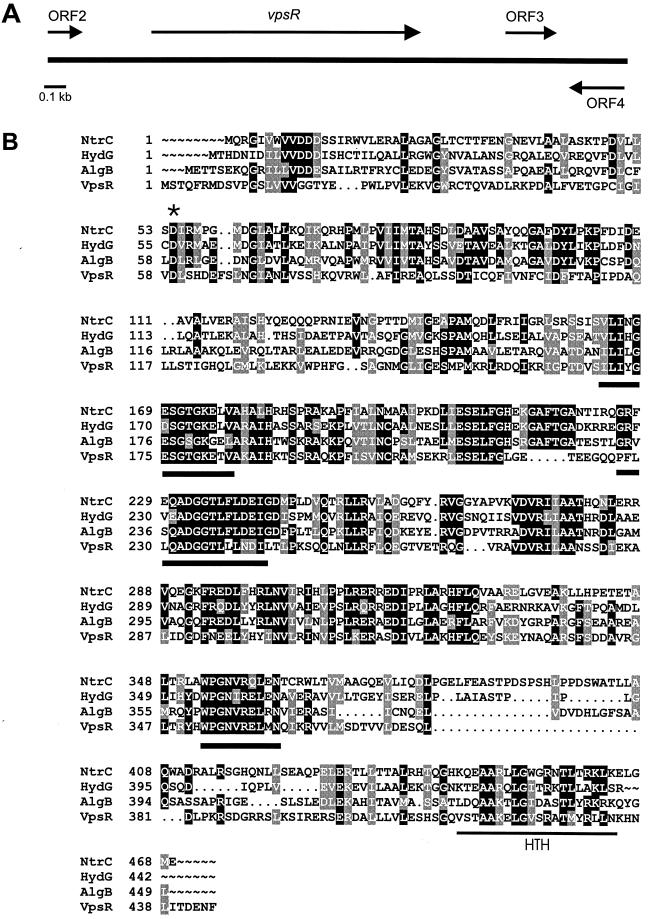
Sequence analysis of the vpsR region was undertaken with specifically designed oligonucleotides using pCos-vpsR as a template. A 2.897-kb region encompassing the insertion site and the flanking regions was sequenced and revealed an apparently full-length vpsR spanning 1,335 nucleotides and located between nucleotides 431 and 1765 of the cloned segment (Fig. 1A). An incomplete ORF was identified 274 bp upstream of the vpsR start codon, in the same orientation, that would specify the carboxy-terminal 51 amino acids of a protein with 87.7% identity and 91.8% similarity to lysS, which encodes an E. coli lysyl-tRNA synthetase (23). Downstream of vpsR and in the same orientation, a third ORF was found between nucleotides 2343 and 2594 of the cloned sequence; it is predicted to encode an 83-amino-acid protein without identifiable homologous sequences in the protein database. Directly downstream of the third ORF, in the opposite orientation, an incomplete ORF was identified; it is predicted to encode the carboxy-terminal 87 amino acids of a protein that exhibits 47.6% identity and 58.1% similarity to the E. coli TAS (tyrosine auxotrophy suppressor) protein (34).
FIG. 1.
VpsR homologues and functional motifs. (A) Map of the vpsR region on the V. cholerae O1 El Tor chromosome. (B) Amino acid sequence alignment of VpsR from V. cholerae O1, NtrC from serovar Typhimurium (P41789), HydG from serovar Typhimurium (P25852), and AlgB from P. aeruginosa (P23747). Amino acids identical in all four sequences are shaded in black. The star denotes a conserved aspartate residue and a putative phosphorylation site. Predicted RpoN interaction sites are indicated by heavy lines, and a C-terminally located helix-turn-helix (HTH) motif is shown by the thin line.
vpsR is predicted to encode a 444-amino-acid, 49.76-kDa protein homologous to HydG (51% similarity, 35.7% identity) from Salmonella enterica serovar Typhimurium (3), AlgB (43.2% similarity, 34.3% identity) from Pseudomonas aeruginosa (11, 37), and NtrC (55.8% similarity, 44.1% identity) from serovar Typhimurium (8) (Fig. 1B). Each of these proteins belongs to the response regulator component of two-component signal transduction systems and possesses an N-terminal domain that can be phosphorylated by a sensor kinase, the other component of this signal transduction system (33). The conserved aspartate residue of VpsR (Asp-59) that is postulated to be phosphorylated is indicated by a star (Fig. 1B). Two other conserved residues, Asp and Lys, typically present within the N-terminal domain of response regulators, are not present in the corresponding region of VpsR.
The central region of VpsR harbors the following consensus sequences shown or postulated to be important for interaction with the alternative sigma-54 factor (Fig. 1B): 1, (L,I,V,M,F,Y) 3xG(D,E,Q)(S,T,E)G(S,T,A,V)GKx2(L,I,V,M,F,Y); and 2, (G,S) x(L,I,V,M,F)x2A(D,N,E,Q,A,S,H)(G,N,E,K)G(S,T,I,M)(L,I,V, M,F,Y)3(D,E)(E,K)(L,I,V,M) (7). These are ATP binding motifs and are located in the N-terminal segment of the central domain. ATP binding promotes open complex formation by a sigma-54 containing RNA polymerase (22, 26). A segment of the vpsR sequence, which would code for amino acids 214 to 356, had been previously identified by Klose et al. as a sigma-54-dependent transcriptional activator (s54act5) through their use of degenerate primers designed to amplify genes coding for this class of transcriptional activators. However, these investigators did not determine the function of the s54act5 gene product (20).
