Abstract
We present the first structure of the human Kir2.1 channel containing both transmembrane domain (TMD) and cytoplasmic domain (CTD). Kir2.1 channels are strongly inward-rectifying potassium channels that play a key role in maintaining resting membrane potential. Their gating is modulated by phosphatidylinositol 4,5-bisphosphate (PIP2). Genetically inherited defects in Kir2.1 channels are responsible for several rare human diseases, including Andersen’s syndrome. The structural analysis (cryo–electron microscopy), surface plasmon resonance, and electrophysiological experiments revealed a well-connected network of interactions between the PIP2-binding site and the G-loop through residues R312 and H221. In addition, molecular dynamics simulations and normal mode analysis showed the intrinsic tendency of the CTD to tether to the TMD and a movement of the secondary anionic binding site to the membrane even without PIP2. Our results revealed structural features unique to human Kir2.1 and provided insights into the connection between G-loop and gating and the pathological mechanisms associated with this channel.
First human Kir2.1 structure reveals the G-loop connection with gating for the loss of function in Andersen’s syndrome mutant.
INTRODUCTION
Inward-rectifier potassium (Kir) channels are a group of integral membrane proteins that selectively control the permeation of K+ (potassium) ions across cell membranes. They are particular in that the channels conduct K+ ions easier in the inward direction (into the cell) than in the outward direction (out of the cell). The small outward K+ current through Kir channels controls the resting membrane potential and membrane excitability, regulates cardiac and neuronal electrical activities, couples insulin secretion to blood glucose levels, and maintains electrolyte balance (1).
All Kir channels are tetramers of identical or related subunits and share characteristic structural features. They have a canonical pore-forming transmembrane domain (TMD) made of two transmembrane helices (M1 and M2) separated by a K+ ion selectivity filter and a large cytoplasmic domain (CTD) containing both N and C termini. The CTD extends the ion conduction pathway and provides docking sites for regulatory ions, proteins, and ligands (2). The inward-rectification mechanism results from a block on the cytoplasmic side of the channels by endogenous polyamines and Mg2+ that plug the channel pore at depolarized potentials, resulting in decreased outward currents. The blockers are then removed from the pore when the K+ ions flow into the cell at hyperpolarized potentials. This voltage-dependent block results in efficient conduction of current only in the inward direction (3–5). In addition to being inwardly rectifying, Kir channels respond to a variety of intracellular messengers that directly control the channel gating, including phosphoinositides (PIPs), G proteins (Kir3 channels), adenosine 5′-triphosphate (Kir6 channels), and changes in pH (Kir1 channels) (1). The Kir family is encoded by 16 genes (KCNJ1 to KCNJ18) and classified in seven subfamilies (Kir1 to Kir7).
To fulfill crucial biological roles, the gating of all eukaryotic Kir channels is directly controlled by regulatory lipid ligands such as PIPs. Different Kir isoforms display distinct specificities for the activating PIPs (2). Kir2.1 channels are selectively activated by the signaling lipid phosphatidylinositol 4,5-bisphosphate (PIP2), while some members (such as Kir3.1/3.4) are less stringent in their PIP sensitivity, and all PIPs indiscriminately activate others such as Kir6.2. PIP2 binds to a “primary site” (6, 7), and its sensitivity is enhanced by the binding of a bulk of anionic lipids (positive allosteric regulators) to a “secondary site” (8, 9). On the other hand, the bacterial KirBac channels are inhibited by PIPs (6, 10).
Kir2.1 is the protein product of the KCNJ2 gene, which has been shown to play a critical role in excitable cells and developmental processes (11). Kir2.1 is expressed at high levels in the heart, skeletal muscle, and neural tissue. The extent of rectification varies among the different subfamilies, ranging from weak (Kir1) to strong (Kir2). Several crystallographic structures of Kir channels with Mg2+ have been published. One study has reported the crystallographic structure of KirBac3.1 channel with an electron density attributed to spermine. However, KirBac3.1 is missing numerous critical spermine-interacting residues (12). The molecular blocking mechanisms of polyamines and Mg2+ are still under scrutiny. KCNJ2 mutations are associated with four disorders, namely, Andersen’s syndrome (AS), atrial fibrillation (AF), short QT syndrome (SQT), and catecholaminergic polymorphic ventricular tachycardia (CPTV) (11, 13–15). While AF- and SQT-associated mutations lead to a gain of channel function, those associated with AS and CPTV are all loss of function mutations. AS is a rare and complex disorder showing variability in patient diagnosis. The most common phenotypes are periodic paralysis, cardiac arrhythmias, and distinctive physical features with development anomalies (16).
It has been proposed that the G-loop is involved in channel modulation by PIP2 and other ligands (17, 18). The G-loop is a conserved motif extending from G300 to Y315, composed predominantly of residues with short hydrophobic side chains that may constrict the pore diameter in some states of the channel (19). Although the role of the G-loop is yet to be fully understood, several structural studies raise the possibility that this motif undergoes relevant conformational changes for channel gating (17–20). However, it is not known how the G-loop gate integrates into channel gating and how it affects the gating kinetics of Kir channels.
To shed light on the structural mechanism of the human Kir channels and the associated rearrangements underlying PIP2, Mg2+, and polyamine-dependent gating, detailed structural information is crucial. Here, we used cryo–electron microscopy (cryo-EM) combined with image analysis to elucidate the structure of a human Kir channel, Kir2.1, at atomic resolution in the closed state. Our findings reveal unique structural features of the human Kir2.1 channel, among other eukaryotic Kir channels. Furthermore, computational investigations [normal mode (NM) analysis and molecular dynamics (MD) simulation] reveal crucial conformational movements, including compaction of the structure and opening movements at the interface between the TMD and CTD, which could facilitate the binding of PIP2. In addition, we provide biophysical and functional data on the Kir2.1 channel [wild type (WT) and the mutant R312H, a mutant responsible for AS disease (21)], and we propose a putative activation mechanism involving the transmission of the signal to the G-loop upon PIP2 binding.
RESULTS
Protein expression and purification of human Kir2.1 WT and R312H
Until recently, the effect of residue R312 on the binding of PIP2 to Kir2.1 was still controversial. The R312Q mutation has been suggested to weaken the interaction of Kir2.1 with PIP2 (22). However, more recent biochemical binding assays have indicated that this mutation does not significantly affect the PIP2 binding (23). To clarify this point, we investigated the affinity of PIP2 to the WT channel and R312H mutant (naturally occurring AS-causing mutation) and the impact of this mutant on the Kir2.1 function. The human Kir2.1 WT channel and its R312H mutant counterpart were recombinantly produced in and purified from the yeast Pichia pastoris. The size exclusion chromatography elution profile of the Kir2.1 WT (fig. S1) shows a prominent elution peak at 11.80 ml, corresponding to its tetrameric quaternary structure. SDS–polyacrylamide gel electrophoresis and Western blot analyses reveal the monomeric form of Kir2.1 at its expected size (~50 kDa; fig. S1). Similar results were obtained with the Kir2.1 R312H mutant.
Overall cryo-EM structure of the human Kir2.1 channel
The structure of the human Kir2.1 channel was resolved by single-particle cryo-EM analysis (Fig. 1A and fig. S2) to an average resolution of 4.3 Å (fig. S3) for the whole protein. The local resolution map shows that some regions, especially in CTD, can reach 3.7 Å resolution, mainly the G-loop region, the PIP2-binding site, residue R312, and its vicinity (fig. S3B). The final structural model contains all typical structural features of Kir channels (Fig. 1, A and B). In the TMD, the outer (M1) and inner (M2) transmembrane helices are observed, in addition to two short helical elements (the slide and the pore helices) and the selectivity filter that contains the K+ channel signature sequence (T-X-G-Y/F-G). The CTD is rich in β strands and encompasses the G-loop in the apex of the cytoplasmic pore. This is the first time that the entire human Kir2.1 channel has been resolved at high resolution; it is also the first cryo-EM structure of a Kir2 channel. This Kir2.1 structure covers an external flexible N-terminal loop (regions 41 to 54) that interacts with CTD and PIP2, which has only been continuously modeled in Kir2.2 PIP2-bound crystal structures (3SPI, 3SPG, and 5KUM) (7, 24). This segment has been shown to be important in the gating process (25).
Fig. 1. Cryo-EM map and atomic structure of the human Kir2.1 channel.
(A) Sharpened cryo-EM map of human Kir2.1 obtained at 4.3-Å resolution (bottom, side, and top views). (B) Side view of human Kir2.1 atomic structure fitted in the cryo-EM map (white surface). The chains A and C are in cartoon and highlight the typical structural features of Kir channels: Outer helix (M1, yellow), inner helix (M2, cyan), pore helix (dark blue), slide helix (red), selectivity filter (magenta), extracellular region (light blue), G-loop (black), and helix bundle crossing (HBC) region. The Sr2+ (green) and K+ (purple) ions inside the pore channel are represented as spheres. Chains B and D are in gray ribbons for clarity. Gray horizontal bars indicate the plasma membrane. The TMD and CTD are indicated. (C) Top view of the human Kir2.1 atomic structure fitted in the cryo-EM map. The four chains (A to D) are in a blue cartoon. The cysteine residues (C122 and C154) involved in the intrasubunit disulfide bridge are highlighted as red sticks. Protein dimensions are shown.
