Abstract
A dissimilatory sulfite reductase (DSR) was purified from the anaerobic, taurine-degrading bacterium Bilophila wadsworthia RZATAU to apparent homogeneity. The enzyme is involved in energy conservation by reducing sulfite, which is formed during the degradation of taurine as an electron acceptor, to sulfide. According to its UV-visible absorption spectrum with maxima at 392, 410, 583, and 630 nm, the enzyme belongs to the desulfoviridin type of DSRs. The sulfite reductase was isolated as an α2β2γn (n ≥ 2) multimer with a native size of 285 kDa as determined by gel filtration. We have sequenced the genes encoding the α and β subunits (dsrA and dsrB, respectively), which probably constitute one operon. dsrA and dsrB encode polypeptides of 49 (α) and 54 kDa (β) which show significant similarities to the homologous subunits of other DSRs. The dsrB gene product of B. wadsworthia is apparently a fusion protein of dsrB and dsrD. This indicates a possible functional role of DsrD in DSR function because of its presence as a fusion protein as an integral part of the DSR holoenzyme in B. wadsworthia. A phylogenetic analysis using the available Dsr sequences revealed that B. wadsworthia grouped with its closest 16S rDNA relative Desulfovibrio desulfuricans Essex 6.
Bilophila wadsworthia is a strictly anaerobic, gram-negative bacterium (2) which belongs to the family Desulfovibrionaceae in the delta subdivision of the Proteobacteria, but does not reduce sulfate (2, 25). B. wadsworthia has been found quite frequently in patients with appendicitis and its complications and is the third most common anaerobic isolate in such infections (11), but can also be isolated from a wide variety of other infections, e.g., biliary tract infection (41), liver abscess (41), and ear infections (39). B. wadsworthia has also been found in the normal fecal flora (2). The organism lacks classical virulence factors like capsules, fimbriae, and extracellular enzymes (2). However, preliminary studies have indicated that B. wadsworthia exerts cytotoxic effects on two cell lines, and endotoxic activity of B. wadsworthia has been described (2, 34).
We recently isolated from a communal sewage plant a strain of B. wadsworthia which utilizes organic sulfonates (e.g., taurine [2-aminoethanesulfonate]) as a carbon source and electron sink. B. wadsworthia respires taurine anaerobically with electrons derived mainly from formate oxidation and oxidation of the taurine carbon (25). Taurine is transaminated to sulfoacetaldehyde (22), which is cleaved to sulfite and an unidentified organic product (K. Denger and A. M. Cook, unpublished). Finally, sulfite is reduced to sulfide by a dissimilatory sulfite reductase (DSR) (6, 25). In addition, sulfite or thiosulfate serves as an electron acceptor for anaerobic respiration with formate as the electron donor in B. wadsworthia (25).
DSR, a key enzyme in dissimilatory sulfate reduction, occurs in all organisms capable of reducing sulfite during anaerobic respiration investigated so far (9, 33). Otherwise, DSRs are rare. An apparently dissimilatory type of sulfite reductase inducible in the presence of sulfite under anoxic conditions has been found in Salmonella enterica serovar Typhimurium, but the function of dissimilatory sulfite reduction by this organism is not clear (16). The sulfite reductase characterized in Clostridium pasteurianum was also proposed to be of the dissimilatory type but differs in its properties from DSRs of sulfate-reducing organisms (12). In contrast to DSRs, assimilatory sulfite reductases are involved in assimilation of sulfate in many organisms.
DSRs are multisubunit enzymes (167 to 225 kDa) that catalyze the six-electron reduction of sulfite to sulfide. They all contain siroheme and [4Fe-4S] prosthetic centers and are classified according to their spectroscopic properties in four major groups (47). Sulfite reductases of the desulfoviridin type are found in Desulfovibrio species (27). Their subunit structure was initially described as α2β2, with a molecular mass of 50 kDa for the α and 40 kDa for the β subunits (28), but a third subunit, γ (11 kDa), was discovered, and an α2β2γ2 structure was proposed for Desulfovibrio vulgaris (Hildenborough), D. vulgaris oxamicus (Monticello), D. gigas, and D. desulfuricans ATCC 27774 (36).
The dsrA (α subunit) and dsrB (β subunit) genes of DSR have been sequenced completely in six organisms: the sulfate-reducing bacterium D. vulgaris (19), the sulfate-reducing archaea Archaeoglobus fulgidus (9) and Archaeoglobus profundus (21), the thermophilic, gram-positive bacterium Desulfotomaculum thermocisternum (21), the sulfur-reducing archaeon Pyrobaculum islandicum (33), and the “reverse sulfite reductase” of the phototrophic Allochromatium vinosum (14). The γ subunit (dsrC) is apparently encoded in a separate locus (18). In all cases except for A. vinosum (14), a third gene, dsrD, encoding a protein of unknown function, was found downstream of dsrB. Recently, Wagner et al. developed a PCR assay for the specific amplification of large parts of the dsrA and dsrB genes, which allows the detection of many organisms capable of dissimilatory sulfate reduction (45).
We report here on purification and properties of the DSR that is involved in energy conservation from taurine metabolism in the anaerobic respiration of B. wadsworthia RZATAU as well as the relationship of the DsrA and DsrB sequences of B. wadsworthia to those of its sulfate-reducing relatives.
MATERIALS AND METHODS
Bacteria and growth conditions.
B. wadsworthia RZATAU (DSM 11045) was routinely grown in batch culture (0.1 or 10 liters) in an anoxic freshwater mineral salts medium containing 12 mM taurine and 80 mM formate (25). Alternatively, 12 mM isethionate (2-hydroxyethanesulfonate) or 12 mM cysteate (2-amino-3-sulfopropionate) plus formate, 12 mM taurine plus 25 mM pyruvate, or 12 mM thiosulfate plus 20 mM dl-lactate was used. B. wadsworthiaT was grown in taurine-plus-formate medium. D. vulgaris was grown in the same salts medium in the presence of 20 mM sulfate and 20 mM dl-lactate. Desulfovibrio sp. strain RZACYSA was grown in salts medium containing 10 mM cysteate and 20 mM dl-lactate (25). The sources of the chemicals and gases (N2 and CO2) used are given elsewhere (25).
