Abstract
Umbelopsis ramanniana is one of the most commonly reported species within the genus and an important oleaginous fungus. The morphology of the species varies remarkably in sporangiospores, columellae and chlamydospores. However, phylogenetic analyses based on ITS and nLSU rDNA had previously shown insufficiency in achieving species level identification in the genus Umbelopsis. In this study, by applying a polyphasic approach involving multi-gene (nSSU, ITS, nLSU, act1, MCM7 and cox1) phylogeny, morphology and maximum growth temperature, U. ramanniana sensu lato was revealed as a polyphyletic group and resolved with five novel taxa, namely U. curvata, U. dura, U. macrospora, U. microsporangia and U. oblongielliptica. Additionally, a key for all currently accepted species in Umbelopsis was also updated.
Keywords: Mucoromycota, Umbelopsidales, Umbelopsidaceae, five new species, taxonomy, molecular phylogeny, maximum growth temperature test
1. Introduction
Umbelopsis ramanniana, a widespread species in the genus Umbelopsis, is a promising oleaginous fungus in biochemistry and biotechnology. The species is well-known for accumulating large amounts of lipids, which makes the species useful in studying the mechanism of lipid biosynthesis [1,2,3] and for the biotransformation of oil [4,5]. Ecologically, U. ramanniana is a typical inhabitant of forest soils [6] and important in biological rehabilitation [7,8,9,10,11]; it is also frequently isolated from rhizospheres of forest plants [12], or as an endophyte of plants [13].
Umbelopsis ramanniana was first described as Mucor ramannianus by Möller [14]. Historically, there has been a long debate about the attribution of the species and position of the genus Umbelopsis in Mucorales [15]. For its similarity with Mortierella isabellina (basionym of U. isabellina), Linnermann [16] transferred the species to Mortierella sect. Pusilla (which was revised to sect. Isabellina by the author in 1969, nom. inval.). Mil’ko [17], however, retained the species in the genus Mucor and introduced a new section Ramannianus. Gams [18] maintained it as a species of Mortierella and proposed the subgenus Micromucor. Von Arx [19] elevated the subgenus to a genus rank as Micromucor in Mucoraceae and treated the species as Micromucor ramannianus. Meyer and Gams [20] combined the genera Umbelopsis and Micromucor based on the results of restriction fragment length polymorphism data of the whole nuclear ribosomal internal transcribed spacer (ITS) region and phylogenetic reconstruction of ITS1; and, therefore, the species was recombined as its current name U. ramanniana.
The species was first reported from pine mycorrhizas collected in Bavaria and Mark Brandenburg of Germany and was proposed with a poorly informative description, i.e., roseate colony, roundish to elongated sporangiospores and two types of chlamydospores [14]. Because of the lack of type information or illustrations, remarkable variations in morphology and biochemistry among strains assigned to this species have been observed in subsequent studies. Consequently, some members with specific features were separated from U. ramanniana. For strains with angular sporangiospores other than ellipsoidal ones in the autonym variety ramanniana, Linnemann [16] introduced a variety angulispora, which was accepted by Chalabuda [21]. Sugiyama et al. [15] found that the ex-neotype strain of the variety angulispora should be identified as U. vinacea and introduced a new species U. angularis for the isolates with angular sporangiospores in the sense of Linnemann. Evans [22] reported that U. ramanniana varied significantly across strains under different cultural conditions and described another variety autotrophica for the strains showing thiamine independence, pale congo-pink colonies and globose sporangiospores as opposed to the thiamine dependence, darker colonies and ellipsoidal sporangiospores of the variety ramanniana. This variety was raised to a species rank as U. autotrophica by Meyer and Gams [20]. A third variety incrustacea was proposed for strains differing from the original variety in the shape and size of sporangiophores, columellae and sporangiospores [21]. This variety is now a doubtful name due to the lack of related strains all over the world [23].
In addition to the variations exhibited by the varieties of U. ramanniana as mentioned above, more morphological, molecular and biochemical variations within the species have been reported. Turner [24] showed that a culture of this species differed from other members in colony color, sporangiophore length, sporangiospore size and chlamydospore shape. Peberdy and Turner [25] pointed out the esterase patterns of different strains varied widely in U. ramanniana, which, however, have little or no correlation with the morphological variations. The genetic divergences on ITS and nuclear large subunit (nLSU) rDNA sequences and chromosomal number and size were observed among isolates of U. ramanniana [15,20,26]. Based on a more comprehensive investigation on variations of sporangiospores, columellae, chlamydospores and sporangiophores, as well as ITS and nLSU rDNA sequences, Ogawa et al. [27,28] proposed that isolates traditionally identified as U. ramanniana were polyphyletic and included at least three morphologically and genetically divergent groups. However, due to little correlation between nuclear rDNA sequences and morphological characteristics, and the lack of robust molecular phylogenies, U. ramanniana has not been redefined and the isolates traditionally identified as this species have not been reclassified.
The present work focuses on re-examining the variation within the species complex and determining the taxonomical status of the cultures in U. ramanniana sensu lato. We collected many more strains of U. ramanniana, especially some well-known ones, such as NRRL 1296, NRRL 5844 and CBS 219.47; performed a comprehensive phylogenetic analysis based on the sequences from six loci; and examined the morphological characters and maximum growth temperatures of the strains. Consequently, a revised circumscription for U. ramanniana is presented here and five new species are recognized from the species complex. Furthermore, combined with our previous studies on Umbelopsis, a key to known species in this genus is also updated.
2. Materials and Methods
2.1. Cultures and Isolation
Details of materials studied are listed in Table S1. Strains from China were isolated by Chen [29] and Wang et al. [23] using the method of Zheng et al. [30] and preserved in the China General Microbial Culture Collection centre, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (CGMCC) and the State Key Laboratory of Mycology, IM, CAS (Um). Others were obtained from the USDA Agricultural Research Culture Collection, Peoria, Illinois, USA (NRRL) and the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands (CBS). Dried cultures for holotypes were deposited in the Herbarium Mycologicum Academiae Sinicae, IM, CAS, Beijing, China (HMAS).
2.2. Media, Cultivation and Morphological Observation
For DNA extraction, strains were cultivated in malt extract (ME: malt extract 2%, peptone 0.1%, and dextrose 2%) for 4–8 days at 20 °C. Isolates were cultivated at 18 °C for 7–14 days on malt extract agar (MEA: malt extract 2% and agar 2%) and cornmeal agar (CMA: cornmeal 2% and agar 2% agar) under natural light for morphophysiological studies [23]. To determine the maximum growth temperature, each strain was tested three times on PDA for 5 days between 25 and 45 °C.
Microscopic observations were conducted with a Zeiss AX10 Imager A2 light microscope using differential interference contrast illumination. Water or Shear’s mounting medium was used for microscopic observation. Description of the sporangial state was based on an integrative observation of all strains within a certain taxon. Capitalized color designations in the descriptions were from Ridgway [31].
2.3. DNA Extraction, Amplification and Sequence Analyses
Total genomic DNA extraction, amplification and sequencing of partial nuclear small subunit (nSSU) rDNA, ITS and D1–D3 region of nLSU rDNA, and the partial γ-actin gene (act1) were conducted according to the protocols described by Wang et al. [23,32]. The partial regions of DNA replication licensing factor (MCM7) and mitochondrial cytochrome c oxidase subunit 1 (cox1) were amplified using the primer pairs Mcm7-8af (or MCM-709f)/MCM7-16r [33,34] and cox1/cox4 [35], respectively. Polymerase chain reaction (PCR) program of the above two loci included an initial denaturation at 94 °C for 5 min, 39 cycles of 94 °C for 1 min, 53 °C for 1 min and 72 °C for 50 s and a final extension of 72 °C for 10 min. DNA sequencing was performed at Majorbio Bio-technology Company Ltd. (Beijing, China) with the PCR primers. Generated sequences were assembled for consensus in Sequencher 4.1.4 (Gene Codes Corp., Ann Arbor, Michigan); then, they were aligned with MAFFT 6.952 [36,37] and optimized manually in BioEdit 7.1.3.0 [38]. Sequence data generated in this study are deposited in GenBank (Table S1).
Phylogenetic analysis based on the nLSU rDNA sequences was performed by using the neighbor-joining (NJ) method executed in MEGA7 [39] with Kimura 2-parameter model. The topology of the trees was assessed by 1000 bootstrap replications. For multi-gene phylogenetic analyses, optimized sequence alignments of nSSU, ITS, nLSU, act1, MCM7 and cox1 were combined with SequenceMatrix1.7.8 [40]. Phylogenetic analyses using the maximum likelihood (ML), maximum parsimony (MP) and strict clock Bayesian inference (BI) algorithms were performed with RAxML8.0.23 [41], MEGA7 [39] and MrBayes v. 3.0b4 [42,43], respectively. The parameters for ML, MP and BI analyses were set following the methods described by Wang et al. [23,32]. Trees were visualized with Figtree [44] and edited in Adobe Illustrator CS4. The node reliability was assessed by no less than 70% of maximum likelihood bootstrap proportion (MLBP) and maximum parsimony bootstrap support value (MPBS) and no less than 95% of Bayesian posterior probability values (BPP) [45].
3. Results
3.1. Molecular Phylogenetic Analyses
In order to investigate the phylogenetic relationships of the strains sequenced in this study with the three subclades recognized by Ogawa et al. [28], the nLSU rDNA sequences of the U. ramanniana and related species determined by Ogawa et al. [28] were retrieved from GenBank and integrated with nLSU sequences of the strains determined in this study for a phylogenetic analysis. The phylogenetic tree (Figure S1) shows that the strains identified as U. ramanniana are polyphyletic and grouped together with U. angularis, U. gibberispora, U. heterosporus, U. swartii, and U. westeae and U. wiegerinckiae, with high bootstrap values. When more strains were added, the three subclades designated by Ogawa et al. [28] were not resolved as three well-supported linages (Figure S1). Six U. ramanniana strains sequenced in this study (CGMCC 3.15777–3.15781 and NRRL 1296) were grouped in a clade together with the strains of subclade I. This clade also includes U. swartii, and U. westeae, but with low bootstrap support (<50%). The strains of the subclade II and subclade III of Ogawa et al. [28] and 13 new isolates employed in this study formed a well-supported (96%) clade together with U. angularis, U. gibberispora, U. heterosporus and U. wiegerinckiae. As mentioned by Ogawa et al. [28], the results indicate that strains of the U. ramanniana complex represent an assemblage of several genetically distinct species, but nLSU rDNA alone is insufficient in resolving the relationships of the strains in this species complex. Therefore, a phylogenetic construction based on multiple genes was performed in this study to clarify cryptic species in the U. ramanniana complex.