The resolved Kir2.1 structure exhibits ion-conduction pore constrictions along the helix bundle crossing region and the G-loop (see following sections). It is 121 Å long and 78 Å wide (Fig. 1). This extended conformation is also observed in the apo-Kir2.2 crystal structure (3JYC) (26) and the apo-Kir3.2 cryo-EM structure (6XIS) (27), both of which are in a closed state. Thus, these data suggest that the human Kir2.1 structure was obtained in the nonconductive closed state. A single disulfide bridge between C122 and C154 residues of the same chain is observed in the outermost region of the TMD, close to the extracellular loops (Fig. 1C). Last, it was possible to accurately map the location of most of the AS-causing mutations in the Kir2.1 structure (fig. S4). Moreover, we studied in detail the impact of one of them, R312H, located outside the PIP2-binding site, on the structure and function of the human Kir2.1 channel (see following sections). The accession codes for these structures are Protein Data Bank (PDB) 7ZDZ and EMD 14678.
Ions’ location in the pore channel
No density consistent with K+ ions or any other ions was observed in the selectivity filter. The cryo-EM structure of apo-Kir3.2 (6XIS) is another structure reported with no K+ ions inside the selectivity filter, despite its slightly larger pore diameter than Kir2.1 (see next section). Nevertheless, the presence of three K+ ions is observed when the same channel is in complex with PIP2 (cryo-EM PIP2-Kir3.2 structure, 6XIT) (27). In most crystallographic structures, four K+ ions are present at the selectivity filter; this includes the crystal structures of apo-Kir2.2, PIP2-Kir2.2 (3JYC and 3SPI, respectively), apo-Kir3.2, PIP2-Kir3.2 (3SYO and 3SYA, respectively), and KirBac3.1 (i.e., 3ZRS) (7, 17, 26, 28). Note that for crystallization assays and cryo-EM sample preparation, the concentration of KCl in the medium is approximately the same (150 to 200 mM).
Three ions (two Sr2+ and one K+; Fig. 2 in blue and green, respectively) are present in the pore below the selectivity filter. The two circular densities, located just below the selectivity filter and slightly below the G-loop, were attributed to Sr2+ ions. These cryo-EM densities show a high magnitude (still persistent at 13σ and 15σ map threshold, respectively) and exhibit higher cross-correlation values (cc = 0.82 and 0.76, respectively) when modeled as Sr2+ ions, compared to when modeled as Mg2+ (0.51 and 0.57) or K+ ions (0.41 and 0.49). The divalent cation Sr2+ mimics Mg2+, a biologically important metal ion blocker of eukaryotic Kir channels (29). The first Sr2+ ion is located below the selectivity filter at equidistance from the Oδ2 of the D172 side chains (~7 Å) and the Oγ1 of the T142 side chains (~6 to 7 Å), the innermost residue of the selectivity filter. The second Sr2+ ion is located below the G-loop at ~8 to 11 Å from Oε1/Oε2 of the E299 side chains and ~9 to 10 Å from Oε1/Oε2 of the E224 side chains. It is closer (~6 Å) to the carbonyl backbone of the A306 in the G-loop region. Sr2+ ions at similar positions have also been found in the crystal structure of chicken Kir2.2 (26), where crystals were grown in the presence of 10 mM Sr2. However, no Sr2+ was added in the sample preparation for cryo-EM analysis, but there are traces of Sr2+ ions in the yeast medium.
Fig. 2. Ions’ location along the pore in the human Kir2.1 channel cryo-EM structure.
Cryo-EM densities modeled as Sr2+ (blue mesh at 5σ) and K+ (green mesh at 2.5σ) ions inside the pore of human Kir2.1 cryo-EM atomic structure (blue ribbon). Insets: Detailed views of the regions where the ions are located. The distances between these ions and the negatively charged atoms of the closest residues (yellow sticks) inside the pore channel are indicated.
A third circular cryo-EM density with a persistent magnitude up to a 6σ map threshold is observed in the outermost region of the CTD. This density was interpreted as a K+ ion (cc = 0.84). This K+ ion is dehydrated and directly coordinated to two D255 residues from opposite chains with typical K-O distances of 2.8 Å (Fig. 2). This confirms that the long pore extends quite far from the cytoplasm (20). A K+ ion in a similar position has been found in the crystal structure of the CTD domain of mouse Kir2.1 R218Q/T309K (2GIX) (30). However, in this structure (2GIX), the K+ ion is hydrated and interacts with the four D255 residues through eight water molecules.
Four constriction points (gates) in the Kir2.1 channel
The analysis of the radius between the van der Waals surfaces of the central ion conduction pore using the HOLE software (31) shows four constriction points located at the selectivity filter and residues I176, M180, and A306 (Fig. 3, A and B). The selectivity filter has a very tight structure (pore radius less than 0.2 Å). Two of the other narrow points, I176 and M180, are located in the TMD immediately below the Sr2+ ion present in this region. These constriction points have a minimum pore radius of 0.77 Å (I176) and 1.05 Å (M180) and close the pore leading to the CTD. Similarly, in the apo-Kir2.2 crystal structure (3JYC) (26), I177, equivalent to I176 in Kir2.1, is also described as a constriction point with a similar pore radius (Fig. 3C). M180 is equivalent to residue Y132 of KirBac3.1 located at the helix bundle crossing, a well-described constriction point in the bacterial channel (12, 32). The fourth constriction point is located at A306 at the apex of the G-loop at the pore entrance into the CTD; this feature has also been observed in the mouse CTD Kir2.1 crystal structure (1U4F) (20). This constriction point is unique to Kir2.1 and is not present in Kir2.2 (3JYC) and Kir3.2 (3SYO) structures (Fig. 3C).
Fig. 3. Pore radius analysis of human Kir2.1 channel structure and constriction points.
(A) Plot of the pore radius between van der Waals surfaces. The constriction points (selectivity filter, I176, M180, and A306) and the position of the two Sr2+ ions are highlighted. (B) Three constriction points below the selectivity filter: I176 (yellow sphere), M180 (orange sphere), and A306 (purple sphere). Chains A and C are in light blue and green cartoons, respectively. Chains B and D are in gray ribbons for clarity. Sr2+ (blue) and K+ (red) ions are represented as spheres. The insets show the zoom of the three constriction points, where the spheres represent the van der Waals surfaces. (C) Comparative analysis of pore radius (stippled surface) between van der Waals surfaces among human Kir2.1, chicken Kir2.2, and mouse Kir3.2 channel structures (light gray ribbons). Along the central ion conduction, red indicates insufficient space for a water molecule, green indicates that one or two water molecules could fit, and blue indicates that many water molecules could fit. Constriction points were considered where the red volume was visualized. Horizontal dashes were placed to compare the constriction points in human Kir2.1, apo-Kir2.2, and apo-Kir3.2 pore channels. Molecular graphics images were prepared with VMD. Pore radius was analyzed using the HOLE software.
Structural comparison between Kir2.1 and Kir2.2 structures
The superposition of Cα atoms of TMD and extracellular region (amino acids 72 to 182) of the four chains of human Kir2.1 channel cryo-EM structure resulted in a root mean square deviation (RMSD) of 1.0 to 1.4 Å (table S1A). The region that presents the most significant deviation is the extracellular region (amino acids 112 to 121), including the C-terminal end of the M1 helix and the extracellular loops, indicating high conformational flexibility (fig. S5A). Moreover, Cα superposition between the four chains of Kir2.1 structure and the crystal structures of Kir2.2 in the absence and presence of PIP2 (3SPI and 3JYC, respectively) resulted in an RMSD of 4.6 to 5.0 Å (table S1A). These high RMSD values are mainly due to deviations observed in extracellular and selectivity filter regions (amino acids 141 to 147) (fig. S5B). This Cα superposition reveals that the selectivity filter in human Kir2.1 is closer to the center of the pore channel compared with the Kir2.2 structures (fig. S5B), resulting in a narrower pore (as shown in the previous section). The superposition of Cα TMD and the extracellular region between the Kir2.2 crystal structures reveals lower RMSD values than the comparison between the Kir2.1 chains (0.8 Å and 1.0 to 1.4 Å, respectively; table S1A). This is mainly due to a smaller deviation in the extracellular region (fig. S5, A and B), suggesting that this region of Kir2.1 channels can present higher conformation flexibility than the same region in Kir2.2.
Superposition between Cα atoms of CTD (amino acids 192 to 364) from the four chains of human Kir2.1 channel cryo-EM structure resulted in RMSD values of 0.9 to 1.6 Å (table S1B). When compared with the crystal structure of mouse Kir2.1 CTD (1U4F), Cα CTD superposition resulted in RMSD values of 1.3 to 1.9 Å (table S1B). In both superpositions, the CTD regions that present the greatest deviation are two flexible loops exposed to the solvent (amino acids 281 to 291 and 330 to 340) and the C-terminal end (amino acids 357 to 364). The G-loop presented low Cα RMSD values in both comparisons (0.4 to 0.7 Å between the four human Kir2.1 chains and 0.5 to 0.7 Å between human and mouse CTD) (table S1C). These data indicate that the G-loop displays a stable backbone conformation in the Kir2.1 CTD structures, although previously described as intrinsically flexible (20).
