Abstract
The MexABM efflux pump exports structurally diverse xenobiotics, utilizing the proton electrochemical gradient to confer drug resistance on Pseudomonas aeruginosa. The MexB subunit traverses the inner membrane 12 times and has two, two, and one charged residues in putative transmembrane segments 2 (TMS-2), TMS-4, and TMS-10, respectively. All five residues were mutated, and MexB function was evaluated by determining the MICs of antibiotics and fluorescent dye efflux. Replacement of Lys342 with Ala, Arg, or Glu and Glu346 with Ala, Gln, or Asp in TMS-2 did not have a discernible effect. Ala, Asn, or Lys substitution for Asp407 in TMS-4, which is well conserved, led to loss of activity. Moreover, a mutant with Glu in place of Asp407 exhibited only marginal function, suggesting that the length of the side chain at this position is important. The only replacements for Asp408 in TMS-4 or Lys939 in TMS-10 that exhibited significant function were Glu and Arg, respectively, suggesting that the native charge at these positions is required. In addition, double neutral mutants or mutants in which the charged residues Asp407 and Lys939 or Asp408 and Lys939 were interchanged completely lost function. An Asp408→Glu/Lys939→Arg mutant retained significant activity, while an Asp407→Glu/Lys939→Arg mutant exhibited only marginal function. An Asp407→Glu/Asp408→Glu double mutant also lost activity, but significant function was restored by replacing Lys939 with Arg (Asp407→Glu/Asp408→Glu/Lys939→Arg). Taken as a whole, the findings indicate that Asp407, Asp408, and Lys939 are functionally important and raise the possibility that Asp407, Asp408, and Lys939 may form a charge network between TMS-4 and TMS-10 that is important for proton translocation and/or energy coupling.
Emergence of infectious agents and neoplastic cells resistant to chemically and functionally diverse chemotherapeutic agents is increasingly problematic in human health. An important factor contributing to this low specific drug resistance is the xenobiotic or drug efflux pump, which exports incoming chemotherapeutic agents across the membranes, thereby lowering the intracellular drug concentration. Xenobiotic efflux pumps seem to be ubiquitous in most, if not all, living organisms from bacteria to mammals, suggesting that the pumps function as a fundamental cellular defense apparatus (19, 28). When xenobiotic efflux pumps are expressed in bacteria, the cells gain resistance to structurally and functionally diverse compounds, including antibiotics and synthetic chemotherapeutic agents (9, 14, 17, 31).
Pseudomonas aeruginosa is a hospital pathogen that often infects immunocompromised patients and exhibits resistance to a broad spectrum of antibiotics. This multiantibiotic resistance is largely attributable to low outer membrane permeability and the function of multidrug efflux pumps (14, 38). Four operons encoding a multidrug transporter have been reported in P. aeruginosa (8, 10, 12, 13, 21, 22). Among these, the MexABM pump is the only one expressed in the wild-type strain (13, 22, 37). Upon mutation of the regulatory gene nalB or mexR, the MexABM pump was overexpressed and the bacterium became more resistant than the wild-type strain to the same spectrum of antibiotics (11, 23, 24, 30). Therefore, the MexABM pump plays a central role in both basal and elevated levels of intrinsic multidrug resistance in P. aeruginosa.
The MexABM pump consists of three subunit proteins. MexB is located in the inner membrane and belongs to the hydrophobe–amphiphile efflux 1 (HAE-1) pump family of the resistance-nodulation-cell division (RND) superfamily (2, 19, 32). MexA is a periplasmic protein anchored to the inner membrane via a fatty acid attached to an amino-terminal cysteine residue that belongs to the membrane fusion protein family. We reported recently that delipidated MexA functions properly without anchoring to the membrane (35). MexM (MexM is a new designation for OprM) is an outer membrane-associated protein which had been assumed to form the antibiotic diffusion path across the outer membrane (15, 16). However, we reported recently that MexM is a periplasmic lipoprotein anchored to the outer membrane (15). Delipidated MexM showed pump function indistinguishable from that of the wild-type protein. Thus, it seems less likely that MexM alone forms the transmembrane xenobiotic exit channel.
