Abstract
Targeted gene disruption efficiency in Acremonium chrysogenum was increased 10-fold by applying the double-marker enrichment technique to this filamentous fungus. Disruption of the mecB gene by the double-marker technique was achieved in 5% of the transformants screened. Mutants T6 and T24, obtained by gene replacement, showed an inactive mecB gene by Southern blot analysis and no cystathionine-γ-lyase activity. These mutants exhibited lower cephalosporin production than that of the control strain, A. chrysogenum C10, in MDFA medium supplemented with methionine. However, there was no difference in cephalosporin production between parental strain A. chrysogenum C10 and the mutants T6 and T24 in Shen's defined fermentation medium (MDFA) without methionine. These results indicate that the supply of cysteine through the transsulfuration pathway is required for high-level cephalosporin biosynthesis but not for low-level production of this antibiotic in methionine-unsupplemented medium. Therefore, cysteine for cephalosporin biosynthesis in A. chrysogenum derives from the autotrophic (SH2) and the reverse transsulfuration pathways. Levels of methionine induction of the cephalosporin biosynthesis gene pcbC were identical in the parental strain and the mecB mutants, indicating that the induction effect is not mediated by cystathionine-γ-lyase.
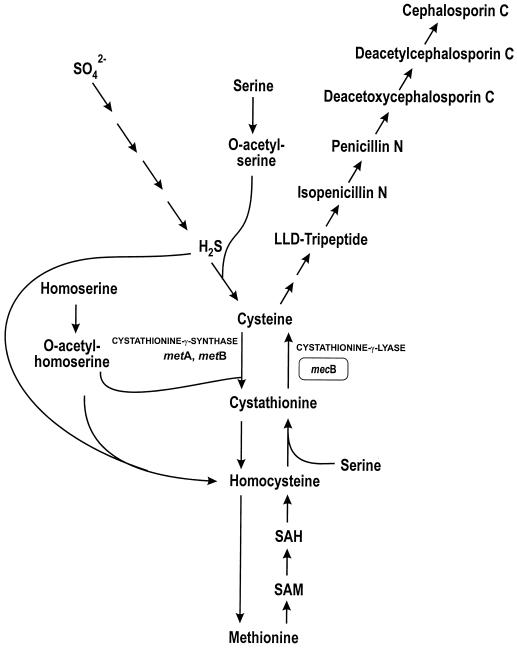
β-Lactam biosynthesis in Acremonium chrysogenum (formerly known as Cephalosporium acremonium) begins with the nonribosomal condensation of the three precursor amino acids l-α-aminoadipic acid, l-cysteine, and l-valine (1, 21, 23). There are two ways to synthesize cysteine in filamentous fungi (35, 26). One pathway, the so-called autotrophic pathway, converts inorganic sulfur to cysteine via serine O-acetyltransferase and O-acetylserine sulfhydrilase (Fig. 1). In the second route, cysteine can be obtained via the reverse transsulfuration pathway, in which the sulfur atom of methionine is transferred to cysteine through S-adenosylmethionine, S-adenosylhomocysteine, homocysteine, and cystathionine as intermediates.
FIG. 1.
Cysteine biosynthesis in A. chrysogenum from sulfate (autotrophic pathway) and from methionine (reverse transsulfuration pathway). The mecB gene encoding cystathionine-γ-lyase is shaded. This gene has been disrupted in transformants T6 and T24 (see the text). In the text, genes designated met belong to the methionine biosynthesis (direct transsulfuration) pathway and those named mec are involved in the reverse transsulfuration pathway. SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine.
The predominant route of the cysteine supply for β-lactam biosynthesis depends on the producer microorganism (6). Cysteine for penicillin biosynthesis in Penicillium chrysogenum is obtained mainly from sulfate reduction (31). In this microorganism, the transsulfuration pathway has been considered to have a minor role in penicillin biosynthesis. On the other hand, the sulfur atom for β-lactam biosynthesis by A. chrysogenum is believed to derive preferentially from methionine via the reverse transsulfuration pathway (3, 24). However, there is no genetic evidence for, or against, this hypothesis.
dl-Methionine is known to stimulate cephalosporin biosynthesis in A. chrysogenum (7, 16). For many years it was unclear whether the stimulatory effect of dl-methionine was due to a precursor effect (one providing cysteine) or to an inducing effect (8, 20, 23). Sawada and coworkers established that methionine increases isopenicillin N synthase, deacetoxycephalosporin C synthase (30), and α-aminoadipyl-cysteinyl-valine synthetase activities (38) in Acremonium cultures. Later, Velasco et al. (35) showed that methionine induces the transcription of three of the four known cephalosporin biosynthetic genes.
Therefore, methionine has presumably a double effect on cephalosporin biosynthesis: (i) it could be the main supplier of cysteine via the reverse transsulfuration pathway and (ii) it has an induction effect on cephalosporin biosynthetic genes.