Like other response regulators, VpsR harbors a helix-turn-helix DNA-binding domain (28), located between amino acids 415 and 434 near the carboxy terminus of the protein (Fig. 1B).
Generation and analysis of vpsR knockout mutants.
To test if vpsR is required for rugose colonial morphology, EPS production, and biofilm formation, it was independently inactivated in the rugose genetic background by using pGP704 harboring a 351-nucleotide internal fragment, which corresponds to the coding sequence for amino acids 148 to 264. Knockouts carrying this mutation, designated vpsR::pGP704, were examined for colonial morphology, EPS production and localization, and biofilm formation.
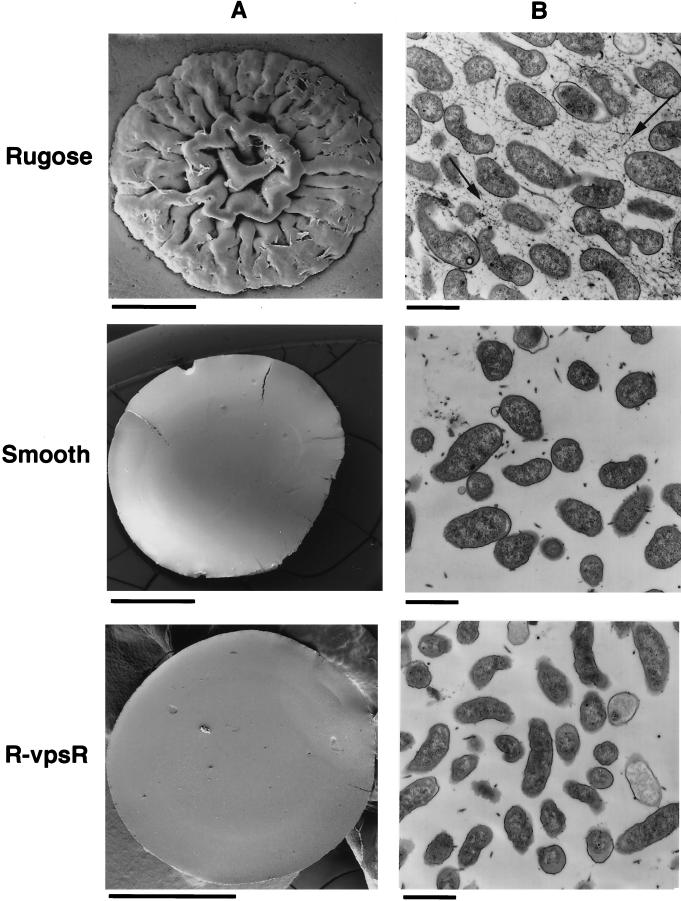
The capacity of V. cholerae O1 to naturally oscillate between two distinctive colony morphotypes—the rugose and smooth colonial variants—led to the identification of the rugose-associated phenotypes described above (39). If vpsR is required for the rugose colonial morphotype, then disruption of this gene in the rugose background should yield phenotypically “smooth” colonies that do not revert to the rugose colonial variant. The nonreverting nature of the mutation was tested by cultivating vpsR::pGP704 under conditions of nutrient limitation that favor reversion of the wild-type smooth variant to the rugose variant (35, 39). This growth condition did not yield any rugose colonies of vpsR::pGP704, indicating that the smooth colonial phenotype of this mutant is stable. To determine if disruption of vpsR in the rugose genetic background resulted in the same colonial morphology as the wild-type smooth variant, colonies formed by vpsR::pGP704 and by the original smooth and rugose colonial variants of the wild-type parent strain were compared by scanning electron microscopy. Figure 2A depicts such a comparison. The colonial morphology of vpsR::pGP704 (denoted R-vpsR in this figure) is dramatically different from the corrugated colonial morphology of the wild-type rugose parent, even though this mutation was made in the genetic background of the wild-type rugose variant. Instead, colonies formed by vspR::pGP704 resemble the featureless, flat colonies of the wild-type smooth variant.
FIG. 2.
Colonial morphology and EPS production by the vpsR mutant. (A) The appearance of agar grown colonies was determined by scanning electron microscopy for the wild-type rugose and smooth variants of V. cholerae O1 El Tor and for the vpsR mutant of the rugose variant (designated R-vpsR). Disruption of vpsR is associated with a change from the rugose to the smooth-colony type. Bars, 1 mm. (B) EPS production by bacteria growing on a cellulose membrane atop nutrient agar was determined by transmission electron microscopic examination of thin sections stained with ruthenium red. A stained matrix, evident between rugose-type bacteria (arrow), is not produced by the vpsR mutant of the rugose variant (designated R-vpsR) or by the wild-type smooth variant. Bars, 1 μm.
One explanation for differences between the colony types depicted in Fig. 2A might be qualitative or quantitative differences in the EPSETr produced by each. To compare the abundance, distribution, and tinctorial qualities of EPS produced by vpsR::pGP704 and the wild-type rugose and smooth variants of the same strain, each bacterial type was grown on a cellophane dialysis membrane placed on the surface of LB agar plates, and the resulting bacterial lawn was subjected to thin-section transmission electron microscopy of ruthenium red-stained samples, a method that preferentially stains acidic polysaccharides. This method led to the comparative features depicted in Fig. 2B. The wild-type rugose variant produces a ruthenium red-positive matrix between well-separated bacteria. In contrast, the wild-type smooth variant and the vpsR::pGP704 mutant were identical; neither produces the ruthenium red-staining interbacterial matrix typical of the rugose variant. EPSETr production by vpsR::pGP704 was also tested by enzyme-linked immunosorbent assay using an EPSETr-specific antiserum, and the result showed that vpsR::pGP704 was EPSETr antigen negative (data not shown). Taken together, these results provide compelling evidence that vpsR is required for EPSETr production.
