Abstract
Arthrobacter sp. strain TAD20, a chitinolytic gram-positive organism, was isolated from the sea bottom along the Antarctic ice shell. Arthrobacter sp. strain TAD20 secretes two major chitinases, ChiA and ChiB (ArChiA and ArChiB), in response to chitin induction. A single chromosomal DNA fragment containing the genes coding for both chitinases was cloned in Escherichia coli. DNA sequencing analysis of this fragment revealed two contiguous open reading frames coding for the precursors of ArChiA (881 amino acids [aa]) and ArChiB (578 aa). ArChiA and ArChiB are modular enzymes consisting of a glycosyl-hydrolase family 18 catalytic domain as well as two and one chitin-binding domains, respectively. The catalytic domain of ArChiA exhibits 55% identity with a chitodextrinase from Vibrio furnissii. The ArChiB catalytic domain exhibits 33% identity with chitinase A of Bacillus circulans. The ArChiA chitin-binding domains are homologous to the chitin-binding domain of ArChiB. ArChiA and ArChiB were purified to homogeneity from the native Arthrobacter strain and partially characterized. Thermal unfolding of ArChiA, ArChiB, and chitinase A of Serratia marcescens was studied using differential scanning calorimetry. ArChiA and ArChiB, compared to their mesophilic counterpart, exhibited increased heat lability, similar to other cold-adapted enzymes.
Chitin, the second most abundant biopolymer in nature next to cellulose, is an insoluble homopolysaccharide composed of β-1,4-linked N-acetylglucosamine (GlcNAc) residues. This polysaccharide is found in fungi, algae, and especially in the exoskeletons of insects and crustaceans. The turnover of chitin in the aquatic biosphere is enormous and mediated by chitinolytic bacteria (37). Chitinases (EC 3.2.1.14) hydrolyze the β-1,4-linkages in chitin, yielding predominantly N-N′-diacetyl chitobiose, which is further degraded by chitobiases to the GlcNAc monomer.
Several chitinases from bacteria have been cloned and expressed in Escherichia coli (6, 7, 18, 35). Furthermore, the structure of two chitinases has been elucidated (16, 23). Based on the amino acid sequence similarity of their catalytic domains, chitinases are classified into two unrelated families in the glycosylhydrolase classification system (15). Family 18 includes chitinases from bacteria, fungi, animals, and certain plants, while family 19 comprises chitinases of plant origin. Bacterial chitinases generally consist of multiple functional domains, such as chitin-binding domains (ChBDs) and fibronectin type III-like domains, linked to the catalytic domain. The importance of the ChBDs in the degradation of insoluble chitin has been demonstrated for several bacterial chitinases (4, 20, 29, 34).
Psychrophilic microorganisms growing at ∼5°C can be found in several permanently cold environments. Psychrophilic enzymes produced by such microorganisms display a high specific activity at low and moderate temperatures and are most often, if not always, associated with high thermosensitivity (14). These properties can be extremely useful for various applications. During the last years, several psychrophilic enzymes have been isolated (11, 13, 24, 25, 31), and the structure of four of them has been elucidated (1, 3, 19, 26).
In this study, we report cloning, sequence, and characterization of the genes coding for the precursors of two chitinases, ArChiA and ArChiB, from the Antarctic, aerobic gram-positive Arthrobacter sp. strain TAD20. The purification and partial characterization of the enzymes from the native strain are also described.
MATERIALS AND METHODS
Bacterial strains, plasmids, and enzymes.
The chitinolytic strain TAD20 was isolated from sea sediments at the Dumont d'Urville Antarctic station (60°40′ S, 40°01′ E) in 1993. It was identified as an Arthrobacter sp. in the Laboratory of Microbiology (Jean Swings) at the University of Ghent (Belgium) by analysis of fatty acid composition and comparison of the profile with the MIDI database. Selection for chitinolytic activity was carried out on Marine agar 2216E (Difco) containing 1% colloidal chitin prepared as described by Hsu and Lockwood (17). E. coli X11-Blue was purchased from Stratagene. Reagents for bacterial media were from Difco. The pSP72 plasmid was purchased from Promega. The enzymes for molecular biology were obtained from Stratagene, Boehringer Mannheim, and Gibco-BRL and used according to the instructions of the manufacturers.
Effect of temperature on growth of Arthrobacter sp. strain TAD20 and enzyme secretion.