The mecB gene of A. chrysogenum has recently been cloned and characterized in our laboratory (19). This gene encodes a cystathionine-γ-lyase activity which splits cystathionine into cysteine and α-ketobutyrate. Several years ago it was proposed that cystathionine-γ-lyase is required for cephalosporin biosynthesis (34). In order to study the influence of the transsulfuration pathway on cephalosporin biosynthesis, mecB was inactivated by a novel disruption technique in this fungus and the effect of this inactivation on cephalosporin production was studied. Results show unequivocally that the splitting of cystathionine is essential for high-level production of cephalosporin but that reverse transsulfuration is not the only pathway for cysteine formation for cephalosporin since the mecB-disrupted mutants were still able to produce cephalosporin.
MATERIALS AND METHODS
Microbial strains.
A. chrysogenum C10 (ATCC 48278), a strain producing high levels of cephalosporin C, released by Panlabs (6, 28), was used for mecB inactivation experiments. Aspergillus nidulans C47 (mecB cysB) and M63 (mecB) were obtained from A. Paszewski and used as controls to test for methionine auxotrophy following gene disruption. Escherichia coli ESS2231, a β-lactam-supersensitive strain, was used for routine bioassays. E. coli DH5α competent cells were used in routine DNA manipulations.
Media and growth conditions for cephalosporin production.
A. chrysogenum was grown in seed medium (32) for 48 h; 10 ml of seed culture was used to inoculate Shen's defined production (MDFA) medium (32) with or without supplementation with dl-methionine (3 g/liter). The cultures were incubated in triple-baffled flasks (500 ml; Bellco) containing 100 ml of medium at 25°C in a rotary shaker. Samples were taken every 24 h, and levels of cephalosporin antibiotics were determined by bioassays against E. coli ESS2231.
A. chrysogenum transformation and isolation of genomic DNA.
Transformation of A. chrysogenum protoplasts was carried out as described previously (12). Transformants were selected in tryptic soy agar (Difco) with sucrose (10.3%), supplemented with hygromycin at 30 μg/ml. Genomic DNA of A. chrysogenum was isolated as described previously (12).
Site-directed mutagenesis.
In vitro mutagenesis was performed with a Quickchange mutagenesis kit (Stratagene, La Jolla, Calif.) by following the manufacturer's instructions. Oligonucleotides used in the formation of an EcoRI site in the mecB gene were Ia (5′-CCTTATGTGCAGAATTCGCTCGACCTC) and Ib (5′-GAGGTCGAGCGAATTCTGCACATAAGG).
Southern blotting and hybridization.
Three-microgram samples of genomic DNAs from A. chrysogenum C10 and its transformants were digested with EcoRI and separated in 0.8% agarose gel. The gel was blotted onto Hybond-N membrane (Amersham Pharmacia Biotech) as described by Sambrook et al. (29). Digoxigenin-alkaline phosphatase labeling, hybridization, and detection were done with a Genius kit (Boehringer Mannheim) according to the manufacturer's protocol. Hybridizations were performed at 68°C, and the blots were washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate for 5 min at room temperature and twice with 0.1× SSC–0.1% sodium dodecyl sulfate for 15 min at 68°C.
Cell extracts and cystathionine-γ-lyase activity assay.
Frozen mycelia (500 mg) collected every 24 h were ground in a mortar with liquid nitrogen and resuspended in 100 mM sodium phosphate buffer (pH 7.3) containing 1 mM EDTA and 0.1 mM pyridoxal 5′-phosphate. Cell debris was removed by centrifugation at 15,000 × g for 20 min at 4°C. Cystathionine-γ-lyase activity was assayed by determining the conversion of l-cystathionine into l-cysteine and α-ketobutyrate. The reaction mixture consisted of 4 mM l-cystathionine, 5.5 × 10−2 mM pyridoxal 5′-phosphate, 7 mM EDTA, 2 mM dithiothreitol, and 0.1 mg of protein in a final volume of 0.5 ml. The reaction mixtures were incubated for 30 min at 30°C, and the reactions were stopped by the addition of 1 ml of Gaitonde's reagent and boiled for 5 min. The precipitated proteins were removed by centrifugation, and the amount of cysteine in 1 ml of supernatant was determined by using the acid ninhydrin assay. This assay is highly specific and gives essentially no reaction with cystathionine or methionine (10). Total protein in cell extracts was determined by the Bradford method with ovalbumin as a standard.
RESULTS
Gene disruption in A. chrysogenum by the double-marker technique.
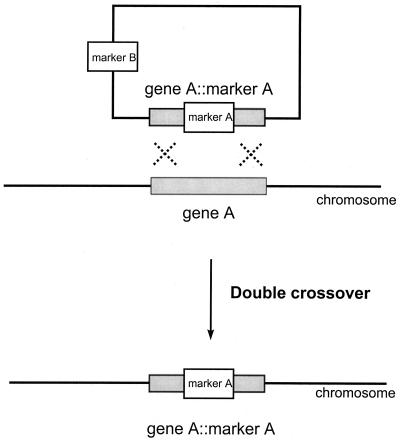
Transforming DNA is integrated into the genome of A. chrysogenum at a very low efficiency, mainly via heterologous recombination (14, 36). The frequency of gene disruption in A. chrysogenum is one event every 200 transformants screened (36). Since target inactivation of the mecB gene, which encodes cystathionine-γ-lyase, would not generate any easily detectable phenotype (i.e., auxotrophy, resistance, color), we tried a technique to enrich the proportion of gene disruption events when inactivation is targeted to nonselectable genes (18). This technique, which has not been applied before to filamentous fungi, is based on the use of two selectable markers (Fig. 2). Marker A has two functions: (i) to inactivate the desired gene and (ii) to serve as the transformation marker. When double recombination takes place, the second marker (marker B) is lost. Thus, transformants are selected as marker A resistant and then screened for marker B sensitivity.