Preparation of cell extracts and enzyme purification.
Cells for the purification of DSR were harvested and stored as described elsewhere (23). Preparation of crude extracts by disruption of suspended cells in a French pressure cell, removal of membrane particles by ultracentrifugation, and precipitation of DNA with streptomycin sulfate are detailed elsewhere (17, 23).
DSR from B. wadsworthia RZATAU was purified from the cytosolic fraction of the crude extract in a two-step protocol involving anion-exchange and gel filtration chromatography. The 2.5-fold-diluted crude extract was applied to a Mono Q column (HR 10/10; Pharmacia) equilibrated with 20 mM MOPS (morpholinepropanesulfonic acid, pH 6.5) at a flow rate of 2 ml min−1. Proteins were eluted with an increasing linear gradient to 1 M Na2SO4 and collected in 5-ml fractions. DSR was identified by its green color, and the identity was confirmed spectroscopically and by red fluorescence under alkaline conditions (37). Enrichment of the protein was monitored by the A630/A280 ratio (purity index derived from reference 9; defined as A630/A280 × 10−3). DSR eluted at about 130 mM Na2SO4 in one fraction and had a purity index of 174. Concentrated protein was loaded onto a Superose 12 column (HR 10/30; Pharmacia) equilibrated at a flow rate of 0.4 ml min−1 with 50 mM MOPS (pH 6.5) containing 150 mM Na2SO4, and 0.5-ml fractions were collected. The molecular masses of the proteins used to calibrate the column are described elsewhere (23). The purity index of the purified DSR was estimated to be 249.
The presence of DSR in crude extract of B. wadsworthia RZATAU grown with different substrates was detected qualitatively by its red fluorescence (37).
Sulfite reductase activity.
Formation of sulfide from taurine was investigated in cell extracts from B. wadsworthia RZATAU grown with taurine plus formate. The assay was performed in 100 mM potassium phosphate buffer (pH 7.0) containing 5 mM taurine, 5 mM pyruvate, 0.5 mM NAD+, 0.1 mM pyridoxal-5′-phosphate, 0.1 mM thiamine pyrophosphate, and 20 mM formate under anoxic conditions in 16-ml serum bottles closed with butyl rubber septa. The center of the bottle contained a tube with a filter soaked with cadmium(II) acetate (10%, wt/wt) and NaOH (10%, wt/wt). The reaction was started by addition of 0.5 mg of protein, and the bottles were incubated on a shaker at 30°C. Sulfide was determined in aliquots of the reaction mixture or in the filter by the formation of methylene blue (5).
Gel electrophoresis and N-terminal sequence analysis.
Proteins were separated by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (SDS–12% PAGE) according to the method of Laemmli (20) or of Schägger and Jagow (38) and subsequently stained with Coomassie brilliant blue G-250 (35). Blotting and N-terminal sequencing were done as described elsewhere (23).
Analytical methods.
Protein concentrations were determined by the method of Bradford (4) with bovine serum albumin as the standard. UV-visible (UV/VIS) spectra were recorded by an Uvikon 922 spectrophotometer (Kontron). Taurine and alanine were quantified by high-pressure liquid chromatography after derivatization with 2,4-dinitrofluorobenzene (10, 26). The G+C content of B. wadsworthiaT was determined by the German Culture Collection (Braunschweig, Germany) in a sample of 0.8 g (wet weight) of cells.
Isolation of nucleic acids and DNA amplification procedures.
Total DNA was prepared from stationary-phase cultures of B. wadsworthia RZATAU (0.5 liter), D. desulfuricans (0.5 liter), or Desulfovibrio sp. strain RZACYSA (0.2 liter) by the cetyltrimethylammonium bromide precipitation method (1). The primer pair DSR1F and DSR4R was used to amplify by PCR a 1.9-kb DNA fragment encoding most of the α and β subunits of DSR (45). PCR was performed as described previously (45) except for the buffer [2.25 mM MgCl2, 50 mM Tris-HCl (pH 9.2), 14 mM (NH4)2SO4, 10% dimethyl sulfoxide] and for the Taq polymerase (MBI) in a Master Cycler gradient thermocycler (Eppendorf). Washed and concentrated cells of B. wadsworthia RZATAU were also added directly to the PCR mixtures instead of purified DNA as the template.
PCR amplification of the adenosine phosphosulfate (APS) reductase genes with degenerate primer sets (wh53, wh54, and wh62) involved the thermal profile described by Hipp et al. (14). The reaction buffer described above for the DSR primer set was used. DNAs from D. desulfuricans and from Desulfovibrio sp. strain RZACYSA were used as positive controls.
DNA sequencing and analysis.
The nucleotide sequence of the 1.9-kb PCR product from the dsr region was determined by cycle sequencing and primer walking using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit and an ABI 377 DNA sequencer (GATC GmbH). We then used adaptor-ligated PCR to obtain the complete sequences of the genes encoding the α and β subunits of DSR. Genomic DNA was digested with different restriction enzymes and ligated to an adaptor of known sequence (Universal Genome Walker kit; Clontech). Nested PCR was done with the Advantage Tth polymerase mix (Clontech) and primer deduced from specific dsrA or dsrB gene sequences in combination with the adaptor primer. The dsr primer sequences for amplification of the upstream region were CAT GCA CGG TTC CAC CGG CGA CAT CGT GCT (primary PCR) and GGA GCC ACG GAG TTC CCA AAT GTC GCA CAG (nested PCR). To amplify the 3′ end of dsrB, two adaptor-ligated PCRs were done successively using primers GCG CCG TGC ACT GCT CCG ACA TCG GTA TCG (primary PCR) and GGT ATC CAC CGC AAG CCT CCG ATG ATC GAC (nested PCR) for the first and AGG ACT TCC TTG AAC TCT TCC CCA CCA AGG (primary PCR) and GTC CGT GCT CGT GTC CGA AGA GTC TCT GGA (nested PCR) for the second PCR. Upstream of the 1.9-kb sequence, a 1.8-kb amplification product was obtained, while downstream a 0.9-kb DNA fragment was amplified and sequenced. Sequence alignments were done using ClustalX (44).
