Abstract
Current knowledge of methicillin-resistant Staphylococcus aureus (MRSA) colonisation in relation to epidemiological characteristics is incomplete. We conducted a cross-sectional study at an acute-care tertiary infectious diseases hospital of MRSA isolates identified through routine surveillance from January 2009 to December 2011. We randomly selected 205 MRSA isolates (119 inpatients) from 798 isolates (427 inpatients) for molecular profiling using multilocus sequence typing. Multilevel multinomial logistic regression was used to estimate odds ratio (OR) assessing the predilection of MRSA strains for anatomic sites, and associations of strains with human immunodeficiency virus (HIV) infection. The most frequent sequence types (STs) were 239, 22 and 45. The proportion of ST22 increased over the sampling period, replacing ST239 as the dominant lineage. However, ST239 remained the most prevalent among HIV-seropositive individuals who were six times more likely to be colonised with this strain than non-HIV patients (adjusted OR (aOR) 6.44, 95% confidence interval (CI) 1.94–21.36). ST45 was >24 times more likely to be associated with perianal colonisation than in the nares, axillae and groin sites (aOR 24.20, 95% CI 1.45–403.26). This study underlines the clonal replacement of MRSA in Singapore as previously reported but revealed, in addition, key strain differences between HIV-infected and non-infected individuals hospitalised in the same environment.
Key words: Epidemiology, hospital microbiology, hospital-acquired (nosocomial) infections, methicillin-resistant S. aureus (MRSA), molecular epidemiology
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) has remained a significant threat for healthcare-associated infections worldwide since its discovery in Britain in the 1960s but first appeared in Singapore in the early 1980s [1]. MRSA infection imposes an additional burden on healthcare systems owing to significant morbidity and mortality, length of hospital stay and hospital costs [2]. Prior colonisation with MRSA is a major risk factor for subsequent infection [3]. An earlier study in Singapore reported that 14% of 840 patients newly colonised with MRSA identified on active surveillance developed infection within a median time of 22 days; 72% of infected patients developed infection within 60 days of the date MRSA colonisation was detected [4]. The risk of subsequent infection appears to be augmented by colonisation at multiple body sites [5].
The most prevalent MRSA isolates reported worldwide are sequence types (STs) 239, ST22, ST5 and ST8, as determined by multilocus sequence typing (MLST) [6]. A whole-genome sequencing study of the population structure of MRSA in Singapore hospitals spanning three decades till year 2010 clearly showed that, alongside clinical practice and antibiotic usage, competition between clones played an important role in driving the evolution of strain populations [7]. Since around 2000, the fluoroquinolone-resistant ST22 replaced ST239 corresponding to the time when fluoroquinolones were widely used globally in the healthcare setting [8]. However, to date, there has been limited information on the colonisation distribution of specific MRSA strains by anatomic sites and the influence of human immunodeficiency virus (HIV) infection on colonisation of such strain types.
We report here the clinical epidemiological characteristics and MRSA STs using MLST patterns, and their correlation with the anatomic sites of colonisation and HIV status of patients attending a tertiary infectious disease referral centre in Singapore over a 3-year period.
Methods
Setting
A cross-sectional study was conducted at the Communicable Disease Centre (CDC), of the Tan Tock Seng Hospital (TTSH), Singapore, which is a 100-bed adult acute-care public hospital and a tertiary referral centre for HIV infection and other infectious diseases; it also provides tertiary inpatient care for dermatological diseases. The CDC introduced MRSA screening surveillance in January 2009. As previously described, this programme requires all inpatients to be screened for MRSA colonisation with three separate samples from anatomic sites: throat, perianal and combined pooled swab from the nares, axillae and groin (NAG) [9, 10]. MRSA-colonised inpatients were isolated as per standard procedure and the study period ran from 1 January 2009 to 31 December 2011.
MRSA isolates
Swab samples were inoculated on selective chromogenic agar plates (MRSASelect, BioRad, France) and incubated at 37 °C for 18–28 h. Growth of pink or mauve colonies was presumptively identified as MRSA and stored in deep freeze. During the study period, 798 MRSA isolates from 427 patients were obtained and a group of MRSA-positive patients was randomly selected proportionate to the number of MRSA patients detected in each calendar quarter for a period of 12 calendar quarters, for subsequent molecular profiling.