FIG. 2.
Scheme of the double-marker enrichment technique for targeted gene disruption. Marker A is the transformation marker. Marker B is the enrichment marker (see the text). Transformants having gene A disrupted were selected as marker A resistant and marker B sensitive.
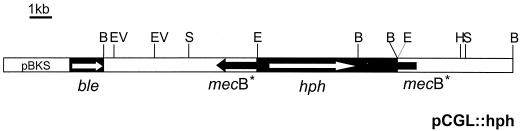
To apply the double-marker technique, we constructed plasmid pCGL::hph (Fig. 3), which contains as marker A the hygromicin resistance gene (hph) under the control of the gpd gene promoter (27) and as marker B the phleomycin resistance (ble) under the control of the pcbC gene promoter (subcloned from pC43) (13).
FIG. 3.
Physical map of the pCGL::hph plasmid constructed for replacement of the mecB gene. The plasmid contains as marker A the hygromycin resistance cassette (hph) and as marker B the phleomycin resistance cassette (ble) (see the text for details of the resistance cassettes). B, BamHI; EV, EcoRV; S, SstI; E, EcoRI; H, HindIII.
A 7.7-kb BamHI DNA fragment containing the mecB gene was inserted into the BamHI site of pC43, which carries the ble gene (resistance to phleomycin). An EcoRI site was introduced in nucleotides 606 to 611 of the mecB gene by in vitro mutagenesis (see Materials and Methods), generating the pCGL plasmid. Finally, a hygromycin resistance (hph) cassette subcloned from pAN7-1E was inserted in the EcoRI position of the pCGL plasmid, generating the construct pCGL::hph (Fig. 3). This plasmid was used in the transformation of A. chrysogenum C10.
Southern blot analysis of transformants sensitive to phleomycin and resistant to hygromycin.
About 600 transformants resistant to hygromycin were selected by transformation of A. chrysogenum C10 with pCGL::hph and screened for resistance to phleomycin. Ninety-nine out of 600 transformants were sensitive to phleomycin and resistant to hygromycin. There are two possibilities to explain the phenotype of these phleomycin-sensitive, hygromycin-resistant transformants: (i) phleomycin resistance is lost because double recombination took place or (ii) the transforming DNA is integrated into the genome through the phleomycin resistance cassette, thus losing its function.
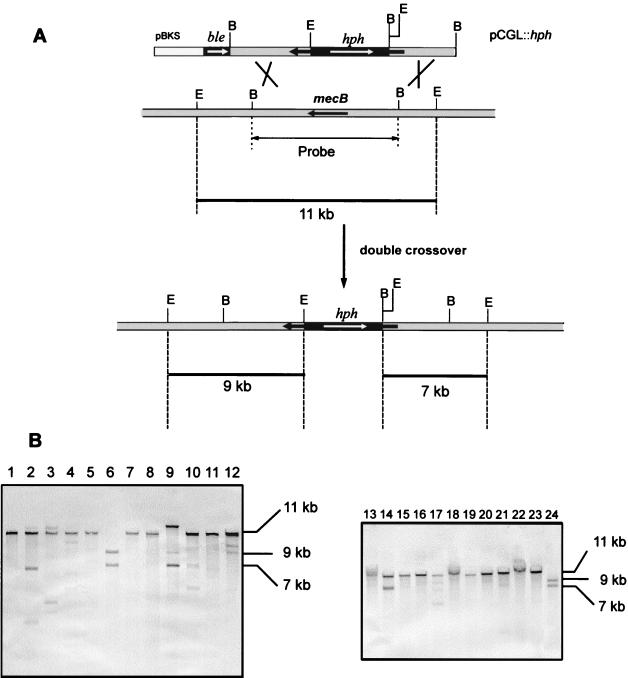
To identify the transformants in which the endogenous mecB gene had been replaced by the inactivated gene, Southern blot analysis was performed in 35 Hygr Phls transformants. As shown in Fig. 4A, the genomic DNA of A. chrysogenum should give hybridization with an 11-kb EcoRI band when the 7.7-kb BamHI fragment is used as a probe. When double recombination takes place within the homologous mecB region, the ble gene (conferring phleomycin resistance) is lost and the 11-kb hybridization band should change into two hybridization bands of 9 and 7 kb.
FIG. 4.