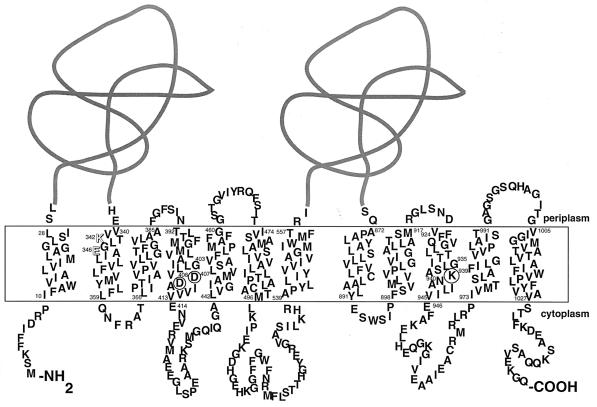
MexB consists of 12 transmembrane segments (TMS) with an inside orientation of both amino and carboxyl termini and has two large hydrophilic periplasmic domains between TMS-1 and -2, as well as TMS-7 and -8 (see Fig. 1) (5). Like many other polytopic membrane proteins, this pump protein was also found to have internal tandem repeats (1, 5, 19, 25, 29). Its transmembrane domains contain five charged amino acid residues, K342 and E346 (TMS-2), D407 and D408 (TMS-4), and K939 (TMS-10) (see Fig. 1). D407, D408, and K939 may be located at about the same level in the membrane and in the corresponding positions in the two repeats (see Fig. 1) (5). Since charged amino acid residues in TMS were observed to play important roles in proton translocation, substrate transport and/or protein biogenesis in many transporters, including multidrug transporters; it is likely that these charged residues in MexB also play important functional roles (3, 4, 6, 20, 26, 27, 33, 34). To obtain insight into this issue, we employed site-directed mutagenesis of all five charged residues and analyzed the function of MexB mutants.
FIG. 1.
Schematic representation of the two-dimensional structure of MexB. The membrane topology and the extramembrane loops were drawn on the basis of results reported previously (5). Two charged residues in TMS-2 are shown as white letters, and three charged residues in TMS-4 and TMS-10 are circled.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains used for site-directed mutagenesis were BMH 71-18 [mutS thi supE Δ(lac-proAB) mutS::Tn10 F′ proA+B+ lacIq ZΔM15] and JM109 [e14− (McrA−) endA1 recA1 gyrA96 thi hsdR17 (rK−mK+) relA1 supE44 λ− Δ(lac-proAB) F′ traD36 proA+B+ lacIq ZΔM15]. We tested the function of MexB mutants in P. aeruginosa TNP071 lacking mexB, a derivative of PAO4290 (37). Plasmid pBSK-mexB carrying wild-type mexB was used as the template for site-directed mutagenesis. Low-copy-number shuttle vector pMMB67EH and its derivative pMEXB1 with wild-type mexB were used to subclone and express mexB-encoded mutant proteins in TNP071 (36).
Site-directed mutagenesis.
Mutants were constructed in vitro with the GeneEditor site-directed mutagenesis system (Promega) using plasmid pBSK-mexB. The sequences of the mutagenic primer used are available upon request. The transformant selected by GeneEditor Antibiotic-Selection-Mix was purified and confirmed to carry the expected mutation by restriction mapping and DNA sequencing. For most of the mutants, an about 1.5-kb BsiwI-MfeI fragment was sequenced to make sure that there was no additional mutation and replaced with the BsiwI-MfeI fragment in pMEXB1. All of mutant mexB in plasmid pMEX-K939R, pMEX-K939A, pMEX-K939E, or pMEX-K939D was sequenced, and the about 3.9-kb EcoRI-XhoI fragment containing all of mexB was subcloned into pMMB67EH treated with EcoRI and SalI.
Double mutants (pMEX-D407K/K939D, pMEX-D408K/K939D, pMEX-D407E/K939R, pMEX-D408E/K939R, pMEX-D407A/K939A, and pMEX-D408A/K939A) and a triple mutant (pMEX-D407E/D408E/K939R) were constructed by replacing the mutant BsiwI-MfeI fragment with the wild-type fragment as described above.
Determination of MICs of antibiotics.
MICs of antibiotic were determined by the agar double-dilution method in the presence of 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Plates were incubated at 37°C for 18 h.
Fluorescent dye extrusion experiment.