Molecular profiling
Genomic DNA was extracted using the EZ1 DNA Tissue Kit (Qiagen Inc., Hilden, Germany) following the manufacturer's instructions. Indexed sequencing libraries were prepared using Covaris fragmentation and Illumina TruSeq DNA/RNA v2 kit (Illumina, Inc., San Diego, CA, USA) following the manufacturer's recommended protocol. Sequencing was done on an Illumina HiSeq2500 Rapid using paired-end 2 × 76 base pair reads. MLST was run through an enhanced Short Read Sequencing Typing 2 (SRST2) [11] of 0.5-kb fragments from seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi and yqiL) as described previously [12].
Data collection and quantitative variables
Data collection included demographic, epidemiological and clinical information of patients, principally Charlson's comorbidities index (CCI) [13] and HIV infection status (see Table 1). The screening year of MRSA was also used as a surrogate index to estimate the degree of strain evolution over time. Based on the ST, an MRSA clone from one of three screening swabs was defined as single site colonisation, and if present in more than one site, was deemed multi-site colonisation. This was further categorised as single strain colonisation where the same ST was cultured from the body sites, and different strains otherwise. All the subject identifiers were anonymised from the dataset for analysis.
Table 1.
Demographic and clinical characteristics of patients
| Characteristics | Total patients (n = 119) |
Single site (n = 61) |
Multiple sites (n = 58) | P1 | P2 | ||
|---|---|---|---|---|---|---|---|
| Total multiple sites (n = 58) | Same strains (n = 33) | Different strains (n = 25) | |||||
| Age, years | 0.24a | 0.27a | |||||
| Mean ± s.d. | 60 ± 19.2 | 58 ± 18.1 | 63 ± 20.3 | 60 ± 20.8 | 66 ± 19.4 | ||
| Range | 19–94 | 21–91 | 19–94 | 19–94 | 19–91 | ||
| Age >65 years | 55 (46.2) | 24 (39.3) | 31 (53.5) | 16 (48.5) | 15 (60.0) | 0.12 | 0.38 |
| Male | 82 (68.9) | 42 (68.9) | 40 (69.0) | 23 (69.7) | 17 (68.0) | 0.99 | 0.89 |
| Ethnic | 0.88 | 0.52 | |||||
| Chinese | 91 (76.5) | 47 (77.0) | 44 (75.9) | 24 (72.7) | 20 (80.0) | ||
| Non-Chinese | 28 (23.5) | 14 (23.0) | 14 (24.1) | 9 (27.3) | 5 (20.0) | ||
| Smoking | 24 (20.2) | 19 (31.2) | 5 (8.6) | 2 (6.1) | 3 (12.0) | <0.01 | 0.43 |
| Alcohol abuse | 4 (3.4) | 3 (4.9) | 1 (1.7) | 0 (0.0) | 1 (4.0) | 0.62b | 0.25b |
| CCI, median (IQR) | 4 (1–7) | 6 (1–7) | 2 (1–6) | 4 (1–6) | 1 (0–6) | 0.15c | 0.30c |
| CCI > 3 | 62 (52.1) | 36 (59.0) | 26 (44.8) | 18 (54.6) | 8 (32.0) | 0.12 | 0.09 |
| HIV positive | 52 (43.7) | 33 (54.1) | 19 (32.8) | 12 (36.4) | 7 (28.0) | 0.02 | 0.50 |
| Diabetes mellitus | 23 (19.3) | 11 (18.0) | 12 (20.7) | 8 (24.2) | 4 (16.0) | 0.71 | 0.44 |
| Myocardial infarction | 3 (2.5) | 1 (1.6) | 2 (3.5) | 1 (3.0) | 1 (4.0) | 0.61b | 1.00b |
| Congestive heart failure | 8 (6.7) | 5 (8.2) | 3 (5.2) | 2 (60.1) | 1 (4.0) | 0.72b | 1.00b |
| Cerebrovascular disease | 22 (18.5) | 9 (14.8) | 13 (22.4) | 7 (21.2) | 6 (24.0) | 0.28 | 0.80 |
| Chronic pulmonary disease | 13 (10.9) | 9 (14.8) | 4 (6.9) | 3 (9.1) | 1 (4.0) | 0.17 | 0.63 |
| Chronic liver disease | 17 (14.3) | 6 (9.8) | 11 (19.0) | 6 (18.2) | 5 (20.0) | 0.16 | 1.00b |
| Renal diseases | 15 (12.6) | 6 (9.8) | 9 (15.5) | 6 (18.2) | 3 (12.0) | 0.35 | 0.72b |
| Solid malignancy | 6 (5.0) | 2 (3.3) | 4 (6.9) | 2 (6.1) | 2 (8.0) | 0.43b | 1.00 |
| Haematologic malignancy | 2 (1.7) | 1 (1.6) | 1 (1.7) | 1 (3.0) | 0 (0.0) | 1.00b | 1.00b |
| Hospitalisation in the past 1 year | 77 (64.7) | 38 (62.3) | 39 (67.2) | 26 (78.8) | 13 (52.0) | 0.57 | 0.03 |
| Invasive procedures during hospitalisationd | 69 (58.0) | 36 (59.0) | 33 (56.9) | 18 (54.6) | 15 (60.0) | 0.82 | 0.68 |
| Primary discipline of admission | |||||||
| Infectious disease | 59 (49.6) | 38 (64.4) | 21 (35.6) | 12 (57.1) | 9 (42.9) | <0.01 | 0.98 |
| Dermatology | 60 (50.4) | 23 (38.3) | 37 (61.7) | 21 (56.8) | 16 (43.2) | ||
| Year of screening | |||||||
| 2009 | 55 (46.2) | 27 (44.3) | 28 (48.3) | 13 (39.4) | 15 (60.0) | 0.28 | 0.34 |
| 2010 | 30 (25.2) | 19 (31.1) | 11 (19.0) | 7 (21.2) | 4 (16.0) | ||
| 2011 | 34 (28.6) | 15 (24.6) | 19 (32.7) | 13 (39.4) | 6 (24.0) | ||
CCI, Charlson's comorbidity index; HIV, human immunodeficiency virus; IQR, interquartile range; s.d., standard deviation.