Multi-gene phylogenetic analyses were carried out based on the sequence data of six loci generated in this study or retrieved from GenBank (Table S1). A total of 86 isolates representing all currently accepted taxa of Umbelopsis were analyzed with Mortierella minutissima and M. verticillata as outgroups. Six sequence alignments (including gaps) were obtained with 1667 characters in nSSU, 723 in ITS, 1013 in nLSU, 820 in act1, 1032 in MCM7 and 1742 in cox1, respectively. The final combined sequence matrix consists of 6996 characters, including 4915 constant, 777 parsimony uninformative and 1304 parsimony informative characters. The most parsimonious tree resulted in a tree length of 4454 steps, consistency index (CI) of 0.655, retention index (RI) of 0.929 and rescaled consistency index (RC) of 0.608. For the Bayesian inference, the GTR + I + G model was selected for nSSU, act1 and MCM7, GTR + G model for ITS and nLSU and HKY + G model for cox1, respectively. The ML, MP and BI analyses produced similar trees with nodes supported by high bootstrap values. The BI phylogeny tree (Figure 1) is presented with BI, ML and MP bootstrap values indicated along branches.
Figure 1.
Phylogenetic tree for Umbelopsis based on a combined data matrix comprised alignments of nSSU, ITS, nLSU, act1, MCM7 and cox1 generated from Bayesian analyses with Mortierella as outgroups. Values above the branches represent significant Bayesian posterior probability values (BPP ≥ 0.95), and values below the branches are maximum likelihood bootstrap proportion (MLBP ≥ 70%) and maximum parsimony bootstrap support values (MPBS ≥ 70%). Branches in bold indicate strong support (MLBP: 100%, MPBS: 100%, BPP: 1.00). Missing or weakly supported nodes (MLBP < 70%, MPBS < 70% or BPP < 0.95) are denoted by a minus sign “−”. The bar at the lower left indicates 0.02 expected changes per site. The new species are highlighted in green and blue. The sporangiospores of novel species established in this study are illustrated on the right side of the tree (scale bar = 5 µm) and correlated with each clade of the U. ramanniana complex using the same clade numbers. U. = Umbelopsis. T = ex-type strain, ET = ex-epitype strain and HT = ex-holotype strain.
As shown in Figure 1, the strains of the U. ramanniana complex (highlighted in green and blue) can be divided into six well-supported clades (C1–6, Figure 1). Clades C1 and C2 were grouped together with U. angularis, U. gibberispora, U. heterosporus and U. wiegerinckiae, forming a highly supported lineage (BPP: 1.00, MLBP: 99, MPBS: 100). Clades C3 to C6 were grouped together with U. swartii and U. westeae, forming another highly supported lineage (BPP: 1.00, MLBP: 97, MPBS: 100). The clade C1 (BPP: 0.98, MLBP: 80, MPBS < 80), including three isolates (CBS 219.47, CGMCC 3.6647 and CGMCC 3.6648), constituted a grade with U. heterosporus and U. wiegerinckiae. The clade C2 (BPP: 1.00, MLBP: 100, MPBS: 100), including 11 strains, formed a sister clade to U. angularis. Four strains (CGMCC 3.15769, CGMCC 3.15770, CGMCC 3.15771 and CGMCC 3.15782) in the clade C3 formed a strongly supported clade (BPP: 1.00, MLBP: 100, MPBS: 100) in both six-genes and nLSU trees. This clade was grouped together with clades C4 to C6 in six-genes tree (Figure 1), but more closely related to the lineage including clades C1 and C2 in nLSU tree (Figure S1). Although five isolates (CGMCC 3.15777–3.15781) in clade C4 (BPP: 1.00, MLBP: 100, MPBS: 100) and strains NRRL 1296 and NRRL 5844 formed a monophyletic group (BPP: 1.00, MLBP: 100, MPBS: 100) in the six-genes tree, they were divided into three clades for their huge variations in morphology (Figure 1, Table 1) and divergencies in some gene makers. All strains of the clade C4 have an insertion of 1160 bp in the mitochondrial cox1 gene in comparison to NRRL 1296 and NRRL 5844, as well as other Umbelopsis species. This indicates that the clade C4 may possess a unique evolutionary pattern in mitochondrial genes. The two clades represented by strains NRRL 1296 and NRRL 5844 contained one strain each. In the nLSU analysis (Figure S1), the strains NRRL 5844 and YODK 004 formed a strongly supported (84%) clade and were diverged distantly from the cluster including NRRL 1296 and clade C4.
Table 1.
The mainly morphological comparison among Umbelopsis ramanniana complex.
| Species | Colony Diam. (mm) | Sporangial Diam. (µm) | Sporangial Walls DLQ 1 | Collars | Columellae (µm) | Sporangiospores (µm) | Macro-Chlamydospores |
|---|---|---|---|---|---|---|---|
| C1: U. curvata | 43–48 | (11.0–) 15.5–22.5 (–24.5) | rapidly | small but conspicuous | depressed globose (4.0–) 6.0–8.5 (–10) × (3.5–) 5.0–7.0 (–8.5); or subglobose 4.0–6.0 (–7.0) | ellipsoidal and often curving to one side (2.7–) 3.2–4.5 (–5.7) × (1.6–) 2.0–2.4 (–2.8) | abundant on CMA and MEA |
| C2: U. ramanniana | 40–44 | (11.0–) 15.0–23.0 (–29.5) | rapidly | small but conspicuous | ovoid to oblong-ovoid 4.0–6.0 (–8.0) × (4.5–) 5.0–7.0 (–9.5) µm | ellipsoidal 2.4–3.6 (–4.6) × 1.6–2.4 (–2.6) | less abundant on CMA or MEA |
| C3: U. microsporangia | 35–40 | (10.0–) 14.0–18.5 (–22.5) | rapidly | small or no | depressed globose (3.0–) 4.0–6.5 (–10.0) × (2.5–) 3.5–5.5 (–9.0); or subglobose (2.5–) 4.0–6.0 | ovoid to subglobose (2.4–) 2.6–3.2 (–4.0) × 1.8–2.5 (–2.8) | rare on CMA and less abundant on MEA |
| C4: U. dura | 41–45 | (10.0–) 15.0–23.5 (–31.5) | slowly | small or no | roundish conical (2.5–) 3.5–6.0 (–7.0) × (3.5–) 4.5–7.0 (–8.0) | ovoid and sometimes slightly narrowing on one end (2.4–) 2.8–3.3 (–4.6) × (1.6–) 1.9–2.4 (–2.8) | undiscovered on CMA and less abundant on MEA |
| C5: U. oblongielliptica | 60–62 | (11.5–) 15.5–21.5 (–26.0) | rapidly | small but conspicuous | roundish conical 3.5–6.0 × 4–6.5 | oblong-ellipsoidal and sometimes slightly curved to one side (3.2–) 4.0–5.0 (–5.5) × 1.6–2.0 (–2.5) | abundant on CMA and MEA |
| C6: U. macrospora | 46 | (13.5–) 16.0–23.0 (–25.5) | rapidly | small but conspicuous | depressed globose (4.0–) 6.0–8.0 (–10) × (3.5–) 4.0–6.0 (–7.0); or subglobose (3.0–) 5.0–7.0 (–8.0) | oblong-ellipsoidal to ellipsoidal and sometimes slightly curving to one side (3.6–) 4.0–4.9 (–5.5) × (1.7–) 2.4–2.8 (–3.2) | undiscovered on CMA or MEA |
Colony diam. and characteristics of the sporangial state were compared on CMA after 10 days at 18 °C. 1 DLQ = deliquescent.
3.2. Maximum Growth Temperature
A total of 25 strains in the Umbelopsis ramanniana species complex were tested three times for their maximum growth temperature. Detailed results for each culture are presented in Table S2. The maximum growth temperature of those strains varies from 31 °C to 38 °C, which indicates that there may be cryptic species in the species complex. According to above mentioned phylogenetic clades, the maximum growth temperature ranges for the clades C1 to C6 (Figure 1) are 35–36 °C, 33–35 (–36) °C, 31–32 °C, 35 °C, 37 °C and 38 °C, respectively.
3.3. Morphology and Taxonomy
Like in other species of the Umbelopisis, morphology is an important basis for the taxonomy of the U. ramanniana species complex. The following characteristics are of prime importance to the classification of Umbelopsis: colony color, the pattern and length of branches, the type and shape of sporangia, the shape and size of columellae and sporangiospores, and the formation of chlamydospores. In this study, more criteria, such as colony diameter, the deliquescence of sporangial walls and the possession of collars, have been adopted to investigate differences in the U. ramanniana complex. Combined with the results of phylogenetic analyses, the mainly morphological comparison is summarized in Table 1.
Based on the results of the multi-gene phylogeny (Figure 1), morphological comparison (Table 1) and maximum growth temperature test (Table S2), five new species are introduced for the isolates formerly identified as U. ramanniana. These five novel taxa are described here. Meanwhile, U. ramannianus is re-described based on our isolated specimens. Additionally, accompanied by these new members, a diagnostic key for all taxa of Umbelopsis is updated herein.
3.3.1. Umbelopsis curvata Y.N. Wang, X.Y. Liu and R.Y. Zheng, sp. nov.
Fungal Name: FN570527.
Type: The Netherlands, on Lactarius deliciosus, 1947, A. L. van Beverwijk. Holotype HMAS 247509, ex-holotype culture CBS 219.47.
Etymology: curvata referring to the shape of sporangiospores, often curving to one side.
Diagnosis: Umbelopsis curvata (Figure 2) differs from other species by forming ellipsoidal and often curved sporangiospores with (2.7–) 3.2–4.5 (–5.7) × (1.6–) 2.0–2.4 (–2.8) µm.
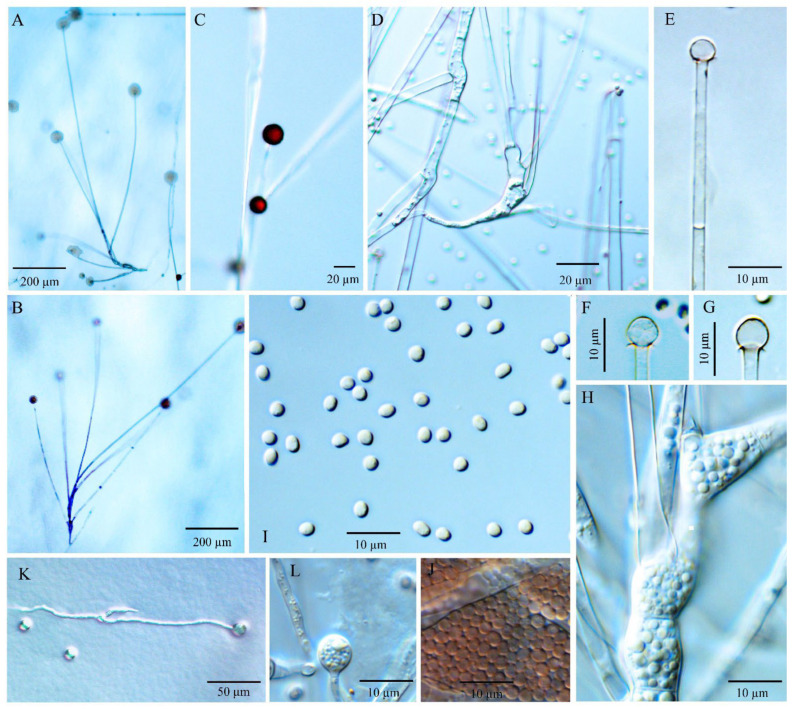
Figure 2.