Phylogeny of DsrA and DsrB.
The phylogenetic analysis (i.e., sequence alignments and treeing) was performed by using the ARB software package (version 2.5b; O. Strunk and W. Ludwig, Technische Universität München, Munich, Germany [http://www.biol.chemie.tu-muenchen.de/pub/ARB/]). Deduced DSR amino acid sequences were fitted manually into an alignment of DSR sequences retrieved from public databases (3) using the Genetic Data Environment (version 2.2) as implemented in the ARB software package. Prior to treeing analysis, amino acid frequency filters (20 to 100% sequence similarity) were generated for a concatenated data set comprising the amino acid sequences of the α (364 positions) and β (238 positions) subunit data sets. Treeing was performed on the concatenated α and β subunit data sets using distance matrix analysis [FITCH (PHYLIP version 3.5) and neighbor-joining (ARB)], parsimony [PROTPARS (PHYLIP version 3.5)], and maximum likelihood [PROTML (PHYLIP version 3.5)] as outlined previously (45). Bootstrap analysis (100 resamplings) was performed using parsimony analysis as implemented in the PHYLIP package.
Nucleotide sequence accession numbers.
The sequences encoding the α and β subunits of DSR from B. wadsworthia RZATAU (accession no. AF269147) and D. desulfuricans (accession no. AF273034) have been deposited in GenBank.
RESULTS
Activity and purification of DSR from B. wadsworthia RZATAU.
In cell extracts of B. wadsworthia RZATAU, 1.6 mM alanine and 80 μM sulfide were formed from taurine (5 mM) and formate (20 mM). However, sulfide was only detectable when trapped from the gas phase with cadmium acetate. In the absence of taurine, negligible amounts of sulfide were formed. We thus presume that there is DSR activity in the extract but that sulfide reacts with other components in the aqueous mixture (13) and that formation of sulfide is underestimated. The enzyme rapidly lost activity, so its purification depended on assaying its physical properties. DSR was detected by fluorescence in crude extracts of cells grown with taurine plus formate or pyruvate, cysteate or isethionate plus formate, thiosulfate plus lactate, or pyruvate as the sole carbon and energy source. We purified DSR from a soluble extract of B. wadsworthia RZATAU grown with taurine and formate. The protein from anion-exchange chromatography was about 90% pure, and apparent homogeneity was obtained by gel filtration chromatography (not shown). Based on the purity index (A630/A280), it can be calculated that DSR was purified 11-fold and represents about 9% of the soluble protein of B. wadsworthia.
Molecular properties.
SDS-PAGE of the protein showed three bands with apparent molecular masses of 53, 49, and 11 kDa (Fig. 1), so DSR seems to be composed of three different subunits. The protein eluted from gel filtration under nondenaturing conditions with an apparent molecular mass of 285 kDa.
FIG. 1.
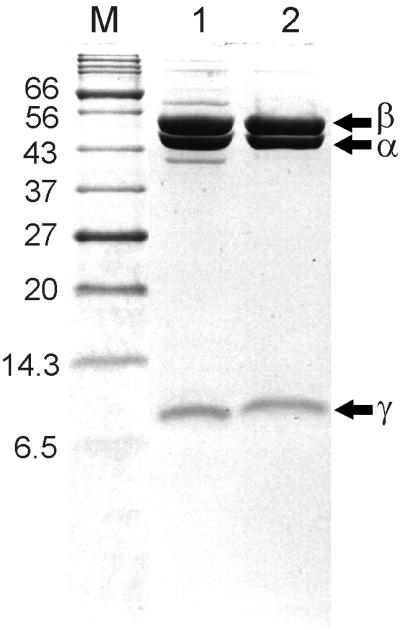
Purification of DSR from B. wadsworthia RZATAU monitored by SDS-PAGE. Protein was separated on an SDS–12% PAGE gel and subsequently stained with Coomassie brilliant blue. Lanes: M, molecular mass standards (in kilodaltons); 1, fraction from Mono Q chromatography (10 μg); 2, fraction from gel filtration on Superose 12 (10 μg).
DSR had a UV/VIS spectrum with absorption maxima at 392, 410, 583, and 630 nm (not shown), characteristic of desulfoviridin (27). The molar extinction coefficients of B. wadsworthia DSR at 392, 410, 583, and 630 nm were estimated to be 5.5 × 105, 5.8 × 105, 1.1 × 105, and 2.0 × 105 M−1 cm−1, respectively. Under alkaline conditions, the enzyme showed red fluorescence, also characteristic of desulfoviridin (37).
The N-terminal sequences of the three subunits were determined by Edman degradation (Table 1). A comparison was done with the amino acid sequence of the subunits of the DSRs from D. vulgaris and D. desulfuricans (Table 1). The N-terminal sequences of the α, β, and γ subunits of the three organisms were each highly conserved. The data indicated, however, that the largest subunit (53 kDa) from B. wadsworthia exhibited similarities to the β subunit from D. vulgaris (40 kDa) and D. desulfuricans (45 kDa), whereas the 49-kDa subunit of the B. wadsworthia enzyme was similar to the α subunits (50 kDa) from the same organisms.
TABLE 1.