Biofilm formation by vpsR::pGP704.
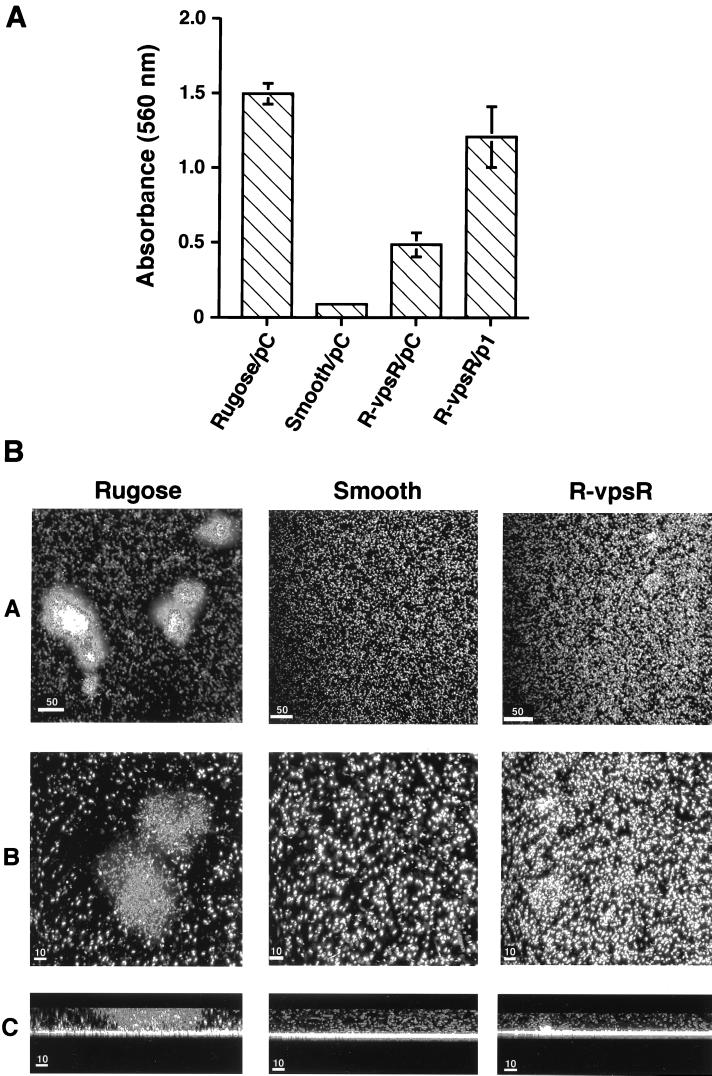
The capacity of the rugose colonial variant to form high-profile biofilms is the rugose-associated property most likely to be important for the environmental persistence of this species in natural aquatic habitats. To determine if biofilm formation by the rugose variant is affected by the vpsR::pGP704 mutation of the rugose parent strain, two assays were performed that monitor different aspects of the biofilm-forming phenotype. Quantitative differences in biofilm-forming capacity were measured by monitoring the intensity of crystal violet staining of a biofilm that formed on the surface of a polyvinyl chloride microtiter well during 24 h of growth in LB medium (27). This assay was used to examine the wild-type rugose and smooth colonial variant, the vpsR::pGP704 mutant, and this mutant complemented with pACYC184 containing a cloned copy of vpsR. To control for possible plasmid effects, each of the other tested strains also contained pACYC184 without the vpsR insert. The biofilm-forming capacity of the wild-type rugose variant was found to be 17-fold greater than the wild-type smooth variant (Fig. 3A). In contrast, the biofilm-forming capacity of the noncomplemented vpsR::pGP704 mutant of the rugose variant was intermediate between the wild-type rugose and smooth variants, being 3-fold less than that of the rugose variant but 5.5-fold greater than that of the smooth variant (Fig. 3A). Furthermore, a time course of biofilm development by the vpsR::pGP704 mutant also exhibited an intermediate biofilm formation phenotype, where the biofilm-forming capability of the vpsR mutant is less than that of the wild-type rugose and more than that of the wild-type smooth strain (data not shown).
FIG. 3.