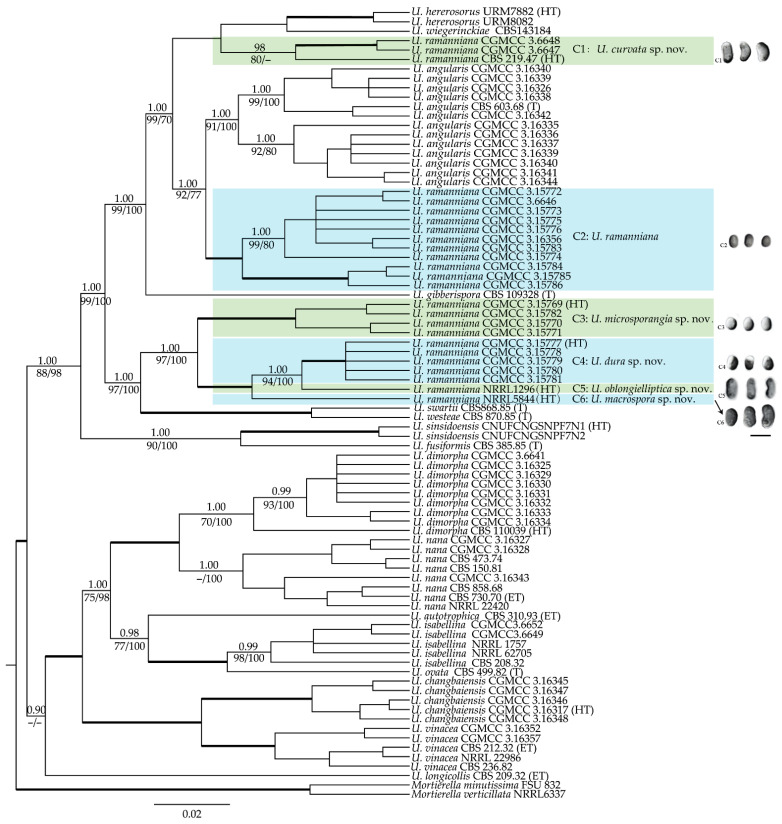
Umbelopsis curvata. (A–C) Branched sporangiophores. (D) Branch point of sporangiophore. (E) Sporangium at tip of sporangiophore. (F–H) Various shapes of collars and columellae at sporangiophore tips after the sporangia have been dissolved. (I, J) Sporangiospores. (K) Macro-chlamydospores. (L) Micro-chlamydospore.
Description: Colonies on MEA reaching 50–60 mm in diam. after 10 days at 18 °C, 2–3 mm high, velvety, slightly zonate, not wrinkled, sometimes forming sectors, at first white, then becoming brownish vinaceous to light russet-vinaceous (Ridgway, Pl. XXXIX) because of abundant sporulation on the obverse side, pinkish cinnamon to cinnamon (Ridgway, Pl. XXIX) on the reverse side; on CMA, reaching 43–48 mm in diam. after 10 days at 18 °C, low, much sparser than on MEA, not zonate, slightly russet, substrate mycelia dense. Sporangiophores abundant, hyaline, smooth, compactly sympodial branching from slightly swollen stalks, (100–) 260–670 (–1000) µm long, (3.5–) 4.0–6.0 (–6.5) µm wide near the base and 2.0–3.5 (–4.0) µm wide near the tip, 2–4 (–6) septate, the uppermost septum at (16.5–) 20–25.5 (–28.5) µm below the columella, often one septum near the base. Sporangia globose, (11.0–) 15.5–22.5 (–24.5) µm in diam., reddish-brown, multi-spored, walls smooth and quickly deliquescent leaving small but conspicuous collars. Columellae hyaline, smooth, small but distinct, mostly depressed globose and (4.0–) 6.0–8.5 (–10) × (3.5–) 5.0–7.0 (–8.5) µm, sometimes subglobose and 4.0–6.0 (–7.0) µm diam. Sporangiospores smooth-walled, ellipsoidal and often curving to one side, (2.7–) 3.2–4.5 (–5.7) × (1.6–) 2.0–2.4 (–2.8) µm, reddish in mass, with small oil droplets. Chlamydospores in substrate hyphae, smooth, containing oil droplets, two types of size: macro-chlamydospores abundant on CMA and MEA, globose to subglobose, (20.0–) 23.5–40.0 (–52.0) µm in diam., yellowish brown, usually intercalary and sometimes at the base of sporangiophore branches, solitary or in short chains; micro-chlamydospores less abundant, subglobose, (4.0–) 5.5–8.0 (–10.5) µm in diam., hyaline, terminal, solitary or in mass. Zygospores unknown.
Maximum growth temperature: 35–36 °C.
Additional strains examined: China, Hubei, Shennongjia, Sister Mountain, dung, 2 Aug 1984, 366a (CGMCC 3.6647) and 373a (CGMCC 3.6648).
Notes: The ex-type culture CBS 219.47 of U. curvata was originally identified as U. ramanniana by Domsch et al. [6] and then illustrated by Meyer and Gams [20]. According to the nLSU phylogeny by Ogawa et al. [28], this culture belonged to subclades III of the U. ramanniana complex. In the present study, multi-gene phylogenetic analyses revealed that U. curvata was distant from the cluster consisting of U. angularis and U. ramannina (Figure 1) and formed a sister cluster to a group that contained U. heterosporus and U. wiegerinckiae. Morphologically, U. curvata differs from U. ramanniana by forming sporangiospores that are larger (3.2–4.5 × 2.0–2.4 µm) and curved; and can be distinguished from U. angularis by not exhibiting angular sporangiospores. Compared to irregular sporangiospores and columellae in U. heterosporus, U. curvata possesses distinct and depressed globose columella and smooth and ellipsoidal sporangiospores. Umbelopsis curvata can be distinguished from U. wiegerinckiae by its pinkish colony and sympodial branching. Additionally, U. curvata is somewhat similar to U. macrospora and U. oblongielliptica in the shape of sporangiospores (ellipsoidal and curving to one side; Table 1), but they are distantly related in molecular phylogeny (Figure 1).
3.3.2. Umbelopsis dura Y.N. Wang, X.Y. Liu and R.Y. Zheng, sp. nov.
Fungal Name: FN570497.
Type: China, Jilin, Changbai Mountain, forest soil mix, 42°10′548″–44°02′274″ N, 126°35′399″–128°55′815″ E, 360–2654 m alt., Jun 2011, Xiao-yong Liu 12448. Holotype HMAS 247505, ex-holotype culture CGMCC 3.15777.
Etymology: dura referring to the permanent nature of sporangial walls, which dissolve slowly and appear stronger than the other species in the U. ramanniana complex.
Diagnosis: Umbelopsis dura (Figure 3) differs from other species due to slowly deliquescent sporangial walls and Indian red to dark vinaceous-brown colonial color.
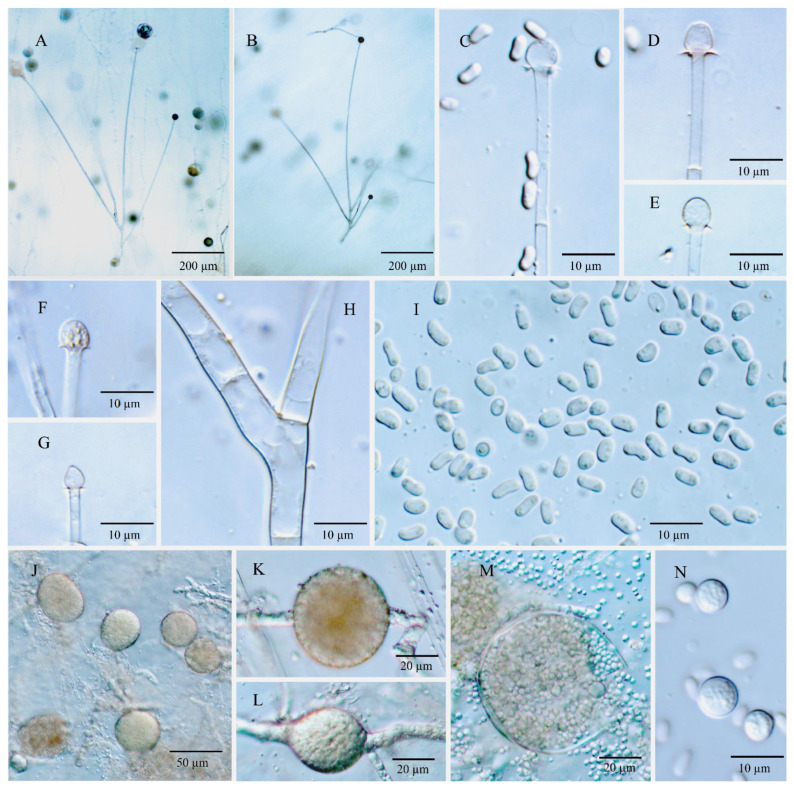
Figure 3.
