Abstract
The ability of antibodies to bind a wide variety of analytes with high specificity and high affinity makes them ideal candidates for therapeutic and diagnostic applications. However, the poor stability and high production cost of antibodies have prompted exploration of a variety of synthetic materials capable of specific molecular recognition. Unfortunately, it remains a fundamental challenge to create a chemically diverse population of protein-like, folded synthetic nanostructures with defined molecular conformations in water. Here we report the synthesis and screening of combinatorial libraries of sequence-defined peptoid polymers engineered to fold into ordered, supramolecular nanosheets displaying a high spatial density of diverse, conformationally constrained peptoid loops on their surface. These polyvalent, loop-functionalized nanosheets were screened using a homogeneous Förster resonance energy transfer (FRET) assay for binding to a variety of protein targets. Peptoid sequences were identified that bound to the heptameric protein, anthrax protective antigen, with high avidity and selectivity. These nanosheets were shown to be resistant to proteolytic degradation, and the binding was shown to be dependent on the loop display density. This work demonstrates that key aspects of antibody structure and function–the creation of multivalent, combinatorial chemical diversity within a well-defined folded structure–can be realized with completely synthetic materials. This approach enables the rapid discovery of biomimetic affinity reagents that combine the durability of synthetic materials with the specificity of biomolecular materials.
Keywords: protein-mimetic materials, multivalent molecular recognition, combinatorial display, two-dimensional nanomaterials, bioinspired polymers
Graphical Abstract:

Antibodies naturally act as molecular recognition elements having high specificity and affinity to their corresponding antigens. While antibodies are nature’s ultimate pathogen detectors, their utility for biomedical applications is restricted by high production cost, low solubility, loss of activity due to improper folding, and difficulties with chemical modification.1,2 To avoid these drawbacks and to emulate these antibodies, various synthetic biomacromolecules have been developed by rational design and combinatorial approaches using natural scaffolds, such as proteins, nucleic acids, and bacterial phages.3,4 These approaches mimic, all in some way, the sophisticated three-dimensional structures of antibody binding, in which multiple, diverse chemical functionalities are presented in precise spatial geometries. However, the process for discovering artificial affinity reagents is laborious due to the requisite iterative hit selection and amplification; moreover, their low stability against proteases and poor binding affinities in the micromolar range limit the utility of these materials. Antibody mimetics discovered by combinatorial approaches have been spotlighted due to their robustness and binding affinities,5,6 but are limited in chemical diversity and their ability to display complex, folded, binding structures. Thus, there is an unmet need for methods enabling the facile and rapid discovery of molecularly defined synthetic antibodies consisting of entirely non-natural, robust materials.
Peptoids are bioinspired and sequence-defined N-substituted glycine polymers. They are ideal as building blocks for constructing protein-like architectures, because their sequence-defined linear chains can reproducibly fold into specific molecular architectures due to assorted long-range inter- and intramolecular noncovalent interactions.7–14 They are efficiently synthesized by the automated solid-phase submonomer method, using diverse and readily available amine synthons, to precisely modulate chain length, monomer sequence, and side chain chemistry. Like proteins, this provides the opportunity to program both folding information and biological activity into the chemical sequence of the polymer chain.15,16 The free-floating 2-dimensional nanosheet is a fascinating example of a folded peptoid-based nanomaterial with atomically defined structure.17 In this work, we mimic the basic structure of the antibody binding site by positioning a diversity of well-defined peptoid loops on the surface of peptoid nanosheets. These nanosheets are made from the self-assembly of peptoid 42-mer strands that contain a central and variable chemically diverse hexameric loop domain, flanked by two fixed nanosheet-forming domains.18,19 Atomic force microscopy (AFM) and protein binding experiments show that these loops are displayed on the nanosheet surface.18,19 Specific binding affinity for target proteins can be encoded by the monomer sequence of the loop-display domain, highlighting its potential as a biologically relevant binding material. Furthermore, nanosheet self-assembly is triggered by interfacial compression of the air–water interface,20 allowing the composition of the loops to be controlled prior to sheet formation. The zwitterionic surface of peptoid nanosheets also makes them an optimal chassis for display of functional loops, since it prevents nonspecific adhesion of biomolecules.21
An important structural feature of loop-functionalized peptoid nanosheets is the opportunity to enable multivalent interactions with substrates due to their high spatial density of loops on the surface.20 The ability of these high-surface-area materials to support multiple binding events simultaneously boosts selectivity and sensitivity for target proteins among various biomacromolecules.22 Furthermore, antibody-like peptoid nanosheets resist degradation by proteases, solvents, and extreme pH and temperature conditions, because they lack peptide bonds in their backbone structure.16,23,24 They can be engineered to employ photoreactive monomers for cross-linking the nanosheet interior, yielding increased chemical and mechanical stability.25 While peptoid nanosheets thus provide valuable chemical and structural properties as affinity reagents, missing from the toolkit is a process for mimicking the recombination and affinity maturation of antibodies, which is critical to the immune system’s ability to obtain specific and high affinity binders to targets. If chemists could develop an in vitro approach for generating specific, high affinity nanosheet binders to unfamiliar targets, these synthetic materials could overcome many of the drawbacks of antibodies.
Herein, we describe a general approach for the rapid discovery of biomimetic affinity reagents by the screening of combinatorial libraries of chemically diverse, loop-functionalized peptoid nanosheets. The discovery cycle comprised four major steps: the chemical synthesis of loop-containing peptoids (loopoids), the supramolecular assembly of loopoid nanosheets, the screening of the loopoid nanosheets against various proteins for binding activity (hits), and hit validation (Figure 1). First, the loopoid strands, which are based on a block-patterned 36-mer nanosheet-forming sequence, with the insertion of six monomers in the middle to form a conformationally rigidified surface-displayed loop, were synthesized robotically using the automated solid phase submonomer method. Drawing upon a set of 20 chemically diverse peptoid monomers, we produced a library of 256 individual loopoid strands. We then employed an autopipetting robot to initiate the supramolecular assembly of nanosheets in a high-throughput, parallel manner from the loopoids in multiwell plates. The assembled loopoid nanosheets were then used directly in a variety of target protein binding assays based on a Förster resonance energy transfer (FRET) assay.20 On the basis of this automated process for synthesis and screening, we discovered a nanosheet binder for anthrax protective antigen [(PA63)7], a ring-shaped homoheptameric toxin-related protein. On the basis of the chemical properties of the hit loop sequences, we identify rules for understanding the specific binding of loopoid nanosheets to (PA63)7. We also demonstrated the importance of multivalency in hit interactions by biolayer interferometry, electron microscopy, and fluorescence imaging and FRET analyses.
Figure 1.

Combinatorial discovery of loop-functionalized peptoid nanosheets as chemically diverse, biomimetic affinity reagents. The hierarchical assembly creates polyvalent chemical diversity within a conformationally constrained, folded context.
RESULTS AND DISCUSSION
Design of Loopoid Libraries.