Biofilm-forming phenotype of the vpsR mutant. (A) Quantitative comparison of biofilm-forming capacity by the wild-type rugose and smooth colonial variants of V. cholerae O1 El Tor and by the vpsR mutant of the rugose variant. Biofilms were quantified for the following four variants of the same strain: the wild-type rugose and smooth colonial morphotypes, each carrying the control plasmid pACYA184 (pC), and the vpsR mutant of the rugose morphotype (R-vpsR) harboring either the control plasmid (pC) or a complementing plasmid (p1), which contains a wild-type copy of vpsR on pACYC184. Biofilm quantification was carried out by growing these variants overnight in LB broth at 30°C in polyvinyl chloride microtiter plates. The wells were washed, the attached bacteria were stained with crystal violet, the stained film was solubilized in 95% ethanol, and the intensity of crystal violet staining was then monitored by absorbance spectroscopy at 595 nm. Disruption of vpsR in the rugose variant, denoted R-vpsR, caused a threefold reduction in biofilm-forming capacity compared to the wild-type rugose parent strain. Complementation of R-vpsR with p1 restored most of the wild-type biofilm phenotype of the rugose variant. The biofilm-forming capacity of R-vpsR was significantly greater than that of the wild-type smooth variant, indicating that biofilm-forming genes, which do not require vpsR, are expressed by the rugose variant. (B) Topographical features of biofilm development by the rugose and smooth wild-type colonial morphotypes and by the vpsR mutant of the rugose variant (denoted R-vpsR). Each of the three tested strains, carrying a plasmid that constitutively expresses the green fluorescent protein, was incubated for 24 h in borosilicate glass-bottom chambers containing LB broth. The chambers were then emptied, and the glass surface was examined with a scanning confocal microscope using 488- and 510-nm excitation and emission wavelengths, respectively. Horizontal (xy) projected images of each of the strains are shown at low and high magnification in rows A and B, respectively. Biofilms formed by the rugose variant contain large circumscribed aggregates of adherent bacteria. In contrast, only individual bacteria of the smooth variant interact with the glass surface. The adherence pattern of R-vpsR is intermediate between the rugose and smooth patterns, showing both small aggregates and individual glass-bound bacteria. Image reconstruction led to the sagittal (xz) view of the same biofilms shown in row C. The high-profile biofilm (>10 μm) produced by the rugose variant contains a mushroom-shaped pillar that is not present in the low-profile biofilms (<10 μm) produced by the wild-type smooth variant or the R-vpsR mutant of the rugose variant. Bars: 50 μm, row A; 10 μm, rows B and C.
Complementation of this mutant with an intact copy of vpsR increased its biofilm-forming capacity to nearly the same level as the wild-type rugose parent strain (Fig. 3A). These results demonstrated that vpsR is required for some but not all of the biofilm-forming capacity of the rugose variant. However, to determine if differences in the biofilm-forming capacity of the vpsR::pGP704 and the wild-type strains were due to differences in the growth rate, both were cultivated in liquid medium. No differences in the growth rate or final cell density were observed (data not shown). Thus, the decreased capacity of vpsR::pGP704 strain to form a biofilm is not due to an altered growth rate.
The same strains were studied by a second assay that provided comparative information about the topographic and architectural features of the biofilms produced by these variants on a glass surface. To this end, a plasmid carrying a gene that constitutively expresses the green fluorescent protein (GFP) was introduced into each of these variants. The biofilms they formed after 24 h of incubation at 30°C were compared using scanning confocal laser microscopy to obtain horizontal (xy) and sagittal (xz) images (Fig. 3B). Under these conditions, the rugose variant produces a well-developed biofilm containing multicellular, 75-μm high pillars and fluid-filled columns. Horizontal projections of the rugose strain showed prominent islands of bacterial aggregates. In contrast, the smooth variant produces a low-profile, 6-μm biofilm lacking pillars, columns, or islands of bacterial aggregates. The vpsR::pGP704 mutant of the rugose variant also formed a low-profile 7- to 10-μm biofilm, similar in depth to the biofilm formed by the wild-type smooth colonial variant. However, in contrast to the smooth variant, the vpsR::pGP704 biofilm contained more bacteria per unit area, and in some areas the bacteria coalesced into poorly developed or temporally delayed islands (Fig. 3B). Taken together, these results show that vpsR is required for normal biofilm development by the rugose colonial variant, but they also point to the probable existence of genes that are independent of vpsR which contribute to the complete biofilm phenotype by this colonial variant.
Transcriptional regulation of vps biosynthesis genes vpsA and vpsL by vpsR.
Sequence analysis of the 30.7-kb region that contains vps biosynthesis genes showed that these genes are organized into two clusters of 11.4 and 6.6 kb harboring 11 and 6 vps genes, respectively (unpublished data). Because VpsR was found to be homologous to several previously described response regulators (Fig. 1B), we reasoned that it might control the expression of vps biosynthesis genes within one or both of these clusters. To investigate this possibility, the first gene of each cluster was selected for gene expression analysis, since each had been shown to be required for EPSETr production by transposon insertion mutagenesis (39). These genes are described below.
vpsA, the first gene in the first vps region, is predicted to encode a 356-amino-acid, 39.2-kDa protein with 73.8% similarity and 66.0% identity to E. coli WecB, which is required for bacteriophage N4 adsorption and encodes UDP–N-acetylglucosamine 2-epimerase function (19). It is also highly homologous (71.2% similarity and 64.6% identity) to EpsC of Pseudomonas solanacearum, which is required for the production of the exopolysaccharide 1 (EPS1) virulence factor (14). vpsL, the first gene in the second vps cluster, is predicted to encode a 464-amino-acid, 52.9-kDa protein that is homologous to members of a large enzyme family that catalyze the transfer of glucosyl-1-phosphate from UDP glucose to polyprenolphosphate—the first step in the biosynthesis of lipid intermediates for polysaccharide synthesis. Homologous family members of VpsL include: WcaJ of E. coli (32) (56.0% similarity and 45.6% identity), AmsG (2) of Erwinia amylovora (39.7 and 30.5%), RfbP (18) of serovar Typhimurium (39.5 and 28.5%), and ExoY (25) of Rhizobium meliloti (48.5 and 38.8%).