Arthrobacter sp. strain TAD20 was cultivated in nutrient broth (Difco) supplemented with 0.1% colloidal chitin in order to measure enzyme secretion at various temperatures. TAD20 was cultured aerobically at 4°C under vigorous shaking in 500-ml Erlenmeyer flasks containing 100 ml of medium. Growth was monitored by turbidity (optical density) measurements at 580 nm. Assays of chitinase were carried out using 0.1 mM p-nitrophenyl N,N′-diacetylchitobiose (pNP-chitobiose) (Sigma) using conditions described under the enzyme assay section. Activities are expressed as micromoles of substrate hydrolyzed per milliliter of sample.
Microorganism cultivation.
The Antarctic strain was cultivated in shake cultures for 5 days at 5°C in 3 liters of medium consisting of 5 g of Bactotryptone, 1 g of yeast extract, 33 g of sea salts, and 1 g of colloidal chitin (pH 7.3) per liter (17) to induce secretion of the enzymes.
Enzyme purification.
After centrifugation of the cell culture at 11,000 × g for 15 min, the supernatant was concentrated up to 400 ml and dialyzed against 20 mM Tris-HCl (pH 6.5) using a Minitan tangential flow ultrafiltration unit (Millipore) fitted with PTCGC membranes (10-kDa cutoff). The sample was then loaded on a QFF-Sepharose column (2 by 20 cm) equilibrated in the abovementioned buffer. The flowthrough was dialyzed against 20 mM Tris-HCl (pH 8), loaded on another QFF-Sepharose column (2 by 20 cm) equilibrated in 20 mM Tris-HCl (pH 8), and eluted with an NaCl linear gradient (0 to 200 mM, 250 to 250 ml). ArChiA active fractions were pooled and stored at 5°C. ArChiB active fractions in the flowthrough were concentrated to 10 ml, applied on a Sephacryl S-200 column (2.5 by 100 cm), and eluted with 20 mM Tris-HCl–100 mM NaCl (pH 7.5). Active fractions corresponding to pure chitinase B (± 82 mg) were pooled and stored at 5°C. Under these conditions, the enzymes were stable for at least 3 months.
Cloning and sequencing.
All general techniques used were described by Sambrook et al. (27). Genomic DNA of Arthrobacter sp. strain TAD20 was digested with EcoRI, and the resulting fragments were ligated to EcoRI-cleaved and alkaline phosphatase-treated pSP72. The recombinant plasmids were used to transform E. coli XL1-Blue competent cells. Transformants were grown at 18°C on Luria-Bertani (LB) agar plates containing 100 μg of ampicillin/ml.
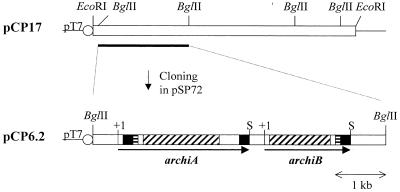
Mature colonies were transferred on nylon membranes (Amersham Life Science), set down on a paper (Wattman), and wetted with a solution of 0.01 mM 4-methylumbelliferyl N,N′-diacetylchitobiose (4-MU-chitobiose), 4-methylumbelliferyl N,N′,N′-triacetylchitotriose (4-MU-chitotriose) (Sigma), and 20 mM HEPES (pH 7.5). Hydrolysis of these substrates results in the liberation of fluorescent 4-methylumbelliferone. Three identical positive clones carrying a 17-kb insert (termed pCP17) were identified by a fluorescent halo when visualized under UV light. A 6.2-kb BglII fragment subcloned from pCP17 was found to carry the DNA coding for ArChiA and ArChiB. It was ligated with pSP72 digested by BglII to give pCP6.2.
The nucleotide sequence of the 6.2-kb fragment was determined by a subcloning strategy and by gene walking with custom sequencing primers using the dideoxy chain termination method (28) on denatured double-stranded DNA templates with an ALF DNA sequencer (Pharmacia) and fluorescein-labeled primers.
Enzyme assay.
Chitinolytic activity was routinely assayed at 25°C using 0.1 mM pNP-chitobiose (Sigma) as the substrate for ArChiA and Serratia marcesens chitinase A (SmChiA) and 0.1 mM pNP-chitotriose (p-nitrophenyl-N,N′,N"-triacetylchitotriose) (Sigma) as the substrate for ArChiB in 20 mM HEPES (pH 7.5). Activities were recorded in a thermostated Uvicon 860 spectrophotometer (Kontron) and calculated on the basis of an extinction coefficient for p-nitrophenol of 14,700 (M × cm)−1 at 405 nm. For comparative studies, preliminary experiments were performed to determine the effect of pH, buffer composition, and monovalent and divalent ions on the activity and stability of ArChiA, ArChiB, and SmChiA. The thermal stability of the enzymes was measured by incubating the enzymes at 50°C at a concentration of 10 μg/μl in the corresponding optimal buffer for stability, i.e., 20 mM bis-Tris (pH 6.1) for ArChiA, 20 mM HEPES (pH 7.3) for ArChiB, and 20 mM HEPES (pH 6.6) for SmChiA. Aliquots were taken at different times, and enzyme activity was measured using the routine assay conditions described above.