Values are no. (%) unless otherwise indicated.
Single site colonisation was defined as detection of an MRSA clone from one of three screening swabs (nares, axillae, groin pooled swab (NAG), perianal and throat).
Multiple sites colonisation was defined as detection of MRSA clone in a minimum of two out of three screening swabs (nares, axillae, groin pooled swab (NAG), perianal and throat).
Multiple sites colonisation was further categorised into same strain where same ST was cultured from body sites of each patient, and different strains otherwise; and a combination of both same and different strains was named as total multiple sites.
P1, statistical test between single site and total multiple sites.
P2, statistical test between same strains and different strains.
Two-sample t test.
Fisher's exact test.
Mann–Whitney U test.
Invasive procedures included surgery, nasogastric tube, chest tube, intravenous line insertions and urinary catheterisation.
Statistical methods
Characteristics of patients were described with frequencies and percentages for categorical variables, mean and standard deviation (s.d.), or median and interquartile range (IQR) for continuous and ordinal variables, respectively. Pearson's χ2 test or Fisher's exact test for categorical variables and unpaired two-sample t test or Mann–Whitney U test for continuous variables were used for bivariate inference methods. A multilevel multivariable multinomial logistic regression with random intercepts was constructed using PROC GLIMMIX in SAS (version 9.4, SAS Institute Inc., Cary, NC, USA) to assess the association between different MRSA STs and anatomic sites, and HIV status. The odds ratios (OR) with 95% confidence intervals (CI) from regression analyses were determined and statistical significance threshold was set at P < 0.05; all reported P values were two-tailed. Statistical analyses comparing differences in continuous and categorical variables using two-sample t test, Mann–Whitney U test, χ2 and Fisher's exact tests were performed with Stata 13.1 (College Station, TX, USA: StataCorp LP).
Ethics approval and STROME-ID statement
The study was approved by the Domain Specific Review Board (DSRB) of the National Healthcare Group, Singapore (DSRB – 2011/01655), and reported in accordance with the Strengthening the Reporting of Molecular Epidemiology for Infectious Diseases (STROME-ID) statement [14].