The diverse antigenic specificity of antibodies is achieved by the chemical diversity (i.e., combination of 20 amino acids) within the structural context of complementarity-determining regions (CDR). To replicate the binding specificity of antibodies to their corresponding antigens, the loop-functionalized nanosheet is an ideal scaffold because of the opportunity to display chemically diverse moieties within a conformationally constrained loop domain projected from the nanosheet polar surface. For these displayed loops, we identified a set of 7 hydrophilic (P) and 13 hydrophobic (H) peptoid monomers possessing distinct chemical properties similar to natural amino acids (Figure 2a). Combinatorial loop libraries were designed to contain specific combinations of H and P monomers to avoid extremes of hydrophobicity or charge within the loop domains. These combinations were selected using design rules based on monomer chemical properties, the feasibility of polypeptoid synthesis, and its capacity to fold into nanosheets (Figure 2b). We fixed the length of the loop domain to be six monomers in order to protrude far enough from the surface of nanosheets for efficient protein binding.20 A peptoid loop six monomers in length is expected to have conformational rigidity, as has been shown in similarly sized peptoid macrocycles.26 There are 64 possible amphiphilic HP patterns of loop sequence in a 6-mer by considering only the H and P property of monomers. Our previous studies determined that the nanosheet formation mechanism involves a key intermediate where the peptoids must adsorb to the air–water interface to form an ordered monolayer.18 Hence, the overall water solubility and surface activity must be balanced to promote self-assembly and the correct folding of loops on the surface of nanosheets. For this reason, we eliminated sequences with alternating H and P monomers, sequences with ≥3 H monomers, and sequences with ≥2 large H monomers per sequence. Employing these rules, we randomly selected 12 hexameric HP patterns. Chemical diversity was introduced into the loop sequence by classifying each monomer according to their primary and secondary chemical characteristics: aromatic, small aliphatic, large aliphatic, large hydrophobic, cationic, anionic, hydrogen bond donor/acceptor, and heterocycle. Considering the chemical characteristics of these monomers, the 12 initial HP patterns were enumerated to a set of 256 specific sequences with a bias toward minimizing the number of different monomers within the same sequence. In this way the chemical distinctness of each loop could be maximized, making it easier to later identify key chemical properties essential for molecular recognition (Supporting Information, Table S1).
Figure 2.
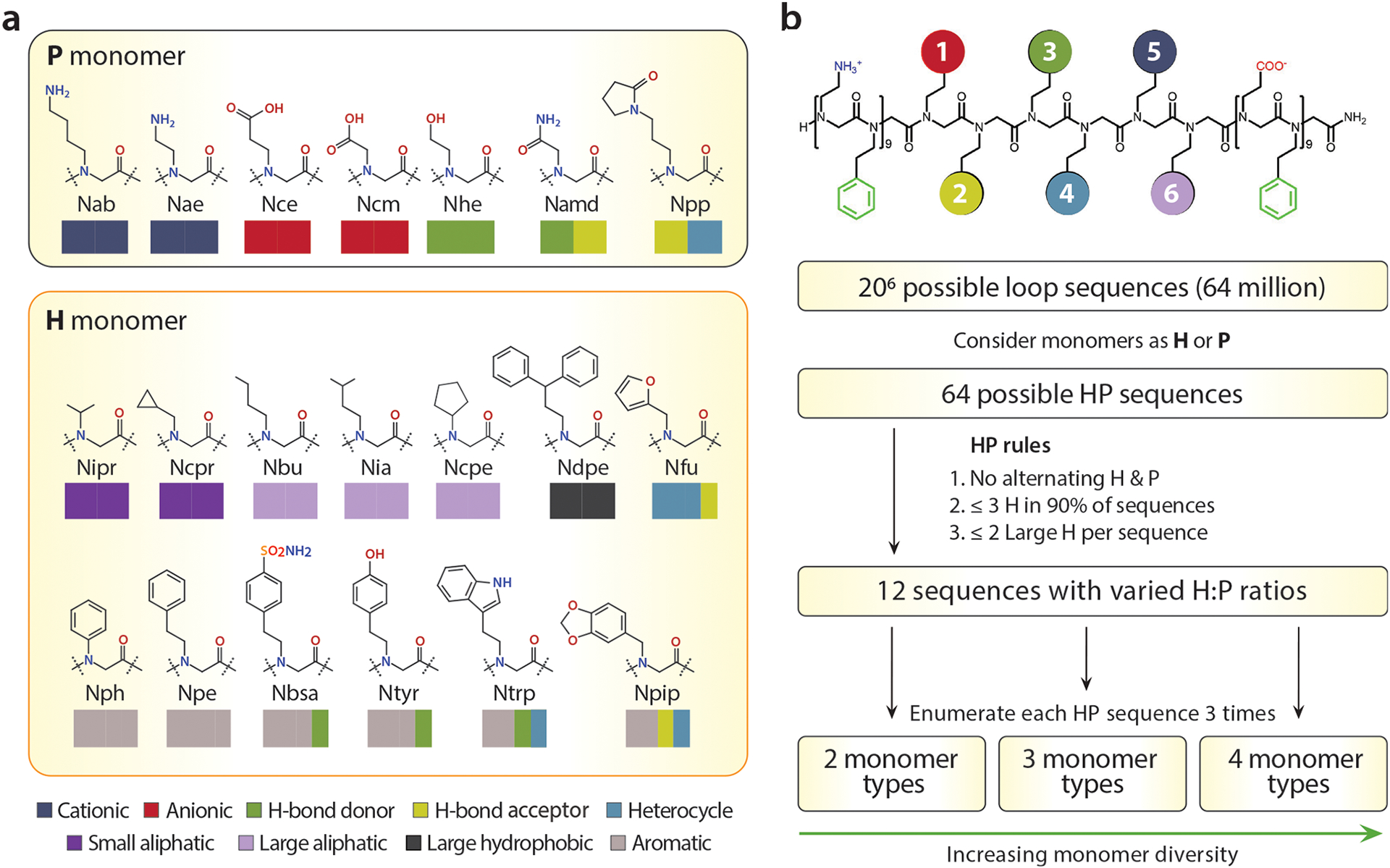
Design of the chemically diverse binding loops, built from a basis of 20 efficiently incorporated monomers. (a) P (hydrophilic) and H (hydrophobic) amine monomers exhibiting various properties. Colored rectangles represent primary and secondary properties of monomers. All abbreviations of monomers are included in Supporting Information. (b) Design algorithm for creating a maximally diverse set of loop insert sequences, while avoiding extremes of hydrophobicity.
Screening of Loopoid Library and Hit Validation.