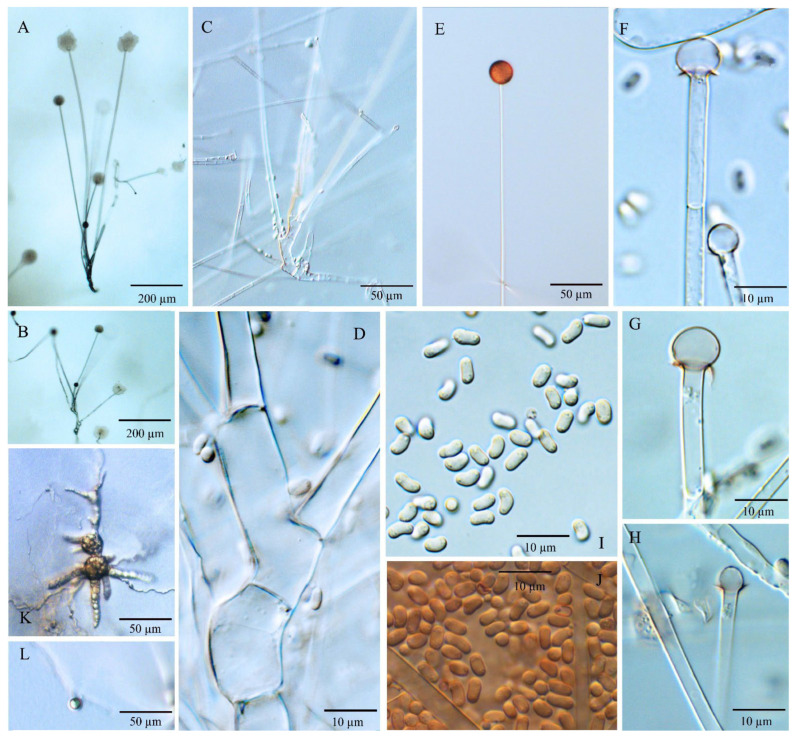
Umbelopsis dura. (A) The main branching pattern of sporangiophore. (B) Branch point of sporangiophore. (C) Sporangium at tip of sporangiophore. (D–H) Various shapes of collars and columellae at sporangiophore tips after the sporangia have been dissolved. (I) Sporangiospores. (J,K) Micro-chlamydospores. (L) Macro-chlamydospore.
Description: Colonies on MEA reaching 50–52 mm in diam. after 10 days at 18 °C, 1 mm high, velvety, zonate, not wrinkled, sometimes forming sectors, at first white, then becoming Indian red (Ridgway, Pl. XXVII) to dark vinaceous-brown (Ridgway, Pl. XXXIX) because of abundant sporulation on the obverse side, pinkish cinnamon to sayal brown (Ridgway, Pl. XXIX) on the reverse side; on CMA, reaching 41–45 mm in diam. after 10 days at 18 °C, low, much sparser than on MEA, zonate, slightly russet, substrate mycelia dense. Sporangiophores abundant, hyaline, smooth, compactly sympodial branching from slightly swollen stalks, (80–) 166–315 (–600) µm long, (2.0–) 4.0–6.0 (–8.0) µm wide near the base and (1.5–) 2.0–4.0 (–5.0) µm wide near the tip, 2–3 (–4) septate, the uppermost septum at (20.0–) 30.5–37.5 (–50.0) µm below the columella, often one septum near the base. Sporangia globose, (10.0–) 15.0–23.5 (–31.5) µm in diam., reddish-brown, multi-spored, walls smooth and slowly deliquescent leaving small or no collars. Columellae hyaline, smooth, small but distinct, depressed globose and (3.5–) 4.0–8.0 (–10.0) × (2.5–) 3.0–7.0 (–9.0) µm, subglobose and (3.0–) 4.0–6.0 (–8.0) µm in diam., or roundish conical and (2.5–) 3.5–6.0 (–7.0) × (3.5–) 4.5–7.0 (–8.0) µm. Sporangiospores smooth-walled, ovoid and sometimes slightly narrowing on one end, (2.4–) 2.8–3.3 (–4.6) × (1.6–) 1.9–2.4 (–2.8) µm, reddish in mass, with small oil droplets. Chlamydospores in substrate hyphae, smooth, containing oil droplets, two types of size: macro-chlamydospores undiscovered on CMA and less abundant on MEA, globose to subglobose, (14.0–) 16.0–23.5 (–27.5) µm in diam., yellowish brown, intercalary, solitary; micro-chlamydospores abundant, subglobose, (3.0–) 5.0–7.5 µm in diam., hyaline, terminal, single or in mass. Zygospores unknown.
Maximum growth temperature: 35 °C.
Additional strains examined: China, Jilin, Changbai Mountain, forest soil mix, 42°10′548″–44°02′274″ N, 126°35′399″–128°55′815″ E, 360–2654 m alt. Jun 2011, Xiao-yong Liu 12,448 CGMCC 3.15778, CGMCC 3.15779, CGMCC 3.15780 and CGMCC 3.15781.
Notes: Umbelopsis dura can be separated from all other Umbelopsis species in the cox1 gene by a significant insertion. Phylogenetic inferences show that this species is closely related to U. oblongielliptica. However, they can be easily distinguished by the characteristics of the growth rate (41–45 mm vs. 60–62 mm in diam. on CMA after 10 days at 18 °C; Table 1), the deliquescence of sporangial walls (slowly vs. quickly) and the shape and size of sporangiospores (ovoid and 2.8–3.3 × 1.9–2.4 µm vs. oblong-ellipsoidal and 4.0–5.0 × 1.6–2.0 µm; Table 1). Although the colony colors of U. dura and U. angularis are somewhat similar, the two species differ in their sporangiospores shape and are distantly related in molecular phylogeny (Figure 1).
3.3.3. Umbelopsis macrospora Y.N. Wang, X.Y. Liu and R.Y. Zheng, sp. nov.
Fungal Name: FN570526.
Type: United Kingdom, associated with Pinus sylvestris. Holotype HMAS 247507, ex-holotype culture NRRL 5844.
Etymology: macrospora referring to the size of its sporangiospores, bigger than other species in the U. ramanniana complex.
Diagnosis: Umbelopsis macrospora (Figure 4) is recognized by forming oblong-ellipsoidal to ellipsoidal sporangiospores with (3.6–) 4.0–4.9 (–5.5) × (1.7–) 2.4–2.8 (–3.2) µm. In addition, it is similar to U. oblongielliptica, but differs in the absence of macro-chlamydospore and the growth rate for colonies.
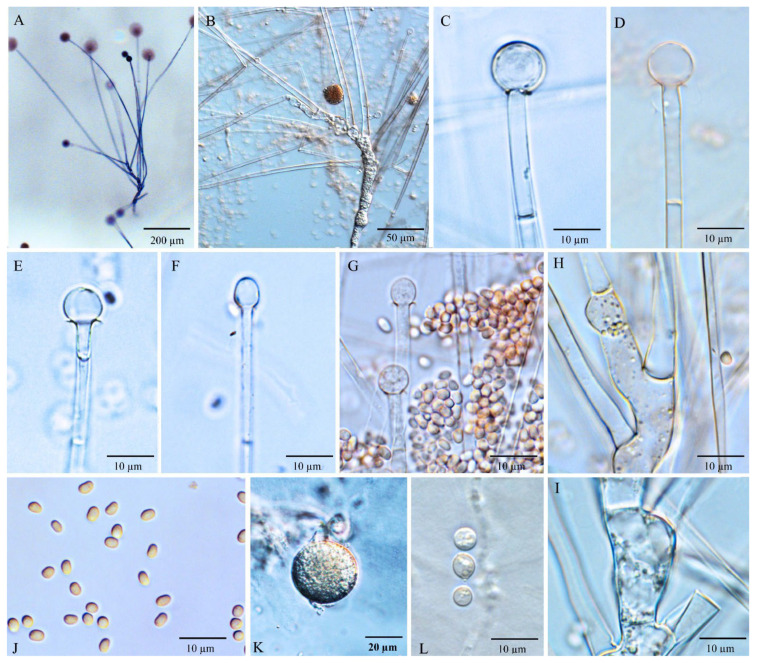
Figure 4.
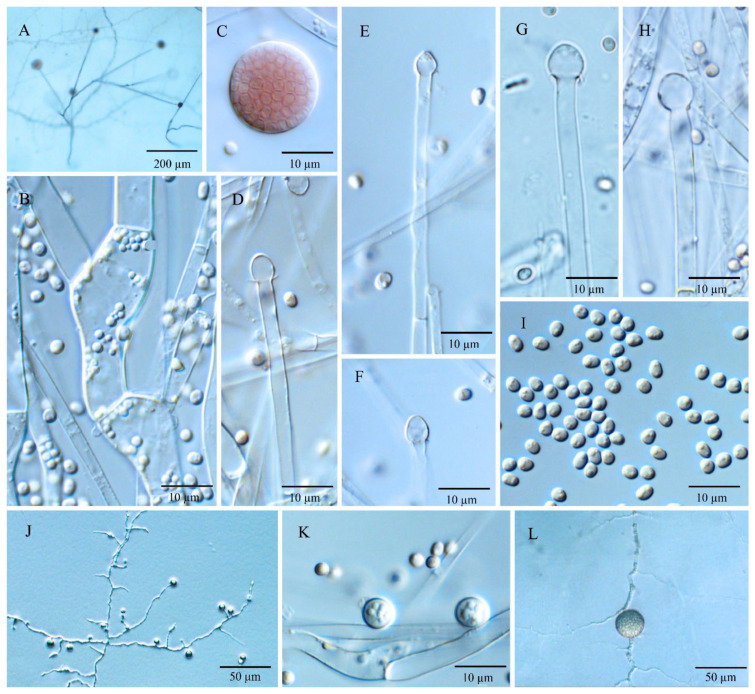
Umbelopsis macrospora. (A,B) Branched sporangiophores. (C) Sporangium at tip of sporangiophore. (D) Branch point of sporangiophore. (E–I) Various shapes of collars and columellae at sporangiophore tips after the sporangia have been dissolved. (J,K) Sporangiospores. (L) Micro-chlamydospores.
Description: Colonies on MEA reaching 62 mm in diam. after 10 days at 18 °C, 1–2 mm high, velvety, zonate, not wrinkled, without sectors, at first white, then becoming russet-vinaceous to vinaceous-brown (Ridgway, Pl. XXXIX) because of abundant sporulation on the obverse side, pinkish cinnamon to cinnamon (Ridgway, Pl. XXIX) on the reverse side; on CMA, reaching 46 mm in diam. after 10 days at 18 °C, low, much sparser than on MEA, not zonate, slightly russet, substrate mycelia dense. Sporangiophores abundant, hyaline, smooth, compactly sympodial branching from slightly swollen stalks, (100–) 215–685 (–900) µm long, (3.5–) 4.3–6.3 (–7.5) µm wide near the base and 2.0–4.0 µm wide near the tip, 3–4 (–6) septate, the uppermost septum at 24.5–39.5 µm below the columella, one septum near the base. Sporangia globose, (13.5–) 16.0–23.0 (–25.5) µm in diam., reddish-brown, multi-spored, walls smooth and quickly deliquescent leaving small but conspicuous collars. Columellae hyaline, smooth, small but distinct, mostly depressed globose and (4.0–) 6.0–8.0 (–10) × (3.5–) 4.0–6.0 (–7.0) µm, sometimes subglobose and (3.0–) 5.0–7.0 (–8.0) µm in diam. Sporangiospores smooth-walled, oblong-ellipsoidal to ellipsoidal and sometimes slightly curving to one side, (3.6–) 4.0–4.9 (–5.5) × (1.7–) 2.4–2.8 (–3.2) µm, reddish in mass, with or without small oil droplets. Chlamydospores in substrate hyphae, smooth, containing oil droplets, one type of size: macro-chlamydospore undiscovered on CMA or MEA; micro-chlamydospores abundant, subglobose, (3.0–) 6.0–8.0 (–10.0) µm in diam., hyaline, terminal, single or in mass. Zygospores unknown.