Quantitative determination of pump function was carried out by the real-time fluorescent dye extrusion method as described elsewhere (18). Briefly, cells were grown at 37°C for 2 h while rotating at 200 rpm after 100-fold dilution of a fully grown preculture and incubated in the presence of 2 mM IPTG for an additional 2 h. Cells were harvested by centrifugation, washed once with 100 mM NaCl–50 mM sodium phosphate buffer (pH 7.0), and suspended to an A600 of 0.1 in the same solution containing 0.05% glycerol. Fluorescence intensity was measured with a JASCO FP-777 spectrofluorometer. The excitation and emission wavelengths for 2-(4-dimethylaminostyryl)-1-ethylpyridinium (DMP) were set to 480 and 560 nm, respectively, and those for ethidium-bromide (EtBr) were 520 and 590 nm, respectively. Slit widths for excitation and emission for DMP were both 10 nm, and those for EtBr were 5 and 10 nm, respectively.
Expression of mutant proteins.
The inner membrane fraction was prepared as described elsewhere (13). Sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and Western blotting were carried out as described previously. The antibody raised against MexB was used to probe mutant proteins (37).
DNA sequencing.
Nucleotide sequence were determined using the ABI PRISM Dye Terminator Cycle Sequencing Core Kit with ampliTaq DNA polymerase, FS.
RESULTS
MexB has five charged amino acid residues in α-helical TMS. They are D407 and D408 in TMS-4, K939 in TMS-10, and K342 and E346 in TMS-2 (Fig. 1). A total of 29 (19 single, 9 double, and 1 triple) mutant proteins were constructed.
Antibiotic susceptibility of cells carrying mutant MexB.
To evaluate the function of mutant MexB, we determined the MICs of the negatively charged antibiotics aztreonam, novobiocin, and nalidixic acid and the neutral antibiotic chloramphenicol. For mutants lacking the MexB subunit, the MICs of the antibiotics were 4 to 16 times lower than those for the wild-type strain, confirming previous results (36). Introduction of the wild-type mexB gene in plasmid pMEXB1 into the strain lacking chromosomal mexB restored antibiotic resistance to the level of the wild-type strain (Table 1). However, higher antibiotic resistances were not attained as more MexB was expressed from the plasmid. This probably is due to limiting of other pump subunits, such as MexA and/or MexM. Overproduction of a full set of MexABM from the chromosome of a nalB mutant or from a plasmid resulted in MICs of antibiotics severalfold higher than those for the wild-type strain (10, 30).
TABLE 1.
MICs of antibiotics for cells carrying mutant MexB
| Strain | MIC (mg/liter)a
|
|||
|---|---|---|---|---|
| Aztreonam | Novobiocin | Nalidixic acid | Chloramphenicol | |
| Wild-type PAO4290 | 6.25 | >1,600 | 100 | 50 |
| PAO4290ΔB | 0.39 | 100 | 25 | 12.5 |
| PAO4290ΔB/pMMB67EH | 0.39 | 100 | 25 | 12.5 |
| PAO4290ΔB/pMEXB1 | 6.25 | >1,600 | 100 | 100 |
| PAO4290ΔB/pMEX-D407N | 0.39 | 100 | 25 | 12.5 |
| PAO4290ΔB/pMEX-D407A | 0.39 | 100 | 25 | 12.5 |
| PAO4290ΔB/pMEX-D407K | 0.39 | 100 | 25 | 12.5 |
| PAO4290ΔB/pMEX-D407E | 0.78 | 200 | 25 | 12.5∼25 |
| PAO4290ΔB/pMEX-D408N | 0.39 | 100 | 25 | 12.5 |
| PAO4290ΔB/pMEX-D408A | 0.39 | 100 | 25 | 12.5 |
| PAO4290ΔB/pMEX-D408K | 0.39 | 100 | 25 | 12.5 |
| PAO4290ΔB/pMEX-D408E | 3.12 | 400 | 50 | 25∼50 |
| PAO4290ΔB/pMEX-K939A | 0.39 | 100 | 12.5 | 12.5 |
| PAO4290ΔB/pMEX-K939E | 0.39 | 100 | 12.5 | 12.5 |
| PAO4290ΔB/pMEX-K939D | 0.39 | 50 | 12.5 | 12.5 |
| PAO4290ΔB/pMEX-K939R | 6.25 | 1,600 | 100 | 50 |
