Abstract
The phototrophic bacterium Rhodobacter capsulatus is able to reduce 2,4-dinitrophenol (DNP) to 2-amino-4-nitrophenol enzymatically and thus can grow in the presence of this uncoupler. DNP reduction was switched off by glutamine or ammonium, but this short-term regulation did not take place in a draTG deletion mutant. Nevertheless, the target of DraTG does not seem to be the nitrophenol reductase itself since the ammonium shock did not inactivate the enzyme. In addition to this short-term regulation, ammonium or glutamine repressed the DNP reduction system. Mutants of R. capsulatus affected in ntrC or rpoN exhibited a 10-fold decrease in nitroreductase activity in vitro but almost no DNP activity in vivo. In addition, mutants affected in rnfA or rnfC, which are also under NtrC control and encode components involved in electron transfer to nitrogenase, were unable to metabolize DNP. These results indicate that NtrC regulates dinitrophenol reduction in R. capsulatus, either directly or indirectly, by controlling expression of the Rnf proteins. Therefore, the Rnf complex seems to supply electrons for both nitrogen fixation and DNP reduction.
The industrial production and abusive use of dyes, explosives, herbicides, pesticides, and drugs result in the release of nitroaromatic compounds into the environment (26). These xenobiotic compounds are resistant to oxygenolytic reactions since the nitroaromatic ring is rendered impervious to electrophilic attack, especially in the case of polynitroaromatics. Therefore, microorganisms have developed reductive pathways that facilitate the metabolism of these recalcitrant compounds. The process may began with reduction of the aromatic ring (5, 18) or with reduction of the nitro group to the corresponding amino or hydroxylamino derivatives that can be assimilated upon release of ammonium and hydroxyaromatic adducts (11, 24).
Under light and anaerobiosis, Rhodobacter capsulatus cometabolizes the uncoupler 2,4-dinitrophenol (DNP) by reducing it to 2-amino-4-nitrophenol, which is almost stoichiometrically accumulated in the medium (2). The reaction is catalyzed by a cytosolic and homodimeric Flavin mononucleotide-linked, 54-kDa nitrophenol reductase (NPR) (3). Once DNP is consumed, the cells began to grow by fixing the dinitrogen dissolved in the medium.
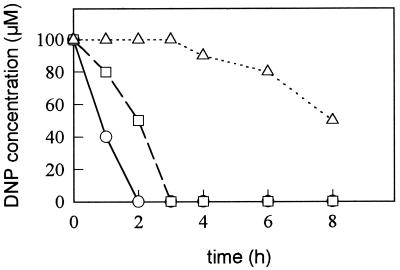
The reduction of DNP in R. capsulatus is repressed by ammonium since the process does not takes place in ammonium-grown cells in the presence of chloramphenicol (2). To assess if the reduction of DNP is activated in N2-fixing cultures, cells cultured as previously described (2) with glutamate (nitrogenase-derepressing conditions), ammonium (negative control), and glutamate plus DNP (positive control) as nitrogen sources were transferred to media with DNP. DNP consumption and NPR activity were determined as published elsewhere (3). As expected, the maximal rate of DNP photoreduction was observed in cells previously cultured with DNP (Fig. 1), showing a NPR activity of 3.2 ± 0.5 mU mg−1. The consumption of DNP by these cells was independent of the presence of chloramphenicol (not shown). If the cells were precultured with glutamate, the rate of DNP reduction was lower (Fig. 1). The NPR activity of extracts from these cells was 0.3 ± 0.02 mU mg−1. In this case, the addition of chloramphenicol inhibited DNP uptake and reduction, which completely ceased after 2 to 3 h incubation (not shown). In contrast, the ammonium-grown cells started to photoreduce DNP after a period of 3 h (Fig. 1), and NPR activity was very low (0.03 ± 0.01 mU mg−1). In this case, DNP reduction does not take place in the presence of chloramphenicol (2). These results suggest that the pathway for DNP metabolism (transport and reduction) is completely repressed in the presence of ammonium, whereas under nitrogenase-derepressing conditions (with glutamate or dinitrogen) the pathway is fully activated only if the substrate DNP is present. The activating effect of DNP was dependent on de novo protein synthesis and can be explained taking into account the lability of the enzyme in the absence of DNP (6). This conclusion was corroborated by staining the diaphorase activity of the NPR protein in nondenaturing pore-gradient polyacrylamide gel electrophoresis and by two-dimensional gel electrophoresis (not shown).