PIP2 binding interaction with Kir2.1 WT and mutant R312H
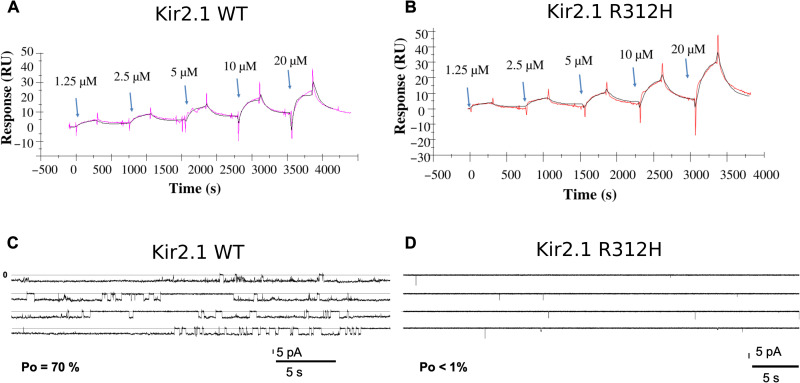
The association and dissociation rate constants, kon and koff, respectively, and the dissociation constant (KD) of the Kir2.1 WT and R312H mutant were determined directly from surface plasmon resonance (SPR) experiments and the fitted curves using CM5 sensor chips. Kir2.1 WT was immobilized at 3900 to 6500 RU (response units) levels on CM5 sensor chips. Sequential injections of PIP2 ranged from 0.15 to 20 μM (Fig. 4A). The KD, derived from koff/kon, was determined to be 3.2 × 10−6 ± 0.7 × 10−6 M. The average kon and koff are 1.8 × 10−3 ± 1.1 × 10−3 Ms−1 and 5.2 × 10−3 ± 3.5 × 10−3 s−1, respectively. Kir2.1 R312H was immobilized to 4000- to 4130-RU level on CM5 sensor chips. Sequential injections of PIP2 ranged from 0.15 to 20 μM. The derived KD was determined to be 3.7 × 10−5 ± 1.2 × 10−5 M with an average kon of 4.5 × 10−2 ± 2.3 × 10−2 ms−1 and koff of 1.4 × 10−2 ± 4.5 × 10−2 s−1 (Fig. 4B). R312H showed a 10-fold difference in affinity compared to WT. However, both have affinities in the micromolar range, which corresponds to generally low binding affinity. This indicates that the R312H mutation slightly modifies the KD but does not impede the binding.
Fig. 4. Biophysical and functional assays on Kir2.1 WT and R312H mutant.
(A and B) Comparative binding kinetics of PIP2 on Kir2.1 WT (A) and R312H mutant (B) immobilized on a CM5 chip. Five sequential injections of PIP2 are shown, 1.25 to 20 μM, at 25 μl/5 min. In color, experimental data; in black, fitted curve. The single-cycle kinetics assays were carried out in triplicate; only one replica is shown. The SPR response is expressed in response units relative to the time in seconds. (C and D) Electrophysiological recordings of Kir2.1 WT (C) and R312H (D) reconstituted in lipid bilayers in the presence of 1% PIP2 at −100 mV. Kir2.1 WT shows an open probability (Po) of 70%, and Kir2.1 R312H has a Po of less than 1%.
Functional studies on Kir2.1 WT and mutant R312H
The purified proteins (Kir2.1 WT and R312H) were investigated in reconstituted planar lipid bilayers. The electrophysiological recordings of Kir2.1 WT and R312H mutant in the absence of PIP2 showed no channel activity (data not shown). However, recordings in the presence of 1% PIP2 at −100 mV revealed an open probability (Po) of 70% for the WT form (Fig. 4C) and less than 1% for the Kir2.1 R312H mutant (Fig. 4D). These activities were all silenced by applying 100 μM BaCl2 to the upper chamber.
Interaction network between R312, H221, and E303 residues
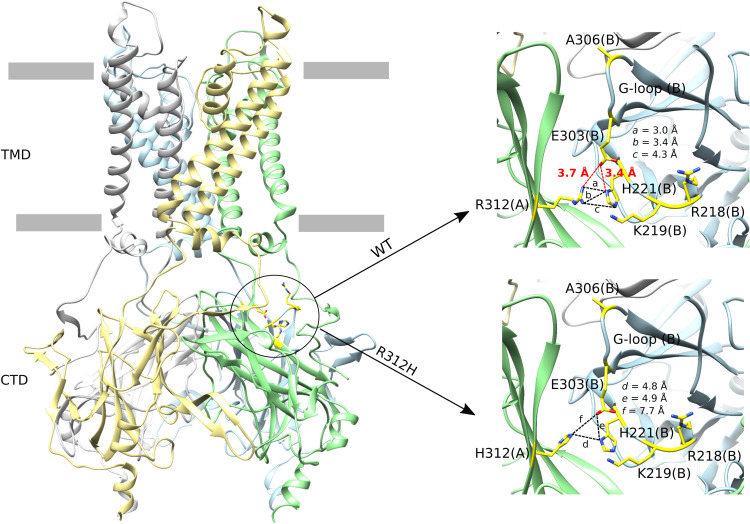
In the human Kir2.1 channel structure, residue R312 is involved in an intersubunit interaction network with residues H221 and E303 of the clockwise-adjacent chain (top view from the extracellular side) (Fig. 5 and fig. S6). Except for chain C, the oxygen atoms of the R312 side chain can establish intersubunit contacts at distances of 3 to 3.4 Å with the oxygen atoms of the H221 imidazole ring of the adjacent chain, although both are positively charged. Computational approaches have demonstrated that arginine can form strong like-charged contact pairs with histidine, which are not weakened by the unfavorable electrostatic interaction between the side chains (33). Moreover, except for chain B, R312 can establish intersubunit salt bridges (3.4 to 3.7 Å) with E303 of the adjacent chain, a residue present in the G-loop. E303 can also establish intrasubunit salt bridges (3.4 Å) with H221 near residues K219 and R218. These two critical residues have been previously reported to control the binding of PIP2 to the Kir2.1 channel, with the mutation R218Q abolishing such a binding (23, 34). Figure S7 highlights the cryo-EM density map quality for H221, E303, and R312 in the four Kir2.1 chains, with a local resolution of 3.7 Å (fig. S3).
Fig. 5. Interaction network between R312, H221, and E303 residues in the human Kir2.1 channel cryo-EM structure.
Cartoon representation of the human Kir2.1 channel cryo-EM structure highlighting (inside the circle) the location of the R312 residue. The insets show the intersubunit interaction network of R312 in the WT (top) and that of H312 in the R312H mutant (bottom) between chains A and B. The chains A, B, C, and D are shown in green, blue, gray, and yellow, respectively. Residues R218, K219, H221, E303, and A306 are represented as yellow sticks. The chains are indicated in parenthesis. Salt bridge distances are highlighted in red dashes and the other distances are highlighted in black dashes.
The R312H mutation was modeled in the human Kir2.1 channel to understand its implication in the interaction network described above. Arginine was changed to histidine using Chimera software, followed by local energy minimization. The analysis of the interaction network distances showed that the nitrogen atoms of the H312 imidazole ring were further away from the nitrogen atoms of the H221 imidazole ring of the adjacent chain than the distances observed in the WT structure (Fig. 5 and fig. S6). Furthermore, all salt bridges involving E303 are lost because the distances between Oε2 of E303 and nitrogen atoms from H312 and H221 increased to 4.9 to 9.0 Å. These data suggest that R312 helps coordinate the position of H221 and E303 side chains to maintain the salt bridges established by E303 with the two other residues. The mutation R312H abolishes this particular network of interactions.
Investigation of global motions of the Kir2.1 channel by NM analysis
The first Kir2.2 channel crystal structures revealed significant structural differences between the apo and PIP2-bound structures. In the apo-Kir2.2 crystal structure, the protein is observed in an extended conformation, and the linker between CTD and TMD is disordered in the outer region (amino acids 57 to 70). On the other hand, in the PIP2-bound structures, this linker adopts a helical conformation, and the CTD approaches the membrane’s inner surface, translating toward the TMD by 6 Å (7, 26). Similar results about PIP2-induced channel compaction were observed in Kir3.2 cryo-EM structures (27). Furthermore, this compact structure has also been associated with a gating conformational change (35). However, it is uncertain that PIP2 binding could be responsible for the movement of CTD toward TMD since the crystal structures of Kir2.2 (5KUM) and Kir3.2 (3SYO and 3SYP) have been obtained in the compact form without PIP2 (17). This suggests that large conformational movements are involved in the gating of this channel. These large-scale conformational movements generally take place on nanosecond to microsecond time scale, often not reached in classical MD simulations. Low-frequency NM analysis is a relevant tool to overcome this limitation and characterize these conformational movements and has been successfully used to describe structural aspects of several proteins (36, 37). NM analysis was performed to assess whether the lowest frequency modes would describe the channel’s compaction/extension movement in the absence of PIP2. We calculated the first five lowest frequency modes as they are sufficient to get the overall collective motions (38).