The first hurdle to discovering a nanosheet affinity reagent is to effectively convert the library of individual peptoid chains into loop-functionalized nanosheet libraries. Conventionally, this supramolecular assembly process has been accomplished by the lateral compression of peptoid monolayers formed at the air/water or oil/water interface.27,28 A device to rotate a glass vial containing milliliter volumes of peptoid-dissolved solutions from vertical to horizontal was designed to induce a 3-fold change of interfacial area for obtaining high-quality peptoid nanosheets. However, the vial-rocking method is too difficult to parallelize for high-throughput library generation and screening and operates at the milliliter scale, which results in an unnecessary waste of material and makes materials cost prohibitively high as the library size increases. To address this challenge, we established a pipetting method for effective preparation of nanosheets that can be performed in multiwell plates with an automated pipetting robot. Withdrawing an aqueous solution by pipetting creates an air/water interface inside the pipet tip. Due to the conical geometry of the tip, the interfacial area changes depending on the volume of liquid aspirated (Figure S1). By repeating aspiration and dispense cycles, surface compression comparable to the vial rocking method could be achieved at much smaller volume scales (~10 μL) (Figure 3a). The quality (size, thickness, and uniformity) of nanosheets synthesized by the pipetting method were in line with that prepared by vial rocking (Figure 3b and Figure S2). Attractive aspects of the pipetting method are that it is readily automated, can be performed in a multiwell plate format, and produces nanosheets at concentrations and in buffers suitable for direct use in subsequent protein-binding screens.
Figure 3.

Automated production and screening of a loop-functionalized peptoid nanosheet library for specific binding to (PA63)7. (a) Schematic illustration of peptoid nanosheet formation by interfacial compression through robotic pipetting. (b) Fluorescence microscopic images of pipetting-induced peptoid nanosheets. All scale bars represent 100 μm. (c) (PA63)7 specificity score of various loopoid nanosheets from screening process. The detail of the calculation of specificity score described in Supporting Information. (d) Chemical structure of loop domain of L034 and TYWWLD nanosheet. (e) Validation of (PA63)7 binding specificity of L034 nanosheet by fluorescent microscopy. All scale bars represent 100 μm.
To evaluate the protein-binding capability of each loop-displayed nanosheet, we developed a screening method based on our previously developed homogeneous FRET binding assay.20 Nanosheet-forming solutions were mixed with octadecyl rhodamine (OR), which anchors its aliphatic tail in the sheet’s hydrophobic core, and serves as the FRET donor. These OR-labeled sheets yield a FRET signal upon binding of Alexa Fluor 647 (AF647)-conjugated target proteins on the nanosheet surface (Figure S3). By analyzing fluorescence spectra of control (loopless), biotinylated, and globotriosylated nanosheets in the absence and presence of protein, a positive FRET signal was generated by a binding event of nanosheets to their corresponding targets (Figure S4). Spectral scanning of the surface of the globotriosylated nanosheet by confocal microscopy verified that the FRET signal originated from the protein binding onto the nanosheet surface (Figure S5). To estimate the degree of binding for the selection of hits, the FRET ratio was calculated based on curve fitting to prevent potential errors caused by noise in the fluorescence spectra. We found that this method could satisfactorily identify high-FRET hits similar to the conventional FRET ratio measurements (~ IAcceptor/IDonor) (Figure S6). Analysis of the fitting-corrected ratio for 10 samples of each loopless, biotinylated, and globotriosylated nanosheets in the presence of Shiga toxin 1 subunit B (STX 1B) (Figure S7), revealed a Z-factor of approximately 0.73, indicating that this assay is sufficient for high-throughput screening.29
We next applied our automated screening platform to discover loop-functionalized nanosheets capable of selective binding to anthrax protective antigen (PA63)7 which is a homoheptameric toxin-related protein. To gauge binding specificity, FRET values of each loopoid nanosheet were obtained against fluorescently labeled (PA63)7 and other multimeric control proteins, STX 1B and streptavidin, and the ratiometric score for binding specificity was calculated. From the parallel screening of a nanosheet library of 60 variants, sample L034 nanosheet was found to have the highest specificity score for (PA63)7 (Figure 3c).
Upon successful identification of a hit for (PA63)7, we scaled up the preparation of several nanosheet candidates to the milliliter scale using the vial-rocking method to allow for more stringent validation and characterization of binding affinity.28 This seamless transition from microscale screening to macro-scale production highlights the utility of this platform for rapid affinity reagent development. After scale-up, we examined the consistency of (PA63)7 binding specificity to the L034 nanosheet by the FRET assay and by fluorescence microscopy. As a positive control, we created a nanosheet that displays a loop containing a known (PA63)7 binding motif composed of a peptide hexamer, TYWWLD, previously discovered by phage-display methods30,31 (Figure 3d). Fluorescence microscope images and the FRET assay indicate that only (PA63)7 was selectively captured by the L034 nanosheet as well as the specific binding by TYWWLD nanosheet (Figure 3e, Figures S8 and S9). This result demonstrates the ability to rapidly discover antibody mimetics from microscale screening of combinatorial libraries, and immediate scale up of hits to the milligram scale for further testing and use.
Binding Specificity: Chemical Properties and Multi-valency of Loop-Functionalized Nanosheets.
The (PA63)7 binding hit, L034, has a loop insert sequence of Namd-Ntyr-Ntyr-Ntyr-Ntyr-Namd, which consists of two small polar carboxamidomethyl (Namd) side chains, and four aromatic hydroxyphenethyl (Ntyr) side chains (Figure 3d). Interestingly, this falls into the same HP family as the positive control loop (TYWWLD), where there are four central hydrophobic groups flanked by two polar groups (i.e., PHHHHP). Additionally, both contain three aromatic residues in the hydrophobic loop region, allowing us to consider the impact of specific chemical features of the loop on (PA63)7 recognition (Table S2). Given the predominance of hydrophobic side chains in both our hit and the positive control sequence, we first examined the binding of other hydrophobic library members. There were five other loop-functionalized nanosheets with the identical PHHHHP pattern that did not recognize (PA63)7 (Figure S10) indicating that aromatic richness is the major factor for (PA63)7 recognition. We also explored the importance of multiple aromatic derivatives within the loop domain by designing an all-peptoid analogue (rationally designed peptoid, Rtoid) inspired from the known TYWWLD peptide sequence, which has three aromatic groups (Figure S11). The Rtoid nanosheets exhibited strong binding, indicating that the aromatic side chains are playing a dominant role in binding. Motivated by these observations, we plotted the (PA63)7 specific binding ability against the number of aromatic monomers within the loop region (Figure 4a), which revealed that ≥3 aromatic monomers were necessary but insufficient for binding. A secondary chemical property, a hydrogen-bonding donor, is also required for binding. Additional secondary properties such as a hydrogen bonding acceptor or chirality were not required.
Figure 4.