FIG. 1.
Time course of DNP photoreduction by R. capsulatus cultured with different nitrogen sources. Cells grown with ammonium-acetate (▵), glutamate-acetate (□) and glutamate-DNP-acetate (○) were washed and incubated with DNP-acetate. DNP concentration was determined in aliquots taken from the cultures at the times indicated, 2-amino-4-nitrophenol produced was almost stoichiometric with the DNP consumed (not shown). Data are from a single experiment; two independent experiments yielded essentially the same results.
The regulation pattern of DNP reduction by ammonium resembles that described for nitrogen fixation in R. capsulatus. The expression of Nif (nitrogen fixation) proteins in R. capsulatus is enhanced by a high intracellular [2-oxoglutarate]/[glutamine] ratio, which activates a cascade mechanism that includes the NtrC-NtrB (NifR1-NifR2) two-component regulatory system, which activates its target genes (e.g., nifA) in concert with RNA polymerase containing the housekeeping ς70 factor (9). Expression of nif genes is then activated by NifA, which interacts with RNA polymerase harboring ς54 (RpoN) (15, 16, 17, 19). The Ntr system detects the oscillations of the glutamine concentration inside the cells, so that the irreversible inhibition of glutamine synthetase by L-methionine-SR-sulfoximine (MSX) prevents the inorganic nitrogen metabolism from being regulated by ammonium (1, 10).
To study the regulatory links between nitrogen fixation and DNP photoreduction, we examined some mutants affected in nitrogen fixation (19).
The R. capsulatus mutants used were defective in the nitrogenase structural genes (nifHDK), in regulatory components (ntrC and poN), and in genes encoding proteins involved in electron transport to nitrogenase (nifF, rnfF, rnfA, rnfC, and orf14) (13, 25). Deletion of the nitrogenase structural genes did not affect DNP photoreduction (Table 1). NifF is a flavodoxin required for electron transfer to nitrogenase under iron deprivation (27). The molecular properties of this protein resemble those of NPR (3), but nifF mutants showed an NPR+ phenotype (Table 1). In addition, NPR did not react against NifF antibodies (data not shown), indicating that NifF and NPR are two different proteins. The rnf cluster consists of two operons, both essential for nitrogen fixation in the light (25) or dark (23). One rnf operon (orf14, fdxC, fdxN, rnfF, and orf10) does not seem to be involved in supplying electrons to NPR since mutants with insertion mutations in orf14 (with the interposon integrated in both directions) and in rnfF reduced DNP to the same extent as the wild type (Table 1). By contrast, strains mutated in two genes of the other rnf operon (rnfA and rnfC) were almost unable to reduce DNP in vivo and showed a constitutive NPR activity around 10-fold lower than that of the wild-type strain (Table 1). The ntr regulatory ntrC and rpoN mutants showed a phenotype similar to that of the rnfA and rnfC mutants (Table 1). All mutants affected in the reduction of DNP in vivo became more sensitive to DNP, showing a lag phase of 85 h in the presence of the uncoupler (not shown). The residual DNP reduction observed in vivo in the rnf and ntr mutants (20% of the wild-type level) could be due to unspecific nitroreductases similar to those found in other bacteria (7).
TABLE 1.
Effect of ammonium on metabolism of DNP and NPR activities in R. capsulatus wild-type and mutant strains.
| R. capsulatus Strain | Genotype | NPR activity (mU mg−1)
|
% DNP consumeda | |
|---|---|---|---|---|
| −NH4+ | +NH4+ | |||
| E1F1 | Wild type | 2.8 ± 0.3 | 0.04 ± 0.01 | 100 ± 10 |
| B10S | Wild type | 2.9 ± 0.3 | 0.03 ± 0.01 | 100 ± 14 |
| B10S mutantb | nifHDK | 3.0 ± 0.2 | 0.04 ± 0.02 | 100 ± 15 |
| ntrC | 0.4 ± 0.2 | 0.3 ± 0.2 | 20 ± 14 | |
| rpoN | 0.20 ± 0.01 | 0.20 ± 0.02 | 18 ± 10 | |
| nifF | 2.5 ± 0.2 | 0.02 ± 0.01 | 100 ± 12 | |
| rnfF | 2.4 ± 0.1 | 0.03 ± 0.02 | 100 ± 11 | |
| rnfA | 0.15 ± 0.01 | 0.17 ± 0.02 | 21 ± 15 | |
| rnfC | 0.20 ± 0.01 | 0.20 ± 0.01 | 23 ± 13 | |
| orf14 | 2.5 ± 0.1 | 0.03 ± 0.02 | 100 ± 15 | |
Cells cultured in the presence of DNP were harvested by centrifugation in the mid-log phase and resuspended in medium with DNP; 100% corresponded to 0.2 mM DNP consumed in 6 h by the wild type. The data represent averages of four independent experiments.