Maximum growth temperature: 38 °C.
Notes: The only strain NRRL 5844 of U. macrospora was received as U. ramanniana. Previously, the strains NRRL 5884 and NRRL 1296 (the ex-hotype of U. oblongielliptica) were widely used as U. ramanniana in the phylogenetic analyses. However, Nagy et al. [26] reported that the chromosomal banding patterns in those two cultures are significantly diverse in orthogonal field alternation gel electrophoreses (OFAGE) and contour clamped homogeneous electric field gel electrophoreses (CHEF). In the present study, multi-gene phylogenetic inferences show that U. macrospora is basal to the U. oblongielliptica/U. dura cluster (BPP: 1.00; MLBP: 94; MPBS: 100). However, it was separated from this cluster in the nLSU tree (Figure S1). This species is morphologically similar to U. oblongielliptica but differs in the growth rate (46 mm vs. 60–62 mm in diam. after 10 days at 18 °C), sporangiospore size (4.0–4.9 × 2.4–2.8 µm vs. 4.0–5.0 × 1.6–2.0 µm) and macro-chlamydospores (undiscovered vs. abundant). Considering the significant divergences in the nLSU tree (Figure S1), morphological characteristics and chromosomal banding patterns demonstrated by Nagy et al. [26], those two isolates were treated as different species.
According to the results of nLSU phylogeny (Figure S1), strain YODK 004 seems like a second specimen of U. macrospora. However, the sporangiospores size of this culture is 2.2 ± 0.05 × 1.6 ± 0.05 µm, which is smaller than NRRL 5844 (Ogawa et al. 2005). To determine whether this strain belongs to the species, it is necessary to further confirm its morphological characteristics and sequences of other gene loci.
3.3.4. Umbelopsis microsporangia Y.N. Wang, X.Y. Liu and R.Y. Zheng, sp. nov.
Fungal Name: FN570525.
Type: China, Jilin, Changbai Mountain, forest soil mix, 42°10′548″–44°02′274″ N, 126°35′399″–128°55′815″ E, 360–2654 m alt., Jun 2011, Xiao-yong 12448. Holotype HMAS 247506, ex-holotype culture CGMCC 3.15769.
Etymology: microsporangia referring to its smaller sporangia than other species in the U. ramanniana complex.
Diagnosis: Umbelopsis microsporangia (Figure 5) differs from other species due to a relatively low maximum growth temperature of 31–32 °C. It is also characterized by smaller sporangia (10.0–) 14.0–18.5 (–22.5) µm.
Figure 5.
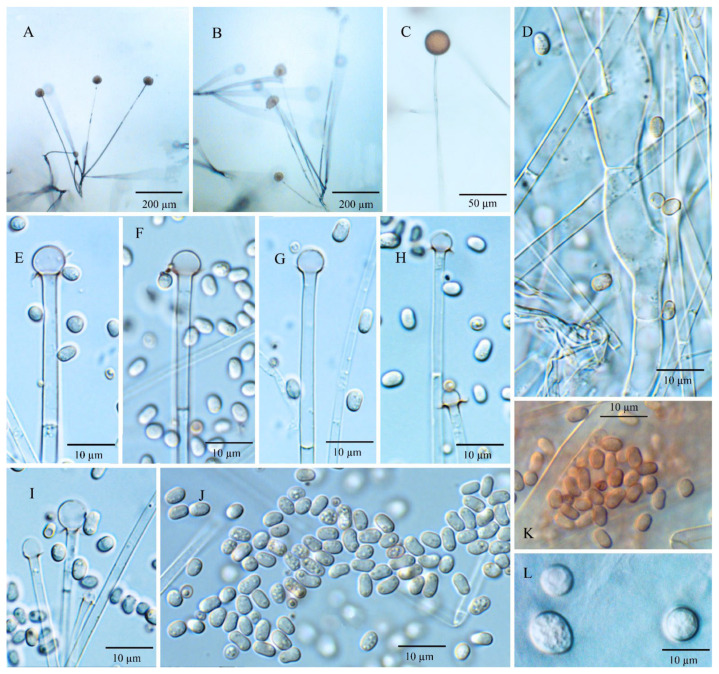
Umbelopsis microsporangia. (A,B) Branched sporangiophores. (C) Sporangium at tip of sporangiophore. (D,H) Branch point of sporangiophore. (E–G) Various shapes of collars and columellae at sporangiophore tips after the sporangia have been dissolved. (I,J) Sporangiospores. (K,L) Micro-chlamydospores.
Description: Colonies on MEA reaching 50–55 mm in diam. after 10 days at 18 °C, 1 mm high, velvety, slightly zonate, not wrinkled, often forming sectors, at first white, then becoming brownish vinaceous (Ridgway, Pl. XXXIX) because of abundant sporulation on the obverse side, light pinkish cinnamon to pinkish cinnamon (Ridgway, Pl. XXIX) on the reverse side; on CMA, reaching 35–40 mm in diam. after 10 days at 18 °C, low, much sparser than on MEA, not zonate, slightly russet, substrate mycelia dense. Sporangiophores abundant, hyaline, smooth, compactly sympodial branching from slightly swollen stalks, (80–) 135–510 (–745) µm long, (2.0–) 3.5–5.0 (–7.0) µm wide near the base and (1.5–) 2.0–3.5 µm wide near the tip, 2–3 (–4) septate, the uppermost septum at (17.5–) 21.0–32.0 (–39.0) µm below the columella, one septum near the base. Sporangia globose, (10.0–) 14.0–18.5 (–22.5) µm in diam., reddish-brown, multi-spored, walls smooth and quickly deliquescent leaving small or no collars. Columellae hyaline, smooth, small but distinct, mostly depressed globose and (3.0–) 4.0–6.5 (–10.0) × (2.5–) 3.5–5.5 (–9.0) µm, sometimes subglobose and (2.5–) 4.0–6.0 µm in diam. Sporangiospores smooth-walled, ovoid to subglobose, uniform in size, (2.4–) 2.6–3.2 (–4.0) × 1.8–2.5 (–2.8) µm, reddish in mass, smooth-walled, without small oil droplets. Chlamydospores in substrate hyphae, smooth, containing oil droplets, two types of size: macro-chlamydospores rare on CMA and less abundant on MEA, globose to subglobose, (9.0–) 16.0–26.0 (–39.5) µm in diam., yellowish brown, usually intercalary and sometimes at the base of sporangiophore branches, solitary; micro-chlamydospores abundant, subglobose, (3.0–) 6.0–8.5 (–12.5) µm in diam., hyaline, terminal, single or in mass. Zygospores unknown.
Maximum growth temperature: 31–32 °C.
Additional strains examined: China, Jilin, Changbai Mountain, forest soil mix, 42°10′548″–44°02′274″ N, 126°35′399″–128°55′815″ E, 360–2654 m alt. Jun 2011, Xiao-yong Liu 12448. CGMCC 3.15770, CGMCC 3.15771 and CGMCC 3.15782.
Notes: Umbelopsis microsporangia formed a strongly supported clade (BPP: 1.00; MLBP: 100%; MPBS: 100%) and occurred as a sister clade of a polytomy consisting of U. dura, U. oblongielliptica and U. macrospora (Figure 1). Morphologically, U. microsporangia produces sporangiospores that are smaller than U. oblongielliptica and U. macrospora (2.6–3.2 × 1.8–2.5 µm vs. 4.0–5.0 × 1.6–2.0 µm and 4.0–4.9 × 2.4–2.8 µm, respectively; Figure 1 and Table 1) and can be distinguished from U. dura by its brownish vinaceous colony color and rapidly deliquescent sporangia. Furthermore, the maximum growth temperature range of the U. microsporangia is 31–32 °C, which is noticeably lower than those of the above mentioned three species (35–38 °C; Table S2).
3.3.5. Umbelopsis oblongielliptica Y.N. Wang, X.Y. Liu and R.Y. Zheng, sp. nov.
Fungal Name: FN570524.
Type: USA, Wisconsin, substrate unknown. Holotype HMAS 247508, ex-holotype culture NRRL 1296.
Etymology: oblongielliptica referring to the shape of sporangiospores.
Diagnosis: Umbelopsis oblongielliptica (Figure 6) differs from other species by forming oblong-ellipsoidal sporangiospores (3.2–) 4.0–5.0 (–5.5) × 1.6–2.0 (–2.5) µm. This species is also characterized by the rapid colony extension (up to 64–70 mm in diameter after 10 d at 18 °C on MEA) and the production of abundant macro-chlamydospores on the substrate of both CMA and MEA.
Figure 6.
Umbelopsis oblongielliptica. (A,B) Branched sporangiophores. (C–G) Various shapes of collars and columellae at sporangiophore tips after the sporangia have been dissolved. (H) Branch point of sporangiophore. (I) Sporangiospores. (J–M) Various macro-chlamydospores. (K). Mature macro-chlamydospores, (L). Immature macro-chlamydospores, (M). Broken macro-chlamydospore spilling oil droplets. (N). Micro-chlamydospores.
Description: Colonies on MEA reaching 64–70 mm in diam. after 10 days at 18 °C, 1 mm high, velvety, zonate, wrinkled or not, sometimes forming sectors, at first white, then becoming russet-vinaceous (Ridgway, Pl. XXXIX) because of abundant sporulation on the obverse side, pinkish cinnamon to brown (Ridgway, Pl. XXIX) on the reverse side; on CMA, reaching 60–62 mm in diam. after 10 days at 18 °C, low, much sparser than on MEA, not zonate, slightly russet, substrate mycelia dense. Sporangiophores abundant, hyaline, smooth, compactly sympodial branching from slightly swollen stalks, (120–) 270–475 (–980) µm long, 3.5–7.0 µm wide near the base and (1.8–) 2.0–4.0 µm wide near the tip, 3–4 (–6) septate, the uppermost septum at 17.5–27.5 µm below the columella, often one septum near the base. Sporangia globose, (11.5–) 15.5–21.5 (–26.0) µm in diam., reddish-brown, multi-spored, walls smooth and quickly deliquescent leaving small but conspicuous collars. Columellae hyaline, smooth, small but distinct, mostly depressed globose and (4.5–) 6.0–8.5 (–10) × (3.0–) 5.0–7.0 (–8.5) µm, sometimes subglobose and 5.0–7.0 µm in diam., occasionally roundish conical and 3.5–6.0 × 4.0–6.5 µm. Sporangiospores smooth-walled, oblong-ellipsoidal and sometimes slightly curved to one side, (3.2–) 4.0–5.0 (–5.5) × 1.6–2.0 (–2.5) µm, reddish in mass, with or without small oil droplets. Chlamydospores in substrate hyphae, smooth, containing oil droplets, two types of size: macro-chlamydospores abundant on CMA and MEA, globose to subglobose, (15.5–) 28.0–39.5 (–47.5) µm in diam., yellowish brown, intercalary, solitary or in chains; micro-chlamydospores abundant, subglobose, (3.0–) 4.0–8.0 (–13.0) µm in diam., hyaline, terminal, single or in mass. Zygospores unknown.