Characterization of selective (PA63)7 binding behavior of L034 nanosheet by monomeric chemical property and multivalent configuration. (a) Relationship of the number of aromatic monomers within loop domain to (PA63)7 specificity score. Arrows indicate loop sequences with aromatic monomers possessing the secondary property as hydrogen bonding donor (black arrows) or others (orange arrows). (b) Competitive binding assay with TYWWLD hexamer as a competitor. (c) Molecular structure of (PA63)7 (PDB 1TZO) and L034 nanosheet. Colored regions in (PA63)7 represent binding site for TYWWLD ligand. L034 nanosheet structure shows the spatial configuration and interspacing of loops (purple) on the backbone chains (green). (d) Degree of (PA63)7 binding as a function of the shortest average loop separation on the nanosheet. (e) Electron microscopic images of B36 (i.e., loopless), TYWWLD and L034 nanosheet with bound (PA63)7.
The specific binding affinity of loops containing aromatic and hydrogen-bond donor properties hints that they interact with a specific site of (PA63)7. By performing competitive binding experiments of a L034 or TYWWLD nanosheet in the presence of the free TYWWLD hexamer as a competitor (Figure 4b), the half maximal inhibitory concentration (IC50) value of the L034 nanosheet of 78 μM is similar to that of the TYWWLD nanosheet (98 μM), indicating they interact with the same binding site of (PA63)7. A computational docking simulation of the TYWWLD peptide in a loop conformation binding to (PA63)7 provided insight into the molecular interactions involved. The central YWW residues are in close contact (≤4.5 Å) to P184, F202, F203, S204, and P205 which are known to form a hydrophobic binding pocket in (PA63)7.32 Phenylalanine facilitates an interaction with aromatic peptide residues via π–π interactions. In addition, proline is known to preferentially interact with electron-rich aromatics, such as tyrosine and tryptophan, by CH-π interactions in contrast to electron-poor aromatics and nonaromatics.33 Taken together, we envisioned that a loop domain comprising many electron-rich aromatic functionalities can recognize the hydrophobic pocket of (PA63)7 via π–π and CH-π interactions.
From a multivalent binding point of view, the spatial arrangement of binding loops on the surface of the peptoid nanosheet plays a key role in its specificity. The distance between loops in the direction of the chain is approximately 5.1 nm, which is close to the second shortest distance, 5.4 nm, between (PA63)7 binding sites, as shown in the simulated structure of the L034 nanosheet (Figure 4c). According to the model, the hydrophobic binding pocket of (PA63)7 covers several loops on the sheet surface, indicating that simultaneous multiple binding events, that is, multivalent binding, may occur. Typically, multivalency leads to nonlinear binding responses such as superselectivity and high binding affinity.22 We explored the effect of loop display density on (PA63)7 binding by diluting the TYWWLD and L034 loop strands with varying amounts of a loopless nanosheet-forming peptoid. This results in a series of coassembled nanosheets with different interloop spacings. The degree of binding exhibited a nonlinear response versus loop spacing (Figure 4d). The threshold distance of both nanosheets is ~3.8 nm, which is close to the shortest distance between binding sites of (PA63)7. This suggests that the close loop spacing is required for cooperative binding and higher affinity. The multivalent interaction between loops and (PA63)7 was also supported by direct observation via transmission electron microscopy which revealed that the central pore of (PA63)7 was vertically aligned on the surface of both the TYWWLD and L034 nanosheets (Figure 4e).
Binding Performance of Loop-Functionalized Nano-sheets: Binding Affinity and Proteolytic Stability.
To evaluate the feasibility of our identified affinity reagents for practical use, we evaluated their binding performance using biolayer interferometry (BLI) according to our previously developed method.34 We transferred a loop-functionalized peptoid monolayer onto the hydrophobic surface of the optical fiber by Langmuir–Schaefer deposition, exposing loop domains that can capture target proteins. BLI allows real-time, label-free monitoring of a bound protein based on interference patterns formed by reflection of white light from a functional surface, allowing the measurement of binding affinity. Both L034 and TYWWLD monolayers immediately captured and tightly immobilized (PA63)7 on their surface in accordance with association and dissociation stages. In contrast, there was no binding response with the loopless peptoid monolayer (Figure 5a). Analysis of the binding curves of L034 and TYWWLD revealed that these loop-functionalized monolayers had single-digit nanomolar binding affinity, 2.0 nM and 1.6 nM, respectively, (Table S3), which is comparable to the KD of recombinant antibodies and natural complementary components (i.e., edema factor and lethal factor) for (PA63)7.30,35,36 The binding affinity of these materials is strikingly high in comparison with the known micromolar binding of the monovalent TYWWLD ligand.30 Taken together, these results indicate a significant enhancement of binding affinity due to the high avidity of designed and engineered multimeric structures.37
Figure 5.

Binding performance and proteolytic stability of L034 nanosheet. (a) Binding response of B36 (i.e., loopless), TYWWLD, and L034 monolayer-deposited optical fiber against (PA63)7 by using the biolayer interferometric technique. Area before and after dotted line represents association and dissociation phase, respectively. (b) AFM images of TYWWLD and L034 nanosheets before and after protease treatment. Scale bars represent 1 μm. (c) Quantification of the thickness upon protease treatment calculated from AFM images. Each thickness was estimated by averaging three AFM measurements. (d) Fluorescence microscopic images of loopoid nanosheets before and after protease treatment. All images were obtained in the presence of (PA63)7 under TR (yellow) and Cy5 (red) filter. Scale bars represent 100 μm. (e) Quantification of the proteolytic stability of TYWWLD and L034 nanosheet by FRET-based assay. L034 nanosheet retains binding affinity after protease treatment, demonstrating resistance to proteolysis.
Beyond their exceptional binding performance, these materials hold promise as a robust affinity reagent scaffold due to the chemical stability and durability of peptoids against proteases.38 To demonstrate this, we monitored the proteolytic stability of the L034 nanosheet by atomic force microscopy. While the all-peptoid L034 nanosheet maintained loop architectures after protease treatment, the initially rough surface of the peptide-containing TYWWLD nanosheet smoothed (Figure 5b). Additionally, the thickness of the L034 nanosheet did not change, whereas the TYWWLD nanosheet thinned from 4.7 to 3.4 nm after protease treatment (Figure 5c). These results indicated that while the peptide loops of the TYWWLD nanosheet were eliminated through proteolytic degradation, the all-peptoid L034 nanosheet has high proteolytic resistance. We further studied the influence of proteolytic degradation on the binding activity of the L034 and TYWWLD nanosheets using multicolor fluorescence imaging, with the Texas Red (TR) filter for nanosheets and the Cy5 filter for the (PA63)7 protein as yellow and red color, respectively (Figure 5d). The orange color of the TYWWLD nanosheet incubated with (PA63)7 turned to yellow after protease treatment, indicating that the removal of TYWWLD loops led to the loss of binding affinity to (PA63)7. The L034 nanosheet, however, remained orange in the merged fluorescence image after the incubation of protease, indicating that they retained (PA63)7 binding activity. We corroborated these results with our FRET assay, where proteolytic degradation of TYWWLD loops also demonstrated loss of (PA63)7 binding in contrast to negligible change in FRET signal from the L034 nanosheet (Figure 5e). The durability of these peptoid constructs against proteolysis highlights a major advantage of this synthetic system.