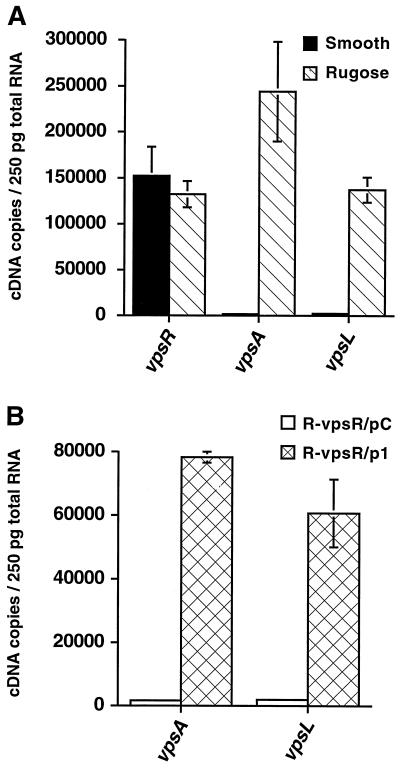
To estimate the abundance of vpsA, vpsL, and vpsR mRNA in smooth and rugose colonial variants, total RNA was isolated from exponentially growing smooth and rugose cultures and reverse transcribed using random hexamers, and the resulting cDNAs were used for real-time PCR using the corresponding TaqMan PCR primer-probe set. A standard curve for each of the genes was generated using different amounts of chromosomal DNA and the respective TaqMan PCR primer-probe set; this standard curve was then used to measure message abundance by reference to the Ct value for each gene. Figure 4A, which depicts the calculated message abundance for the each of the three tested genes in exponentially growing rugose and smooth variants, shows low levels of vpsA mRNA (1.5 × 103 cDNA copies/250 pg of RNA) and vpsL mRNA (2.3 × 103 cDNA copies/250 pg of RNA) in the smooth colonial variant. Even though these levels were low, samples subjected to RT contained higher amounts of template molecules than the untreated samples. Thus, vpsA and vpsL are transcribed during exponential growth of the smooth variant, although at low “basal levels.” In comparison, the message abundances of vpsA and vpsL in the rugose variant were dramatically greater, 2.3 × 105 and 1.3 × 105 copies/250 pg of RNA, respectively. Therefore, high levels of vpsA and vpsL mRNA are correlated with the rugose colonial morphotype and accordingly with EPSETr production and biofilm formation. In contrast, the abundance of the vpsR message was similar in both colonial variants (Fig. 4A).
FIG. 4.
Transcriptional activity of vpsR and of the EPSETr biosynthetic gene cluster. (A) vpsR, vpsA, and vpsL mRNA abundance in wild-type smooth and rugose colonial variants. Standard curves for the vpsR, vpsA, and vpsL genes were generated by plotting the Ct value against the input DNA concentration, as described in Materials and Methods. Message abundance in exponentially growing wild-type smooth and rugose strains was calculated using the corresponding equations from the standard curves and the Ct values obtained after RT of total RNA, followed by real-time PCR amplification. vpsA and vpsL of the EPSETr biosynthetic gene cluster are strongly expressed in the rugose variant, but they are also expressed at low, basal levels in the smooth variant. Smooth and rugose variants expressed vpsR at equivalent levels. (B) Complementation of the vpsR mutant restores vpsA and vpsL expression. The vpsR mutant of the rugose colonial variant (denoted R-vpsR) was complemented with either a control plasmid (pC) or the control plasmid containing a wild-type copy of vpsR (p1). vpsA and vpsL mRNA abundances were determined during exponential growth for the complemented (R-vpsR/p1) and noncomplemented (R-vpsR/pC) mutant. Message abundance is expressed as the number of cDNA copies detected in 250 pg of total RNA.
These results show that changes in vpsR expression, at the transcriptional level, are not involved in the regulated expression of vpsA and vpsL. However, this finding is still compatible with the possibility that vpsR is required for the expression of these genes because this class of response regulators is activated by a posttranslational event, namely, phosphorylation of a critical Asp residue by the sensor kinase. Therefore, to further test if vpsR is required for the transcriptional regulation of the EPSETr biosynthetic gene cluster, the abundance of vpsA mRNA and vpsL mRNA was determined for the vpsR mutant of the rugose colonial variant. As shown in Fig. 4B, the expression of vpsA and vpsL is dramatically lower in the vpsR regulatory mutant. However, expression levels were partially restored upon complementation. The partial complementation could be due to the fact that the expression of the normal chromosomal copy of vpsR might differ from the expression of an episomal copy of the same gene. These results indicate that vpsR likely functions as a positive transcriptional regulator of vpsA and vpsL and therefore acts to positively regulate EPSETr production and biofilm formation as well.
Comparison of vps gene expression by V. cholerae O1 El Tor strains A1552 and N16961 during biofilm formation.