Maximum growth temperature: 37 °C.
Notes: The only culture NRRL 1296 of U. oblongielliptica was received as U. ramanniana. It is phylogenetically closely related to U. dura (BPP: 1.00; MLBP: 100%; MPBS: 100%) and U. macrospora (Figure 1), but morphologically differs from the former species by forming oblong-ellipsoidal sporangiospores (4.0–5.0 × 1.6–2.0 µm; Table 1), and the later one by producing abundant chlamydospores on MEA and narrower sporangiospores (1.6–2.0 µm vs. (1.7–) 2.4–2.8 (–3.2) µm in width; Table 1).
3.3.6. Umbelopsis ramanniana (Möller) W. Gams, Mycol. Res. 107(3): 349 (2003)
-
≡
Mucor ramannianus Möller, Z. Forst- u. Jagdw. 35: 330 (1903)
-
≡
Mortierella ramanniana (Möller) Linnem., Mucor.-Gatt. Mortierella Coem.: 19 (1941)
-
≡
Micromucor ramannianus (Möller) Arx, Sydowia 35: 19 (1984)
Diagnosis: Umbelopsis ramanniana (Figure 7) is recognized by brownish vinaceous to light russet-vinaceous colonies, small but distinct columellae and ellipsoidal sporangiospores with 2.4–3.6 (–4.6) × 1.6–2.4 (–2.6) µm.
Figure 7.
Umbelopsis ramanniana. (A,B) Branched sporangiophores. (C–G) Various shapes of collars and columellae at sporangiophore tips after the sporangia have been dissolved. (H,I) Branch point of sporangiophore. (J) Sporangiospores. (K) Macro-chlamydospores. (L) Micro-chlamydospores.
Description: Colonies on MEA reaching 48–56 mm in diam. after 10 days at 18 °C, 1–2 (–3) mm high, velvety, zonate, not wrinkled, without sectors, at first white, then becoming brownish vinaceous to light russet-vinaceous (Ridgway, Pl. XXXIX) because of abundant sporulation on the obverse side, pinkish cinnamon to cinnamon (Ridgway, Pl. XXIX) on the reverse side; on CMA, reaching 40–44 mm in diam. after 10 days at 18 °C, low, much sparser than on MEA, not zonate, slightly russet, substrate mycelia dense. Sporangiophores abundant, hyaline, smooth, compactly sympodial branching from slightly swollen stalks, (100–) 275–745 (–1275) µm long, (3.0–) 4.0–6.0 (–8.0) µm wide near the base and (1.5–) 2.0–4.0 µm wide near the tip, 2–4 (–7) septate, the uppermost septum at (9.0–) 16.0–26.0 (–37.0) µm below the columella, one septum near the base. Sporangia globose, (11.0–) 15.0–23.0 (–29.5) µm in diam., reddish-brown, multi-spored, walls smooth and quickly deliquescent leaving small but conspicuous collars. Columellae hyaline, smooth, small but distinct, mostly subglobose and (4.0–) 6.0–8.0 (–10.0) µm in diam., sometimes depressed globose and (4.0–) 6.0–10.0 (–13.0) × (3.5) 5.0–8.5 (–12.0) µm, or ovoid to oblong-ovoid and 4.0–6.0 (–8.0) × (4.5–) 5.0–7.0 (–9.5) µm. Sporangiospores smooth-walled, ellipsoidal, 2.4–3.6 (–4.6) × 1.6–2.4 (–2.6) µm, reddish in mass, without small oil droplets. Chlamydospores in substrate hyphae, smooth, containing oil droplets, two types of size: macro-chlamydospores less abundant on CMA or MEA, globose to subglobose, (12.0–) 16.0–30.0 (–44.0) µm in diam., yellowish brown, usually intercalary and seldom at the base of sporangiophore branches, solitary; micro-chlamydospores sparse, subglobose, (3.5–) 6.0–8.0 (–10.0) µm in diam., hyaline, terminal, single. Zygospores unknown.
Maximum growth temperature: 33–35 (–36) °C.
Strains examined: China, Jilin, Changbai Mountain, forest soil mix, 42°10′548″–44°02′274″ N, 126°35′399″–128°55′815″ E, 360–2654 m alt., Jun 2011, Xiao-yong Liu (CGMCC 3.15772, CGMCC 3.15773, CGMCC 3.15774, CGMCC 3.15775 and CGMCC 3.15776); China, Hubei, Shennongjia, Jiuhuping, from Gastrodia elata, 1984, 377a (CGMCC 3.6646); China, Tibet, Lulang, rotten cloth, 5 Aug 2009, Xue-wei Wang (CGMCC 3.15783); China, Hubei, Shennongjia, Nanyinzhai, soil under Fargesia spathacea, Jul 1984, 402b (CGMCC 3.16356); China, Fujian, Wuyishan National Nature Reserve, wild fruits fell on the ground, 27°44′414″ N, 117°41′113″ E, 1232 m alt., 20 Jun 2012, Ya-wing Wang 12,954 (CGMCC 3.15784 and CGMCC 3.15785); China, Fujian, Wuyishan City, soil in grove of bamboo, 20 Jun 2012, Ya-ning Wang 12,983 (CGMCC 3.15786).
Notes: The strains of clade C2 in the multi-gene phylogenetic tree (Figure 1) were characterized by producing ellipsoidal spores, small but distinct columellae and two types of chlamydospores, which is compatible with Möller’s description for U. ramanniana [14]. Moreover, they formed a sister lineage to U. angularis, which was a variant of U. ramanniana in the sense of Linnermann [15,16]. As a result, the strains in clade C2 were regarded as representative of the U. ramanniana in this study.
Umbelopisis ramanniana can be distinguished from U. angularis by having ellipsoidal sporangiospores and a lighter colony color. Umbelopisis curvata (C1, Figure 1) can be distinguished from U. ramanniana (C2, Figure 1) by its columellae shape and the shape and size of sporangiospores (Table 1). In addition, slight morphological variations were observed in several isolates of this clade. The sporangiophores of isolates CGMCC 3.6646 and CGMCC 3.15774 are shorter than other isolates, which may be intraspecific variations caused by differences in geographical environment.
In this species complex, remarkable variations were found in the monophyletic cluster consisting of U. microsporangia, U. dura, U. oblongielliptica and U. macrospora (C3, C4, C5 and C6, respectively; Figure 1). Umbelopisis microsporangia and U. dura produce smaller ovoid sporangiospores; however, some of them are oblong-ellipsoidal to ellipsoidal in U. oblongielliptica and U. macrospora (Figure 1 and Table 1). Compared to U. microsporangia and other taxa in the U. ramanniana complex, the deliquescence of sporangia walls in U. dura is slower. Moreover, the colony diam. after 10 days at 18 °C in the clade U. microsporangia is 35–40 mm, which is 41–45 mm in U. dura. Regarding their morphology, two single strain clades C5 and C6 can be easily distinguished from each other by their colony diam., the shape and size of columellae, sporangiospores size and macro-chlamydospores (abundant vs. undiscovered). Therefore, we treated them as different taxa U. oblongielliptica and U. macrospora.
