Abstract
Primary cilia are specialized cell-surface organelles that mediate sensory perception and, in contrast to motile cilia and flagella, are thought to lack motility function. Here, we show that primary cilia in human and mouse pancreatic islets exhibit movement that is required for glucose-dependent insulin secretion. Islet primary cilia contain motor proteins conserved from those found in classic motile cilia, and their three-dimensional motion is dynein-driven and dependent on adenosine 5′-triphosphate and glucose metabolism. Inhibition of cilia motion blocks beta cell calcium influx and insulin secretion. Human beta cells have enriched ciliary gene expression, and motile cilia genes are altered in type 2 diabetes. Our findings redefine primary cilia as dynamic structures having both sensory and motile function and establish that pancreatic islet cilia movement plays a regulatory role in insulin secretion.
Motile primary cilia in pancreatic islets control insulin secretion through glucose-controlled movement.
INTRODUCTION
Cilia are slender, hair-like projections that mediate cellular interactions with the surrounding environment. There are two main types of vertebrate cilia: primary cilia that are solitary, present on most cells of the body, and specialize in sensory and signaling function (1) and motile cilia that are typically present in large numbers per cell in select tissues and beat in coordinated waves (2). Fundamentally, these two cilia types are distinguished by their capacity for movement. Classic motile cilia have a “9 + 2” axonemal microtubule structure, composed of a ring of nine outer microtubule doublets and two central microtubule singlets, with interconnecting motor protein complexes such as dynein arms and radial spokes (2, 3). In contrast, conventional primary cilia have a “9 + 0” configuration of microtubules (4, 5) and are considered immotile due to the lack of central microtubules and motor accessories.
There are exceptions to this dichotomous cilia classification, most notably, the embryonic nodal cilia, which are “9 + 0” primary cilia that have both motility and signaling capacity (6–8). In other tissues, primary cilia can bend passively under external forces such as fluid flow (9), where cilia mechanosensing regulates cellular homeostasis via ciliary and cytoplasmic [Ca2+]i (10–12). Primary cilia can also exhibit spontaneous fluctuations from internal actin-myosin forces generated at the cell cortex (13). However, to date, active dynein-driven motility in primary cilia has not been reported. Recent three-dimensional (3D) ultrastructural studies have shown that primary cilia architecture can differ extensively from the classic “9 + 0” configuration, owing to microtubule shifts along the length of the cilium that result in outer microtubules rotating into the center of the axoneme (14–16). These findings suggest that primary cilia microtubules are dynamic and that the structural distinction between primary and motile cilia may not be absolute.
To examine the possibility that primary cilia may have motility and the functional significance of their movement, we used multimodal imaging to study cilia structure and behavior in human and mouse pancreatic islets. Islet cells express primary cilia that regulate glucose-stimulated hormone secretion via G protein–coupled receptor (GPCR) and other specialized signaling pathways (17–20). Defects in ciliary function manifest in metabolic disorders such as obesity and type 2 diabetes in human ciliopathy syndromes (21–23). We show here that, contrary to assumption, islet primary cilia are motile by using the same dynein machinery as classic motile cilia, and that beta cell cilia motility is required for glucose-stimulated calcium influx and insulin release. Our studies identify cilia motility as a core beta cell function that, when impaired, leads to reduced ability by the beta cell to respond to glucose and to secrete insulin. These findings raise the possibility that cilia dysmotility may be a pathway to islet dysfunction and offers a new therapeutic target for human metabolic diseases.
RESULTS
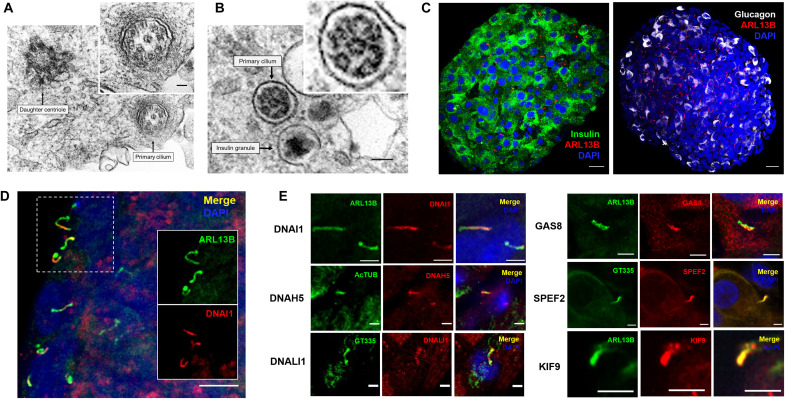
We examined the structure and distribution of primary cilia in human donor islets using complementary imaging modalities (Fig. 1). Ultrastructural examination by transmission electron microscopy (TEM) of solitary beta cell cilia from two individual human donors showed characteristics consistent with primary cilia but deviated from the expected 9 + 0 microtubule arrangement (Fig. 1, A and B). Instead, we detected eight outer microtubule doublets with a central microtubule doublet or singlet. Electron densities decorating the microtubules resemble inner and outer dynein arms and radial spokes but were not preserved clearly for definitive labeling. These cilia cross sections are internal, approximately 0.3 μm in diameter, similar to an insulin granule (Fig. 1B), and can be found juxtaposed to a daughter centriole several micrometers away (Fig. 1A).
Fig. 1. Human islets contain motile primary cilia.
(A) Transmission electron microscopy (TEM) of a beta cell primary cilium and daughter centriole from a healthy 40-year-old female human donor. Inset: cilium cross section showing outer and inner microtubules decorated with unidentified electron densities. Inset scale bar, 50 nm. (B) TEM of a beta cell cilium from a healthy 55-year-old male human donor, showing similar outer/inner microtubule arrangements. Scale bar, 100 nm. (C) Primary cilia distribution on beta and alpha cells in human islets. Insulin (green, left), glucagon (gray, right), cilia (red), and DAPI (blue). Scale bar, 20 μm. (D) Low-magnification view of a human islet showing dynein (DNAI1) expression in the primary cilia of multiple cells. Scale bar, 5 μm. (E) High-magnification views of primary cilia on human islet cells showing endogenous expression of motile proteins, including dynein arm components (DNAI1, DNAH5, and DNALI1), nexin regulatory protein (GAS8), and central pair-associated proteins (SPEF2 and KIF9). Reference cilia markers are shown in green, including ARL13B (ADP ribosylation factor-like GTPase), AcTUB (acetylated alpha tubulin), and GT335 (polyglutamylated tubulin). Nuclei are labeled with DAPI in blue. Scale bars, 2 μm.
Consistent with electron microscopy findings, we detected abundant primary cilia in both beta and alpha cells by immunofluorescence in healthy human donor islets (Fig. 1C). Targeted immunostaining showed strong endogenous expression and ciliary localization of key motile cilia proteins (Fig. 1, D and E). These include dynein arm components dynein axonemal intermediate chain DNAI1, light intermediate chain DNALI1, and heavy chain DNAH5 (24–26), which are distributed continuously but non-uniformly along the length of the cilium; growth arrest–specific protein GAS8 that forms the nexin-dynein regulatory complex (N-DRC) (27, 28) and is detected in the axonemal lumen; and the spermatogenesis and flagellar assembly protein SPEF2 (29, 30) and kinesin family member KIF9 (31, 32), which are known to associate with the central pair and were found in the axoneme and base. The differential enrichment of these proteins in different ciliary compartments likely reflects their unique roles in motile cilia and flagellar assembly and function.