| PAO4290ΔB/pMEX-D407E/D408E | 0.39 | 100 | 25 | 12.5 |
| PAO4290ΔB/pMEX-D407E/D408E/K939R | 0.78 | 100∼200 | 25 | 12.5∼25 |
| PAO4290ΔB/pMEX-D407E/K939R | 0.78 | 200 | 25 | 25 |
| PAO4290ΔB/pMEX-D407K/K939D | 0.39 | 100 | 25 | 12.5 |
| PAO4290ΔB/pMEX-D407A/K939A | 0.39 | 100 | 25 | 12.5 |
| PAO4290ΔB/pMEX-D408E/K939R | 3.12 | 400 | 50∼100 | 25∼50 |
| PAO4290ΔB/pMEX-D408K/K939D | 0.39 | 100 | 12.5 | 12.5 |
| PAO4290ΔB/pMEX-D408A/K939A | 0.39 | 100 | 12.5 | 12.5 |
| PAO4290ΔB/pMEX-D407R/D408R | 0.39 | 100 | 25 | 12.5 |
| PAO4290ΔB/pMEX-K342A | 6.25 | 1,600 | 100 | 50 |
| PAO4290ΔB/pMEX-K342R | 6.25 | 1,600 | 100 | 100 |
| PAO4290ΔB/pMEX-K342E | 6.25 | 1,600 | 100 | 100 |
| PAO4290ΔB/pMEX-E346Q | 6.25 | 1,600 | 100 | 50 |
| PAO4290ΔB/pMEX-E346A | 6.25 | 1,600 | 100 | 100 |
| PAO4290ΔB/pMEX-E346D | 6.25 | 1,600 | 100 | 100 |
| PAO4290ΔB/pMEX-E346K | 1.56 | 400 | 50 | 25 |
| PAO4290ΔB/pMEX-E346K/K342E | 3.12 | 400∼800 | 50∼100 | 25∼50 |
MICs of antibiotics were determined by the agar dilution method, in which the plate was incubated for 18 h at 37°C. The MICs shown are representative results of several repeated experiments.
D407 or D408 (TMS-4) mutants and K939 (TMS-10) mutants.
For all of the mutants harboring plasmids encoding the mutations D407N, D407A, D407K, D408N, D408A, and D408K, the MICs of the antibiotics were indistinguishable from those for the mutant lacking the MexB subunit (Table 1). The mutant with mutation D407E also exhibited dramatically decreased antibiotic resistance, and the MICs of aztreonam and novobiocin were only twice that resulting from the mexB deletion. However, replacement of D408 with Glu resulted in MICs of antibiotics two to eight times higher than those for the mexB mutant. Substitution of Ala, Glu, or Asp for K939 caused total loss of antibiotic resistance. The mutant with the mutation K939R conserved the resistance indistinguishably from the wild-type strain.
Double and triple mutations.
When D407 and D408 were replaced with Glu simultaneously, pump function was totally abolished. Furthermore, we replaced K939 with Arg in the D407E/D408E background. Interestingly, the triple mutation D407E/D408E/K939R restored the MIC of aztreonam to that for the D407E or D407E/K939R mutant. This result was confirmed repeatedly with three independent constructions. The D407A/K939A and D408A/K939A double mutants completely lost antibiotic resistance. Again, for the D407K/K939D and D408K/K939D mutants the MICs of the antibiotics were similar to those for the strain lacking MexB. The D407E/K939R mutant also exhibited dramatically decreased antibiotic resistance, similar to the D407E single mutant. However, for the D408E/K939R mutant, the MICs of the antibiotics were two to eight times as high as those for the mexB mutant, similar to the D408E single mutant.
K342 or E346 (TMS-2) mutants.
We replaced K342 with Arg, Ala, or Glu and E346 with Gln, Ala, or Asp. The MICs of the antibiotics for each of these mutants were comparable to those for the wild-type strain, suggested that K342 and E346 are replaceable. However, the E346K mutant exhibited antibiotic resistances two to four times lower than those of the wild-type strain. A positive charge in this position was somewhat unfavorable to pump function. The MICs for the K342E/E346K double mutant were between those of the wild-type strain and the E346K mutant, but the presence of two Glu residues at both positions did not affect the antibiotic susceptibilities. One possibility is that K342 and E346 are located in the hydrophilic surface.