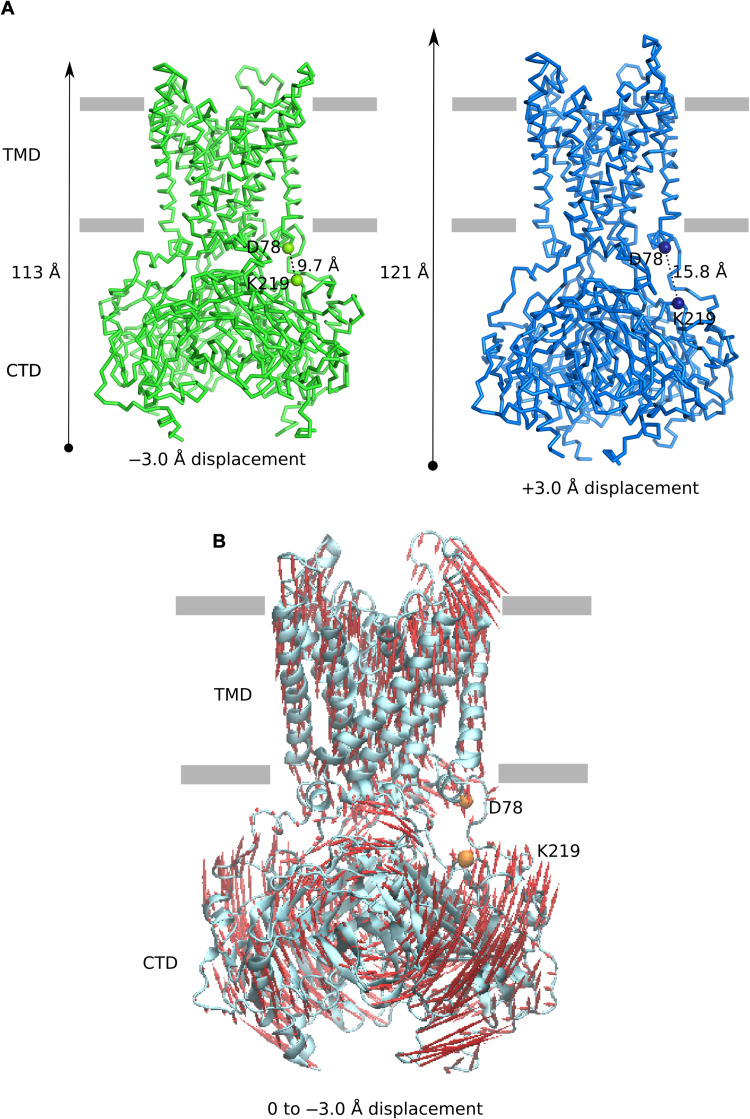
The longitudinal length of Kir2.1 (fig. S8, left) was evaluated by the Cα-Cα distance between the most distal points of the structure: residues K117 (in the extracellular loop) and E363 (lowest part of the C-terminal region). Tethering of the CTD to the TMD was assessed by the Cα-Cα distance between D78, located in the slide helix at the interface between the membrane and the CTD, and K219, located in the CTD; the latter has been identified as a PIP2 binding controller, being close to H221 (2). In the extended structure of the Kir2.1 channel obtained here, the protein is seen to be 121 Å long and has a D78/K219 Cα-Cα distance of ~15 Å (fig. S8, left). After the initial energy minimization necessary for the all-atom NM calculation (constituting the reference structure), a slight decrease in the protein’s longitudinal length and a slightly closer distance between the CTD and TMD were observed (fig. S8, center). To better analyze the motions corresponding to the five lowest frequency modes, ±3.0 Å structural displacements were achieved along each of them. Then, the absolute value of the longitudinal length difference between the two extremes of the displaced structures along each of the mode vectors and the CTD-TMD tethering length were calculated.
Mode 5 was the only NM for which the Cα K117/E363 and D78/K219 distances varied cooperatively by at least 6 Å between the displaced structures at +3.0 and −3.0 Å (fig. S8, right). A displacement of −3.0 Å along the mode 5 from the energy-minimized reference structure leads to a compaction of the entire protein down to 113 Å and a reduction of Cα-Cα D78/K219 down to 9.7 Å (Fig. 6). On the other hand, the reverse +3-Å displacement leads to an extension up to 121 Å and to an increase in the Cα-Cα D78/K219 distance up to 15.8 Å, which are values close to those observed in the cryo-EM structure (Fig. 6A). These data show that NM5 was able to recover and describe the extension/compression conformational motion of the Kir2.1 channel even without PIP2 (Fig. 6A and movie S1).
Fig. 6. Description of extension/compression conformational movement of the human Kir2.1 channel using NM analysis.
(A) Ribbon representation of the human Kir2.1 cryo-EM structure after displacement of −3.0 Å (green; compacted structure) and +3.0 Å (blue; expanded structure) along mode 5. The longitudinal length and the Cα-Cα distance between residues D78 and K219 (represented by spheres) are indicated in angstroms. (B) Description of the compression movement on the cartoon representation of the human Kir2.1 structure (cyan). Red arrows show Cα displacement from the reference structure toward the −3.0 Å compacted structure along the mode 5. Orange spheres represent D78 and K219. Gray horizontal bars indicate plasma membrane boundaries.
Modes 1 and 2 describe a swing motion that results in a lateral opening of the area located at the interface between CTD and TMD in adjacent chains (Fig. 7, fig. S9, and movies S2 and S3). Mode 1 describes a swing motion between chains B and C in this interface area and chains A and D (Fig. 7A, fig. S9A, and movie S2). The extent of this motion was evaluated by the Cα-Cα distance between R82 and K219 from adjacent chains [R82(B)/K219(C) and R82(D)/K219(A)] (fig. S9A). A displacement of +3.0 Å along the mode 1 from the reference structure leads to an increase in Cα R82(B)/K219(C) distance of 5.1 Å, and the reverse −3.0 Å displacement leads to an increase in Cα R82(D)/K219(A) distance of 1.8 Å (fig. S9A). K187 is a residue that interacts directly with a PIP2 phosphate group (7) and is located in the central region of the inner loop between TMD and CTD. We observed a significant increase in the solvent-accessible surface area (SASA) of the residue K187 from chains B and D by 10.0 and 20.0 Å2, respectively, after applying ±3.0 Å displacements along the NM1 (fig. S9A). Mode 2 describes the same swing motion as mode 1 but between chains A and B and chains C and D (Fig. 7B, fig. S9B, and movie S3). Likewise, the Cα R82/K219 distances increase in the ±3.0 Å displaced structures along the NM2 [by 2.1 and 5.8 Å for R82(A)/K219(B) and R82(C)/K219(D) distances, respectively] and the SASA of the K187 residues [by 13.7 and 15.3 Å2 for K187(A) and K187(C), respectively] (fig. S9B and movie S3). In conclusion, modes 1 and 2 may describe an inherent movement of opening the PIP2-binding site in the four possible PIP2-binding sites available in the Kir2.1 structure. The residues involved in putative PIP2 binding for Kir2.x (R80, W81, R82, K182, K185, K187, K188, R189, R218, and K219) (2) are shown in movies S2 and S3. Last, the motions referring to modes 3 and 4 can be seen in movies S4 and S5; we do not address them in detail.
Fig. 7. Description of the opening of the PIP2-binding site of the human Kir2.1 channel using NM analysis.
(A and B) Description of the conformational movement on the cartoon representation of the human Kir2.1 structure. Chains A, B, C, and D are in cyan, orange, green, and yellow, respectively. Red arrows show Cα displacement from the reference structure toward the displacement of +3.0 and −3.0 Å along the modes 1 (A) and 2 (B).
To identify the different conformational states described by NM analysis in our cryo-EM dataset, we followed the protocol described by Scheres (39). The compression/extension conformations were observed when the particles from class 3 of the second three-dimensional (3D) classification round (blue box; fig. S2) were submitted to 3D classification using local angular searches (fig. S10). This 3D classification provided two classes (classes 2 and 6; fig. S10), which after 3D refinement, resulted in maps with size lengths of 120 and 113 Å (fig. S10), values similar to those observed along the ±3.0 Å structural displacements in NM5 (Fig. 6). Concerning the lateral opening at the interface between CTD and TMD similar to that described by modes 1 and 2, we noticed that class 3 from the first round of 3D classification presented already this feature (red box; fig. S2). This was confirmed when we submitted the particles of this class to a 3D classification using local angular searches. In all classes resulting from this 3D classification, it was possible to observe the lateral opening of the structure (fig. S11). This interesting conformation can lead to increased accessibility of the PIP2-binding site, as described by the NM1 and NM2 (Fig. 7).