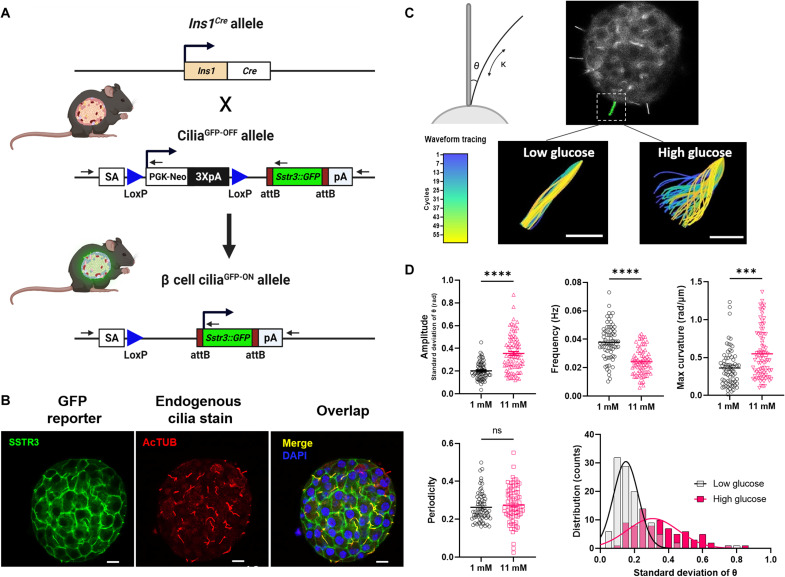
Next, we tested whether primary cilia exhibit motility behavior in live islets. Cilia movement was observed by video microscopy in intact human islets labeled with green cilia cADDis (cAMP difference detector in situ) or SiR (silicon rhodamine)-tubulin, but imaging was suboptimal due to high background fluorescence and lack of cell specificity by the biosensors (movies S1). To specifically study beta cell cilia dynamics, we generated a beta cell–targeted fluorescent cilia reporter mouse by crossing the Ins1-Cre and somatostatin receptor 3 (SSTR3)–green fluorescent protein (GFP) lines (33, 34) (Fig. 2A). The Ins1-Cre strain was chosen for its efficient and selective beta cell recombination without leaky expression in the central nervous system (33). Homozygous beta cell cilia GFP mice are fertile and produce normal size healthy litters, expressing constitutive green fluorescence in the primary cilia of beta cells. Fluorescence is robust and withstands prolonged live imaging and fixation (Fig. 2B). Live imaging of Ins1-Cre cilia reporter islets revealed spontaneous movement of beta cell cilia throughout the islet. Cilia at low glucose displayed slow and variable oscillations, with wave periods on the order of 20 to 30 s (movies S2). This contrasts with the fast and regular beating of conventional motile cilia with periods in the 0.1- to 0.3-s range (35–37). In addition to planar beating, beta cell cilia displayed a lasso-like 3D rotation, whose trajectory was less amenable to tracing as it moved in and out of imaging focus and required a z-stack to capture. We prioritized preserving temporal resolution of the videos and thus focused on the more clear planar view that captured cilia in profile. Raising ambient glucose from 1 to 11 mM stimulated cilia movement by nearly a twofold increase in amplitude as measured by SD of angle of deflection (θ), representing the transverse variance from the cilium base to a reference central axis (Fig. 2C, movie S2, quantitation Fig. 2D). Movement is most dynamic in the distal tip of the beta cell cilium, a region that is continually remodeled (38) and represents a specialized compartment enriched in GPCRs and other signaling molecules (20, 39, 40). High glucose induced greater axonemal bend and larger tip displacement, up to 10 μm, which exceeds that expected from thermal bending due to Brownian motion or actin-mediated internal forces (13, 41). Cilia beating frequency was greater in low glucose conditions as the cilia traversed shorter distances. All cilia waveform parameters, including curvature, frequency, periodicity, and amplitude, displayed increased distribution range under high glucose conditions, reflecting heterogeneity in beta cell responses. These observations suggest that active cilia motility may have evolved to aid dynamic interactions between the cell and its environment and possibly to augment the detection of nutrient cues.
Fig. 2. Beta cell cilia exhibit glucose-dependent motility.
(A) Beta cell cilia reporter mice were generated by crossing INS1-Cre with SSTR3-GFPOFF strains. (B) SSTR3-GFP protein is enriched in beta cell cilia with detectable expression also in the plasma membrane. Colocalization of SSTR3 fluorescence reporter (green) with antibody-stained acetylated alpha tubulin (cilia marker, red) is seen as yellow in the merged image. Non–beta cell cilia are red only. Nuclei are labeled in DAPI (blue). Scale bar, 10 μm. (C) Beta cell cilia exhibit glucose-dependent motility. Schematic shows measurement of theta (θ) as the angle of deflection and kappa (κ) as the curvature of the cilia axoneme. Video microscopy captures primary cilia movement on murine beta cells in response to glucose (movie S2), with representative traces shown in colored panels corresponding to the temporal heatmap where the starting frame of the ciliary wave is depicted in blue and the end in yellow. Scale bar, 5 μm. (D) Cilia waveform analysis showing increased ciliary curvature, reduced frequency, and increased amplitude at high glucose (movie S2); n = 80 per condition. ***P < 0.001 and ****P < 0.0001. Histogram shows cilia amplitude distribution at 1 mM low glucose (black) versus 11 mM high glucose (magenta).
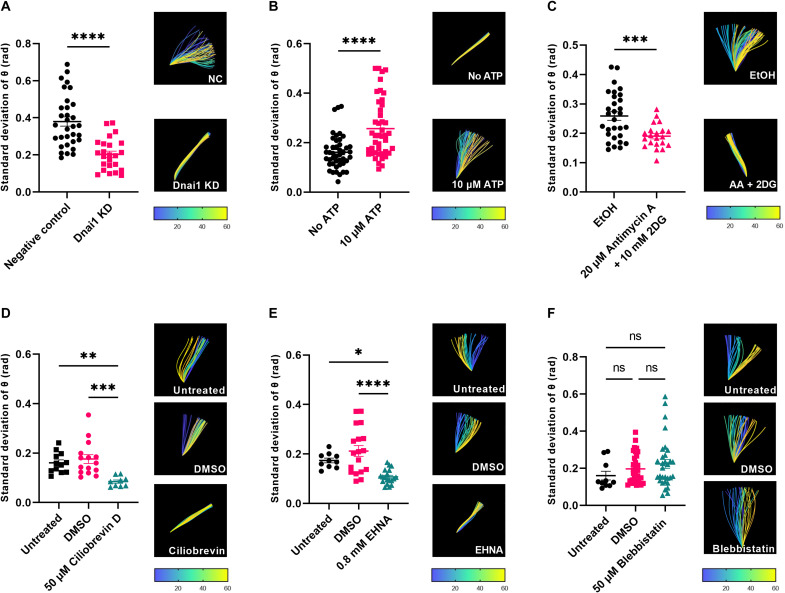
To test the role of specific motor proteins in beta cell cilia motility, we used targeted genetic deletion of axonemal dynein Dnai1 in cilia reporter mouse islets. Dynein knockdown resulted in abrogated cilia motion, suggesting that motility is dynein dependent (Fig. 3A and movie S3). To test energy requirements for ciliary motion, islets were cultured with no glucose stimulation with or without 10 μM supplemental adenosine 5′-triphosphate (ATP), a concentration established to induce ciliary beating of motile cilia (42, 43). Exogenous ATP induced a near 60% increase in mean cilia amplitude, demonstrating a stimulatory effect (Fig. 3B and movie S4). Conversely, ATP depletion using antimycin A and 2-deoxy-d-glucose profoundly inhibited cilia movement (Fig. 3C and movie S5). These results suggest that beta cell cilia movement depends on a microtubule dynein–driven mechanism and requires ATP as energy source. Dynein knockdown studies were corroborated by pharmacologic inhibitors including ciliobrevin D, a small-molecule inhibitor of dynein adenosine triphosphatase (44, 45), at 50 μM, effectively inhibiting cilia movement after 1 hour of treatment (Fig. 3D and movie S6), and erytho-9-(2-hydroxynonyl) adenine (EHNA) (46) at 0.8 mM (Fig. 3E and movie S7). In contrast, inhibition of the actin-associated motor myosin II by blebbistatin did not obviously impair cilia movement (Fig. 3F and movie S8).