Results
Characteristics of study population
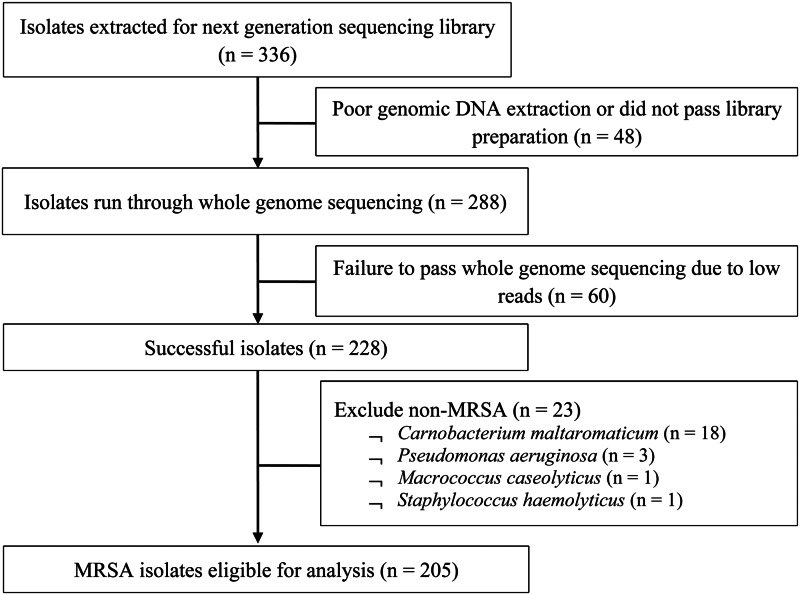
From 2009 to 2011, a total of 205 out of 336 MRSA isolates (61%) from 119 patients aged from 19 to 94 years (mean 60 ± 19.2 years) were eligible for analysis (Fig. 1). There was no statistically significant difference between MRSA-positive patients who were selected into the study and those who were not, in terms of age, gender and ethnicity. Table 1 presents the demographics and clinical characteristics of the study population; nearly half (46.2%) were admitted in 2009 with the remaining patients almost equally represented (25.2% and 28.6%) in the successive years. A significantly higher proportion of multi-site colonisation was observed in dermatological inpatients (single 38.3% vs. multi 61.7%, P < 0.01) but a lower proportion were HIV-positive (single 54.1% vs. multi 32.8%, P = 0.02). There were no significant differences between single-site and multi-site colonisation according to gender, ethnicity, prior hospitalisation, invasive procedures, year of screening and median CCI.
Fig. 1.
Flow diagram of MRSA isolates selection for study.
Of those subjects colonised at multiple sites, 57% (n = 33/58) grew the same strain at different sites, and those with a history of hospitalisation in the past year were significantly more likely to be colonised with the same strain (same 78.8% vs. different 52.0%, P = 0.03). No significant differences in other covariates were observed between the two groups.
Distribution of MRSA STs
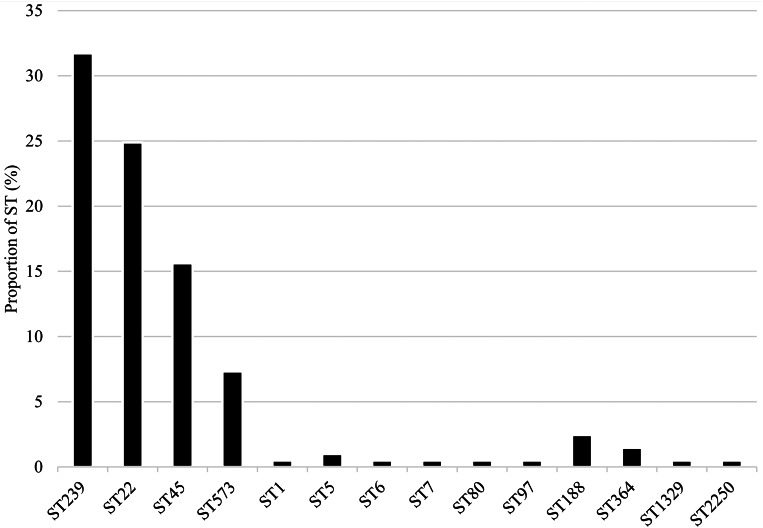
Fourteen STs were identified among MRSA isolates from NAG sites (n = 79; 38.5%), perianal (n = 66; 32.2%) and throat (n = 60; 29.3%) samples. The most prevalent were ST239 (n = 65; 31.7%), ST22 (n = 51; 24.9%), ST45 (n = 31; 15.1%) and ST573 (n = 15; 7.3%) (Fig. 2).
Fig. 2.
Distribution of MRSA sequence types.