Quantitative determination of fluorescent dye accumulation.
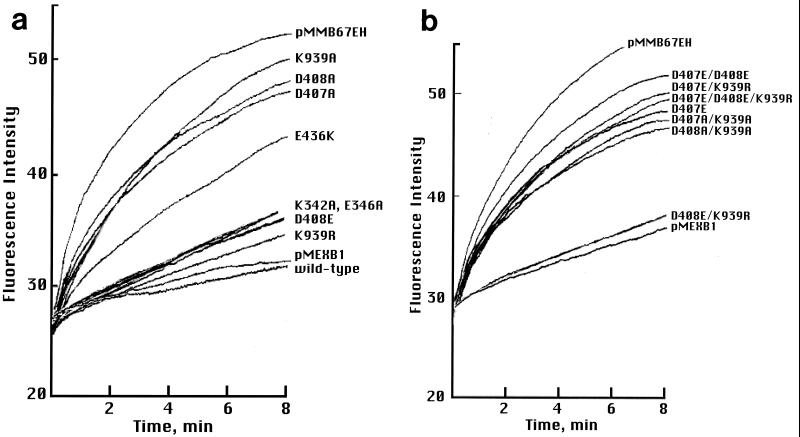
In order to determine the substrate-exporting activity of cells carrying mutant MexB, we quantified the intracellular accumulation of the hydrophobic fluorescent dye DMP and the hydrophilic dye EtBr. The mutant lacking MexB rapidly accumulated the fluorescent dye, indicating that the pump was unable to extrude the dye (18) (Fig. 2a). Both the wild-type strain and the strain carrying pMEXB1 showed lower accumulation curves. Mutants with D407A, D408A, and K939A accumulated the dye in a manner similar to that of the mexB mutant, suggesting that transport activity was abolished in these mutants (Fig. 2a). The mutant with K939R accumulated DMP slowly, in a manner similar to that of the wild-type strain (Fig. 2a). This result confirmed that the uphill efflux activity was preserved in the mutant with K939R and, therefore, the resistance of this mutant to antibiotics is attributable to a functional efflux pump.
FIG. 2.
Fluorescent dye extrusion experiments with cells carrying mutant MexB. Protein was expressed in the presence of 2 mM IPTG for 2 h, harvested, washed, and suspended in buffer solution at an A600 of 0.1. One milliliter of this suspension was mixed with 75 μM DMP, and the fluorescence intensity was recorded immediately. pMEXB1 and pMMB67EH represent experiments using cells expressing wild-type MexB from a plasmid and those carrying the plasmid only, respectively.
The D408E or D408E/K939R mutant showed pump activity close to that of the wild-type strain (Fig. 2a and b). The D407E, D407E/K939R and D407E/D408E/K939R mutants showed DMP accumulation curves similar to that of the D407E/D408E mutant (Fig. 2b), although the MICs of the antibiotics for these mutants showed a little difference. There was no significant difference in extrusion capability between hydrophilic EtBr and hydrophobic DMP in the functional and partially functional mutants (data not shown). In addition, these mutants showed no change in the pattern of resistance to the antibiotics tested.
The K342A, K342R, K342E, E346A, E346Q, and E346D mutants all showed DMP accumulation curves similar to that of the wild-type strain (Fig. 2a), consistent with the MIC results.
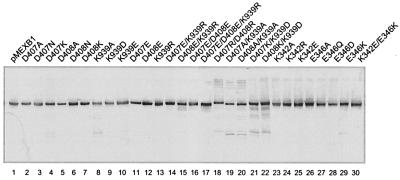
Expression of the mutant protein.
All of the mutant proteins appeared in the inner membrane in amounts comparable to that of the wild-type MexB (Fig. 3). Most of the mutants with a single amino acid substitution exhibited a distinct protein band regardless of pump function (Fig. 3). Besides the major band in K939 mutants, a small amount of breakdown products was detectable regardless of whether the mutants were functional (Fig. 3, lane 15) or dysfunctional (Fig. 3, lanes 8, 10, and 19 to 22), suggesting that a small fraction of these mutant proteins was unstable upon proteolysis. However, this is probably not a major cause of pump dysfunction. In addition, a D407R/D408R mutant had a little lower mobility relative to the others, suggesting that substitution of Arg for both Asp residues may cause a conformational change (Fig. 3, lane 18).