Fig. 3. Beta cell cilia motility requires dynein, ATP, and glucose.
(A) Cilia movement in mouse beta cells is inhibited by targeted dynein knockdown, as shown in mean waveform amplitude (SD of cilium tangent angle, θ, rad). Representative wave traces are shown for each condition; temporal heatmap denoting the starting and ending frames of ciliary wave traces. (B) ATP supplementation without glucose promotes cilia movement. (C) ATP depletion using antimycin A and 2-deoxy-d-glucose (2DG) inhibits cilia movement. (D) Dynein chemical inhibition by ciliobrevin D blocks cilia movement. DMSO-treated islet cilia exhibited unperturbed motility; untreated islet cilia are shown as an additional control. (E) EHNA inhibits cilia movement compared to DMSO-treated and untreated cilia. (F) The actin-myosin inhibitor blebbistatin does not significantly affect ciliary beat amplitude. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; n = 10 to 50 traces per condition, pooled from three or more independent experiments. Corresponding movies S3 to S8.
Building on these findings, we next tested the physiologic relevance of dynein-mediated cilia motility in islet function. Dynein inhibition was achieved using both molecular and pharmacologic approaches as for cilia waveform studies. As calcium is a crucial messenger in beta cells underlying glucose-stimulated insulin secretion (GSIS), we tested the effect of axonemal dynein knockdown on islet intracellular calcium dynamics. Glucose normally induces a large rise in cytosolic [Ca2+]i in both mouse and human islet beta cells, measurable by fluorescent calcium sensors such as Calbryte 590 AM. Lentiviral short hairpin–mediated (shRNA) knockdown of DNAI1 in human islets led to a flattening of Ca2+ flux during the glucose response (Fig. 4A). The delay in Ca2+ flux corresponded to a nearly 50% decrease in 1-hour insulin secretion, as measured by static GSIS assay (Fig. 4B). Mouse islets exhibited a similar reaction to dynein inhibition, where both targeted mouse Dnai1 knockdown and chemical inhibition by ciliobrevin D reduced both the time onset and amplitude of the calcium influx (fig. S1 and movies S9 and S10). Treatment of human islets with ciliobrevin D reduced insulin secretion by more than 50% in multiple healthy donors (fig. S2), with an average fold induction of 19.2 in dimethyl sulfoxide (DMSO) versus 9.3 in ciliobrevin D. Human islets exhibited a range of insulin secretion in both baseline GSIS and response to ciliobrevin D, which was expected given donor heterogeneity. Ciliobrevin treatment had minimal cytotoxicity on intact islets when limited to short-term exposures, and its inhibitory effect on insulin secretion was partly reversed by the potassium channel blocker tolbutamide (fig. S3), suggesting that ciliobrevin-mediated disruption of stimulus-secretion coupling is upstream of membrane depolarization. Together, these results demonstrate that cilia motility is conserved between mouse and human islets and that insulin secretory function is at least in part dependent on cilia motility.
Fig. 4. Cilia motility is required for glucose-stimulated insulin secretion.
(A) Dual-color live imaging showing GFP-labeled transduced beta cells in whole islets labeled with Calbryte dye. Still images show control versus dynein shRNA islets reporting calcium activity after exposure to 11 mM glucose. Three representative regions of interest (ROIs) per islet are shown. Calbryte, red; cilia, green. Scale bar, 20 μm. Calcium response to glucose is delayed and dampened by ciliobrevin D, as shown by fluorescence intensity tracings and areas under the curve (AUCs). Traces are composite of three ROIs and representative of islets examined in three independent experiments. (B) Dynein knockdown in human islets results in reduced DNAI1 gene expression and diminished glucose-stimulated insulin secretion (GSIS); data are plotted from two independent knockdown experiments. Statistical significance was analyzed by Student’s t test and two-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (C) Gene set enrichment analysis (GSEA) of a published human RNA-seq study (47) showing differential expression of motile cilia genes in beta cells versus acinar cells. Red-blue color bar represents the gene order in beta cells (red, left) to acinar cells (blue, right). Each black line above the color bar represents a high-confidence ciliary gene in the reference gene set CiliaCarta (48). More black lines are enriched toward the left, showing greater frequency of ciliary gene detection in beta cells. (D) Increased expression of select motile cilia genes in human T2D beta cells. Z-transformed normalized average expression of ciliary genes in alpha and beta cells from healthy controls and patients with T2D. Source data from a published single-cell RNA-seq study (49).
Last, to explore the physiologic role of motile primary cilia in human islets and potential link to metabolic disease, we examined ciliary gene expression in islet transcriptome studies from both healthy donors and individuals with type 2 diabetes. Gene set enrichment analysis (GSEA) of a previously published RNA sequencing (RNA-seq) study (47) using fluorescence-activated cell sorting–sorted human pancreatic cells from 28 healthy donors across the age span (1 to 66 years) showed enrichment of cilia genes in islet beta cells compared to acinar cells. The reference gene set used was CiliaCarta, a validated compendium of ciliary genes (48) [Fig. 4C, normalized enrichment score (NES) 2.59]. We then performed gene expression analysis on another published single-cell RNA-seq dataset from human pancreatic islets obtained from either healthy individuals or those with type 2 diabetes (49); reference gene list was manually curated from literature review including genes with known ciliary function such as dynein and other core components of motile cilia. When comparing average expression in alpha and beta cells separated by donor disease status, we observed that a number cilia motility genes are more highly expressed in beta cells than alpha cells and higher in diabetic donors than healthy controls, including axonemal dynein and central pair assembly factors DNAH5, DNAAF2, SPAG16, and others (Fig. 4D).
DISCUSSION
The primary cilium is often called the cellular antenna for its prominent role in sensory perception and signaling. Here, we show that the antenna is capable of active movement and that this facilitates islet beta cell nutrient sensing and hormone secretion. Motility of beta cell primary cilia is stimulated by glucose, the main metabolic signal for pancreatic islets. We find that islet primary cilia contain axonemal dynein motors and associated regulatory protein complexes including N-DRC, radial spoke proteins, and central microtubule-associated proteins. Cilia waveform studies confirm that axonemal movement is driven by dynein via ATP hydrolysis, the classic force-generating system that confers cilia and flagellar motility. Our ex vivo imaging show that beta cell cilia can be quite dynamic in their movement with tip displacements up to 10 micrometers or the entire length of a cilium. These movement patterns are exaggerated on the islet periphery in isolated islets, an ex vivo observation that should be validated in more physiological settings, e.g., pancreatic slices or whole pancreas by intravital imaging. As dictated by islet cytoarchitecture, the surface of the islet may have cilia projecting outward into the exocrine-endocrine interface, while most of the islet cilia may be directed inward to the canalicular space between adjacent cells (39, 50, 51), an important region where glucose is extracted from the interstitial fluid between arterial-venous blood flow and where paracrine hormones, nutrient cues, and signaling molecules are present in high concentrations. In the relatively enclosed islet environment where cells themselves are nonmotile, having motile cilia would increase the range of signal detection and cell-cell communication, which may contribute to a more effective whole-islet response to extracellular cues. Consistent with this idea, the mode of primary cilia motility appears to be an irregular, exploratory pattern, a combination of the planar beating exhibited by conventional motile cilia and rotational movement of nodal cilia, such that it allows each beta cell to sample its surroundings for sensory stimuli. Similar nonperiodic, 3D, wave-like motion provides exploratory function in other biological contexts, for example, the hunting behavior of the unicellular predator Lacrymaria olor (52). Of note, in addition to ciliary movement as purposeful sensory surveillance, our data are equally compatible with increased motility as a consequence of increased rate of axonemal transport, especially given the apparent lack of directionality to the movement of these cilia. This possibility may be tested by restricting cilia movement, but not axonemal transport, e.g., by embedding islets in agarose before live imaging. Clarifying between these mechanisms and characterizing the complete 3D waveform (53) in beta cell primary cilia will be a priority of future research.