Investigation by MD simulations of local motions of residues K64 and R67
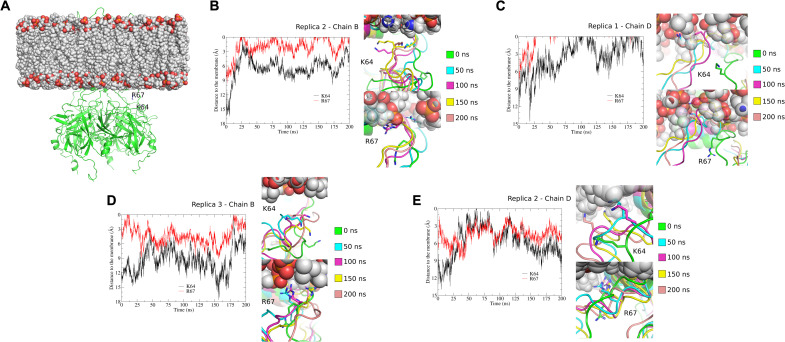
Residues K64 and K219 have been identified as constituting a secondary binding site, directly interacting with nonspecific anionic phospholipids (PL−). These positive allosteric regulators increase the PIP2 sensitivity by 10- to 100-fold (8, 9, 24). We studied the local motion of K64 (at the N-terminal end of the slide helix) by carrying out MD simulations on the cryo-EM structure of Kir2.1 (extended conformation) embedded in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid bilayer for 200 ns in triplicate. Snapshots of the K64 position during MD simulations show that this residue can approach the membrane significantly in most of the chains and replicas. Moreover, for three of the chains, we observed that K64 can enter the membrane (Fig. 8, fig. S12, and movie S6). These data support the previously proposed movement of K64 tethering to the membrane (9, 24), which would be required for the PL− binding to this secondary binding site. In addition, the dynamics of R67 (in the vicinity of K64 and closer to the membrane) were also examined. Snapshots of the R67 position along the MD simulations show that this residue can penetrate the membrane for most of the chains and therefore gets significantly closer to the membrane than K64 residue (Fig. 8, fig. S12, and movie S5). The analysis of the distances between the K64 and R67 residues to the phosphate groups of the inner membrane shows that these residues move toward the membrane (Fig. 8 and fig. S12). Furthermore, this analysis also shows that K64 and R67 have similar trajectory profiles and may have inherent coupled local motions. Last, the Cα RMSD and Cα root mean square fluctuations (RMSFs) from the MD simulations are shown in fig. S13. All three replicas exhibited Cα RMSD variation of less than 0.5 Å in the last 50 ns of the MD simulations (fig. S13A). Cα RMSF analysis shows that extracellular loops have the highest RMSF values in all four chains of the human Kir2.1 structure (fig. S13B), indicating a high level of backbone flexibility. On the other hand, G-loop exhibits low Cα RMSF values, indicating a low level of backbone flexibility in this region.
Fig. 8. Tethering of K64 and R67 to the membrane displayed in MD simulations.
(A) Human Kir2.1 cryo-EM structure and membrane (POPC bilayer) system used in MD simulations. The Kir2.1 channel structure is represented in a green cartoon, and the residues K64 and R67 of chain B are highlighted. (B to E) Trajectory analysis of the K64 and R67 residues along the 200 ns of MD simulations in some representative chains of the three replicates. In each of these panels, snapshots along MD simulations are shown: 0 ns (green), 50 ns (cyan), 100 ns (magenta), 150 ns (yellow), and 200 ns (salmon). K64 and R67 residues are highlighted in sticks. (B) to (E) show the distances between the K64 and R67 residues to the plane of the phosphate groups from the inner membrane. To calculate these distances, the inner membrane was centered at Z = 0. The 0 value represents the membrane level. (B) Chain B, replica 2; (C) chain D, replica 1; (D) chain B, replica 3; (E) chain D, replica 2.
DISCUSSION
Conducting ions and blocking ions compete for sites in the pore
Strong rectification of Kir2.1 has been attributed to two principal electronegative regions: D172 in the M2 helix (40) and E224/E299 below the G-loop (41). The arrangement of residues is essential for blockade because they create negatively charged rings in the channel pore, which is crucial for the blocking and rectification of Kir2.1 channels (4). Mutations of these residues decreased the affinity for blockers such as Mg2+ and polyamines and reduced the intensity of inward rectification (1, 42). Our data support these findings, as one of the Sr2+-blocking ions is located in the TMD, below the selectivity filter near D172, and in the CTD near E224 and E299 just below the G-loop (Fig. 2). Each site contains several negatively charged carboxyl groups that create a strong electric field that easily accommodates multivalent cations. Sr2+ ions are used in electrophysiological experiments to promote the blockade of Kir2.1 channels (43). The Sr2+ blocker just below the selectivity filter is stabilized not only with the negatively charged ring made of D172 but also with T142 (conserved in all Kir members) located at the same distance as D172 (~6 to 7 Å), which could help stabilize Sr2+ (Fig. 2). However, this site is too wide (diameter of ~14 Å) to ensure the direct coordination of a dehydrated ion in the center. The Sr2+ ions at the center of the site should most likely interact with D172 and T142 through bridging water molecules. The second Sr2+, just below the G-loop, is stabilized by the negatively charged residues E224 and E299. In addition, the backbone carbonyl group of A306 is closer to the Sr2+ than the other negatively charged groups (Fig. 2). A306 is conserved only in the Kir2 subfamily (the strongest rectifiers) and KirBac (1.1 and 3.1). However, again, the pore diameter is too large (diameter of ~13 Å) to allow direct coordination; presumably, bridging with water molecules seems important. The presence of blocking ions could influence the conformation of the pore and, in particular, the radial size of the selectivity filter, which is very narrow (less than 0.5 Å in diameter; Fig. 3) and cannot accommodate a K+ ion whose diameter is 2.67 Å when dehydrated (44).
Another di-aspartate cluster (D255/D259) has been described as important in determining the extent of inward rectification of the Kir2.1 channel (20). Multivalent blocking ions can occupy this site. For instance, the chicken Kir2.2 channel structure shows an Sr2+ ion in this position (D256) when the crystals were grown in the presence of Sr2+ (26). In our case, we identified one K+ ion at this position, which suggests that conducting and blocking ions may compete for the same sites in the pore. The high concentration of KCl present in the buffer for cryo-EM analysis (150 mM) probably favors the occupation of this site.
Inherent local and large-amplitude motions of the human Kir2.1 channel structure may describe the mechanism of CTD tethering to TMD
It has been suggested that the PIP2 binding displaces the CTD by approximately 6 Å in Kir2 and Kir3 channels: The CTD becomes tethered to the TMD, resulting in a compacted shape of the channel (from 120 to 114 Å longitudinally; 3SPI and 6XIT) (7, 27). However, some apo-Kir2.2 and apo-Kir3.2 crystal structures exhibited a compact conformation even in the absence of PIP2 (3SYO and 5KUM) (17, 24). The fifth lowest-frequency NM describes the extension/compression conformational motion of the human Kir2.1 channel (Fig. 6 and movie S1) and shows that the Kir2.1 longitudinal length can vary from 121 Å (extended form) to 113 Å (compact form) as the CTD moves closer to the TMD in the compact form of the channel (Fig. 6 and movie S1). These conformation states were also found on cryo-EM experimental data (fig. S10). These findings demonstrate that extension/compression movement is a natural large-amplitude motion of the Kir2.1 channel and that its compact form can be achieved even in the absence of PIP2. In addition, the PIP2-binding sites can be more accessible due to a swinging movement that results in a lateral opening at the interface between CTD and TMD described in modes 1 and 2 (Fig. 7, fig. S9, and movies S2 and S3). Protein conformations presenting this lateral opening with various amplitudes were identified in our cryo-EM study (fig. S11).
Using MD simulations, we then investigated the motions of residue K64, which is part of the secondary binding site (9, 24). We recovered its tethering motion to the membrane and demonstrated that R67, in a coupled local movement with K64, can penetrate the membrane (Fig. 8, fig. S12, and movie S5). Note that the conformation of Kir2.1 remained in its extended form along the MD simulations. The movement of K219, also considered part of the secondary binding site (9) and pointed out to be PIP2 binding controller (2), toward the membrane is noticed during NM analysis (large movements; Fig. 6 and movie S1) but not in MD simulations.
Together, our observations support the intrinsic Kir2.1 compaction and an inherent opening of the PIP2-binding site in the absence of PIP2. The former large conformational movement could possibly be initiated by the local movements of K64 and R67 residues into the membrane. Therefore, PIP2 binding would not induce the compact conformation but would instead stabilize the compressed state.
Central role of the G-loop in the gating and mechanism for releasing the constriction point
Previous studies have supported a functional role of the G-loop region. For instance, endogenous microRNAs could physically bind to Kir2.1 at the G-loop, modulating cardiac electrophysiology (45). In addition, two residues in the G-loop, E303 (20) and C311 (46), are important for channel conductance and gating.
The crystal structure of the CTD domain of mouse Kir2.1 R218Q/T309K (2GIX) (30) indicated a possible role of two salt bridges, E303/R312 and R218/T309, for G-loop gating. MD simulations from Li et al. (47) suggest that G-loop gating may be regulated by three pairs of weak electrostatic interactions (E303-H221, H221-R189, and E303-R312).
Previous studies by Pegan et al. (20) describe the G-loop as an intrinsically flexible region. Our structural comparison of the human and mouse Kir2.1 shows that the G-loop backbone exhibits a similar conformation in both structures. In addition, MD simulations indicate that the G-loop adopts a rigid structure in the closed conformation without PIP2 (one of the lowest RMSF values; fig. S13). Both mouse and human Kir2.1 structures exhibit a constriction point in the G-loop at residue A306. Although we showed that the G-loop is rigid in the absence of PIP2, it must undergo a conformational change to release the constriction point and gate.
Here, we demonstrated by SPR experiments on Kir2.1 WT and its R312H mutation counterpart that residue R312 does not significantly alter the affinity of human Kir2.1 for PIP2. Nonetheless, electrophysiological recordings showed that the Kir2.1 R312H mutant channels could not open despite binding PIP2. These data indicated that binding of PIP2 to Kir2.1 alone is insufficient to induce channel opening, emphasizing the requirement for structural signal transmission of PIP2-binding at gating.