Replacement of mecB by an inactive copy using the double-marker technique and molecular analysis of the transformants. (A) Disruption of mecB with the pCGL::hph plasmid. The 11-kb EcoRI fragment in the genome is changed into two fragments of 9 and 7 kb B, BamHI; E, EcoRI. (B) Southern blot hybridization of EcoRI-digested genomic DNA (two Southern blots are shown) using as the probe the 7.7-kb BamHI fragment containing mecB. Lane 1, A. chrysogenum C10; lanes 2 to 24 transformants T2 to T24. The sizes of the hybridization bands are indicated on the right.
In order to study the genotypes of the transformants, the DNAs of 35 randomly selected Hygr Phls transformants were digested with EcoRI, blotted onto a nylon membrane, and hybridized with a 7.7-kb BamHI fragment as the probe. Southern hybridization results showed that the parental strain, A. chrysogenum C10 (Fig. 4B, lane 1), hybridized with an 11-kb band that was also present in most of the transformants. However, transformants T6 (Fig. 4B, lane 6) and T24 (Fig. 4B, lane 24) did not contain the 11-kb hybridization band, showing instead the predicted change for a gene disruption event (hybridizing bands of 9 and 7 kb).
Transformants T6 and T24 with the disrupted mecB gene lack cystathionine-γ-lyase activity.
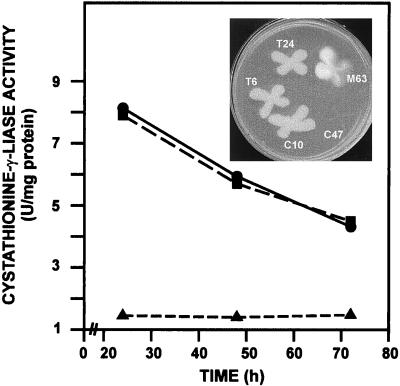
To study if transformants T6 and T24 had lost cystathionine-γ-lyase activity (encoded by mecB), fermentation in defined production medium was performed (see Materials and Methods) using strains A. chrysogenum C10, the transformants T6 and T24 (with a disrupted mecB region), and transformant T99 (unmodified mecB region). Results showed (Fig. 5) a typical profile of a primary metabolism enzyme for A. chrysogenum C10; a very similar level of cystathionine-γ-lyase activity was obtained with transformant T99. However, transformants T6 and T24 showed no detectable cystathionine-γ-lyase activity. These results indicated that those transformants that suffered the canonical recombination process lost the mecB gene in the gene replacement process. These results confirmed that there is a single copy of the mecB gene in A. chrysogenum, as was proposed before on the basis of Southern hybridization evidence (19).
FIG. 5.
Specific cystathionine-γ-lyase activity in A. chrysogenum C10 (●), mutant T99 (■), and mutants T6 and T24 (▴); the last two mutants showed no activity. (Inset) Growth of the T6 and T24 mutants in minimal Czapek medium. The parental strain, A. chrysogenum C10, is also a prototroph. Aspergillus nidulans C47 (mecB cysB), used as control, is unable to grow in minimal medium since it lacks both cysteine biosynthesis pathways, whereas Aspergillus nidulans M63 (mecB) behaves like the A. chrysogenum transformants T6 and T24.
MecB-inactivated mutants of A. chrysogenum are not auxotrophs.
Transformants T6 and T24, deficient in cystathionine-γ-lyase activity, lack the reverse transsulfuration pathway, but they are not auxotrophs, i.e., they are able to synthesize cysteine from sulfate and convert cysteine to methionine to meet the methionine requirement of the cell (Fig. 5, inset). The control Aspergillus nidulans strain C47 (mecB cysB), a double auxotroph defective in the autotrophic as well as in the reverse transsulfuration pathway, was unable to grow in minimal Czapek medium. These results indicate that cystathionine-γ-lyase (catalyzing the splitting of cystathionine in the reverse transsulfuration pathway) is different from cystathionine-γ-synthase, which catalyzes the formation of cystathionine from cysteine and O-acetylserine in direct transsulfuration (conversion of cysteine to methionine); mutants lacking cystathionine-γ-synthase are known to be methionine auxotrophs, and this is not the case with transformants T6 and T24.
Inactivation of the mecB gene decreases cephalosporin production in methionine-supplemented cultures.