Ours is the first demonstration of dynein-controlled motility and the presence of motor accessory components in primary cilia. Primary cilia have been found to move in the context of mechanosensing and actin forces (11, 13, 54), while our discovery of motile primary cilia containing dynein challenges the conventional dichotomy of primary (immotile) versus motile cilia. Our findings support a more permissive view where primary cilia not only might be considered as sensory dominant but also have motile function. Correspondingly, motile cilia have been shown as not only motile dominant but also capable of sensory and signaling function (55–57). There has been mounting evidence of primary cilia structural deviations from the classic “9 + 0” paradigm, where microtubule arrangements can evolve from “9 + 0” to “8 + 1” to “7 + 2,” depending on location along the axoneme and is likely cell type dependent (14, 15, 58). Our current study reveals “8 + 1” microtubule patterns in human beta cell primary cilia that may also contain dynein arms and radial spokes, although the immunostaining data could benefit from additional validation using RNA interference (RNAi) knockdown of endogenous motile components. These structures had not been highlighted in prior studies likely because their detection is critically dependent on fixation and staining protocol (58, 59) and can be missed because of sampling. While orthogonally positioned centriole pairs are frequently found in our TEM islet studies, cilia cross sections were rare observations whose detection depends on sample preservation, meticulous searching, and luck. With the advance of focused ion beam scanning electron microscopy (FIB-SEM) and tomography techniques, we expect to see more whole-cilia structures that demonstrate the dynamic 3D organization of microtubules and distribution of motor complexes from base to tip (15, 16).
On the basis of the cross-species conservation of cilia motility and the strong link between cilia movement and insulin secretion in both human and mouse islets, we conclude that cilia motility is an essential aspect of beta cell function. This notion is consistent with the observation that cilia motility genes are enriched in human beta cells, and their expression was altered in type 2 diabetes, a disorder often characterized by insulin hypersecretion. The demonstration of motility gene enrichment in beta cells across the human age span strongly suggests a role of islet cilia motility on human health, although we present only limited bioinformatics analyses here and would suggest a more comprehensive future study of cilia gene expression in human islets to compare disease-correlated changes across multiple T2D datasets. In terms of expression and control of motile primary cilia in islets, we speculate that there exist multiple levels of heterogeneity: different microtubule arrangements and, thus, mechanical stability and robustness of motion among different cilia, mosaicism with motile and nonmotile primary cilia on different cells within a tissue, or inducible switch between nonmotile and motile cilia depending on environmental cues and transcriptional state of the cell. In all of these scenarios, having motile cilia likely augments intercellular communication as neighboring primary cilia can physically interact (60), which may serve to transduce signals between cells, and future studies might delineate a functional role of cilia motility in mediating paracrine or juxtacrine signaling. Whether motility in beta cell primary cilia induces mechanotransduction and calcium signaling through transient receptor potential (TRPV) and polycystin channels (61) is another open question. Altered expression in motile cilia genes in human T2D suggests a potential connection between cilia motility and islet function that may provide a new therapeutic target in diabetes.
MATERIALS AND METHODS
Generation of beta cell cilia GFP mice
INS1-Cre mice from the Jackson Laboratory (JAX #026801) were crossed with SSTR3 GFP mice (34) provided by the Yoder laboratory at UAB. Mice at weaning were genotyped by a commercial vendor (Transnetyx) and fed a standard rodent diet (PicoLab Mouse Diet S053, 13.2% calories from fat). Both male and female mice were used for experiments between 2 and 6 months of age. Animals were maintained in accordance with Institutional Animal Care and Use Committee regulations at the Washington University School of Medicine, approval #20190188.
Human islets
Intact islets from cadaveric nondiabetic human donors were obtained from the NIDDK Integrated Islet Distribution Program (IIDP) or purchased commercially from Prodo Laboratories Inc. Upon arrival, islets were visually inspected, washed twice in islet media [RPMI-1640, 10% FBS, 11 mM glucose, 1% (v/v) penicillin/streptomycin, and 20 mM Hepes], and cultured overnight before experiments. Islets from both male and female donors were used. Human donor islet use adhered to institutional review board (IRB) guidelines. All references to “low” and “high” glucose in insulin secretion and imaging experiments correspond to 1 and 11 mM glucose, respectively.
Electron microscopy
Human islet cilia were examined using electron microscopy. Intact isolated islets were received from IIDP, recovered overnight in islet culture media, fixed in Karnovsky’s solution (3% glutaraldehyde, 1% paraformaldehyde), then secondarily fixed in osmium tetroxide, dehydrated in alcohol, embedded in resin, and polymerized at 90°C for 48 hours. Ultrathin sections of 90-nm thickness were cut, stained with uranyl acetate, and imaged with a JEOL 1200 EX. Different human samples showed batch-dependent variability in staining quality and contrast.
Live cilia motility imaging
Islets were isolated from beta cell cilia GFP mice using an established protocol (62) and recovered for 24 hours in islet media at 37°C and 5% CO2. On imaging day, islets were treated for 1 hour with inhibitors and vehicle control, including ciliobrevin D (50 μM in DMSO; Sigma-Aldrich, 250401), EHNA (0.8 mM in DMSO, Cayman Chemical 13352), ATP (10 μM in dH2O; Sigma-Aldrich, 660-30), antimycin A (20 μM in EtOH; Sigma-Aldrich, A8674), 2-deoxy-d-glucose [10 mM in phosphate-buffered saline (PBS); Sigma-Aldrich, D8375], and blebbistatin (50 μM in DMSO, Sigma 203389). Inhibitor treatments were performed at 1 mM glucose except antimycin A and 2-deoxy-d-glucose studies, which had no glucose supplementation. Treatment conditions were well tolerated by cells as determined by viability assays (Live/Dead viability/cytotoxicity kit, Invitrogen L3224). For human islet cilia live imaging, SiR-Tubulin (Cytoskeleton CY-SC002) and green cilia-targeted cADDis sensors (Montana Molecular D0201G) were used to label cilia in intact human islets. After treatment or labeling, islets were washed twice with KRBH and transferred to 35-mm glass-bottom imaging dishes. Live cilia motility images were recorded using an inverted Zeiss LSM880 fluorescence microscope using a 63× oil immersion objective. Each experiment was independently performed by two to four laboratory members to ensure reproducible findings. Imaging parameters varied among experiments and researchers but generally included 60 to 120 cycles, 7 to 12 slices (for z-stack images only), 1 to 2 Airy units (pinhole size), 512 × 512 pixel size, 16-bit depth, 0.26 to 0.44 μm/pixel, and up to 660 s duration.