Sequence analysis.
Similarity searches were performed with updated versions of the Blast (2) and the Fasta (22) programs, using the facilities of the Greek EMBnet Node. The Genedoc program (http://www.psc.edu/biome/genedoc) was used for editing and shadowing the sequence alignment.
Other methods.
Protein concentration was measured using the bicinchoninic acid protein assay reagent (Pierce). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was run as described by the supplier of the electrophoresis equipment (Hoeffer Scientific Instruments). Isoelectric focusing was run on a Fast System Separation and Control Unit from Pharmacia using gels with a 3-10 pH gradient. The NH2-terminal amino acid sequence of ArChiA and ArChiB was determined using a pulsed liquid-phase protein sequenator (Applied Biosystems 477A) equipped with an on-line 120A phenylthiohydantoin analyzer. C-terminal amino acid sequences were obtained on a Procise 494CT sequencer (Applied Biosystems, Perkin-Elmer Division) equipped for alkylthiohydantion analysis.
Differential scanning calorimetry (DSC) measurements were performed using a MicroCal MCS-DSC instrument at a scan rate of 60 K h−1 and under a nitrogen pressure of 2 atm. Samples were dialyzed overnight against the appropriate buffer, the dialysate being used in the reference cell and for buffer base line determination. Protein concentration after dialysis was ∼4 mg/ml for ArChiA and ArChiB and ∼2 mg/ml for SmChiA. Buffers used were those ensuring optimal stability for the enzymes, as previously described.
Denaturation curves for ArChiA and ArChiB were analyzed using MicroCal Origin software (version 2.9).
Nucleotide sequence accession numbers.
The nucleotide sequences of ArchiA and ArchiB have been deposited and assigned accession numbers AJ250585 and AJ250586, respectively, in the EMBL database.
RESULTS
Effect of temperature on enzyme secretion.
Arthrobacter strain TAD20 was grown at 4, 17, and 24°C. Chitinase was assayed as described in Materials and Methods. Highest activity was observed in supernatants of cultures incubated at 4°C, similar to other enzymes from psychrophilic bacteria (10). Chitinase activity could not be detected when TAD20 was grown at 17 or 24°C.
Cloning and sequencing of archiA and archiB.
The structural genes for ArChiA and ArChiB were cloned from a genomic library of the Antarctic bacterium Arthrobacter sp. strain TAD20. In order to prevent thermal denaturation of the cloned product, E. coli transformants were grown at 18°C. Three thousand transformants were transferred on nylon membranes and screened for activity on a mixture of 4-MU-chitobiose and 4-MU-chitotriose. Three identical clones carrying a 17-kb insert (pCP17) were detected by the appearance of a fluorescent halo when exposed to UV light.
From subcloning, a plasmid containing a 6.2-kb fragment (pCP6.2) and conferring chitinase activity on both 4-MU-chitobiose and 4-MU-chitotriose substrates to transformed E. coli cells was isolated (Fig. 1) and sequenced on both strands. The nucleotide sequence revealed two open reading frames of 2,640 and 1,731 bp, coding for ArChiA (archiA) and ArChiB (archiB), respectively. Upstream of the ATG codon of those genes, a short sequence has been identified that may function as a Shine-Dalgarno ribosome-binding site; typical transcription initiation sequences were not identified (Fig. 2). Downstream of the stop codons of archiA and archiB are short inverted repeats which are putative terminators. Upstream of archiA and archiB are inverted repeat sequences which are potential sites for protein binding (21).
FIG. 1.
Cloning of archiA and archiB. The black boxes in archiA and archiB correspond to ChBDs, the boxes with horizontal streaks correspond to Pro/Thr-rich regions, and the boxes with diagonal streaks correspond to the catalytic domains. The circles labeled pT7 show T7 promoter. S, stop codon.
FIG. 2.
Flanking sequences of archiA and archiB. The boxed sequences are inverted repeats that are potential sites for DNA-binding proteins. The Shine-Dalgarno sequence (SD) and the putative terminators are indicated.
ArChiA and ArChiB peptide sequence analysis.