CONCLUSION
We have described an approach to generate 2D antibody mimetic materials with specific protein binding using a purely synthetic approach. The method uses high-throughput screening of combinatorial libraries of non-natural, sequence-defined peptoid polymers folded into protein-like architectures. The selective binding affinity of the L034 nanosheet hit to (PA63)7 sensitively depends on the chemical characteristics of the surface-displayed loops. Furthermore, the spatially dense display of surface loops enables high binding avidity that significantly improves binding affinity to the target protein via multivalent interactions. This is a major advantage of these non-natural sheets which have a massive binding surface area relative to antibodies, and make them particularly well-suited to engage with large, multivalent targets. Moreover, their non-natural peptoid chemistry makes them robust to proteolytic attack as demonstrated by detailed analysis using AFM, fluorescence imaging, and FRET binding assay. Robustness is a valuable property of using non-natural backbone chemistry, allowing screening in less stringently purified biofluids and discovery of affinity reagents that operate under harsher and more practical use cases. This work advances on several key aspects of antibody mimetic materials, demonstrating a rational and scalable strategy for generating and screening large chemical libraries, generation of materials with high and selective binding affinity via multivalency, and the ability to immediately scale up production of hits by simple batch synthesis approaches that do not involve animals. Screening capacity is limited only by the fluidic approach for mixing and testing the combinatorial library members. Using established microfluidic miniaturization approaches, screening hundred-million-member libraries should be feasible, allowing multiple loopoid strands to be combined prior to nanosheet assembly to access enormous chemical diversity akin to the natural immune system. Combined, these properties should allow rapid and effective discovery of robust recognition elements for pathogens (e.g., toxin proteins, viruses, and cells) important to many biomedical applications such as sensing, diagnostics, and therapeutics.
MATERIALS AND METHODS
Instrumentation.
Fluorescence data were collected with a BioTek Synergy H1 microplate reader. Fluorescence microscopy was performed on an Olympus IX81 inverted microscope, and confocal FRET experiments were conducted on a Zeiss LSM710 microscope equipped with a spectral detector. Confocal image spectra were collected from images by image thresholding across all channels and background subtraction through ImageJ. Biolayer interferometry measurements were conducted on the Octet Red384 system from ForteBio.
Loopoid Library Synthesis and Purification.
Methods for the solid-phase synthesis and purification of peptoid polymers were previously reported by our group and others.15 Submonomer peptoid synthesis was automated using a Protein Technologies Symphony X or AAPPTEC Apex 396 peptide synthesizers to allow for the parallel synthesis of up to 24 different loopoid sequences at a time. β-Alanine tert-butyl ester (Nce) and glycine tert-butyl ester (Ncm) were prepared from the HCl salt using 1 M aq NaOH and extracted with DCM before preparation of 1 M solutions in DMF. Glycinamide HCl (Namd) was neutralized in DMF by adding an equimolar amount of KOH (50% w/w aq solution) and filtered by centrifugation immediately before use. Protected ethanolamine (Nhe) was prepared according to published procedures.39 All other amines were purchased from commercial suppliers and used without further purification. Peptoid loopoids were prepared using a Rink amide resin (200 mg) from Protein Technologies with a standard amine loading of 0.64 mmol/g. Standard peptoid coupling proceeded in two steps, initial bromoacetylation with bromoacetic acid (BrAA, 0.8 M) and N,N-diisopropylcarbodiimide (DIC, 0.8 M) in N,N-dimethylformamide (DMF) for 2.5 min. Subsequent displacement with amine (1 M) in DMF lasted for 5 min. Anilines, and other low nucleophilicity amines, were bromoacetylated for 20 min and their displacement lasted for 1 h using amine (2 M) solutions with added potassium iodide (1 M).40
The TYWWLD loopoid strand was prepared on a low loading (200 mg, 0.2 mmol/g) Rink amide resin from Protein Technologies to counter aggregation. Peptoid coupling proceeded with the standard reagents using 1 h displacement and bromoaceylation times. Amino acids in the loop were coupled twice with HCTU (0.4 M), N-methylmorpholine (0.4 M), and Fmoc-protected amino acids (0.4 M) in DMF for 1 h and then deprotected with 4-methylpiperidine (20% v/v) for 1 h. The TYWWLD hexamer was prepared on standard Rink amide resin using DIC (0.8 M) as coupling agent with 1 h coupling and deprotection times.
Crude loopoids were cleaved from the resin using 95% TFA/2.5% H2O/2.5% TIPS while mixing on a rotary shaker for 30 min. The cleaved peptoid was filtered and resin was washed with DCM before concentration of the combined organics on a Biotage V10 evaporator. The loopoids were purified by reverse-phase HPLC using a Waters XBridge Prep C18 (5 μm, 19 × 100 mm) column. The typical gradient was 35–65% over 30 min with H2O+0.1% TFA as solvent A and ACN+0.1% TFA as solvent B. The pure fractions were combined and concentrated with a Biotage V10 evaporator before dissolving with Milli-Q water and lyophilizing to produce a typical yield of 15–30 mg. Purified loopoids were evaluated by analytical reverse-phase HPLC and MALDI-TOF mass spec to meet a standard of ≥85% purity for inclusion in the library screening.
Loop-Functionalized Nanosheet Assembly and Screening.
A library of loopoid nanosheets was prepared with a robotic liquid handling system (Biomek FX, Beckman Coulter) as follows. Each well of 96-well plates (polypropylene, round-bottom; Greiner Bio-One, cat. no. 655209) was filled with 247.5 μL of 5 μM OR (octadecyl rhodamine B chloride; ThermoFisher, cat. no. O246) in 10 mM Tris buffer (pH 8.0). 2.5 μL of 2 mM loopoid stock solution in 2:1 (v/v) DMSO/water was added to each well to make a final loopoid concentration of 20 μM. The robotic pipettor with a 96-channel head (AP96) was programmed to perform 3:1 interfacial area compression inside a P250 pipet tip as follows (Figure S1; Beckman Coulter, cat. no. 717252): (a) aspirate 200 μL of loopoid solution; (b) 200 cycles of dispense 160 at 16 μL/s flow rate, aspirate 160 at 32 μL/s with a delay time of 10 s after the aspiration; (c) dispense 200 μL. Then, 50 μL of nanosheet-containing solutions were transferred to the plates prefilled with 50 μL of Alexa 647-conjugated substrate protein solutions and mixed with the 96-channel pipet head. After 30 min incubation at room temperature, the fluorescence intensities and/or spectra of both donor and acceptor fluorophores were obtained with a plate reader (Infinite M200 PRO, Tecan; excitation at 550 nm, donor emission at 590 nm, isosbestic point at 645 nm, acceptor emission at 675 nm) and analyzed to calculate FRET ratios. Fluorescence images of produced nanosheets were acquired with an automated microscope (EVOS FL Auto, Life Technologies) using the RFP light cube (Ex. 531/40 nm, Em. 593/40 nm) with a raster scan mode, and the images from the same well were stitched together with the manufacturer provided software (PerlScope, revision no. 31201).