Cilia waveform analysis
Cilium traces were obtained using a custom program written in MATLAB (The MathWorks Inc., Natick, MA). Using the first image of a time series, the user manually traces the cilium to obtain length, base coordinates, and base angle; the program then automatically traces the path of the cilium in successive frames, using the following algorithm. The length of the cilium is divided into n segments. For the first segment, a rectangular array of points spanning the cilium width and the length of one segment is rotated through a range of angles about the cilium base. At each angle, the weighted average of the interpolated pixel intensities at all points in the array is calculated. The next point along the cilium is located using this angle, the base coordinate, and the segment length. To improve the robustness of the angle selection, terms are applied to penalize curvature, translational velocity, rotational velocity, and change in curvature per time step. The process is then repeated along successive points along the cilium. When all n + 1 points along the cilium have been identified, the program proceeds to the next image in the sequence. The program requires that any translation of the base of the cilium be removed beforehand (35).
Quantities of interest including period, periodicity, curvature, and amplitude are calculated by postprocessing the raw angle data obtained above. Oscillation period is estimated from the nonzero time delay that maximizes the autocorrelation of the angle data; this delay is found at all points along the cilium and averaged to obtain a single estimate of the period. The mean of the maximum autocorrelation value at each point is used as a measure of periodicity or how similar the beats are to each other. For each frame, a fourth-order polynomial is fit to the angle data and used to reconstruct angle as a continuous function of the arc length, s. The shape of the cilium, as represented by x and y positions along the cilium axis, is reconstructed by integrating the cosine and sine, respectively, of the angle along the length of the cilium. Curvature is calculated as the derivative of the fitted angle function with respect to arc length. Beating amplitude is quantified by taking the SD of all angle data within the middle 80% of the length of the cilium, excluding the base and the tip. The autotrace code is deposited at Zenodo via https://doi.org/10.5281/zenodo.6687921.
Calcium imaging in intact mouse islets
Intracellular calcium was measured using the cell-permeant dye Calbryte 590 AM (AAT Bioquest, CA). Intact islets were incubated in 4 μM Calbryte 590 AM at 37°C. Mouse and human islets were loaded for 15 and 30 min, respectively. For ciliobrevin and tolbutamide experiments, islets were treated for 1 hour with ciliobrevin D (50 μM, DMSO vehicle control) with or without tolbutamide (100 μM; Sigma-Aldrich T0891, EtOH vehicle control) before Calbryte dye loading. Following dye incubation, islets were washed and allowed to recover in KRBH buffer for 10 min. Islets were then transferred to a four-chamber glass-bottom imaging dish and mounted on a climate-controlled stage with 37°C and 5% CO2. Calcium imaging was performed on a Zeiss LSM880 inverted confocal system using a 63× oil immersion objective. Calbryte 590 AM was excited by a 561-nm laser and detected in the range of 571 to 700 nm. Baseline recordings were performed at 1 mM glucose for 5 min, then glucose was added to a final concentration of 11 mM by manual pipetting, and islets were continuously imaged for a total duration of 12 min. Sixteen-bit 512 × 512 pixel images were acquired every 465 ms, which is the frequency best suited to resolve islet Ca2+ dynamics upon glucose stimulation. For two-channel z-stacked time lapse imaging of cilia overlapped with calcium signals, 12-μm-thick islet regions were imaged in seven consecutive planes along the z axis with a total scan time of 3.25 s per cycle. A total of 120 cycles of z-stack time lapse were recorded, and glucose was added on cycle 30 (140 s), while islets were continuously imaged for a total duration of 564 s. Calcium traces were extracted using Calbryte fluorescence integrated intensity (mean gray value) from ROIs in ImageJ software (FIJI) (63). Traces were selected from cells that exhibited minimal motion artifacts and were averaged across biological replicates. Calcium kinetics was time-corrected in Excel and then plotted in Prism GraphPad software (San Diego, CA).
Immunohistochemistry
Isolated islets were washed with PBS, fixed with 4% paraformaldehyde (PFA) for 30 min, and permeabilized with 0.3% Triton X-100 in PBS (PBST) for 30 min at room temperature. After incubation with blocking buffer PBS with 10% normal goat serum for 1 hour at room temperature, islets were incubated for 48 hours at 4°C with primary antibodies diluted in PBST. The next day, islets were washed, incubated overnight at 4°C with secondary antibodies diluted in PBS, and washed again with PBS. 4′,6-Diamidino-2-phenylindole (DAPI) provided nuclear counterstain. Islets were mounted on glass slides with ProLong Gold Antifade Mountant (Thermo Fisher Scientific, P36930) before imaging.
Primary antibodies used for cilia immunostaining include the following: ARL13B (Proteintech, 17711-1-AP; NeuroMab, 75-287), AcTUB (Proteintech, 66200-1-Ig; Sigma-Aldrich, T7451), GFP (Thermo Fisher Scientific; Invitrogen, #A-11122), polyglutamylated tubulin (GT335; AdipoGen, AG-20B-0020), DNAI1 (NeuroMab, 75-372), DNAH5 (Atlas Antibodies, HPA037469), DNALI1 (Atlas Antibodies, HPA028305), GAS8 (Atlas Antibodies, HPA041311), SPEF2 (Atlas Antibodies, HPA040343), and KIF9 (Atlas Antibodies, HPA022033). Secondary antibodies include Alexa Fluor goat anti-mouse and goat anti-rabbit (Invitrogen).
Images were acquired on a Zeiss LSM 880 microscopy with AiryScan detector (Zeiss, Oberkochen, Germany). Plan-Apochromat 63×/1.40 numerical aperture (NA) Oil DIC M27 oil immersion objective was used for all experiments. Images were acquired using an optical magnification of at least 1.8×. XY pixel size was optimized per Zeiss Zen software (11-nm pixel size). Z pixel dimension was set to 250 nm per Z-step. Laser power and Z-ramp (when necessary) for each channel were set independently to equalize signals at the top and bottom of the imaging stack and to occupy the first ∼1/3 of the detector dynamic range. Scan speed was set to 6, scan averages to 2, gain to 700 to 900, digital offset to 0, and digital gain to 1. Confirmation of AiryScan detector alignment was performed before image acquisition and was rechecked with every new slide. Following acquisition, images were 3D Airyscan processed in Zeiss Zen Black.
Insulin secretion
For DNAI1 knockdown experiments, after successful lentiviral infection and incubation in islet media, 10 size-matched, reaggregated human islets were picked from 384-well microplates and equilibrated in the Krebs-Ringer Bicarbonate Hepes (KRBH) buffer [128 mM NaCl, 4.8 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4 7H2O, 2.5 mM CaCl2, 20 mM Hepes, 5 mM NaHCO3, and 0.1% BSA (pH 7.4)] at 2.8 mM glucose for 1 hour at 37°C. Islets were then transferred to the KRBH buffer with 1 or 11 mM glucose, respectively, for 1 hour at 37°C for insulin secretion. Supernatant and islets were collected and stored at −80°C before quantification using the Lumit Insulin Immunoassay Kit (Promega 2021). Secreted insulin levels were normalized to total insulin content of whole islets after acid ethanol extraction. Fold induction was calculated as the ratio of insulin secretion at 11 mM glucose versus 1 mM glucose.