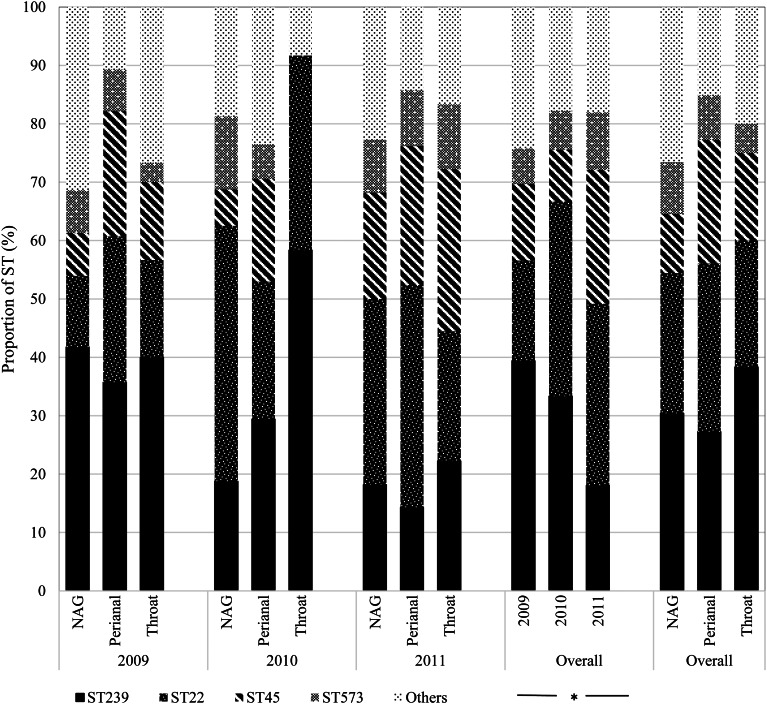
Overall, ST239 (n = 24; 30.4%) and ST22 (n = 19; 24.1%) were the most frequent colonists of NAG sites followed by ST45 (n = 8; 10.1%); the first two types also predominated in the throat (ST239: n = 23; 38.3% and ST22: n = 13; 21.7%). By contrast, ST45 (n = 14; 21.2%) was almost as frequent as ST239 (n = 18; 27.3%) and ST22 (n = 19; 28.8%) in perianal sites (Fig. 3). Over time, ST239 was the most prevalent in 2009 (n = 39; 39.4%), with the distribution of ST239 and ST22 balancing out in 2010 (n = 15; 33.3% each), and ST22 dominating over ST239 in 2011 (n = 19; 31.2% vs. n = 11; 18.0%) (P = 0.04) suggesting that clonal replacement had occurred in the hospital setting (Fig. 3).
Fig. 3.
Temporal distribution of MRSA sequence types by anatomic sites. NAG, nares, axillae, groin pooled swab; ST, sequence type. *Fisher's exact test (P = 0.04).
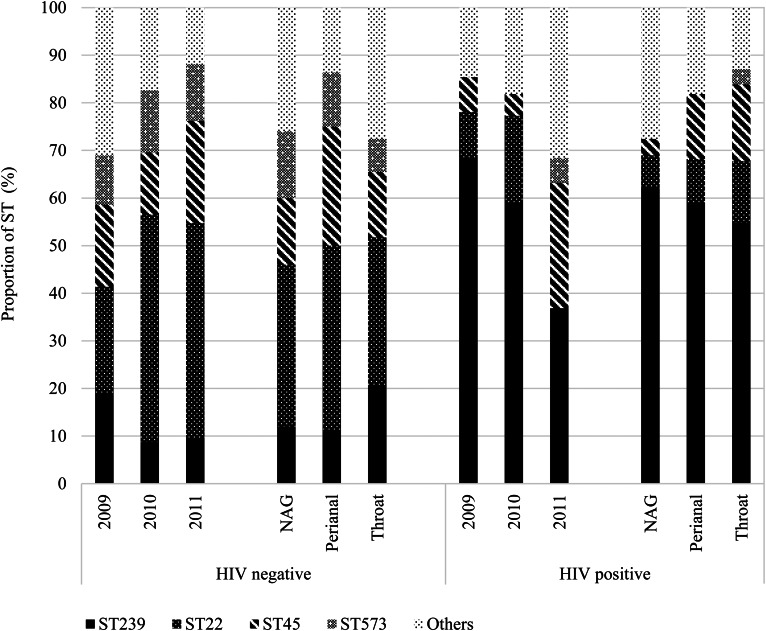
We further stratified the MRSA isolates based on HIV status, including 82 (40.0%) isolates from 52 (43.7%) HIV-positive patients and observed differences in colonisation of STs across the years and between anatomic sites (Fig. 4). Indeed, HIV-positive patients tended to be predominantly colonised with ST239, even in the year 2011 when ST22 was generally more prevalent than ST239.
Fig. 4.
Distribution of MRSA isolates (n = 205) sequence types (STs) by anatomic sites stratified by HIV-seropositive status. HIV, human immunodeficiency virus; NAG, nares, axillae, groin pooled swab; ST, sequence type.
Correlates of colonisation with specific STs
In the two-level multinomial logistic regression analyses, the reference category was other STs. Compared with NAG colonisation, perianal sites were >24 times more likely to harbour ST45 strains after adjustment for screening year (adjusted OR (aOR) 24.20, 95% CI 1.45–403.26; Table 2). Likewise, HIV-positive patients were six times more likely to be colonised with a ST239 strain than HIV-negative individuals (aOR 6.44, 95% CI 1.94–21.36; Table 2).