Alignment of the N-terminal amino acid sequences of DSR subunits (α, β, and γ) determined by Edman degradation from D. vulgaris Hildenborough, D. desulfuricans Essex 6, and B. wadsworthia RZATAUa
| Subunit | Size (kDa) | Organism | Sequence | Reference |
|---|---|---|---|---|
| α | 50 | D. vulgaris | A—K H A T P K L D Q L E S G P W X S F V v d k k | 36 |
| 50 | D. desulfuricans | A—K H A T P L L D Q L E S G P W P S F V | 40 | |
| 49 | B. wadsworthia | A I K H P T P L L D Q L E T G P W P X F V S | This study | |
| β | 40 | D. vulgaris | A F I S S G Y N P E K p m a n | 36 |
| 45 | D. desulfuricans | A F I P T G Y N P X K P M | 40 | |
| 53 | B. wadsworthia | A F V S S G Y N P E K P M E G R I S D I | This study | |
| γ | 11 | D. vulgaris | A E V T Y K G K S F E V D E D G F L L R F D D W | 18 |
| 11 | D. desulfuricans | A E I T Y K G K | 40 | |
| 11 | B. wadsworthia | A E V T Y K G K T F E V D E D G F L L K F D d | This study |
Identifications in lowercase letters are uncertain. X, residue not identified; —, alignment gap.
Nucleotide sequence analysis.
A 1.9-kb DNA region encoding most of the α and β subunits of DSR was amplified by PCR from B. wadsworthia. The sequence showed high similarities to those determined for D. vulgaris and other organisms. This was in contrast to the different sizes of the β subunits (53 kDa rather than 40 to 45 kDa; Table 1), so we amplified and sequenced the complete dsrAB region. A total of 4.3 kb of double-stranded sequence was examined. It comprised three open reading frames, ORF1, dsrA, and dsrB. The gene dsrA (1,317 bp) was identified because the deduced amino acid sequence included the N-terminal sequence that we observed in the α subunit. Similarly, dsrB (1,452 bp) was identified because the N-terminal amino acid sequence of the β subunit corresponded to the deduced sequence.
In contrast to the published value for the G+C content of B. wadsworthiaT (39 to 40 mol% [2]), we now report the G+C content to be 59.2%, so the G+C content of dsrA and dsrB (59.4 and 60.2%, respectively) corresponds to the overall G+C content of the organism. A putative promoter sequence was located 123 nucleotides upstream of the translational start of the dsrA gene, and a putative transcription terminator was found downstream of the stop codon of dsrB. The start codons of dsrA and dsrB are both preceded by a putative ribosome-binding site (not shown). The intergenic distance between the 3′ end of the dsrA and the 5′ end of the dsrB gene comprised 18 nucleotides, analogous to the dsr operons of D. vulgaris and A. fulgidus (19).
The amino acid sequences deduced from dsrA and dsrB consisted of 438 and 483 residues, respectively, but the initiating methionine residue was obviously removed from each protein after synthesis. The derived sizes of the α (49.0 kDa) and β (53.6 kDa) subunits are in good agreement with the data determined for the purified sulfite reductase (49 and 53 kDa, respectively). The isoelectric points calculated for DsrA and DsrB were 5.3 and 6.5, respectively.
The third open reading frame, ORF1 (711 bp), was located at the 5′ end of the 4.3-kb DNA fragment, 618 bp upstream of the translational start codon of dsrA. Parts of the deduced amino acid sequence exhibited similarities to the rare lipoprotein A (rlpA) from Escherichia coli (39% identity) (42). The region between ORF1 and dsrA contained no open reading frames of more than 130 bp in length.
Sequence similarities.
The deduced amino acid sequences of dsrA and dsrB were highly similar to the α and β subunits of DSRs from other sulfate-reducing microorganisms (Table 2). Analysis of partial DSR sequence data (available via the ∼1.9-kb DSR PCR fragment) revealed that DsrA of B. wadsworthia RZATAU was most similar to that of D. desulfuricans Essex 6 on both the nucleotide and deduced amino acid levels (88.3 and 86.0%, respectively; Table 2), whereas DsrB was slightly more similar to that of D. vulgaris (83.6 and 82.3%, respectively; Table 2).
TABLE 2.
Sequence similarities of DSR α and β subunit gene fragments (dsrA and dsrB) and their deduced amino acid sequences (DsrA and DsrB)a
| Strain | Sequence similarity (%)
|
||||||
|---|---|---|---|---|---|---|---|
| B. wadsworthia | D. desulfuricans | D. vulgaris | Desulfovibrio sp. strain PT-2 | D. multivorans | D. sapovorans | D. latus | |
| B. wadsworthia | 100 | 82.4/81.4 | 83.6/82.3 | 80.1/81.1 | 74.7/72.2 | 72.8/70.1 | 71.4/69.2 |
| D. desulfuricans | 88.3/86.0 | 100 | 82.9/81.1 | 82.1/80.7 | 70.6/71.4 | 71.0/68.1 | 69.2/67.8 |
| D. vulgaris | 86.4/83.9 | 84.1/83.3 | 100 | 89.9/86.7 | 75.2/73.8 | 76.1/70.4 | 73.3/70.7 |
| Desulfovibrio sp. strain PT-2 | 80.9/82.0 | 81.6/83.7 | 87.2/87.3 | 100 | 71.8/73.1 | 73.4/71.7 | 71.3/71.4 |
| D. multivorans | 74.4/73.3 | 75.1/73.9 | 77.1/75.3 | 76.7/75.3 | 100 | 85.8/77.8 | 81.2/73.8 |
| D. sapovorans | 73.3/71.3 | 72.5/72.7 | 76.6/74.1 | 73.5/72.9 | 76.3/71.2 | 100 | 83.0/76.5 |
| D. latus | 73.3/70.4 | 70.0/70.2 | 74.4/70.9 | 72.4/70.4 | 78.0/73.5 | 76.5/73.3 | 100 |
Alignment positions of unambiguously determined nucleotides represented in all sequences were used for pairwise comparisons of dsrA (968 positions), dsrB (668 positions), DsrA (322 positions), and DsrB (224 positions). Values for DsrA/dsrA are below the diagonal, and those for DsrB/dsrB are above the diagonal.