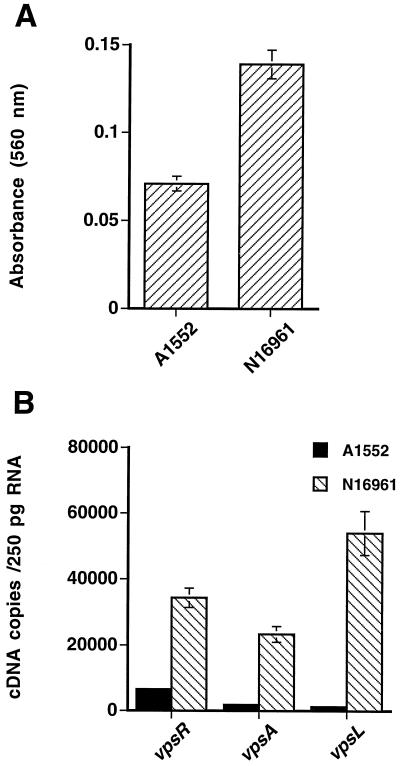
Transposon mutagenesis of V. cholerae O1 N16961 by Watnick and Kolter led to the identification of three classes of genes (pili, flagella, and some of the EPSETr biosynthesis genes previously identified by Yildiz and Schoolnik [39] that contribute to the biofilm phenotype [36]). However, Watnick and colleagues used only the smooth colonial variant in their studies, yet their mutational strategy and biofilm-forming screen were able to identify genes in the EPS biosynthetic cluster. In contrast, the experiments we report here, which were conducted with strain A1552 in a liquid medium, show that vpsR-regulated genes of the EPS biosynthetic cluster are highly expressed in the rugose colonial variant (Fig. 4A). Three possible reasons might account for this apparent discrepancy: first, the expression levels of the vps genes may exhibit strain-specific differences under the same condition of growth; second, the biofilm environment, including contact with the glass or plastic surface, might induce transcription of the vps biosynthetic gene cluster, even in the smooth variant, when compared to growth in a liquid medium; and third, the biofilm microenvironment might induce phase transition from the smooth to the rugose morphotype. To address these issues, biofilms formed by the smooth colonial variants of the two strains after 24 h of incubation at 30°C were quantitatively compared using the crystal violet-based method described for Fig. 3. The results (Fig. 5A) indicate that N16961 forms an approximately twofold-greater biofilm than A1552. To determine if differences between the biofilm phenotype of the two strains are correlated with differences in the expression of vps biosynthesis genes, the abundance of vpsR, vpsA, and vpsL mRNA in biofilms formed in polystyrene petri plates after 24 h of incubation at 30°C was quantified for each of the two strains using real-time PCR. As shown in Fig. 5B, the amounts of vpsR, vpsA, and vpsL message were 5.2-, 12.6-, and 44.7-fold higher, respectively, in N16961 biofilms compared to A1552 biofilms. The differences in expression data are therefore consistent with the differences in biofilm magnitude for the smooth variants of the two strains.
FIG. 5.
Comparison of V. cholerae O1 El Tor strains A1552 and N16961 for biofilm phenotype and vps gene expression. (A) Biofilm-forming phenotypes of the smooth strains A1552 and N16961. Biofilm formation was analyzed by growing the strains in LB broth at 30°C for 24 h in polyvinyl chloride microtiter plates, and then the biofilms were quantified by crystal violet staining. The biofilm-forming capacity of the N16961 strain after 24 h of growth in polyvinyl chloride microtiter plates is twofold higher than that of the A1552 strain. (B) Expression analysis of vps genes in the strains A1552 and N16961. The abundance of vpsR, vpsA, and vpsL messages was quantified by copy number determination using real-time PCR. All three genes were expressed at a higher level in N16961 than in A1552.
To determine if the differences could be due to a possible effect of the biofilm microenvironment that might induce transition from the smooth to the rugose morphotype and thereby be associated with increased expression of the vps genes, cells from the same biofilms were removed, serially diluted, and plated onto LB agar plates. We recovered 108 to 109 cells per biofilm and observed that neither of the strains exhibited conversion from the smooth to the rugose morphotype under the conditions tested. Thus, the difference between the biofilm phenotypes of V. cholerae O1 El Tor A1552 and N16961 depicted in Fig. 5A is most likely due in part to strain-specific variation in the expression of vps genes.
DISCUSSION
We have identified vpsR, a gene encoding a positive transcriptional regulator of EPSETr biosynthetic gene expression and the biofilm-forming phenotype by the rugose colonial variant of V. cholerae O1 El Tor. When mutation of vpsR was undertaken in a rugose background, the colony formed by the resulting mutant resembled the smooth colonial variant. However, the quantitative biofilm assay results depicted in Fig. 3 showed that this mutant exhibits a biofilm-forming capacity that is intermediate between those of the wild-type rugose and smooth variants. EPS biosynthesis mutants of the rugose colonial variants, including vpsA and vpsL mutants, also exhibit an intermediate biofilm phenotype (unpublished data). In view of these results, it seems most likely that the rugose variant selectively expresses not only genes in the vps biosynthetic cluster but also other genes as well. These other genes are not under the control of vpsR, but they nonetheless contribute to the biofilm-forming phenotype of the rugose variant. Thus, the fully manifested three-dimensional biofilm of the rugose variant is likely to be the product of several separate effector genes and their cognate regulators, including vpsR.
VpsR was found to be highly homologous to several response regulators which, in well-characterized two-component signal transduction systems, have been shown to act in conjunction with a cognate sensor kinase protein. Activation of the response regulator typically occurs as a result of sensor kinase-mediated phosphorylation of a conserved Asp residue within the N-terminal segment of the response regulator component. VpsR lacks two conserved residues, Asp and Lys, which are critical for phosphorylation and are present at the N-terminal domain. AlgB and AlgR response regulators, which control alginate production by activating the transcription of algD, do not need to be phosphorylated for their roles in alginate production (24). Therefore, it will be of interest to determine if VpsR requires phosphorylation for its function and if the mode of phosphorylation differs from the typical members of the response regulators.
In well-characterized two-component signal transduction systems, response regulators have been shown to act in conjunction with a cognate sensor kinase protein. Because the response regulator and the sensor kinase are in close proximity to one another in many two-component regulatory systems, we examined ca. 2 kb of the sequence flanking vpsR. However, we failed to locate a gene that might encode the cognate sensor component of VpsR within this sequenced region, nor was a candidate cognate sensor kinase identified among the transposon mutants characterized thus far. Analysis of the annotated genome sequence of V. cholerae O1 may lead to the identification of candidate genes for this sensor kinase (13). VpsR also contains a helix-turn-helix, DNA-binding motif near the C terminus. Thus, it is possible that VpsR directly activates the transcription of vps biosynthetic genes by binding upstream promoter regions within this cluster. Alternatively, VpsR may mediate its effect on vps gene transcription indirectly, by interacting with another positive regulator of vps expression. The results described here are compatible with either possibility.