Table 2.
Multilevel multivariable multinomial models (205 isolates nested within 119 patients) of colonisation with specific methicillin-resistant Staphylococcus aureus (MRSA) sequence types (STs) including ST239, ST22, ST45 and ST573 with other STs as the reference category
| Variables | Total number (%) in analysis | ST239 | ST22 | ST45 | ST573 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Crude OR (95% CI) | Adjusted ORa (95% CI) | Crude OR (95% CI) | Adjusted ORa (95% CI) | Crude OR (95% CI) | Adjusted ORa (95% CI) | Crude OR (95% CI) | Adjusted ORa (95% CI) | ||
| Isolates based on anatomic sites | |||||||||
| NAGb | 79 (38.5) | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| Perianal | 66 (32.2) | 1.83 (0.55–6.17) | 1.91 (0.57–6.41) | 3.81 (0.36–39.90) | 3.69 (0.37–36.56) | 26.55 (1.50–469.47) | 24.20 (1.45–403.26) | 20.01 (0.05–999.99) | 14.74 (0.07–999.99) |
| Throat | 60 (29.3) | 1.93 (0.60–6.23) | 1.85 (0.59–5.79) | 2.94 (0.19–45.18) | 2.63 (0.18–38.69) | 4.12 (0.23–72.86) | 3.75 (0.23–61.97) | 0.85 (0.01–57.98) | 0.79 (0.01–49.75) |
| Isolates based on HIV infection status | |||||||||
| Negative | 123 (60.0) | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| Positive | 82 (40.0) | 5.92 (1.88–18.65) | 6.44 (1.94–21.36) | 0.28 (0.01–14.08) | 0.22 (0.003–14.57) | 0.62 (0.02–19.48) | 0.50 (0.01–24.86) | 0.19 (0.001–347.33) | 0.17 (0.001–553.52) |
CI, confidence interval; HIV, human immunodeficiency virus; NAG, nares, axillae and groin; OR, odds ratio.
Adjusted OR was adjusted for year of screening.
A single pooled swab was taken from nares, axillae and groin (NAG) sites.
Discussion
The 3-year temporal distribution analysis of MRSA clones from 2009 to 2011 in our hospital population showed significant clonal replacement whereby the imported UK-EMRSA-15 clone ST22 gradually became predominant and replacing the ST239 strain; this finding corroborates the conclusions of previous studies in Singapore [7, 15, 16]. The prevalence of the Berlin clone ST45, which is relatively new to Singapore, was also exhibiting a rising trend in an earlier study [15]. ST239 and ST22 have been widely reported to be prevalent hospital-associated clones in many parts of the world [17–19]. Nevertheless, similar to this study, ST239 appears to be in relative decline while ST22 continues to spread in acute-care hospitals in Germany [20] and the Czech Republic [21]. The reasons behind the emergence and replacement of MRSA strains remain unclear but hypothetical suggestions include host preferences, differences in antibiotic resistance, infectivity and ability to cope with environmental stress, and virulence gene complement [20]. Nonetheless, this might have considerable impact on clinical consequences as different clones respond differently to antibiotics and possess varying levels of virulence genes. Notably, ST573 was identified in 7.3% of all isolates and was observed to increase in frequency over the study period. Albeit fairly rare, this type has been recovered in many Asian countries including Singapore [22, 23] and in Australia and Europe [24, 25].
This is one of the few studies, to our knowledge, to examine the specific MRSA strain types associated with HIV infection. Compared with the general population, the prevalence of MRSA and recurrence of infection is significantly higher in HIV-seropositive individuals [26, 27]. Previous studies from our hospital have also highlighted an increased MRSA prevalence among such patients [28, 29]. In terms of specific clonal colonisation, studies from the USA and Europe, for instance, indicate that pulsed-field gel electrophoresis type USA300, corresponding to MLST ST8, is the most frequently colonist among HIV-infected persons [30, 31]. In our study, the hospital-associated clone ST239 was six times more likely to be found in HIV patients. Some clinical studies have suggested that the apparent capability of ST239 for endothelial invasion could have resulted in higher bacteraemia and case-fatality rates than non-ST239 strains [32, 33]. Recent insights have also been gained from in vitro studies suggesting the invasiveness of the ST239 clone might be due to toxin expression and other virulence factors for endothelial inflammation, cell growth and cellular interactions [34]. Due to its apparent increased pathogenicity leading to life-threatening conditions such as bacteraemia, endocarditis and pneumonia [35], its high prevalence in HIV-infected individuals, as found here, poses significant clinical implications and concerns.