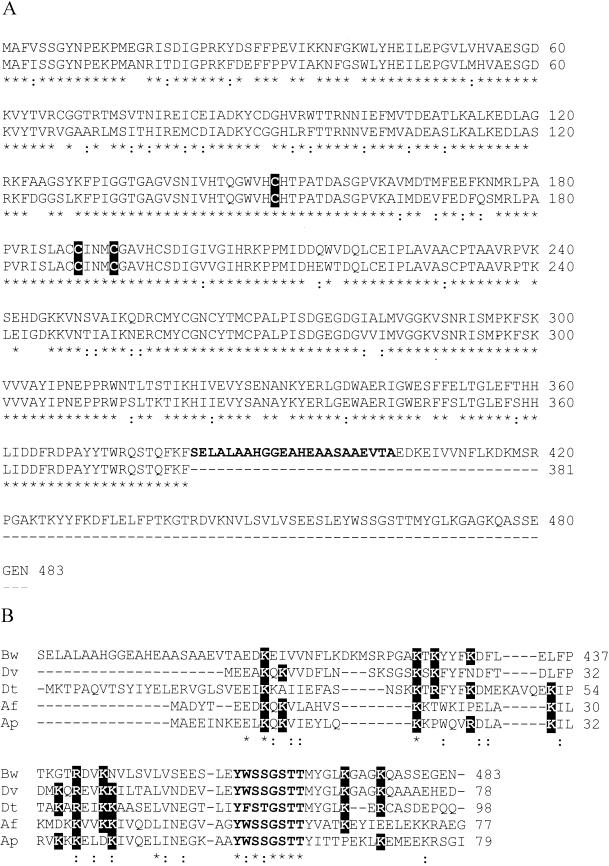
Full amino acid sequences (all positions) of DsrA were 84% identical to DsrA from D. vulgaris (19) and 57% identical to DsrA from A. fulgidus (9). Amino acids 1 to 381 of DsrB exhibited 83% identity to DsrB from D. vulgaris (19) and 56% identity to DsrB from A. fulgidus (9), while the C-terminal amino acids exhibited no similarity to other DsrBs. However, the C-terminal amino acids 405 to 483 of DsrB were 59% identical to DsrD from D. vulgaris (19), 42% identical to DsrD from Desulfotomaculum thermocisternum (21), and 46% identical to DsrD from A. fulgidus (9) (Fig. 2). Amino acids 382 to 404 of the B. wadsworthia DsrB showed similarities neither to other DsrBs nor to other DsrDs. Between the 3′ end of dsrB and the 5′ end of dsrD in D. vulgaris and A. fulgidus are 58 and 41 bp of intergenic sequence, respectively.
FIG. 2.
Sequence comparisons with DSR from B. wadsworthia RZATAU. (A) Amino acid sequences derived from dsrB (upper line) aligned with those of DsrB from D. vulgaris (lower line). The amino acid sequence positions are indicated in the margins. Highly conserved cysteine residues proposed to coordinate siroheme-[4Fe-4S] binding (9) are indicated by boxes. Amino acids which do not show any homologies are in boldface. (B) Alignment of amino acids 405 to 483 of DsrB from B. wadsworthia DSR (Bw), with DsrD from D. vulgaris (Dv), Desulfotomaculum thermocisternum (Dt), A. fulgidus (Af), and A. profundus (Ap). Conserved lysine and arginine residues are indicated by boxes; a conserved stretch of eight amino acids is shown in boldface. ∗, positions with a conserved residue; :, positions with a conservative replacement.
Phylogeny of DsrA and DsrB.
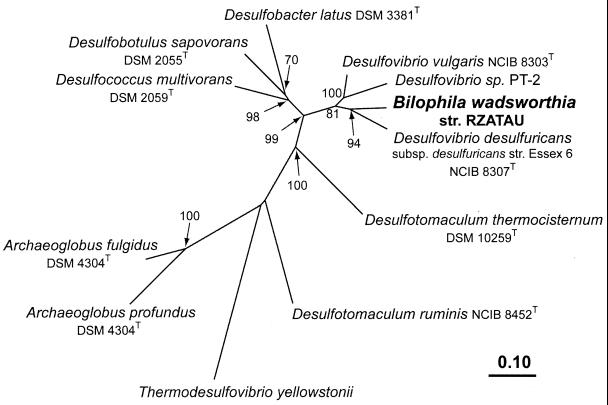
Phylogenetic trees for the DSR α and β subunits (not shown) and for a region representing α and β subunits (Fig. 3) were estimated from deduced amino acid data sets of the ∼1.9-kb PCR product by distance matrix, parsimony, and maximum-likelihood methods. Consistently, B. wadsworthia RZATAU grouped with its closest 16S rDNA relative D. desulfuricans Essex 6 (91.5% 16S rDNA sequence similarity). This tree topology was supported by all treeing algorithms utilized, by high bootstrap scores, and by individual analyses of both subunits as well as analysis of the concatenated α and β subunit data sets.
FIG. 3.
Phylogenetic tree reflecting the relationship of the B. wadsworthia RZATAU DSR to the DSRs from Desulfovibrio sp. and other sulfate-reducing microorganisms (21, 45). Tree topology was estimated by using FITCH distance matrix analysis of the concatenated α and β subunit amino acid data sets (546 positions, including D. vulgaris positions 73 to 421 [DsrA] and 18 to 257 [DsrB]). Parsimony analysis was used to determine bootstrap values with an identical data set. Bootstrap values are only indicated for branches, which were recovered in the majority of bootstrap replicates (>50%). The scale bar indicates the number of expected amino acid substitutions per site per unit of branch length. GenBank accession numbers used: U58114, U58115, U58118 to U58127, M95624, AF071499, AF074396, U16723, AF273034, and AF269147.
Investigation of presence of genes encoding APS reductase.
PCR conditions and primer sets (14) to amplify regions of the apr genes (encoding APS reductase) were applied to investigate the presence of these genes in B. wadsworthia. However, we did not obtain specific PCR fragments of the apr gene in B. wadsworthia, though in the Desulfovibrio control strains the corresponding 1.6- or 2.2-kb products were detected. The presumed absence of the apr genes is also reflected by the fact that B. wadsworthia is not able to reduce sulfate (6).