For ciliobrevin experiments, triplicates of five size-matched human islets per condition were equilibrated in 24-well plates containing 400 μl of KRBH buffer at 1 mM glucose for 1 hour at 37°C. Islets were then transferred to 1 or 11 mM glucose for secretion for 1 hour at 37°C, with or without ciliobrevin D (50 μM, DMSO vehicle control). For rescue experiments, a subset of islets received cotreatment with tolbutamide (100 μM, EtOH vehicle control) during the 1-hour ciliobrevin incubation. Supernatants and islets were collected separately, stored at −80°C until insulin measurements, and performed on Crystal Chem insulin ELISA assay kits according to manufacturer specifications. Secreted insulin was normalized to total protein, which was determined using a Pierce BCA protein assay kit (Thermo Fisher Scientific, 23225) according to the manufacturer’s instructions. Fold induction was calculated as the ratio of insulin secretion at 11 mM glucose versus 1 mM glucose.
RNAi knockdown of dynein in mouse and human islets
Mouse dynein (Dnai1) knockdown was performed using ON-TARGETplus SMART pool small interfering RNA (siRNA). Intact SSTR3-GFP mouse islets were isolated, recovered overnight in islet media, and then partly dissociated using brief treatment (3 to 5 min) of Accutase (Innovative Cell Technologies AT-104). Cells were treated with 1 μM siRNA for 24 hours and then imaged under live-cell conditions. Human DNAI1 knockdown was performed using lentiviral particles generated from Origene (TL313431V) using four unique 29-mer target-specific shRNA and one scramble control (TR30021V), all expressing GFP. After dissociation of human islets using Accutase for 10 to 20 min at 37°C, cells were seeded in microwells or low-cell attachment plates and treated with control and DNAI1 shRNA lentiviral particles at 1 × 106 titer unit/ml for 24 to 48 hours. Cells were further incubated for 96 hours to allow recovery and reaggregation and then used for gene expression and insulin secretion experiments.
Quantitative real-time PCR
After shRNA knockdown, reaggregated human islets were lysed in 350 μl of lysis buffer RA1 (Macherey-Nagel, no. 740961) with β-mercaptoethanol per 100 to 200 islets. RNA was purified with the NucleoSpin RNA Kit (Macherey-Nagel, Germany), and complementary DNA (cDNA) was synthesized using a Thermo Fisher Scientific High-Capacity cDNA reverse transcription kit at 200 ng/20 μl. Quantitative polymerase chain reaction (PCR) was performed in triplicate on a 7900 Step One Plus reverse transcription PCR machine (Applied Biosystems) using 2× PowerSYBR Green PCR Master Mix (Thermo Fisher Scientific, 4367659). Changes in gene expression were quantified using 2−∆∆Ct, and results were normalized to the housekeeping gene GAPDH. The following primer sequences were used: human DNAI1: F: 5′-TCAGCCAAGTCTGGCAAGCACT-3′, R: 5′-GAGTCCAAGACACAATCCTGCC-3′; human GAPDH: F: 5′-GTCTCCTCTGACTTCAACAGCG-3′, R: 5′-ACCACCCTGTTGCTGTAGCCAA-3′.
Bioinformatics
For GSEA, raw fastq files from a published RNA-seq study of isolated pancreatic cells were downloaded from the European Nucleotide Archive (47) (Submission ID SRA167055). Reads were quasi-mapped to the human genome (GRCh38, Ensembl release 106) using Salmon (64). Transcripts were imported and aggregated to the gene level in R using Tximport (65), and differential expression analysis was carried out with Limma-Voom (66). Differential expression testing was performed comparing beta cells to acinar cells, and the t test statistic of that comparison was used to perform GSEA using the FGSEA package (doi: https://doi.org/10.1101/060012). The gene set for comparison was built from the 402 genes in CiliaCarta database (48) with an internal score greater than or equal to 1.
For T2D gene expression analysis, raw read counts from the published single-cell RNA-seq data of human pancreatic islet cells were downloaded from ArrayExpress (Accession: E-MTAB-5061). All data analysis was performed in R using Seurat (version 4.0.5). Cells with at least 1500 expressed genes and at least 15,000 detected unique molecular identifiers were retained for analysis. Counts were log-normalized and scaled, and clustering was performed by UMAP (uniform manifold approximation and projection). Cell clusters were identified by marker gene expression. Genes involved with ciliary function were identified by literature review, and a subset of these was identified with measurable expression and variability across cell types in the dataset. The R code used to generate Fig. 4 (C and D) of the manuscript has been archived at DOI:10.5281/zenodo.6789947 and may also be accessed at https://github.com/mueggeb/Cho-2022-Beta-Cell.
Statistics
Data are presented as means ± SEM. Statistical significance was analyzed by paired Student’s t test (two groups) or ANOVA (more than two groups), *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Sample size and number of replicates for each experiment are indicated in figure legends.
Acknowledgments
We are grateful to the human islet donors and their families. We thank the Yoder laboratory at UAB and the Horani, Brody, Piston, Millman, and Remedi laboratories at Washington University for reagent sharing. We thank X. W. Ng for islet imaging expertise and pilot studies. We thank the Hughes laboratory members and the Washington University cilia research community for data review as well as S. Brody and C. Semenkovich for critical reading of the manuscript. Graphics were created with BioRender.com.
Funding: This work was funded by National Institutes of Health grant DK115795A (J.W.H.), National Institutes of Health grant DK127748 (J.W.H.), U.S. Department of Veterans Affairs Biomedical Laboratory Research and Developmental Service Career Development Award 5IK2BX004909 (B.D.M.), NSF grant CMMI-1633971 (P.V.B.), Doris Duke Charitable Foundation grant DDFRCS (J.W.H.), Diabetes Research Connection (J.W.H.), and NIH grant P30 DK 020579 (Washington University Diabetes Research Center). Human pancreatic islets were provided by the NIDDK-funded Integrated Islet Distribution Program (IIDP) (RRID:SCR_014387) at City of Hope, NIH grant #2UC4DK098085, and the JDRF-funded IIDP Islet Award Initiative. Light microscopy was performed at the Washington University Center for Cellular Imaging (WUCCI), supported by Washington University School of Medicine, The Children’s Discovery Institute of Washington University, St. Louis Children’s Hospital (CDI-CORE-2015-505 and CDI-CORE-2019-813), and the Foundation for Barnes-Jewish Hospital (3770 and 4642). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official view of the U.S. Department of Veterans Affairs or the United States Government.
Author contributions: Conceptualization: J.W.H., J.H.C., and H.F.R. Methodology: J.W.H., J.H.C., Z.A.L., H.F.R., T.U., L.G.W., and P.V.B. Investigation: J.W.H., J.H.C., Z.A.L., L.Z., B.D.M., H.F.R., E.Y.L., T.U., and L.G.W. Visualization: J.W.H., J.H.C., Z.A.L., L.Z., E.Y.L., and B.D.M. Funding acquisition: J.W.H., B.D.M., and P.V.B. Supervision: J.W.H. and P.V.B. Writing—original draft: J.W.H., J.H.C., and Z.A.L. Writing—review and editing: All authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The code used for cilia waveform autotrace is deposited via Zenodo at DOI:10.5281/zenodo.6687921, and the code for RNA-seq analysis is deposited at DOI:10.5281/zenodo.6789947. Transgenic animals will be made available pending scientific review and a completed material transfer agreement. Requests for mice should be submitted to jing.hughes@wustl.edu.