Structural analysis of R312 in the human Kir2.1 cryo-EM structure shows that this residue is involved in a strong network of intersubunit interactions (salt bridges and like-charged contact pairs) with residues H221 and E303 (on G-loop, close to the A306 constriction point), both on the adjacent chain. Our studies highlight the relevance of the structural integrity of E303-mediated salt bridges with R312 and H221 for proper G-loop gating. These findings support the previous work of Pegan et al. (30) and of Li et al. (47) with significant differences: (i) where Li et al. found weak interactions (E303-H221 and E303-R312) on the Kir2.1 modeled structure, we found strong interactions. In addition, they observed the weak interaction H221-R189, which we have not identified because both side chains are far away in our cryo-EM structure (separation distance, >10 Å). (ii) the R218/T309 salt bridge described in Pegan et al.’s work is not seen in our structure; because the distance between the two side chains in the human Kir2.1 channel cryo-EM structure is too large to allow any interaction (>12 Å).
In conclusion, our human Kir2.1 channel cryo-EM structure describes a well-connected interaction network between the PIP2-binding site residues, R218 and K219, and the G-loop region (E303) via residues R312 and H221. Our data suggest that the conformational changes required for the G-loop opening are most likely controlled by PIP2 binding. The replacement of R312 with histidine leads to a complete loss of the interaction network described above. Therefore, the interaction network integrity between subunits seems necessary for the proper allosteric transmission of the signal between R312 and the G-loop of the adjacent subunit upon PIP2 binding, which possibly allows the release of the constriction point on the G-loop. We can then hypothesize a PIP2-dependent G-loop gating mechanism that consists of the following: PIP2 binding triggers local conformational changes in the position of the side and main chains of R218 and K219, which, because of the structural proximity, lead to significant changes in the position of H221, displacing it laterally toward the intracellular medium. This movement would, in turn, cause E303 and R312 of the adjacent chain to move in the same direction, causing the G-loop to open. This, of course, needs to be confirmed by detailed experiments. Last, the results regarding R312H mutation indicate a direct involvement of an AS-causing mutation in the G-loop gating of the Kir2.1 channels.
MATERIALS AND METHODS
Protein expression
Protein expression was handled according to Hartmann et al. (48). Briefly, the synthetic KCNJ2 genes encoding residues 1 to 427 (the whole-sequence UniProt reference P63252) of human Kir2.1 WT and R312H mutant were cloned in a pPIC9K vector upstream of a sequence coding for a PreScission protease cleavage site (LEVLFQGP), followed by a linker of 11 amino acids and a 10His tag. The plasmids were introduced in P. pastoris strain SMD1163 (his4, pep4, and prb1), and the resulting colonies were further analyzed via an in situ Yeastern blot immunoassay to identify the best expressing clones. For protein production, a freshly streaked colony was grown overnight in BMGY [1% (w/v) yeast extract, 2% (w/v) peptone, 1.34% (w/v) yeast nitrogen base without amino acids, 1% (w/v) glycerol, and 0.1 M phosphate buffer at pH 6.0] at 30°C with shaking at 250 rpm until an optical density at 600 nm of 3 to 5. Expression of the recombinant Kir2.1 was induced with methanol by transferring yeast cells in a BMMY medium [1% (w/v) yeast extract, 2% (w/v) peptone, 1.34% (w/v) yeast nitrogen base without amino acids, 0.5% (v/v) methanol, and 0.1 M phosphate buffer at pH 6.0] and incubating them for about 20 hours at 22°C. Cells were harvested by centrifugation, washed in phosphate-buffered saline buffer (pH 7.4), and stored at −80°C until use.
Protein purification
To purify, the Kir2.1 WT and Kir2.1 R312H cells were resuspended in buffer A [50 mM tris-HCl (pH 7.4), 200 mM KCl, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride] and protease inhibitor cocktail tablets (Roche and Merck). Cells were first incubated with 20 U of Zymolyase 20T (Amsbio) per gram of pellet for 15 min at room temperature and then ruptured using a Constant System Cell Disrupter or, better, a FastPrep 24 (MP Biomedicals). Cell debris was removed by a 5-min centrifugation at 4000g at 4°C. The supernatant was collected and centrifuged at 100,000g for 30 min at 4°C to collect crude membranes. The pellet was subsequently resuspended in buffer B [50 mM tris-HCl (pH 7.4) and 150 mM KCl], and the membrane proteins were solubilized by addition of 29.3 mM DDM (1.5% n-dodecyl-d-maltoside, Glycon) for 30 min at room temperature with stirring and then centrifuged at 100,000g for 30 min at 4°C. Supernatant was added to pre-equilibrated [50 mM tris-HCl (pH 7.4), 150 mM KCl, 30 mM imidazole, and 0.03% DDM] 1-ml cobalt affinity resin (TALON, Clontech) at 4°C incubated for 45 min. The protein was eluted with an increasing concentration of imidazole. After the addition of 5 mM dithiothreitol (DTT), the eluted protein was concentrated on 100-kDa cutoff Vivaspin concentrator, filtered through 0.22-μm-pore-size polyethersulfone (PES) membrane, and injected on a Superdex-200 size exclusion chromatography column Increase 10/300 GL (Cytiva) pre-equilibrated with buffer C [20 mM tris-HCl (pH 7.4), 150 mM KCl, 0.03%/0.59 mM DDM, and 1 mM EDTA) using the Äkta Purifier system (Cytiva). Fractions corresponding to the tetramer were pooled, added 2 mM DTT, and concentrated to reach a concentration of about 0.7 to 1 mg/ml.
Surface plasmon resonance
The interaction between Kir2.1 and the lipid PIP2 was characterized by SPR on a Biacore 3000 instrument (Cytiva) controlled by Biacore 3000 Control software v4.1. All biosensor experiments were performed in triplicate at 25°C using the running buffer [20 mM tris-HCl (pH 7.5), 150 mM KCl, 0.05 mM EDTA, and 0.05% DDM]. In all experiments, a flow cell was left blank to be used as a reference for the sensorgrams. Kir2.1 (WT/R312H) was immobilized onto a carboxymethylated dextran (CM5) sensor chip. The activation of CM5 chips and immobilization of the protein steps were done using standard Biacore procedures (flow. 10 μl/min; contact time, 7 min; protein concentration, 25 to 50 nM]. Briefly, the CM5 chip was activated by amine coupling by injecting 0.2 M N-ethyl-N′-(3-dimethylaminopropyl) carbodiimide and 0.05 M N-hydroxysuccinimide. For immobilization, the protein was diluted to 25 to 50 nM using sodium acetate (pH 4.0 to 4.5), and saturation was achieved with 1 M ethanolamine HCl (pH 8.5; Cytiva).
The binding and kinetic assays were performed using single-cycle kinetics. PIP2 was serially diluted in running buffer to working concentrations (0.15 to 20 μM). For each cycle, PIP2 was injected at increasing concentration with a flow of 5 μl/min over both the reference cell and the ligand cell. Before the first PIP2 injection, a running buffer was injected and used as a double reference. Each injection consisted of 300-s contact time with 300-s dissociation time. No regeneration step was done between injections, as these buffers were detrimental to Kir2.1. The data were analyzed using BIA evaluation software 4.1, and kinetic parameters were determined using general fit and the titration kinetics 1:1 binding with drift model. The association and dissociation rate constants, kon and koff, respectively, and the KD were determined directly from the fitted curves.
Functional reconstitution of Kir2.1 WT and R312H mutant in lipid bilayer
Kir2.1 (WT and R312H) protein purified in DDM detergent (0.03%) was added to the upper chamber (150 μl) to a preformed bilayer. The Orbit mini apparatus was used (Nanion, Germany, horizontal planar lipid bilayer system), with two chambers separated by a partition with a 100-μm hole where the lipid bilayer is formed by 1,2-diphytanoyl-sn-glycero-3-phosphocholine (10 to 30 pF). The upper and lower chambers contained 150 mM KCl and 10 mM Mops (pH 7.4). Currents were recorded using Elements Data Reader (Nanion, Germany) and analyzed using Clampfit (Molecular Devices, CA, USA) software, sampled at 100 μs, and filtered at 1.25 kHz. The recording was performed at 24°C using a Nanion temperature control unit.
Sample preparation and cryo-EM data collection
Negatively stained electron microscopy showed good homogeneity and optimal distribution of a K+ channel molecule. After optimizing the cryo grid preparations, quantifoil (1.2/1.3) Cu 300 mesh grids with a thin carbon layer were glow discharged for 25 s before sample freezing. Three microliters of Kir2.1 at 0.6-mg/ml concentration was placed on the grid, blotted for 3.0 s, and flash-frozen in liquid ethane using a Vitrobot Mark III (Thermo Fisher Scientific) operated at 4°C and 100% humidity. The electron microscopy data collection statistics are available in table S2. A total of 9895 micrographs were collected on a Titan Krios G3 microscope operated at 300 kV equipped with a Falcon 3 direct electron detectors and a bioquantum/K3 energy filter (Gatan Inc.). The automation of the data collection was done with the software EPU. Movies were recorded in electron-counting mode using exposures of 4 s dose-fractionated into 36 frames with a dose rate of 1.7139 electrons/Å2, resulting in a total dose of 61.7 electrons/Å. A defocus range of −1.2 to −2.8 μm was used. The pixel size was 0.86 Å per pixel.