The observation that MRSA ST45, which was probably imported [15], exhibited a propensity for the perianal site is unique to our study and is in contrast to a US study which found USA300 (ST8) to be frequently recovered from the perianal site [36]. ST45 has been linked with a small subset of hospital infections in Singapore [15] and so continued surveillance of this type as a colonist and its epidemiology for infection prevention is warranted.
The study was subject to limitations. It was cross-sectional and patients were screened for MRSA on admission as part of the hospital's routine screening programme. Hence, persons who were intermittently colonised might not have been detected, with the potential underestimation of the prevalence of certain strains. Medical information was collected retrospectively; thus, availability of data was limited on certain potential confounders such as prior MRSA infection, health-seeking practise, and sexual orientation and behaviour. We might have underestimated the prior hospitalisation rate since data on admission to other hospitals were not readily available. Nonetheless, the misclassification was likely to be minimal, as the majority of previously hospitalised patients would be admitted to the same hospital for continuity of care. As a combined swab from the NAG was taken, we were unable to differentiate the colonisation pattern of MRSA clones from each of these body sites separately. However, this does not invalidate our study's findings on the predilection of ST45 for perianal colonisation. We did not perform a sample size calculation a priori, and hence our study could have been insufficiently powered to detect small differences in characteristics between strain types. Although multivariable models were used to control for the main confounding variables, residual confounding might still persist.
In summary, our study observed a differing clonal pattern among HIV-infected and non-infected individuals. ST239 remains the dominant strain in HIV-infected patients while ST22 has replaced ST239 in the general patient population. These interesting phenomena would require a longer study period to verify and seek further explanation. Furthermore, we observed that ST45 appeared to have a predilection for perianal colonisation and this potentially has implications for infection prevention and control and warrants further study to determine its clinical significance.
Acknowledgements
Authors acknowledge Ms Linda Kay Lee and Mr Kwa Wit Thun Stephen for their project management and laboratory work, respectively.
Financial support
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Conflict of interest
None.
References
- 1.Esuvaranathan K et al. (1992) A study of 245 infected surgical wounds in Singapore. Journal of Hospital Infection 21, 231–240. [DOI] [PubMed] [Google Scholar]
- 2.Cosgrove SE et al. (2005) The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infection Control and Hospital Epidemiology 26, 166–174. [DOI] [PubMed] [Google Scholar]
- 3.Coello R et al. (1997) Risk factors for developing clinical infection with methicillin-resistant Staphylococcus aureus (MRSA) amongst hospital patients initially only colonized with MRSA. Journal of Hospital Infection 37, 39–46. [DOI] [PubMed] [Google Scholar]
- 4.Balm MN et al. (2013) Progression from new methicillin-resistant Staphylococcus aureus colonisation to infection: an observational study in a hospital cohort. BMC Infectious Diseases 13, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chipolombwe J et al. (2016) Methicillin-resistant Staphylococcus aureus multiple sites surveillance: a systemic review of the literature. Infection and Drug Resistance 9, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stefani S et al. (2012) Methicillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. International Journal of Antimicrobial Agents 39, 273–282. [DOI] [PubMed] [Google Scholar]
- 7.Hsu L-Y et al. (2015) Evolutionary dynamics of methicillin-resistant Staphylococcus aureus within a healthcare system. Genome Biology 16, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holden MT et al. (2013) A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Research 23, 653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Win M-K et al. (2015) Review of a two-year methicillin-resistant Staphylococcus aureus screening program and cost-effectiveness analysis in Singapore. BMC Infectious Diseases 15, 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Win M-K et al. (2013) Evaluation of universal methicillin-resistant Staphylococcus aureus screening using nasal polymerase chain reaction compared with nasal, axilla, and groin and throat and perianal cultures in a hospital setting. Infection Control and Hospital Epidemiology 34, 1335–1337. [DOI] [PubMed] [Google Scholar]
- 11.Inouye M et al. (2014) SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Medicine 6, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enright MC et al. (2000) Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. Journal of Clinical Microbiology 38, 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charlson M et al. (1994) Validation of a combined comorbidity index. Journal of Clinical Epidemiology 47, 1245–1251. [DOI] [PubMed] [Google Scholar]