The NH2-terminal amino acid sequences of the purified ArChiA and ArChiB (ASPSGT and AAPPNTA respectively) allowed us to locate the signal peptidase cleavage site, which fulfills the −3, −1 rule of von Heijne (33); the leader peptides of ArChiA and ArChiB are composed of 34 and 38 amino acid residues, respectively. It also adopts the general pattern of prokaryotic signal sequences, i.e., a positively charged amino terminus followed by a hydrophobic core and a string of polar residues (36). Furthermore, C-terminal amino acid sequence analysis of ArChiA and ArChiB (ILCGK and TGACN, respectively) confirmed the C termini deduced from DNA translation. The deduced primary structures of the mature ArChiA and ArChiB consist of 846 and 539 amino acids with a predicted Mr of 89,415 and 57,123, respectively.
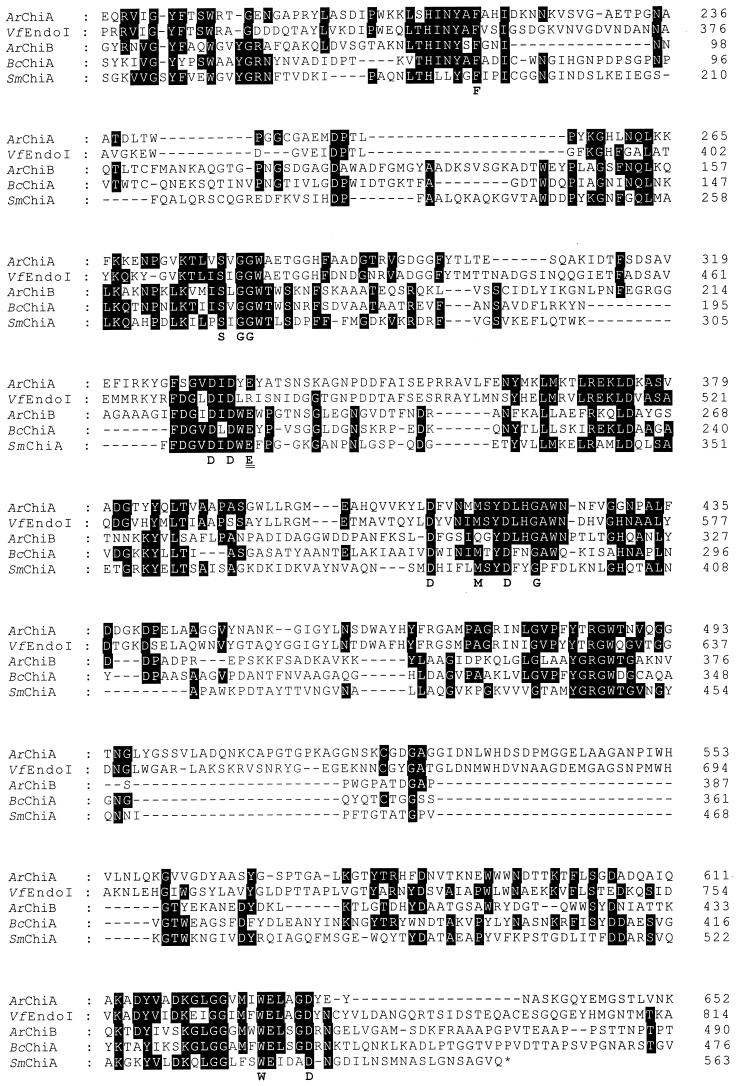
Blast analysis of the amino acid sequence of ArChiA and ArChiB revealed that both enzymes exhibit a catalytic domain as well as two and one ChBDs, respectively (Fig. 1). The catalytic domains of ArChiA (residues 183 to 653) and ArChiB (residues 57 to 473) exhibit 55 and 33% identity, respectively, with homologous regions of the chitodextrinase of Vibrio furnissii (18) and the chitinase A1 of Bacillus circulans WL-12, respectively (35) (Fig. 3). The identity of the catalytic domains of ArChiA and ArChiB with that of the crystallised chitinase A of S. marcencens (SmChiA) (23) is 29% in a 458-amino-acid (aa) overlap for ArChiA and 26.5% in a 475-aa overlap for ArChiB (Fig. 3).
FIG. 3.
Sequence alignment of ArChiA, ArChiB, several bacterial chitinases, and a chitodextrinase. The alignment was constructed using the multiple alignment program (PILEUP) from the Greek EMBnet Node. VfEndoI, chitodextrinase of V. furnissii; BcChia, chitinase A of B. circulans; SmChiA, chitinase A of S. marcescens. Amino acid numbering is indicated on the right. The amino acids under the sequence alignment are the consensus sequence determined from the alignment of five bacterial chitinases as described in the text. The catalytic residue is double underlined.
At the N and C termini of ArChiA and at the C terminus of ArChiB, similar (ChBDs) occur, namely, ChBDA1 (residues 44 to 93) and ChBDA2 (residues 827 to 878) for ArChiA and ChBDB (residues 425 to 578) for ArChiB (Fig. 4).