Cryo-EM data processing
After a visual inspection to remove poor-quality micrographs (containing ice contamination, thick ice, low contrast, and abnormal background), a total of 7188 micrographs were selected for further cryo-EM data processing. The movies were motion-corrected and dose-weighted using MotionCor2 (49), and contrast function parameters were estimated using CTFFIND4 (50). A total of 1,031,472 particles were automated picked using SPHIRE-crYOLO (51) and a trained network of about 2000 particles. The image processing was then performed using RELION (52, 53). The extracted particles were binned four times and subject to one round of 2D classification. A total of 837,808 particles were extracted (no binned) from 45 2D class averages and subjected to 3D initial model ab initio construction using C4 symmetry. Then, the obtained 3D initial model was submitted to three rounds of 3D classification, the first two using C1 symmetry and the last one using C4 symmetry. The best class from the last round of 3D classification contained 63,584 particles. After the creation of mask to masking out the detergent layer, the map resulted from this class was refined using 3D autorefine using C4 symmetry with decreasing values of initial angular sampling and local searches from autosampling, being the final map obtained using 1.8° and 0.5°, respectively. The resolution of the final map is 4.3 Å [gold-standard Fourier shell correlation (FSC) = 0.143]. This final map was subjected to handedness correction and B-factor map sharpening using the Auto-sharpen tool available in the PHENIX software suite (54). Figure S2 shows the complete cryo-EM processing workflow. Cryo-EM data collection information is summarized in table S2.
Model building and refinement
An initial in silico homology model of human Kir2.1 was generated using I-TASSER (55) and the crystal structure of the chicken Kir2.2 channel (3JYC) (26) as a template. For building and refinement of the atomic model, the TMD (regions 55 to 184) of this in silico model was placed into the final sharpened cryo-EM map using the Dock in Map tool available in PHENIX (54). For the CTD (188 to 367 region), the crystal structure of the CTD from mouse Kir2.1 channel (1U4F) (20) was placed into the final cryo-EM map using the same approach. Once the models were placed in the cryo-EM density map, the loops that connect the two domains (regions 185 to 187) and an N-terminal loop (41 to 54 region) absent in the in silico model were manually built using Coot (56). Visual inspection of the cryo-EM density map revealed that it was also possible to manually build a fraction of the N-terminal region that reaches the CTD (regions 41 to 54). After these manual builds, several iterative cycles of refinement using the phenix.real_space_refine in PHENIX with secondary structure restraints and manual adjustments in Coot provided the final cryo-EM model for the human Kir2.1 channel. Refinement statistics are summarized in table S2. All figures were generated using Chimera, ChimeraX, PyMOL, and VMD software.
R312H mutation modeling
The R312H mutation was modeled on the human Kir2.1 channel cryo-EM structure by replacing the arginine residue with the most likely rotamer of histidine in the Chimera software. Next, local energy minimization was performed using Chimera (around 6.5 Å of the modeled histidine, keeping other protein atoms fixed) using 1000 steepest descent steps (0.02-Å step size), 20 conjugate gradient steps (0.02-Å step size), and the AMBER ff14SB force field. After energy minimization, no clashes and bad contacts were observed between the modeled histidine and its vicinity.
NM analysis
The first five internal lowest-frequency modes of the entire Kir2.1 channel structure were obtained by carrying out all-atom NM calculations using a physical force field. The protein was first energy minimized in vacuum to a low-energy gradient of 10−6 kcal × mol × Å−1 by applying 10,000 conjugate gradient steps, followed by 100,000 adopted basis Newton-Raphson steps by using the CHARMM36m force field (57). Electrostatic interactions were calculated with a distance-dependent dielectric constant. The NMs were calculated using the DIMB diagonalization method (58) in CHARMM under the same force field parameters as those described previously for initial energy minimization.
Two outer structures were generated for each mode, one corresponding to a structural displacement of 3.0 Å RMSD along the mode with respect to the minimum energy structure and the other in the inverse direction with the same RMSD value. These outer structures were, in turn, energy minimized by applying an umbrella potential to maintain their positions along the modes. CHARMM’s VMOD module was applied to generate these structures. In addition, intermediate structures between −3.0 and 3.0 Å RMSD were also generated by steps of 0.1 Å to make the movie animations for each mode. The first six NMs were not taken into account as they are rigid-body overall modes (three rotations and three translations). Thus, the five NMs described here refer to modes 7 to 11, but they were renumbered from 1 to 5.
MD simulations
The structure of the human Kir2.1 channel was subjected to MD simulations using NAMD software (59) under the CHARMM36m force field. All preparation steps were performed using CHARMM-GUI server (60). The Kir2.1 channel was embedded in a lipid bilayer containing 176 POPC molecules, with a size of 80 Å in the x and y directions. The complete system has box dimensions of 106.8 Å by 106.8 Å by 165.1 Å and contains 44,276 molecules of water and 246 neutralizing ions. The KCl concentration was 0.15 M. The system was heated and equilibrated in the standard equilibration protocol suggested by CHARMM-GUI developers, gradually decreasing protein and membrane atomic positional restraints during a duration of 2 ns. Last, the production run was performed without any positional restraints for 200 ns in triplicate. The distances between the K64 and R67 residues to the plane of the phosphate groups from the inner membrane were calculated, with the inner membrane centered at Z = 0.
Acknowledgments
We thank J.-M. Guigner for initial electron microscopy images, M. G. S. Costa for discussions, and M. Dezi for preliminary work (IMPMC); Instruct-ERIC facility at IGBMC Strasbourg, France and B. Klaholz; Nanoimaging Core facility (C2RT) at the Institut Pasteur and the help of J.-M. Winter and M. Vos; and Core-facility Molecular Interactions (IBPS) at the Sorbonne University and T. Bouceba.
Funding: This work was supported by AFM-Téléthon #23207 for S.B. and C.V.-B., AFM-Téléthon #23210 for C.A.H.F., Ecole Doctorale ED515 Sorbonne Université for D.Z. and C.F., and EQUIPEX CACSICE ANR-11-EQPX-0008 (C.V.-B.). The project has received funding from the European Union’s Horizon Europe Research and Innovation Program under grant agreement no. 101026386.
Author contributions: Conceptualization: C.V.-B., S.B., and D.P. Project coordination: C.V.-B. Protein expression and purification: D.Z., R.W., V.K., and C.V.-B. SPR: D.Z. Cryo-EM sample preparation and data collection: D.Z., G.P.-A., and C.V.-B. Cryo-EM image analysis and structure determination: C.A.H.F., C.F., and C.V.-B. Computational investigations: C.A.H.F. and D.P. Electrophysiology: R.S. and S.B. Writing—original draft: C.A.H.F., D.Z., and C.V.-B. Writing—review and editing: C.A.H.F., D.Z., C.F., R.W., D.P., S.B., and C.V.-B. Funding acquisition: C.V.-B., S.B., and C.A.H.F.
Competing interests: The authors declare that they have no competing interest.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and the Supplementary Materials. Cryo-EM density maps and coordinates of human Kir2.1 cryo-EM structure have been deposited in the EMDB with ID code EMD-14678 and RCSB PDB with ID code 7ZDZ.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S6
REFERENCES AND NOTES
- 1.Hibino H., Inanobe A., Furutani K., Murakami S., Findlay I., Kurachi Y., Inwardly rectifying potassium channels: Their structure, function, and physiological roles. Physiol. Rev. 90, 291–366 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Fürst O., Mondou B., D’Avanzo N., Phosphoinositide regulation of inward rectifier potassium (Kir) channels. Front. Physiol. 4, 404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nichols C., Lopatin A., Inward rectifier potassium channels. Annu. Rev. Physiol. 59, 171–191 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Baronas V. A., Kurata H. T., Inward rectifiers and their regulation by endogenous polyamines. Front. Physiol. 5, 325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu Z., Mechanism of rectification in inward-rectifier K+ channels. Annu. Rev. Physiol. 66, 103–129 (2004). [DOI] [PubMed] [Google Scholar]