To compare binding specificity quantitatively, we calculate specificity score (S) for each loopoid (i = 1, 2, ⋯, n) and each target protein (j = 1, 2, ⋯, m) from FRET ratio values (Rij) as follows: (1) filter out loopoids with negative FRET ratio values due to background noise (Rij > 0); (2) normalize FRET ratio values for each target protein group by dividing the values with the maximum FRET value from each group ; (3) calculate the relative ratio as defined:
For example, when FRET ratio values for three target proteins (anthrax, shiga, and strep) are available from a binding affinity screen, the anthrax binding specificity score for loopoid i is calculated as
For hit validation, nanosheets were prepared by the conventional vial-rocking method according to the literature.28 Briefly, 1 mL of Tris buffer (10 mM, pH 8.0) containing 20 μM peptoid strands were prepared in a 4 dram glass vial. The vial was tilted by a custom-built machine from horizontal to vertical for 200 repetitions and 10 s horizontal wait times.
(PA63)7 Expression, Purification, and Dye Labeling.
A plasmid for expression of the 83 kDa anthrax protective antigen (PA83; K563C, D425A mutant) cloned into a pD444-ompA vector (ATUM, Newark, CA) with ampicillin resistance, IPTG inducible T5 promoter and ompA signal peptide was a gift from the Pentelute lab at MIT. Similar protocols for purification of (PA63)7 have been described previously.41,42 BL21 (DE3) Escherichia coli (New England Biosciences, Ipswich, MA) transformants were grown from overnight cultures in 0.5 L batches at 37 °C with shaking at 250 rpm to OD 0.6–0.8. Overexpression was induced with 1 mM IPTG at 30 °C and 250 rpm for 3 h. Cells were harvested by centrifugation and stored as pellets at −80 °C until protein purification. Cells were exposed to osmotic shock to extract proteins from the periplasmic space by dispersion in 10 mM bis-tris propane pH 8.9 with 1 mM EDTA and 30% glucose, followed by centrifugation and redispersion into 5 mM MgCl2. A round of ultracentrifugation was used to clarify the supernatant (50 000g, 30 min), then the solution was brought to 10 mM bis-tris propane pH 8.9 with 5 mM TCEP (Buffer A) with concentrated stocks. This solution was applied to a 5 mL HiTrap Q Fast Flow anion exchange column (GE Healthcare Life Sciences, Marlborough, MA) and eluted over 20 column volumes with a 0–60% gradient of Buffer A + 1 M NaCl. The fraction containing PA83 was identified by SDS-PAGE and dialyzed into Buffer A overnight at 4 °C. Cleavage of PA83 to the active PA63 form was done by adding 1:1000 (g/g) trypsin/PA63 for 30 min at room temperature, followed by the addition of a 10 times excess of trypsin inhibitor. It was found that heptamerization of PA63 did not occur until a second round of anion exchange (this time over a 1 mL column) after cleavage to separate PA63 from the 20 kDa cleaved unit. The (PA63)7-containing fraction was dialyzed into phosphate buffered saline pH 7.4 (PBS; 10 mM phosphate buffer, 2.7 mM potassium chloride, and 137 mM sodium chloride) overnight at 4 °C.
Alexa Fluor 647 succinimidyl ester (ThermoFisher Scientific, Waltham, MA) was conjugated to solvent-exposed lysine residues of (PA63)7. The protein in PBS pH 7.4 solution was made basic with 1 M sodium bicarbonate pH 9.0, and dye was dissolved in dry DMSO. Ten molar equivalents of the dye (relative to heptamer concentration obtained by UV–vis absorbance at 280 nm using a measured extinction coefficient of 4.39 × 105 M−1 cm−1) was added to the protein solution while stirring vigorously, and the reaction was allowed to proceed for 1 h. The reaction was quenched with 0.2 M tris(hydroxymethyl)-aminomethane hydrochloride pH 7.5 and the protein was purified as a heptamer via gel filtration with a Superdex 200 Increase 10/300 GL column (GE Healthcare Life Sciences, Marlborough, MA). Final protein concentrations were measured by UV–vis, and samples were stored in PBS pH 7.4 at 4 °C.
Scanning Transmission Electron Microscope Sample Preparation.
Nanosheet samples were combined 1:1 with 1 mM (PA63)7 in PBS pH 7.4. The solution was drop-cast onto copper TEM grids (ultrathin carbon with lacey carbon film, 400 mesh, Ted Pella, Redding, CA) and allowed to incubate for 10 min. After wicking away the sample solution and washing twice with water, an organotungsten negative stain (Nano-W, Nanoprobes, Yaphank, NY) was applied for 5 min. The stain was wicked away and the samples were allowed to air-dry overnight before imaging at 30 kV in STEM mode on a Zeiss Gemini Ultra-55 analytical FE-SEM.
Transmission Electron Microscope Sample Preparation.
Negative staining EM specimen preparation: PA63 Heptamer sample was prepared by optimized negative staining method as described.43–45 In brief, the sample was diluted to 0.01 mg/mL with Dulbecco’s phosphate buffered saline (DPBS). An aliquot (~4 μL) of sample was placed on a glow-discharged thin-carbon-coated 200 mesh copper grid (CF200-Cu-UL, Electron Microscopy Sciences, Hatfield, PA 19440, USA). After ~1 min incubation, excess solution was blotted with filter paper. Then, the grid was stained by 1% (w/v) uranyl formate (UF) on Parafilm. The grid was dried with nitrogen.
EM Data Acquisition and Image Processing.
The negative staining EM specimen was examined on a Zeiss Libra 120 Plus TEM (Carl Zeiss NTS, Oberkochen, Germany) operating at 120 kV with 20 eV in-column energy filtering at room temperature. The micrographs were acquired by a Gatan UltraScan 4Kx4K CCD at 80 000× magnification (each pixel of the micrographs corresponded to 1.48 Å) under near Scherzer focus (0.1 μm) and defocus of 0.4 μm.
Protease Treatment of Peptoid Nanosheets.
For the observation of the proteolytic stability of a loop-functionalized nanosheet, a fresh stock solution of 20 mg/mL proteinase K, 4 mM CaCl2 was prepared in DI H2O. A 500 mL nanosheet solution was incubated with proteinase K at 60 °C, and agitated on a rotary shaker at 300 rpm overnight. Final concentration is 0.25 mg/mL of Proteinase K, 4 mM CaCl2, and 50 mM Tris pH 7.5 buffer. After incubation with Proteinase K, nanosheets were dialyzed overnight, using a 1 mL Spectra-Por Float-A-Lyzer device with 100 kDa MWCO to remove Proteinase K. The dialyzed samples were transferred onto the surface of freshly cleaved mica to observe morphology by AFM.