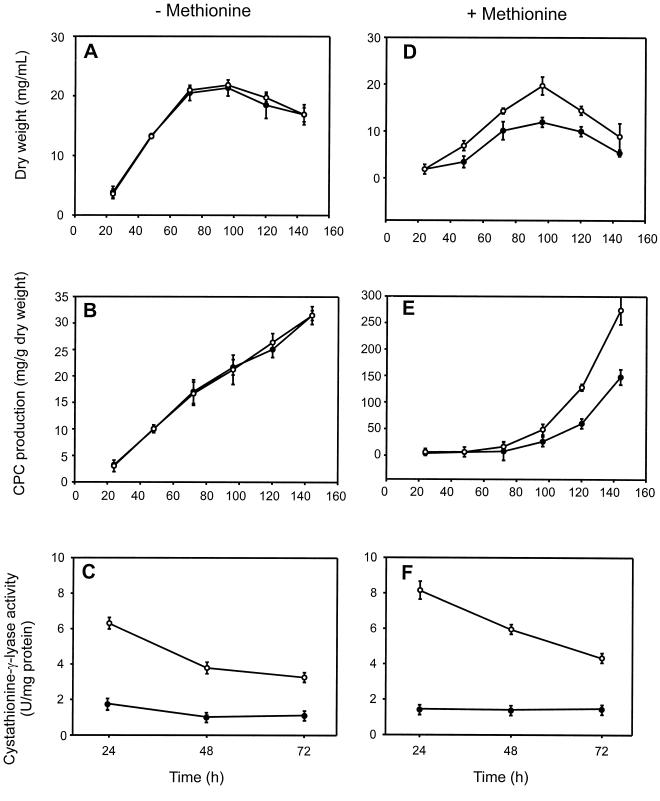
The effect of inactivation of the mecB gene on cephalosporin biosynthesis was studied in cultures of the transformants T6 and T24 in triplicate flasks, and the same fermentation was repeated three times. Fermentations to evaluate the effect of inactivation of mecB were performed in MDFA medium with (3 g/liter) and without dl-methionine. Results showed that in medium without methionine (Fig. 6A to C), the growth behavior of the inactivated strains (T6 and T24) and their specific levels of cephalosporin production were similar to those of the control strain, A. chrysogenum C10. The lack of cystathionine-γ-lyase activity (Fig. 6C) did not affect growth in MDFA medium.
FIG. 6.
Growth kinetics (A and D), cephalosporin production (B and E), and cystathionine-γ-lyase activity (C and F) in A. chrysogenum C10 (○) and mutant T6 or T24 (●) in MDFA medium without methionine (left) and in MDFA medium supplemented with 3 g of dl-methionine per liter (right). Note the different scales in panels B and E. The increase in cephalosporin C (CPC) production (E) is due to the induction of cephalosporin biosynthesis by methionine. Results are the averages of three determinations from triplicate flasks. Vertical bars indicate the standard deviations from the averages using nine determinations.
However, in cultures supplemented with methionine, the behavior was different (Fig. 6D to F). Mutants T6 and T24 exhibited a smaller growth extent than the parental strain A. chrysogenum C10, i.e., the lack of cystathionine-γ-lyase results in less growth than in the parental strain C10, probably because methionine reduces the synthesis of cysteine through the autotrophic pathway, which is the only remaining route for cysteine formation in the mecB mutants. The specific production of cephalosporin was decreased by 40 to 50% in the mutants T6 and T24 compared with that in the control strain, A. chrysogenum C10. The differences in cephalosporin biosynthesis correlate with the lack of cystathionine-γ-lyase activity observed in these mutants (Fig. 6C and F). In A. chrysogenum C10, the cystathionine-γ-lyase activity was slightly higher in cultures grown with methionine than in those without methionine, and the same happened with cephalosporin biosynthesis (see below). However, the T6 and T24 mutants showed no significant cystathionine-γ-lyase activity with or without methionine, thus excluding the possibility of the existence of a second silent mecB gene that might be triggered by dl-methionine in mecB mutants. Both mecB mutants behaved identically.
dl-Methionine still induces cephalosporin gene expression in mecB-disrupted mutants.
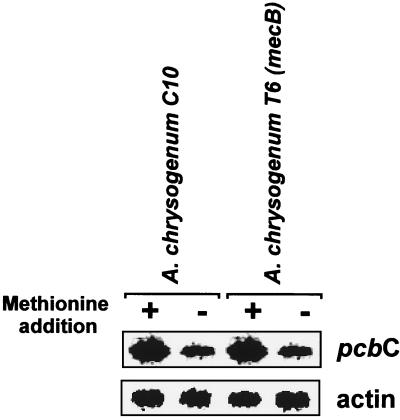
It is well known that exogenous dl-methionine induces the transcription of cephalosporin C biosynthetic genes (35). It was unclear, however, whether the methionine induction effect could be mediated by α-ketobutyrate (a product of cystathionine splitting by cystathionine-γ-lyase) or by the cystathionine-γ-lyase protein itself acting as a regulatory protein. To determine if the methionine induction effect was still present in mecB-disrupted mutants lacking cystathionine-γ-lyase activity, Northern blot analysis was performed using RNAs extracted from mycelia of A. chrysogenum C10 and mutant T6, which were grown for 48 h in MDFA without and with methionine (3 g/liter). Results showed (Fig. 7) that the induction effect on expression of the A. chrysogenum pcbC gene was still present in the strain lacking cystathionine-γ-lyase activity. These results indicate that cystathionine-γ-lyase is not involved in cephalosporin gene expression. The involvement of α-ketobutyrate in cephalosporin induction cannot be ruled out since α-ketobutyrate may still be formed in the cell by other catabolic pathways.
FIG. 7.
Methionine stimulation of transcription of the pcbC gene in the parental strain A. chrysogenum C10 and the A. chrysogenum T6 mutant, disrupted in mecB. Northern hybridizations were performed with a 0.8-kb NcoI/KpnI fragment containing the actin gene of Aspergillus nidulans and a 1.8-kb SalI/BamHI fragment that contains the pcbC gene of A. chrysogenum.