- 6.D’Avanzo N., Cheng W. W. L., Doyle D. A., Nichols C. G., Direct and specific activation of human inward rectifier K+ channels by membrane phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 285, 37129–37132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen S. B., Tao X., MacKinnon R., Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 477, 495–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng W. W. L., D’Avanzo N., Doyle D. A., Nichols C. G., Dual-mode phospholipid regulation of human inward rectifying potassium channels. Biophys. J. 100, 620–628 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S.-J., Wang S., Borschel W., Heyman S., Gyore J., Nichols C. G., Secondary anionic phospholipid binding site and gating mechanism in Kir2.1 inward rectifier channels. Nat. Commun. 4, 2786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rohacs T., Lopes C. M. B., Jin T. H., Ramdya P. P., Molnar Z., Logothetis D. E., Specificity of activation by phosphoinositides determines lipid regulation of Kir channels. Proc. Natl. Acad. Sci. U.S.A. 100, 745–750 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plaster N. M., Tawil R., Tristani-Firouzi M., Canun S., Bendahhou S., Tsunoda A., Donaldson M. R., Iannaccone S. T., Brunt E., Barohn R., Clark J., Deymeer F., George A. L. Jr., Fish F. A., Hahn A., Nitu A., Ozdemir C., Serdaroglu P., Subramony S. H., Wolfe G., Fu Y. H., Ptacek L. J., Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen’s syndrome. Cell 105, 511–519 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Clarke O. B., Caputo A., Hills A. P., Vandenberg J. I., Smith B. J., Gulbis J. M., Domain reorientation and rotation of an intracellular assembly regulate conduction in Kir potassium channels. Cell 141, 1018–1029 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Xia M., Jin Q., Bendahhou S., He Y., Larroque M.-M., Chen Y., Zhou Q., Yang Y., Liu Y., Liu B., Zhu Q., Zhou Y., Lin J., Liang B., Li L., Dong X., Pan Z., Wang R., Wan H., Qiu W., Xu W., Eurlings P., Barhanin J., Chen Y., A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem. Biophys. Res. Commun. 332, 1012–1019 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Priori S. G., Pandit S. V., Rivolta I., Berenfeld O., Ronchetti E., Dhamoon A., Napolitano C., Anumonwo J., di Barletta M. R., Gudapakkam S., Bosi G., Badiale M. S., Jalife J., A novel form of short QT syndrome (SQT3) is caused by a mutation in the KCNJ2 gene. Circ. Res. 96, 800–807 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Tester D. J., Arya P., Will M., Haglund C. M., Farley A. L., Makielski J. C., Ackerman M. J., Genotypic heterogeneity and phenotypic mimicry among unrelated patients referred for catecholaminergic polymorphic ventricular tachycardia genetic testing. Heart Rhythm 3, 800–805 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Andersen E. D., Krasilnikoff P. A., Overvad H., Intermittent muscular weakness, extrasystoles, and multiple developmental anomalies. A new syndrome? Acta Paediatr Scand. 60, 559–564 (1971). [DOI] [PubMed] [Google Scholar]
- 17.Whorton M. R., MacKinnon R., Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell 147, 199–208 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whorton M. R., MacKinnon R., X-ray structure of the mammalian GIRK2-βγ G-protein complex. Nature 498, 190–197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishida M., Cadene M., Chait B., Mackinnon R., Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 26, 4005–4015 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pegan S., Arrabit C., Zhou W., Kwiatkowski W., Collins A., Slesinger P. A., Choe S., Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification. Nat. Neurosci. 8, 279–287 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Sacconi S., Simkin D., Arrighi N., Chapon F., Larroque M. M., Vicart S., Sternberg D., Fontaine B., Barhanin J., Desnuelle C., Bendahhou S., Mechanisms underlying Andersen’s syndrome pathology in skeletal muscle are revealed in human myotubes. Am. J. Physiol. Cell Physiol. 297, C876–C885 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Lopes C. M. B., Zhang H., Rohacs T., Jin T., Yang J., Logothetis D. E., Alterations in conserved Kir channel-PIP2 interactions underlie channelopathies. Neuron 34, 933–944 (2002). [DOI] [PubMed] [Google Scholar]
- 23.D’Avanzo N., Lee S.-J., Cheng W. W. L., Nichols C. G., Energetics and location of phosphoinositide binding in human Kir2.1 channels. J. Biol. Chem. 288, 16726–16737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S.-J., Ren F., Zangerl-Plessl E.-M., Heyman S., Stary-Weinzinger A., Yuan P., Nichols C. G., Structural basis of control of inward rectifier Kir2 channel gating by bulk anionic phospholipids. J. Gen. Physiol. 148, 227–237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An H.-L., Lü S.-Q., Li J.-W., Meng X.-Y., Zhan Y., Cui M., Long M., Zhang H.-L., Logothetis D. E., The cytosolic GH loop regulates the phosphatidylinositol 4,5-bisphosphate-induced gating kinetics of Kir2 channels. J. Biol. Chem. 287, 42278–42287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao X., Avalos J. L., Chen J. Y., MacKinnon R., Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 angstrom resolution. Science 326, 1668–1674 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niu Y., Tao X., Touhara K. K., MacKinnon R., Cryo-EM analysis of PIP2 regulation in mammalian GIRK channels. eLife 9, e60552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bavro V. N., De Zorzi R., Schmidt M. R., Muniz J. R. C., Zubcevic L., Sansom M. S. P., Vénien-Bryan C., Tucker S. J., Structure of a KirBac potassium channel with an open bundle crossing indicates a mechanism of channel gating. Nat. Struct. Mol. Biol. 19, 158–163 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuda H., Saigusa A., Irisawa H., Ohmic conductance through the inwardly rectifying K-channel and blocking by internal Mg2+. Nature 325, 156–159 (1987). [DOI] [PubMed] [Google Scholar]
- 30.Pegan S., Arrabit C., Slesinger P. A., Choe S., Andersen’s syndrome mutation effects on the structure and assembly of the cytoplasmic domains of Kir2.1. Biochemistry 45, 8599–8606 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S. P., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. Model. 14, 354–360 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Fagnen C., Bannwarth L., Oubella I., Forest E., De Zorzi R., de Araujo A., Mhoumadi Y., Bendahhou S., Perahia D., Vénien-Bryan C., New structural insights into Kir channel gating from molecular simulations, HDX-MS and functional studies. Sci. Rep. 10, 8392 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heyda J., Mason P. E., Jungwirth P., Attractive interactions between side chains of histidine-histidine and histidine-arginine-based cationic dipeptides in water. J. Phys. Chem. B 114, 8744–8749 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Fuerst O., Nichols C. G., Lamoureux G., D’Avanzo N., Identification of a cholesterol-binding pocket in inward rectifier K+ (Kir) channels. Biophys. J. 107, 2786–2796 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zangerl-Plessl E.-M., Lee S.-J., Maksaev G., Bernsteiner H., Ren F., Yuan P., Stary-Weinzinger A., Nichols C. G., Atomistic basis of opening and conduction in mammalian inward rectifier potassium (Kir2.2) channels. J. Gen. Physiol. 152, e201912422 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mouawad L., Perahia D., Motions in hemoglobin studied by normal mode analysis and energy minimization: Evidence for the existence of tertiary T-like, quaternary R-like intermediate structures. J. Mol. Biol. 258, 393–410 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Bahar I., Lezon T. R., Yang L.-W., Eyal E., Global dynamics of proteins: Bridging between structure and function. Annu. Rev. Biophys. 39, 23–42 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaynak B. T., Krieger J. M., Dudas B., Dahmani Z. L., Costa M. G. S., Balog E., Scott A. L., Doruker P., Perahia D., Bahar I., Sampling of protein conformational space using hybrid simulations: A critical assessment of recent methods. Front. Mol. Biosci. 9, 832847 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheres S. H. W., Processing of structurally heterogeneous Cryo-EM data in RELION. Methods Enzymol. 579, 125–157 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Lu Z., MacKinnon R., Electrostatic tuning of Mg2+ affinity in an inward-rectifier K+ channel. Nature 371, 243–246 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Kubo Y., Murata Y., Control of rectification and permeation by two distinct sites after the second transmembrane region in Kir2.1 K+ channel. J. Physiol. 531, 645–660 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujiwara Y., Kubo Y., Functional roles of charged amino acid residues on the wall of the cytoplasmic pore of Kir2.1. J. Gen. Physiol. 127, 401–419 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uchimura N., Cherubini E., North R., Inward rectification in rat nucleus accumbens neurons. J. Neurophysiol. 62, 1280–1286 (1989). [DOI] [PubMed] [Google Scholar]
- 44.Moldenhauer H., Díaz-Franulic I., González-Nilo F., Naranjo D., Effective pore size and radius of capture for K+ ions in K-channels. Sci. Rep. 6, 19893 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang D., Wan X., Dennis A. T., Bektik E., Wang Z., Costa M. G., Fagnen C., Venien-Bryan C., Xu X., Hund T. J., Mohler P. J., Laurita K. R., Deschenes I., Fu J.-D., MicroRNA biophysically modulates cardiac action potential by direct binding to ion channel. Circulation 143, 1597–1613 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garneau L., Klein H., Parent L., Sauvé R., Contribution of cytosolic cysteine residues to the gating properties of the Kir2.1 inward rectifier. Biophys. J. 84, 3717–3729 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J., Xiao S., Xie X., Zhou H., Pang C., Li S., Zhang H., Logothetis D. E., Zhan Y., An H., Three pairs of weak interactions precisely regulate the G-loop gate of Kir2.1 channel. Proteins 84, 1929–1937 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.L. Hartmann, V. Kugler, R. Wagner, in Heterologous Expression of Membrane Proteins: Methods and Protocols, 2nd Edition, I. MusVeteau, Ed. (Humana Press Inc., Totowa, 2016), vol. 1432, pp. 143–162; www.webofscience.com/wos/woscc/summary/d2fa6128-1783-46fe-bf75-ff2c82819c51-2dfd5b3c/relevance/1. [Google Scholar]
- 49.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D., Quentin D., Roderer D., Tacke S., Siebolds B., Schubert E., Shaikh T. R., Lill P., Gatsogiannis C., Raunser S., SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J., Lindahl E., Scheres S. H., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liebschner D., Afonine P. V., Baker M. L., Bunkoczi G., Chen V. B., Croll T. I., Hintze B., Hung L.-W., Jain S., McCoy A. J., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., Richardson J. S., Richardson D. C., Sammito M. D., Sobolev O. V., Stockwell D. H., Terwilliger T. C., Urzhumtsev A. G., Videau L. L., Williams C. J., Adams P. D., Macromolecular structure determination using x-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y., The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang J., MacKerell A. D. Jr., CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perahia D., Mouawad L., Computation of low-frequency normal modes in macromolecules: Improvements to the method of diagonalization in a mixed basis and application to hemoglobin. Comput. Chem. 19, 241–246 (1995). [DOI] [PubMed] [Google Scholar]
- 59.Phillips J., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R., Kale L., Schulten K., Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 and S2
Movies S1 to S6