Supplementary Materials
This PDF file includes:
Figs. S1 to S3
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S10
REFERENCES AND NOTES
- 1.Pazour G. J., Witman G. B., The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 15, 105–110 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Satir P., Heuser T., Sale W. S., A structural basis for how motile cilia beat. Bioscience 64, 1073–1083 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gui M., Ma M., Sze-Tu E., Wang X., Koh F., Zhong E. D., Berger B., Davis J. H., Dutcher S. K., Zhang R., Brown A., Structures of radial spokes and associated complexes important for ciliary motility. Nat. Struct. Mol. Biol. 28, 29–37 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto M., Kataoka K., Electron microscopic observation of the primary cilium in the pancreatic islets. Arch. Histol. Jpn. 49, 449–457 (1986). [DOI] [PubMed] [Google Scholar]
- 5.Mitchison H. M., Valente E. M., Motile and non-motile cilia in human pathology: From function to phenotypes. J. Pathol. 241, 294–309 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., Kido M., Hirokawa N., Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Takeda S., Yonekawa Y., Tanaka Y., Okada Y., Nonaka S., Hirokawa N., Left-right asymmetry and kinesin superfamily protein KIF3a: New insights in determination of laterality and mesoderm induction by KIF3A(−/−) mice analysis. J. Cell Biol. 145, 825–836 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corbit K. C., Aanstad P., Singla V., Norman A. R., Stainier D. Y. R., Reiter J. F., Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018–1021 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Praetorius H. A., Spring K. R., Bending the MDCK cell primary cilium increases intracellular calcium. J. Membr. Biol. 184, 71–79 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Pazour G. J., San Agustin J. T., Follit J. A., Rosenbaum J. L., Witman G. B., Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr. Biol. 12, R378–R380 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Delling M., DeCaen P. G., Doerner J. F., Febvay S., Clapham D. E., Primary cilia are specialized calcium signalling organelles. Nature 504, 311–314 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruhlen R., Marberry K., The chondrocyte primary cilium. Osteoarthr. Cartil. 22, 1071–1076 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Battle C., Ott C. M., Burnette D. T., Lippincott-Schwartz J., Schmidt C. F., Intracellular and extracellular forces drive primary cilia movement. Proc. Natl. Acad. Sci. U.S.A. 112, 1410–1415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun S., Fisher R. L., Bowser S. S., Pentecost B. T., Sui H., Three-dimensional architecture of epithelial primary cilia. Proc. Natl. Acad. Sci. U.S.A. 116, 9370–9379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiesel P., Alvarez Viar G., Tsoy N., Maraspini R., Gorilak P., Varga V., Honigmann A., Pigino G., The molecular structure of mammalian primary cilia revealed by cryo-electron tomography. Nat. Struct. Mol. Biol. 27, 1115–1124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu C. S., Pang S., Shtengel G., Müller A., Ritter A. T., Hoffman H. K., Takemura S., Lu Z., Pasolli H. A., Iyer N., Chung J., Bennett D., Weigel A. V., Freeman M., van Engelenburg S. B., Walther T. C., Farese R. V., Lippincott-Schwartz J., Mellman I., Solimena M., Hess H. F., An open-access volume electron microscopy atlas of whole cells and tissues. Nature 599, 147–151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerdes J. M., Christou-Savina S., Xiong Y., Moede T., Moruzzi N., Karlsson-Edlund P., Leibiger B., Leibiger I. B., Östenson C.-G., Beales P. L., Berggren P.-O., Ciliary dysfunction impairs beta-cell insulin secretion and promotes development of type 2 diabetes in rodents. Nat. Commun. 5, 5308 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Volta F., Scerbo M. J., Seelig A., Wagner R., O’Brien N., Gerst F., Fritsche A., Häring H.-U., Zeigerer A., Ullrich S., Gerdes J. M., Glucose homeostasis is regulated by pancreatic β-cell cilia via endosomal EphA-processing. Nat. Commun. 10, 5686 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes J. W., Cho J. H., Conway H. E., DiGruccio M. R., Ng X. W., Roseman H. F., Abreu D., Urano F., Piston D. W., Primary cilia control glucose homeostasis via islet paracrine interactions. Proc. Natl. Acad. Sci. U.S.A. 117, 8912–8923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu C.-T., Hilgendorf K. I., Bevacqua R. J., Hang Y., Demeter J., Kim S. K., Jackson P. K., Discovery of ciliary G protein-coupled receptors regulating pancreatic islet insulin and glucagon secretion. Genes Dev. 35, 1243–1255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hildebrandt F., Benzing T., Katsanis N., Ciliopathies. N. Engl. J. Med. 364, 1533–1543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaghloul N. A., Katsanis N., Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J. Clin. Invest. 119, 428–437 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kluth O., Stadion M., Gottmann P., Aga H., Jähnert M., Scherneck S., Vogel H., Krus U., Seelig A., Ling C., Gerdes J., Schürmann A., Decreased expression of cilia genes in pancreatic islets as a risk factor for type 2 diabetes in mice and humans. Cell Rep. 26, 3027–3036.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Guichard C., Harricane M. C., Lafitte J. J., Godard P., Zaegel M., Tack V., Lalau G., Bouvagnet P., Axonemal dynein intermediate-chain gene (DNAI1) mutations result in situs inversus and primary ciliary dyskinesia (Kartagener syndrome). Am. J. Hum. Genet. 68, 1030–1035 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rashid S., Breckle R., Hupe M., Geisler S., Doerwald N., Neesen J., The murine Dnali1 gene encodes a flagellar protein that interacts with the cytoplasmic dynein heavy chain 1. Mol. Reprod. Dev. 73, 784–794 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Olbrich H., Häffner K., Kispert A., Völkel A., Volz A., Sasmaz G., Reinhardt R., Hennig S., Lehrach H., Konietzko N., Zariwala M., Noone P. G., Knowles M., Mitchison H. M., Meeks M., Chung E. M. K., Hildebrandt F., Sudbrak R., Omran H., Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left–right asymmetry. Nat. Genet. 30, 143–144 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Lewis W. R., Malarkey E. B., Tritschler D., Bower R., Pasek R. C., Porath J. D., Birket S. E., Saunier S., Antignac C., Knowles M. R., Leigh M. W., Zariwala M. A., Challa A. K., Kesterson R. A., Rowe S. M., Drummond I. A., Parant J. M., Hildebrandt F., Porter M. E., Yoder B. K., Berbari N. F., Mutation of growth arrest specific 8 reveals a role in motile cilia function and human disease. PLOS Genet. 12, e1006220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bower R., Tritschler D., Vanderwaal K., Perrone C. A., Mueller J., Fox L., Sale W. S., Porter M. E., The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol. Biol. Cell 24, 1134–1152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sironen A., Kotaja N., Mulhern H., Wyatt T. A., Sisson J. H., Pavlik J. A., Miiluniemi M., Fleming M. D., Lee L., Loss of SPEF2 function in mice results in spermatogenesis defects and primary ciliary dyskinesia. Biol. Reprod. 