DISCUSSION
Gene disruption is a technique difficult to apply to filamentous fungi (9). However, there are significant differences among filamentous fungi. Some of them are able to efficiently integrate homologous DNA into the genome, as is the case with Aspergillus nidulans (33, 37), Ascobolus immersus (11), and Cochiobolus heterostrophus (15). On the other hand, with other filamentous fungi such as A. chrysogenum (14, 36), Podospora anserina (2), and Penicillium chrysogenum (5), the frequency of homologous recombination is very low (usually lower than 1 in 100 transformants).
Only two cases of targeted gene disruption have been described for A. chrysogenum. Hoskins et al. (14) inactivated the pcbAB gene by the single-crossover integration approach. However, this technique generates direct repeats in the genome and it has been shown to be highly unstable in the genomes of filamentous fungi (4). Waltz and Kück (36) studied the frequency of gene disruption by double crossover in A. chrysogenum and established that only in 0.5% of the transformants was the targeted gene inactive.
In this work, we have adapted the gene disruption enrichment method developed for mouse embryo-derived stem cells (18) based on the use of two selection markers. This technique resulted in obtaining 5% gene disruption (one event in every 20 transformants); thus, the expected gene disruption efficiency (0.5%) was increased 10-fold. The use of this enrichment method in our laboratory has resulted in very high efficiencies (30% of gene disruption events) for the inactivation of other A. chrysogenum genes (G. Liu, J. Casqueiro, and J. F. Martín, unpublished results).
In Saccharomyces cerevisiae and filamentous fungi, cysteine is synthesized by two pathways (25, 26) (Fig. 1). The l-cysteine molecule required for the biosynthesis of the ACV tripeptide in cephalosporin-producing A. chrysogenum strains was proposed to be synthesized mainly from methionine via the reverse transsulfuration pathway through homocysteine and cystathionine (Fig. 1) (23). Treichler et al. (34) at CIBA-Geigy suggested that cystathionine-γ-lyase, the enzyme that splits cystathionine into cysteine and α-ketobutyrate, is a key enzyme for cephalosporin biosynthesis. An A. chrysogenum mutant isolated by random mutagenesis and defective in cystathionine-γ-lyase activity showed a drastic reduction in cephalosporin biosynthesis (34). However, this mutant might have contained a second mutation affecting cephalosporin production and, unfortunately, the CIBA-Geigy mutant is not available for further studies (J. Heim, personal communication). In this article, we have inactivated the mecB gene, which encodes cystathionine-γ-lyase activity in A. chrysogenum (19). The inactivation of the mecB gene does not affect cephalosporin biosynthesis when the sulfur source is sulfate, i.e., cysteine for cephalosporin biosynthesis may be synthesized from either sulfate or methionine. Addition of methionine stimulates cephalosporin biosynthesis in the parental strain A. chrysogenum C10. But when mecB is inactivated (mutants T6 and T24), there is a clearly lower production of cephalosporin, indicating that the supply of cysteine from methionine via the reverse transsulfuration pathway is required for high-level production of cephalosporin. A correlation between the positive effect of methionine and cystathionine-γ-lyase activity in A. chrysogenum C10 was observed (Fig. 6). The cystathionine-γ-lyase activity is higher in the presence of methionine (as previously described by Zanca [38]), in agreement with a direct supply of cysteine from the reverse transsulfuration pathway. The growth of mutants T6 and T24 is retarded in methionine-supplemented medium, probably because in the presence of methionine the utilization of sulfate is depressed (17). Sulfate utilization is needed for cysteine biosynthesis in these mecB mutants because they cannot synthesize cysteine through the reverse transsulfuration pathway. Complementation tests of the mecB mutants would be desirable to confirm this hypothesis.
dl-Methionine greatly stimulates cephalosporin biosynthesis (16, 20) by inducing expression of the cephalosporin biosynthesis genes pcbAB, pcbC, and cefEF (30, 35). Disruption of the mecB gene did not affect the induction of pcbC by methionine, i.e., induction of the cephalosporin genes is not mediated by a putative regulatory mechanism exerted by the cystathionine-γ-lyase protein. The induction mechanism may be triggered by methionine itself or by a catabolite derived from methionine, e.g., α-ketobutyrate. α-Ketobutyrate (a product of cystathionine splitting) cannot be formed in mecB mutants. However, the involvement of α-ketobutyrate as an inducer in A. chrysogenum cannot be ruled out since it may still be formed by methionine-deaminases similar to those that occur in Aspergillus nidulans (R. E. Cardoza and J. F. Martín, unpublished results), although their presence in A. chrysogenum has not yet been demonstrated.
In conclusion, targeted inactivation of the mecB gene shows that methionine has two positive effects on cephalosporin biosynthesis; one of them, the supply of cysteine from the reverse transsulfuration pathway, required for high-level cephalosporin biosynthesis, is catalyzed by cystathionine-γ-lyase.
ACKNOWLEDGMENTS
This work was supported by a grant of the CICYT (BIO97-0289-C02-01).