3.3.7. Key to Species of Umbelopsis
| 1. | Sporangia sometimes with neck-like base | Umbelopsis longicollis |
| – | Sporangia without neck-like base | 2 |
| 2. | Colonies drab grey when the sporangia are produced heavily | 3 |
| – | Colonies roseate to almost white when the sporangia are produced heavily | 4 |
| 3. | Sporangia obovate to ovate | U. ovata |
| – | Sporangia globose to subglobose | U. isabellina |
| 4. | With two kinds of sporangia, single- and multi-spored | U. dimorpha |
| – | With one kind of sporangia, single- or multi-spored | 5 |
| 5. | Colony color white to pale pinkish; sporangia single-spored with 4.5–8.0 µm in diam.; sporangiospores globose with the same size of sporangia | U. nana |
| – | Colony color roseate to pale pinkish; sporangia multi-spored larger than 10 µm; sporangiospore globose to oblong-ellipsoidal far smaller than sporangia | 6 |
| 6. | Sporangiospores globose, maximum growth temperature 40 °C | U. autotrophica |
| – | Sporangiospores subglobose, ovoid to oblong-ellipsoidal, angular or irregular; maximum growth temperature no more than 39 °C | 7 |
| 7. | Sporangiophores no more than 100 µm in length | 8 |
| – | Sporangiophores mainly more than 100 µm in length | 10 |
| 8. | Colonies reaching 20–21 mm in diam. on MEA after 7 days at 20 °C; sporangiospores globose, ellipsoidal or angular | U. sinsidoensis |
| – | Colonies reaching more than 30 mm in diam. on MEA after 7 days at 20 °C; sporangiospores angular | 9 |
| 9. | Colonies russet; sporangiophores umbellately branched from distinct vesicles on agar surface, (16–) 23–63 (−80) µm in length; columellae small but distinct 3.2–5.5 × 1.6–3.8; without micro-chlamydospores | U. changbaiensis |
| – | Colonies light russet-vinaceous pinkish; sporangiophores simply branched from slightly swollen stalks, (27.7–) 47.4–94.8 µm in length; columellae absent or slightly convex up to 1 µm diam.; with micro-chlamydospores | U. vinacea |
| 10. | Sporangia fusiform; without columella | U. fusiformis |
| – | Sporangia globose to subglobose; with distinct columellae | 11 |
| 11. | Columellae distinct to inconspicuous and variable in shape and size; sporangiospores variable in shape and size up to 3.5–5 × 5–11 µm | U. heterosporus |
| – | Columellae distinct and uniform; sporangiospores mostly uniform and no more than 6 µm in diam | 12 |
| 12. | Sporangiophores mainly umbellately branched from swollen portion of the subtended stalk | U. wiegerinckiae |
| – | Sporangiophores sympodial branched from slightly swollen stalks | 13 |
| 13. | Sporangiospores angular | U. angularis |
| – | Sporangiospores not angular | 14 |
| 14. | Sporangiospores with appendage | 15 |
| – | Sporangiospores smooth | 17 |
| 15. | Sporangiophores straight, sporangiospores without a narrowed end, walls thickened unilaterally | U. gibberispora |
| – | Sporangiophores sometimes recurved; sporangiospores with a narrowed end, walls thickened or not | 19 |
| 16. | Sporangiospores tear-shaped with 4.0–6.0 ×2.0–2.5 µm | U. swartii |
| – | Sporangiospores oval to clavate with distinctively thickened wall at the narrower end with 4.0–6.0–8.0 × 2.5–3.5 µm | U. westeae |
| 17. | Colony color deep roseate, Indian red to dark vinaceous-brown; sporangial walls slowly deliquescent | U. dura |
| – | Colony color roseate to pinkish, brownish vinaceous to light russet-vinaceous; sporangial walls quickly deliquescent | 18 |
| 18. | Sporangiospores ovoid to ellipsoidal no curved, mostly 2.4–3.5 µm in length | 19 |
| – | Sporangiospores ellipsoidal to oblong-ellipsoidal and sometimes slightly curved to one side, mainly 3.5–4.9 µm in length | 20 |
| 19. | Colony diam. on CMA reaching 35–40 mm in diam. after 10 days at 18 °C; maximum growth temperature 31–32 °C; columellae mostly depressed globose (3.0–) 4.0–6.5 (–10.0) × (2.5–) 3.5–5.5 (–9.0) µm | U. microsporangia |
| – | Colony diam. on CMA reaching 40–44 mm in diam. after 10 days at 18 °C, maximum growth temperature higher than 32 °C; columellae mostly subglobose (4.0–) 6.0–8.0 (–10.0) µm | U. ramanniana |
| 20. | Colony diam. on CMA reaching 60–62 mm in diam. after 10 days at 18 °C; colonies 1 mm in high; sporangiophores(120–) 270–475 (–980) µm in length | U. oblongielliptica |
| – | Colony diam. on CMA reaching 43–48 mm in diam. after 10 days at 18 °C; colonies up to 2 or 3 mm; sporangiophores commonly more than 500 µm in length | 21 |
| 21. | Sporangiospores oblong-ellipsoidal to ellipsoidal, (3.6–) 4.0–4.9 (–5.5) × (1.7–) 2.4–2.8 (–3.2) µm; macro-chlamydospore not formed on CMA and MEA | U. macrospora |
| – | Sporangiospores ellipsoidal (2.7–) 3.2–4.5 (–5.7) × (1.6–) 2.0–2.4 (–2.8) µm in size; macro-chlamydospore abundant on CMA and MEA | U. curvata |
4. Discussion
The noticeable variations of Umbelopsis ramanniana have been described by the morphological characteristics of their sporangiospores shape, columella size and chlamydospore production, and by their ubiquitous ecological distribution by many studies [22,24,27,28]. However, it was difficult to make a clear delimitation as those morphological characteristics vary continuously among the isolates. Previous research indicated that ITS and nLSU rDNA were not sufficient in achieving species-level identification for the genus Umbelopsis [15,20,27,28,46]. In the present study, based on a combined analysis of multi-loci phylogeny (nSSU, ITS and nLSU rDNA, act1, MCM7 and cox1), morphology and maximum growth temperature, isolates previously identified as U. ramanniana were re-examined and divided into six clades, including five morphologically cryptic lineages. They are described herein as novel species, namely U. curvata, U. dura, U. macrospora, U. microsporangia and U. oblongielliptica.
Umbelopsis ramanniana was meagerly described by Möller [14] without illustration or typification. Based on the culture isolated from mycorrhiza of trees in Eberswalde (Brandenburg, Germany; habitation in protolog) by Möller, Lendner [47] provided a more detailed description and illustration of sporangia, columella and chlamydospores for the species. According to the two studies mentioned above, this species produces roseate colonies, ellipsoidal sporangiospores (2.5 × 1.7 µm), small but distinct columellae and two types of chlamydospores. In this study, the isolates in the clade C2 (Figure 1) tend to fit the descriptions of Möller [14] and Lendner [47] best. Moreover, the clade C2 formed a sister linage to U. angularis, which is a closely related species to U. ramanniana in the sense of Linnemann [16]. Therefore, strains in the clade C2 were chosen as the deputies of U. ramanniana, although there may have been some arbitrariness.
Based on the sequences of the nLSU rDNA D1/D2 region, Ogawa et al. [28] examined intraspecific variations of U. ramanniana and pointed out that this species is an assemblage of several genetically distinct species. In the present study, U. ramanniana was polyphyletic in the nLSU tree (Figure S1), which supports the conclusion of Ogawa et al. [28]. When 23 strains of U. ramanniana were added to the subclades identified by Ogawa et al. [28], more divergent clades were recognized in this species complex. However, due to the low bootstrap values in the nLSU tree, the phylogenetic relationships between subclades of U. ramanniana and closely related species were undetermined. As mentioned by Ogawa et al. [27,28], it is more reasonable to make the taxonomic treatment when species relationships are clarified.
In the present study, the six-loci phylogeny of Umbelopsis was reconstructed and obtained strong support values. The U. ramanniana complex is polyphyletic and can be divided into six clades. The first two clades U. curvata (C1) and U. ramanniana (C2) were located in a well-supported lineage that included U. angularis, U. gibberispora, U. heteropsproa and U. wiegerinckiae. The last two species in this lineage are recently reported, of which, U. wiegerinckiae is characterized by forming umbellate sporangiophores, and U. heterosporus produces varied sporangiophores and columellae [48,49]. The other four species can be distinguished from each other by their unique sporangiospores. In detail, they are the biggest and unilaterally thickened oblong-ellipsoidal sporangiospores in U. gibberispora, bigger and oblong-ellipsoidal with slight curved to one side in U. curvata, smaller and ellipsoidal without any appendage in U. ramanniana and smallest and polygonal in U. angularis. Variations of sporangiospores seem to confirm the hypothesis that tight sporangial walls physically limit the free expansion of sporangiospores and consequently several kinds of shape of sporangiospores could evolve [15]. Moreover, it appears that the tighter sporangial walls seem to result in the deeper sporangial and colonial color. The color of colonies is pale vinaceous in U. gibberispora, brownish vinaceous to light russet-vinaceous in U. curvata and U. ramanniana, and etruscan red to prussian red in U. angularis. The acquisition of a tight sporangial wall should be a main evolution strategy for taxa in this lineage, which supports the hypothesis that relatively few mutations are required to determine a tight sporangial wall [15]. Hence, the differentiation of the lineage is probably a recent event in the evolution of the Umbelopsis. The other four clades U. dura (C4), U. macrospora (C6), U. microsporangia (C3) and U. oblongiellptica (C5) were clustered together with U. swartii and U. westeae. These species formed another well-supported lineage. In this lineage, the sporangiospore shape of the above mentioned six species also varies from appendage to smooth and subglobose to oblong-ellipsoidal. In detail, the sporangiospores are subglobose to ovoid in U. microsporangia, ovoid in U. dura, ellipsoidal to oblong-ellipsoidal with irregularly slight curved to one side in U. macrospora and U. oblongielliptica, and appendaged in U. swartii and U. westeae. It is suggested that variations in sporangiospores shape and size are acquired independently in those two lineages. Moreover, U. dura and U. microsporangia possess a similar sporangiospore shape and size but differ in their maximum growth temperature. Additionally, the maximum growth temperature increased gradually from 31–32 °C to 38 °C in the clades U. microsporangia (C3) to U. macrospora (C6) (Table S2). It is indicated that at least two strategies occurred in the evolution of the lineage of U. swartii through to U. microsporangia.
As shown in previous studies, the ITS barcode was not sufficient in achieving species level identification in Umbelopsis [27,32]. Secondary barcodes, usually protein-coding genes, have been introduced for species discrimination [50,51]. The extensively used loci, such as beta-tubulin (ßtub), translation elongation factor 1-alpha (TEF1), the largest subunit of RNA polymerase II (RPB1) and the second largest subunit of RNA polymerase II (RPB2), were tested, but resulted in multiple copies [32]. The present study preliminarily suggests that the MCM7 should be a suitable secondary barcode, but this needs to be further studied.
5. Conclusions
The multi-gene (nSSU, ITS, nLSU, act1, MCM7 and cox1) phylogeny were proved to be reliable indications of taxon differentiation for the Umbelopsis ramanniana complex. Five species are newly described from the species complex: U. curvata, U. dura, U. macrospora, U. microsporangia and U. oblongielliptica. Those species can be distinguished by morphological traits in combination with the speed of growth and their maximum growth temperature.
Acknowledgments
CBS and NRRL are acknowledged for supplying cultures for this study. Thanks are due to Yi-jian Yao, Paul Kirk and David Hawksworth for their suggestions on nomenclature, and Feng-yan Bai for the critical revision of the manuscript. We also thank Hong-Mei Liu, Mei-Lin Lv and Zhen Song of this research group for taking care of the cultures and generating some molecular phylogenetic data.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8090895/s1, Table S1: Mortierella and Umbelopsis cultures used in phylogenetic analyses; Table S2: Maximum growth temperature (°C) of strains of Umbelopsis ramanniana complex; Figure S1: Phylogenetic tree based on nLSU rDNA D1/D2 region inferred from neighbor-joining (NJ) analysis for Umbelopsis ramanniana and related species.