85, 690–701 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenzie C. W., Lee L., Genetic interaction between central pair apparatus genes CFAP221, CFAP54, and SPEF2 in mouse models of primary ciliary dyskinesia. Sci. Rep. 10, 12337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yokoyama R., O’toole E., Ghosh S., Mitchell D. R., Regulation of flagellar dynein activity by a central pair kinesin. Proc. Natl. Acad. Sci. U.S.A. 101, 17398–17403 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyata H., Shimada K., Morohoshi A., Oura S., Matsumura T., Xu Z., Oyama Y., Ikawa M., Testis-enriched kinesin KIF9 is important for progressive motility in mouse spermatozoa. FASEB J. 34, 5389–5400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorens B., Tarussio D., Maestro M. A., Rovira M., Heikkilä E., Ferrer J., Ins1(Cre) knock-in mice for beta cell-specific gene recombination. Diabetologia 58, 558–565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Connor A. K., Malarkey E. B., Berbari N. F., Croyle M. J., Haycraft C. J., Bell P. D., Hohenstein P., Kesterson R. A., Yoder B. K., An inducible CiliaGFP mouse model for in vivo visualization and analysis of cilia in live tissue. Cilia 2, 8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayly P. V., Lewis B. L., Kemp P. S., Pless R. B., Dutcher S. K., Efficient spatiotemporal analysis of the flagellar waveform of Chlamydomonas reinhardtii. Cytoskeleton (Hoboken). 67, 56–69 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith C. M., Djakow J., Free R. C., Djakow P., Lonnen R., Williams G., Pohunek P., Hirst R. A., Easton A. J., Andrew P. W., O’Callaghan C., ciliaFA: A research tool for automated, high-throughput measurement of ciliary beat frequency using freely available software. Cilia 1, 14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oltean A., Schaffer A. J., Bayly P. V., Brody S. L., Quantifying ciliary dynamics during assembly reveals stepwise waveform maturation in airway cells. Am. J. Respir. Cell Mol. Biol. 59, 511–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chien A., Shih S. M., Bower R., Tritschler D., Porter M. E., Yildiz A., Dynamics of the IFT machinery at the ciliary tip. eLife 6, e28606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Burght S. N., Rademakers S., Johnson J.-L., Li C., Kremers G.-J., Houtsmuller A. B., Leroux M. R., Jansen G., Ciliary tip signaling compartment is formed and maintained by intraflagellar transport. Curr. Biol. 30, 4299–4306.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Pala R., Alomari N., Nauli S. M., Primary cilium-dependent signaling mechanisms. Int. J. Mol. Sci. 18, 2272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battle C., Broedersz C. P., Fakhri N., Geyer V. F., Howard J., Schmidt C. F., MacKintosh F. C., Broken detailed balance at mesoscopic scales in active biological systems. Science 352, 604–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshitsugu M., Rautiainen M., Matsune S., Nuutinen J., Ohyama M., Effect of exogenous ATP on ciliary beat of human ciliated cells studied with differential interference microscope equipped with high speed video. Acta Otolaryngol. 113, 655–659 (1993). [DOI] [PubMed] [Google Scholar]
- 43.Hayashi T., Kawakami M., Sasaki S., Katsumata T., Mori H., Yoshida H., Nakahari T., ATP regulation of ciliary beat frequency in rat tracheal and distal airway epithelium. Exp. Physiol. 90, 535–544 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Firestone A. J., Weinger J. S., Maldonado M., Barlan K., Langston L. D., O’Donnell M., Gelfand V. I., Kapoor T. M., Chen J. K., Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature 484, 125–129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roossien D. H., Miller K. E., Gallo G., Ciliobrevins as tools for studying dynein motor function. Front. Cell. Neurosci. 9, 252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouchard P., Penningroth S. M., Cheung A., Gagnon C., Bardin C. W., Erythro-9-[3-(2-hydroxynonyl)]adenine is an inhibitor of sperm motility that blocks dynein ATPase and protein carboxylmethylase activities. Proc. Natl. Acad. Sci. U.S.A. 78, 1033–1036 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arda H. E., Li L., Tsai J., Torre E. A., Rosli Y., Peiris H., Spitale R. C., Dai C., Gu X., Qu K., Wang P., Wang J., Grompe M., Scharfmann R., Snyder M. S., Bottino R., Powers A. C., Chang H. Y., Kim S. K., Age-dependent pancreatic gene regulation reveals mechanisms governing human β-cell function. Cell Metab. 23, 909–920 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Dam T. J. P., Kennedy J., van der Lee R., de Vrieze E., Wunderlich K. A., Rix S., Dougherty G. W., Lambacher N. J., Li C., Jensen V. L., Leroux M. R., Hjeij R., Horn N., Texier Y., Wissinger Y., van Reeuwijk J., Wheway G., Knapp B., Scheel J. F., Franco B., Mans D. A., van Wijk E., Képès F., Slaats G. G., Toedt G., Kremer H., Omran H., Szymanska K., Koutroumpas K., Ueffing M., Nguyen T.-M. T., Letteboer S. J. F., Oud M. M., van Beersum S. E. C., Schmidts M., Beales P. L., Lu Q., Giles R. H., Szklarczyk R., Russell R. B., Gibson T. J., Johnson C. A., Blacque O. E., Wolfrum U., Boldt K., Roepman R., Hernandez-Hernandez V., Huynen M. A., CiliaCarta: An integrated and validated compendium of ciliary genes. PLOS ONE 14, e0216705 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Segerstolpe Å., Palasantza A., Eliasson P., Andersson E.-M., Andréasson A.-C., Sun X., Picelli S., Sabirsh A., Clausen M., Bjursell M. K., Smith D. M., Kasper M., Ämmälä C., Sandberg R., Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aughsteen A. A., The ultrastructure of primary cilia in the endocrine and excretory duct cells of the pancreas of mice and rats. Eur. J. Morphol. 39, 277–283 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Gan W. J., Zavortink M., Ludick C., Templin R., Webb R., Webb R., Ma W., Poronnik P., Parton R. G., Gaisano H. Y., Shewan A. M., Thorn P., Cell polarity defines three distinct domains in pancreatic β-cells. J. Cell Sci. 130, 143–151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coyle S. M., Flaum E. M., Li H., Krishnamurthy D., Prakash M., Coupled active systems encode an emergent hunting behavior in the unicellular predator Lacrymaria olor. Curr. Biol. 29, 3838–3850.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen J. N., Rassmann S., Stüven B., Jurisch-Yaksi N., Wachten D., CiliaQ: A simple, open-source software for automated quantification of ciliary morphology and fluorescence in 2D, 3D, and 4D images. Eur. Phys. J. E. 44, 18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Narita K., Kawate T., Kakinuma N., Takeda S., Multiple primary cilia modulate the fluid transcytosis in choroid plexus epithelium. Traffic 11, 287–301 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Shah A. S., Ben-Shahar Y., Moninger T. O., Kline J. N., Welsh M. J., Motile cilia of human airway epithelia are chemosensory. Science 325, 1131–1134 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bloodgood R. A., Sensory reception is an attribute of both primary cilia and motile cilia. J. Cell Sci. 123, 505–509 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Mao S., Shah A. S., Moninger T. O., Ostedgaard L. S., Lu L., Tang X. X., Thornell I. M., Reznikov L. R., Ernst S. E., Karp P. H., Tan P., Keshavjee S., Alaiwa M. H. A., Welsh M. J., Motile cilia of human airway epithelia contain hedgehog signaling components that mediate noncanonical hedgehog signaling. Proc. Natl. Acad. Sci. U.S.A. 115, 1370–1375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caspary T., Larkins C. E., Anderson K. V., The graded response to sonic hedgehog depends on cilia architecture. Dev. Cell 12, 767–778 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Behnke O., Forer A., Evidence for four classes of microtubules in individual cells. J. Cell Sci. 2, 169–192 (1967). [DOI] [PubMed] [Google Scholar]
- 60.Ott C., Elia N., Jeong S. Y., Insinna C., Sengupta P., Lippincott-Schwartz J., Primary cilia utilize glycoprotein-dependent adhesion mechanisms to stabilize long-lasting cilia-cilia contacts. Cilia 1, 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nauli S. M., Pala R., Kleene S. J., Calcium channels in primary cilia. Curr. Opin. Nephrol. Hypertens. 25, 452–458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li D.-S., Yuan Y.-H., Tu H.-J., Liang Q.-L., Dai L.-J., A protocol for islet isolation from mouse pancreas. Nat. Protoc. 4, 1649–1652 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soneson C., Love M. I., Robinson M. D., Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., Smyth G. K., limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S3
Movies S1 to S10