FIG. 4.
Sequence alignment of the ChBDs of ArChiA (ChBDA1 and ChBDA2) and ArChiB (ChBDB) and of Alteromonas sp. strain O-7 chitinase A (ChBDaltso). The alignment was constructed using the multiple alignment program (PILEUP) from the Greek EMBnet Node. Amino acid numbering is indicated on the right. The amino acids under the sequence alignment are the consensus sequence determined from the alignment of putative bacterial ChBDs as described in the text. @, aromatic residue.
Characterization of ArChiA and ArChiB.
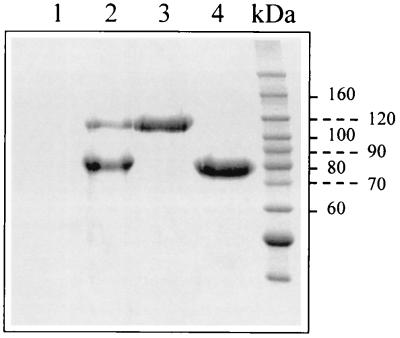
The apparent molecular masses on SDS-PAGE of the purified ArChiA (±110,000 Da) and ArChiB (±80,000 Da) (Fig. 5) were higher than those the estimated one from the DNA sequence translation (89,415 and 57,123 Da, respectively). The isoelectric points of ArChiA and ArChiB are 5.7 and 8.1, respectively. No ion was found to increase the activity of these enzymes, and they retain 100% of their activity in the presence of a 10 mM EDTA solution. Optimal buffer for activity was 20 mM HEPES (pH 7.3 to 8) for ArChiA and ArChiB. No ion was found to increase the stability of the enzymes from Arthrobacter or S. marcescens. Optimal buffer for stability was 20 mM bis-Tris (pH 6.1) for ArChiA and 20 mM HEPES (pH 7.3) for ArChiB.
FIG. 5.
SDS-PAGE of ArChiA and ArChiB secreted by Arthrobacter sp. strain TAD20. Lanes 1 and 2, 20 μl of the supernatants of cultures grown in the absence of chitin (lane 1) or in the presence of 0.1% colloidal chitin (lane 2). All cultures (10 ml) were grown at 5°C under identical conditions in a medium containing 2 g of yeast extract, 5 g of tryptone, and 33 g of sea salts per liter (pH 7.2). Colloidal chitin was prepared as described by Hsu and Lockwood (17). Lanes 3 and 4, purified ArChiA and ArChiB, respectively. The gel was stained using Coomassie brilliant blue.
Thermal stability.
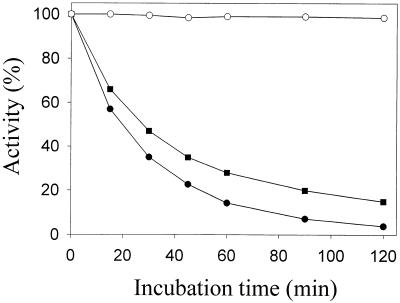
The denaturation curves of the psychrophilic and mesophilic enzymes were recorded at 50°C in the corresponding optimal buffers for stability (Fig. 6), showing that the psychrophilic chitinases are less stable than their mesophilic counterpart, a common characteristic of cold-adapted enzymes.
FIG. 6.
Thermal stability of the chitinases of Arthrobacter sp. strain TAD20 and the mesophilic chitinase A of S. marcescens. ArChiA (●), ArChiB (■), and SmChiA (○) were incubated at 50°C for the indicated periods of time in 20 mM bis-Tris (pH 6.1) for ArChiA, 20 mM HEPES (pH 7.3) for ArChiB, and 20 mM HEPES (pH 6.6) for SmChiA.
DSC.
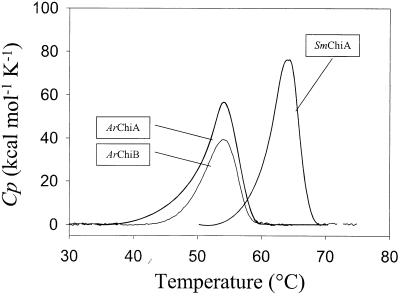
The denaturation curves of ArChiA, ArChiB, and SmChiA show single peaks with apparent Tms of 54.3, 54, and 64.2°C, respectively (Fig. 7). Calculation of the areas under the heat absorption peaks determined the calorimetric denaturation enthalpy (ΔHcal) of ArchiB (415 kcal/mol), ArChiB (270 kcal/mol), and SmChiA (449 kcal/mol).
FIG. 7.