Supplementary Material
ACKNOWLEDGMENTS
This project was funded by the DARPA Fold F(x) program. Portions of this work were conducted at the Molecular Foundry at Lawrence Berkeley National Laboratory, which is supported by the Office of Science, Office of Basic Energy Sciences, U.S. Department of Energy under Contract No. DEAC02-05CH11231. Pacific Northwest National Laboratory (PNNL) is a multiprogram national laboratory operated for Department of Energy by Battelle under Contracts No. DE-AC05-76RL01830. The authors thank R. Garcia for help with peptoid synthesis.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.9b07498.
Design and synthesis of the peptoid library, microscale formation of nanosheet libraries, nanosheet characterization, FRET assay validation and binding data, analysis of binding specificity (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Bradbury AR; Sidhu S; Dubel S; McCafferty J Beyond Natural Antibodies: The Power of In Vitro Display Technologies. Nat. Biotechnol 2011, 29, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Holliger P; Hudson PJ Engineered Antibody Fragments and the Rise of Single Domains. Nat. Biotechnol 2005, 23, 1126–1136. [DOI] [PubMed] [Google Scholar]
- (3).Haussner C; Lach J; Eichler J Synthetic Antibody Mimics for the Inhibition of Protein-Ligand Interactions. Curr. Opin. Chem. Biol 2017, 40, 72–77. [DOI] [PubMed] [Google Scholar]
- (4).Binz HK; Amstutz P; Pluckthun A Engineering Novel Binding Proteins from Nonimmunoglobulin Domains. Nat. Biotechnol 2005, 23, 1257–1268. [DOI] [PubMed] [Google Scholar]
- (5).Mahon CS; Fulton DA Mimicking Nature with Synthetic Macromolecules Capable of Recognition. Nat. Chem 2014, 6, 665–672. [DOI] [PubMed] [Google Scholar]
- (6).Kodadek T Synthetic Receptors with Antibody-Like Binding Affinities. Curr. Opin. Chem. Biol 2010, 14, 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Elgersma RC; Mulder GE; Kruijtzer JAW; Posthuma G; Rijkers DTS; Liskamp RMJ Transformation of the Amyloidogenic Peptide Amylin(20–29) into Its Corresponding Peptoid and Retropeptoid: Access to Both an Amyloid Inhibitor and Template for Self-Assembled Supramolecular Tapes. Bioorg. Med. Chem. Lett 2007, 17, 1837–1842. [DOI] [PubMed] [Google Scholar]
- (8).Murnen HK; Rosales AM; Jaworski JN; Segalman RA; Zuckermann RN Hierarchical Self-Assembly of a Biomimetic Diblock Copolypeptoid into Homochiral Superhelices. J. Am. Chem. Soc 2010, 132, 16112–16119. [DOI] [PubMed] [Google Scholar]
- (9).Nam KT; Shelby SA; Choi PH; Marciel AB; Chen R; Tan L; Chu TK; Mesch RA; Lee BC; Connolly MD; Kisielowski C; Zuckermann RN Free-Floating Ultrathin Two-Dimensional Crystals from Sequence-Specific Peptoid Polymers. Nat. Mater 2010, 9, 454–460. [DOI] [PubMed] [Google Scholar]
- (10).Hebert ML; Shah DS; Blake P; Turner JP; Servoss SL Tunable Peptoid Microspheres: Effects of Side Chain Chemistry and Sequence. Org. Biomol. Chem 2013, 11, 4459–4464. [DOI] [PubMed] [Google Scholar]
- (11).Sun J; Jiang X; Lund R; Downing KH; Balsara NP; Zuckermann RN Self-Assembly of Crystalline Nanotubes from Monodisperse Amphiphilic Diblock Copolypeptoid Tiles. Proc. Natl. Acad. Sci. U. S. A 2016, 113, 3954–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Jin H; Ding Y-H; Wang M; Song Y; Liao Z; Newcomb CJ; Wu X; Tang X-Q; Li Z; Lin Y; Yan F; Jian T; Mu P; Chen C-L Designable and Dynamic Single-Walled Stiff Nanotubes Assembled from Sequence-Defined Peptoids. Nat. Commun 2018, 9, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Jin H; Jiao F; Daily MD; Chen Y; Yan F; Ding Y-H; Zhang X; Robertson EJ; Baer MD; Chen C-L Highly Stable and Self-Repairing Membrane-Mimetic 2D Nanomaterials Assembled from Lipid-Like Peptoids. Nat. Commun 2016, 7, 12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Xuan S; Jiang X; Spencer RK; Li NK; Prendergast D; Balsara NP; Zuckermann RN Atomic-Level Engineering and Imaging of Polypeptoid Crystal Lattices. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 22491–22499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zuckermann RN; Kerr JM; Kent SBH; Moos WH Efficient Method for the Preparation of Peptoids [Oligo(N-Substituted Glycines)] by Submonomer Solid-Phase Synthesis. J. Am. Chem. Soc 1992, 114, 10646–10647. [Google Scholar]
- (16).Knight AS; Zhou EY; Francis MB; Zuckermann RN Sequence Programmable Peptoid Polymers for Diverse Materials Applications. Adv. Mater 2015, 27, 5665–5691. [DOI] [PubMed] [Google Scholar]
- (17).Robertson EJ; Battigelli A; Proulx C; Mannige RV; Haxton TK; Yun L; Whitelam S; Zuckermann RN Design, Synthesis, Assembly, and Engineering of Peptoid Nanosheets. Acc. Chem. Res 2016, 49, 379–389. [DOI] [PubMed] [Google Scholar]
- (18).Olivier GK; Cho A; Sanii B; Connolly MD; Tran H; Zuckermann RN Antibody-Mimetic Peptoid Nanosheets for Molecular Recognition. ACS Nano 2013, 7, 9276–9286. [DOI] [PubMed] [Google Scholar]
- (19).Zhu L; Zhao Z; Cheng P; He Z; Cheng Z; Peng J; Wang H; Wang C; Yang Y; Hu Z Antibody-Mimetic Peptoid Nanosheet for Label-Free Serum-Based Diagnosis of Alzheimer’s Disease. Adv. Mater 2017, 29, 1700057. [DOI] [PubMed] [Google Scholar]
- (20).Battigelli A; Kim JH; Dehigaspitiya DC; Proulx C; Robertson EJ; Murray DJ; Rad B; Kirshenbaum K; Zuckermann RN Glycosylated Peptoid Nanosheets as a Multivalent Scaffold for Protein Recognition. ACS Nano 2018, 12, 2455–2465. [DOI] [PubMed] [Google Scholar]
- (21).Jiang SY; Cao ZQ Ultralow-Fouling, Functionalizable, and Hydrolyzable Zwitterionic Materials and Their Derivatives for Biological Applications. Adv. Mater 2010, 22, 920–932. [DOI] [PubMed] [Google Scholar]
- (22).Martinez-Veracoechea FJ; Frenkel D Designing Super Selectivity in Multivalent Nano-Particle Binding. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 10963–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Sun J; Zuckermann RN Peptoid Polymers: A Highly Designable Bioinspired Material. ACS Nano 2013, 7, 4715–4732. [DOI] [PubMed] [Google Scholar]