VpsR is highly homologous with NtrC of serovar Typhimurium (Fig. 1B), and most such homologues act in concert with the alternative sigma factor RpoN. However, the expression of vpsA and vpsL were not altered in an rpoN-null mutant generated in the rugose variant (data not shown). Further, analysis of the upstream regions of vpsA and vpsL did not reveal the TGGCACn4TTTGCA sequences recognized by sigma-54 (7). Thus, like AlgB of P. aeruginosa, a response regulator that harbors a RpoN-binding domain (11, 37), VpsR does not require RpoN to activate vps gene transcription.
Complementation studies reported in this study were performed with the vpsR gene cloned from the smooth form of V. cholerae O1 El Tor. At present the mechanism of smooth-to-rugose transition and vice versa is not known. However, our observations suggest that vpsR is not the gene functioning as the switch. Instead, VpsR may be the target of the switch.
Biofilm formation and decay must be a tightly controlled system if it is to confer a selective advantage in natural aquatic habitats that are subject to changes in a variety of physicochemical parameters, including nutrient availability, salinity, and sheer force generated by flowing water. For the system to be adequately responsive, flexible, and robust, some environmental signals should trigger biofilm formation, whereas others should inhibit this process and initiate biofilm dissolution. Thus, depending on the signal, processes would be initiated that change the ratio of the sessile to the planktonic populations of the species. We predict that VpsR will be only one of several regulators of this process as is the case for the much more thoroughly studied alginate-dependent biofilm system of P. aeruginosa. Alginate biosynthesis by mucoid strains of this species requires two response regulators (AlgB and AlgR) (6, 11, 37), an alternative sigma factor (AlgT) (38), and a ribbon-helix-helix DNA-binding protein (AlgZ) (1). Further, the development of a mature biofilm architecture containing pillars of alginate-embedded bacteria that are separated by water-containing channels requires quorum sensing mediated by the lasI product, N-(3-oxododecanoly)-l-homoserine (5). The V. cholerae O1 biofilm system is likely to be equally complex. For example, studies by Watnick and Kolter of the complex biofilm-forming system of this species (36) led them to propose a three-step model of biofilm development: first, mannose-sensitive hemagglutinin type IV pili and flagella facilitate the attachment of free-swimming bacteria to a surface; second, the flagella cause attached bacteria to spread across the surface; and third, EPS is secreted (36), providing the extracellular matrix of the mature biofilm's three-dimensional structure. The work presented here highlights another level of complexity by pointing out that different V. cholerae O1 El Tor isolates can vary significantly with respect to the third proposed step of biofilm development, vps gene expression. V. cholerae O1 El Tor, strain A1552, the prototype strain used in our study, was isolated in Peru at the beginning of the recent South American outbreak, where epidemic cholera had been absent for over a century. In contrast, strain N16961, studied by Watnick and Kolter, was isolated in India, where cholera has long been endemic. Based on the comparative analysis of the biofilm-forming phenotypes of these strains depicted in Fig. 5A, the smooth variant of N16961 is a better biofilm former than the smooth variant of A1552 and correspondingly exhibits more vpsA and vpsL expression. Strain-to-strain variation in the biofilm phenotype further compounded by intrastrain differences between the rugose and smooth-colony types. If the biofilm phenotype is important for the survival of V. cholerae O1 El Tor in an aquatic habitat, then differences of the kind exhibited by strains A1552 and N16961 could cause the latter to be more environmentally fit than the former. If so, then it is possible that adaptation to different geographic locations and, in turn, acclimation to distinct microenvironments could have led to the observed differences in this important phenotype.
ACKNOWLEDGMENTS
We thank Melissa Carter (CSU Hayward) for help with the scanning electron microscopy, Nafisa Ghori (Stanford) for help with the transmission electron microscopy, Susan Palmieri for help with the CSLM, Lee Kozar for his assistance with sequence analysis of the vps gene cluster, Denise Monack for providing pACYC-GFP, and TIGR for early release of vps coding sequence.
This work is supported by NIH grant RO1-AI43422.