We thank A. Paszewski for providing Aspergillus nidulans strains M63 and C47 and M. Mediavilla, B. Martín, R. Barrientos, and M. Corrales for excellent technical assistance.
REFERENCES
- 1.Aharonowitz Y, Cohen G, Martín J F. Penicillin and cephalosporin biosynthetic genes: structure, organization, regulation and evolution. Annu Rev Microbiol. 1992;46:461–496. doi: 10.1146/annurev.mi.46.100192.002333. [DOI] [PubMed] [Google Scholar]
- 2.Brygoo Y, Debuchy R. Transformation by integration in Podospora anserina. I. Methodology and phenomenology. Mol Gen Genet. 1985;200:128–131. [Google Scholar]
- 3.Caltrider P G, Niss H F. Role of methionine in cephalosporin synthesis. Appl Microbiol. 1966;14:746–753. doi: 10.1128/am.14.5.746-753.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casqueiro J, Bañuelos O, Gutiérrez S, Hijarrubia M J, Martín J F. Intrachromosomal recombination between direct repeats in Penicillium chrysogenum: gene conversion and deletion events. Mol Gen Genet. 1999;261:994–1000. doi: 10.1007/s004380051048. [DOI] [PubMed] [Google Scholar]
- 5.Casqueiro J, Gutiérrez S, Bañuelos O, Hijarrubia M J, Martín J F. Gene targeting in Penicillium chrysogenum: disruption of the lys2 gene leads to penicillin overproduction. J Bacteriol. 1999;181:1181–1188. doi: 10.1128/jb.181.4.1181-1188.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demain A L. Strain exchange between industry and academia. ASM News. 1983;49:431. [Google Scholar]
- 7.Demain A L, Wolfe S. Biosynthesis of cephalosporins. Dev Ind Microbiol. 1987;27:175–182. [Google Scholar]
- 8.Drew S, Demain A L. Effect of primary metabolites on secondary metabolism. Annu Rev Microbiol. 1977;31:343–356. doi: 10.1146/annurev.mi.31.100177.002015. [DOI] [PubMed] [Google Scholar]
- 9.Fincham J R S. Transformation in fungi. Microbiol Rev. 1989;53:148–170. doi: 10.1128/mr.53.1.148-170.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaitonde M K. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem J. 1967;104:627–633. doi: 10.1042/bj1040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goyon C, Faugeron G. Targeted transformation of Ascobolus inmersus and de novo methylation of the resulting duplicated sequences. Mol Cell Biol. 1989;9:2818–2827. doi: 10.1128/mcb.9.7.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutiérrez S, Díez B, Montenegro E, Martín J F. Characterization of the Cephalosporium acremonium pcbAB gene encoding α-aminoadipyl-cysteinil-valine synthetase, a large multidomain peptide synthetase: linkage to the pcbC gene as a cluster of early cephalosporin biosynthetic genes and evidence of multiple functional domains. J Bacteriol. 1991;173:2354–2365. doi: 10.1128/jb.173.7.2354-2365.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutiérrez S, Marcos A T, Casqueiro J, Kosalková K, Fernández F J, Velasco J, Martín J F. Transcription of the pcbAB, pcbC and penDE genes of Penicillium chrysogenum AS-P-78 is repressed by glucose and the repression is not reversed by alkaline pHs. Microbiology. 1999;145:317–324. doi: 10.1099/13500872-145-2-317. [DOI] [PubMed] [Google Scholar]
- 14.Hoskins J A, O'Callaghan N, Queener S W, Cantwell C A, Wood J S, Chen V J, Skatrud P L. Gene disruption of the pcbAB gene encoding ACV synthetase in Cephalosporium acremonium. Curr Genet. 1990;18:523–530. doi: 10.1007/BF00327023. [DOI] [PubMed] [Google Scholar]
- 15.Keller N P, Bergstrom G C, Yoder O C. Mitotic stability of transforming DNA is determined by its chromosomal configuration in the fungus Cochiobolus heterostrophus. Curr Genet. 1991;19:227–233. [Google Scholar]
- 16.Komatsu K-I, Mizuno M, Kodaira R. Effect of methionine on cephalosporin C and penicillin N production by a mutant of Cephalosporium acremonium. J Antibiot. 1975;28:881–888. doi: 10.7164/antibiotics.28.881. [DOI] [PubMed] [Google Scholar]
- 17.Lewandwska M, Paszewski A. Sulphate and methionine as sulphur sources for cysteine and cephalosporin C synthesis in Cephalosporium acremonium. Acta Microbiol Pol. 1988;37:17–26. [PubMed] [Google Scholar]
- 18.Mansour S L, Thomas K R, Capecchi M R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 19.Marcos, A. T., K. Kosalková, R. E. Cardoza, F. Fierro, S. Gutiérrez, and J. F. Martin. Characterization of the reverse transsulfuration mecB gene of Acremonium chrysogenum encoding a functional cysthationine-γ-lyase. Mol. Gen. Genet, in press. [DOI] [PubMed]