- 14.Field N et al. (2014) Strengthening the Reporting of Molecular Epidemiology for Infectious Diseases (STROME-ID): an extension of the STROBE statement. The Lancet Infectious Diseases 14, 341–352. [DOI] [PubMed] [Google Scholar]
- 15.Teo J et al. (2013) ST22 and ST239 MRSA duopoly in Singaporean hospitals: 2006–2010. Epidemiology and Infection 141, 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow A et al. (2017) MRSA transmission dynamics among interconnected acute, intermediate-term, and long-term healthcare facilities in Singapore. Clinical Infectious Diseases 64(Suppl_2), S76–S81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campanile F et al. (2009) Hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) in Italy. Annals of Clinical Microbiology and Antimicrobials 8, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song J-H et al. (2011) Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. Journal of Antimicrobial Chemotherapy 66, 1061–1069. [DOI] [PubMed] [Google Scholar]
- 19.Johnson AP et al. (2001) Dominance of EMRSA-15 and-16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS). Journal of Antimicrobial Chemotherapy 48, 143–144. [DOI] [PubMed] [Google Scholar]
- 20.Albrecht N et al. (2011) Clonal replacement of epidemic methicillin-resistant Staphylococcus aureus strains in a German university hospital over a period of eleven years. PLoS ONE 6, e28189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melter O et al. (2006) Emergence of EMRSA-15 clone in hospitals throughout the Czech Republic. Euro Surveillance 11, E060803–E060806. [DOI] [PubMed] [Google Scholar]
- 22.Wijaya L, Hsu L and Kurup A (2006) Community-associated methicillin-resistant Staphylococcus aureus: overview and local situation. Annals of the Academy of Medicine 35, 479–486. [PubMed] [Google Scholar]
- 23.Hsu L-Y et al. (2006) Establishment of ST30 as the predominant clonal type among community-associated methicillin-resistant Staphylococcus aureus isolates in Singapore. Journal of Clinical Microbiology 44, 1090–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coombs GW et al. (2006) Methicillin-resistant Staphylococcus aureus clones, Western Australia. Emerging Infectious Diseases 12, 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karden-Lilja M et al. (2007) Panton-Valentine leukocidin genes and staphylococcal chromosomal cassette mec types amongst Finnish community-acquired methicillin-resistant Staphylococcus aureus strains, 1997–1999. European Journal of Clinical Microbiology and Infectious Diseases 26, 729–733. [DOI] [PubMed] [Google Scholar]
- 26.Shet A et al. (2009) Colonization and subsequent skin and soft tissue infection due to methicillin-resistant Staphylococcus aureus in a cohort of otherwise healthy adults infected with HIV type 1. Journal of Infectious Diseases 200, 88–93. [DOI] [PubMed] [Google Scholar]
- 27.Drapeau CM et al. (2007) Role of previous hospitalization in clinically-significant MRSA infection among HIV-infected inpatients: results of a case-control study. BMC Infectious Diseases 7, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villacian J et al. (2004) Prevalence of and risk factors for nasal colonization with Staphylococcus aureus among human immunodeficiency virus-positive outpatients in Singapore. Infection Control and Hospital Epidemiology 25, 438–440. [DOI] [PubMed] [Google Scholar]
- 29.Kyaw WM et al. (2012) Prevalence of and risk factors for MRSA colonization in HIV-positive outpatients in Singapore. AIDS Research and Therapy 9, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shadyab A and Crum-Cianflone N (2012) Methicillin-resistant Staphylococcus aureus (MRSA) infections among HIV-infected persons in the era of highly active antiretroviral therapy: a review of the literature. HIV Medicine 13, 319–332. [DOI] [PubMed] [Google Scholar]
- 31.Ferreira DdC et al. (2014) Methicillin-resistant Staphylococcus aureus in HIV patients: risk factors associated with colonization and/or infection and methods for characterization of isolates – a systematic review. Clinics 69, 770–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgeworth JD et al. (2007) An outbreak in an intensive care unit of a strain of methicillin-resistant Staphylococcus aureus sequence type 239 associated with an increased rate of vascular access device-related bacteremia. Clinical Infectious Diseases 44, 493–501. [DOI] [PubMed] [Google Scholar]
- 33.Wang S-H et al. (2012) Methicillin-resistant Staphylococcus aureus sequence type 239-III, Ohio, USA, 2007–2009. Emerging Infectious Diseases 18, 1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grundmeier M et al. (2010) Staphylococcal strains vary greatly in their ability to induce an inflammatory response in endothelial cells. Journal of Infectious Diseases 201, 871–880. [DOI] [PubMed] [Google Scholar]
- 35.Hidron AI et al. (2010) Methicillin-resistant Staphylococcus aureus in HIV-infected patients. Infection and Drug Resistance 3, 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popovich KJ et al. (2014) Anatomic sites of colonization with community-associated methicillin-resistant Staphylococcus aureus. Infection Control and Hospital Epidemiology 35, 1192–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]