- (24).Miller SM; Simon RJ; Ng S; Zuckermann RN; Kerr JM; Moos WH Comparison of the Proteolytic Susceptibilities of Homologous L-Amino Acid, D-Amino Acid, and N-Substituted Glycine Peptide and Peptoid Oligomers. Drug Dev. Res 1995, 35, 20–32. [Google Scholar]
- (25).Flood D; Proulx C; Robertson EJ; Battigelli A; Wang S; Schwartzberg AM; Zuckermann RN Improved Chemical and Mechanical Stability of Peptoid Nanosheets by Photo-Crosslinking the Hydrophobic Core. Chem. Commun 2016, 52, 4753–4756. [DOI] [PubMed] [Google Scholar]
- (26).Shin SBY; Yoo B; Todaro LJ; Kirshenbaum K Cyclic Peptoids. J. Am. Chem. Soc 2007, 129, 3218–3225. [DOI] [PubMed] [Google Scholar]
- (27).Robertson EJ; Olivier GK; Qian M; Proulx C; Zuckermann RN; Richmond GL Assembly and Molecular Order of Two-Dimensional Peptoid Nanosheets through the Oil-Water Interface. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 13284–13289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Sanii B; Kudirka R; Cho A; Venkateswaran N; Olivier GK; Olson AM; Tran H; Harada RM; Tan L; Zuckermann RN Shaken, Not Stirred: Collapsing a Peptoid Monolayer to Produce Free-Floating, Stable Nanosheets. J. Am. Chem. Soc 2011, 133, 20808–20815. [DOI] [PubMed] [Google Scholar]
- (29).Zhang JH; Chung TDY; Oldenburg KR A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screening 1999, 4, 67–73. [DOI] [PubMed] [Google Scholar]
- (30).Mourez M; Kane RS; Mogridge J; Metallo S; Deschatelets P; Sellman BR; Whitesides GM; Collier RJ Designing a Polyvalent Inhibitor of Anthrax Toxin. Nat. Biotechnol 2001, 19, 958–961. [DOI] [PubMed] [Google Scholar]
- (31).Patke S; Boggara M; Maheshwari R; Srivastava SK; Arha M; Douaisi M; Martin JT; Harvey IB; Brier M; Rosen T; Mogridge J; Kane RS Design of Monodisperse and Well-Defined Polypeptide-Based Polyvalent Inhibitors of Anthrax Toxin. Angew. Chem., Int. Ed 2014, 53, 8037–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Joshi A; Kate S; Poon V; Mondal D; Boggara MB; Saraph A; Martin JT; McAlpine R; Day R; Garcia AE; Mogridge J; Kane RS Structure-Based Design of a Heptavalent Anthrax Toxin Inhibitor. Biomacromolecules 2011, 12, 791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zondlo NJ Aromatic-Proline Interactions: Electronically Tunable CH/π Interactions. Acc. Chem. Res 2013, 46, 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Murray DJ; Kim JH; Grzincic EM; Kim SC; Abate AR; Zuckermann RN Uniform, Large-Area, Highly Ordered Peptoid Monolayer and Bilayer Films for Sensing Applications. Langmuir 2019, 35, 13671–13680. [DOI] [PubMed] [Google Scholar]
- (35).Maynard JA; Maassen CB; Leppla SH; Brasky K; Patterson JL; Iverson BL; Georgiou G Protection against Anthrax Toxin by Recombinant Antibody Fragments Correlates with Antigen Affinity. Nat. Biotechnol 2002, 20, 597–601. [DOI] [PubMed] [Google Scholar]
- (36).Mohamed N; Clagett M; Li J; Jones S; Pincus S; D’Alia G; Nardone L; Babin M; Spitalny G; Casey L A High-Affinity Monoclonal Antibody to Anthrax Protective Antigen Passively Protects Rabbits before and after Aerosolized Bacillus Anthracis Spore Challenge. Infect. Immun 2005, 73, 795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Strauch EM; Bernard SM; La D; Bohn AJ; Lee PS; Anderson CE; Nieusma T; Holstein CA; Garcia NK; Hooper KA; Ravichandran R; Nelson JW; Sheffler W; Bloom JD; Lee KK; Ward AB; Yager P; Fuller DH; Wilson IA; Baker D Computational Design of Trimeric Influenza-Neutralizing Proteins Targeting the Hemagglutinin Receptor Binding Site. Nat. Biotechnol 2017, 35, 667–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Miller SM; Simon RJ; Ng S; Zuckermann RN; Kerr JM; Moos WH Proteolytic Studies of Homologous Peptide and N-Substituted Glycine Peptoid Oligomers. Bioorg. Med. Chem. Lett 1994, 4, 2657–2662. [Google Scholar]
- (39).Zuckermann RN; Martin EJ; Spellmeyer DC; Stauber GB; Shoemaker KR; Kerr JM; Figliozzi GM; Goff DA; Siani MA; Simon RJ; Banville SC; Brown EG; Wang L; Richter LS; Moos WH Discovery of Nanomolar Ligands for 7-Transmembrane G-Protein-Coupled Receptors from a Diverse N-(Substituted)Glycine Peptoid Library. J. Med. Chem 1994, 37, 2678–2685. [DOI] [PubMed] [Google Scholar]
- (40).Proulx C; Yoo S; Connolly MD; Zuckermann RN Accelerated Submonomer Solid-Phase Synthesis of Peptoids Incorporating Multiple Substituted N-Aryl Glycine Monomers. J. Org. Chem 2015, 80, 10490–10497. [DOI] [PubMed] [Google Scholar]
- (41).Wigelsworth DJ; Krantz BA; Christensen KA; Lacy DB; Juris SJ; Collier RJ Binding Stoichiometry and Kinetics of the Interaction of a Human Anthrax Toxin Receptor, Cmg2, with Protective Antigen. J. Biol. Chem 2004, 279, 23349–23356. [DOI] [PubMed] [Google Scholar]
- (42).Christensen KA; Krantz BA; Collier RJ Assembly and Disassembly Kinetics of Anthrax Toxin Complexes. Biochemistry 2006, 45, 2380–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Zhang L; Song J; Newhouse Y; Zhang S; Weisgraber KH; Ren G An Optimized Negative-Staining Protocol of Electron Microscopy for Apoe4 Popc Lipoprotein. J. Lipid Res 2010, 51, 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Rames M; Yu Y; Ren G Optimized Negative Staining: A High-Throughput Protocol for Examining Small and Asymmetric Protein Structure by Electron Microscopy. J. Visualized Exp 2014, No. e51087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Zhang L; Song J; Cavigiolio G; Ishida BY; Zhang S; Kane JP; Weisgraber KH; Oda MN; Rye KA; Pownall HJ; Ren G Morphology and Structure of Lipoproteins Revealed by an Optimized Negative-Staining Protocol of Electron Microscopy. J. Lipid Res 2011, 52, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