Thermal unfolding of the chitinases of Arthrobacter sp. strain TAD20 and the mesophilic chitinase A of S. marcescens. Microcalorimetric records of ArChiA, ArChiB, and SmChiA. Experiments were performed in 20 mM bis-Tris (pH 6.1) for ArChiA, 20 mM HEPES (pH 7.3) for ArChiB, and 20 mM HEPES (pH 6.6) for SmChiA.
DISCUSSION
In this report, we describe for the first time the cloning, sequencing, and characterization of two chitinase genes from the Antarctic marine strain Arthrobacter sp. strain TAD20. This strain secretes mainly two chitinases, ArChiA (∼10 mg/liter) and ArChiB (∼40 mg/liter) in response to chitin induction (Fig. 5). A single 17-kb chromosomal DNA fragment of Arthrobacter sp. strain TAD20 containing the genes coding for the precursors of ArChiA and ArChiB was cloned in E. coli using a mixture of 4-MU-chitobiose and 4-MU-chitotriose in order to detect positive clones which appeared fluorescent under UV light. Attempts to screen the transformed cells on plates containing colloidal chitin (32) were unsuccessful, possibly due either to the fact that the cloned chitinases are not secreted by E. coli or that their expression level is under the detection limit (12).
Upstream of archiA and archiB, inverted repeat sequences were identified (Fig. 2). The marked differences between these sequences provide a good indication that archiA and archiB are regulated independently and that the mode of their regulation is different.
The archiA and archiB genes encode the precursors of modular chitinases composed of an N-terminal signal peptide, a catalytic domain of the glycosyl-hydrolase family 18, as well as two and one ChBDs, respectively (Fig. 1).
Chitinases follow the general acid-base catalytic mechanism (8), and from sequence comparison with SmChiA, Glu-335 of ArChiA and Glu-230 of ArChiB are predicted to be the catalytic residues, acting as proton donors (23, 30).
The N-terminal signal sequences of ArChiA (34 aa) and ArChiB (38 aa) are relatively long, a common characteristic of enzymes secreted by gram-positive organisms (35). The catalytic domain of ArChiA exhibits the best homology (55% identity) with a chitodextrinase of V. furnissii. However, ArChiA, in contrast to chitodextrinase, is active on insoluble chitin (18). Furthermore, ArChiA carries two ChBDs, while the chitodextrinase carries none, which is in support of the observed difference in substrate specificity (34). The catalytic domain of ArChiB exhibits the best homology (33% identity) with chitinase A of B. circulans and a low homology (26% identity) with ArChiA (Fig. 3).
The ChBDs of ArChiA and ArChiB are 50 to 60 residues long, similar to other ChBDs, and exhibit good homology (50% identity) with the ChBD of chitinase A from Alteromonas sp. (Fig. 4) (32). Sequence alignment of this ChBD with ChBDA1, ChBDA2, and ChBDB revealed a consensus sequence which appears to be quite well conserved in several bacterial ChBDs (4, 34). Furthermore, Trp and Tyr residues are conserved, suggesting that these aromatic side chains might be involved in the stacking against the pyranosyl rings of N-acetylglucosamine residues in chitin (5). Finally, ChBDA1 and ChBDB are linked to the catalytic domain via a long Pro/Thr-rich region, while ChBDA2 is linked via a 9-aa (815 to 823) sequence containing six glycines, both regions acting as flexible hinges.
The native ArChiA and ArChiB were purified to homogeneity employing conventional chromatographic techniques. The optimum pH was 7.3 to 8 for both enzymes. The apparent molecular masses of ArChiA and ArChiB as determined by SDS-PAGE were 110 and 80 kDa, respectively, higher than those estimated from the deduced amino acid sequence (Fig. 5). However, C-terminal amino acid analysis for both enzymes confirmed the C termini deduced from DNA translation.
Increased flexibility related to increased heat lability has been proposed to be the main structural feature of cold-adapted enzymes, allowing conformational changes necessary to reach the transition state enabling catalysis (14). The results obtained by thermal denaturation (Fig. 6) and DSC (Fig. 7) demonstrate that under optimal conditions, ArChiA and ArChiB are less stable than their mesophilic counterpart SmChiA. The psychrophilic enzymes exhibited remarkable thermal lability. Following incubation of ArChiA and ArChiB at 50°C for 60 min, the enzymes retained 18 and 30% of their original activity, respectively, while SmChiA retained almost 100% of the original activity. DSC denaturation curves show that, compared to SmChiA, the psychrophilic chitinases have a lower apparent Tm (54.3 and 54 for ArChiA and ArChiB, respectively, and 64.2 for SmChiA) as well as a significant lower calorimetric denaturation enthalpy per mole of residue. The nonsymmetrical shape of the denaturation curves is probably the result of a multitransition process combined with an aggregation phenomenon. For this reason, deconvolution of the denaturation curves into symmetrical components was not attempted.