- 20.Martín J F, Aharonowitz Y. Regulation of the biosynthesis of β-lactam antibiotics. In: Demain A L, Solomon N A, editors. Antibiotics containing the β-lactam structure, part I. Berlin, Germany: Springer-Verlag; 1983. pp. 229–254. [Google Scholar]
- 21.Martín J F, Casqueiro J, Kosalková K, Marcos A T, Gutiérrez S. Penicillin and cephalosporin biosynthesis: mechanism of carbon catabolite regulation of penicillin production. Antonie Leeuwenhoek. 1999;75:21–31. doi: 10.1023/a:1001820109140. [DOI] [PubMed] [Google Scholar]
- 22.Martín J F, Gutiérrez S, Demain A L. β-Lactams. In: Anke T, editor. Fungal biotechnology. Weinheim, Germany: Chapman and Hall GmbH; 1997. pp. 91–127. [Google Scholar]
- 23.Nüesch J, Heim J, Treichler H J. The biosynthesis of sulfur-containing β-lactam antibiotics. Annu Rev Microbiol. 1987;41:51–57. doi: 10.1146/annurev.mi.41.100187.000411. [DOI] [PubMed] [Google Scholar]
- 24.Nüesch J, Treichler H J, Liersch M. The biosynthesis of cephalosporin C. In: Vanek Z, Hostalek Z, Cudlin J, editors. Genetics of industrial microorganisms. Vol. 2. Prague, Czechoslovakia: Academia; 1973. pp. 309–334. [Google Scholar]
- 25.Ono B, Hazu T, Yoshida S, Kawato T, Shinoda S, Brzvwczy J, Paszewiski A. Cysteine biosynthesis in Saccharomyces cerevisiae: a new outlook on pathway and regulation. Yeasts. 1999;15:389–397. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1365::AID-YEA468>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 26.Paszewski A, Brzywczy J, Natorff R. Sulphur metabolism. Prog Ind Microbiol. 1994;29:299–319. [PubMed] [Google Scholar]
- 27.Punt P J, Oliver R P, Dingemanse M A, Pouwels P H, van den Hondel C A J J. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56:117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- 28.Ramos F R, López-Nieto J M, Martín J F. Coordinate increase of isopenicillin N synthetase, isopenicillin N epimerase and deacetoxycephalosporin C synthetase in a high cephalosporin-producing mutant of A. chrysogenum and simultaneous loss of the three enzymes in a non-producing mutant. FEMS Microbiol Lett. 1986;35:123–127. [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Sawada Y, Konomi T, Solomon N A, Demain A L. Increase in activity of β-lactam synthetases after growth of Cephalosporium acremonium with methionine or norleucine. FEMS Microbiol Lett. 1980;9:281–284. [Google Scholar]
- 31.Segal I H, Johnson J M. Intermediates in inorganic sulphate utilization by Penicillium chrysogenum. Arch Biochem Biophys. 1963;103:216–226. doi: 10.1016/0003-9861(63)90398-9. [DOI] [PubMed] [Google Scholar]
- 32.Shen Y Q, Wolfe S, Demain A L. Levels of isopenicillin N synthetase and deacetoxycephalosporin C synthetase in Cephalosporium acremonium producing high and low levels of cephalosporin C. Bio/Technology. 1986;4:61–64. [Google Scholar]
- 33.Tilburn J, Scazzochio C, Taylor G G, Zabicky-Zissman J H, Lockington R A, Davies R W. Transformation by integration in Aspergillus nidulans. Gene. 1983;26:205–221. doi: 10.1016/0378-1119(83)90191-9. [DOI] [PubMed] [Google Scholar]
- 34.Treichler H J, Liersch M, Nüesch J, Dobeli H. Role of sulfur metabolism in cephalosporin C and penicillin biosynthesis. In: Sebek O K, Laskin A I, editors. Genetics of industrial microorganisms. Washington, D.C.: American Society for Microbiology; 1979. p. 97. [Google Scholar]
- 35.Velasco J, Gutiérrez S, Fernández F J, Marcos A T, Arenós C, Martín J F. Exogenous methionine increases levels of mRNAs transcribed from pcbAB, pcbC, and cefEF genes, encoding enzymes of the cephalosporin biosynthetic pathway in Acremonium chrysogenum. J Bacteriol. 1994;176:985–991. doi: 10.1128/jb.176.4.985-991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walz M, Kück U. Targeted integration into the Acremonium chrysogenum genome: disruption of the pcbC gene. Curr Genet. 1993;24:421–427. doi: 10.1007/BF00351851. [DOI] [PubMed] [Google Scholar]
- 37.Yelton M M, Hamer J E, Timberlake W E. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci USA. 1984;81:1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zanca D. Regulación de la biosíntesis de la penicilina Ny de la cefalosporina C en cepas de Cephalosporium acremonium. Ph.D. thesis. Salamanca, Spain: University of Salamanca; 1982. [Google Scholar]
- 39.Zhang J Y, Banko G, Wolfe S, Demain A L. Methionine induction of ACV synthetase in Cephalosporium acremonium. J Ind Microbiol. 1987;2:251–255. [Google Scholar]