Previously described (24).
Since the expression of rnf genes is dependent on the Ntr system, DNP photoreduction is regulated either directly (by controlling the expression of NPR) or indirectly (through expression of the rnf genes). We propose an indirect control because relatively high in vitro NPR activities could be detected in crude extracts of ntr and rnf mutant strains (Table 1). Nevertheless, the NPR activity in the ntr and rnf mutants cultured in the presence of DNP was significantly lower than that observed in the wild-type strain. This fact could be explained taking into account that the NPR of R. capsulatus is very unstable in vitro when it became exposed to blue light in the absence of NADPH (6). Therefore, mutants affected in electron supply may be unable to maintain the enzyme in an appropriate reduced state to avoid photoinactivation.
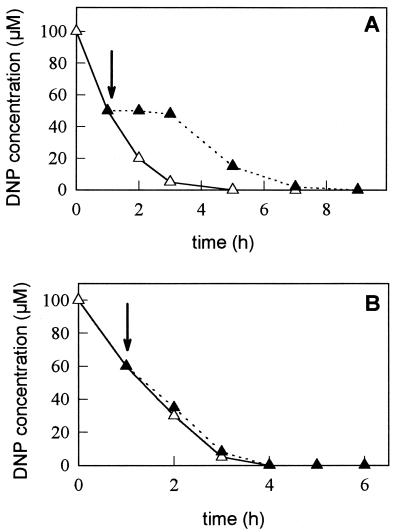
In addition to the long-term effect of ammonium on the reduction of DNP described above, ammonium addition causes a rapid and reversible inhibition of DNP photoreduction in R. capsulatus (2) (Fig. 2A). A similar inhibition pattern was observed by glutamine addition, but other amino acids such as glutamate did not inhibit DNP metabolism (not shown). This effect is achieved probably not by ammonium per se since (i) in phototrophic bacteria, ammonium shock increases the glutamine concentration inside the cells, which activates regulatory processes that inhibit nitrogen fixation (14) and nitrate assimilation (10), (ii) MSX relieved the ammonium suppression of DNP metabolism (4), and (iii) DNP reduction was rapidly and reversibly inhibited by addition of either ammonium or glutamine. This short-term inhibition of DNP metabolism resembles the effect described for nitrate transport or nitrogen fixation, which are switched off by ammonium addition in Rhodospirillaceae (8, 14, 27, 28). Nitrogen fixation in these bacteria is short-term regulated by ammonium by reversible ADP-ribosylation of the nitrogenase, catalyzed by the DraT and DraG proteins (12, 22). To test if the short-term effect of ammonium on DNP metabolism was mediated by the DraTG system that regulates nitrogenase activity in R. capsulatus, we analyzed a mutant defective in draTG (20) and found that ammonium did not repress DNP reduction in this mutant (Fig. 2B). Therefore, both nitrogen fixation and DNP reduction are regulated on the posttranslational level via DraTG (Fig. 2). In contrast to nitrogenase, which is modified at residue Arg102 of the NifH protein and thus is inactivated, the activity of NPR itself is not suppressed. It is interesting that an R. capsulatus strain carrying an NifH-Arg102 substitution mutant still showed ammonium switch-off (21), indicating a second, not yet identified target for DraT-dependent regulation. Since nitrogen fixation and DNP reduction share the rnf-encoded electron transfer system, one of the Rnf proteins might be this target.
FIG. 2.
Effect of ammonium on DNP metabolism in R. capsulatus wild-type (A) and draTG double-deletion mutant (B) strains. Cells grown with 0.1 mM DNP were harvested by centrifugation and resuspended in fresh medium with 10 mM acetate and 0.1 mM DNP. The flasks were incubated under light at 30°C, and DNP concentration was determined. At the time indicated by arrows, half of each culture was made 2 mM in NH4Cl (filled symbols), whereas the other half remained under the same growth conditions (open symbols). Data are representatives of three different experiments.