Author Contributions
Conceptualization, Y.-N.W., X.-Y.L. and R.-Y.Z.; methodology, software, validation, formal analysis, investigation resources and data curation, Y.-N.W.; writing—original draft preparation, Y.-N.W.; writing—review and editing, X.-Y.L. and R.-Y.Z.; visualization, Y.-N.W.; supervision, R.-Y.Z.; project administration, R.-Y.Z. and Y.-N.W.; funding acquisition, Y.-N.W. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequence data are available in NCBI GenBank following the accession numbers in the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by National Natural Science Foundation of China, grant number 31600022.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kamisaka Y., Yokochi T., Nakahara T., Suzuki O. Characterization of the diacylglycerol acyltransferase activity in the membrane fraction from a fungus. Lipids. 1993;28:583–587. doi: 10.1007/BF02536050. [DOI] [PubMed] [Google Scholar]
- 2.Pillai M.G., Certik M., Nakahara T., Kamisaka Y. Characterization of triacylglycerol biosynthesis in subcellular fractions of an oleaginous fungus, Mortierella ramanniana var. angulispora. Biochim. Biophys. Acta. 1998;1393:128–136. doi: 10.1016/S0005-2760(98)00069-1. [DOI] [PubMed] [Google Scholar]
- 3.Mysyakina I.S., Sergeeva Y.E., Bokareva D.A. Lipid composition of the spores of zygomycetous and ascomycetous fungi during cessation of the exogenous dormancy state. Microbiology. 2018;87:51–59. doi: 10.1134/S0026261718010125. [DOI] [Google Scholar]
- 4.Özşen Ö., Kıran İ., Dağ İ., Atlı Ö., Çiftçi G.A., Demirci F. Biotransformation of abietic acid by fungi and biological evaluation of its metabolites. Process Biochem. 2017;52:130–140. doi: 10.1016/j.procbio.2016.09.022. [DOI] [Google Scholar]
- 5.Lardizabal K., Effertz R., Levering C., Mai J., Pedroso M.C., Jury T., Aasen E., Gruys K., Bennett K. Expression of Umbelopsis ramanniana DGAT2A in seed increases oil in soybean. Plant Physiol. 2008;148:89–96. doi: 10.1104/pp.108.123042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domsch K.H., Gams W., Anderson T.-H. Compendium of Soil Fungi. Academic Press; London, UK: 1980. [Google Scholar]
- 7.Parshikov I.A., Freeman J.P., Lay J.O., Beger R.D., Williams A.J., Sutherland J.B. Regioselective transformation of ciprofloxacin to N-acetylciprofloxacin by the fungus Mucor ramannianus. FEMS Microbiol. Lett. 1999;177:131–135. doi: 10.1111/j.1574-6968.1999.tb13723.x. [DOI] [PubMed] [Google Scholar]
- 8.Parshikov I.A., Freeman J.P., Lay J.O., Beger R.D., Williams A.J., Sutherland J.B. Microbiological transformation of enrofloxacin by the fungus Mucor ramannianus. Appl. Environ. Microbiol. 2000;66:2664–2667. doi: 10.1128/AEM.66.6.2664-2667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo J., Jeon J., Kim S.D., Kang S., Han J., Hur H.G. Fungal biodegradation of carbofuran and carbofuran phenol by the fungus Mucor ramannianus: Identification of metabolites. Water Sci. Technol. 2007;55:163–167. doi: 10.2166/wst.2007.051. [DOI] [PubMed] [Google Scholar]
- 10.Kang S.-I., Kang S.-Y., Hur H.-G. Identification of fungal metabolites of anticonvulsant drug carbamazepine. Appl. Microbiol. Biotechnol. 2008;79:663. doi: 10.1007/s00253-008-1459-5. [DOI] [PubMed] [Google Scholar]
- 11.Murphy C.D. Microbial degradation of fluorinated drugs: Biochemical pathways, impacts on the environment and potential applications. Appl. Microbiol. Biotechnol. 2016;100:2617–2627. doi: 10.1007/s00253-016-7304-3. [DOI] [PubMed] [Google Scholar]
- 12.Summerbell R.C. Root endophyte and mycorrhizosphere fungi of black spruce, Picea mariana, in a boreal forest habitat: Influence of site factors on fungal distributions. Stud. Mycol. 2005;53:121–145. doi: 10.3114/sim.53.1.121. [DOI] [Google Scholar]
- 13.Ikeda H., Fukuda T., Yokoyama J. Endophytic fungi associated with a holoparasitic pant, Balanophora japonica (Balanophoraceae) Am. J. Plant Sci. 2016;7:152–158. doi: 10.4236/ajps.2016.71016. [DOI] [Google Scholar]
- 14.Möller A. Untersuchungen über ein- und zweijährige Kiefern im märkischen Sandbode. Z. Forst Jagdwes. 1903;35:257–321. [Google Scholar]
- 15.Sugiyama M., Tokumasu S., Gams W. Umbelopsis gibberispora sp. nov. from Japanese leaf litter and a clarification of Micromucor ramannianus var. angulisporus. Mycoscience. 2003;44:217–226. doi: 10.1007/S10267-003-0105-4. [DOI] [Google Scholar]
- 16.Linnemann G. Die Mucorineen-Gattung Mortierella Coemans. Gustav Fischer; Jena, Germany: 1941. [Google Scholar]
- 17.Mil’ko A.A. Opredeltiel’ Mukoral’nykh Gribov [Key to the identification of Mucorales] Naukova Dumka; Kyiv, Ukraine: 1974. [Google Scholar]
- 18.Gams W. Some new or noteworthy species of Mortierella. Persoonia. 1976;9:111–140. [Google Scholar]
- 19.Von Arx J. On Mucoraceae s. str. and other families of the Mucorales. Sydowia. 1984;35:10–26. [Google Scholar]
- 20.Meyer W., Gams W. Delimitation of Umbelopsis (Mucorales, Umbelopsidaceae fam. nov.) based on ITS sequence and RFLP data. Mycol. Res. 2003;107:339–350. doi: 10.1017/S0953756203007226. [DOI] [PubMed] [Google Scholar]
- 21.Chalabuda T. Systematics of the family Mortierellaceae. Novosti Sist. Nizsh. Rast. 1968;1968:120–131. [Google Scholar]
- 22.Evans E.H. Studies on Mortierella ramanniana: I. Relationship between morphology and cultural behaviour of certain isolates. Trans. Br. Mycol. Soc. 1971;56:201–216. doi: 10.1016/S0007-1536(71)80031-1. [DOI] [Google Scholar]
- 23.Wang Y.N., Liu X.Y., Zheng R.Y. Umbelopsis changbaiensis sp. nov. from China and the typification of Mortierella vinacea. Mycol. Prog. 2014;13:657–669. doi: 10.1007/s11557-013-0948-9. [DOI] [Google Scholar]
- 24.Turner M. Studies in the genus Mortierella: I. Mortierella isabellina and related species. Trans. Br. Mycol. Soc. 1963;46:262–272. doi: 10.1016/S0007-1536(63)80082-0. [DOI] [Google Scholar]
- 25.Peberdy J.F., Turner M. The esterases of Mortierella ramanniana in relation to taxonomy. J. Gen. Microbiol. 1968;51:303–312. doi: 10.1099/00221287-51-2-303. [DOI] [PubMed] [Google Scholar]
- 26.Nagy Á., Pesti M., Galgóczy L., Vágvölgyi C. Electrophoretic karyotype of two Micromucor species. J. Basic Microbiol. 2004;44:36–41. doi: 10.1002/jobm.200310272. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa Y., Suda A., Kusama-Eguchi K., Watanabe K., Tokumasu S. Intraspecific groups of Umbelopsis ramanniana inferred from nucleotide sequences of nuclear rDNA internal transcribed spacer regions and sporangiospore morphology. Mycoscience. 2005;46:343–351. doi: 10.1007/S10267-005-0257-5. [DOI] [Google Scholar]
- 28.Ogawa Y., Sugiyama M., Hirose D., Kusama-Eguchi K., Tokumasu S. Polyphyly of intraspecific groups of Umbelopsis ramanniana and their genetic and morphological variation. Mycoscience. 2011;52:91–98. doi: 10.1007/S10267-010-0074-3. [DOI] [Google Scholar]
- 29.Chen F.-J. The genus Mortierella Coemans in China. Graduate University of Chinese Academy of Sciences; Beijing, China: 1986. [Google Scholar]
- 30.Zheng R.-Y., Chen G.-Q., Huang H., Liu X.-Y. A monograph of Rhizopus. Sydowia. 2007;59:273–372. [Google Scholar]
- 31.Ridgway R. Color Standards and Color Nomenclature. Published by the Author; Washington, DC, USA: 1912. [Google Scholar]
- 32.Wang Y.-N., Liu X.-Y., Zheng R.-Y. Umbelopsis longicollis comb. nov. and the synonymy of U. roseonana and U. versiformis with U. nana. Mycologia. 2015;107:1023–1032. doi: 10.3852/14-339. [DOI] [PubMed] [Google Scholar]
- 33.Schmitt I., Crespo A., Divakar P.K., Fankhauser J.D., Herman-Sackett E., Kalb K., Nelsen M.P., Nelson N.A., Rivas-Plata E., Shimp A.D., et al. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia. 2009;23:35–40. doi: 10.3767/003158509X470602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tretter E.D., Johnson E.M., Wang Y., Kandel P., White M.M. Examining new phylogenetic markers to uncover the evolutionary history of early-diverging fungi: Comparing MCM7, TSR1 and rRNA genes for single- and multi-gene analyses of the Kickxellomycotina. Persoonia. 2013;30:106–125. doi: 10.3767/003158513X666394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molitor C., Inthavong B., Sage L., Geremia R.A., Mouhamadou B. Potentiality of the cox1 gene in the taxonomic resolution of soil fungi. FEMS Microbiol. Lett. 2010;302:76–84. doi: 10.1111/j.1574-6968.2009.01839.x. [DOI] [PubMed] [Google Scholar]
- 36.Katoh K., Kuma K.-I., Toh H., Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katoh K., Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 38.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 39.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaidya G., Lohman D.J., Meier R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27:171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- 41.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 43.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Page R.D.M. TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 45.Alfaro M.E., Zoller S., Lutzoni F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Biol. Evol. 2003;20:255–266. doi: 10.1093/molbev/msg028. [DOI] [PubMed] [Google Scholar]
- 46.Mahoney D., Gams W., Meyer W., Starink-Willemse M. Umbelopsis dimorpha sp. nov., a link between U. vinacea and U. versiformis. Mycol. Res. 2004;108:107–111. doi: 10.1017/S0953756203008876. [DOI] [PubMed] [Google Scholar]
- 47.Lendner A. Materiaux pour la Flore Cryptogamique Suisse. Volume III K.-J. Wyss; Bern, Switzerland: 1908. Les Mucorinées de La Suisse. [Google Scholar]
- 48.Crous P.W., Wingfield M.J., Burgess T.I., Carnegie A.J., Hardy G.S.J., Smith D., Summerell B.A., Cano-Lira J.F., Guarro J., Houbraken J. Fungal Planet description sheets: 625–715. Persoonia. 2017;39:270. doi: 10.3767/persoonia.2017.39.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan H.-S., Lu X., Dai Y.-C., Hyde K.D., Kan Y.-H., Kušan I., He S.-H., Liu N.-G., Sarma V.V., Zhao C.-L. Fungal diversity notes 1277–1386: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2020;104:1–266. doi: 10.1007/s13225-020-00461-7. [DOI] [Google Scholar]
- 50.Buyck B., Kauff F., Eyssartier G., Couloux A., Hofstetter V. A multilocus phylogeny for worldwide Cantharellus (Cantharellales, Agaricomycetidae) Fungal Divers. 2014;64:101–121. doi: 10.1007/s13225-013-0272-3. [DOI] [Google Scholar]
- 51.Xu J. Fungal DNA barcoding. Genome. 2016;59:913–932. doi: 10.1139/gen-2016-0046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data are available in NCBI GenBank following the accession numbers in the manuscript.