The values of the denaturation enthalpy per residue are 490, 501, and 907 cal (mol of residue)−1 for ArChiA, ArChiB, and SmChiA, respectively. Although these values are small, possibly due to aggregation phenomena, they are comparable to those found for the psychrophilic α-amylase from Alteromonas haloplanctis [525 cal (mol of residue)−1] and the mesophilic α-amylase from Bacillus amyloliquefaciens [1,008 cal (mol of residue)−1] (9).
The relationship between stability, specific activity, and flexibility for ArChiA and ArChiB is now under study in our laboratory.
REFERENCES
- 1.Aghajari N, Feller G, Gerday C, Haser R. Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 1998;7:564–572. doi: 10.1002/pro.5560070304. . (Erratum, 7:1481.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller E, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez, M., J. P. Zeelen, V. Mainfroid, F. Rentier-Delrue, J. A. Martial, L. Wyns, R. K. Wierenga, and D. Maes. Triose-phosphate isomerase (TIM) of the psychrophilic bacterium Vibrio marinus. Kinetic and structural properties. J. Biol. Chem. 273:2199–2206. [DOI] [PubMed]
- 4.Blaak H, Schrempf H. Binding and substrate specificities of a Streptomyces olivaceoviridis chitinase in comparison with its proteolytically processed form. Eur J Biochem. 1995;229:132–139. doi: 10.1111/j.1432-1033.1995.tb20447.x. [DOI] [PubMed] [Google Scholar]
- 5.Brun E, Moriaud F, Gans P, Blackledge M, Barras F, Marion D. Solution structure of the cellulose-binding domain of the endoglucanase Z secreted by Erwinia chrysanthemi. Biochemistry. 1997;36:16074–16086. doi: 10.1021/bi9718494. [DOI] [PubMed] [Google Scholar]
- 6.Brurberg M B, Nes I F, Eijsink V G. Comparative studies of chitinases A and B from Serratia marcescens. Microbiology. 1996;142:1581–1589. doi: 10.1099/13500872-142-7-1581. [DOI] [PubMed] [Google Scholar]
- 7.Chernin L S, De la Fuente L, Sobolev V, Haran S, Vorgias C E, Oppenheim A B, Chet I. Molecular cloning, structural analysis, and expression in Escherichia coli of a chitinase gene from Enterobacter agglomerans. Appl Environ Microbiol. 1997;63:834–839. doi: 10.1128/aem.63.3.834-839.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 9.Feller G, d'Amico D, Gerday C. Thermodynamic stability of a cold-active alpha-amylase from the Antarctic bacterium Alteromonas haloplanctis. Biochemistry. 1999;38:4613–4619. doi: 10.1021/bi982650+. [DOI] [PubMed] [Google Scholar]
- 10.Feller G, Narynx E, Arpigny J L, Zekhini Z, Swings J, Gerday C. Temperature dependence of growth, enzyme secretion and activity of psychrophilic Antarctic bacteria. Appl Microbiol Biotechnol. 1994;41:477–479. [Google Scholar]
- 11.Feller G, Zekhnini Z, Lamotte-Brasseur J, Gerday C. Enzymes from cold-adapted microorganisms: the class C beta-lactamase from the Antarctic psychrophile Psychrobacter immobilis A5. Eur J Biochem. 1997;244:186–191. doi: 10.1111/j.1432-1033.1997.00186.x. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs R L, McPherson S A, Drahos D J. Cloning of a Serratia marcescens gene encoding chitinase. Appl Environ Microbiol. 1986;51:504–509. doi: 10.1128/aem.51.3.504-509.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galkin A, Kulakova L, Ashida H, Sawa Y, Esaki N. Cold-adapted alanine dehydrogenases from two Antarctic bacterial strains: gene cloning, protein characterization, and comparison with mesophilic and thermophilic counterparts. Appl Environ Microbiol. 1999;65:4014–4020. doi: 10.1128/aem.65.9.4014-4020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerday C, Aittaleb M, Arpigny J L, Baise E, Chessa J P, Garsoux G, Petrescu I, Feller G. Psychrophilic enzymes: a thermodynamic challenge. Biochim Biophys Acta. 1997;1342:119–131. doi: 10.1016/s0167-4838(97)00093-9. [DOI] [PubMed] [Google Scholar]
- 15.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollis T, Monzingo A F, Bortone K, Ernst S, Cox R, Robertus J D. The X-ray structure of a chitinase from the pathogenic fungus Coccidioides immitis. Protein Sci. 2000;9:544–551. doi: 10.1110/ps.9.3.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu S C, Lockwood J L. Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl Microbiol. 1975;29:422–426. doi: 10.1128/am.29.3.422-426.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keyhani N O, Roseman S. The chitin catabolic cascade in the marine bacterium Vibrio furnissii: molecular cloning, isolation, and characterization of a periplasmic chitodextrinase. J Biol Chem. 1996;271:33414–33424. doi: 10.1074/jbc.271.52.33414. [DOI] [PubMed] [Google Scholar]