In conclusion, both nitrogen fixation and DNP reduction in R. capsulatus need the low level reduction power provided by the Rnf system. The Ntr control of rnf expression could explain the long-term (transcriptional) regulation of DNP reduction by ammonium. In addition, we found that the short-term regulatory effect of ammonium on DNP reduction depended on the DraTG system. Since NPR itself was shown not to be the target of ADP-ribosylation, we speculated that either the electron transfer system encoded by rnf or the DNP uptake system may be controlled by DraTG.
Acknowledgments
We acknowledge financial support from DGICYT (grant PB 95 0554 CO2 02) and PAI (grant CVI 0117) (Spain) and the Alexander von Humboldt Foundation (Germany). R.B. acknowledges financial support from the MEC (Contratos de Incorporación de Doctores y Tecnólogos).
We thank Alexander Yakunin and Patrick Hallenbeck for providing the antibodies against NifF and C. Moreno-Vivián for critically reading the manuscript.
REFERENCES
- 1.Arp D J, Zumft W G. l-Methionine-SR-sulfoximine as a probe for the role of glutamine synthetase in nitrogenase switch-off by ammonia and glutamine in Rhodopseudomonas palustris. Arch Microbiol. 1983;134:17–22. doi: 10.1007/BF00429400. [DOI] [PubMed] [Google Scholar]
- 2.Blasco R, Castillo F. Light-dependent degradation of nitrophenols by the phototrophic bacterium Rhodobacter capsulatus E1F1. Appl Environ Microbiol. 1992;58:690–695. doi: 10.1128/aem.58.2.690-695.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco R, Castillo F. Characterization of a nitrophenol reductase from the phototropic bacterium Rhodobacter capsulatus E1F1. Appl Environ Microbiol. 1993;59:1774–1778. doi: 10.1128/aem.59.6.1774-1778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco R, Castillo F. Characterization of 2,4-dinitrophenol uptake by Rhodobacter capsulatus. Pestic Biochem Physiol. 1997;58:1–6. [Google Scholar]
- 5.Blasco R, Moore E, Wray V, Pieper D, Timmis K, Castillo F. 3-Nitroadipate, a metabolic intermediate for mineralization of 2,4-dinitrophenol by a new strain of a Rhodococcus species. J Bacteriol. 1999;181:149–152. doi: 10.1128/jb.181.1.149-152.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasco R, Aparicio P J, Castillo F. Photoinactivation of the detoxifying enzyme nitrophenol reductase from Rhodobacter capsulatus. Arch Microbiol. 1995;163:248–253. [Google Scholar]
- 7.Brian D W, McCalla D R, Leeksma M, Laneuville P. Type I nitroreductases of Escherichia coli. Can J Microbiol. 1981;27:81–86. doi: 10.1139/m81-013. [DOI] [PubMed] [Google Scholar]
- 8.Caballero F J, Moreno-Vivián C, Castillo F, Cárdenas J. Nitrite uptake system in photosynthetic bacterium Rhodopseudomonas capsulata E1F1. Biochim Biophys Acta. 1986;848:16–23. [Google Scholar]
- 9.Cullen P J, Bowman W C, Foster-Harnett D, Reilly S C, Kranz R G. Translational activation by an NtrC enhancer-binding protein. J Mol Biol. 1998;278:903–914. doi: 10.1006/jmbi.1998.1745. [DOI] [PubMed] [Google Scholar]
- 10.Dobao M M, Martínez-Luque M, Moreno-Vivián C, Castillo F. Effect of carbon and nitrogen metabolism on nitrate reductase activity of Rhodobacter capsulatus E1F1. Can J Microbiol. 1994;40:645–650. [Google Scholar]
- 11.He Z, Spain J C. Studies of the catabolic pathway of degradation of nitrobencene by Pseudomonas pseudoalcaligenes JS45: removal of the amino group from 2-aminomuconic semialdehide. Appl Environ Microbiol. 1997;63:4839–4843. doi: 10.1128/aem.63.12.4839-4843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jouanneau Y, Roby C, Meyer C M, Vignais P M. ADP-ribosylation of dinitrogenase reductase in Rhodobacter capsulatus. Biochemistry. 1989;28:6524–6530. [Google Scholar]