REFERENCES
- 1.Baynham P J, Wozniak D J. Identification and characterization of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol Microbiol. 1996;22:97–108. doi: 10.1111/j.1365-2958.1996.tb02659.x. [DOI] [PubMed] [Google Scholar]
- 2.Bugert P, Geider K. Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora. Mol Microbiol. 1995;15:917–933. doi: 10.1111/j.1365-2958.1995.tb02361.x. [DOI] [PubMed] [Google Scholar]
- 3.Chopra A K, Peterson J W, Prasad R. Cloning and sequence analysis of hydrogenase regulatory genes (hydHG) from Salmonella typhimurium. Biochim Biophys Acta. 1991;1129:115–118. doi: 10.1016/0167-4781(91)90224-a. [DOI] [PubMed] [Google Scholar]
- 4.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 5.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 6.Deretic V, Dikshit R, Konyecsni W M, Chakrabarty A M, Misra T K. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J Bacteriol. 1989;171:1278–1283. doi: 10.1128/jb.171.3.1278-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drummond M, Whitty P, Wootton J. Sequence and domain relationships of ntrC and nifA from Klebsiella pneumoniae: homologies to other regulatory proteins. EMBO J. 1986;5:441–447. doi: 10.1002/j.1460-2075.1986.tb04230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flashner Y, Weiss D S, Keener J, Kustu S. Constitutive forms of the enhancer-binding protein NtrC: evidence that essential oligomerization determinants lie in the central activation domain. J Mol Biol. 1995;249:700–713. doi: 10.1006/jmbi.1995.0330. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher M. Bacterial attachment in aquatic environments: a diversity of surfaces and adhesion strategies. In: Fletcher M, editor. Bacterial adhesion: molecular and ecological diversity. New York, N.Y: Wiley-Liss; 1996. pp. 1–24. [Google Scholar]
- 10.Gibson U E, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg J B, Dahnke T. Pseudomonas aeruginosa AlgB, which modulates the expression of alginate, is a member of the NtrC subclass of prokaryotic regulators. Mol Microbiol. 1992;6:59–66. doi: 10.1111/j.1365-2958.1992.tb00837.x. [DOI] [PubMed] [Google Scholar]
- 12.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 13.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haf D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White W O, Salzberg S T, Smith H O, Colwell R R, Mekalanos J J, Venter J C, Fraser C M. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, Schell M. Molecular characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation at a single promoter. Mol Microbiol. 1995;16:977–989. doi: 10.1111/j.1365-2958.1995.tb02323.x. [DOI] [PubMed] [Google Scholar]
- 15.Islam M S, Drasar B S, Bradley D J. Long-term persistence of toxigenic Vibrio cholerae O1 in the mucilaginous sheath of a blue-green alga, Anabaena variabilis. J Trop Med Hyg. 1990;93:133–139. [PubMed] [Google Scholar]
- 16.Islam M S, Drasar B S, Bradley D J. Survival of toxigenic Vibrio cholerae O1 with a common duckweed, Lemna minor, in artificial aquatic ecosystems. Trans R Soc Trop Med Hyg. 1990;84:422–424. doi: 10.1016/0035-9203(90)90345-f. [DOI] [PubMed] [Google Scholar]
- 17.Islam M S, Drasar B S, Sack R B. The aquatic flora and fauna as reservoirs of Vibrio cholerae: a review. J Diarrhoeal Dis Res. 1994;12:87–96. [PubMed] [Google Scholar]
- 18.Jiang X M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 19.Kiino D R, Licudine R, Wilt K, Yang D H, Rothman-Denes L B. A cytoplasmic protein, NfrC, is required for bacteriophage N4 adsorption. J Bacteriol. 1993;175:7074–7080. doi: 10.1128/jb.175.21.7074-7080.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klose K E, Novik V, Mekalanos J J. Identification of multiple sigma 54-dependent transcriptional activators in Vibrio cholerae. J Bacteriol. 1998;180:5256–5259. doi: 10.1128/jb.180.19.5256-5259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolter R, Losick R. One for all and all for one. Science. 1998;280:226–227. doi: 10.1126/science.280.5361.226. [DOI] [PubMed] [Google Scholar]
- 22.Kustu S, Santero E, Keener J, Popham D, Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leveque F, Plateau P, Dessen P, Blanquet S. Homology of lysS and lysU, the two Escherichia coli genes encoding distinct lysyl-tRNA synthetase species. Nucleic Acids Res. 1990;18:305–312. doi: 10.1093/nar/18.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma S, Selvaraj U, Ohman D E, Quarless R, Hassett D J, Wozniak D J. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J Bacteriol. 1998;180:956–968. doi: 10.1128/jb.180.4.956-968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller P, Keller M, Weng W M, Quandt J, Arnold W, Puhler A. Genetic analysis of the Rhizobium meliloti exoYFQ operon: ExoY is homologous to sugar transferases and ExoQ represents a transmembrane protein. Mol Plant-Microbe Interact. 1993;6:55–65. doi: 10.1094/mpmi-6-055. [DOI] [PubMed] [Google Scholar]
- 26.North A K, Klose K E, Stedman K M, Kustu S. Prokaryotic enhancer-binding proteins reflect eukaryote-like modularity: the puzzle of nitrogen regulatory protein C. J Bacteriol. 1993;175:4267–4273. doi: 10.1128/jb.175.14.4267-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 28.Pabo C A, Sauer R T. Protein DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Singleton F, Attwell C, Jangi M, Colwell R. Influence of salinity, nutrient concentration and temperature on growth and survival of Vibrio cholerae in the aquatic environment. In: Kuwahara S, Pierce N, editors. Advances in research on cholera and related diarrheas. Tokyo, Japan: JTJ Scientific Publisher; 1983. [Google Scholar]
- 31.Singleton F L, Attwell R W, Jangi M S, Colwell R R. Influence of salinity and organic nutrient concentration on survival and growth of Vibrio cholerae in aquatic microcosms. Appl Environ Microbiol. 1982;43:1080–1085. doi: 10.1128/aem.43.5.1080-1085.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevenson G, Andrianopoulos K, Hobbs M, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timms A R, Bridges B A. Reversion of the tyrosine ochre strain Escherichia coli WU3610 under starvation conditions depends on a new gene tas. Genetics. 1998;48:1627–1635. doi: 10.1093/genetics/148.4.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wai S N, Mizunoe Y, Takade A, Kawabata S I, Yoshida S I. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl Environ Microbiol. 1998;64:3648–3655. doi: 10.1128/aem.64.10.3648-3655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watnick P I, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wozniak D J, Ohman D E. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J Bacteriol. 1991;173:1406–1413. doi: 10.1128/jb.173.4.1406-1413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wozniak D J, Ohman D E. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yildiz F H, Schoolnik G K. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]