DISCUSSION
DSR was purified from B. wadsworthia as an inactive protein which we initially identified by its spectral and fluorescence properties and confirmed by sequence homologies. No other green protein was detected in separated extracts (H. Laue, unpublished results). We detected three subunits, α (49 kDa), β (53 kDa), and γ (11 kDa) (Fig. 2), and we interpret the native structure to be α2β2γn (n ≥ 2), as suggested for D. desulfuricans Essex 6 (40). The function of the 11-kDa γ subunit that was copurified in most sulfite reductases (36) except that from A. fulgidus (9) is not clear at present. Whereas Pierik et al. reported that the γ subunit was tightly associated in the DSR of D. vulgaris (36), it was shown that in D. desulfuricans Essex 6, this subunit can be separated during gel filtration, which indicated a less tight association of DsrC with the α and β subunits in that organism (40). The presence and position of the putative promoter 123 nucleotides upstream of the translational start of the dsrA gene and putative termination sequences downstream of the stop codon of dsrB indicate that dsrA and dsrB constitute a single transcription unit and that the genes are coordinately expressed.
The order and sequences of the dsrAB genes of DSR from bacteria and archaea are highly conserved. Whereas in all organisms sequenced so far except A. vinosum, the operon consists of dsrA, dsrB, and dsrD, in B. wadsworthia there are only two genes, dsrA and dsrB. Apparently, the dsrB gene product of B. wadsworthia is a fusion of dsrB and dsrD. A function for DsrD has not been detected in earlier biochemical work (19); however, the presence of DsrD as an integral part of the DSR holoenzyme in B. wadsworthia as a DsrB-DsrD fusion suggests a possible involvement in DSR function. This hypothesis is further corroborated by the high degree of conservation of DsrD sequences (Fig. 2) among the microorganisms sequenced so far, which suggests an essential role of this protein in dissimilatory sulfite reduction in general. Because of its high content and significant conservation of Lys residues among DsrDs, a function as a sulfite-binding protein was proposed (19), but spectroscopic analysis of DsrD indicated that it bound neither sulfite nor sulfide (15). Alignment of DsrD from five different organisms shows in addition to the conserved Lys residues, a short, remarkably conserved stretch of eight amino acids (Y W/F S S/T G S T T) (Fig. 2) (21). Recently, DsrD from D. vulgaris Hildenborough has been crystallized, and preliminary results concerning the crystal structure have been described (32). The forthcoming high-resolution three-dimensional structure may provide a clue to the function of DsrD.
The stretch of 23 residues between the sequences homologous to dsrB and dsrD in the DSR of B. wadsworthia revealed no homologies to these genes. The presence of several small residues like alanine (nine residues) and glycine (two residues) may indicate a function as a linker between the DsrB and DsrD domains, possibly necessary to allow correct assembly.
DSRs usually contain two sirohemes and additional iron-sulfur clusters, possibly four of the [4Fe-4S] type (9). By sequence alignment of different siroheme [4Fe-4S]-binding proteins, Dahl et al. identified highly conserved cysteine-containing clusters (C-X5-C)-Xn-(C-X3-C) proposed to coordinate siroheme-[4Fe-4S] binding (9). This arrangement was also present in the predicted DsrA and DsrB amino acid sequence from B. wadsworthia, which indicates the same content of siroheme. Analogous to DsrB from A. fulgidus (9) and D. vulgaris (19), the first Cys residue of this motif in DsrB is replaced by a Thr residue (Fig. 2), so that the α subunit is most likely to bind siroheme, with no binding of the β subunit. Crane et al. showed that based on the crystallographic structure of E. coli sulfite reductase hemoprotein (8), key residues important for stability and function of siroheme-containing sulfite and nitrite reductases are clustered in five homology regions, H1 to H5 (7). As has been shown for the α and β subunits of DSR from D. vulgaris (7), the corresponding subunits of the B. wadsworthia enzyme also contained conserved sequences belonging to the homology regions.
The codon usage of the dsr genes of B. wadsworthia is very similar to the codon usage of the dsr genes of D. vulgaris, e.g., the arginine codons in B. wadsworthia are, as in D. vulgaris (19), almost exclusively CGT or CGC (41 of 43 codons). This is also reflected by the nearly identical G+C content of these genes (60.4% for D. vulgaris [19] and 59.8% for B. wadsworthia). B. wadsworthia differs in the prevalence of GAA for glutamate (65 of 70 codons) from D. vulgaris, which utilizes the codons GAA and GAG (36 and 24 of 60 codons, respectively).
The similar codon usage is perhaps not surprising, because B. wadsworthia is phylogenetically a member of the Desulfovibrionaceae, and its nearest defined neighbor is D. desulfuricansT (25). The phylogeny is based on the similarity of the 16S rDNA sequences, which is not reflected in the published G+C contents of these bacteria, 59% for D. desulfuricansT (46) and 40% for B. wadsworthiaT (2). The latter value was obtained when cells had to be harvested from plates and worked up for melting points (2), whereas the recent discovery of ready growth in liquid culture facilitates high growth yields of metabolically active cells (22, 23), from which DNA was separated and hydrolyzed, and the monomers were subjected to chromatographic separation and determination. The newly obtained value (59.2%) for the G+C content corresponds to the G+C content of the B. wadsworthia genes sequenced in this work and elsewhere (22, 23) and is consistent with the values for the family Desulfovibrionaceae, e.g., 59% for D. desulfuricans and 66% for D. vulgaris (46).
One might speculate that B. wadsworthia, which does not reduce sulfate, was once a sulfate reducer and that the capacity for sulfate reduction was lost. The coupling of putative sulfite generation from organosulfonates with energy conservation involving DSR is well known in Desulfovibrio spp. (24, 29, 30) as it is in B. wadsworthia (25). What probably distinguishes most Desulfovibrio spp. from B. wadsworthia is clinical importance (2, 11, 39, 41), though recent papers suggest pathogenic roles for some Desulfovibrio spp. (31, 43).
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (H.L.) and the Max Planck Society (M.F.).