- 13.Jouanneau Y, Jeong H S, Hugo N, Meyer C, Willison J C. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus. Characterization of 2 membrane-bound iron-sulfur proteins. Eur J Biochem. 1998;251:54–64. doi: 10.1046/j.1432-1327.1998.2510054.x. [DOI] [PubMed] [Google Scholar]
- 14.Kanemoto R H, Ludden P W. Effect of ammonia, darkness and phenazine methosulfate on whole-cell nitrogenase activity and Fe protein modification in Rhodospirillum rubrum. J Bacteriol. 1984;158:713–720. doi: 10.1128/jb.158.2.713-720.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kranz R G, Cullen P J. Regulation of nitrogen fixation genes. In: Blankenship R E, et al., editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1191–1208. [Google Scholar]
- 16.Kranz R G, Foster-Hartnett D. Transcriptional regulatory cascade of nitrogen-fixation genes in anoxygenic photosynthetic bacteria: oxygen- and nitrogen-responsive factors. Mol Microbiol. 1990;4:1793–1800. doi: 10.1111/j.1365-2958.1990.tb02027.x. [DOI] [PubMed] [Google Scholar]
- 17.Kustu S, Santero E, Keerner J, Popham D, Weiss D. Expression of ς54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenke H, Pieper D-H, Bruhn C, Knackmuss H-J. Degradation of 2,4-dinitrophenol by two Rhodococus strains, HL 24-1 and HL 24-2. Appl Environ Microbiol. 1992;58:2928–2932. doi: 10.1128/aem.58.9.2928-2932.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masepohl B, Klipp W. Organization and regulation of genes encoding the molybdenum nitrogenase and the alternative nitrogenase in Rhodobacter capsulatus. Arch Microbiol. 1996;165:80–90. [Google Scholar]
- 20.Masepohl B, Krey R, Klipp W. The draTG gene region of Rhodobacter capsulatus is required for post-translational regulation of both the molybdenum and the alternative nitrogenase. J Gen Microbiol. 1993;139:2667–2675. doi: 10.1099/00221287-139-11-2667. [DOI] [PubMed] [Google Scholar]
- 21.Pierrard J, Ludden P W, Roberts G P. Posttranslational regulation of nitrogenase in Rhodobacter capsulatus: existence of two independent regulatory effect of ammonium. J Bacteriol. 1993;175:1358–1366. doi: 10.1128/jb.175.5.1358-1366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pope M R, Murrell S A, Ludden P W. Covalent modification of the iron protein of nitrogenase from Rhodospirillum rubrum by adenosine diphosphoribosylation of a specific Arg residue. Proc Natl Acad Sci USA. 1985;82:3173–3177. doi: 10.1073/pnas.82.10.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeki K, Kumagai H. The rnf gene products in Rhodobacter capsulatus play an essential role in nitrogen fixation during anaerobic DMSO-dependent growth in the dark. Arch Microbiol. 1998;169:464–467. doi: 10.1007/s002030050598. [DOI] [PubMed] [Google Scholar]
- 24.Schenzle A, Lenke H, Spain L C, Knackmuss H-J. Chemoselective nitro group reduction and reductive declhorination initiate degradation of 2-chloro-5-nitrophenol by Ralstonia eutropha JMP 134. Appl Environ Microbiol. 1999;65:2317–2323. doi: 10.1128/aem.65.6.2317-2323.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmehl M, Jahn A, Meyer A, Hennecke S, Masephol B, Schuppler M, Marxer M, Oelze J, Klipp W. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol Gen Genet. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 26.Spain J C. Biodegradation of nitroaromatic compounds. Annu Rev Microbiol. 1995;49:523–555. doi: 10.1146/annurev.mi.49.100195.002515. [DOI] [PubMed] [Google Scholar]
- 27.Yakunin A F, Gennaro G, Hallenbeck P C. Purification and properties of a nif-specific flavodoxin from the photosynthetic bacterium Rhodobacter capsulatus. J Bacteriol. 1993;175:6775–6780. doi: 10.1128/jb.175.21.6775-6780.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zumft W G, Castillo F. Regulatory properties of the nitrogenase from Rhodopseudomonas palustris. Arch Microbiol. 1978;117:53–60. doi: 10.1007/BF00689351. [DOI] [PubMed] [Google Scholar]