- 19.Kim S Y, Hwang K Y, Kim S H, Sung H C, Han Y S, Cho Y. Structural basis for cold adaptation: sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum. J Biol Chem. 1999;274:11761–11767. doi: 10.1074/jbc.274.17.11761. [DOI] [PubMed] [Google Scholar]
- 20.Morimoto K, Karita S, Kimura T, Sakka K, Ohmiya K. Cloning, sequencing, and expression of the gene encoding Clostridium paraputrificum chitinase ChiB and analysis of the functions of novel cadherin-like domains and a chitin-binding domain. J Bacteriol. 1997;179:7306–7314. doi: 10.1128/jb.179.23.7306-7314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pabo C O. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 22.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 23.Perrakis A, Tews I, Dauter Z, Oppenheim A B, Chet I, Wilson K S, Vorgias C E. Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure. 1994;2:1169–1180. doi: 10.1016/s0969-2126(94)00119-7. [DOI] [PubMed] [Google Scholar]
- 24.Rina M, Caufrier F, Markaki M, Mavromatis K, Kokkinidis M, Bouriotis V. Cloning and characterization of the gene encoding PspPI methyltransferase from the Antartic psychrotroph Psychrobacter sp. strain TA137: predicted interactions with DNA and organization of the variable region. Gene. 1997;197:353–360. doi: 10.1016/s0378-1119(97)00283-7. [DOI] [PubMed] [Google Scholar]
- 25.Rina M, Pozidis C, Mavromatis K, Tzanodaskalaki M, Kokkinidis M, Bouriotis V. Alkaline phosphatase from the Antarctic strain TAB5. Properties and psychrophilic adaptations. Eur J Biochem. 2000;267:1230–1238. doi: 10.1046/j.1432-1327.2000.01127.x. [DOI] [PubMed] [Google Scholar]
- 26.Russell R J, Gerike U, Danson M J, Hough D W, Taylor G L. Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium. Structure. 1998;6:351–361. doi: 10.1016/s0969-2126(98)00037-9. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svitil A L, Kirchman D L. A chitin-binding domain in a marine bacterial chitinase and other microbial chitinases: implications for the ecology and evolution of 1,4-beta-glycanases. Microbiology. 1998;144:1299–1308. doi: 10.1099/00221287-144-5-1299. [DOI] [PubMed] [Google Scholar]
- 30.Tews I, Perrakis A, Oppenheim A, Dauter Z, Wilson K S, Vorgias C E. Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat Struct Biol. 1996;3:638–648. doi: 10.1038/nsb0796-638. [DOI] [PubMed] [Google Scholar]
- 31.Tsigos I, Velonia K, Smonou I, Bouriotis V. Purification and characterization of an alcohol dehydrogenase from the Antarctic psychrophile Moraxella sp. TAE123. Eur J Biochem. 1998;254:356–362. doi: 10.1046/j.1432-1327.1998.2540356.x. [DOI] [PubMed] [Google Scholar]
- 32.Tsujibo H, Orikoshi H, Tanno H, Fujimoto K, Miyamoto K, Imada C, Okami Y, Inamori Y. Cloning, sequence, and expression of a chitinase gene from a marine bacterium, Altermonas sp. strain O-7. J Bacteriol. 1993;175:176–181. doi: 10.1128/jb.175.1.176-181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Von Heijne G. Signal sequences: the limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe T, Ito Y, Yamada T, Hashimoto M, Sekine S, Tanaka H. The roles of the C-terminal domain and type III domains of chitinase A1 from Bacillus circulans WL-12 in chitin degradation. J Bacteriol. 1994;176:4465–4472. doi: 10.1128/jb.176.15.4465-4472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe T, Suzuki K, Oyanagi W, Ohnishi K, Tanaka H. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J Biol Chem. 1990;265:15659–15665. [PubMed] [Google Scholar]
- 36.Watson M. Compilation of published signal sequences. Nucleic Acids Res. 1984;12:5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu C, Bassler B L, Roseman S. Chemotaxis of the marine bacterium Vibrio furnissii to sugars: a potential mechanism for initiating the chitin catabolic cascade. J Biol Chem. 1993;268:9405–9409. [PubMed] [Google Scholar]