M. Claros (Leipzig) kindly made B. wadsworthiaT available. We thank K. Sulger (Konstanz) for providing purified desulfoviridin from D. vulgaris, L. Cobianchi (Konstanz) for N-terminal sequencing, B. Wagner (Marburg) for excellent technical assistance, and J. Fritz-Steuber (Zürich), U. Schumacher (Tübingen), and C. Kisker (Stony Brook) for valuable discussions.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley; 1987. [Google Scholar]
- 2.Baron E J, Summanen P, Downes J, Roberts M C, Wexler H, Finegold S M. Bilophila wadsworthia, gen. nov. and sp. nov., a unique gram-negative anaerobic rod recovered from appendicitis specimens and human faeces. J Gen Microbiol. 1989;135:3405–3411. doi: 10.1099/00221287-135-12-3405. [DOI] [PubMed] [Google Scholar]
- 3.Benson O D A, Karsch-Mizrachi I, Lipman D J, Ostell J, Rapp B A, Wheeler D L. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Cline J D. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- 6.Cook A M, Laue H, Junker F. Microbial desulfonation. FEMS Microbiol Rev. 1999;22:399–419. doi: 10.1111/j.1574-6976.1998.tb00378.x. [DOI] [PubMed] [Google Scholar]
- 7.Crane B R, Getzoff E D. The relationship between structure and function for the sulfite reductases. Curr Opin Struct Biol. 1996;6:744–756. doi: 10.1016/s0959-440x(96)80003-0. [DOI] [PubMed] [Google Scholar]
- 8.Crane B R, Siegel L M, Getzoff E D. Sulfite reductase structure at 1.6 Å: evolution and catalysis for reduction of inorganic anions. Science. 1995;270:59–67. doi: 10.1126/science.270.5233.59. [DOI] [PubMed] [Google Scholar]
- 9.Dahl C, Kredich N M, Deutzmann R, Trüper H G. Dissimilatory sulphite reductase from Archaeoglobus fulgidus: physico-chemical properties of the enzyme and cloning, sequencing and analysis of the reductase genes. J Gen Microbiol. 1993;139:1817–1828. doi: 10.1099/00221287-139-8-1817. [DOI] [PubMed] [Google Scholar]
- 10.Denger K, Laue H, Cook A M. Anaerobic taurine oxidation: a novel reaction by a nitrate-reducing Alcaligenes sp. Microbiology (Reading UK) 1997;143:1919–1924. doi: 10.1099/00221287-143-6-1919. [DOI] [PubMed] [Google Scholar]
- 11.Finegold S, Jousimies-Somer H. Recently described clinically important anaerobic bacteria: medical aspects. Clin Infect Dis. 1997;25:S88–S93. doi: 10.1086/516237. [DOI] [PubMed] [Google Scholar]
- 12.Harrison G, Curle C, Laishley E J. Purification and characterization of an inducible dissimilatory type sulfite reductase from Clostridium pasteurianum. Arch Microbiol. 1984;138:72–78. doi: 10.1007/BF00425411. [DOI] [PubMed] [Google Scholar]
- 13.Heunisch G W. Stoichiometry of the reaction of sulfites with hydrogen sulfide ion. Inorg Chem. 1976;16:1411–1413. [Google Scholar]
- 14.Hipp W M, Pott A S, Thum-Schmitz N, Faath I, Dahl C, Trüper H G. Towards the phylogeny of APS reductases and sirohaem sulfite reductases in sulfate-reducing and sulfur-oxidizing prokaryotes. Microbiology (Reading UK) 1997;143:2891–2902. doi: 10.1099/00221287-143-9-2891. [DOI] [PubMed] [Google Scholar]
- 15.Hittel D S, Voordouw G. Overexpression, purification and immunodetection of DsrD from Desulfovibrio vulgaris Hildenborough. Antonie van Leeuwenhoek. 2000;77:271–280. doi: 10.1023/a:1002449227469. [DOI] [PubMed] [Google Scholar]
- 16.Huang C J, Barret E L. Sequence analysis and expression of the Salmonella typhimurium asr operon encoding production of hydrogen sulfide from sulfite. J Bacteriol. 1991;173:1544–1553. doi: 10.1128/jb.173.4.1544-1553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Junker F, Leisinger T, Cook A M. 3-Sulphocatechol 2,3-dioxygenase and other dioxygenases (EC 1.13.11.2 and EC 1.14.12.-) in the degradative pathways of 2-aminobenzenesulphonic, benzenesulphonic and 4-toluenesulphonic acids in Alcaligenes sp. strain O-1. Microbiology (Reading UK) 1994;140:1713–1722. doi: 10.1099/13500872-140-7-1713. [DOI] [PubMed] [Google Scholar]
- 18.Karkhoff-Schweizer R R, Bruschi M, Voordouw G. Expression of the γ-subunit genes of desulfoviridin-type dissimilatory sulfite reductase and of the α- and β-subunit genes is not coordinately regulated. Eur J Biochem. 1993;211:501–507. doi: 10.1111/j.1432-1033.1993.tb17576.x. [DOI] [PubMed] [Google Scholar]
- 19.Karkhoff-Schweizer R R, Huber D P W, Voordouw G. Conservation of the genes for dissimilatory sulfite reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus allows their detection by PCR. Appl Environ Microbiol. 1995;61:290–296. doi: 10.1128/aem.61.1.290-296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Larsen O, Lien T, Birkeland N K. Dissimilatory sulfite reductase from Archaeoglobus profundus and Desulfotomaculum thermocisternum: phylogenetic and structural implication from gene sequences. Extremophiles. 1999;3:63–70. doi: 10.1007/s007920050100. [DOI] [PubMed] [Google Scholar]
- 22.Laue H, Cook A M. Biochemical and molecular characterization of taurine:pyruvate aminotransferase from the anaerobe Bilophila wadsworthia. Eur J Biochem. 2000;267:6847–6848. doi: 10.1046/j.1432-1033.2000.01782.x. [DOI] [PubMed] [Google Scholar]
- 23.Laue H, Cook A M. Purification, properties and primary structure of alanine dehydrogenase involved in taurine metabolism in the anaerobe Bilophila wadsworthia. Arch Microbiol. 2000;174:162–167. doi: 10.1007/s002030000190. [DOI] [PubMed] [Google Scholar]