FIG. 3.
Western blotting analysis of mutant MexB. Sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis was run with 10 μg of inner membrane protein prepared by solubilizing the crude membrane fraction with 0.8% lauryl sarcosine. The total envelope fraction of the D407E/D408E/K939R mutant (10 μg) was applied. Protein was blotted onto a polyvinylidene difluoride membrane and visualized with an antibody raised against MexB.
DISCUSSION
The MexB subunit of the MexABM efflux pump exports an extremely broad range of substrates, including stereochemically and electrically unrelated xenobiotics. The only properties shared by the substrate compounds are hydrophobicity and amphiphilicity (16, 17). Phylogenic analysis showed that most RND transporters have a transmembrane topology similar to that of MexB and extrude the substrate utilizing the proton motive force (5, 19, 29, 32). Recent in vitro reconstitution studies of two members of this family have confirmed that RND transporters are proton-substrate antiporters (4, 38). However, the mechanism by which the pump couples cellular energy, substrate selection, and transport remained elusive.
We ran a multiple alignment of the full-length amino acid sequences of 39 proteins that belong to the heavy metal efflux and HAE-1 subsets of the RND superfamily. G403XXXD407XXXXXXE414 in TMS-4 of MexB is strictly conserved in all of the sequences aligned. These residues are predicted to line up on the same face of the α-helical TMS, and their strict conservation suggests that they play an important role(s) in a proton translocation that is shared by the family of proteins (Fig. 1). D408 in TMS-4 and K939 in TMS-10 in the MexB protein are only conserved in the HAE-1 family.
Ala replacement at position 407, 408, or 939 inactivated the pump function without decreasing the expression of mutant proteins, indicating that these conserved charges located in the central region of TMS play important roles in pump function. Highly conserved residue E414 is two turns away from D407 and may be located at the boundary of the cytoplasmic side of TMS-4. This residue might also play an important role and should be further investigated. Substitutions of Asn or Lys for D407 and D408 lead to complete loss of extrusion activity. Replacement of K939 with Glu or Asp also inactivated pump function. D407 could not be replaced with Glu, in which the carboxyl-containing side chain is one methylene longer, suggesting that side-chain length at position 407 is important. Pump function was preserved when D408 was replaced with Glu or K939 was replaced with Arg, suggesting that positions 408 and 939 required a negative and a positive charge, respectively. The volume of the side chain may not be essential. The fact that these two positions could tolerate larger side chains also suggested that TMS-4 and TMS-10 are flexible. It was reported that D408 of Ralstonia eutropha CzcA, a member of the RND family, corresponding to D407 of MexB, is essential for proton translocation (4). The role of D407 in MexB might be similar to that of D408 in CzcA of R. eutropha. Thus, it is likely that conserved D407, D408, and K939 in MexB are involved in proton translocation and/or energy coupling.
The D408E/K939R double mutant, where the charge pair was conserved, showed significant pump function, while the D407E/K939R mutant exhibited only marginal function, similar to that of the D407E single mutant. The D407E/D408E double mutant totally lost pump function, but significant activity was restored by replacing K939 with Arg. The D407A/K939A and D408A/K939A double neutral mutants and mutants in which charged residues D407 and K939 or D408 and K939 were interchanged also totally lost pump function. All of these data support the idea that D407, D408, and K939 play an important role(s) in pump function.
Amphiphilic TMS-4 and TMS-10 contained important charged residues, two negative charges in TMS-4, and one positive charge plus one polar residue (N940) in TMS-10. It is possible that these important charges form ionic interactions to stay in the hydrophobic membrane environment. There is no functional restoration upon charge reversal substitution or double neutral mutation. Given that three charges form a charge network, all of the experimental data are not against this assumption. The use of approaches such as site-directed cysteine cross-linking or engineered divalent metal binding sites in conjunction with electron paramagnetic resonance would be a way to study the geometrical relationship between these sites and the interaction of TMS-4 and TMS-10 (7).
ACKNOWLEDGMENTS
We are indebted to Ronald Kaback for critical reading of the manuscript. Thanks are also due to Michael Ehrmann for stimulating discussion.
This work was supported in part by grants from the Ministry of Education, Science, Sport and Culture of Japan, the Ministry of Health and Welfare of Japan under the Microbiological Resistance Program, the Japan Society for Promotion of Science, and the Tokai University School of Medicine Research Project. L.G. was the recipient of a Tokai University School of Medicine research fellowship.
REFERENCES
- 1.Chen C J, Chin J E, Ueda K, Clark D P, Pastan I, Gottesman M M, Roninson I B. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell. 1986;47:381–389. doi: 10.1016/0092-8674(86)90595-7. [DOI] [PubMed] [Google Scholar]
- 2.Dong Q, Mergeay M. Czc/cnr efflux: a three-component chemiosmotic antiport pathway with a 12-transmembrane-helix protein. Mol Microbiol. 1994;14:185–187. doi: 10.1111/j.1365-2958.1994.tb01278.x. [DOI] [PubMed] [Google Scholar]
- 3.Edgar R, Bibi E. A single membrane-embedded negative charge is critical for recognizing positively charged drugs by the Escherichia coli multidrug resistance protein MdfA. EMBO J. 1999;18:822–832. doi: 10.1093/emboj/18.4.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg M, Pribyl T, Juhnke S, Nies D H. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J Biol Chem. 1999;274:26065–26070. doi: 10.1074/jbc.274.37.26065. [DOI] [PubMed] [Google Scholar]
- 5.Guan L, Ehrmann M, Yoneyama H, Nakae T. Membrane topology of the xenobiotic-exporting subunit, MexB, of the MexA,B-OprM extrusion pump in Pseudomonas aeruginosa. J Biol Chem. 1999;274:10517–10522. doi: 10.1074/jbc.274.15.10517. [DOI] [PubMed] [Google Scholar]
- 6.Gupta S S, DeWitt N D, Allen K E, Slayman C W. Evidence for a salt bridge between transmembrane segments 5 and 6 of the yeast plasma-membrane H+-ATPase. J Biol Chem. 1998;273:34328–34334. doi: 10.1074/jbc.273.51.34328. [DOI] [PubMed] [Google Scholar]
- 7.Kaback H R, Voss J, Wu J. Helix packing in polytopic membrane proteins: the lactose permease of Escherichia coli. Curr Opin Struct Biol. 1997;7:537–542. doi: 10.1016/s0959-440x(97)80119-4. [DOI] [PubMed] [Google Scholar]
- 8.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J-C. Cheracterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 9.Ma D, Cook D N, Hearst J E, Nikaido H. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 10.Maseda H, Yoneyama H, Nakae T. Assignment of the substrate-selective subunits of the MexEF-OprN multidrug efflux pump of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2000;44:658–664. doi: 10.1128/aac.44.3.658-664.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2000;44:2242–2246. doi: 10.1128/aac.44.9.2242-2246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:415–417. doi: 10.1128/aac.43.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreshed S R M, Lei Y, Yoneyama H, Nakae T. Expression of genes associated with antibiotic extrusion in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1995;210:356–362. doi: 10.1006/bbrc.1995.1669. [DOI] [PubMed] [Google Scholar]
- 14.Nakae T, Yoshihara E, Yoneyama H. Multiantibiotic resistance caused by active drug extrusion in hospital pathogens. J Infect Chemother. 1997;3:173–183. doi: 10.1007/BF02490031. [DOI] [PubMed] [Google Scholar]
- 15.Nakajima A, Sugimoto Y, Yoneyama H, Nakae T. Localization of the outer membrane subunit OprM of resistance-nodulation-cell division family multicomponent efflux pump in Pseudomonas aeruginosa. J Biol Chem. 2000;275:30064–30068. doi: 10.1074/jbc.M005742200. [DOI] [PubMed] [Google Scholar]
- 16.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikaido H. Multiple antibiotic resistance and efflux. Curr Opin Microbiol. 1998;1:516–523. doi: 10.1016/s1369-5274(98)80083-0. [DOI] [PubMed] [Google Scholar]
- 18.Ocaktan A, Yoneyama H, Nakae T. Use of fluorescence probes to monitor function of the subunit proteins of the MexA-MexB-OprM drug extrusion machinery in Pseudomonas aeruginosa. J Biol Chem. 1997;272:21964–21969. doi: 10.1074/jbc.272.35.21964. [DOI] [PubMed] [Google Scholar]