- 24.Laue H, Denger K, Cook A M. Fermentation of cysteate by a sulfate-reducing bacterium. Arch Microbiol. 1997;168:210–214. doi: 10.1007/s002030050502. [DOI] [PubMed] [Google Scholar]
- 25.Laue H, Denger K, Cook A M. Taurine reduction in anaerobic respiration of Bilophila wadsworthia RZATAU. Appl Environ Microbiol. 1997;63:2016–2021. doi: 10.1128/aem.63.5.2016-2021.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laue H, Field J A, Cook A M. Bacterial desulfonation of the ethanesulfonate metabolite of the chloroacetanilide herbicide metazachlor. Environ Sci Technol. 1996;30:1129–1132. [Google Scholar]
- 27.Lee J-P, Peck H D. Purification of the enzyme reducing bisulfite to trithionate and its identification as desulfoviridin. Biochem Biophys Res Commun. 1971;45:583–589. doi: 10.1016/0006-291x(71)90457-8. [DOI] [PubMed] [Google Scholar]
- 28.Lee J-P, LeGall J, Peck H D. Isolation of assimilatory- and dissimilatory-type sulfite reductases from Desulfovibrio vulgaris. J Bacteriol. 1973;115:529–542. doi: 10.1128/jb.115.2.529-542.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lie T J, Leadbetter J R, Leadbetter E R. Metabolism of sulfonic acids and other organosulfur compounds by sulfate-reducing bacteria. Geomicrobiol J. 1998;15:135–149. [Google Scholar]
- 30.Lie T J, Pitta T, Leadbetter E R, Godchaux III W, Leadbetter J R. Sulfonates: novel electron acceptors in anaerobic respiration. Arch Microbiol. 1996;166:204–210. doi: 10.1007/s002030050376. [DOI] [PubMed] [Google Scholar]
- 31.Loubinoux J, Mory F, Pereira I A C, Le Faou A E. Bacteremia caused by a strain of Desulfovibrio related to the provisionally named Desulfovibrio fairfieldensis. J Clin Microbiol. 2000;38:931–934. doi: 10.1128/jcm.38.2.931-934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizuno N, Hittel D S, Miki K, Voordouw G, Higuchi Y. Preliminary X-ray crystallographic study of DsrD protein from the sulfate-reducing bacterium Desulfovibrio vulgaris. Acta Crystallogr Sect D Biol Crystallogr. 2000;56:754–755. doi: 10.1107/s0907444900004327. [DOI] [PubMed] [Google Scholar]
- 33.Molitor M, Dahl C, Molitor I, Schäfer U, Speich N, Huber R, Deutzmann R, Trüper H G. A dissimilatory sirohaem-sulfite reductase-type protein from the hyperthermophilic archaeon Pyrobaculum islandicum. Microbiology (Reading UK) 1998;144:529–541. doi: 10.1099/00221287-144-2-529. [DOI] [PubMed] [Google Scholar]
- 34.Mosca A, D'Alagni M, Del Prete R, De Michele G P, Summanen P H, Finegold S M, Miragliotta G. Preliminary evidence of endotoxic activity of Bilophila wadsworthia. Anaerobe. 1995;1:21–24. doi: 10.1016/s1075-9964(95)80379-3. [DOI] [PubMed] [Google Scholar]
- 35.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie brilliant blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 36.Pierik A J, Duyvis G, van Helvoort J M L M, Wolbert R B G, Hagen W R. The third subunit of desulfoviridin-type dissimilatory sulfite reductases. Eur J Biochem. 1992;205:111–115. doi: 10.1111/j.1432-1033.1992.tb16757.x. [DOI] [PubMed] [Google Scholar]
- 37.Postgate J R. A diagnostic reaction of Desulphovibrio desulphuricans. Nature (London) 1959;183:481–482. doi: 10.1038/183481b0. [DOI] [PubMed] [Google Scholar]
- 38.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 39.Schumacher U K, Bless D, Plinkert P K, Laue H, Werner H. Bilophila wadsworthia in ear infections: report of three cases. Anaerobe. 1999;5:371–372. [Google Scholar]
- 40.Steuber J, Arendsen A F, Hagen W R, Kroneck P M H. Molecular properties of the dissimilatory sulfite reductase from Desulfovibrio desulfuricans (Essex) and comparison with the enzyme from Desulfovibrio vulgaris (Hildenborough) Eur J Biochem. 1995;233:873–879. doi: 10.1111/j.1432-1033.1995.873_3.x. [DOI] [PubMed] [Google Scholar]
- 41.Summanen P H, Jousimies-Somer H, Manley S, Bruckner D, Marina M, Goldstein E J C, Finegold S M. Bilophila wadsworthia isolates from clinical specimens. Clin Infect Dis. 1995;20(Suppl. 2):S279–S282. doi: 10.1093/clinids/20.supplement_2.s210. [DOI] [PubMed] [Google Scholar]
- 42.Takase I, Ishino F, Wachi M, Kamata H, Doi M, Asoh S, Matsuzawa H, Ohta T, Matsuhashi M. Genes encoding two lipoproteins in the leuS-dacA region of the Escherichia coli chromosome. J Bacteriol. 1987;169:5692–5699. doi: 10.1128/jb.169.12.5692-5699.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tee W, Dyall-Smith M, Woods W, Eisen D. Probable new species of Desulfovibrio isolated from a pyogenic liver abscess. J Clin Microbiol. 1996;34:1760–1764. doi: 10.1128/jcm.34.7.1760-1764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner M, Roger A J, Flax J L, Brusseau G A, Stahl D A. Phylogeny of dissimilatory sulfite reductase supports an early origin of sulfate respiration. J Bacteriol. 1998;180:2975–2982. doi: 10.1128/jb.180.11.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. IV. New York, N.Y: Springer; 1992. pp. 3352–3378. [Google Scholar]
- 47.Widdel F, Hansen T A. The dissimilatory sulfate- and sulfur-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. I. New York, N.Y: Springer; 1992. pp. 583–624. [Google Scholar]