- 19.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux system. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pebay-Peyroula E, Rummel G, Rosenbusch J P, Landau E M. X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases. Science. 1997;277:1676–1681. doi: 10.1126/science.277.5332.1676. [DOI] [PubMed] [Google Scholar]
- 21.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T. Overexpression of the mexC-mexD-oprJ efflux operion in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 22.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D E, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rella M, Haas D. Resistance of Pseudomonas aeruginosa PAO to nalidixic acid and low levels of β-lactam antibiotics: mapping of chromosomal genes. Antimicrob Agents Chemother. 1982;22:242–249. doi: 10.1128/aac.22.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubin R A, Levy S B, Heinrikson R L, Kezdy F J. Gene duplication in the evolution of the two complementing domains of gram-negative bacterial tetracycline efflux proteins. Gene. 1990;87:7–13. doi: 10.1016/0378-1119(90)90489-e. [DOI] [PubMed] [Google Scholar]
- 26.Sahin-Tóth M, Karlin A, Kaback H R. Unraveling the mechanism of the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 2000;97:10729–10732. doi: 10.1073/pnas.200351797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahin-Tóth M, le Coutre J, Kharabi D, le Maire G, Lee J C, Kaback H R. Characterization of Glu126 and Arg144, two residues that are indispensable for substrate binding in the lactose permease of Escherichia coli. Biochemistry. 1999;38:813–819. doi: 10.1021/bi982200h. [DOI] [PubMed] [Google Scholar]
- 28.Saier M H, Jr, Paulsen I T, Sliwinski M K, Pao S S, Skurray R A, Nikaido H. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 1998;12:265–274. doi: 10.1096/fasebj.12.3.265. [DOI] [PubMed] [Google Scholar]
- 29.Saier M H, Jr, Tam R, Reizer A, Reizer J. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol. 1994;11:841–847. doi: 10.1111/j.1365-2958.1994.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 30.Saito K, Yoneyama H, Nakae T. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol Lett. 1999;179:67–72. doi: 10.1111/j.1574-6968.1999.tb08709.x. [DOI] [PubMed] [Google Scholar]
- 31.Sharom F J. The P-glycoprotein efflux pump: how does it transport drugs? J Membr Biol. 1997;160:161–175. doi: 10.1007/s002329900305. [DOI] [PubMed] [Google Scholar]
- 32.Tseng T T, Gratwick K S, Kollman J, Park D, Nies D H, Goffeau A, Saier M H., Jr The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 33.Yamaguchi A, Akasaka T, Ono N, Someya Y, Nakatani M, Sawai T. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by transposon Tn10. Roles of the aspartyl residues located in the putative transmembrane helices. J Biol Chem. 1992;267:7490–7498. [PubMed] [Google Scholar]
- 34.Yerushalmi H, Schuldiner S. An essential glutamyl residue in EmrE, a multidrug antiporter from Escherichia coli. J Biol Chem. 2000;275:5264–5269. doi: 10.1074/jbc.275.8.5264. [DOI] [PubMed] [Google Scholar]
- 35.Yoneyama H, Maseda H, Kamiguchi H, Nakae T. Function of the membrane fusion protein, MexA, of the MexA, B-OprM efflux pump in Pseudomonas aeruginosa without an anchoring membrane. J Biol Chem. 2000;275:4628–4634. doi: 10.1074/jbc.275.7.4628. [DOI] [PubMed] [Google Scholar]
- 36.Voneyama H, Ocaktan A, Gotoh N, Nishino T, Nakae T. Subunit swapping in the Mex-extrusion pumps in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1998;244:898–902. doi: 10.1006/bbrc.1998.8351. [DOI] [PubMed] [Google Scholar]
- 37.Yoneyama H, Ocaktan A, Tsuda M, Nakae T. The role of mex-gene products in antibiotic extrusion in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1997;233:611–618. doi: 10.1006/bbrc.1997.6506. [DOI] [PubMed] [Google Scholar]
- 38.Zgurskaya H I, Nikaido H. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]